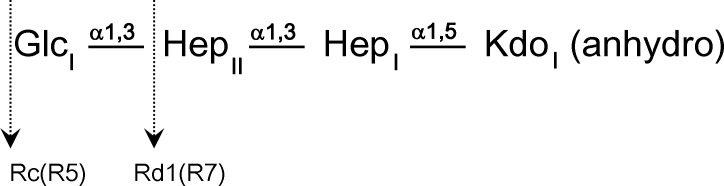Abstract
The crystal structures of a biologically and therapeutically active recombinant homotrimeric fragment of native human SP-D (hSP-D) complexed with the inner core oligosaccharide of the Salmonella enterica sv Minnesota rough strains R5 and R7 (rough mutant chemotypes Rc and Rd1) have been determined. The structures reveal that hSP-D specifically and preferentially targets the LPS inner core via the innermost conserved Hep-Kdo pair with the flexibility for alternative recognition when this preferred epitope is not available for binding. Hep-Kdo binding is achieved through calcium dependent recognition of the heptose dihydroxyethyl side chain coupled with specific interactions between the Kdo and the binding site flanking residues Arg343 and Asp325 with evidence for an extended binding site for LPS inner cores containing multiple Kdo residues. In one subunit of the R5-bound structure this preferred mode of binding is precluded by the crystal lattice and oligosaccharide is bound through the terminal inner core glucose. The structures presented here thus provide unique multiple insights into the recognition and binding of bacterial LPS by hSP-D. Not only is it demonstrated that hSP-D targets the highly conserved LPS proximal inner core Hep-Kdo motif, but also that hSP-D can recognise either terminal or non-terminal sugars and has the flexibility and versatility to adopt alternative strategies for bacterial recognition, utilising alternative LPS epitopes when the preferred inner core Hep-Kdo disaccharide is not available for binding.
Introduction
The innate immune protein human surfactant protein D (hSP-D) is a collagenous C-type lectin prototypically associated with the lung surfactant but now identified throughout the human body [1]. SP-D recognises a wide range of pathogens, through their surface carbohydrate arrays, leading to rapid deactivation and clearance through activation of phagocytic cells [2–4].
Numerous studies of the binding of simple sugars to both hSP-D and rfhSP-D, a recombinant homotrimeric native fragment comprised of the neck region plus three carbohydrate recognition domains (CRDs) which exhibits significant biological and therapeutic activity in vivo and in vitro [5–10] have been carried out both in solution and through 3-dimensional structural studies. Binding studies which demonstrate affinity for simple sugars such as glucose, mannose, maltose, heptose and inositol [11–13] have been confirmed crystallographically (see for example [14–18]). In all cases recognition is through CRD Ca-dependent binding of the terminal monosaccharide through a mannose-type equatorial hydroxyl pair O3' and O4' or a stereochemically equivalent pair.
A number of studies of gram-negative bacteria, including Escherichia coli, Salmonella enterica serovar Minnesota and Haemophilus influenzae [19,20] have reported that lipopolysaccharide (LPS) on the bacterial surface mediates hSP-D binding with LPS recognition initially thought to be via terminal carbohydrate residues through carbohydrate recognition domain Ca-dependent binding of a mannose-type equatorial hydroxyl pair on the terminal monosaccharide [14–16]. Our recent structural definition of the recognition of H. influenzae Eagan strains inner core LPS by a recombinant fragment of native hSP-D (rfhSP-D) [6,7,21] revealed that recognition occurs by calcium-dependent binding of the non-terminal heptose in the proximal inner core, with the Kdo positioned to interact with the binding site flanking residues Asp325 and Arg343 [15,16]. Furthermore, a recent study by Reinhardt and colleagues has identified that a number of synthetic oligosaccharide cores can be bound by wildtype hSP-D with increased affinities for heptose-rich cores and a clear preference for a terminal Hep-Kdo motif compared to Hep alone [22].
Salmonella enterica is a gram-negative bacterium that causes widespread outbreaks of gastroenteritis and bacteraemia in humans, with the most severe S. enterica Typhi infections causing enteric fever (reviewed by Coburn et al. [23]). Serovar Minnesota has been identified as the causative agent in a number of gastroenteritis outbreaks and, more recently, urosepsis in a Crohn’s patient [24–26]. The LPS oligosaccharide can be divided into the core oligosaccharides, containing 3-deoxy-D-manno-oct-2-ulosonic acid (Kdo), L-glycero-D-manno-heptose (Hep), α-D-glucose (Glc), α-D-galactose (Gal) and N-acetyl-D-glucosamine (GlcNAc), and the antigen-specific O-oligosaccharides that extend in repeats away from the bacterial cell surface [27]. In Salmonella species the inner and, to a large extent, the outer core oligosaccharides are a conserved epitope [28].
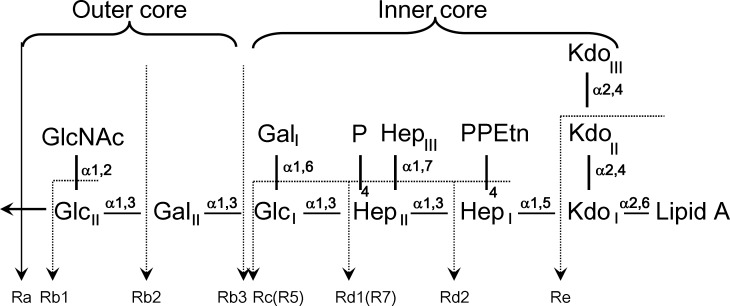
Rough mutants of S. enterica Minnesota lack the O-antigen, leaving the core sugar residues exposed to the immune system leading to the possibility of recognition by SP-D in a similar manner to that reported for H. influenzae Eagan 4A [21]. While the LPS core is largely conserved, mutations in the synthetic enzymes responsible for assembling the LPS can result in truncated oligosaccharide cores such as those of strains R5 and R7, equivalent to the Rc and Rd1 phenotypes (Fig 1), demonstrated through lectin blotting and agglutination-inhibition studies to be recognised by hSP-D [19]. The R5 and R7 rough strain LPS exhibit similar structures with the R5 extended by an extra glucose (GlcI) bound to HepII. The R5 strain arises from a single mutation in the phosphorylation step of HepI, inhibiting and impacting on the phosphorylation of HepII and the addition of GalI and GalII to GlcI. Phosphorylation of HepI and HepII initiates the extension of GalI and GalII from GlcI, producing the Rb2 strain [30–33].
Fig 1. Core LPS structure of Salmonella enterica rough mutant strains.
The LPS phenotypes (Ra, Rb1, Rb2, Rb3, Rc, Rd1, Rd2, and Re) and the mutant strains R5 and R7 are shown (modified from Mansfield and Forsythe, 2001 [29]). Glc: glucose; GlcNAc: N-acetyl-glucosamine; Gal: galactose; Hep: L-D-Heptose; P: phosphate; PEtn: phosphoethanolamine; Kdo: 3-deoxy-D-manno-oct-2-ulosonic acid.
While the H. influenzae Eagan 4A LPS [21] includes a truncated inner core with a single Kdo and a single Hep with a single Glc extension, the S. enterica Minnesota strains R5 and R7provide the opportunity to investigate, through crystal structure analysis, host recognition of more complex natural LPS structures with inner cores containing multiple Kdo and Hep alongside a terminal Glc in the R5 strain. Here we show that hSP-D recognition of Salmonella enterica sv Minnesota rough strains R5 and R7 (rough mutant chemotypes Rc and Rd1) LPS occurs preferentially through the inner core Hep-Kdo, with hSP-D having the flexibility and versatility to target the terminal glucose of R5 when the preferred epitope is not accessible for binding.
Materials and methods
S. enterica Minnesota R5 and R7 oligosaccharide preparation
The oligosaccharide (delipidated) portions of Salmonella enterica sv. Minnesota R5 (Enzo Lifesciences-ALX-581-017-L002) and R7 (Enzo Lifesciences-ALX-581-018-L002) mutants were prepared under mild hydrolysis conditions with 2% acetic acid, in accordance with the method previously published by Masoud et al. [34] and our previously published procedures [21]. For the R5 LPS the molecular weight was calculated as ≈ 3036 Da by the addition of all glycoconjugates and Lipid A. The lipid A portion accounted for ≈ 1800 Da [35], making up 60% of the LPS. 2 mg of R5 LPS were initially hydrolysed, yielding ≈0.7 mg of the oligosaccharide representing 88% hydrolysis efficiency. The R5 oligosaccharide was dissolved in deionised water to give a KdoI-HepI-HepII-GlcI concentration of ≈16 mM (accounting for the beta elimination of KdoII-KdoII) for storage and for use in crystal soaking. The same procedure was followed for the R7 LPS providing a stock solution of ≈18mM for storage and for use in crystal soaking and cryoprotection preparation.
Generation and purification of a recombinant fragment of hSP-D
A recombinant homotrimeric fragment of human SP-D (rfhSP-D) was expressed and purified as described previously [5,6,14]. Each chain contains 177 amino acids (Gly179 to Phe355) comprising a short collagen region with eight Gly-X-Y repeats, the α-helical coiled coil neck region and the globular CRD. The contaminating level of endotoxin present in the rfhSP-D preparation was minimized [6] using a Polymyxin B column (Detoxi-Gel, Pierce, UK) to remove endotoxin followed by assaying with the QCL-1000 Limulus amoebocyte lysate system (BioWhittaker, Walkersville, MD) according to manufacturer's instructions. The assay was linear over a range of 0.1–1.0 EU/ml (10 EU = 1 ng of endotoxin).
Crystallisation, ligand soaking and cryoprotection
The endotoxin treated rfhSP-D at 8.8 mg/ml in PBS was used for crystallisation both directly (for the R7 studies), following addition of CaCl2 to give 10mM, and after dialysis (for the R5 studies) into 50 mM tris(hydroxymethyl)aminomethane (Tris), 10 mM CaCl2, 150 mM NaCl, 0.02% NaN3 at pH7.4. Protein concentrations used were 7.7–8.9 mg/ml. Native crystals of rfhSP-D were grown in sitting drops using screens around conditions previously reported [14]. Those for soaking with the delipidated R5 LPS were grown in precipitant buffer 0.1 M tris pH 8.0, 16% polyethylene glycol (PEG) 6000, and for the delipidated R7 LPS in 0.1 M Tris pH 7.0, 16% PEG 10000. Cryoprotectant buffers were prepared as previously described [21] to provide increasing concentrations of MPD (2,4-methylpentane diol) in precipitant buffer with ligand added to provide final concentrations of ≈8 mM and ≈13 mM in the crystallisation drop for R5 and R7 respectively. The crystals were soaked with ligand and prepared for cryocooling by successive addition of cryoprotectant containing increasing concentrations of MPD (5–15% for R7; 5–20% for R5) in 2 μl aliquots over three minutes followed by a final exchange of 6–8 μl of the drop with 6–8 μl of the final MPD cryoprotectant solution (15% MPD for R7; 20% MPD for R5). Crystals were flash-frozen at 100 K after a total of 24 minutes (R5) or 8 minutes (R7) soaking with ligand and cryoprotectant buffer.
Data collection, structure solution and refinement
Data for the rfhSP-D R5 and R7 complexes were collected at 100 K at Diamond Light Source, Oxfordshire, on beamlines I04-1 and I03, respectively, using Dectris Pilatus 6M hybrid photon counting detectors. Integrated intensities were calculated using the program MOSFLM [36]. The 1.6 Å native structure of rfhSP-D (PDB ID 1PW9 [14]) was used as a starting model for modelling both the rfhSP-D/R5 and rfhSP-D/R7 structures. Electron density maps were calculated using the CCP4 suite of programs including AIMLESS, TRUNCATE, SORTMTZ and, where necessary, REINDEX [37]. Model building was completed using COOT [38] and refined using maximum likelihood refinement in REFMAC5 [39], available as part of the CCP4 package, in alternating rounds. Ligand topology and parameter files were generated using ProDRG [40] following fitting into the density. Final refinement statistics are presented in Table 1 and the quality of the final model was confirmed using the MolProbity web server [41]. The coordinates and structure factors for the R5-bound (PDB ID 5OXS) and R7-bound (PDB ID 5OXR) structures have been deposited with the Protein Data Bank. All main and side-chain stereochemical parameters are within accepted limits or better, with more than 98% of residues in favoured regions of the Ramachandran plot with no outliers. Molecular figures were generated using CCP4mg molecular graphics software [42] and the PyMOL Molecular Graphics System Version 1.4 (Schrödinger, LLC, 2011).
Table 1. Data collection and processing.
| Data collection | rfhSP-D/R7 | rfhSP-D/R5 |
|---|---|---|
| Synchrotron station | DLS I03 | DLS I04-1 |
| Wavelength (Å) | 0.97625 | 0.92819 |
| Space group | P21 | P21 |
| Cell dimensions | a = 55.33 Å, b = 108.14 Å, c = 55.67 Å, β = 91.82⁰ | a = 55.68 Å, b = 108.51 Å, c = 56.17 Å, β = 92.89⁰ |
| Resolution range (Å) | 55.3–1.75 (1.78–1.75) | 56.10–1.65 (1.68–1.65) |
| Observations | 143,142 (7,653) | 230,306 (10,955) |
| Unique reflections | 61,250 (3,299) | 74,199 (3,629) |
| Completeness (%) | 93.2 (91.2) | 93.0 (93.2) |
| Rmerge a | 0.066 (0.359) | 0.054 (0.397) |
| Mean (I/σ(I)) | 7.3 (2.2) | 11.4 (2.5) |
| Refinement | ||
| Protein atoms | 3483 | 3537 |
| Residues chain A | 205–355 | 204–355 |
| Residues chain B | 204–355 | 203–355 |
| Residues chain C | 205–355 | 203–355 |
| Water molecules | 479 | 477 |
| Other molecules | ||
| Subunit | A B C | A B C |
| Calcium ions | 3 3 3 | 2 2 1 |
| Oligosaccharide | - 1 1 | 1 1 1 |
| Rwork b (%) | 17.3 | 16.5 |
| Rfree c (%) | 19.6 | 18.7 |
| r.m.s.d. bond length (Å) | 0.010 | 0.010 |
| r.m.s.d. bond angle (°) | 1.40 | 1.32 |
| Average B-values (Å2) | ||
| Protein | 23.1 | 22.6 |
| Water | 34.2 | 34.9 |
| Other hetero-atoms | 31.1 | 40.2 |
| PDB ID | 5OXR | 5OXS |
| Ramachandran plot values d (%) | ||
| Favoured | 97.8 | 97.8 |
| Outliers | 0.0 | 0.0 |
Figures in parentheses refer to the highest resolution bin
a Rmerge = ∑h∑j |Ih,j—Ih| /∑h∑j |Ih,j|, where Ih,j is the jth observation of reflection h and Ih is the mean of the j measurements of reflection h.
b Rwork = ∑h ||Foh|—|Fch|| /∑h |Foh| where Foh and Fch are the observed and calculated structure factor amplitudes, respectively, for the reflection h.
c Rfree is equivalent to Rwork for a randomly selected subset (5%) of reflections not used in the refinement.
d Defined according to Molprobity
Results
The trimeric rfhSP-D structures comprising three carbohydrate recognition domains (CRDs) and the α-helical coiled-coil neck region have been refined to 1.65 Å and 1.75 Å resolution for the R5-bound and R7-bound structures respectively. In all but one of the CRDs the structures reveal oligosaccharide ligand in the Ca1 binding pocket. No ligand is present in subunit A of the R7-bound structure while the mode of oligosaccharide recognition in subunit A of the R5-bound structure differs from the Hep-Kdo binding seen in subunits B and C of both structures.
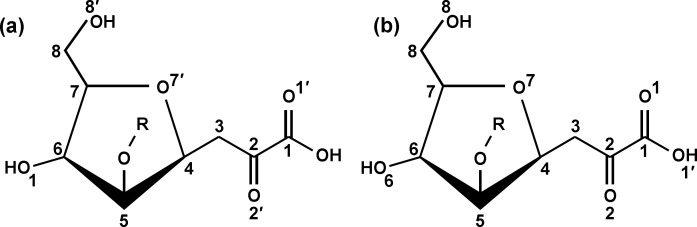
The bound oligosaccharide is clearly defined in the electron density of both structures; Kdo-HepI-HepII for the R7 and Kdo-HepI-HepII-Glc1 for R5 (Fig 2). The Kdo is in the anhydro form (Fig 3) reported by Shrive and co-workers [21] in the rfhSP-D–H. influenzae Eagan 4A structure (pdb entry 4E52) but here the 5-membered ring and all substituents are fully defined in the electron density (Fig 4) except in subunit A of the R5-bound structure where LPS binding is not via HepI and the terminal anhydro Kdo is poorly defined. The Kdo electron density clearly shows the 2-oxobutanoic acid sidechain off C4 (Fig 3B) in a beta conformation with respect to the glycosidic bond in both structures. KdoII and KdoIII (Fig 1) are not present in the electron density.
Fig 2. Structure of the product of mild acid hydrolysis of S. enterica Minnesota rough mutants R5 and R7.
Glc: glucose; Hep: L-D-Heptose; Kdo: 3-deoxy-D-manno-oct-2-ulosonic acid.
Fig 3. Structure of the 4,7 closure furanoid derivative (anhydro Kdo) of Kdo following LPS delipidation by mild acid hydrolysis and β-elimination of the Kdo O4 substituent, reported by Shrive and co-workers (PDB ID 4E52, ligand KD5).
Anhydro Kdo is proposed to be a racemic mixture (Auzanneau et al., 1991 [43]) with the 2-oxobutanoic acid side chain at C4 both alpha and beta to the glycosidic bond (at C5). (a) Original Kdo numbering retained. (b) Numbering according to pdb entry 4E52 (alpha) and used for the structure here.
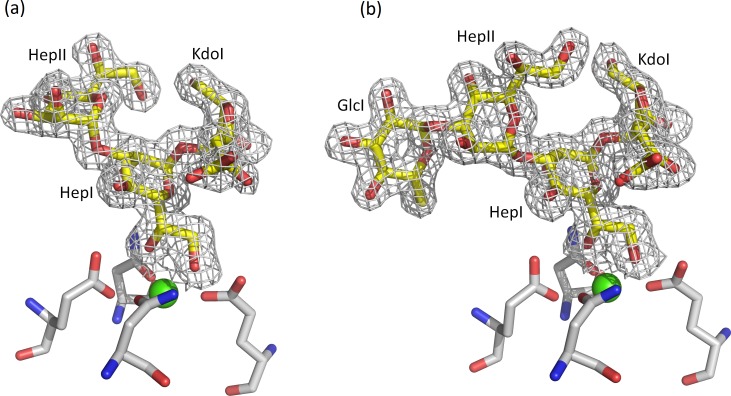
Fig 4. Electron density for the Salmonella enterica oligosaccharides bound to rfhSP-D (subunit B) with HepI in the Ca1 binding site.
The calcium ion is in green and the four calcium and ligand coordinating residues Glu321, Asn323, Glu329 and Asn341 are shown in cpk. (a) R7 oligosaccharide KdoI (anhydro)-HepI-HepII (b) R5 oligosaccharide KdoI (anhydro)-HepI-HepII-Glc1. Maps are 2mFo-DFc contoured at 1σ.
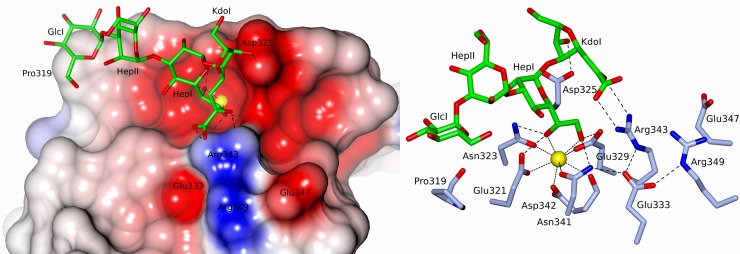
In subunits B and C, in both structures, recognition of the oligosaccharide by rfhSP-D (Fig 5 and Table 2) is primarily via protein and Ca1 coordination of the 6'OH and 7'OH of the dihydroxyethyl side chain of HepI. Further HepI interactions with the binding pocket in these four subunits include those routinely associated with carbohydrate recognition by hSP-D [21].
Fig 5. S. enterica R5 oligosaccharide bound to rfhSP-D (subunit C) with the inner core HepI in the Ca1 binding site.
KdoI (anhydro) interacts with both Asp325 and Arg343. Similar binding is observed in subunit B and in subunits B and C of the R7-bound structure which lacks the terminal glucose GlcI. Calcium coordinating bonds are represented by dots and protein-ligand interactions by dashes. The interactions of Glu333 are also indicated.
Table 2. Calcium and ligand binding distances (Å).
| Atom 1 | Atom 2 | R5 | R7 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | A | B | C | |||||
| Ca1 | Glu | 321 | OE1 | 2.63 | 2.61 | 2.57 | 2.65 | 2.60 | 2.64 | |
| Asn | 323 | OD1 | 2.46 | 2.42 | 2.47 | 2.49 | 2.44 | 2.38 | ||
| Glu | 329 | OE1 | 2.42 | 2.49 | 2.44 | 2.51 | 2.41 | 2.34 | ||
| Asn | 341 | OD1 | 2.50 | 2.44 | 2.44 | 2.37 | 2.43 | 2.44 | ||
| Asp | 342 | OD1 | 2.47 | 2.38 | 2.40 | 2.35 | 2.34 | 2.28 | ||
| Asp | 342 | O | 2.61 | 2.55 | 2.62 | 2.54 | 2.51 | 2.51 | ||
| HepI | O6' | - | 2.34 | 2.34 | - | 2.36 | 2.37 | |||
| O7' | - | 2.38 | 2.34 | - | 2.35 | 2.34 | ||||
| GlcI | O3' | 2.38 | - | - | - | - | - | |||
| O4' | 2.36 | - | - | - | - | - | ||||
| HepI | O6' | Glu | 321 | OE2 | - | 2.67 | 2.61 | - | 2.63 | 2.52 |
| Asn | 323 | ND2 | - | 3.01 | 2.92 | - | 3.03 | 2.93 | ||
| O7' | Glu | 329 | OE2 | - | 2.66 | 2.65 | - | 2.64 | 2.57 | |
| Asn | 341 | ND2 | - | 2.98 | 3.04 | - | 2.94 | 3.07 | ||
| GlcI | O3' | Glu | 321 | OE2 | 2.67 | - | - | - | - | - |
| Asn | 323 | ND2 | 2.76 | - | - | - | - | - | ||
| O4' | Glu | 329 | OE2 | 2.69 | - | - | - | - | - | |
| Asn | 341 | ND2 | 3.00 | - | - | - | - | - | ||
| KdoI | O1 | Arg | 343 | NH2 | - | 3.09 | 2.87 | - | - | - |
| KdoI | O2 | Arg | 343 | NH1 | - | 3.09 | 3.26 | - | 3.10 | 3.06 |
| KdoI | O6 | Asp | 325 | OD2 | - | 2.80 | 2.56 | - | 2.76 | 2.69 |
| GlcI | O6' | Arg | 343 | NH1 | 3.30 | - | - | - | - | - |
| GlcI | O6' | Arg | 343 | NH2 | 3.22 | - | - | - | - | - |
| HepI | O4' | Arg | 343 | NH2 | 3.06 | - | - | - | - | - |
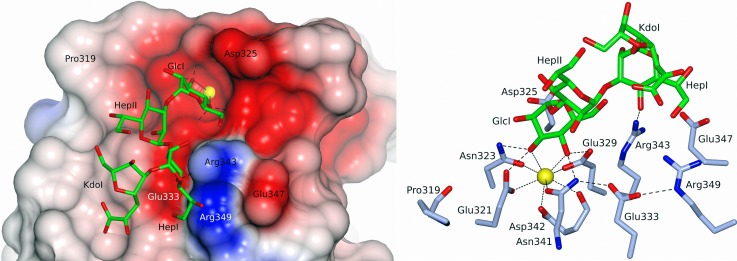
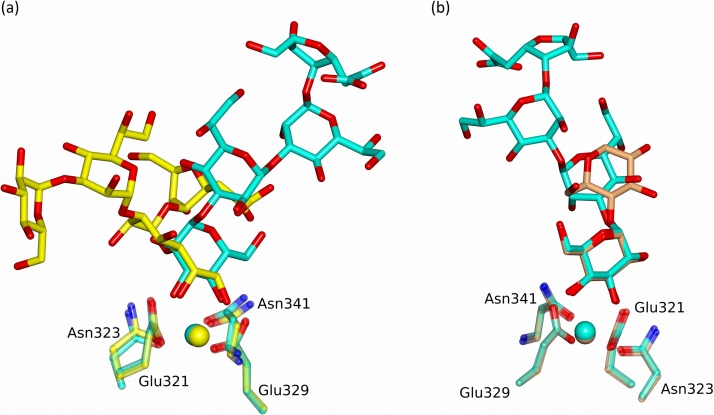
In contrast to the Ca1-HepI binding in the B and C subunits of both structures, subunit A of the R5-bound structure reveals an alternative mode of LPS recognition with calcium-dependent binding of the terminal glucose GlcI of R5, with the remaining R5 oligosaccharide chain HepII-HepI-Kdo extending over Arg343 and the SP-A (surfactant protein A), SP-D conserved Arg349 and Glu333 (Fig 6). The relative positions of the LPS for the two alternative binding modes are shown in Fig 7A where least-squares fitting of the main chain CRD in the two cases reveals the completely different LPS orientations despite the conserved Ca1-OH interactions. The position of the Ca1-bound glucose GlcI and its interactions with Ca1 and the binding pocket closely mirror those reported in a variety of structures including the rfhSP-D maltose bound structure [14] as shown in Fig 7B. The Ca1 coordinates to the terminal glucose GlcI through 3'OH (2.38 Å) and 4'OH (2.36 Å). Protein coordination to both calcium (via Glu321, Asn323, Glu329, Asn341) and the calcium-bound Glc1 hydroxyls (via Glu321, Asn323, Glu329, Asn341, Asp342) follows the established pattern for ligand-bound SP-D structures (Table 2). Calcium Ca3, the more weakly coordinated of the three calcium ions Ca1, Ca2, Ca3 [15], is absent throughout the R5-bound structure. Ca2 is only weakly defined in subunit B of the R5-bound structure and is absent in subunit C where the calcium (Ca2 and Ca3) coordinating loop between Asp297 and Tyr304 is rearranged and a water molecule replaces Ca2.
Fig 6. S. enterica R5 oligosaccharide bound to rfhSP-D (subunit A) with the terminal glucose GlcI in the Ca1 binding site.
HepI interacts with Arg343. No ligand was observed in subunit A of the R7-bound structure due to crystal lattice constraints. Calcium coordinating bonds are represented by dots and protein-ligand interactions by dashes. The interactions of Glu333 are also indicated.
Fig 7.
(a) Superposition of subunit A (GlcI bound) in cyan and subunit B (HepI bound) in yellow of the R5 LPS-bound structure following a least-squares fit of the CRD protein main chain atoms. (b) Superposition of subunit A (GlcI bound) in cyan of the R5 LPS-bound structure and subunit A (Glc1 bound) in coral of the maltose-bound structure [14] following a least-squares fit of the CRD protein main chain atoms.
In both structures and for both modes of binding (Ca1 coordination of HepI or GlcI) additional direct interactions between the protein and bound oligosaccharide are dominated by the binding-pocket flanking residues Arg343 and Asp325 (Table 2). For both ligands where HepI is coordinated to Ca1 (subunits B and C, both structures) the Kdo (anhydro) interacts with both binding site flanking residues, forming hydrogen bonds with Asp325 through Kdo O6 (2.56–2.80 Å) and with Arg343 through the extended 2-oxobutanoic acid sidechain of the anhydro Kdo (Fig 5 and Table 2). In subunit A of the R5-bound structure where GlcI is coordinated to Ca1, additional direct interaction between protein and oligosaccharide again involves Arg343 (Fig 6) which forms weak hydrogen bonds to both GlcI 6'OH and O4' of HepI (Table 2). The direct interactions between protein and oligosaccharide ligand in both structures and all subunits where oligosaccharide ligand is present are supplemented by extensive water molecule networks linking the ligand to the protein.
Discussion
The structures of rfhSP-D, a biologically active recombinant native fragment of hSP-D, bound to inner core structures of S. enterica Minnesota LPS from deep rough mutant strains reveal SP-D targeting of the proximal inner core Hep-Kdo motif through calcium-dependent binding of the (non-terminal) Hep supplemented by interaction of both Hep and Kdo with the binding site flanking residues Arg343 and Asp325. The structures also provide a definitive demonstration for the first time that hSP-D has the versatility to recognise alternative LPS epitopes when the Hep-Kdo inner core unit is inaccessible. This flexibility and versatility of LPS recognition is supplemented by evidence of an extended recognition surface that interacts with the LPS inner core oligosaccharide. The Kdo structure itself provides the first direct confirmation that mild hydrolysis removal of the lipid A from intact LPS exhibiting 4-KdoI substituted KdoII is accompanied by the elimination of KdoII from KdoI C4 [44,45] in a manner entirely equivalent to the β-elimination of phosphate from KdoI C4 through mild acid hydrolysis [21]. This is of some significance in the interpretation of mass spectrometry studies of bacterial LPS structure.
In all subunits where the carbohydrate binding site is openly accessible in the crystal (subunits B and C, both structures), HepI is coordinated to calcium Ca1 as reported for the H. influenzae Eagan 4A –rfhSP-D structure [21] with KdoI extending between the two binding-site flanking residues, Asp325 and Arg343, both of which interact directly with the bound HepI-Kdo (anhydro) unit (Fig 5). Superposition of the anhydro Kdo furanose rings together with HepI from all these subunits reveals an essentially conserved position of both rings resulting in very similar Asp325 –anhydro Kdo O6 interactions across the structures (2.56–2.80 Å), but significant variations in the Arg343 –Kdo interactions (2.87–3.26 Å) (Table 2). This suggests an important flexibility inherent in the Arg343 –Kdo interaction. Combined with the well documented ability of Arg343 to respond to different ligands through conformational change in the Arg343 side chain [15], this reinforces the importance of the Arg343—ligand interaction as a flexible determinant of specificity.
The R5 bound structure (Fig 6) reveals for the first time that hSP-D has the flexibility and versatility to recognise and bind alternative LPS epitopes if the HepI-Kdo core motif is not accessible or, in the physiological context, is shielded by extended oligosaccharides in the outer core and beyond [21]. In subunit A access to the carbohydrate binding site is restricted with the crystal packing precluding binding of either the R5 or R7 oligosaccharide via the HepI-Kdo pair. The crystal packing does however allow Ca1 binding of the terminal glucose of the longer R5 oligosaccharide, which binds to the SP-D subunit A binding pocket with the oligosaccharide ligand (Fig 6) oriented in a direction roughly orthogonal to the ligand when HepI is bound to Ca1 (Fig 7) as in subunits B and C of both structures (Fig 5). Interestingly, despite the completely different position of HepI with respect to the protein structure, Arg343 remains fundamental to the recognition of the LPS, stabilising the interaction through an Arg343-HepI hydrogen bond (Fig 6 and Table 2) and perhaps contributing to directing the glucose into the binding site [15,16]. While this novel structural demonstration that hSP-D can adopt alternative binding mechanisms for the same LPS is a fortuitous artefact of crystallisation, it clearly shows not only alternative recognition strategies for the same LPS but also the ability to recognise more than just the inner core heptose and Kdo residues in natural LPS ligands. This potentially provides an explanation for the extraordinary ability of hSP-D to recognise such a broad range of gram-negative bacteria by utilising Ca1-dependent binding of not just inner core HepI but also of multiple types of saccharide residue in the LPS [13,21]. The extent of the LPS, and the variety of different carbohydrate residues displayed on bacterial surfaces, provides the opportunity for versatility of recognition by the carbohydrate recognition domain.
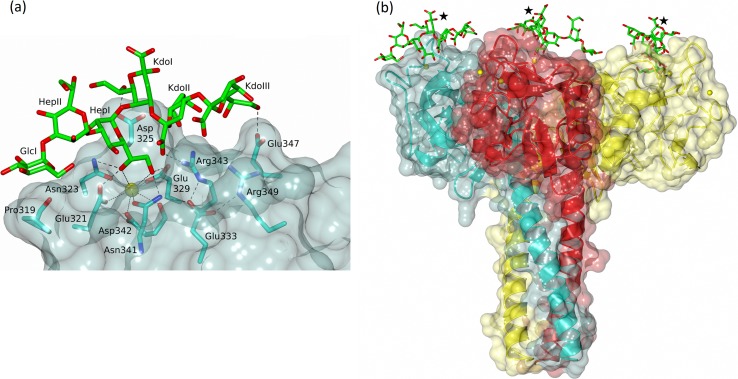
The full S. enterica sv. Minnesota R5 and R7 LPS oligosaccharides (Fig 1) are known to be recognised by hSP-D [19]. Positioning the full R5 oligosaccharide, including KdoII, KdoIII and a full 6-membered KdoI ring, in the R5-bound structure (subunit C) by overlaying HepI reveals that KdoI may be oriented such that the dihydroxyethyl side chain (7'OH, 8'OH) off C6 interacts either with Asp325 or Arg343. In the orientation shown in Fig 8, KdoII sits between KdoI and the conformationally flexible side chain of Arg343 with both KdoII and KdoIII proximal to the SP-D conserved Glu347 and the SP-A, SP-D conserved Arg349 and Glu333. KdoI O7' interacts with Asp325, KdoII O1' with Arg343, and KdoIII O8' with Glu347. The lipid A linked to KdoI O2' extends out from the CRD and the trimer overall. Salmonella enterica mutants with more extended cores and which are recognised by SP-D [19] can also be accommodated in this orientation. For the Rb3 mutant for example (Fig 1), the sugars linked to HepII and Glc1 all extend away from the CRD. This strongly suggests an extended inner core binding site with recognition of the Hep-Kdo motif accompanied by additional binding of inner cores with multiple Kdo structures such as those of Pseudomonas aeruginosa, Escherichia coli and Klebsiella pneumoniae which are known to be recognised by rfhSP-D [46].
Fig 8. Model of complete S. enterica R5 oligosaccharide bound to rfhSP-D with the inner core HepI in the Ca1 binding site.
KdoI (intact) is positioned to interact with Asp325, with KdoII and KdoIII interacting with Arg343 and Glu347 respectively. (a) subunit C, R5-bound structure (b) the rfhSP-D trimer with R5 similarly placed in each subunit and the link to lipid A from KdoI O2' indicated by ★.
Previous studies of the hSP-D/LPS interaction have shown the importance of Asp325 and Arg343 as oligosaccharide ligand selection mediators, flanking the binding site and directing calcium recognition [13–17], alongside the targeting of the LPS proximal inner core. The structures presented here combine and extend these key elements, revealing the preferred inner core Hep-Kdo binding mechanism alongside a flexibility to recognise alternative epitopes, to provide unique insights into the recognition and binding of a wide variety of gram negative bacteria by hSP-D.
Acknowledgments
We thank Diamond Light Source for access to beamlines I03 and I04-1 (proposal numbers MX8359, MX10369, MX14692) that contributed to the results presented here. Help from the beamline scientists at the Diamond Light Source is gratefully acknowledged.
Data Availability
The coordinates and structure factors for the R5-bound (PDB ID 5OXS) and R7-bound (PDB ID 5OXR) structures have been deposited with the Protein Data Bank.
Funding Statement
This work was supported by Diamond Light Source awards MX8359, MX10369, MX14692 (TJG, AKS). Diamond Light Source had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Ujma S, Horsnell WGC, Katz A, Clark HW, Schäfer G. Non-Pulmonary Immune Functions of Surfactant Proteins A and D. J Innate Immun. 2017;9:3–11. doi: 10.1159/000451026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reid KBM. Interactions of surfactant protein D with pathogens, allergens and phagocytes. Biochim Biophys Acta 1998;1408:290–5. [DOI] [PubMed] [Google Scholar]
- 3.LeVine AM, Whitsett JA. Pulmonary collectins and innate host defense of the lung. Microbes Infect. 2001;3:161–6. [DOI] [PubMed] [Google Scholar]
- 4.Crouch EC, Wright JR. Surfactant proteins A and D and pulmonary host defence. Annu Rev Physiol. 2001;63:521–4. doi: 10.1146/annurev.physiol.63.1.521 [DOI] [PubMed] [Google Scholar]
- 5.Madan T, Kishore U, Singh M, Strong P, Clark HW, Hussain EM et al. Surfactant proteins A and D protect mice against pulmonary hypersensitivity induced by Aspergillus fumigatus antigens and allergens. J Clin Invest. 2001;107:467–75. doi: 10.1172/JCI10124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singh M, Madan T, Waters P, Parida SK, Sarma PU, Kishore U. Protective effects of a recombinant fragment of human surfactant protein D in a murine model of pulmonary hypersensitivity induced by dust mite allergens. Immunology Letters 2003;86:299–307. [DOI] [PubMed] [Google Scholar]
- 7.Clark HW, Reid KBM. Structural requirements for SP-D function in vitro and in vivo: Therapeutic potential of recombinant SP-D. Immunobiology 2002;205:619–31. doi: 10.1078/0171-2985-00159 [DOI] [PubMed] [Google Scholar]
- 8.Madan T, Kishore U, Singh M, Strong P, Hussain EM, Reid KBM et al. Protective role of surfactant protein D in a murine model of Invasive Pulmonary Aspergillosis. Infect Immun. 2001b;69:2728–31. doi: 10.1128/IAI.69.4.2728-2731.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hickling TP, Bright H, Wing K, Gower D, Martin SL, Sim RB et al. A recombinant trimeric surfactant D carbohydrate recognition domain inhibits respiratory syncytial virus infection in vitro and in vivo. Eur J Immunol. 1999;29:3478–84. doi: 10.1002/(SICI)1521-4141(199911)29:11<3478::AID-IMMU3478>3.0.CO;2-W [DOI] [PubMed] [Google Scholar]
- 10.Strong P, Townsend P, Mackay R, Reid KBM, Clark HW. A recombinant fragment of human SP-D reduces allergic responses in mice sensitised to house dust mite allergens. Clin Exp Immunol. 2003;134:181–7. doi: 10.1046/j.1365-2249.2003.02281.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kishore U, Wang J-Y, Hoppe HJ, Reid KBM. The α-helical neck region of human lung surfactant protein D is essential for the binding of the carbohydrate recognition domains to lipopolysaccharides and phospholipids. Biochem J. 1996;318:505–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crouch E, Tu Y, Briner D, McDonald B, Smith K, Holmskov U, Hartshorn K. Ligand specificity of human surfactant protein D: expression of a mutant trimeric collectin that shows enhanced interactions with influenza A virus. J Biol Chem. 2005;280(17):17046–56. doi: 10.1074/jbc.M413932200 [DOI] [PubMed] [Google Scholar]
- 13.Wang H, Head J, Kosma P, Brade H, Müller-Loennies S, Sheikh S et al. Recognition of Heptoses and the Inner Core of Bacterial Lipopolysaccharides by Surfactant Protein D. Biochemistry 2008;47:710–20. doi: 10.1021/bi7020553 [DOI] [PubMed] [Google Scholar]
- 14.Shrive AK, Tharia HA, Strong P, Kishore U, Burns I, Rizkallah PJ et al. High-resolution structural insights into ligand binding and immune cell recognition by human lung surfactant protein D. J Mol Biol. 2003;331:509–23. [DOI] [PubMed] [Google Scholar]
- 15.Shrive AK, Martin C, Burns I, Paterson JM, Martin JD, Townsend JP et al. Structural Characterisation of Ligand-Binding Determinants in Human Lung Surfactant Protein D: Influence of Asp325. J Mol Biol. 2009;394:776–88. doi: 10.1016/j.jmb.2009.09.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crouch E, Hartshorn K, Horlacher T, McDonald B, Smith K, Cafarella T et al. Recognition of mannosylated ligands and influenza a virus by human surfactant protein D: Contributions of an extended site and residue 343. Biochemistry 2009;48:3335–45. doi: 10.1021/bi8022703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crouch E, McDonald B, Smith K, Roberts M, Mealy T, Seaton B et al. Critical role of Arg/Lys343 in the species-dependent recognition of phosphatidylinositol by pulmonary surfactant protein D. Biochemistry 2007;46:5160–9. doi: 10.1021/bi700037x [DOI] [PubMed] [Google Scholar]
- 18.Crouch E, McDonald B, Smith K, Cafarella T, Seaton B, Head J. Contributions of phenylalanaine 335 to ligand recognition by human surfactant protein D. J Biol Chem. 2006a;281:18008–14. [DOI] [PubMed] [Google Scholar]
- 19.Kuan SF, Rust K, Crouch E. Interactions of Surfactant Protein-D with Bacterial Lipopolysaccharides—Surfactant Protein-D is an Escherichia-Coli Binding-Protein in Bronchoalveolar Lavage. Journal of Clinical Investigation 1992;90:97–106. doi: 10.1172/JCI115861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.LeVine AM, Whitsett JA, Gwozdz JA, Richardson TR, Fisher JH, Burhans MS et al. Distinct Effects of Surfactant Protein A or D Deficiency During Bacterial Infection on the Lung. J Immunol. 2000;165:3934–40. [DOI] [PubMed] [Google Scholar]
- 21.Clark HW, Mackay R-M, Deadman ME, Hood DW, Madsen J, Moxon ER et al. Crystal structure of a complex of Surfactant Protein D and Haemophilus influenzae lipopolysaccharide reveals shielding of core structures in SP-D resistant strains. Infect Immun. 2016;84:1585–92. doi: 10.1128/IAI.01239-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reinhardt A, Wehle M, Geissner A, Crouch EC, Kang Y, Yang Y et al. Structure binding relationship of human surfactant protein D and various lipopolysaccharide inner core structures. J Struct Biol. 2016;195:387–95. doi: 10.1016/j.jsb.2016.06.019 [DOI] [PubMed] [Google Scholar]
- 23.Coburn B, Grassl G, Finlay BB. Salmonella, the host and disease: A brief review. Immunol Cell Biol. 2007;85:112–18 doi: 10.1038/sj.icb.7100007 [DOI] [PubMed] [Google Scholar]
- 24.McCracken DA. Salmonella minnesota Infection in Northamptonshire. J R Sanit Inst 1954;74:1091–4. [DOI] [PubMed] [Google Scholar]
- 25.Borbolla L, Guerra Chabau A, Prieto E, Valledor T [Meningitis caused by Salmonella minnesota: case report] Translated from Spanish. Rev Cubana Pediatr. 1956;10:611–18. [PubMed] [Google Scholar]
- 26.Steinebrunner N, Sandig C, Zimmermann S, Stremmel W, Eisenbach C, Mischnik A. Salmonella enterica serovar Minnesota urosepsis in a patient with Crohn’s disease in the absence of recent or current gastrointestinal symptoms. J Med Microbiol. 2013;62:1360–2. doi: 10.1099/jmm.0.059352-0 [DOI] [PubMed] [Google Scholar]
- 27.Holst O. Structure of the lipopolysaccharide core region, in Bacterial Lipopolysaccharides: Structure, Chemical Synthesis, Biogenesis and Interaction with Host Cells (Knirel Y. A., and Valvani M. A., eds) pp. 21–39, 2011. Springer, Vienna. [Google Scholar]
- 28.Holst O, Brade H. Structural studies of the core region of the lipopolysaccharide from Salmonella minnesota strain R7 (rough mutant chemotype Rd1). Carbohydr Res. 1991;219:247–51. [DOI] [PubMed] [Google Scholar]
- 29.Mansfield LP, Forsythe SJ. Demonstration of the Rb-1 lipopolysaccharide core structure in Salmonella strains with the monoclonal antibody M105. Journal of Medical Microbiology 2001;50(4):339–44. doi: 10.1099/0022-1317-50-4-339 [DOI] [PubMed] [Google Scholar]
- 30.Muhlradt P. Biosynthesis of Salmonella lipopolysaccharide. Studies on the transfer of glucose, galactose, and phosphate to the core in a cell free system. Eur J Biochem. 1971;18(1):20–7. [DOI] [PubMed] [Google Scholar]
- 31.Schnaitman CA, Klena JD. Genetics of lipopolysaccharide biosynthesis in enteric bacteria. Microbiological Reviews 1993;57:655–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jousimies H, Mäkelä PH. Genetic analysis of Salmonella minnesota R mutants with defects in the biosynthesis of the lipopolysaccharide core. J Bacteriol. 1974;119(3):753–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raetz CHR, Whitfield C. Lipopolysaccharide Endotoxins. Annu Rev Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Masoud H, Altman E, Richards JC, Lam JS. General strategy for structural analysis of the oligosaccharide region of lipooligosaccharides. Structure of the oligosaccharide component of Pseudomonas aeruginosa IATS serotype 06 mutant R5 rough-type lipopolysaccharide. Biochemistry 1994;33:10568–78. [DOI] [PubMed] [Google Scholar]
- 35.Que NLS, Lin S, Cotter RJ, Raetz CRH. Purification and Mass Spectrometry of Six Lipid A Species from the Bacterial Endosymbiont Rhizobium etli. J Biol Chem. 2000;275(36):28006–16. doi: 10.1074/jbc.M004008200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leslie AGW. Recent changes to the MOSFLM package for processing film and image plate data Joint CCP4 and ESF-EACMB Newsletter on Protein Crystallography 26. 1992. Daresbury Laboratory, Warrington, UK. [Google Scholar]
- 37.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. 2011;D67:235–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and Development of Coot. Acta Crystallogr. 2010;D66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murshudov GN, Vagin AA, Dodson EJ. Refinement of Macromolecular Structures by the Maximum-Likelihood method. Acta Crystallogr. 1977;D53:240–55. [DOI] [PubMed] [Google Scholar]
- 40.Schuettelkopf AW, van Aalten DMF. PRODRG–a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr. 2004;D60:1355–63. [DOI] [PubMed] [Google Scholar]
- 41.Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. 2010;D66:12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McNicholas S, Potterton E, Wilson KS, Noble MEM. Presenting your structures: the CCP4mg molecular-graphics software. Acta Crystallogr. 2011;D67:386–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Auzanneau F-I, Charon D, Szabó L. Phosphorylated sugars. Part 27. Synthesis and reactions, in acid medium, of 5-O-substituted methyl 3-deoxy-α-D-manno-oct-2-ulopyranosidonic acid 4-phosphates. J Chem Soc Perkin Trans 1. 1991;509–17. [Google Scholar]
- 44.Yethon JA, Vinogradov E, Perry MB, Whitfield C. Mutation of the Lipopolysaccharide Core Glycosyltransferase Encoded by waaG Destabilizes the Outer Membrane of Escherichia coli by Interfering with Core Phosphorylation. Journal of Bacteriology 2000;182(19):5620–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Olsthoorn MA, Haverkamp J, Thomas-Oates JE. Mass Spectrometric Analysis of Klebsiella pneumoniae ssp. pneumoniae Rough Strain R20 (O1−:K20−) Lipopolysaccharide Preparations: Identification of Novel Core Oligosaccharide Components and Three 3-Deoxy-D-manno-oct-2-ulopyranosonic Artifacts. J Mass Spectrom. 1999;34:622–36. doi: 10.1002/(SICI)1096-9888(199906)34:6<622::AID-JMS814>3.0.CO;2-V [DOI] [PubMed] [Google Scholar]
- 46.Lim BL, Wang JY, Holmskov U, Hoppe HJ, Reid KBM. Expression of the carbohydrate recognition domain of lung surfactant protein D and demonstration of its binding to lipopolysaccharides of gram-negative bacteria. Biochem Biophys Res Commun. 1994;202(3):1674–80. doi: 10.1006/bbrc.1994.2127 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The coordinates and structure factors for the R5-bound (PDB ID 5OXS) and R7-bound (PDB ID 5OXR) structures have been deposited with the Protein Data Bank.