Melanoma is a paradigm of cancers with an inherent potential for lymph node colonization, a process generally preceded by neo-lymphangiogenesis1, 2, 3. However, sentinel lymph node removal does not necessarily extend overall survival of melanoma patients4, 5. Moreover, lymphatic vessels become collapsed and dysfunctional as melanomas progress6, 7. Therefore, it is unclear whether (and how) lymphangiogenesis contributes to visceral metastasis. Soluble and vesicle-associated proteins secreted by tumors and/or their stroma have been proposed to condition premetastatic sites in this disease8, 9, 10, 11, 12, 13, 14. Still, the identity and prognostic impact of lymphangiogenic mediators remain elusive2, 14. Moreover, the understanding of lymphangiogenesis (in melanoma and other tumor types) has been challenged by the paucity of mouse models for live imaging of distal pre-metastatic niches15. Injectable lymphatic tracers have been developed7, but their limited diffusion precludes whole-body imaging at visceral sites16. Vegfr3 is an attractive “lymphoreporter”17 as its expression is strongly downregulated in normal adult lymphatic endothelial cells, but gets activated under pathological situations such as inflammation and cancer17, 18. Here, we exploit this inducibility of Vegfr3 to engineer mouse melanoma models for whole-body imaging of metastasis generated by human cells, clinical biopsies or endogenously deregulated oncogenic pathways. This strategy revealed an early induction of distal premetastatic niches uncoupled to lymphangiogenesis at primary lesions. Analyses of the melanoma secretome and validation in clinical specimens uncovered the growth factor MIDKINE as a systemic inducer of neo-lymphangiogenesis that defines patient prognosis. This newly identified role of MIDKINE was found linked to a paracrine activation of the mTOR pathway in lymphatic endothelial cells. These data support the use of Vegfr3-reporters as a “MetAlert” discovery platform for drivers and inhibitors of metastasis.
To generate animal models for whole-body imaging of neolymphangiogenesis in melanoma progression, we exploited the Flt4tm1.1Sgo (Vegfr3EGFPLUC) mice17, a knock-in system whereby an EGFP-Luciferase fusion cassette is coupled to the transcription of Vegfr3 (Fig. 1a). These Vegfr3 reporters (herein referred to as Vegfr3Luc for simplicity) were crossed into immune deficient Foxn1nu (nu/nu) mice to assess human melanoma cell lines and patient-derived xenografts (PDX). Crosses to Tyr::CreERT2;BrafV600E or to Tyr::CreERT2;BrafV600·;Ptenlox/lox immune competent strains19 were performed to monitor benign nevi or malignant melanomas. These genetically engineered mouse models (GEMM) were maintained in black-coat or albino backgrounds to characterize pigmented or amelanotic lesions (Fig. 1a).
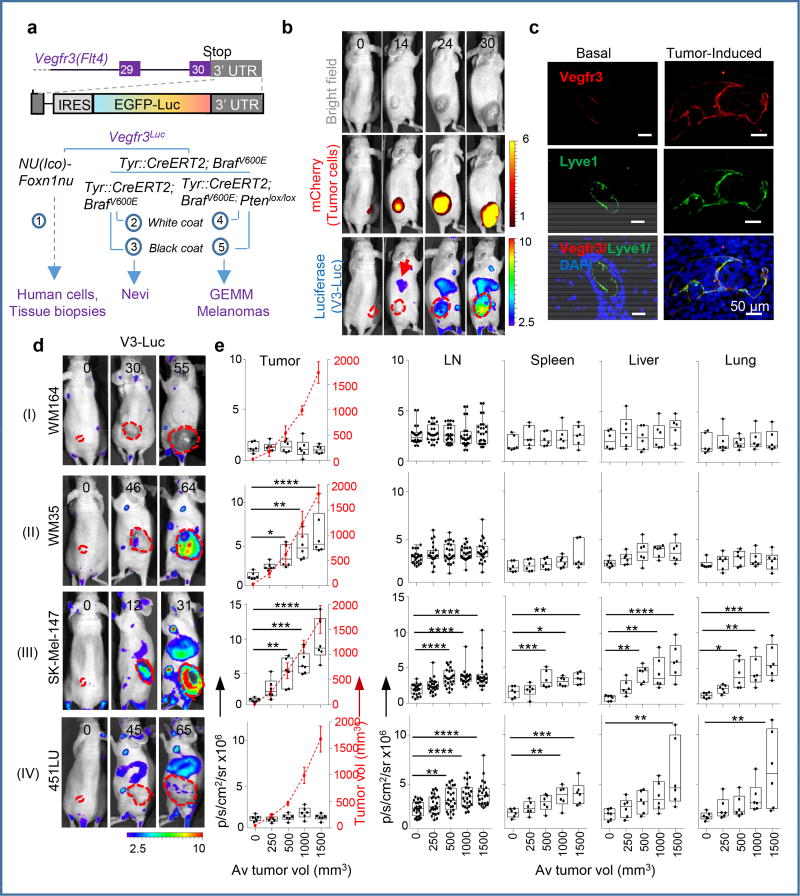
Figure 1. Vegfr3Luc reporter mice for whole-body analysis of benign and malignant melanocytic lesions.
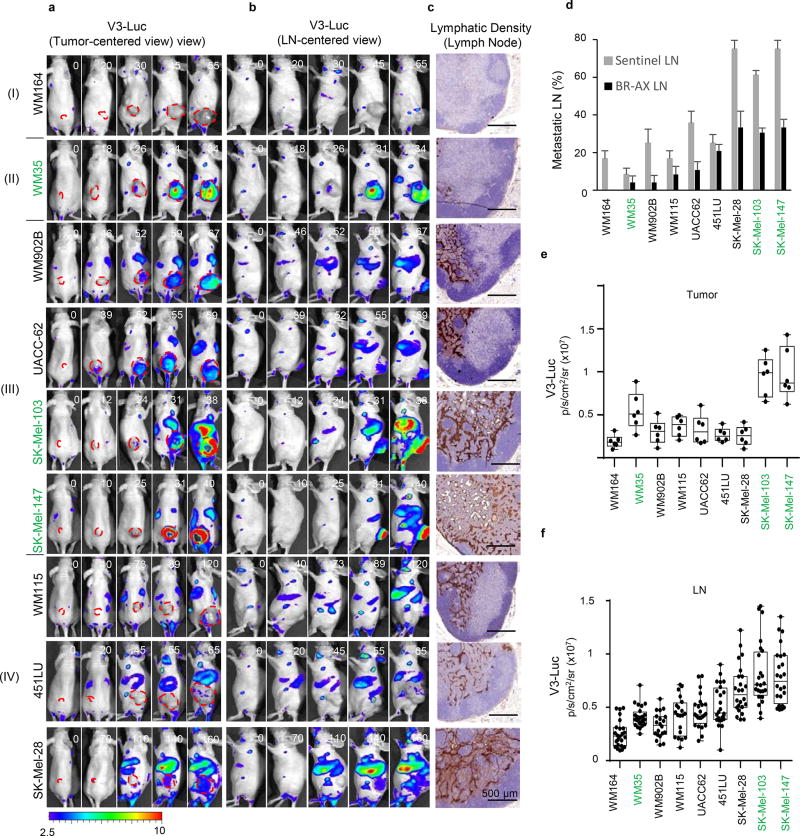
a, Vegfr3Luc mouse models in this study. b, Xenografts of mCherry-SK-Mel-147 imaged as indicated. c, Confocal immunomicroscopy of Vegfr3 (red) and Lyve1 (green) in normal skin or in xenografts by SK-Mel-147. d, Four main patterns (I–IV) of V3-Luc emission identified by whole-body bioluminescence of xenografts of the indicated melanoma cell lines. Numbers represent days upon implantation and dotted lines tumor area. e, V3-Luc at the indicated locations and tumor sizes. Average tumor volumes are indicated in the X-axis, and visualized with a dotted red line for simplicity. Data are mean ± SD (6 mice per condition, 24 LN per cell line). One-way Anova/Dunnett's correction for multiple comparisons. Fluorescence: p/s/cm2/sr ×109; bioluminescence: p/s/cm2/sr ×106. See Source Data for V3-Luc quantifications in panel (e).
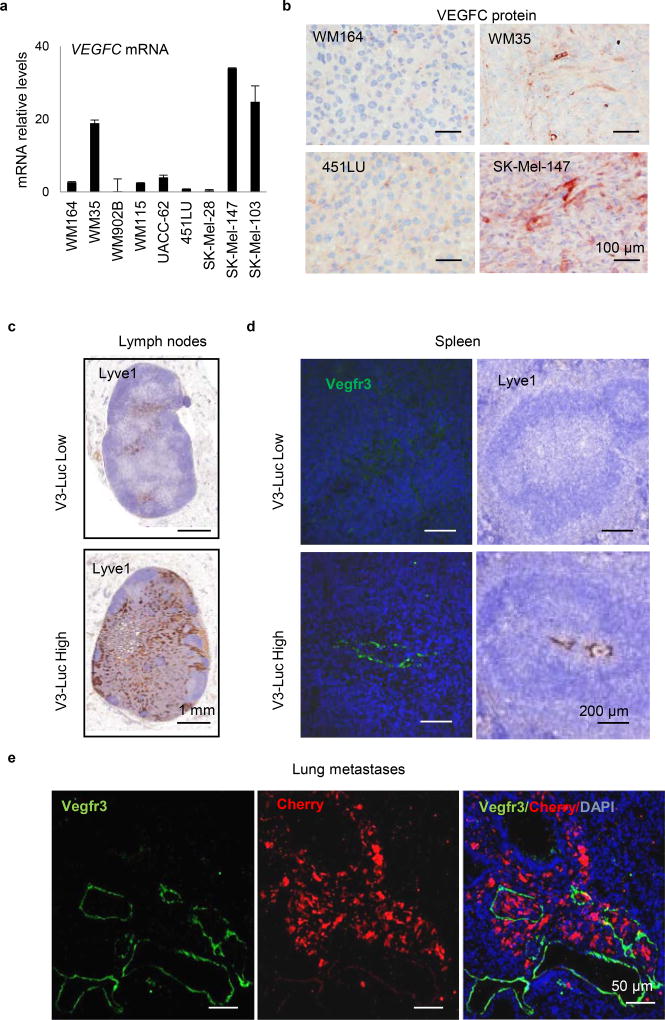
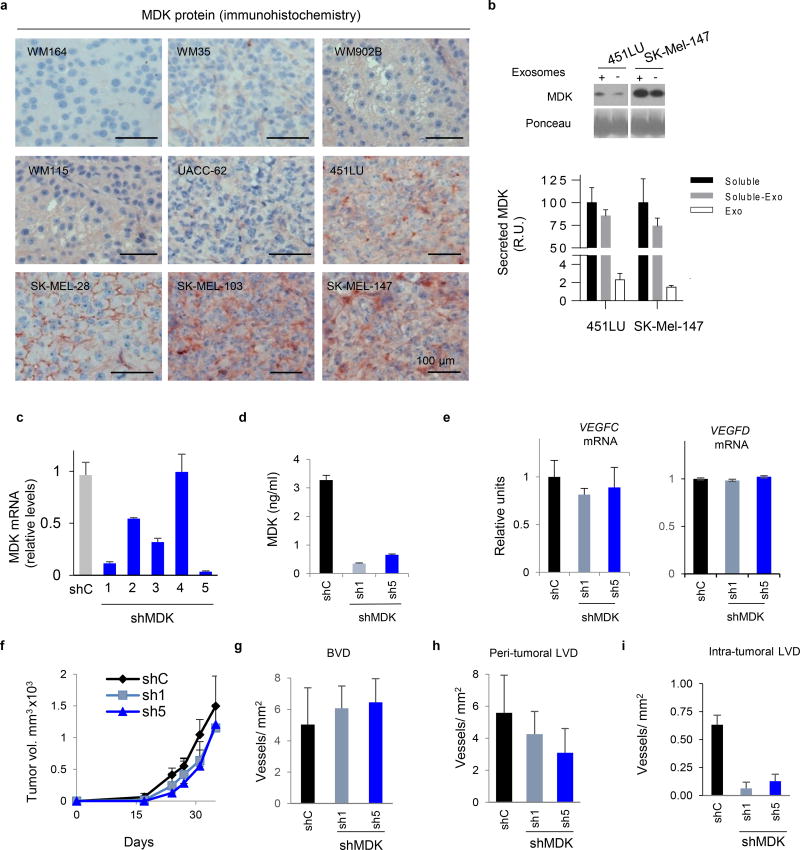
Vegfr3Luc nu/nu mice were used for the generation of subcutaneous xenografts by a panel of 9 melanoma cell lines with different levels of VEGFC (Extended Fig. 1a,b), a main known driver of lymphangiogenesis3. This set of cell lines also recapitulated the most frequent melanoma-associated alterations in BRAF, NRAS, PTEN and p53 (Table S1). Prior implantation, cells were transduced with mCherry for dual fluorescence/bioluminescence imaging of tumor growth and the subsequent emission of Vegfr3-associated luciferase (V3-Luc), respectively (see examples in Fig. 1b). Histological staining for Vegfr3 in Lyve1-positive lymphatic endothelial cells (LEC) confirmed neo-lymphangiogenesis (see Fig. 1c for examples in the skin, and Extended Figs. 1c–e, for lymph nodes (LN), spleen and lungs, respectively).
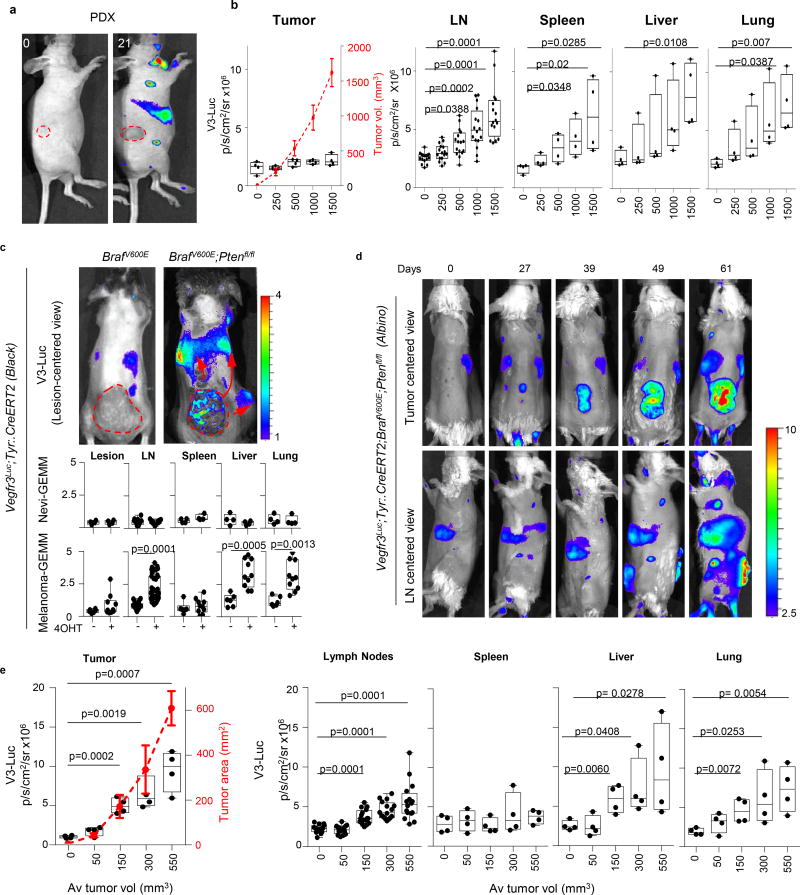
As summarized in Fig. 1d, quantified in Fig. 1e, and presented in additional detail in Extended Fig. 2a–c, and Supplementary Information, we identified 4 major patterns of tumor-driven neolymphangiogenesis; with no V3-Luc at any site (Pattern I); restricted to the primary tumor (II); occurring locally and distally (III); and the most unexpected, with or highly delayed tumoral V3-Luc, and yet, a potent systemic emission (IV). Interestingly, only patterns III and IV (i.e. including distal V3-Luc) were found to significantly increase lymphatic vessel density at lymph nodes (LN) and result in efficient nodal metastasis (Extended Figs. 2c–f). These patterns did not to correlate with BRAF, NRAS, p53 or PTEN status (not shown). Moreover, tumoral VEGFC was neither sufficient, nor essential for the activation of distal V3-Luc and nodal metastases (see Fig. 1a and Fig. S2, where VEGFC-positive cell lines are labeled in green). PDX from human skin metastases further confirmed early and systemic induction of V3-Luc uncoupled from prior tumoral lymphangiogenesis (Extended Fig. 3a,b). Systemic V3-Luc was also obvious in GEMM melanomas (but not in nevi), both in pigmented amelanotic backgrounds (Extended Figs. 3c–e).
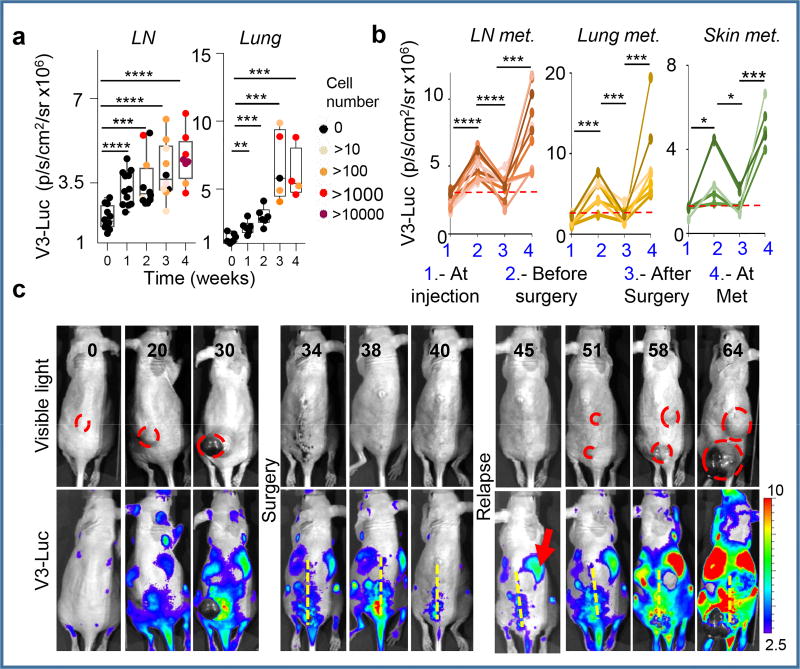
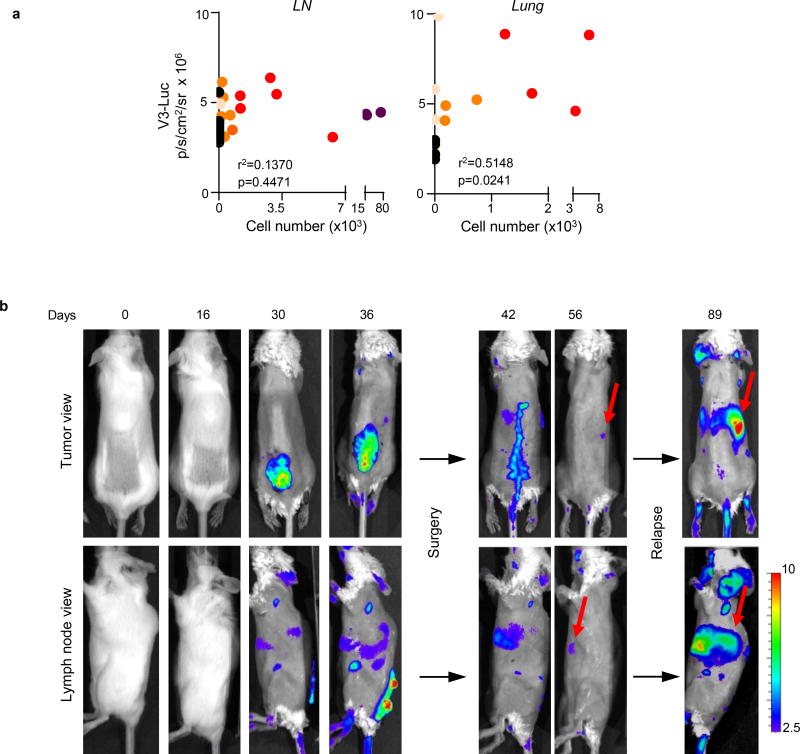
Tumor burden estimated by quantitative RT-PCR at different anatomical locations and time points after tumor cell implantation revealed a significant systemic activation of V3-Luc prior to tumor cell colonization (see black dots in Fig. 2a,b for quantitative analyses of in LN and lungs). Therefore, these data support V3-Luc positive sites as pre-metastatic niches. V3-Luc imaging was then analyzed prior and post-surgical removal of xenografts of human melanoma cells (SK-Mel-147; Fig. 2c), or GEMM-melanomas (Extended Fig. 4). In both cases, excision of primary lesions led to a marked reduction of V3-Luc. Metastatic relapses were found at later time points (involving skin, LN and lung metastases), invariably preceded by V3-Luc emission (see for SK-Mel-147 in Fig. 2d). Therefore early systemic luciferase emission in Vegfr3Luc mice reflects long-distance signals fueled by the primary lesions.
Figure 2. Vegfr3Luc mice reveal pre-metastatic niches.
a, V3-Luc emission by xenografts of mCherry-SK-Mel-147 in sentinel LN and lungs. Colored dots correspond to tumor cell burden defined by RT-PCT. b, Quantification of V3-Luc emission by SK-Mel-147-mCherry prior and after surgical resection of the cutaneous lesions. LN (n=9), lung metastases (n=7) and skin metastases (n=5). t-Test. c, representative whole-body imaging of experiments as in (b). See Source Data for V3-Luc quantifications in panels (a,b).
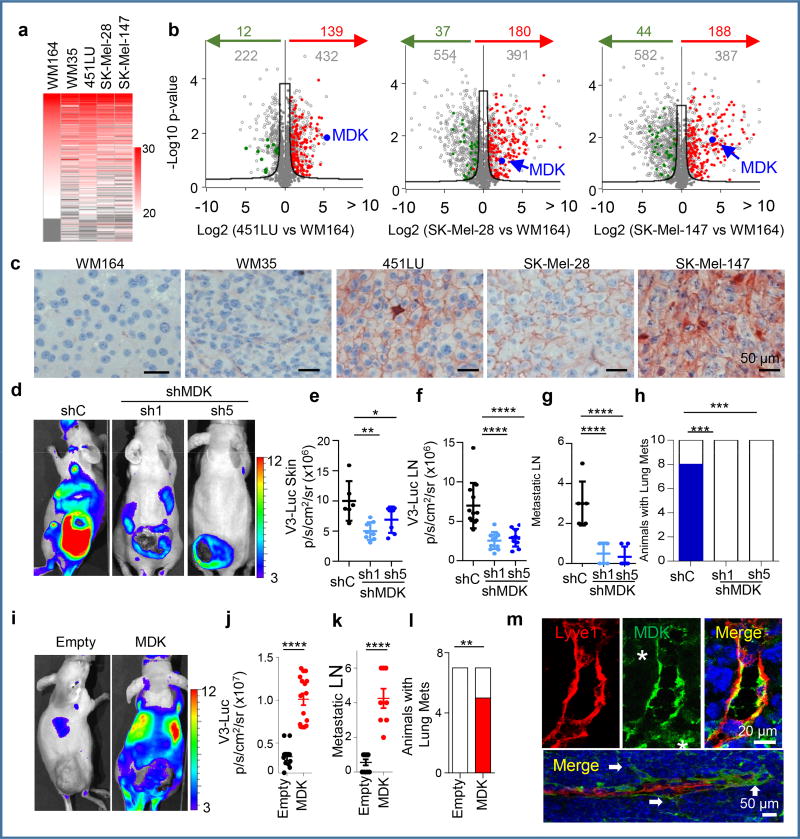
Proteomic analyses were then performed to identify tumor-secreted drivers of distal lymphangiogenesis/metastasis. Exosome cargo was then defined by label-free Liquid Chromatography–Mass Spectrometry (Fig. 3a) in representative examples of the 4 V3-Luc emission patterns described above. Candidates were filtered for statistical significance, selective expression in cell lines with systemic V3-Luc emission, no previous lymphangiogenic roles and no links to melanoma (see Online Methods). Top-scoring in this strategy was the heparin binding factor MIDKINE (MDK, marked in blue in the Volcano plots of Fig. 3b; see additional secreted proteins in Table S2). We found particularly encouraging the fact that MDK is secreted by other cancer types20, 21, 22, 23, and has been proposed to mediate metastasis by its mitogenic, pro-inflammatory and angiogenic functions20, 23, 24, 25, but with no connection to lymphovascular expansion.
Figure 3. Proteomic analyses identify MIDKINE as new pro-lymphangiogenic/pro-metastatic factor.
a, LFQ (Label Free Quantitative) expression of exosome cargo identified by LC/MS-MS in the indicated cell lines. b, Volcano plots showing exosomal proteins (grey dots) differentially expressed with respect to the non-lymphangiogenic WM164. Proteins upregulated or downregulated vs. the poorly-metastatic WM35 are depicted in red or green, respectively. The hyperbolic black curve separates statistically-significant regulated proteins as defined by two-sample Student’s t-Test, FDR=0.05; S0=0.8. c, Immunological detection of MDK (pink) in tumor xenografts. d, V3-Luc emission by SK-Mel-147-mCherry transduced with control or MDK shRNA(1),(5), with quantifications for cutaneous lesions in (e) and for sentinel and brachial LN in (f). g, Metastatic LN (mCherry positive) of animals in (e). Data correspond to mean ± SD (n≥ 6 mice/condition). One-way ANOVA/Dunnett's correction for multiple comparison. h, Lung metastases (blue) after tumor removal. Fisher's exact test, n=10. i, V3-Luc emission by MDK overexpression in the otherwise negative WM164. Scale, p/s/cm2/sr (×106). j, Relative nodal V3-Luc emission at the indicated conditions. The number of metastatic LN per animal is shown in panel (k). t-Test. i, Lung metastases (mCherry positive; red) in MDK GoF studies (generated by overexpressing MDK in the negative WM164). Fisher's exact test, n=7. m, Immunomicrographs showing MDK (green) accumulated at areas of neo-lymphangiogenesis and at sites of lymphatic sprouting in LN (arrows). Asterisks mark stromal cells. See Source Data for V3-Luc quantifications of panels (e–h, j–l) and Supplemental Figure 1 for uncropped blots.
Immunohistochemical analyses confirmed MDK expression in highly metastatic melanoma xenografts (Fig. 3c and Extended Fig. 5a). Importantly, although we had found MDK as an exosome cargo, immune-based analyses (ELISA) and immunoblotting after exosome depletion showed a 40–60 fold higher expression of MDK in the soluble fraction of aggressive melanoma cells (Extended Fig. 5b). Therefore we proceeded with the analysis of soluble MDK in the secretome of melanoma cells.
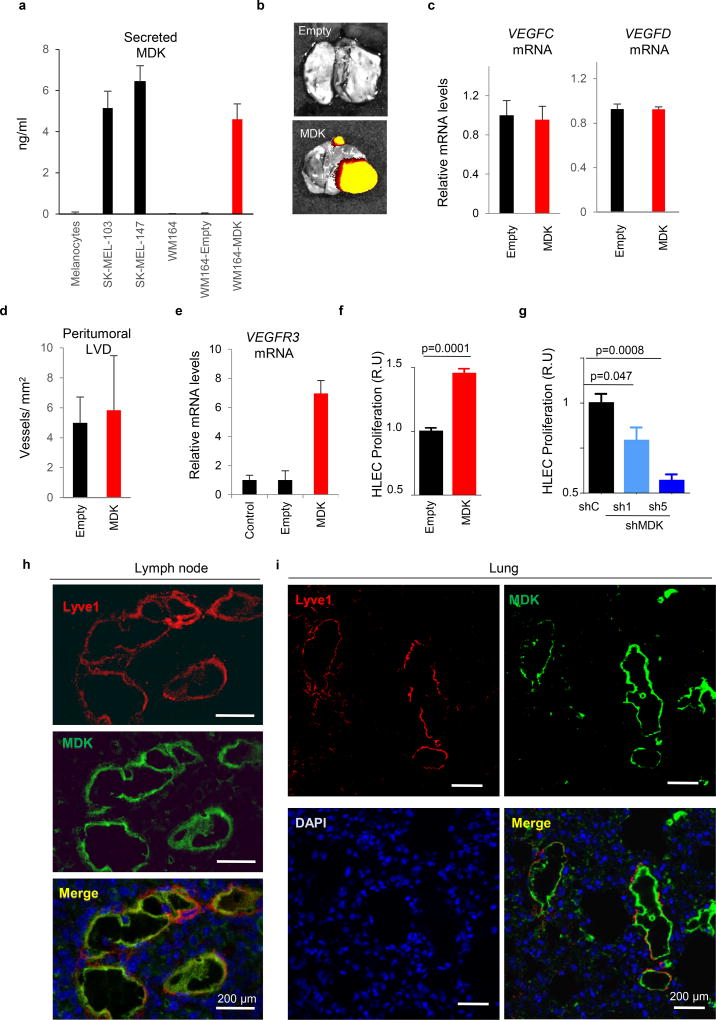
Loss- and gain-of-function studies (LoF and GoF) were performed to manipulate the endogenous levels of MDK. For LoF, 5 shRNAs were tested. Of those, qRT-PCR and ELISA identified shMDK(1) and (5) as the most potent blockers of MDK expression and secretion (Extended Fig. 5c,d). None of these shRNAs affected the intracellular VEGFC or VEGFD mRNA levels (Extended Fig. 5e). Moreover, MDK depletion was not found to significantly affect the growth and the blood vessel density of melanoma xenografts (Extended Fig. 5f,g). This is in contrast to mitogenic20, 24 and angiogenic roles of MDK in other settings26, 27. Instead, in melanoma xenografts, MDK shRNAs decreased lymphatic vessel density (LVD) and reduced tumoral V3-Luc (Extended Fig. 5 h,i, and Fig. 3e, respectively). Nevertheless, the most obvious effect of MDK downregulation was found systemically, with a marked inhibition of bioluminescence and metastasis to LNs (Fig. 3d–g) and visceral organs (Fig 3h). In turn, GoF (restoring MDK expression; Extended Fig. 6a) was sufficient to convert the non-lymphangiogenic WM164 into a potent inducer of systemic V3-Luc (Fig. 3i,j), favoring metastatsis to LN (Fig. 3k) and distal sites (shown for lungs in Fig. 3l and Extended Fig. 6b), without affecting VEGFC/D mRNA, nor the growth and vascularization of the cutaneous lesions (Extended Figs 6c,d, and data not shown).
Further supporting a pro-lymphangiogenic role of MDK, ectopic expression of this protein in LEC induced Vegfr3 mRNA (see qRT-PCR analyses in Extended Fig. 6e). Moreover, conditioned media from MDK-expressing melanoma cells enhanced LEC proliferation, while media from LoF analyses had an inhibitory effect (Extended Figs. 6f,g). In addition, confocal immunofluorescence in MDK-GoF demonstrated that MDK accumulated at sites of nodal neolymphangiogenesis in vivo (see Fig. 3m for colocalization with Lyve1-LEC in LN, including tip cells of sprouting lymphatic vessels; and Extended Figure 6h for additional detail). MDK enrichment in lymphatic vessels could also be detected in visceral organs such as lungs or liver before tumor cell colonization (Extended Fig. 6i and data not shown).
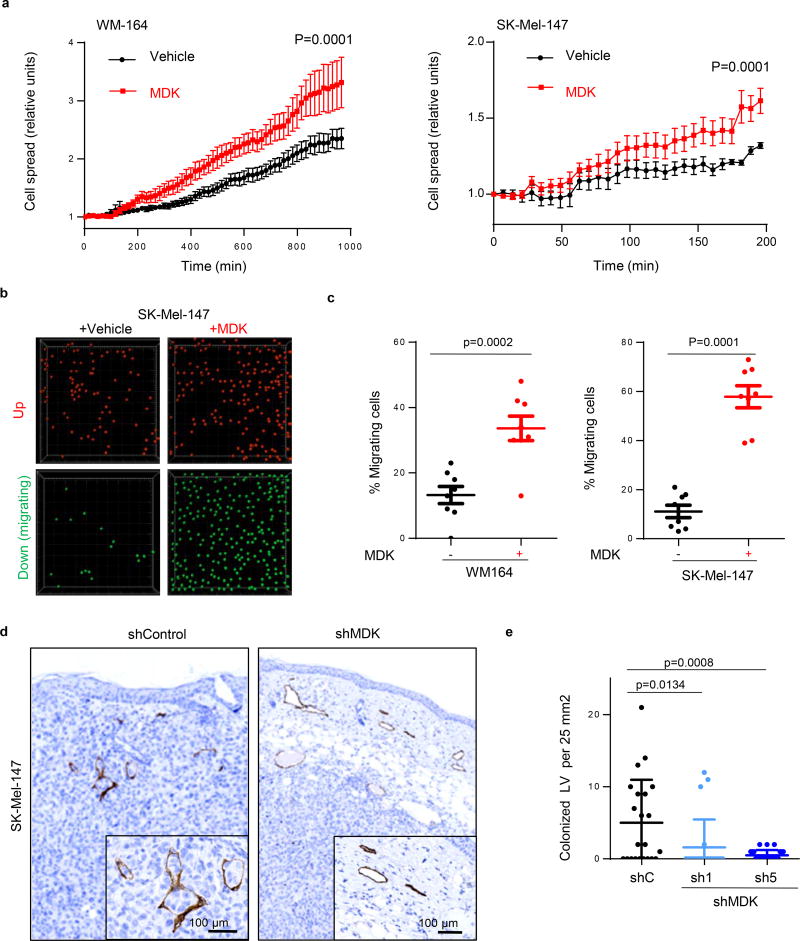
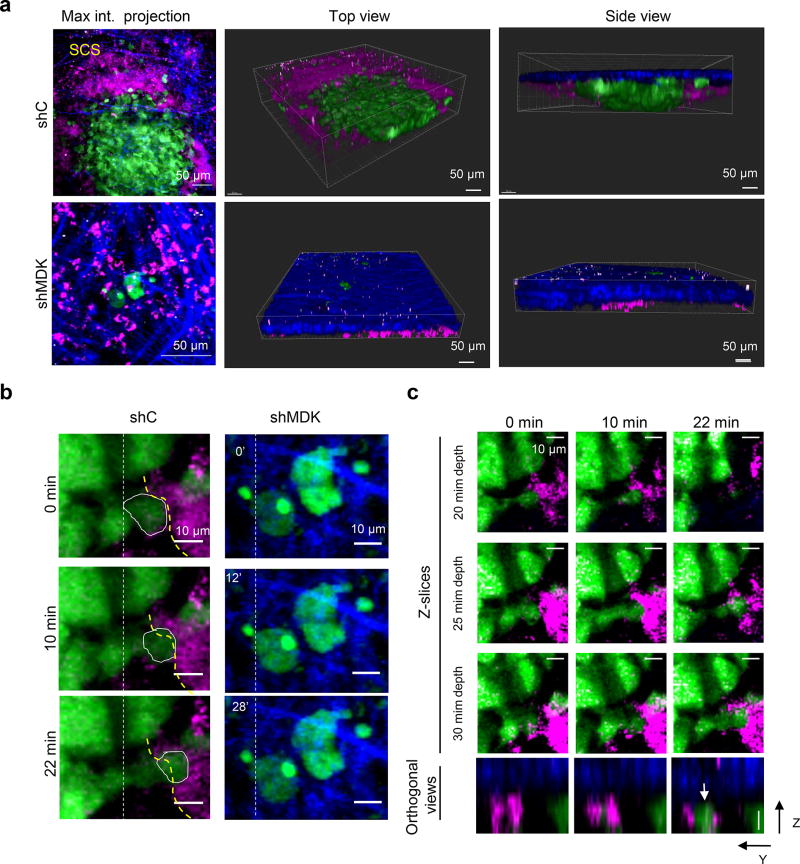
Conceptually, increased lymphatic vessel density would not be expected to promote metastasis per se, unless it is coupled to an effective intra/extravasation of tumor cells. Time-lapse videomicroscopy revealed that incubating LEC with MDK increased the adhesion of WM164 or SK-Mel-147 (Videos S1 and S2, respectively; see quantifications in Extended Fig. 7a), and their transmigration properties (Extended Fig. 7b,c). This inhibitory effect of MDK shRNA on the intravasation of melanoma cells through LEC was confirmed histologically (Extended Fig. 7d,e). Moreover, the prometastatic ability of MDK was further reinforced by intravital imaging of melanoma cells invading the subcapsular sinus at sentinel lymph nodes (Extended Fig. 8; see Methods for detailed explanation of the imaging procedures).
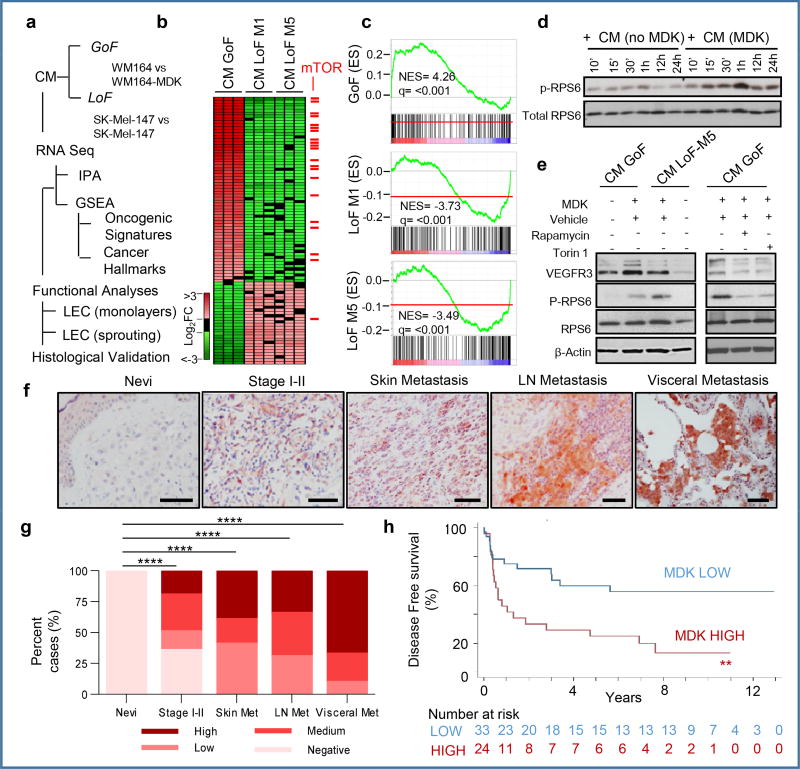
RNA sequencing was performed in LECs incubated with conditioned media from GoF and LoF studies, to identify downstream effectors of MDK (Fig. 4a). Genes with opposing regulation in these GoF and LoF are shown as heatmaps in Fig. 4b. Ingenuity Pathway Analyses (IPA) and gene set enrichment analyses (GSEA) identified multiple Cancer Hallmarks and Oncogenic Signatures as putative downstream effectors of MDK (Table S3, Table S4). These included the mTOR pathway as one of most enriched MDK-associated signaling cascades (Fig 4b,c).
Figure 4. MIDKINE activates the mTOR pathway and is an indicator of poor prognosis in melanoma.
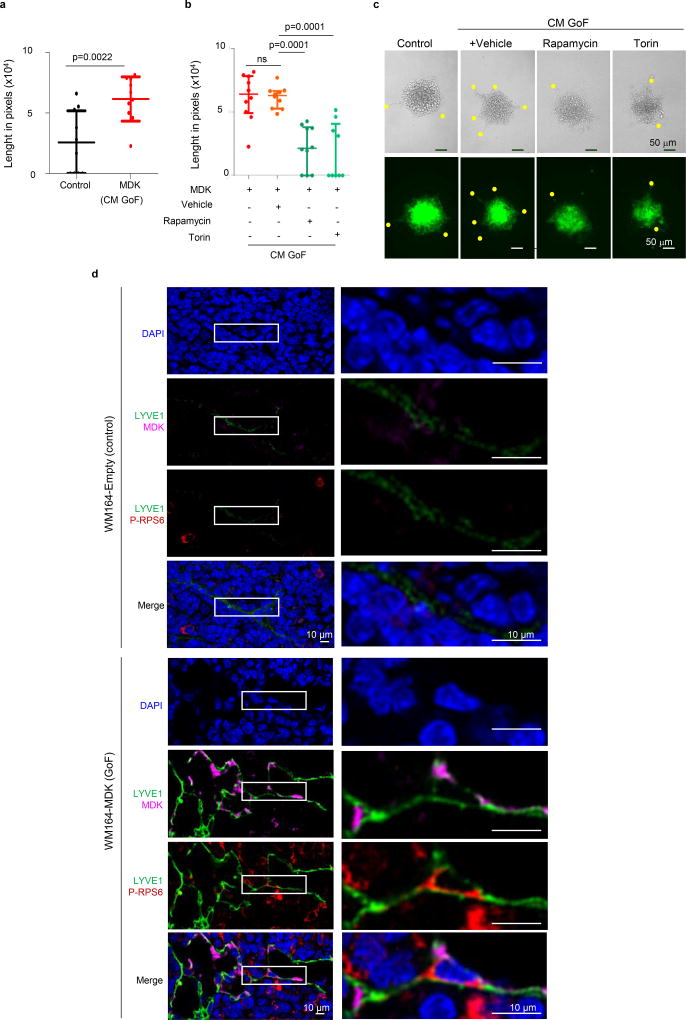
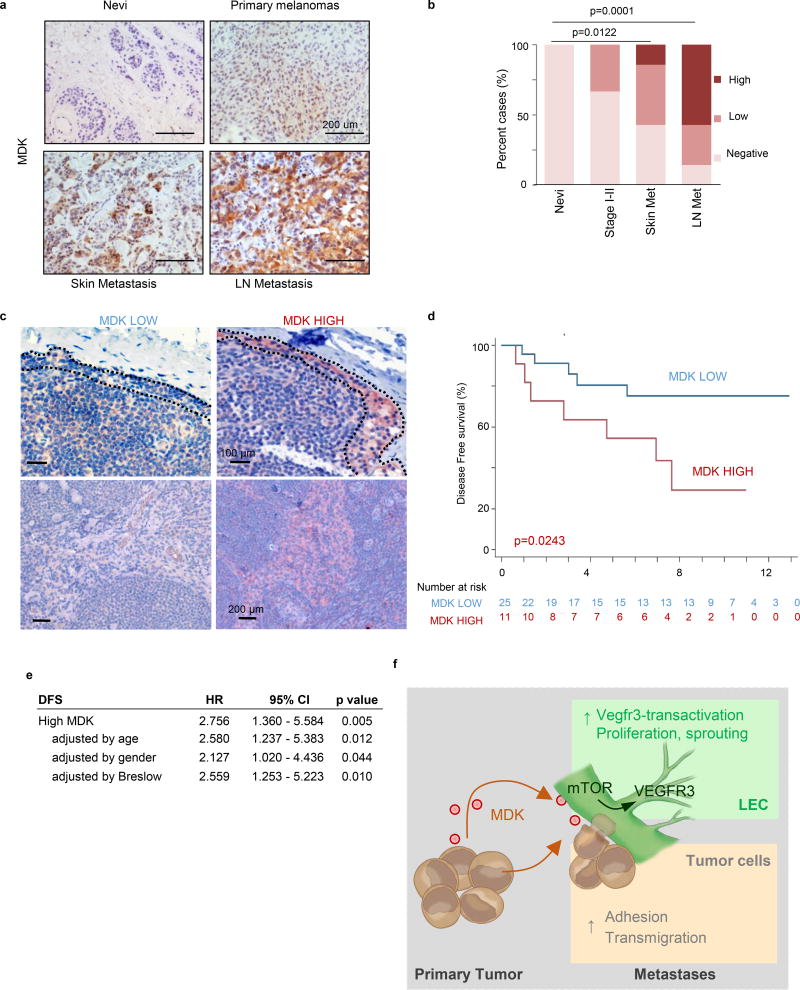
a, Identification of downstream effectors of MDK in LEC by RNA-seq and the corresponding validation analyses. b, Heatmap of deregulated genes (fold change >1.5), with mTOR pathway-associated factors in red. c, Enrichment score plots for mTOR-related pathways. d, Time-course analyses of RPS6 phosphorylation in LEC incubated with CM from control or MDK-GoF analyses. e, Visualization by immunoblotting of the impact of conditioned media (CM-GoF or CM-LoF) on VEGFR3, and total and phosphorylated RPS6 in human LEC. When indicated, LECs were incubated with the mTOR inhibitors rapamycin (1 µM) or torin 1 (10 nM). f, MDK immunohistochemistry (pink) in paraffin sections of the indicated human biopsies, with data quantified in (g). h, Kaplan-Meier survival curves of melanoma patients (Stage II–IV) classified on the basis of low and high-MDK expression in the corresponding sentinel LN. Fisher's exact test.
The mTOR pathway was intriguing because in addition to its long-known contribution to tumor cell growth and motility, it has been found to modulate lymphangiogenesis28, 29, although with no previous link to MDK. The conditioned media (CM) of MDK-expressing melanoma cells promoted the phosphorylation of the mTOR target RPS6 in LEC (Fig 4d). MDK-induced phospho-RPS6 was accompanied by VEGFR3 induction (Fig. 4e) and LEC sprouting (Extended Fig. 9a). The dependency on MDK and mTOR in these LEC-associated effects were confirmed by CM from MDK-depleted cells and the use of the mTOR inhibitors torin 1 and rapamycin (Figs. 4e and Extended Figs. 9b,c). Moreover, histological analyses in Vegfr3Luc xenografts demonstrated mTOR pathway activation (phospho-RPS6) in vivo, at sites of MDK-induced neolymphangiogenesis (Extended Fig. 9d). Together, these results identify a functional role of MDK in melanoma as a mechanistic link between two key pro-tumorigenic processes: neolymphangiogenesis and mTOR signaling.
A retrospective series of human benign nevi and melanomas (confirmed a tumor-associated expression of MDK (see two cohorts in Fig. 4f,g and Extended Fig. 10a,b). Moreover, an independent series of sentinel LN of 62 patients at Stage II–III melanoma showed that patients with high nodal MDK expression (Extended Fig. 10c) have a significantly poor disease free survival (DFS, p=0.0034; Fig. 4h), even in cases with tumor-negative sentinel LN (p=0.0243; Extended Fig. 10d). Multivariate analyses confirmed that the correlation between high MDK expression and reduced DFS is independent on the age and gender of the patient and importantly, on the thickness (Breslow depth) of the primary lesion (Extended Fig. 10e).
The immunocompetent and immunodeficient Vegfr3-“lymphoreporter” melanoma mouse models generated in this study have uncovered distinct patterns of metastatic progression that can bypass the requirement of intra- or peri-tumoral lymphangiogenesis. Live imaging coupled to loss and gain of function studies revealed unexpected roles of MDK on LEC via paracrine effects of the mTOR pathway on VEGFR3 expression (see model in Extended Fig. 10f). Moreover, the physiological relevance of MDK was validated by multivariate analysis of disease-free survival of melanoma patients. We anticipate that the genome-wide proteomic RNA-seq data presented here, with modulation of pro-inflammatory molecules in melanoma cells and LEC, may open new areas of investigation. For example, the accumulation of MDK at the LN subcapsular sinus (Extended Figs. 8, 10c), may affect functions of this structure as a physical and immunological barrier to tumor cell dissemination30. In this context, it will be interesting to determine whether MDK (and mTOR) cooperate with other factors we found enriched in the secretome of metastatic melanoma cells. As the Vegfr3Luc reporters in this study are not limited to melanoma, they may represent a versatile “metastasis-alert” platform for gene discovery and drug testing in a broad spectrum of malignant diseases.
METHODS
Mouse breeding and induction of nevi and melanomas in Vegfr3Luc GEMM
The Vegfr3Luc nu/nu immunodeficient mice and the four Tyr:CreERT2 immunocompetent strains (Vegfr3Luc; BrafV600E and Vegfr3Luc; BrafV600E; PTENlox/lox) generated in albino- and black-coat animals are summarized in Fig. 1a. Strains used in this study for the crosses are as follows: Vegfr3Luc (Flt4tm1.1Sgo), initially engineered in CD-1;129Sv;C57BL6/J-albino background17; nu/nu mice (Crl:NU(Ico)Foxn1nu); Tyr::CreERT2/1Lru (ref31); BrafCA (Braftm1Mmcm)19 and Ptentm2Mak (ref32). Albino and black coat animals were bred independently. Mice were genotyped as previously described19, 31, 32. Nevi and melanomas in the Tyr::CreERT2 strains were induced in 14 week-old mice by topical treatment with 5 µl of 5 mM 4-hydroxy-tamoxifen as previously described19. Animals were maintained in an inbreed background and genotyped as described by the manufactures. All experiments with mice were performed in accordance with protocols approved by the Institutional Ethics Committee of the CNIO and the Instituto de Salud Carlos III (see below for maximum size of the tumor lesions).
Cell culture
Human primary melanocytes were isolated from neonatal foreskins33 and maintained in Medium 254 supplemented with melanocyte growth factors (HMG-1) containing 10 ng/ml phorbol 12-myristate 13-acetate (Invitrogen, Carlsbad, CA). Melanoma cells34 were cultured in DMEM (Invitrogen) supplemented with 10% FBS, 100 µg/mL of penicillin and 100 µg/mL of streptomycin. See also supplementary Table 1 for a list of melanoma cells used and their genetic background. Human LN endothelial cells (HLEC) Innoprot (Derio – Bizkaia, Spain) were cultivated in Clonetics™ EGM™-2 MV BulletKit™ (Lonza, Basel Switzerland) following manufactured instructions. HLEC were used within 6 passages in culture. Al the melanoma cell lines have been authenticated using GenePrint® 10 Loci Service and all the cell lines were tested for mycoplasma contamination routinely.
Gene silencing and overexpression by lentiviral transduction
MDK silencing was performed by lentiviral-driven expression of shRNAs, with pLKO-constructs purchased from Sigma (St. Louis, MO): MDK-sh1 (Clone ID NM_002391.3-505s21c1); MDK-sh2 (238s21c1); MDK-sh3 (337s21c1); MDK-sh4 (421s21c1); MDK-sh5 (621s21c1). Non-Target shRNA (CAACAAGATGAAGAGCACCAA) was used as control. Infected cells were selected by incubation with puromycin (1µg/mL) and MDK downregulation was determined by protein immunoblotting, ELISA or qRT-PCR (see below). For the overexpression of human MDK, ORF lentiviral expression vector pReceiver-Lv105-A0792 (MDK) and the corresponding empty vector were purchased from Genecopoeia (Rockville, MD). Viruses were produced in 293FT cells and infections were performed as previously described35. When indicated, cells were stably infected with mCherry pLV-puro lentiviral vectors as reported before35.
Non-invasive imaging of tumor growth and neo-lymphangiogenesis in vivo by biolumiscence and fluorescence optical imaging
For the analyses of xenografts from human melanoma cells, subcutaneous implantations were performed with mCherry labeled cells (1–5×106 cells) in the flank of 14-week old Vegfr3Luc nu/nu mice. PDX were generated from freshly isolated biopsies of skin metastases. Tumor growth was monitored every two days by caliper measurement of the two orthogonal large and small external diameters (a, b), and estimated using the formula V=a × b2 × 0.52. Non-invasive imaging of mCherry fluorescence or V3-Luc bioluminiscence were performed using an IVIS-SPECTRUM imaging system (Perkin Helmer, Baesweiler Germany). Animals were anesthetized with isoflurane and injected intraperitoneally with 150 mg/kg luciferin (Perkin Helmer). Sequential images were obtained after luciferin injection and the maximum light emission was determined for each animal as previously described17. Photons emitted from specific regions were quantified using Living Image software (Caliper Life Sciences). In vivo dual fluorescence/bioluminescence imaging was performed at different time points after cell implantation, until tumors reached an average final size of 1500 mm3 or an average diameter of 1.5 cm (set to follow ethical procedures for humane handling of animals36). Post hoc analyses were then performed in representative cell lines for an in depth quantification of V3-Luc emission at the tumor, inguinal and brachial lymph nodes and in visceral organs (spleen, liver and lung), corresponding to animals with defined tumor sizes (i.e. 0, 250, 500, 1000 and 1500 mm3), to control for the varied growth rates of the different cell lines.
The IV parterns of V3-Luc emission identified from the human melanoma cell lines tested as xenografts in Vegfr3Luc nu/nu mice were as follows: Pattern I (e.g. WM164) corresponded to the expected behavior of low VEGFC expressing xenografts, namely, no signs of V3-Luc emission at any anatomical site (Fig. 1d,e, top panels), not altered lymphatic density (Extended Fig. 2c) and infrequent metastatic colonization of sentinel LN, as determined by fluorescence-based imaging (Extended Fig. 2d). Pattern II was exemplified by WM35, a VEGFC expressing cell line (Extended Fig. 1c,d). In this case, a potent induction of V3-Luc was detected at the tumor site, but curiously, with minor distal luciferase emission, and no significant nodal metastatic potential (second row in Fig. 1d, e; Extended Fig. 2c,d). Pattern III was constituted by metastatic cell lines (WM902B, UACC62, SK-Mel-103 and SK-Mel-147) with local and distal activation of Vegfr3Luc (third row in Fig. 1c,d; Extended Fig. 2a,b), and nodal metastases (Extended Figure 2d), but with no correlation to VEGFC mRNA expression (Extended Fig. 1c). Pattern IV was even more intriguing, as it corresponded to cell lines (WM115, 451LU and SK-Mel-28) that did not express significant VEGFC and induced minimal or highly delayed tumoral bioluminescence, but still promoted a marked V3-Luc emission at LN and subsequently at spleen, liver and lung (bottom row in Fig. 1d,e; Extended Fig. 2d–f).
Immunocompetent Vegfr3Luc-GEMM nevi and melanoma were imaged following similar techniques and procedures as for Vegfr3Luc nu/nu mice, with animals euthanized when tumor areas covered a maximum 700 mm2 of the mice. Sample sizes were chosen to ensure statistical significance. No randomization or blind analyses were performed. All experiments with mice were performed in accordance with protocols approved by the Institutional Ethics Committee of the CNIO and the Instituto de Salud Carlos III36.
Protein immunobloting and histological analyses of gene expression in mouse tumors
For Western blotting, primary and secondary antibodies were diluted in 5% milk TBS-T incubated overnight at 4°C or 1 hour at RT. Primary antibodies used were: MIDKINE (sc-1398, Santa Cruz), VEGFR3 (Millipore), RPS6 (5G10, Cell Signaling, Danvers, MA), P-RPS6 (Ser240/244; #2215, Cell Signaling) and Actin (Sigma-Aldrich, A5441). HRP-conjugated secondary antibodies used were anti-mouse and anti-rabbit (GE Healthcare) or anti-goat (Jackson Immunoresearch).
For analyses of tissue architecture and expression of lymphangiogenic markers, tissues were fixed in formalin and embedded in paraffin. Sections were prepared for hematoxylin-and-eosin (H&E) staining. For immunostaining, 3 µm paraffin sections were deparaffinized and placed in PBS. Endogenous peroxidase activity was quenched with 3% hydrogen peroxide. Sections were incubated with blocking solution (DAKO, Copenhagen, Denmark) to reduce non-specific staining. Incubation with the indicated antibodies (see below) was then followed by staining with the appropriate biotinylated secondary antibodies (DAKO). Tissue sections were then incubated with Envision-Peroxidase Rabbit Ready-to-Use (DAKO) using diaminobenzidine (DAB) as a substrate for the peroxidase reaction. Sections were finally counterstained with hematoxylin (Anatech, Battle Creek, MI) and mounted with permanent mounting medium. For MDK and VEGFC histological analyses, Ultravision ONE Detection System (RTU, Thermo Scientific, Waltham, MA) was used following manufacturer’s protocols. For immunofluorescence-based analyses, tissue sections were deparaffinized, incubated overnight with primary antibodies at 4 °C in a humidified chamber and then rinsed and incubated with fluorescent secondary antibodies for 1 hour at room temperature. Nuclei were counterstained with Prolong Gold + DAPI (Invitrogen, concentration 5µg/mL) 20 minutes before imaging. Primary antibodies used were: hVEGFR3 (MAB3757, Millipore), mVegfr3 (AF743, R&D Systems), mLyve-1 (ab14917, Millipore), hLYVE-1 (ab36993, Abcam), CD-31(ab28364, Abcam), MIDKINE (sc-1398, Santa Cruz), VEGFC (NB110-61022, NB110-61022), RPS6 (5G10, Cell Signaling, Danvers, MA), P-RPS6 (Ser240/244; #2215, Cell Signaling, Danvers, MA) CD169 (clone 3D6.112; AbD Serotec), MCSF antibody (ab183316, Abcam).
Assessment of metastatic tumor cell burden in Vegfr3Luc mice
1×106 mCherry-labeled SK-Mel-147 melanoma cells were implanted subcutaneously in Vegfr3Luc nu/nu immunodeficient mice. Animals were randomized in 4 groups for weekly analyses/collection. At the desired times, V3-Luc emission was measured as described above. Sentinel lymph nodes and lungs were collected and samples were preserved in RNAlater® (ThermoFisher, Cat. AM7020). RNA and DNA purification from tissue samples were performed using AllPrep DNA/RNA Mini Kit (Qiagen, Cat. 80204). cDNA preparations were performed using the High-Capacity cDNA Reverse Transcription Kit (ThermoFisher, Cat. 4368814). Quantification of melanoma cells was performed by qRT-PCR, using primers for mCherry and 18S as described in the qRT-PCR section. Standard curves for the estimation of cell number were performed using defined numbers of SK-Mel-147-mCherry cells.
Quantitative real-time reverse transcription-PCR (qRT-PCR)
RNA purification from melanoma tissue samples and qRT-PCR were performed using the following primers:
| hVEGFC | hVEGFC-F | TGCCAGCAACACTACCACAG |
| hVEGFC-R | GTGATTATTCCACATGTAATTGGTG | |
| hVEGFD | hVEGFD-F | GGAGGAAAATCCACTTGCTG |
| hVEGFD-R | GCAACGATCTTCGTCAAACA | |
| hVEGFR3 (Flt4) | hVEGFR3-F | CAAGAAAGCGGCTTCAGGTA |
| hVEGFR3-R | GCAGAGAAGAAAATGCTGACG | |
| MDK | MDK-F | CCTGCAACTGGAAGAAGGAG |
| MDK-R | CTGGCACTGAGCATTGTAGC | |
| mCherry | mCherry-F | CCCGCCGACATCCCCGACTA |
| mCherry-R | GGGTCACGGTCACCACGCC | |
| 18s | 18s-F | TTGGAGGGCAAGTCTGGTG |
| 18s-R | CCGCTCCCAAGATCCAACTA |
HLEC proliferation assay
To define the impact of MDK-overexpressing melanoma cells on the growth of HLEC, conditioned media was collected from WM164 cells infected with control (empty vector) or MDK-expressing lentiviruses. Melanoma cells were cultured in DMEM supplemented with 10% of FBS until they reach 80% of confluence. Medium was then changed to EGM™-2 MV for 24 hours. This conditioned medium (CM) was collected, centrifuged for 5 minutes at 1500 rpm and filtered throught a 0.45 µm pore membrane before use. CM was then added to human LEC (HLEC) 24 h after plating (6 well plates, 2×105/per well) for a 5-day analysis of proliferation rate using the Promega’s CellTiter MTS cell proliferation assay kit. Conversely, growth of HLEC was performed on cells pre-conditioned with media from SK-Mel-147 expressing shC or shMDK(1) and shMDK(5) to determine inhibitory effects after MDK depletion.
HLEC sprouting assay
HLECs were cultured in EGM™-2 MV medium (Lonza). Hanging drop cultures were generated 24 h after incubation with Cell Tracker (Molecular Probes, Life Technologies, Madrid, Spain), using 1000 cells per drop (25 µl). This was performed in EBM-2 medium containing 20% Methocel. Sphere formation was checked by microscopy 24 h thereafter. Spheres were then collected in PBS supplemented with 10% of FBS, centrifuged for 3 minutes at 200 rpm, and resuspended in the desired EGM™-2 MV conditional medium obtained as described before. When indicated, spheres were treated with 1 µM rapamycin or 10 nM torin 1 and embedded in type I collagen. Pictures were collected 24 hours thereafter. Number and length of the sprouts were quantified using Image J software.
Melanoma cell attachment and transmigration through HLEC
HLEC were seeded in a 24-well glass bottom plate and then incubated at 37 °C with 5% CO2. At confluency, media was refreshed in the absence or presence of 500 µg/ml recombinant human MDK (Peprotech, Rocky Hill, NJ) for a 16 hours incubation. 105 mCherry labeled SK-Mel-147 or WM164 cells were then seeded on top of the pre-conditioned HLEC cells for immediate time-lapse imaging (fluorescence videomicroscopy using a wide-field microscope coupled with a CCD camera). For SK-Mel-147 cells, images were captured at 2.33 min intervals for a total of 200 minutes, and for WM164 every 3.33 min (total of 965.7 minutes). Videos were assembled using ImageJ software, which was also used to quantify melanoma cell surface (mCherry-positive areas) at the indicated acquisition times. Results are represented as the average and SEM of the fold-induction of the mCherry-covered area per frame, analyzed in 4 different wells (n>200 cells per time point and condition). t-Test analyses were performed on time-matched observations to define statistical differences in the spreading capacity of melanoma cells on control- vs MDK-conditioned HLEC.
Transmigration analyses were performed on 8 µm pore Boyden chambers containing confluent HLEC monolayer (preconditioned for 24 h with empty media or with media contained 500 µg/ml recombinant human MDK). 105 mCherry labeled SK-Mel-147 or WM164 cells were seeded on top of these pre-treated HLEC cells using DMEM medium without growth factors. As chemo-attractant, complete DMEM media was added to the bottom of the plates. Cells were incubated in these conditions for 16 h for subsequent fixation in 4% paraformaldehyde. Chambers were then scanned using a Leica SP5-multifoton confocal microscope (Leica Microsystems, Weztlar, Germany). The number of melanoma-mCherry cells in the upper part (non-migrating) and lower part (migrating) of the chamber was quantified using the Imaris image analysis software (Bitplane, Zurich, Switzerland). t-Test was performed on matched observations using Graphpad Prism software.
Exosome purification
Exosomes were prepared essentially as described37. Briefly, cells were cultured in media supplemented with 10% exosome-depleted FBS (FBS, Hyclone). FBS was depleted of bovine exosomes by ultracentrifugation at 100,000g overnight. Supernatant fractions collected from 24 h cell cultures were pelleted by centrifugation at 500g for 10 min. The supernatant was centrifuged at 20,000g for 20 min. Exosomes were then harvested by centrifugation at 100,000g for 70 min. The exosome pellet was resuspended in 13 ml of PBS and collected by ultracentrifugation at 100,000g for 70 min. The exosome producing cells were trypsinized, counted and stored for proteomic analysis.
ELISA-based analysis of MDK secretion
For the ELISA estimation of soluble or exosome-contained MDK, cells were incubated for 24 h in DMEM-10% FBS before collection. The conditioned media was then divided in two fractions, one for elimination of cell debris by centrifugation for 13000 rpm for 10 min, and another to deplete exosomes by ultracentrifugation at 100000g for 14 h. MDK expression in these soluble and exosome-cleared conditions was analyzed by ELISA (Human MIDKINE ELISA Development Kit 900-K190; Prepotech). Readings were obtained for three independent biological replicates. To define MDK concentration in exosomes, these were lysed with ELISA extraction buffer 75 mM Tris pH 7.0; 150 mM NaCl; 1 mM EGTA; 1mM EDTA and 1% Triton 100; 0.5% Sodium deoxycholate and protease and phosphatase inhibitors.
LC-MS/MS analysis of exosome cargo
For exosome profiling, samples were digested by means of the standard FASP (Filter Aided Sample Preparation) protocol38. Briefly, samples were resuspended in UT buffer (8 M urea in 100 mM Tris-HCl, pH=8.01). Proteins were then reduced with 10 mM DTT, alkylated using 50 mM IAA for 20 min in the dark. Proteins were digested with Lys-C (Wako, Neuss, Germany) during 6 hours (1:50). Finally, samples were diluted in 50 mM ammonium bicarbonate to reduce the urea concentration to 1 M, and were subsequently digested with Trypsin (Promega, Madison, WI; 1:100 sample concentration, overnight at 37 °C). Resulting peptides were desalted and using micro-columns filled with Poros Oligo R3 beads (Life Technologies). Samples were dried and dissolved in 30 µL of 0.1% formic acid (FA). Peptides were separated by RP chromatography using a nanoLC Ultra system (Eksigent, Dublin, CA), directly coupled with a LTQ-Orbitrap Velos instrument (Thermo) via nanoESI (ProxeonBiosystem, Waltham, MA). Peptides were loaded onto a Reprosil-Pur C18 column (3 µm, 400×0.075 mm; Dr. Maisch, Ammerbuch-Entringen Germany), with a trapping column (Prot Trap Column 0.3×10 mm, ReproSil C18-AQ, 5 µm), for 10 min with a flow rate of 2.5 µL/min of loading buffer (0.1% FA). Elution was performed with a 120 min linear gradient (buffer A: 2% ACN, 0.1%FA; buffer B: 100% ACN, 0.1%FA) at 300 nL/min. Peptides were directly electrosprayed into the mass spectrometer using a PicoTip emitter (360/20 OD/ID µm tip ID 10 µm, New Objective) at 1.4 kV spray voltage with a heated capillary temperature of 325°C and S-Lens of 60%. Mass spectra were acquired in a data-dependent manner, with an automatic switch between MS and MS/MS scans using a top 10 method. MS spectra were acquired with a resolution of 60000 (FWHM) at 400 m/z in the Orbitrap, scanning a mass range between 350 and 1500 m/z. Peptide fragmentation was performed using collision-induced dissociation (CID), set at 35%. Label-free quantification was performed in MaxQuant (1.5.1.2) using default settings and further statistical analysis of the quantitative data was conducted with Perseus (1.5.1.6). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD002703. The proteomic analysis resulted in the identification and quantification of a total of 1,705 exosomal proteins (Fig. 3a). A statistical filter (cut off of FDR=0.05 and a threshold S0=0.8) was applied to identify factors enriched with respect to the non-metastatic and non-lymphangiogenic WM164. A second filter was used to eliminate factors shared with cell line WM35 (with lymphangiogenesis restricted mostly to cutaneous lesions). This approach identified 139, 180 and 188 candidates for long range-acting metastatic factors in 451LU, SK-Mel-28 and SK-Mel-147, respectively, as summarized in the Volcano Plots of Fig. 3b (see Table S2). The upper 20% of these proteins were then scored on the basis of three criteria: (i) being common to all cell lines that activate distal lymphangiogenesis, (ii) with described functions associated with metastasis but not previously linked to lymphangiogenesis, and (iii) understudied in melanoma. The highest ranked protein in this analysis was the heparin binding factor MIDKINE/Neurite growth-promoting factor 2 (MDK, marked in blue in Fig. 3b)
RNA sequencing (RNA-Seq) and bioinformatic analyses
RNA sequencing was performed by the CNIO Genomics Unit. 1 µg of total RNA from the samples was used. Average sample RNA Integrity Number was 9.75 (Agilent 2100 Bioanalyzer). PolyA+ fraction was purified and randomly fragmented, converted to double stranded cDNA and processed through subsequent enzymatic treatments of end-repair, dA-tailing, and ligation to adapters as in Illumina's "TruSeq Stranded mRNA Sample Preparation Part # 15031047 Rev. D" kit (this kit incorporates dUTP during 2nd strand cDNA synthesis, which implies that only the cDNA strand generated during 1st strand synthesis is eventually sequenced). Adapter-ligated library was completed by PCR with Illumina PE primers (8 cycles). The resulting purified cDNA library was applied to an Illumina flow cell for cluster generation and sequenced on an Illumina HiSeq2500 by following manufacturer's protocols. 50bp single-end sequenced reads were analyzed with the nextpresso pipeline (http://bioinfo.cnio.es/nextpresso/), as follows: sequencing quality was checked with FastQC v0.11.0 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Reads were aligned to the human genome (GRCh37/hg19) with TopHat-2.0.10 39 using Bowtie 1.0.040 and Samtools 0.1.1941, allowing 2 mismatches and 20 multihits. Transcripts quantification and differential expression were calculated with Cufflinks 2.2.139, using the human GRCh37/hg19 transcript annotations from https://ccb.jhu.edu/software/tophat/igenomes.shtml. GSEAPreranked42 was used to perform gene set enrichment analysis of the described gene signatures on a pre-ranked gene list, setting 1000 gene set permutations. Only those gene sets with significant enrichment levels (FDR q-value < 0.25) were finally considered.
Intravital multiphoton microscopy
1×106 SK-Mel-147 cells labeled with GFP and transduced with control shRNA (shC) or MDK shRNA(5) were injected subcutaneously in 6 week-old female nude mice. Intravital imaging of the inguinal lymph node adjacent to the tumor was performed 3 weeks after tumor cell injection. To label the inguinal lymph node, 20 microliters of 155 kDa TRITC-dextran was injected intradermally at the lateral tail base on the tumor side. Animals were anesthetized with 5% isofluorane and the lymph node was exposed by skin flap surgery as previously described43. Imaging was performed using an Olympus FV1000 multiphoton microscope with a 25× 1.05 NA water immersion objective as previously described44 and adapted for the lymph node. Specifically, we focused on the subcapsular sinus, the outer area of lymph nodes that serves as a main barrier to tumor cell entry45, 46. Briefly, a Coherent Chameleon Vision II laser was tuned to 880 nm for excitation of GFP and TRITC-dextran. Collagen was visualized by second harmonic generation. 3D images were reconstructed with Imaris (Bitplane). 4D images were acquired at a 5 µm step-size ranging from 100–150 µm in depth from the lymph node surface with a 2 minute time interval for 30 minutes. Individual Z-slices are provided at different time points displayed in Figure S7 C (3 different planes containing the cell volume, starting at 20 microns depth from lymph node capsule with 5 microns step size) showing no Z-drift. Orthogonal Z-Y views are also provided to visualize the three dimensional location of the cell in relation to the lymphatic vessel over time. Images were analyzed in Image J.
As shown in Extended Fig. 8a (upper panels) MDK expressing SK-Mel-147 cells were found in clusters invading the subcapsular sinus at sites of a large expansion of the vasculature (see dextran-labeled areas). This was in contrast to individual or small cell clusters of MDK deficient SK-Mel-147 cells found blocked at the border of the subcapsular sinus on the collagen enrich area (Extended Fig. 8a, bottom panels). Snap shots in Extended Fig. 8b,c further illustrate the impact of MDK deficiency in the ability of melanoma cells to move towards the vasculature. Therefore, these data provide additional mechanistic explanation as to why modulating the endogenous levels of MDK in melanoma cell lines not only affects their pro-lymphangiogenic capability, but ultimately their metastatic potential.
MIDKINE expression and metastatic risk in human clinical biopsies
MDK levels were estimated by two independent pathologists. LN and paraffin-embedded blocks were collected from a retrospective series of 62 melanoma patients (21–85 years old, with a median age of 65; Stage II–III at diagnosis). Gender distribution was 66.1% female and 33.9% male. Immunohistochemistry was performed in 3 µm sections of primary lesions or sentinel lymph nodes as indicated and MDK-specific staining was scored as negative, low or high taking into account the intensity of its expression. Survival curves were estimated with Kaplan-Meier product-limit method and compared using logrank test. Hazard ratios (HR) were calculated using Cox regression and adjusted with univariate and multivariate models for Breslow depth, age or gender. Disease Free Survival (DFS) was defined as the time interval between diagnosis and the development of the earliest metastasis detected at any anatomical site. DFS was considered censored for patients who did not present metastases at last follow-up. . Human tumor biopsies were obtained from the i+12 Biobank (RD09/0076/00118) of the Hospital 12 Octubre, the biobank from the Hospital Clínic de Barcelona, and the Spanish National Biobank Network, with the corresponding informed consent and ethical protocols approved by their Clinical Investigation Ethical Committees.
Statistical analyses
Cell proliferation and tumor growth curves were analyzed by one way and two-way ANOVA with Dunnett’s multiple comparation text (mixed model) considering matching among the measures at different time points. P-values were indicated in each figure for statistical significant comparisons (p<0.05). For the analysis of proteomic data, LFQ values were log2 transformed and missing data were imputed based on the observed normal distributions (width value = 0.3 and down-shift = 2). Then, a two-sample Student’s Test was performed using the following settings and filters in the Perseus software: 250 randomizations, FDR = 0.05 and fudge factor S0 = 0.8. For simplicity, only significant p values are shown in all experiments.
Data Availability Statement
The mass spectrometry proteomics data have been deposited in the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD002703. RNA-Seq data have been deposited in NCBI's Gene Expression Omnibus with accession number GSE94549. Source data are provided for Figs 1e; 2a,b; 3e–l and Extended Figs 1d; 2d–f; 3b–e; 4a, 5a,c–h,I; 6b–e; 7a,c,e; 9a,b. All other data that support the findings of this study are available from the authors upon reasonable request.
Extended Data
Extended Data Figure 1. Non invasive imaging of neo-lymphangiogenesis in the Vegfr3Luc reporter mice.
a, qRT-PCR analysis of VEGFC mRNA levels of the indicated melanoma cell lines. Graphs average ± SD of 3 biological replicates. Data were normalized to mRNA levels in the poorly metastatic WM164. b, Immunostaining for VEGFC (pink) in xenografts of the indicated melanoma cell lines. c, Histological staining of Lyve1 (brown) in lymph nodes with low or high V3-Luc emission. d, Costaining of Vegfr3 (green) and Lyve1 LEC (brown) in the spleen of mice identified as low and high V3-Luc emitters in vivo. e, Lung metastases of animals as in (Fig 1c) imaged by immunofluorescence for the detection of Vegfr3 (green) and melanoma cells (red).
Extended Data Figure 2. Assessment of metastatic potential of melanoma cell lines in Vegfr3Luc mice.
a–b, Tumor-associated bioluminiscence driven by subcutaneous implantation of the indicated cell lines (labeled with mCherry) in Vegfr3Luc nu/nu mice, with images centered on the tumor (a) or showing the lateral flank of the mice to visualize sentinel-inguinal and brachial LN, as well as signal from spleen liver and lung (b). Four main patterns (I–IV) of Vegfr3Luc emission identified by are shown. Red lines mark the tumor area as detected by mCherry emission (data not shown). c, Detection of the lymphatic marker Lyve1 in histological sections of sentinel LN of xenografts generated as in (a), and euthanized when primary tumors reached 1500 mm3. d, Tumor-positive LN (Sentinel, Brachial and Axillar) in xenografts of the indicated cell lines. e, Relative V3-Luc signal in subcutaneous xenografts generated by the indicated cell lines. The average signal in sentinel and brachial LN are shown in (f). Data correspond to mean ± SE (minimum 6 mice per condition, 24 lymph nodes per cell line).
Extended Data Figure 3. Whole-body analysis of neo-lymhangiogenesis induced in immunodeficient and immunocompetent genetically engineered mouse models (GEMM).
a, Imaging of luciferase induction by a patient-derived xenograft isolated from a skin melanoma metastasis and implanted in Vegfr3Luc nu/nu mice. Animals were imaged at different time points to show distal lymph nodes induced prior detection of V3-Luc at the subcutaneous lesion. Dashed red lines mark tumor area. Scale p/s/cm2/sr (×106). b, Quantification of the luciferase signal in tumor, LN (sentinel and brachial sites), spleen, liver and lung, at the defined average tumor sizes indicated in the X-axis (visualized with a dotted red line for simplicity). Specific tumor volumes are indicated in the right Y-axis. Bioluminescence data correspond to mean ± SD (4 mice per condition, 16 lymph nodes per PDX). Statistical analysis: t-test. c, Comparative imaging of V3-Luc emission by benign nevi and cutaneous melanomas generated respectively in Tyr::CreERT2-inducible BrafV600E or BrafV600E; Ptenlox/lox mice (black coat background), imaged 4 weeks after topical administration of 4-OH-tamoxifen. Arrows mark systemic activation of luciferase. (Bottom), Quantification of luciferase signal in tumor, sentinel and brachial LN, spleen, liver and lung. Units: p/s/cm2/sr (×106). Data correspond to mean ± SD (4 mice for nevi and 10 animals for melanoma). Statistical analysis: One-way Anova with Dunnett's test for multiple corrections. d, Luciferase emission induced by cutaneous melanomas generated in immunocompetent Vegfr3Luc; BrafV600E; Ptenlox/lox (albino), visualized at the indicated times after topical administration of 4-OH-tamoxifen. e, Quantification of the luciferase signal in the indicated organs, as defined in the Methods section. The average tumor size in each point is indicated by a red doted line and in the x-axis. Data correspond to mean ± SD (4 mice per condition). Statistical analysis: t-test.
Extended Data Figure 4. Analysis of metastatic potential in Vegfr3Luc mice.
a, Pearson correlation analyses of the luciferase signal vs cell burden, corresponding to data presented in Fig. 2a. Shown are lymph nodes or lungs with luciferase signal over the background. b, V3-Luc emission in immunocompetent BrafV600E;Ptenlox/lox (albino) mice at the indicated times prior and after surgical removal of the primary cutaneous melanoma. Note the reduction of tumor-driven Vegfr3-Luc signal particularly in visceral sites, and the reactivation at later time points (black arrows), marking metastatic relapse. Fur was removed to ease in the imaging.
Extended Data Figure 5. MDK lost of function (LoF) impairs melanoma –induced lymphangiogenesis and metastasis.
a, Detection by MDK protein expression by immunohistochemistry in xenografts of the indicated cell lines. b, Secreted MDK in the indicated cell lines. Upper panel shows immunoblots of conditioned media containing (+) or being depleted of exosomes (-). Lower panel shows ELISA-based quantification of MDK in conditioned media of the indicated cell lines contained or being depleted for exosomes (black and grey bars, respectively). MDK levels in the purified exosome fraction (white bars) are indicted also as a reference. Data were normalized to levels in the complete soluble fraction (black), and correspond to mean ± SD of 3 biological replicates. c, Depletion of MDK mRNA detected by qPCR of SK-Mel-147 cells infected with lentiviruses coding for control or MDK shRNA (1) or (5). Data correspond to mean ± SD of 3 biological replicates. d, Impact of the indicated shRNAs in MDK secretion, analyzed by ELISA. e, q-RT-PCR analysis of VEGFC (left panel) and VEGFD (right panel) mRNA expression in SK-Mel-147 expressing control of MDK shRNA (1) or (5). Data correspond to mean ± SD of 3 biological replicates. f, Lack of significant impact of MDK shRNA (shown for shRNA (1) and (5)) on the growth of SK-Mel-147 as subcutaneous xenografts in Vegfr3Luc nu/nu mice. Data correspond to mean ± SD of 6 mice per condition. g, Blood vessel density (BLV) defined by IHC staining for CD31 in xenografts of SK-Mel-147 expressing control or MDK shRNA (1) or (5). h–i, Peritumoral or intratumoral lymphatic vessel density (LVD), respectively, determined by histological staining for Lyve1 in lesions as in (f). Data correspond to mean ± SD of 3 tumors per experimental condition.
Extended Data Figure 6. MDK accumulates at sites of neo-lymphangiogenesis.
a, Relative expression of secreted MDK in WM164 infected with control (empty) or lentiviruses expressing MDK cDNA (right), analyzed in parallel with respect to basal levels in primary melanocytes and the indicated cell lines (left). Data correspond to biological triplicates measured by ELISA, using purified recombinant protein as a reference for quantification. b, Representative images of lung metastases identified by mCherry fluorescence. c, qRT-PCR analysis of VEGFC (left panel) and VEGFD (right panel) mRNA showing minimal changes in control vs MDK-overexpressing WM164 cells. d, Assessment of peritumoral lymphatic vessel density (Lyve1 staining) in consecutive histological sections of xenografts generated by WM164 expressing MDK (and their parental controls). e, Increased VEGFR3 mRNA in MDK-expressing HLEC, determined by qRT-PCR. Data in graphs a-d correspond to mean ± SD of 3 biological replicates. f, Proliferative capacity of human LEC (HLEC) incubated with conditioned media from control- or MDK-overexpressing WM164 (left panels, red). Statistical analysis: t-Test. The converse analysis, namely, depletion of MDK in SK-Mel-147 is shown in g, (blue). Data are represented as the relative mean ± SD of three biological replicates. Statistical analysis: One-way ANOVA. h, Dual immunofluorescence analysis to visualize MDK (green) and lymphatic vessels (Lyve1, red) in Vegfr3Luc positive lymph nodes of mice implanted with WM164-expressing MDK. The equivalent analysis in lungs is shown in i.
Extended Data Figure 7. MIDKINE enhances the ability of melanoma cells to interact with and migrate through human LEC.
a, Attachment and spreading of mCherry-labeled WM164 or SK-MEL-147 melanoma cell lines on a confluent monolayer of human LEC (HLEC) preincubated for 16 hours with 500µg/ml recombinant MDK. Statistical analysis: t-Test. (See Extended Data Videos S1 and S2 for live imaging of the spreading capacity of WM164 and SK-Mel-147 melanoma cells respectively in control vs MDK-treated HLEC cells). b, Migration of SK-Mel-147 through a confluent layer of HLEC in the absence or presence of recombinant MDK (500 µg/ml). Pictures show representative images of cells retained (up, pseudocolored in red) or transmigrating (down, green) through the HLEC layer defined by confocal videomicroscopy as described in the Methods section. c, Quantification of the percentage of migrating WM-164 or SK-MEL-147 cells in conditions as in (c). Statistical analysis: t-test. d, Histological sections of xenografts generated by SK-Mel-147 expressing control or MDK shRNA and stained for the lymphatic cell marked Lyve-1 (brown) to visualize tumor cell intravasation. e, Quantification of the number of lymphatic vessels colonized by melanoma cells in (d) per field of 25 mm2 analyzed. Data correspond to mean ± SD of 3 biological replicates (with a minimum of 6 fields analyzed per tumor).
Extended Figure 8. Intravital multiphoton imaging of lymph node metastases from melanoma cells expressing or lacking MDK.
a, Maximum projection, top views or side views of lymph nodes from representative mice three weeks after implantation of SK-Mel-147 labeled with GFP and transduced with control shRNA (shC, upper panels) or MKD shRNA (shMDK, bottom panels). Tumor cells are visualized by green fluorescence, and collagen fibers by second harmonic generation (blue). Vessels at the subcapsular sinus (SCS) are stained with 155KD dextran (magenta). Note the horizontal and vertical growth of shC cells (i.e. metastasis expressing MDK), and the dissorganization of the surrounding SCS. Instead, shMDK cells were identified as single cells or rare micrometastases. Scale bar 50 microns b, Still images showing the differential intralesional motility of the indicated SK-Mel-147 cell populations from lesions in (a). The yellow dotted lines are shown to visualize the border of a dextran-labeled vessel in the context of a highly motile shC-melanoma cell (encircled with a solid white line). Vertical dotted white lines are included as reference to track cell movements. Scale bar 10 microns. c, Z-slices at different time points from the time images in (b). Bottom panels correspond to orthogonal Z-Y views at the indicated time points to show the three dimensional location of the cell (circled in b and marked with an arrow in c) with respect to the lymphatic vasculature. Scale bar 10 microns. Blue: Second harmonic generation, collagen; green: tumor cells; magenta: lymphatic vasculature.
Extended Data Figure 9. Modulation of lymphangiogenic features by MDK via the mTOR pathway.
a, Sprouting ability of collagen-embedded LEC (stained with CellTracker green for fluorescence imaging). Data correspond to the length of sprouts generated when LEC colonies were incubated with conditioned media (CM) from WM164 or from WM164-MDK. b, Length of sprouts of collagen-embedded LEC colonies in the absence or presence of mTOR inhibitors. Statistical analysis: t-Test; n=10. c, Micrographs to visualize LEC-sprouting assays in panels a, b. Yellow dots mark the end of sprouts. d, Immunofluorescence micrographs showing the expression of MDK (pink), lymphatic vessels (LYVE1, green) and p-RPS6 (red) in lymph nodes of a MDK Gain of Function (GoF) setting, whereby Vegfr3Luc mice were implanted subcutaneously with WM164 and its isogenic derivative WM164-MDK. Images were collected 2 weeks after implantation. White boxes mark magnified areas shown up the bottom.
Extended Data Figure 10. MDK expression in human melanoma specimens.
a, MDK expression shown by immunostaining (brown) in an alternative cohort of paraffin-embedded sections of representative examples of human benign nevus, stage I–II melanoma specimens and skin and lymph node metasases. b, MDK levels in the indicated melanocytic lesions scored as negative, low and high as indicated in the Methods section. Statistical analysis: Chi-square t-test; n=20 per category. c, Micrographs of low- and high-MDK expressing sentinel LN, focusing on subcapsular sinus (upper panels) and medullary areas (bottom panels). d, Kaplan-Meier survival curves of melanoma patients as in (fig 4h) considering only stage I–II (sentinel lymph node negative) patients classified on the basis of low and high-MDK expression in the corresponding sentinel LN. The number of patients at risk is shown for each time point. e, Multivariate analysis of the prognostic value of MDK expression on disease-free survival (DFS) showing independency with age, gender and Breslow depth. f, Proposed mode of action of MDK in the induction of lymphovascular-niches and metastasis in melanoma.
Supplementary Material
Time lapse video microscopy of mCherry-labeled WM164 melanoma cells (red) plated on top of a monolayer of HLEC preincubated for 24h with vehicle (left) or 500ug/ml recombinant MDK (right). at 3.33 min intervals for a total of 965.7 minutes to visualize adhesion and spreading of melanoma cells as a function of changes in the size and area of mCherry-emitting cells.
Time lapse video-microscopy of the metastatic SK-Mel-M17 cells labeled in red with mCherry to assay for increases in cell spreading and adhesion to HLEC preincubated for 24h with vehicle (left) or 500ug/ml recombinant MDK (right). Images were captured at 2.33 min intervals for a total of 200 minutes.
Acknowledgments
The authors thank the colleagues at the CNIO Melanoma Group, particularly to Albano Tenaglia for technical assistance at early stages of this study; José A Esteban (CBMSO, Spain) for critical reading of this manuscript; the i+12 Biobank of the Hospital 12 de Octubre (Madrid, Spain) for help with histological analyses and for providing patient biopsies; Lionel Larue (INSERM; France) and Martin McMahon (UCSD, USA) for Tyr:CreERT2 and inducible BrafCA mouse strains, respectively; the Animal Facility Unit and the Histopathology Core of CNIO for assistance. M.S.S. is funded by grants from the Spanish Ministry of Economy and Innovation (projects SAF2014-56868-R), the Asociación Española Contra el Cáncer (AECC), the Worldwide Cancer Research, and by Team Science and Established Investigator Awards by the Melanoma Research Alliance. The CNIO Proteomics Unit belongs to ProteoRed, PRB2-ISCIII, supported by grant PT13/0001. N.I. and J.M. are funded by SAF2013-45504-R (MINECO). J.M. is also supported by Ramon y Cajal Programme (MINECO) RYC-2012-10651. J.L.R-P and P.O-R are funded by grants FIS 2014/173711/02568 and FIS 11/17592014/01784, respectively, from the Spanish Ministry of Health. F.M. is funded by the AMIT Project/CDTI/CENIT Programme (MICINN), S.O. by SAF2013-44866-R (MINECO), and J.J.B-C. by an NCI K22CA196750 grant and the TCI Young Scientist Cancer Research Award JJR Fund (P30 CA196521). J. Di M. is recipient of a postdoctoral fellowship from the ARC Foundation and E.R-F from “Fundación Científica de la Asociación Española Contra el Cáncer”. D.C-W is a recipient of a predoctoral fellowship from Fundación La Caixa, and M.C-A and X.C. by the Immutrain Marie Skłodowska-Curie ITN Grant.
Footnotes
AUTHOR CONTRIBUTIONS
M.S.S. and D.O. conceived and designed all expression studies, mechanistic analyses and prognostic evaluations in this work. S.O. conceived, designed and developed the Vegfr3Luc (Flt4tm1.1Sgo) and Vegfr3Luc nu/nu mouse models (Strain 1 of this study), contributed to experimental design, discussed data and revised the manuscript. I.M-C generated the Vegfr3Luc immunocomptetent allele and the Vegfr3Luc nu/nu Strain 1, and contributed to their initial characterization. D.O. performed all the spatio-temporal studies of neo-lymphangiogenesis, as well as LoF and GoF studies of MDK in vivo. D.C-W contributed to mechanistic and histological analyses of lymphangiogenesis, helped with in vivo experiments and revised the manuscript. E.R-F, P.O.R. and J.L.R-P performed the analysis of patient prognosis (with bioinformatic aid from D.L. and S.P). P.P, M.C-A, and X.C contributed to functional analyses of lymphangiogenesis. N.I. and J.M. performed LC/MS-MS. F.M. helped with whole-body image analyses, D.M. with time-lapse confocal microscopy and M. Cañamero with histological evaluations. O.G and M. Cifdaloz performed RNA-seq. T.G.C. was in charge of animal breeding and genotyping. J.J.B-C., C.M and J. di M. performed intravital microscopy. J.S, L.O., J.M-T, E.C., T.G.C and D.A-C contributed with technical assistance. The manuscript was written by M.S.S and D.O., revised by S.O and D.A-C., and approved by all authors. M.S.S. supervised the project.
Competing financial interests. The authors have no competing financial interests
References
- 1.Zheng W, Aspelund A, Alitalo K. Lymphangiogenic factors, mechanisms, and applications. J Clin Invest. 2014;124:878–887. doi: 10.1172/JCI71603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stacker SA, et al. Lymphangiogenesis and lymphatic vessel remodelling in cancer. Nat Rev Cancer. 2014;14:159–172. doi: 10.1038/nrc3677. [DOI] [PubMed] [Google Scholar]
- 3.Karaman S, Detmar M. Mechanisms of lymphatic metastasis. J Clin Invest. 2014;124:922–928. doi: 10.1172/JCI71606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Akkooi AC, Eggermont AM. Melanoma: MSLT-1-SNB is a biomarker, not a therapeutic intervention. Nat Rev Clin Oncol. 2014 doi: 10.1038/nrclinonc.2014.64. [DOI] [PubMed] [Google Scholar]
- 5.Morton DL, et al. Final trial report of sentinel-node biopsy versus nodal observation in melanoma. N Engl J Med. 2014;370:599–609. doi: 10.1056/NEJMoa1310460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Padera TP, et al. Lymphatic metastasis in the absence of functional intratumor lymphatics. Science. 2002;296:1883–1886. doi: 10.1126/science.1071420. [DOI] [PubMed] [Google Scholar]
- 7.Proulx ST, et al. Use of a PEG-conjugated bright near-infrared dye for functional imaging of rerouting of tumor lymphatic drainage after sentinel lymph node metastasis. Biomaterials. 2013;34:5128–5137. doi: 10.1016/j.biomaterials.2013.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaplan RN, et al. VEGFR1-positive haematopoietic bone marrow progenitors initiate the pre-metastatic niche. Nature. 2005;438:820–827. doi: 10.1038/nature04186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Demirsoy S, Martin S, Maes H, Agostinis P. Adapt, Recycle, and Move on: Proteostasis and Trafficking Mechanisms in Melanoma. Front Oncol. 2016;6:240. doi: 10.3389/fonc.2016.00240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peinado H, et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med. 2012 doi: 10.1038/nm.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoshino A, et al. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527:329–335. doi: 10.1038/nature15756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coelho P, Almeida J, Prudencio C, Fernandes R, Soares R. Effect of Adipocyte Secretome in Melanoma Progression and Vasculogenic Mimicry. J Cell Biochem. 2016;117:1697–1706. doi: 10.1002/jcb.25463. [DOI] [PubMed] [Google Scholar]
- 13.Obenauf AC, et al. Therapy-induced tumour secretomes promote resistance and tumour progression. Nature. 2015;520:368–372. doi: 10.1038/nature14336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Y, Cao X. Characteristics and Significance of the Pre-metastatic Niche. Cancer Cell. 2016;30:668–681. doi: 10.1016/j.ccell.2016.09.011. [DOI] [PubMed] [Google Scholar]
- 15.Eklund L, Bry M, Alitalo K. Mouse models for studying angiogenesis and lymphangiogenesis in cancer. Mol Oncol. 2013;7:259–282. doi: 10.1016/j.molonc.2013.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sevick-Muraca EM, Kwon S, Rasmussen JC. Emerging lymphatic imaging technologies for mouse and man. J Clin Invest. 2014;124:905–914. doi: 10.1172/JCI71612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martinez-Corral I, et al. In vivo imaging of lymphatic vessels in development, wound healing, inflammation, and tumor metastasis. Proc Natl Acad Sci U S A. 2012;109:6223–6228. doi: 10.1073/pnas.1115542109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petrova TV, et al. VEGFR-3 expression is restricted to blood and lymphatic vessels in solid tumors. Cancer Cell. 2008;13:554–556. doi: 10.1016/j.ccr.2008.04.022. [DOI] [PubMed] [Google Scholar]
- 19.Dankort D, et al. Braf(V600E) cooperates with Pten loss to induce metastatic melanoma. Nat Genet. 2009;41:544–552. doi: 10.1038/ng.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muramatsu T, Kadomatsu K. Midkine: an emerging target of drug development for treatment of multiple diseases. Br J Pharmacol. 2014;171:811–813. doi: 10.1111/bph.12571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yao J, Li WY, Gao SG. The advances of Midkine with peripheral invasion in pancreatic cancer. Am J Cancer Res. 2015;5:2912–2917. [PMC free article] [PubMed] [Google Scholar]
- 22.Jones DR. Measuring midkine: the utility of midkine as a biomarker in cancer and other diseases. Br J Pharmacol. 2014;171:2925–2939. doi: 10.1111/bph.12601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kishida S, Kadomatsu K. Involvement of midkine in neuroblastoma tumourigenesis. Br J Pharmacol. 2014;171:896–904. doi: 10.1111/bph.12442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kadomatsu K, Kishida S, Tsubota S. The heparin-binding growth factor midkine: the biological activities and candidate receptors. J Biochem. 2013;153:511–521. doi: 10.1093/jb/mvt035. [DOI] [PubMed] [Google Scholar]
- 25.Jono H, Ando Y. Midkine: a novel prognostic biomarker for cancer. Cancers (Basel) 2010;2:624–641. doi: 10.3390/cancers2020624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weckbach LT, et al. Midkine acts as proangiogenic cytokine in hypoxia-induced angiogenesis. Am J Physiol Heart Circ Physiol. 2012;303:H429–438. doi: 10.1152/ajpheart.00934.2011. [DOI] [PubMed] [Google Scholar]
- 27.Choudhuri R, Zhang HT, Donnini S, Ziche M, Bicknell R. An angiogenic role for the neurokines midkine and pleiotrophin in tumorigenesis. Cancer Res. 1997;57:1814–1819. [PubMed] [Google Scholar]
- 28.Luo Y, et al. Rapamycin inhibits lymphatic endothelial cell tube formation by downregulating vascular endothelial growth factor receptor 3 protein expression. Neoplasia. 2012;14:228–237. doi: 10.1593/neo.111570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patel V, et al. Decreased lymphangiogenesis and lymph node metastasis by mTOR inhibition in head and neck cancer. Cancer Res. 2011;71:7103–7112. doi: 10.1158/0008-5472.CAN-10-3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pucci F, et al. SCS macrophages suppress melanoma by restricting tumor-derived vesicle-B cell interactions. Science. 2016;352:242–246. doi: 10.1126/science.aaf1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yajima I, et al. Spatiotemporal gene control by the Cre-ERT2 system in melanocytes. Genesis. 2006;44:34–43. doi: 10.1002/gene.20182. [DOI] [PubMed] [Google Scholar]
- 32.Marino S, et al. PTEN is essential for cell migration but not for fate determination and tumourigenesis in the cerebellum. Development. 2002;129:3513–3522. doi: 10.1242/dev.129.14.3513. [DOI] [PubMed] [Google Scholar]
- 33.Denoyelle C, et al. Anti-oncogenic role of the endoplasmic reticulum differentially activated by mutations in the MAPK pathway. Nat Cell Biol. 2006;8:1053–1063. doi: 10.1038/ncb1471. [DOI] [PubMed] [Google Scholar]
- 34.Soengas MS, et al. Inactivation of the apoptosis effector Apaf-1 in malignant melanoma. Nature. 2001;409:207–211. doi: 10.1038/35051606. [DOI] [PubMed] [Google Scholar]
- 35.Tormo D, Alonso-Curbelo D, Soengas MS. Cytosolic delivery of dsRNA triggers MDA-5 mediated autonomous cell death in malignant melanomas. Clinical and Translational Oncology. 2009;11:39–42. [Google Scholar]
- 36.Workman P, et al. Guidelines for the welfare and use of animals in cancer research. British Journal of Cancer. 2010;102:1555–1577. doi: 10.1038/sj.bjc.6605642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peinado H, et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med. 2012;18:883–891. doi: 10.1038/nm.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wisniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 39.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature protocols. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome biology. 2009;10 doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics (Oxford, England) 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Das S, et al. Tumor cell entry into the lymph node is controlled by CCL1 chemokine expressed by lymph node lymphatic sinuses. The Journal of experimental medicine. 2013;210:1509–1528. doi: 10.1084/jem.20111627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Patsialou A, et al. Intravital multiphoton imaging reveals multicellular streaming as a crucial component of in vivo cell migration in human breast tumors. Intravital. 2013;2:e25294. doi: 10.4161/intv.25294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Willard-Mack CL. Normal structure, function, and histology of lymph nodes. Toxicol Pathol. 2006;34:409–424. doi: 10.1080/01926230600867727. [DOI] [PubMed] [Google Scholar]
- 46.Pucci F, et al. SCS macrophages suppress melanoma by restricting tumor-derived vesicle-B cell interactions. Science. 2016;352:242–246. doi: 10.1126/science.aaf1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Time lapse video microscopy of mCherry-labeled WM164 melanoma cells (red) plated on top of a monolayer of HLEC preincubated for 24h with vehicle (left) or 500ug/ml recombinant MDK (right). at 3.33 min intervals for a total of 965.7 minutes to visualize adhesion and spreading of melanoma cells as a function of changes in the size and area of mCherry-emitting cells.
Time lapse video-microscopy of the metastatic SK-Mel-M17 cells labeled in red with mCherry to assay for increases in cell spreading and adhesion to HLEC preincubated for 24h with vehicle (left) or 500ug/ml recombinant MDK (right). Images were captured at 2.33 min intervals for a total of 200 minutes.
Data Availability Statement
The mass spectrometry proteomics data have been deposited in the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD002703. RNA-Seq data have been deposited in NCBI's Gene Expression Omnibus with accession number GSE94549. Source data are provided for Figs 1e; 2a,b; 3e–l and Extended Figs 1d; 2d–f; 3b–e; 4a, 5a,c–h,I; 6b–e; 7a,c,e; 9a,b. All other data that support the findings of this study are available from the authors upon reasonable request.