Abstract
Background: This study evaluated the psychometric properties of a new, comprehensive measure of knowledge about genomic sequencing, the University of North Carolina Genomic Knowledge Scale (UNC-GKS). Methods: The UNC-GKS assesses knowledge in four domains thought to be critical for informed decision making about genomic sequencing. The scale was validated using classical test theory and item response theory in 286 adult patients and 132 parents of pediatric patients undergoing diagnostic whole exome sequencing (WES) in the NCGENES study. Results: The UNC-GKS assessed a single underlying construct (genomic knowledge) with good internal reliability (Cronbach’s α = 0.90). Scores were most informative (able to discriminate between individuals with different levels of genomic knowledge) at one standard deviation above the scale mean or lower, a range that included most participants. Convergent validity was supported by associations with health literacy and numeracy (rs = 0.41–0.46). The scale functioned well across subgroups differing in sex, race/ethnicity, education, and English proficiency. Discussion: Findings supported the promise of the UNC-GKS as a valid and reliable measure of genomic knowledge among people facing complex decisions about WES and comparable sequencing methods. It is neither disease- nor population-specific, and it functioned well across important subgroups, making it usable in diverse populations.
Keywords: genomic sequencing, knowledge, whole exome sequencing, informed decision making
Rapidly evolving genetic testing practices have begun to include panels of dozens of genes in some clinical scenarios, or even more comprehensive tests ranging from thousands of genes to the entire genome or exome (e.g., whole exome sequencing [WES]).1 Genome-scale tests (“genomic sequencing”) are more complex than single gene tests in important ways. For instance, they yield a wide spectrum of potential results, many of which have uncertain meaning. Providing informed consent for genomic sequencing requires that people have knowledge that includes, but goes beyond, knowledge needed for informed consent for single gene testing.2,3 People with greater knowledge of the nature of genes and their effects on health, how genes are inherited in families, and the potential benefits, harms, and limitations of genomic sequencing are better equipped than their less knowledgeable peers to make informed decisions about undergoing sequencing, comprehend the meaning and limitations of their results, and take appropriate actions on learning these results.4,5 Yet many people currently offering genomic sequencing have inadequate knowledge and misconceptions about basic genetics2,6 and are unfamiliar with genomic sequencing. The unique and complex issues raised by genome-scale tests are not well covered by existing knowledge measures. Having a validated, comprehensive measure of genomic knowledge could help identify knowledge gaps and reduce the chance that people’s decisions and responses to genomic sequencing are based on false assumptions, unrealistic expectations, or misconceptions.
To meet the need for a valid way to assess genomic knowledge, we conducted the present study to evaluate a new measure, the University of North Carolina Genomic Knowledge Scale (UNC-GKS). We conceptualized “genomic knowledge” as encompassing four domains: the structure and function of genes, how they are inherited, their relation to health, and potential benefits, harms, and limitations of WES—a sequencing method that identifies variants in the subset of the genome that encodes the genes. These domains are based on our clinical experiences with patients offered sequencing and reflect a pragmatic approach that is highly relevant to typical research applications of genomic sequencing. The domains are also consistent with a framework discussed by Smerecnik and colleagues7 that includes awareness knowledge (knowing that there are genetic risk factors for disease), how-to knowledge (knowing how those risk factors influence risk for developing a disease), and principles knowledge (knowledge of pathways through which genes are theorized to influence health). Our comprehensive definition allows evaluation of the extent to which people have a basic working knowledge they can use to evaluate pros and cons, risks, uncertainties, and alternatives to genomic sequencing.
The UNC-GKS was also designed to address several limitations in existing measures. First, many existing knowledge measures focus on testing for mutations in a single gene8 or are specific to a particular disease.9,10 However, genome-scale tests, in addition to becoming increasingly common, raise unique and complex issues not well covered by these kinds of existing measures. Moreover, because the UNC-GKS is general rather than disease-specific, scores can be compared across populations affected by different diseases. The UNC-GKS can also be used in populations affected by diseases for which disease-specific measures do not exist. A second issue addressed by the UNC-GKS relates to the sociodemographic diversity of most patient populations. Given recognized subgroup differences in knowledge and views of genomic sequencing,11,12 it is often useful to compare knowledge scores across subgroups in a study’s sample. However, these comparisons are only informative if observed subgroup knowledge differences reflect real differences rather than measurement artifacts. It is rare to see formal analyses investigating the psychometric functioning of a knowledge measure across different subgroups. Finally, some existing knowledge measures assess agreement or disagreement with statements about genetics or genomics13—a common approach for measuring beliefs or attitudes that may, but does not necessarily, correspond to knowledge. Studies in educational testing and knowledge assessment more often use items with multiple-choice or true/false response options that can be unambiguously scored as correct or incorrect.14,15
Accordingly, our goal was to develop a measure that met the following criteria: 1) it covers domains of knowledge relevant to the complex decision contexts created by genome-scale sequencing; 2) it applies to a broad range of contexts and populations rather than being specific to a particular disease or population; 3) it has adequate validity and reliability across important sociodemographic subgroups; and 4) it uses a true/false response scale. No existing measures meet these criteria. We note that a recently introduced measure of basic knowledge about genetics and genetic causes of disease16 used true/false response options; however, it was evaluated in a sample with little diversity, and analyses did not examine the psychometric functioning of its items across subgroups, so it is unclear whether the measure can assess knowledge with similar validity and reliability across important subgroups. Moreover, it does not include items about genomic sequencing and implications of potential sequencing results. The UNC-GKS includes those types of items, and we examined its item functioning across subgroups varying by sex, race/ethnicity, education, and English language proficiency. Consequently, the present study was expected to yield a new tool for research with diverse populations offered genomic sequencing.
Methods
Participants
Participants were enrolled in the North Carolina Clinical Genomic Evaluation by Next-generation Exome Sequencing (NCGENES) study, which is investigating the performance and best use of WES in the diagnosis and clinical care of patients with suspected genetic disorders.3 Adult and pediatric patients evaluated at a UNC-affiliated hospital or at Vidant Medical Center (Greenville, NC) were eligible for NCGENES if they had symptoms or an illness with a possible genetic etiology (as determined by the referring physician) and if they were in one of the following diagnostic groups: hereditary cancers, cardiovascular disorders (mainly cardiomyopathies), neurodevelopmental disorders, congenital disorders, retinopathies, or selected other disorders (e.g., mitochondrial disorders). Adult patients and parents of pediatric patients sequenced in NCGENES completed study measures including the UNC-GKS. Participants also included a small sample of guardians of adult patients whose cognitive or physical functioning precluded completion of study procedures; however, none were included in the present sample. We recruited 418 participants (286 adult patients, 132 parents) for the present study’s sample between August 2012 and December 2014. All completed study measures in English.
Procedures
Eligible individuals were contacted by study staff to schedule a study visit and then were mailed an appointment letter, consent and HIPAA forms, educational brochures designed for the study, and an intake questionnaire that included the UNC-GKS. Potential participants then met with a certified genetic counselor who obtained informed consent for sequencing. At this meeting, consenting participants returned their completed intake questionnaire, completed health literacy measures, and had their blood drawn. Data for the present study came from the intake questionnaire, the literacy measures, and UNC Hospitals chart abstraction. Prior to completing the UNC-GKS, participants had the opportunity to read the mailed educational brochures, which provided an overview of genomic sequencing and potential sequencing results. They had not yet received the more specific, personalized information provided during informed consent procedures. Their intake UNC-GKS scores therefore may not reflect the level of genomic knowledge in the general public; instead, these scores approximate knowledge likely to be found in candidates for sequencing. The institutional review boards of the University of North Carolina and Vidant Medical Center approved the study protocols.
Measures
Development of UNC-GKS
The UNC-GKS was developed in an iterative process that gathered feedback on measure domains and on item content and clarity from a team that included certified genetic counselors and medical geneticists with extensive clinical experience educating patients, behavioral scientists with formal training in communication and measure development, and others with and without genetics expertise. This team identified four key domains for the measure: the structure and function of genes, how they are inherited in families, their relation to health, and strengths and limitations of WES. We viewed the latter domain as a potentially separate module that future users could adapt or replace for other sequencing contexts (e.g., newborn or population screening). We reviewed existing measures and adapted or drafted knowledge items with the goal of ensuring good content validity across the four domains and cohesion across the underlying construct of genomic testing knowledge.17 Some items within each domain specifically addressed misconceptions that could affect informed decision making (e.g., “A mother and daughter who look alike are more genetically similar than a mother and daughter who do not look alike”). We used genetic terms that participants were exposed to throughout the study (e.g., in consent procedures, brochures, and counseling). Because the term gene variant was especially important, the instructions included the following reminder:
We are using the term gene variant to mean a version of a gene. Sometimes two people have the same version of a gene (they have the same gene variant) and other times two people have different versions of a gene (they have different gene variants).
The resulting measure includes 25 items framed as statements and uses the following response categories: true, false, and not sure/don’t know (with the latter provided to minimize guessing). The statements and correct answers appear in Table 1. We scored correct responses as 1 and incorrect responses and not sure/don’t know responses as 0.
Table 1.
UNC Genomic Knowledge Scale Items
| Content Area | Item |
|---|---|
| Genes | 1. Genes are made of DNA. |
| 2. Genes affect health by influencing the proteins our bodies make. | |
| 3. All of a person’s genetic information is called his or her genome. | |
| 4. A person’s genes change completely every 7 years.* | |
| 5. The DNA in a gene is made of four building blocks (A, C, T, and G). | |
| 6. Everyone has about 20,000 to 25,000 genes. | |
| Genes and health | 7. Gene variants can have positive effects, harmful effects, or no effects on health. |
| 8. Most gene variants will affect a person’s health.* | |
| 9. Everyone who has a harmful gene variant will eventually have symptoms.* | |
| 10. Some gene variants have a large effect on health while others have a small effect. | |
| 11. Some gene variants decrease the chance of developing a disorder. | |
| 12. Two unrelated people with the same genetic variant will always have the same symptoms.* | |
| How genes are inherited in families | 13. Genetic disorders are always inherited from a parent.* |
| 14. If only one person in the family has a disorder it can’t be genetic.* | |
| 15. Everyone has a chance for having a child with a genetic disorder. | |
| 16. A girl inherits most of her genes from her mother while a boy inherits most of his genes from his father.* | |
| 17. A mother and daughter who look alike are more genetically similar than a mother and daughter who do not look alike.* | |
| 18. If a parent has a harmful gene variant, all of his or her children will inherit it.* | |
| 19. If one of your parents has a gene variant, your brother or sister may also have it. | |
| Whole exome sequencing | 20. Whole exome sequencing can find variants in many genes at once. |
| 21. Whole exome sequencing will find variants that cannot be interpreted at the present time. | |
| 22. Whole exome sequencing could find that you have a high risk for a disorder even if you do not have symptoms. | |
| 23. Your whole exome sequencing may not find the cause of your disorder even if it is genetic. | |
| 24. The gene variants that whole exome sequencing can find today could have different meanings in the future as scientists learn more about how genes work. | |
| 25. Whole exome sequencing will not find any variants in people who are healthy.* |
Note: Correct answer to the items is true unless followed by an asterisk (*).
Sociodemographic and Medical Characteristics
Sociodemographic variables came from clinical records or self-report in the intake questionnaire. They included participant sex, race/ethnicity, educational attainment, and annual household income. Nominating clinicians reported patients’ diagnosis or symptoms; this information was supplemented and confirmed during the informed consent session.
English Proficiency
We used 3-item subscale of the Cultural Identity Scale18 to assess English proficiency in speaking, reading, and writing. Responses, ranging from 1 (Poor) to 4 (Excellent), were summed to create a single score for which higher scores indicated greater proficiency (Cronbach’s α = 0.90).
Health Literacy
Study staff assessed general health literacy and genetics-related health literacy in person using the 66-item Rapid Estimate of Adult Literacy Measure19 and the 8-item Rapid Estimate of Adult Literacy–Genetics.6 For both scales, we created scores by summing the number of words a participant pronounced correctly; words that a participant pronounced incorrectly or skipped were not counted. Higher scores indicate greater general health literacy and genetics-related health literacy, respectively. REALM raw scores can be used to categorize people as having low health literacy (scores of 0–44, less than or equal to sixth-grade reading level), marginal literacy (scores of 45–60, seventh- to eighth-grade reading level), and functional health literacy (scores of 61–66, ninth-grade or higher reading level).19 REAL-G scores of three or less have been interpreted as indicating low genetics-related health literacy (less than or equal to sixth-grade level).6
Numeracy
We measured subjective numeracy with a validated 3-item version of the Subjective Numeracy Scale.20–22 Items assess perceived numerical aptitude and preference for numbers on a scale from 1 (Not at all good/helpful) to 6 (Extremely good/helpful). Summing responses yields a single score for which higher scores indicated stronger preference for numerical over textual information and greater perceived ability to perform mathematical tasks (Cronbach’s α = 0.89). We measured objective numeracy with a validated measure that presents three arithmetic problems testing the use of proportions, fractions, and percentages.23 Summing correct responses yields an objective numeracy score.
Data Analysis
We examined the psychometric properties, factor structure (to evaluate the assumption that all items reflect a single underlying construct—in this case, genomic knowledge), and convergent validity (to evaluate the scale’s association with conceptually related variables) of the UNC-GKS. We also conducted item response theory (IRT) analyses that offer more in-depth information than classical test theory methods, including evaluation of variation in item performance (differential item functioning) across demographic subgroups.
Item-Level Descriptive Statistics
First, we examined the proportion of participants correctly answering each UNC-GKS item to evaluate whether items were too easy (ceiling effects, indicated by >90% of participants answering them correctly) or too hard (floor effects, indicated by >90% of participants answering them incorrectly). Second, we computed inter-item tetrachoric correlations24 to evaluate whether UNC-GKS items were positively associated with each other, as we would expect. Third, we evaluated whether responses to each item were consistent with the sum of the responses to the remaining items by examining whether item-total correlations were positive. Negative or low item-total correlations indicate items that may need to be reworded or discarded.
Factor Analyses
Before completing IRT analyses, we checked the assumption that the measure was unidimensional by conducting a confirmatory factor analysis of the inter-item tetrachoric correlation matrix using the Mplus25“weighted least squares with robust standard errors, mean- and variance-adjusted” algorithm. We evaluated model fit using the root mean square error of approximation (RMSEA; acceptable if <0.05),26 the Tucker-Lewis index (TLI; acceptable if >0.95),27 the comparative fix index (CFI; acceptable if >0.95),28 and residual correlations between items via modification indices. Large modification indices (>10) reveal possible local dependence for sets of items, indicating possible violation of the local dependence assumption of IRT. Local dependencies indicate content redundancy or similar wording between two or more items and may suggest additional factors exist in the scale.
IRT Analyses
We evaluated performance of the UNC-GKS items by fitting one-, two-, and three-parameter logistic IRT models (1PL, 2PL, and 3PL, respectively) using the software program IRTPRO.29 The 1PL model30 characterizes each item by a single parameter—the difficulty parameter, b, which indicates the level of genetic knowledge at which there is a 50% chance of answering the item correctly (i.e., how difficult the item is). The 2PL model31 estimates both b and an additional parameter, the discrimination parameter, a, which reflects the degree to which item responses are associated with the latent construct being measured (how effectively an item discriminates between individuals with higher versus lower genomic knowledge). The 3PL model31 estimates the a and b parameters and an additional parameter, c, which accounts for guessing. We chose the best fitting model by examining chi-square tests of the likelihood ratio for each model pair, then examined this model’s goodness of fit to the data using Orlando and Thissen’s S-χ2 statistic,32,33 for which a nonsignificant result indicates adequate model fit at the item level (i.e., how well each item fits the model). We controlled for multiple comparisons using the Benjamini-Hochberg procedure.34,35
In addition to items flagged for potential local dependence in the confirmatory factor analysis, we used the IRT-based local dependence statistic36 to identify items that were excessively related after controlling for the underlying construct (genomic knowledge)—an undesirable characteristic. Values >10 indicate substantial local dependence. We then conducted an additional check on the dimensionality of the data by estimating a bifactor IRT model in which each locally dependent set of items was specified as a second-order factor. Violations of local dependence were deemed negligible if the variance accounted for by first-order or general factor (common variance)37–40 was at least 0.85.
Next, we examined differential item functioning (DIF), which enables evaluation of whether items behave differently across subgroups after holding the underlying construct (genomic knowledge) constant.41 It detects a form of measurement bias that occurs when people in different groups with the same level of the underlying construct have a different probability of getting a particular score on a scale. DIF may indicate that attributes other than those the scale is intended to measure are affecting responses. In the present study, we examined DIF across sex, race/ethnicity, education, and English proficiency groups. For each item, we used a logistic regression model to evaluate whether item responses were associated with group membership after controlling for participants’ IRT score on the UNC-GKS. Uniform DIF (of a similar magnitude across the range of the underlying construct) was evaluated with a likelihood ratio test comparing a logistic regression model with one predictor (IRT score) to a model with both IRT score and an additional predictor (group membership); this approach allowed us to evaluate whether, after controlling for overall level of genomic knowledge, one group was more likely than the other to answer the item correctly. Nonuniform DIF (for which magnitude may differ across the range of the underlying construct) was evaluated with a likelihood ratio test comparing a model with both predictors (IRT score and group membership) to a model that also included their interaction term. This model allowed us to evaluate whether an item provided better measurement of genomic knowledge for one group versus another. We used the Benjamini-Hochberg procedure to make inferential decisions in multiple comparisons.
According to common practice, we planned to drop items if they did not fit well, substantially violated local dependence, or functioned differently for key groups. The remaining items would then be used to calibrate a final IRT model to use in subsequent analyses.
IRT Scoring and Reliability
We computed IRT scores for the UNC-GKS based on the parameters from the final IRT model. These scores are relative to the population of this sample, assuming a normal distribution with a mean of 0 and standard deviation of 1. To be more easily interpretable, we scaled the IRT scores to the T-score metric with a mean of 50 and a standard deviation of 10. Analysts typically compute IRT scaled scores based on response patterns, essentially weighting item responses by their IRT a parameters so that items more strongly related to the underlying construct have a greater impact on the score. However, analysts also often use summed scores because they do not require special software. To enable practical use of scaled scores, we computed a scoring table to convert summed scores to expected scaled scores. We also computed a scoring table for a 19-item version of the UNC-GKS that omitted the WES items, for use when those items are not needed. The 19-item version was scored using the 19 IRT parameters from the 25-item calibration so that scores would be on the same scale and comparable.
Next, we used the IRT test information function (TIF) to examine the precision of scale scores—the extent to which an estimate of genomic knowledge at a given scale score is reasonably close to the true value. Given that these scores estimate individuals’ genomic knowledge, greater precision improves the scale’s ability to distinguish between individuals with different levels of genomic knowledge in addition to providing other useful information. The TIF sums information functions for each individual item into a single function. Greater test “information” indicates greater precision.42 TIF is depicted in a graphical format where the amount of information is plotted against the latent construct (here, genomic knowledge) to show how well the test estimates the construct over the full range of individuals’ ability or knowledge. The areas of greatest measurement precision are indicated by the highest points of the curve.
Classical Test Theory Reliability
We evaluated internal consistency reliability of the 25 UNC-GKS items by computing Cronbach’s coefficient α.43 Ideal α values are at least 0.70,44 indicating a set of items that are strongly related to one another.
Convergent Validity
We calculated Pearson correlations between the UNC-GKS scale score and the REAL-G, REALM, and subjective and objective numeracy scales to evaluate convergent validity, or the extent to which measures that should be associated with each other are in fact associated. We predicted that the UNC-GKS would correlate positively with genetics-related literacy and general health literacy, because individuals with greater ability to obtain, process, and understand health information should be more able to learn the domains of information assessed by the UNC-GKS. We also predicted positive correlations between the UNC-GKS and both measures of numeracy because the ability to reason and apply numerical concepts influences ability to learn these domains of information.45
Results
Sample
The final sample included 286 adult patients and 132 parents (Table 2). Three quarters were women and 17% were racial/ethnic minorities. Participants’ mean age was 47 years (range 17–84 years). Nearly 20% of participants had not attended college; just over half had a college degree. The median annual household income category was $60,000 to $74,999. Nearly 13% of the sample had marginal or worse general health literacy and 6% had low genetic literacy. About 18% reported less than “excellent” proficiency in speaking, writing, and/or reading English.
Table 2.
Participant Characteristics (N = 418)
| n (%) | Mean (SD) | |
|---|---|---|
| Role | ||
| Adult patient | 286 (68.4) | |
| Parent of pediatric patient | 132 (31.6) | |
| Age (years)a | 46.5 (14.3) | |
| Sex | ||
| Female | 315 (75.4) | |
| Male | 103 (24.6) | |
| Ethnicity | ||
| Non-Hispanic | 391 (93.5) | |
| Hispanic | 19 (4.5) | |
| Missing | 8 (1.9) | |
| Race | ||
| White | 345 (82.5) | |
| Non-White | 71 (17.0) | |
| Missing | 2 (0.5) | |
| Education | ||
| Less than high school | 28 (6.7) | |
| High school graduate | 52 (12.4) | |
| Some college | 88 (21.1) | |
| Associates degree or vocational program | 69 (16.5) | |
| 4-Year college degree | 108 (25.8) | |
| Graduate degree | 71 (17.0) | |
| Missing | 2 (.5) | |
| Income ($) | ||
| <30,000 | 107 (25.6) | |
| 30,000-59,999 | 83 (19.9) | |
| 60,000-89,999 | 84 (20.1) | |
| 90,000-104,999 | 17 (4.1) | |
| >105,000 | 97 (23.2) | |
| Missing | 30 (7.2) | |
| Clinical group | ||
| Hereditary cancers | 100 (23.9) | |
| Cardiovascular disorders | 46 (11.0) | |
| Neurodevelopmental disorders | 112 (26.8) | |
| Congenital disorders | 32 (7.7) | |
| Other | 128 (30.6) | |
| General health literacy | 63.0 (6.7) | |
| Functional (9th grade and above) | 358 (85.6) | |
| Marginal (7th or 8th grade) | 44 (10.5) | |
| Low (6th grade and below) | 12 (2.9) | |
| Missing | 4 (1.0) | |
| Genetics-related health literacy | 7.1 (1.6) | |
| High (above 6th grade) | 384 (91.9) | |
| Low (6th grade and below) | 23 (5.5) | |
| Missing | 11 (2.6) | |
| Objective numeracy | 1.7 (1.0) | |
| Subjective numeracy | 4.6 (1.3) | |
Ages for participating parents of pediatric patients were not collected early in the study; therefore, descriptive statistics for participant age are based on all adult patients and 27 of the 132 participating parents.
Scale and Item Descriptive Statistics
The number of missing responses was minor, ranging from 2 to 8 of 418 participants per item. The mean proportion of participants correctly answering each item was 0.73 (SD = 0.12) with a range of 0.48 to 0.89 across the items. Figure 1 shows the distribution of responses for each item, revealing items for which knowledge and uncertainty were highest and lowest. Items from the four domains assessed by the measure (the structure and function of genes, how they are inherited, their relation to health, and the strengths and limitations of WES) were well distributed across the range of items answered correctly, incorrectly, or for which there was uncertainty. There were no floor or ceiling effects; thus, no items were too easy or too hard for this sample. All correlations among items were positive and statistically significant, with a mean of r = 0.46 (SD = 0.15) and range of 0.10 to 0.90. Similarly, all item-total correlations were positive and of medium to large magnitude, with a mean of r = 0.49 (SD = 0.11) and range of 0.29 to 0.68. Thus, item-level statistics did not identify any items as candidates for revision or removal and subsequent analyses considered all 25 items. Distributions of the summed score for the 19 and 25 item versions of the UNC-GKS appear in Figures 2 and 3. Scores are skewed to the left, indicating that participants correctly answered most items.
Figure 1.
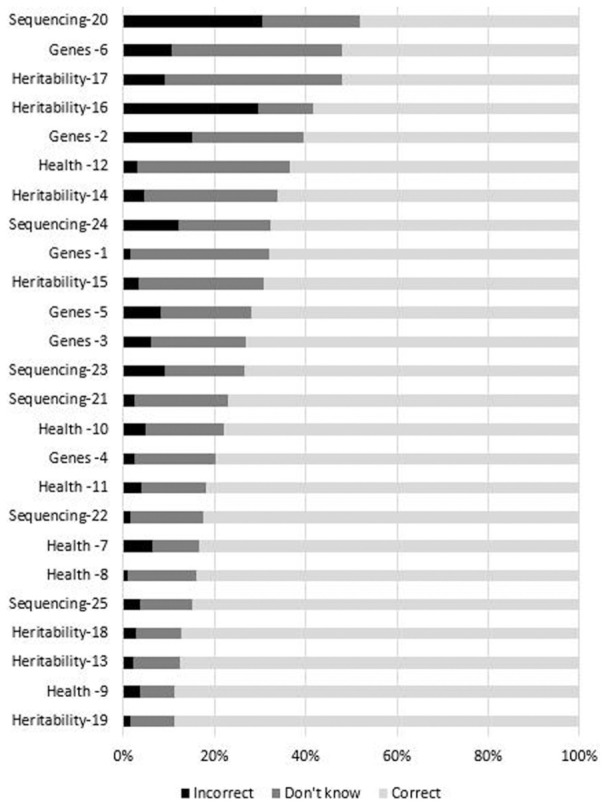
Item response distributions for the University of North Carolina Genomic Knowledge (UNC-GKS)
Figure 2.
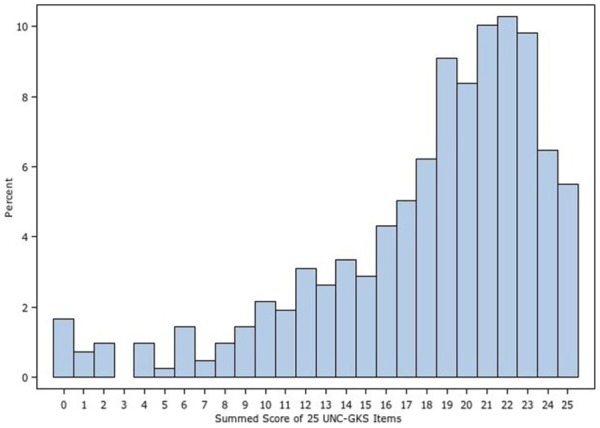
Summed score distribution for the 25-item University of North Carolina Genomic Knowledge (UNC-GKS)
Figure 3.
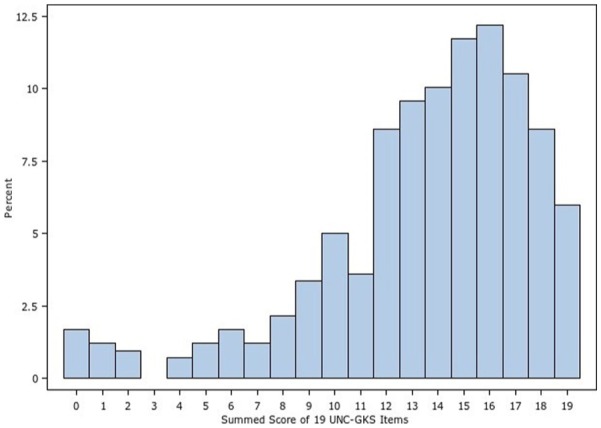
Summed score distribution for the 19-item University of North Carolina Genomic Knowledge (UNC-GKS)
Factor Analyses
The one-factor confirmatory factor analysis model fit the data well (χ2 = 476.4, df = 275, P < 0.001; RMSEA = 0.04; CFI = 0.96; TLI = 0.95), indicating that the items represented a single underlying construct. The standardized factor loadings were positive, statistically significant, and of moderate to large magnitude (0.40–0.95); thus, all items were associated with the underlying construct of genomic knowledge (Table 4). Two modification indices indicated possible local dependence between Items 5 and 6, and between Items 9 and 12. Fitting a model that allowed the errors for these two item pairs to correlate led to good fit (χ2 = 456.0, df = 273, P < 0.001; RMSEA = 0.04; CFI = 0.96; TLI = 0.96) with a statistically significant improvement in fit over the simpler model ( = 20.4, df = 2, P < 0.001). However, the residual correlations were not large (r = 0.33 for Items 5 and 6, r = 0.42 for Items 9 and 12) and the items’ text did not suggest content redundancy so we retained them for further evaluation.
Table 4.
Table for Converting Summed Scores to Item Response Theory (IRT)–Scaled T-Scores for the University of North Carolina Genomic Knowledge Scale (UNC-GKS)
| 25-Item UNC-GKS |
19-Item UNC-GKS |
|||
|---|---|---|---|---|
| Summed Score | T-Score | Standard Error | T-Score | Standard Error |
| 0 | 21.7 | 5.0 | 21.9 | 5.1 |
| 1 | 24.3 | 4.6 | 24.6 | 4.7 |
| 2 | 26.7 | 4.2 | 27.1 | 4.3 |
| 3 | 28.8 | 3.8 | 29.4 | 4.0 |
| 4 | 30.6 | 3.5 | 31.4 | 3.8 |
| 5 | 32.2 | 3.2 | 33.3 | 3.6 |
| 6 | 33.7 | 3.0 | 35.1 | 3.5 |
| 7 | 35.0 | 2.8 | 36.8 | 3.5 |
| 8 | 36.2 | 2.7 | 38.4 | 3.5 |
| 9 | 37.3 | 2.6 | 40.1 | 3.5 |
| 10 | 38.4 | 2.5 | 41.8 | 3.6 |
| 11 | 39.5 | 2.4 | 43.5 | 3.7 |
| 12 | 40.6 | 2.4 | 45.4 | 3.8 |
| 13 | 41.6 | 2.4 | 47.3 | 4.0 |
| 14 | 42.7 | 2.5 | 49.4 | 4.2 |
| 15 | 43.9 | 2.6 | 51.8 | 4.5 |
| 16 | 45.1 | 2.7 | 54.4 | 4.9 |
| 17 | 46.3 | 2.9 | 57.4 | 5.4 |
| 18 | 47.7 | 3.1 | 61.0 | 5.9 |
| 19 | 49.3 | 3.4 | 65.0 | 6.5 |
| 20 | 51.1 | 3.7 | ||
| 21 | 53.1 | 4.1 | ||
| 22 | 55.5 | 4.5 | ||
| 23 | 58.2 | 5.1 | ||
| 24 | 61.6 | 5.7 | ||
| 25 | 65.4 | 6.3 | ||
Note: This table can be used to convert the summed count of a participant’s correct responses (summed scores) to item response theory scaled scores in the T-score metric, allowing easy comparison across studies and populations. These T-scores were calculated using this study’s sample, a population mean of 50, and standard deviation of 10. To convert a score, find a participant’s summed score in column 1 and follow the row to determine the IRT-scaled T-score and standard error for the 25- and 19-item versions of the UNC-GKS.
Item Response Theory Analyses
The UNC-GKS items varied in their ability to discriminate between participants with differing amounts of genomic knowledge, as indicated by the better fit of the 2PL rather than the 1PL model ( = 221.37, df = 24, P < 0.001). The 2PL model fit the items well; no items exhibited misfit or local dependence (P < 0.05). Participants were not likely to have been guessing when they answered “true” or “false,” given that the 3PL model did not fit better than the 2PL model ( = 2.92, df = 24, P = 1.00); participants who were unsure of their responses were likely selecting “not sure/don’t know.” The S-χ2 and LD statistics appear in Table 3.
Table 3.
Results of the One-Factor Confirmatory Factor Analysis Model and Two-Parameter Logistic Item Response Theory Model for University of North Carolina Genomic Knowledge Scale items
| CFA Model |
2PL IRT Model
Parameters |
2 PL IRT Model Item Fit |
|||||
|---|---|---|---|---|---|---|---|
| Item Number | Λ | a | SE | b | SE | S-χ 2 | df |
| 1 | 0.55 | 0.92 | 0.18 | −2.56 | 0.43 | 21.55 | 18 |
| 2 | 0.67 | 1.72 | 0.23 | −0.83 | 0.11 | 20.82 | 16 |
| 3 | 0.60 | 1.27 | 0.18 | −0.94 | 0.14 | 20.83 | 19 |
| 4 | 0.59 | 1.19 | 0.17 | −0.84 | 0.14 | 18.69 | 18 |
| 5 | 0.64 | 1.56 | 0.21 | −0.49 | 0.10 | 14.78 | 15 |
| 6 | 0.52 | 0.96 | 0.15 | −0.10 | 0.12 | 15.86 | 17 |
| 7 | 0.75 | 1.80 | 0.28 | −1.66 | 0.17 | 26.53 | 17 |
| 8 | 0.58 | 1.35 | 0.19 | 0.09 | 0.10 | 15.16 | 15 |
| 9 | 0.67 | 1.42 | 0.20 | −0.69 | 0.11 | 19.87 | 15 |
| 10 | 0.84 | 1.98 | 0.30 | −1.52 | 0.15 | 13.01 | 15 |
| 11 | 0.57 | 1.26 | 0.18 | −0.07 | 0.10 | 18.99 | 16 |
| 12 | 0.78 | 1.82 | 0.26 | −1.21 | 0.13 | 18.69 | 15 |
| 13 | 0.40 | 0.59 | 0.13 | −0.62 | 0.22 | 18.27 | 19 |
| 14 | 0.70 | 1.51 | 0.22 | −1.44 | 0.17 | 17.43 | 18 |
| 15 | 0.48 | 0.77 | 0.14 | −1.48 | 0.27 | 23.4 | 20 |
| 16 | 0.65 | 1.40 | 0.20 | −1.19 | 0.15 | 22.67 | 17 |
| 17 | 0.53 | 0.94 | 0.15 | −0.53 | 0.14 | 14.25 | 18 |
| 18 | 0.81 | 1.72 | 0.26 | −1.59 | 0.17 | 17.5 | 16 |
| 19 | 0.77 | 1.80 | 0.26 | −1.40 | 0.15 | 18.06 | 17 |
| 20 | 0.74 | 2.20 | 0.29 | −0.55 | 0.09 | 16.71 | 13 |
| 21 | 0.73 | 2.01 | 0.27 | −0.53 | 0.09 | 9.28 | 14 |
| 22 | 0.95 | 4.90 | 0.90 | −0.90 | 0.07 | 16.04 | 9 |
| 23 | 0.90 | 3.58 | 0.56 | −0.84 | 0.08 | 6.1 | 12 |
| 24 | 0.94 | 3.80 | 0.63 | −1.01 | 0.09 | 13.93 | 11 |
| 25 | 0.86 | 2.91 | 0.42 | −0.79 | 0.08 | 17.36 | 12 |
Note: CFA = confirmatory factor analysis; 2PL IRT = two-parameter logistic item response theory; SE = standard error; df = degrees of freedom.
To further evaluate the residual correlations found in the confirmatory factor analysis model between Items 5 and 6 and Items 9 and 12, we estimated a bifactor 2PL model with each of these item pairs loading on a second-order factor in addition to the overall genomic knowledge factor. Explained common variance for this model was 0.91; thus, most of the variance in the items was attributable to the overall factor. This finding supported our decision to retain these items and led us to consider the data to be “essentially unidimensional,” meaning that there were no other meaningful underlying dimensions. Thus, these four items could be considered part of the overall factor.
DIF detection analyses, estimated with logistic regression models, did not reveal any statistically significant parameters (P < 0.05) for uniform or nonuniform DIF. That is, after controlling for level of genomic knowledge, the scale items did not function differently in terms of their difficulty or how related they were to genomic knowledge when answered by individuals who differed on each of the key demographic groups of sex, race/ethnicity, education, and English proficiency. The measure performed comparably across these groups.
Our analytic plan called for items to be dropped prior to calibration of a final IRT model if they did not fit well, substantially violated local dependence, or functioned differently for key groups. Because no items were flagged for removal using these criteria, the original 2PL calibration was the final IRT model used for scoring. As shown in Table 3, the difficulty (b) parameters for the items ranged from −2.56 to 0.09; higher b parameters indicate more difficult items. The discrimination (a) parameters indicated that all items were highly related to the underlying construct of genomic knowledge and that they were able to discriminate between individuals with different levels of genomic knowledge. Items specific to WES (Items 20–25) had a parameters >2.0 and thus were particularly good at discriminating between these individuals, perhaps because the items covered information that was relatively unfamiliar to participants, many of whom had not known about WES prior to the study.
Item Response Theory Scoring and Reliability
We examined score precision (TIF) and reliability for both forms of the scale (see Figure 4). For the 25-item UNC-GKS, precision and reliability was especially high for T-scores between 32 and 49, and reliability was good (≥ 0.70) for all scores below 61 (approximately one standard deviation above the mean). For the 19-item UNC-GKS, precision and reliability was highest for T-scores between 36 and 40, and reliability was good (≥0.70) for all T-scores below 60. Thus, reliability was above accepted cutoffs for individuals who correctly answered ≤23 items on the 25-item UNC-GKS or ≤17 items on the 19-item UNC-GKS. Table 4 provides a scoring translation table for converting summed scale scores to T-scores based on the final (2PL) IRT model using the overall sample as a reference group. The correlation between T-scores resulting from the two versions of the scale was 0.97.
Figure 4.
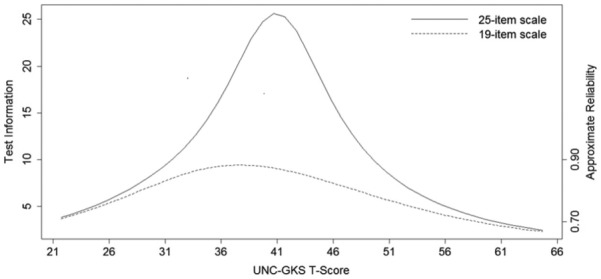
Item response theory test information and reliability for the University of North Carolina Genomic Knowledge (UNC-GKS) T-scores.
Classical Test Theory Reliability
Internal consistency reliability was high (Cronbach’s α = 0.90) for the 25-item UNC-GKS and for the 19-item UNC-GKS (α = 0.86).
Convergent Validity
As expected, the UNC-GKS correlated moderately and positively with general health literacy, genetics-related health literacy, and objective and subjective numeracy (Table 5), providing evidence of convergent validity.
Table 5.
Pearson Correlations Between University of North Carolina Genomic Knowledge (UNC-GKS) Scale Score and Other Measures
| Scale | Correlation With UNC-GKS | 95% CI | n |
|---|---|---|---|
| Genetics-related health literacy | 0.46* | 0.38-0.53 | 407 |
| Health literacy | 0.40* | 0.31-0.47 | 410 |
| Subjective numeracy | 0.43* | 0.34-0.52 | 412 |
| Objective numeracy | 0.41* | 0.32-0.48 | 418 |
P < 0.001.
Discussion
In order to make informed decisions about accepting or declining sequencing, understand their sequencing results, and decide on appropriate ways to apply these results, people need knowledge about the structure and function of genes, how genes are inherited in families, effects of genes on health, and potential benefits, harms, and limitations of sequencing.4,5 Our study used rigorous methods to evaluate the psychometric properties of the UNC-GKS, which assesses these domains of knowledge. Findings indicate that all of the scale’s items measured genomic knowledge well, and taken together, they cover a broad range of critical genomic knowledge. Thus, the UNC-GKS can generate a single score representing a person’s genomic knowledge.
Importantly, findings indicated that the UNC-GKS measures genomic knowledge in the same way regardless of people’s sociodemographic background. That is, different subgroup scores indicate different levels of genomic knowledge rather than differences in the way people in certain subgroups understood scale items or differences in their response patterns that are unrelated to their level of genomic knowledge. As such, the UNC-GKS is valid for a variety of subgroups and researchers can use it with diverse populations. For instance, group differences in UNC-GKS scores could be used to advance understanding of disparities in genomic sequencing.
Furthermore, UNC-GKS scores were most informative below one standard deviation above the scale’s mean. Thus, the scores are precise for most individuals, with slightly less reliability, although still well within an acceptable range for those who answer all items correctly or those who give one incorrect answer (i.e., scores at one standard deviation above the scale’s mean or higher). These high-scoring individuals may have less need for educational interventions, so this psychometric property of the UNC-GKS does not detract from its usefulness. In sum, the measure is reliable and especially sensitive for identifying people with varying degrees of low to moderately high genomic knowledge—the group in greatest need of education and decision support when offered genomic sequencing.
We created two versions of the UNC-GKS: a 25-item version that includes items assessing knowledge about strengths and limitations of WES (the context for which we developed the scale) and a 19-item version that excludes these WES-specific items. T-scores were comparable for these two forms—an advantage for researchers and clinicians who use only the first 19 items because they are not offering WES, or they wish to develop a new set of items specific to the type of testing they are using. Our team has developed and will validate versions of the scale that substitute the WES items with new items relevant to the application of genomic sequencing to newborn screening and to screening adults for highly penetrant mutations that confer risk for treatable or preventable diseases.
Moreover, findings indicated that participants were not guessing, likely as a positive result of including the “not sure/don’t know” response option. This response option also allows evaluation of the extent to which individuals are unsure of responses versus having been forced to indicate that each item is true or false, without being able to indicate uncertainty. Being able to evaluate uncertainty may have practical utility in some applications. For instance, items with high unsure/don’t know responses may represent specific areas that should be targeted with educational efforts.
Limited literacy and numeracy—basic abilities necessary for seeking, comprehending, and using health information46,47—is a prevalent problem and one that is highly relevant for researchers and clinicians who need to educate individuals about genomic sequencing. Both were associated with lower scores on the UNC-GKS. These associations have been identified as important in the context of genetic testing and genomic sequencing,45 and they have contributed to calls to increase educational efforts necessary for making scientific knowledge about genetics and genomics more broadly accessible.4
Additional research will be needed to evaluate additional aspects of validity, including other correlates of genomic knowledge measured by the UNC-GKS, group differences in genomic knowledge that may contribute to disparities in decision making and decision outcomes, and whether UNC-GKS scores identify people at risk for experiencing poor psychosocial or medical outcomes after testing. Research using this measure may also help guide development of psychoeducational interventions to address knowledge deficits, targeting clinicians (e.g., training in communicating effectively about genomic sequencing), patients and research participants (e.g., more useful and accessible educational resources), or both.
Future studies should also address limitations of the present study. Some of our subgroups were smaller than a suggested subgroup size of 100 for DIF analyses.48 This rule of thumb is an estimate, and given the distribution of our item responses, we believe our results to be reliable. Yet a more diverse sample would enable more in-depth examination of differential item functioning across subgroups, including the potential for more difficult or complex items to function poorly in some subgroups. We note that we did not find evidence for this type of problem, perhaps in part because our items were iteratively reviewed by measurement and clinical experts. However, it would be valuable to recruit a larger proportion of underserved minority groups, individuals with less than a high school education, and those with low health literacy. In addition, the research was conducted primarily at an academic medical center in a single geographic area. Future research should evaluate the measure in community-based samples and other geographic areas. Finally, the scale is likely to be too long for some purposes, and validation of a briefer form could increase its usefulness.
Conclusion
This study used a rigorous approach to demonstrate that the UNC-GKS is a promising tool for advancing research on people’s decisions about and responses to genomic sequencing. The scale has promising psychometric properties across different sociodemographic subgroups, making it appropriate for research in diverse populations. In addition, it covers domains of knowledge considered by a multidisciplinary team of experts to be critical for informed decision making about genomic sequencing, which is rapidly replacing single gene testing in some populations. Moreover, the scale is not disease or population specific, making it usable across a wide range of populations facing complex decisions involving next generation genomic sequencing.
Acknowledgments
We would like to thank Kristy Lee and Kate Foreman for piloting and providing helpful feedback on the scale.
Footnotes
None of the authors have conflicts of interest to disclose.
The study sponsor was not involved in study design, data collection, analysis, or interpretation of data; writing this report; or the decision to submit the report for publication. This research was supported by the National Human Genome Research Institute of the National Institutes of Health under award number U01HG006487 (PIs: James P. Evans, Jonathan S. Berg, Karen E. Weck, Kirk C. Wilhelmsen, and Gail E. Henderson). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- 1. Warman Chardon J, Beaulieu C, Hartley T, Boycott KM, Dyment DA. Axons to exons: the molecular diagnosis of rare neurological diseases by next-generation sequencing. Curr Neurol Neurosci Rep. 2015;15(9):64. [DOI] [PubMed] [Google Scholar]
- 2. Bernhardt BA, Roche MI, Perry DL, Scollon SR, Tomlinson AN, Skinner D. Experiences with obtaining informed consent for genomic sequencing. Am J Med Genet A. 2015;167A(11):2635–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Roche MI, Berg JS. Incidental findings with genomic testing: implications for genetic counseling practice. Curr Genet Med Rep. 2015;3(4):166–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Secretary’s Advisory Committee on Genetics, Health, and Society. Genetics Education and Training. Washington, DC: Department of Health & Human Services; 2011. [Google Scholar]
- 5. Lautenbach DM, Christensen KD, Sparks JA, Green RC. Communicating genetic risk information for common disorders in the era of genomic medicine. Annu Rev Genomics Hum Genet. 2013;14:491–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Erby LH, Roter D, Larson S, Cho J. The rapid estimate of adult literacy in genetics (REAL-G): a means to assess literacy deficits in the context of genetics. Am J Med Genet. 2008;146A(2):174–81. [DOI] [PubMed] [Google Scholar]
- 7. Smerecnik CM, Mesters I, de Vries NK, de Vries H. Educating the general public about multifactorial genetic disease: applying a theory-based framework to understand current public knowledge. Genet Med. 2008;10(4):251–8. [DOI] [PubMed] [Google Scholar]
- 8. Green MJ, Biesecker BB, McInerney AM, Mauger D, Fost N. An interactive computer program can effectively educate patients about genetic testing for breast cancer susceptibility. Am J Med Genet. 2001;103(1):16–23. [DOI] [PubMed] [Google Scholar]
- 9. Richman AR, Tzeng JP, Carey LA, Retèl VP, Brewer NT. Knowledge of genomic testing among early-stage breast cancer patients. Psychooncology. 2011;20(1):28–35. [DOI] [PubMed] [Google Scholar]
- 10. Scuffham TM, McInerny-Leo A, Ng SK, Mellick G. Knowledge and attitudes towards genetic testing in those affected with Parkinson’s disease. J Community Genet. 2014;5(2):167–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Singer E, Antonucci T, Van Hoewyk J. Racial and ethnic variations in knowledge and attitudes about genetic testing. Genet Test. 2004;8(1):31–43. [DOI] [PubMed] [Google Scholar]
- 12. Haga SB, Barry WT, Mills R, et al. Public knowledge of and attitudes toward genetics and genetic testing. Genet Test Mol Biomarkers. 2013;17(4):327–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kaphingst KA, Facio FM, Cheng MR, et al. Effects of informed consent for individual genome sequencing on relevant knowledge. Clin Genet. 2012;82(5):408–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Frisbie DA. Multiple choice versus true-false—comparison of reliabilities and concurrent validities. J Educ Meas. 1973;10(4):297–304. [Google Scholar]
- 15. Ebel RL, Frisbie DA. Essentials of Educational Measurement. 5th ed. Englewood Cliffs, NJ: Prentice Hall; 1991. [Google Scholar]
- 16. Fitzgerald-Butt SM, Bodine A, Fry KM, et al. Measuring genetic knowledge: a brief survey instrument for adolescents and adults. Clin Genet. 2016;89(2):235–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. DeVellis RF. Scale Development: Theory and Applications. Vol 26 2nd ed.Newbury Park, CA: Sage; 2003. [Google Scholar]
- 18. Félix-Ortiz M, Newcomb MD, Myers H. 1994. A multidimensional measure of cultural identity for Latino and Latina adolescents. Hisp J Behav Sci. 1994;16(2):99–115. [Google Scholar]
- 19. Davis TC, Crouch MA, Long SW, et al. Rapid assessment of literacy levels of adult primary care patients. Fam Med. 1991;23(6):433–5. [PubMed] [Google Scholar]
- 20. Fagerlin A, Zikmund-Fisher BJ, Ubel PA, Jankovic A, Derry HA, Smith DM. Measuring numeracy without a math test: development of the Subjective Numeracy Scale. Med Decis Making. 2007;27(5):672–80. [DOI] [PubMed] [Google Scholar]
- 21. McNaughton C, Wallston KA, Rothman RL, Marcovitz DE, Storrow AB. Short, subjective measures of numeracy and general health literacy in an adult emergency department. Acad Emerg Med. 2011;18(11):1148–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. McNaughton CD, Cavanaugh KL, Kripalani S, Rothman RL, Wallston KA. Validation of a short, 3-item version of the subjective numeracy scale. Med Decis Making. 2015;35(8):932–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schwartz LM, Woloshin S, Black WC, Welch HG. The role of numeracy in understanding the benefit of screening mammography. Ann Intern Med. 1997;127(11):966–72. [DOI] [PubMed] [Google Scholar]
- 24. Wirth RJ, Edwards MC. Item factor analysis: current approaches and future directions. Psychol Methods. 2007;12(1):58–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Muthén LK, Muthén BO. Mplus User’s Guide. 7th ed.Los Angeles, CA: Muthén & Muthén; 1998–2012. [Google Scholar]
- 26. Steiger JH. Structural model evaluation and modification—an interval estimation approach. Multivariate Behav Res. 1990;25(2):173–80. [DOI] [PubMed] [Google Scholar]
- 27. Tucker LR, Lewis C. Reliability coefficient for maximum likelihood factor-analysis. Psychometrika. 1973;38(1):1–10. [Google Scholar]
- 28. Bentler PM. Comparative fit indexes in structural models. Psychol Bull. 1990;107(2):238–46. [DOI] [PubMed] [Google Scholar]
- 29. Cai L, Thissen D, du Toit SHC. IRTPRO for Windows. Lincolnwood, IL: Scientific Software International; 2011. [Google Scholar]
- 30. Thissen D. Marginal maximum likelihood estimation for the one-parameter logistic model. Psychometrika. 1982;47:201–14. [Google Scholar]
- 31. Birnbaum AS. Some latent trait models and their use in inferring an examinee’s ability. In: Lord FM, Novick MR, eds. Statistical Theories of Mental Test Scores. Reading, MA: Addison-Wesley; 1968. p 392–479. [Google Scholar]
- 32. Orlando M, Thissen D. Further investigation of the performance of S-X-2: an item fit index for use with dichotomous item response theory models. Appl Psychol Meas. 2003;27(4):289–98. [Google Scholar]
- 33. Orlando M, Thissen D. Likelihood-based item-fit indices for dichotomous item response theory models. Appl Psychol Meas. 2000;24(1):48–62. [Google Scholar]
- 34. Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate - a practical and powerful approach to multiple testing. J Roy Stat Soc B Method. 1995;57(1):289–300. [Google Scholar]
- 35. Williams VSL, Jones LV, Tukey JW. Controlling error in multiple comparisons, with examples from state-to-state differences in educational achievement. J Educ Behav Stat. 1999;24(1):42–69. [Google Scholar]
- 36. Chen WH, Thissen D. Local dependence indexes for item pairs: using item response theory. J Educ Behav Stat. 1997;22(3):265–89. [Google Scholar]
- 37. Reise SP, Moore TM, Haviland MG. Bifactor models and rotations: exploring the extent to which multidimensional data yield univocal scale scores. J Pers Assess. 2010;92(6):544–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. ten Berge JMF, Socan G. The greatest lower bound to the reliability of a test and the hypothesis of unidimensionality. Psychometrika. 2004;69(4):613–25. [Google Scholar]
- 39. Bentler PM. Alpha, dimension-free, and model-based internal consistency reliability. Psychometrika. 2009;74(1):137–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sijtsma K. On the use, the misuse, and the very limited usefulness of Cronbach’s alpha. Psychometrika. 2009;74(1):107–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Thissen D, Steinberg L, Wainer H. Detection of differential item functioning using the parameters of item response models. In: Holland PW, Wainer H, eds. Differential Item Functioning. Hillsdale, NJ: Erlbaum; 1993. p 67–113. [Google Scholar]
- 42. Thissen D, Wainer H. Test scoring. J Educ Meas. 2002;39(3):265–8. [Google Scholar]
- 43. Cronbach LJ. Coefficient alpha and the internal structure of tests. Psychometrika. 1951;16(3):297–34. [Google Scholar]
- 44. Streiner DL, Norman GR. Health Measurement Scales. A Practical Guide to Their Development and Use. 2nd ed.Oxford, England: Oxford University Press; 1995. [Google Scholar]
- 45. Lea DH, Kaphingst KA, Bowen D, Lipkus I, Hadley DW. Communicating genetic and genomic information: health literacy and numeracy considerations. Public Health Genomics. 2011;14(4–5):279–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Institute of Medicine. Health Literacy: A Prescription to End Confusion. Washington, DC: National Academies Press; 2004. [PubMed] [Google Scholar]
- 47. Kutner M, Greenberg E, Jin Y, et al. The Health Literacy of America’s Adults: Results From the 2003 National Assessment of Adult Literacy (NCES 2006-483). Washington, DC: US Department of Education; 2006. [Google Scholar]
- 48. Lai JS, Teresi J, Gershon R. Procedures for the analysis of differential item functioning (DIF) for small sample sizes. Eval Health Prof. 2005;28(3):283–94. [DOI] [PubMed] [Google Scholar]


