Abstract
Metallo-β-lactamases (MBLs) are a significant clinical problem because they hydrolyze and inactivate nearly all β-lactam containing antibiotics. These “lifesaving drugs” constitute >50% of the available contemporary antibiotic arsenal. Despite the global spread of MBLs, MBL inhibitors have not yet appeared in clinical trials. Most MBL inhibitors target active site zinc ions and vary in mechanism from ternary complex formation to metal ion stripping. Importantly, differences in mechanism can impact pharmacology in terms of reversibility, target selectivity, and structure activity relationship interpretation. This article surveys the mechanisms of MBL inhibitors and describes methods that determine the mechanism of inhibition to guide development of future therapeutics.
Keywords: metallo-β-lactamase, inhibitor, mechanism, spectroscopy
Antibiotic Resistance
Bacterial resistance towards antibiotics is a serious medical problem. In fact, several bacterial strains have been shown to be resistant to all current antibiotics, including “last resort” compounds such as the carbapenems [1, 2]. Efforts to combat resistance include the discovery of new antibiotics and the discovery of inhibitors, which target a protein that is involved in the resistance phenotype. This review describes the latter approach, involving inhibitors of metallo-β-lactamases.
Metallo-β-lactamases (MBLs)
β-Lactamases are bacterial enzymes that hydrolyze β-lactam containing compounds, which are the largest group of clinically-used antibiotics. Currently, there are more than 1,800 β-lactamases, which are classified according to molecular properties and/or amino acid sequences into 4 distinct groups: A, B, C, and D [3]. The A, C, and D β-lactamases utilize an active site serine for catalysis, and there are clinically-used inhibitors, such as clavulanate, sulbactam, avibactam, and tazobactam, that inhibit many of these serine-β-lactamases to restore sensitivity to β-lactam drugs [4]. The group B β-lactamases are called metallo-β-lactamases (MBLs) because they use 1 or 2 zinc ions (Zn(II)) at the active site for catalysis (Figure 1). MBLs (Box 1) can hydrolyze and inactivate all β-lactam containing antibiotics, except monobactams. Importantly, there are currently no drugs or drug combinations, including those used to inhibit serine-β-lactamases, that can effectively counteract MBL-mediated resistance.
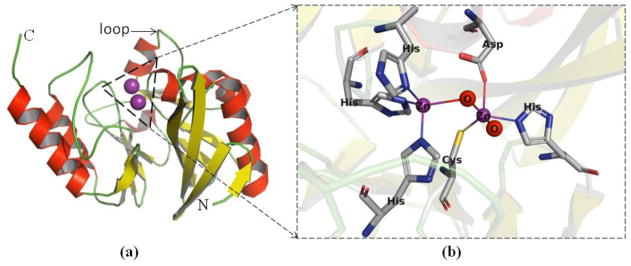
Figure 1. Ribbon diagram of NDM-1.
(a) The αββα tertiary fold with metal binding sites of NDM-1 is colored by secondary structure. (b) A close up view of active-site of NDM-1. Zinc ions (purple) and water molecules (red) are represented as spheres. Labeled residues are shown as sticks and coordination bonds as lines. Structures are rendered using Pymol (version 2.0.6) and the PDB coordinates 3SPU. (Abbreviations in this Figure: C: C terminus, N: N terminus, Zn: Zinc ion, O: water molecule, His: histidine, Cys: cysteine, Asp: aspartic acid.)
Box 1. Metallo-β-lactamases.
The group B β-lactamases (metallo-β-lactamases, MBLs) are further subdivided into three distinct subgroups (B1, B2, and B3) based on the identity of metal binding ligands and number of metal ions bound in the active site (generally two Zn(II) ions bound to the B1 and B3 MBLs and one Zn(II) ion bound to the B2 MBLs) (Figure 1) [61]. The most clinically important MBLs belong to the B1 subgroup: IMPs (IMipenemases), VIMs (Verona ImipeneMases), and NDMs (New Delhi Metallo-β-lactamases). Among these, IMP-1 was reported in 1991 from Pseudomonas aeruginosa, VIM-1 was reported in 1999 from P. aeruginosa, and NDM-1 was reported in 2009 from Klebsiella pneumoniae [62].
Selective pressures continue to drive the evolution of these resistance determinants, with many clinical variants of MBLs emerging: currently there are 53–70 clinical variants of IMP, 46–55 variants of VIM, and 16–21 variants of NDM, according to the Lahey.org (http://www.lahey.org/studies/other.asp#table1) or the NCBI website (http://www.ncbi.nlm.nih.gov/protein/). As with the serine β-lactamases, some of the clinical MBL variants show improvements in stability, selectivity and/or kinetic properties [5–8]. For example, MBLs from 2 Aeromonads require only 1 Zn(II) for full catalytic activity and preferentially hydrolyze carbapenems [9]. However, recent studies have revealed an additional adaptation peculiar to the MBLs. Clinical variants of NDM have evolved improved activity at low zinc concentrations, presumably in response to selective pressures placed by zinc scarcity at infection sites [7, 8]. Additionally, MBL genes are often encoded on mobile elements and some MBL-encoding plasmids also carry resistance genes for different classes of antibiotics, greatly increasing the difficulty of effectively treating bacterial infections [10].
Mechanisms of MBL Inhibition
The lack of effective treatments to overcome MBL-mediated resistance is an unmet clinical need driving the development of MBL inhibitors. Several recent reviews have described progress in this area, often grouping inhibitors by structural similarities [11–14]. Here, we suggest an alternative categorization that may also be useful: mechanism of inhibition. We surveyed a sampling of MBL inhibitors reported in the literature and found that examples of most structural categories of MBL inhibitors can be grouped into one of 4 different mechanisms (Table 1): (1) inhibition through metal ion binding, (2) inhibition through covalent bond formation, (3) allosteric inhibitors, or (4) inhibitors with uncharacterized mechanisms (Table 1 and Figure 2).
Table 1.
Four mechanisms of inhibition by MBL inhibitors.a
| Mechanism of inhibition | Inhibitor name | References | |
|---|---|---|---|
| Metal ion binding inhibitorsb | Metal ion binding inhibitors that form ternary (MBL:Zn(II): inhibitor) complexes | (1) 1,2,4-triazole-3-thiones | [63] |
| (2) captopril and analogs | [12] | ||
| (3) cyclic boronates | [14] | ||
| (4) bisthiazolidines | [12] | ||
| (5) mercaptoacetate | [11] | ||
| (6) mercaptocarboxylate | [11] | ||
| (7) mercaptophosphonates | [64] | ||
| (8) aminophtalic acid derivatives | [13] | ||
| (9) disubstituted succinic acids | [12] | ||
| (10) biphenyl tetrazoles | [12] | ||
| (11) tiopronin | [12] | ||
| (12) pyridine-2,4-dicarboxylic acids | [13] | ||
| (13) thiomandelic acids, other thiol containing derivatives | [12, 65] | ||
| (14) rhodanine hydrolysis products | [12] | ||
| (15) triazolythioacetamide analog | [14] | ||
| (16) sulfonamides | [12] | ||
| (17) tricyclic natural products | [12] | ||
| (18) benzophenone | [14] | ||
| (19) 3-oxoisoindoline-4-carboxylates (also called isoquinolines; note these do not directly coordinate Zn(II) but bind very close) | [14] | ||
| (20) dipicolinic acid derivatives | [29] | ||
| (21) benzyl thiol | [66] | ||
| (22) cyclobutanone penem analog | [58] | ||
| Metal stripping inhibitors that result in a metal:inhibitor complex and an MBL with reduced amounts of Zn(II) bound | (23) EDTAc | [12] | |
| (24) 1,10-phenanthroline | [13] | ||
| (25) picolinic acids | [13] | ||
| (26) TPENd | [12] | ||
| (27) NODAGAe | [12] | ||
| (28) NOTAf | [12] | ||
| (29) DOTAg | [12] | ||
| (30) 1,4,7-triazonane-1,4,7-tris (carboxylodithioate) analogs | [67] | ||
| (31) AMAh | [12, 27] | ||
| Covalent inhibitors | (32) mercaptophenylacetic acid | [12] | |
| (33) moxalactam | [12] | ||
| (34) cephamycins | [12] | ||
| (35) 3-(3-mercaptopropionylsulfanyl)-propionic acid pentafluorophenyl ester | [12] | ||
| (36) cefaclor | [12] | ||
| (37) ebselen | [12] | ||
| (38) formylchromone | [14] | ||
| Allosteric inhibitors | (39) arginine peptides | [12] | |
| (40) camelid nanobody | [11] | ||
| (41) graphene oxide and nanotubes | [68] | ||
| (42) DNA nanoribbon | [69] | ||
| (43) single stranded DNAs | [11] | ||
| Inhibitors with uncharacterized mechanisms | (44) azolylthioacetamides | [12] | |
| (45) several different classes of thionylpeptides | [12] | ||
| (46) hydroxamates and “reverse” hydroxamates | [12, 70] | ||
| (47) maleic acids | [13] | ||
| (48) mitoxantrone | [12] | ||
| (49) triazoles | [12] | ||
| (50) mercaptotriazoles | [12] | ||
| (51) substituted thiosemicarbazides | [12] | ||
| (52) arylsulfonyl hydrazones | [12] | ||
| (53) pyrrole derivatives | [13, 71] | ||
| (54) thioesters | [13] | ||
| (55) thiols | [12] | ||
| (56) trifluoro alcohols and ketones | [11] | ||
| (57) sulfonamides | [12] | ||
| (58) polyketides | [12] | ||
| (59) cystatins | [72] | ||
All of the MBL inhibitors shown are currently for laboratory use only.
Metal ion binding inhibitors can be divided into 2 distinct subgroups, including ternary complex formation and metal ion stripping.
EDTA: ethylenediaminetetraacetic acid.
TPEN: tetrakis(2-pyridylmethyl)ethylenediamine.
NODAGA: 1,4,7-triazacyclononane-1-glutaric acid-4,7-diacetic acid.
NOTA: 1,4,7-triazacyclononane-1,4,7-triacetic acid.
DOTA: 1,4,7,10-tetraazacyclododecane-1,4,7,10-tetraacetic acid.
AMA: aspergillomarasmine A.
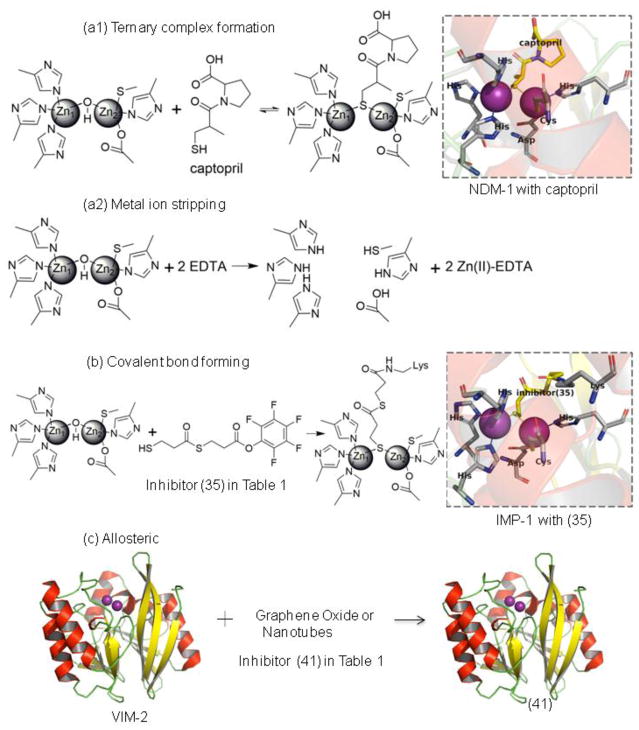
Figure 2. Different mechanisms of inhibition of MBL inhibitors.
(a1) Inhibitor that binds metal ions and forms a ternary (MBL:Zn(II):inhibitor) complex, such as the crystal structure of NDM-1 with captopril (PDB code 4EXS). (a2) Inhibitor that removes metal ions from active sites of MBL. (b) Covalent bond formation, such as the crystal structure of IMP-1 with inhibitor (35) (PDB code 1VGN). (c) Allosteric inhibitor, cartoon structure is VIM-2 (PDB code 5LCA). The binding site for allosteric inhibitors has not been characterized in detail and is shown schematically.
The first mechanism, inhibition through metal ion binding, will be discussed in depth below. The second mechanism, inhibition through covalent bond formation, has a number of potential advantages, including irreversibility, ability to outcompete high concentrations of substrate, duration of action, and ease of target (and off-target) identification. These types of inhibitors have a long history of success exemplified by covalent inhibitors of serine β-lactamases [15]. However, since the catalytic nucleophile (see Glossary) (the zinc-bridging hydroxide) used in MBL catalysis is not covalently attached, these enzymes lack a convenient residue to target for modification. Covalent inhibitors of B1 MBLs are typically reactive compounds (e.g., ebselen, activated esters, aldehydes) that either target the Cys, which is a Zn2 ligand, or an active site Lys that is not conserved within the subfamily. Thiol-containing compounds could also potentially be used to form covalent adducts, provided that the sulfhydryl group is deprotonated in the enzyme active site [16, 17]. However, such reactive compounds are typically non-selective and carry significant toxicity risks including potential immunogenicity of protein-inhibitor adducts [18].
The third mechanism, allosteric modulation of activity, is not easily designed rationally as MBLs are not known to have evolved allosteric control mechanisms. MBL inhibitors discovered in this class are typically macromolecules (e.g., DNA constructs, nanobodies), which are sometimes more difficult than small molecules to characterize and manufacture [19]. Most notably, significant sequence differences found outside of MBL active sites suggest that targeting an exosite may not yield a broad-spectrum inhibitor.
MBL inhibitors in the fourth category, those with uncharacterized mechanisms, are often only characterized using IC50 values to estimate binding potency, with no further mechanistic characterization or an assumed active-site binding [20]. We posit that this extent of characterization is insufficient to accurately guide inhibitor development. Since IC50 values of time-dependent or irreversible inhibitors, including metal-stripping inhibitors (see below) vary with time, kinetic and binding components of inhibition are comingled, complicating structure-activity relationship interpretations [21]. The presence of a functional group with a propensity to bind metal ions is not sufficient evidence to conclude that an inhibitor binds at the active site of MBLs, and differing mechanisms of inhibition have implications in terms of target selectivity, pharmacology, and suitability for drug development.
The most widespread strategy used by MBL inhibitors is that of the first mechanism, metal ion binding, and we focus on this mechanism for the remainder of the article.
Mechanism of Metal Ion Binding
Utilization of metal-binding inhibitors to overcome MBL-mediated resistance is unsurprising as the active site metal center of MBLs is their defining feature and constitutes the most conserved region of this diverse family of enzymes [12]. Metal binding inhibitors function within two limiting mechanisms: 1) metal ion stripping, where the inhibitor either actively removes the active-site metal ions from the enzyme or sequesters metal ions that exited the active site; or 2) ternary complex formation, where the inhibitor binds to the metal ions and the surrounding protein residues, preventing antibiotics from binding.
The first mechanism used by metal-binding inhibitors, metal ion stripping, has the longest precedent. The original MBL report, more than 50 years ago, included EDTA treatment to remove zinc and abrogate activity [22, 23]. MBL inhibition by the natural product aspergillomarasmine A (AMA) is a more recent example of a compound demonstrated to inhibit NDM and VIM enzymes and to restore the ability of meropenem to counter VIM and NDM producing infections in mice, through the same metal stripping mechanism [24–27]. This strategy however has potential drawbacks. MBLs that weakly bind Zn(II) will be inhibited easier, but some clinical variants bind zinc more tightly, suggesting that resistance to such inhibitors is already emerging [7, 8]. Conversely, MBLs that tightly bind Zn(II) have an affinity for metal ion cofactors more similar to host metalloproteins, so inhibitors capable of stripping these metal ions are predicted to have significant off target effects. The potency of metal-stripping agents would be expected to vary with exogenous zinc concentrations, which can vary between infection sites [7]. Once metal ions are removed, some MBLs can readily rebind zinc and recover activity, but some cannot, resulting in irreversible inhibition that can complicate the interpretation of structure-activity data [27]. Finally, optimizing the potency of metal-stripping inhibitors maximizes the inhibitor:metal ion complex and is not specific for any particular enzyme, so more effective metal stripping agents would be expected to have decreased selectivity. It is difficult to envision how to generate selectivity for an inhibitor to a particular active site if the inhibitor does not reside in the active site (e.g., dissociates upon metal stripping). Inhibitors that utilize a metal stripping mechanism can serve as useful tool compounds and can guide development of MBL inhibitors that utilize metal binding as one element of forming stable ternary complexes.
In contrast are the metal-binding inhibitors that work though the other limiting mechanism, formation of an MBL:zinc:inhibitor ternary complex. One of the most common inhibitors that uses this mechanism is D- or L-captopril, which coordinates to both zinc active-site zinc ions by replacing the bridging hydroxide with a sulfur group on the inhibitor (Figure 2(a1)) and also makes binding interactions with surrounding amino acids to stabilize the ternary complex [28]. In fact, there are now several thiol-containing compounds that are thought to inhibit MBLs in the same way [16, 17]. There are a number of MBL inhibitor examples where metal-inhibitor contacts are optimized along with protein-inhibitor contacts to maximize the ternary complex [11–14]. This type of metalloenzyme inhibition mechanism is more commonly used in the optimization of drug lead compounds [16]. Unlike metal-stripping inhibitors, optimization of ternary complex formation is expected to lead to an increase, rather than a decrease, of inhibitor selectivity because the ternary complex forms in the first place due to specific enzyme:inhibitor interactions.
While these two limiting mechanisms describe the extremes, a subset of metal-binding inhibitors have properties of both mechanisms. A recent report of dipicolinic acid-based MBL inhibitors illustrates how lead optimization can be used to drive ternary complex formation and minimize metal-stripping when guided by a mechanistic analysis of how inhibition is achieved [29]. Due to the impact of inhibition mechanism on reversibility, target selectivity, sensitivity to exogenous zinc concentrations, and interpretation of structure activity relationships, we stress the importance of understanding these mechanisms and provide some examples below to guide MBL inhibitor development.
Mechanism of inhibition by metal binding inhibitors
One of the most straightforward experiments to address mechanism of inhibition of metal binding inhibitors is equilibrium dialysis studies (Figure 3(a)) [29]. In these studies, the MBL is incubated with the inhibitor, and the resulting solution is dialyzed versus multiple changes of buffer. The metal content of the MBL is determined by using atomic absorption (AA) spectroscopy, inductively-coupled plasma with atomic emission (ICP-AES) or mass spectrometry (ICP-MS) detection, or one of the colorimetric/fluorimetric assays for zinc. The concentrations of MBL and inhibitor necessary for this experiment are dictated by the detection limits of the technique used to determine the metal content (micromolar for AA, ICP-AES, and most colorimetric assays and 10–100 nanomolar for ICP-MS). Inhibitors that exhibit a metal stripping mechanism of inhibition will show a reduction of metal content in the presence of inhibitor, while those inhibitors that exhibit a metal coordination mechanism of inhibition will show no loss of metal. It is possible that some metal-targeted inhibitors will remove some of the metal in the MBLs at elevated concentrations of inhibitor. Inhibitor redesign efforts would optimize enzyme-inhibitor contacts to anchor the compound in the active site [11–14].
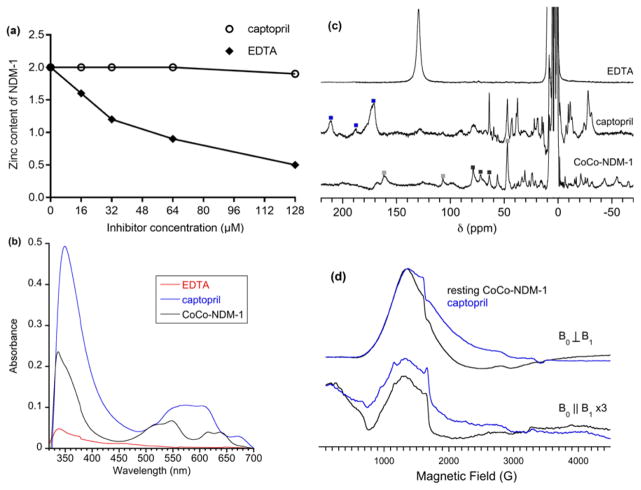
Figure 3. NDM-1 inhibitor studies.
(a) Equilibrium dialysis study of the zinc content of NDM-1 (8 μM) after dialysis with various concentrations of captopril (circles) and EDTA (solid rhombuses). Incubation of NDM-1 with captopril results in no loss of zinc, while incubation of NDM-1 with EDTA results in reduction of zinc content. (b) UV-Vis spectroscopy of CoCo-NDM-1 with 2 equivalent EDTA (red line), CoCo-NDM-1 with 2 equivalent captopril (blue line), and CoCo-NDM-1 (black line). Incubation of CoCo-NDM-1 with EDTA results in significant reduction of absorbance at 320–350 nm (which indicates that cysteine is bound to Co(II)) and at 500–650 nm (d-d transitions) (which reveals information about the number of atoms bound to the Co(II) and often the identity of the atoms bound). Incubation of CoCo-NDM-1 with captopril results in an increased absorbance at 320–350 nm and a shift of absorbance at 500–650 nm. (c) 300 MHz 1H NMR spectroscopy of CoCo-NDM-1 with 2 equivalent EDTA (top), CoCo-NDM-1 with 3 equivalent captopril (center) and CoCo-NDM-1 (bottom). Resonances assigned to metal 1 site are marked with black squares. Resonances to metal 2 site are marked with gray squares. Resonances indicative of a ternary (NDM-1:Co(II):captopril) complex are marked with blue squares. The loss of peaks in the NMR spectrum of CoCo-NDM-1 with EDTA indicates the inhibitor removes Co(II) from the active site. The shifted peaks in the NMR spectrum of CoCo-NDM-1 with captopril indicates the binding of the inhibitor to the metal ions in the active site of the protein. (d) X-band EPR spectroscopy of CoCo-NDM-1 (black lines) and CoCo-NDM-1 with 1 equivalent captopril (blue lines). Standard perpendicular mode spectra are on top, and parallel mode spectra (scaled 3-fold) are shown below. The EPR spectra of CoCo-NDM-1 before and after captopril binding shows that the atoms bound to Co(II) in the two enzymes are different. In particular, the peak at 800 G in the bottom figure (parallel mode) demonstrates that a different atom bridges the Co(II) ions: in the resting enzyme the bridging atom is oxygen from water and in the CoCo-NDM-1/captopril complex, the bridging atom is sulfur from captopril.
Another technique to probe inhibitor mechanisms is native-state electrospray ionization mass spectrometry (ESI-MS) [30–32]. This technique involves the use of micromolar concentrations of MBL and inhibitor and often requires a tight association (micromolar) of the inhibitor with the MBL, since MS requires that the complex be ionized and vaporized in a manner that preserves a native-like folded state [32]. ESI-MS has been used to screen thiol-based inhibitor libraries, ascertain Zn(II) binding to MBLs, and probe the presence of ternary (MBL:zinc:inhibitor) complexes [30, 33]. An advantage of this technique is that peaks can be obtained for a ternary complex, an enzyme-inhibitor complex in the absence of metal ions, and apo MBL (MBL without bound zinc) simultaneously. However, this technique does not indicate whether the inhibitors coordinate directly to metal ions at the active site.
One direct method to probe inhibition mechanisms is to determine the three -dimensional structure of an MBL-inhibitor complex. There are many examples in the Protein Databank of MBL:metal:inhibitor complexes (75 unique structures as of early 2018), supporting the ternary complex formation mechanism of inhibition (compounds (1) to (22) in Table 1). X-ray crystallography requires that well diffracting crystals of the enzyme-inhibitor complex be obtained, which usually requires millimolar concentrations of enzyme. This requirement has complicated the universal use of this crystallography for MBL inhibitor characterization. Solution NMR structures require millimolar quantities of isotopically-labeled protein (13C, 15N, 2H) and high-field NMR spectrometers (>600 MHz) [34]. The size of the MBL is also important for solution NMR structures, as enzymes >30 kDa are difficult to probe using this technique [34]. The catalytic domains of many recombinant MBLs are <30 kDa, enabling the use of this technique, although higher oligomers can cause complication. The inability to obtain a structure of a ternary complex does not necessarily indicate a metal stripping mechanism because high concentrations of MBL and inhibitor used in structural determinations could favor a mechanism different from that at more physiologically-relevant concentrations [29].
To address some of the experimental issues experienced with structure determinations, spectroscopic techniques can be used to probe inhibitor mechanisms. In nature, MBLs predominantly bind Zn(II) ions, but there are only two spectroscopic techniques that can be used to interrogate diamagnetic Zn(II) in metalloproteins: (1) 67Zn NMR spectroscopy, which yields limited information and is costly [35], and (2) extended X-ray absorption fine structure (EXAFS) spectroscopy, which requires beam time on a synchrotron and millimolar concentrations of the enzyme [36]. EXAFS studies on Zn(II)-metalloproteins are conducted on frozen samples, so ternary complexes can be trapped by rapid freezing [37]. EXAFS yields information about the number of atoms bound to the Zn(II) ions and the metal-ligand distances [36]. EXAFS studies have been reported on several MBLs with substrate or product bound [36].
A common strategy to overcome the spectroscopically-taciturn Zn(II) is to prepare Co(II)-substituted metalloforms of MBLs. Co(II) is a common surrogate for Zn(II) in Zn-metalloproteins because Co(II) often binds identically to Zn(II) and Co(II) metalloforms are usually catalytically-active [9, 38]. Unlike diamagnetic Zn(II), Co(II), which can be substituted in Zn(II) sites, is paramagnetic (3 unpaired electrons) and can be interrogated by many magnetic resonance techniques. Co(II) MBL metalloforms (NDM, VIM, IMP, L1, CcrA, ImiS, BcII, and many others) have been reported and characterized using 1H NMR, UV-Vis, EPR, and EXAFS spectroscopies [39–46]. For some MBLs, it is even possible to prepare mixed-metal analogs [39, 43, 47] with Co(II) in one particular metal binding site and Zn(II) in the other. These CoZn metalloforms allow for interrogation of one specific metal binding site because only the Co(II)-containing site is observable using selective spectroscopic techniques. Such techniques do not require crystal formation and can be used to detect and characterize ternary complexes. The use of these techniques can reveal inhibition mechanisms of metal binding inhibitors and distinguish between metal stripping and ternary complex formation.
Spectroscopic studies on MBL-inhibitor complexes
There are several methods to prepare Co(II)-substituted MBLs. The most common method is to remove Zn(II) using chelators, such as EDTA or phenanthroline, and then add Co(II) directly to the metal-free enzyme [38, 48]. Some MBLs require alternative methods to prepare the Co(II)-substituted analogs. One method is to over-express and purify the MBL in minimal medium while including Co(II) in the media used for protein over-expression [40]. A common problem with this approach is that Co(II) oxidizes to Co(III), resulting in an inactive MBL. Another method is to prepare apo MBL (metal-free), unfold the MBL using urea or guanidine-HCl, and refold the MBL in the presence of Co(II) [49]. After a Co(II)-metalloform is prepared, it is imperative to use spectroscopic/biochemical techniques to verify metal stoichiometry, catalytic activity, expected coordination number of Co(II), and integrity of the refolded binding site [39, 40, 43, 48]. For each MBL, the optimal method to prepare the Co(II)-substituted analog must be empirically-determined, but there are published methods to guide this effort [40, 48, 49].
The preparation of a Co(II)-MBL metalloform enables use of UV-Vis spectroscopy, a technique that relies on the absorption of UV-Vis radiation by d electrons in Co(II). Most Co(II)-substituted MBLs are pink in color, and two important pieces of information can be obtained by obtaining a UV-Vis spectrum on a Co(II)-substituted MBL (required concentrations are 100 μM to 1 mM). First, an intense peak (extinction coefficients of 500–2,000 M−1cm−1) at 320–350 nm often signifies the presence of a cysteine sulfhydryl bound to Co(II) [39]. If binding of an inhibitor causes the cysteine to dissociate from Co(II) (perhaps by oxidation of the cysteine sulfhydryl or by removal of Co(II) by an inhibitor), this peak will disappear. Alternatively, an increase in peak intensity could indicate an additional thiolate coordination to Co(II) by the inhibitor. Second, another group of peaks (3, perhaps with an additional shoulder) from 500–650 nm can be found for Co(II)-containing MBLs. These peaks arise from d electrons in Co(II) absorbing visible light and are often called d-d or ligand field transitions [39]. The intensities of these peaks often correlate to the number of ligands bound to Co(II), with 4-coordinate Co(II) exhibiting intense peaks (300–500 M−1cm−1), 5-coordinate Co(II) exhibiting intermediate peaks (150 M−1cm−1), and 6-coordinate Co(II) exhibiting weak peaks (30 M−1cm−1) [39]. The binding of an inhibitor can result in an extra ligand bound to the Co(II) resulting in less intense d-d peaks or significantly altered d-d peaks (Figure 3(b), with captopril as the inhibitor). If an inhibitor, for example EDTA, removes Co(II), the resulting Co-EDTA complex is 6-coordinate, and the intensities of the d-d transitions are significantly reduced (Figure 3(b)). Although UV-Vis spectrophotometry is a relatively simple experiment, this technique can offer valuable information about inhibitor binding to the MBLs.
A second technique to probe inhibitor binding to Co(II)-substituted MBLs is 1H NMR spectroscopy [50]. This technique can be used to identify amino acids bound to Co(II) in MBLs. The unpaired electrons on high-spin Co(II) alter the relaxation properties of protons near Co(II), resulting in large shifts in the resonance position of those protons [51]. For example, -CH2- protons on a Cys bound to Co(II) can be shifted to 300–500 ppm relative to the water peak at 4.8 ppm. The presence of such shifted peaks confirms the binding of a Cys to Co(II), as shown in the UV-Vis spectra described above. Histidine groups bound to Co(II) (and by analogy Zn(II)) are readily assigned in the 1H NMR spectra (Figure 1), because N-H protons on the histidine ring usually appear in the 30–100 ppm (relative to the solvent peak) region and are solvent-exchangeable [47]. Therefore, exchanging the solvent water with D2O by dialyzing a Co(II)-MBL metalloform in D2O-containing buffer results in a sample with NMR spectra with fewer NMR peaks. If the missing peaks appeared originally in the 30–100 ppm region, they can usually be assigned to N-H protons on histidines coordinated to Co(II) [50]. Other peaks are typically assigned by comparing the NMR spectrum of the MBL with previously published spectra [39] or with spectra from inorganic model compounds [52].
Once the 1H NMR spectrum of Co(II)-substituted MBL has been obtained and peaks assigned, NMR spectra of the MBL with different concentrations of inhibitor can be obtained. An inhibitor that forms a ternary complex often results in shifted peaks, as can be observed in the spectrum of captopril and NDM-1 (Figure 3(c)). These shifted peaks provide evidence for formation of a ternary complex. On the other hand, an inhibitor that removes Co(II) from the active site results in the loss of shifted 1H resonances from the MBL and the presence of new peaks that can be assigned to a Co(II)-inhibitor complex formed separate from the MBL (see NMR spectrum of CoCo-NDM-1 with EDTA, Figure 3(c)). We note that 1H NMR spectroscopy requires millimolar concentrations of enzyme and inhibitor, and it is possible that at millimolar concentrations a metal stripping mechanism may be utilized that may be less predominant at lower concentrations of inhibitor and enzyme [29].
A third technique using Co(II)-MBL metalloforms to probe inhibitor mechanisms is electron paramagnetic resonance (EPR) spectroscopy [38]. EPR spectroscopy monitors the electron relaxation properties of unpaired electrons in paramagnetic compounds, in the case of Co(II)-MBL metalloforms, the 3 unpaired electrons on high-spin Co(II) ions. The electronic properties of Co(II) are affected by the ligands that are bound and by the presence of any other unpaired electron or nuclear spins in close proximity to the Co(II) [38]. EPR spectra of Co(II)-metalloforms must be obtained at low temperatures (<30 K), and good signal-to-noise often requires protein concentrations between 0.2 and 1 mM [38]. In general, EPR signals for Co(II)-substituted MBLs are broad [29], and there are small changes in the signal upon binding of an inhibitor to the metal ions (Figure 3(d)). However if inhibitors bridge the Co(II) ions in the MBLs, the coupling between the Co(II) ions can change and be detected with EPR spectroscopy when run in the parallel and perpendicular modes [29]. The correct acquisition and interpretation of EPR spectra can be difficult [38, 53], but the technique yields valuable information about inhibitor binding.
The spectroscopic techniques described above offer detailed information about inhibitor binding to the metal center(s) in the MBLs. Nonetheless, these spectroscopic techniques cannot reveal information about inhibitor binding at a distant site. Previous studies have indicated that conserved, non-metal features of MBLs (e.g., a substrate-binding β-hairpin loop in B1 MBLs or a neighboring α-helix in B2 MBLs) can play a role in binding and catalysis [12, 54]. It is likely these features interact with inhibitors to increase binding affinity [12]. Recently, it has been shown that 19F NMR and an MBL (at ca. 50 micromolar concentrations) that was site-specifically labeled with a fluoro compound, could be used to probe the interaction of an inhibitor with the invariant loop [55–58]. Another technique used to probe loop motions is double electron electron resonance spectroscopy (DEER), a pulsed EPR technique, that can detect a change in loop positioning upon inhibitor binding, providing evidence for a ternary complex (Figure 4) [59, 60]. Such techniques provide information on inhibition mechanisms without requiring the preparation of alternative metalloforms and can be used with lower concentrations (micromolar) of enzyme.
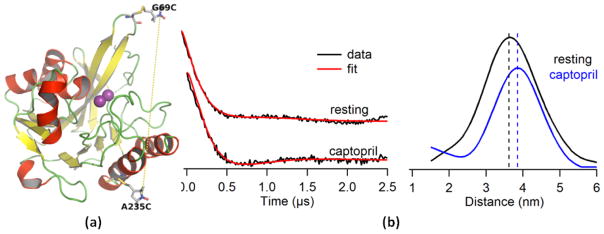
Figure 4. Structure of a site-specifically labeled NDM-1 and DEER spectra of captopril binding to NDM-1.
(a) Cartoon structure of double spin labeled NDM-1 (G69C/A235C). The positions are Gly69 on the hairpin loop and Ala235 on a remote α-helix as optimal sites for MTSL labeling (PDB code: 3ZR9). (b) Q-band DEER spectra of the labeled NDM-1. Time domain traces with corresponding fit of the resting, labeled NDM-1 and of NDM-1 complexed with captopril (left). These fits were included to demonstrate how well the data were fitted by the DEER analysis software. Distance distribution spectra of the resting, labeled NDM-1 (black) and of labeled NDM-1 complexed with captopril (blue) (right). The horizontal lines show the average distance between the introduced spin labels in the two samples.
Concluding Remarks
The rise of metallo-β-lactamase mediated antibiotic resistance threatens the effectiveness of the largest class of antibiotics in clinical use. Therapeutic compounds that counter the activity of these enzymes would represent a significant clinical advance (see Outstanding Questions). We argue here that development of effective MBL inhibitors requires considerations other than just overall potency. The mechanism of inhibition can significantly impact reversibility, target selectivity, and structure activity relationship interpretation and should be integrated into the development of new MBL inhibitors. Several methods highlighted here may help guide the choice of inhibition strategy and the optimization of novel compounds to combat this serious antibiotic resistance threat.
Outstanding Questions Box.
Many reported inhibitors do not inhibit all MBLs. What is the best strategy to develop a broad-spectrum MBL inhibitor?
Many biochemical and structural techniques can elucidate inhibition mechanisms. Should inhibitors with particular mechanisms be prioritized during optimization
Many of the metal targeting compounds inhibit the MBLs by stripping the zinc ion(s) from the active site. Is a metal stripping mechanism appropriate for clinical use?
Highlights.
Clinical resistance to β-lactam containing antibiotics due to bacterial expression of metallo-β-lactamases is becoming more prevalent and problematic.
Metallo-β-lactamase inhibitors have been reported, but the mechanism of inhibition is unknown for many of these inhibitors.
Mechanism of inhibition impacts reversibility, selectivity, and interpretation of structure activity relationships, and should be strongly considered in inhibitor development.
Biochemical and structural studies can be used to demonstrate the mechanism of inhibition, and this information can be used to guide lead optimization.
Acknowledgments
This work was supported in part by the National Institutes of Health (Grants GM111926; GM111926 (to RAB), R01AI100560 (to RAB), R01AI063517 (to RAB), and R01AI072219 (to RAB)), by the National Science Foundation (CHE-1509285 to MWC), and by the Robert A. Welch Foundation (Grant F-1572 to WF). This study was also supported in part by funds and/or facilities provided by the Cleveland Department of Veterans Affairs, Award Number1I01BX001974 (to RAB) from the Biomedical Laboratory Research & Development Service of the VA Office of Research and Development and the Geriatric Research Education and Clinical Center VISN 10 (to RAB). The authors thank Dr. Mahesh Aitha and Dr. Indra Sahu for providing the DEER data shown in Figure 4.
Glossary
- Catalytic nucleophile
enzyme groups that participate in the nucleophilic attack on substrate. In serine-β-lactamases, a serine hydroxide (on the side chain) is the catalytic nucleophile that attacks the β-lactam carbonyl. In MBLs, the bridging hydroxide is thought to be the catalytic nucleophile that attacks the β-lactam carbonyl.
- Exosite
a region of the enzyme that is not the active site. Often exosites are called allosteric sites.
- Inhibitory concentration at half-maximum (IC50)
concentration of an inhibitor that causes 50% inhibition of the enzyme.
- Ternary complex
complex of three distinct entities. In this article, ternary complex refers to a complex of MBL, Zn(II), and inhibitor.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References list
- 1.Rojas LJ, et al. NDM-5 and OXA-181 Beta-Lactamases, a Significant Threat Continues To Spread in the Americas. Antimicrob Agents Chemother. 2017;61:e00454–17. doi: 10.1128/AAC.00454-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen L, et al. Notes from the Field: Pan-Resistant New Delhi Metallo-Beta-Lactamase-Producing Klebsiella pneumoniae - Washoe County, Nevada, 2016. MMWR Morbidity and mortality weekly report. 2017;66:33. doi: 10.15585/mmwr.mm6601a7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fisher JF, et al. Bacterial resistance to beta-lactam antibiotics: compelling opportunism, compelling opportunity. Chem Rev. 2005;105:395–424. doi: 10.1021/cr030102i. [DOI] [PubMed] [Google Scholar]
- 4.Papp-Wallace KM, Bonomo RA. New beta-Lactamase Inhibitors in the Clinic. Infect Dis Clinics North America. 2016;30:441–464. doi: 10.1016/j.idc.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Makena A, et al. Biochemical characterization of New Delhi metallo-beta-lactamase variants reveals differences in protein stability. J Antimicrob Chemother. 2015;70:463–469. doi: 10.1093/jac/dku403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Makena A, et al. Comparison of Verona Integron-Borne Metallo-beta-Lactamase (VIM) Variants Reveals Differences in Stability and Inhibition Profiles. Antimicrob Agents Chemother. 2015;60:1377–1384. doi: 10.1128/AAC.01768-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stewart AC, et al. Clinical Variants of New Delhi Metallo-β-Lactamase Are Evolving To Overcome Zinc Scarcity. ACS Infect Dis. 2017;3:927–940. doi: 10.1021/acsinfecdis.7b00128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bahr G, et al. Clinical Evolution of New Delhi Metallo-beta-Lactamase (NDM) Optimizes Resistance under Zn(II) Deprivation. Antimicrob Agents Chemother. 2018 doi: 10.1128/AAC.01849-01817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meini MR, et al. Overcoming differences: The catalytic mechanism of metallo-beta-lactamases. FEBS Lett. 2015;589:3419–3432. doi: 10.1016/j.febslet.2015.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Potter RF, et al. The rapid spread of carbapenem-resistant Enterobacteriaceae. Drug resistance updates: reviews and commentaries in antimicrobial and anticancer chemotherapy. 2016;29:30–46. doi: 10.1016/j.drup.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fast W, Sutton LD. Metallo-beta-lactamase: inhibitors and reporter substrates. Biochim Biophys Acta. 2013;1834:1648–1659. doi: 10.1016/j.bbapap.2013.04.024. [DOI] [PubMed] [Google Scholar]
- 12.González MM, Vila AJ. An Elusive Task: A Clinically Useful Inhibitor of Metallo-β-Lactamases. In: Supuran CT, Capasso C, editors. Zinc Enzyme Inhibitors: Enzymes from Microorganisms. Springer International Publishing; 2017. pp. 1–34. [Google Scholar]
- 13.McGeary RP, et al. Progress toward inhibitors of metallo-beta-lactamases. Future Med Chem. 2017;9:673–691. doi: 10.4155/fmc-2017-0007. [DOI] [PubMed] [Google Scholar]
- 14.Rotondo CM, Wright GD. Inhibitors of metallo-beta-lactamases. Curr Opin Microbiol. 2017;39:96–105. doi: 10.1016/j.mib.2017.10.026. [DOI] [PubMed] [Google Scholar]
- 15.Drawz SM, Bonomo RA. Three decades of beta-lactamase inhibitors. Clin Microbiol Rev. 2010;23:160–201. doi: 10.1128/CMR.00037-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klingler FM, et al. Approved Drugs Containing Thiols as Inhibitors of Metallo-beta-lactamases: Strategy To Combat Multidrug-Resistant Bacteria. J Med Chem. 2015;58:3626–3630. doi: 10.1021/jm501844d. [DOI] [PubMed] [Google Scholar]
- 17.Buttner D, et al. Challenges in the Development of a Thiol-Based Broad-Spectrum Inhibitor for Metallo-beta-Lactamases. ACS Infect Dis. 2018;4:360–372. doi: 10.1021/acsinfecdis.7b00129. [DOI] [PubMed] [Google Scholar]
- 18.Park BK, et al. Metabolic activation in drug allergies. Toxicology. 2001;158:11–23. doi: 10.1016/s0300-483x(00)00397-8. [DOI] [PubMed] [Google Scholar]
- 19.Leigh R, Eva F. An Introduction to Biologics and Biosimilars. Part II: Subsequent Entry Biologics: Biosame or Biodifferent? Canadian Pharmacists Journal/Revue des Pharmaciens du Canada. 2010;143:184–191. [Google Scholar]
- 20.Xiang Y, et al. Azolylthioacetamides as a potent scaffold for the development of metallo-beta-lactamase inhibitors. Bioorg Med Chem Lett. 2017;27:5225–5229. doi: 10.1016/j.bmcl.2017.10.038. [DOI] [PubMed] [Google Scholar]
- 21.Holdgate GA, et al. Mechanistic enzymology in drug discovery: a fresh perspective. Nature reviews Drug discovery. 2018;17:115–132. doi: 10.1038/nrd.2017.219. [DOI] [PubMed] [Google Scholar]
- 22.Sabath LD, Abraham EP. Zinc as a cofactor for cephalosporinase from Bacillus cereus 569. Biochem J. 1966;98:11c–13c. doi: 10.1042/bj0980011c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pitout JD, et al. Carbapenemase-Producing Klebsiella pneumoniae, a Key Pathogen Set for Global Nosocomial Dominance. Antimicrob Agents Chemother. 2015;59:5873–5884. doi: 10.1128/AAC.01019-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.King AM, et al. Aspergillomarasmine A overcomes metallo-beta-lactamase antibiotic resistance. Nature. 2014;510:503–506. doi: 10.1038/nature13445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Albu SA, et al. Total Synthesis of Aspergillomarasmine A and Related Compounds: A Sulfamidate Approach Enables Exploration of Structure-Activity Relationships. Angew Chem Int Ed Engl. 2016;55:13259–13262. doi: 10.1002/anie.201606657. [DOI] [PubMed] [Google Scholar]
- 26.Koteva K, et al. Total Synthesis and Activity of the Metallo-β-lactamase Inhibitor Aspergillomarasmine A. Angewandte Chemie. 2016;128:2250–2252. doi: 10.1002/anie.201510057. [DOI] [PubMed] [Google Scholar]
- 27.Bergstrom A, et al. Probing the Interaction of Aspergillomarasmine A with Metallo-β-lactamases NDM-1, VIM-2, and IMP-7. ACS Infect Dis. 2017 doi: 10.1021/acsinfecdis.1027b00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brem J, et al. Structural Basis of Metallo-beta-Lactamase Inhibition by Captopril Stereoisomers. Antimicrob Agents Chemother. 2015;60:142–150. doi: 10.1128/AAC.01335-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen AY, et al. Dipicolinic Acid Derivatives as Inhibitors of New Delhi Metallo-β-lactamase-1. J Med Chem. 2017;60:7267–7283. doi: 10.1021/acs.jmedchem.7b00407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lienard BM, et al. Dynamic combinatorial mass spectrometry leads to metallo-beta-lactamase inhibitors. J Med Chem. 2008;51:684–688. doi: 10.1021/jm070866g. [DOI] [PubMed] [Google Scholar]
- 31.De Vriendt K, et al. Monitoring the zinc affinity of the metallo-beta-lactamase CphA by automated nanoESI-MS. J Am Soc Mass Spectrom. 2006;17:180–188. doi: 10.1016/j.jasms.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 32.Selevsek N, et al. Studies on ternary metallo-beta lactamase-inhibitor complexes using electrospray ionization mass spectrometry. J Am Soc Mass Spectrom. 2006;17:1000–1004. doi: 10.1016/j.jasms.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 33.Lienard BM, et al. Structural basis for the broad-spectrum inhibition of metallo-beta-lactamases by thiols. Org Biomol Chem. 2008;6:2282–2294. doi: 10.1039/b802311e. [DOI] [PubMed] [Google Scholar]
- 34.Cuniasse P, et al. Structures of biomolecular complexes by combination of NMR and cryoEM methods. Curr Opin Struct Biol. 2017;43:104–113. doi: 10.1016/j.sbi.2016.12.008. [DOI] [PubMed] [Google Scholar]
- 35.Coutsolelos AG, Spyroulias GA. 67ZnNMR, a Tool for Coordination Chemistry Problems. In: Marek ZRaI., editor. PATAI’S Chemistry of Functional Groups. John Wiley & Sons, Ltd; 2009. pp. 147–161. [Google Scholar]
- 36.Tierney DL, Schenk G. X-ray absorption spectroscopy of dinuclear metallohydrolases. Biophys J. 2014;107:1263–1272. doi: 10.1016/j.bpj.2014.07.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pievo R, et al. A rapid freeze-quench setup for multi-frequency EPR spectroscopy of enzymatic reactions. ChemPhysChem. 2013;14:4094–4101. doi: 10.1002/cphc.201300714. [DOI] [PubMed] [Google Scholar]
- 38.Bennett B. EPR of Cobalt-Substituted Zinc Enzymes. In: Hanson G, Berliner L, editors. Metals in Biology: Applications of High-Resolution EPR to Metalloenzymes. Springer; New York: 2010. pp. 345–370. [Google Scholar]
- 39.Yang H, et al. Spectroscopic and mechanistic studies of heterodimetallic forms of metallo-beta-lactamase NDM-1. J Am Chem Soc. 2014;136:7273–7285. doi: 10.1021/ja410376s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aitha M, et al. Biochemical, mechanistic, and spectroscopic characterization of metallo-beta-lactamase VIM-2. Biochemistry. 2014;53:7321–7331. doi: 10.1021/bi500916y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Griffin DH, et al. Structural and kinetic studies on metallo-beta-lactamase IMP-1. Biochemistry. 2011;50:9125–9134. doi: 10.1021/bi200839h. [DOI] [PubMed] [Google Scholar]
- 42.Aitha M, et al. Probing substrate binding to the metal binding sites in metallo-beta-lactamase L1 during catalysis. Medchemcomm. 2016;7:194–201. doi: 10.1039/C5MD00358J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Periyannan GR, et al. Sequential binding of cobalt(II) to metallo-beta-lactamase CcrA. Biochemistry. 2006;45:1313–1320. doi: 10.1021/bi051105n. [DOI] [PubMed] [Google Scholar]
- 44.Sharma N, et al. Conformational changes in the metallo-beta-lactamase ImiS during the catalytic reaction: an EPR spectrokinetic study of Co(II)-spin label interactions. J Am Chem Soc. 2008;130:8215–8222. doi: 10.1021/ja0774562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Llarrull LI, et al. Evidence for a dinuclear active site in the metallo-beta-lactamase BcII with substoichiometric Co(II). A new model for metal uptake. J Biol Chem. 2007;282:30586–30595. doi: 10.1074/jbc.M704613200. [DOI] [PubMed] [Google Scholar]
- 46.Lisa MN, et al. A general reaction mechanism for carbapenem hydrolysis by mononuclear and binuclear metallo-beta-lactamases. Nature Commun. 2017;8:538. doi: 10.1038/s41467-017-00601-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hu Z, et al. Role of the Zn1 and Zn2 sites in metallo-beta-lactamase L1. J Am Chem Soc. 2008;130:14207–14216. doi: 10.1021/ja8035916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hawk MJ, et al. Differential binding of Co(II) and Zn(II) to metallo-beta-lactamase Bla2 from Bacillus anthracis. J Am Chem Soc. 2009;131:10753–10762. doi: 10.1021/ja900296u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hu Z, et al. Folding strategy to prepare Co(II)-substituted metallo-beta-lactamase L1. Anal Biochem. 2008;378:177–183. doi: 10.1016/j.ab.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bertini I, et al. Perspectives in paramagnetic NMR of metalloproteins. Dalton Trans. 2008:3782–3790. doi: 10.1039/b719526e. [DOI] [PubMed] [Google Scholar]
- 51.Bertini I, et al. Nuclear magnetic resonance of paramagnetic metalloproteins. Chem Rev. 1993;93:2833–2932. [Google Scholar]
- 52.Craig WR, et al. Substituent Effects on the Coordination Chemistry of Metal-Binding Pharmacophores. Inorg Chem. 2017;56:11721–11728. doi: 10.1021/acs.inorgchem.7b01661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baum RR, et al. Paramagnetic Resonance of High-Spin Co(II) in Biologically-Relevant Environments: Models to Metalloproteins. In: Hanson G, Berliner L, editors. Future Directions in Metalloprotein and Metalloenzyme Research. Springer International Publishing; 2017. pp. 33–54. [Google Scholar]
- 54.Aitha M, et al. Conformational dynamics of metallo-beta-lactamase CcrA during catalysis investigated by using DEER spectroscopy. J Biol Inorg Chem. 2015;20:585–594. doi: 10.1007/s00775-015-1244-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rydzik AM, et al. Monitoring conformational changes in the NDM-1 metallo-beta-lactamase by 19F NMR spectroscopy. Angew Chem Int Ed Engl. 2014;53:3129–3133. doi: 10.1002/anie.201310866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brem J, et al. Studying the active-site loop movement of the Sao Paolo metallo-beta-lactamase-1. Chemical science. 2015;6:956–963. doi: 10.1039/c4sc01752h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Abboud MI, et al. (19) F-NMR Reveals the Role of Mobile Loops in Product and Inhibitor Binding by the Sao Paulo Metallo-beta-Lactamase. Angew Chem Int Ed Engl. 2017;56:3862–3866. doi: 10.1002/anie.201612185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Abboud MI, et al. Cyclobutanone Mimics of Intermediates in Metallo-beta-Lactamase Catalysis. Chemistry - A Eur J. 2018 doi: 10.1002/chem.201705886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Aitha M, et al. Investigating the position of the hairpin loop in New Delhi metallo-beta-lactamase, NDM-1, during catalysis and inhibitor binding. J Inorg Biochem. 2016;156:35–39. doi: 10.1016/j.jinorgbio.2015.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jeschke G. DEER Distance Measurements on Proteins. Annu Rev Phys Chem. 2012;63:419–446. doi: 10.1146/annurev-physchem-032511-143716. [DOI] [PubMed] [Google Scholar]
- 61.Crowder MW, et al. Metallo-beta-lactamases: novel weaponry for antibiotic resistance in bacteria. Acc Chem Res. 2006;39:721–728. doi: 10.1021/ar0400241. [DOI] [PubMed] [Google Scholar]
- 62.Mojica MF, et al. B1-Metallo-beta-Lactamases: Where Do We Stand? Curr Drug Targets. 2016;17:1029–1050. doi: 10.2174/1389450116666151001105622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sevaille L, et al. 1,2,4-Triazole-3-thione Compounds as Inhibitors of Dizinc Metallo-β-lactamases. ChemMedChem. 2017;12:972–985. doi: 10.1002/cmdc.201700186. [DOI] [PubMed] [Google Scholar]
- 64.Skagseth S, et al. Metallo-beta-lactamase inhibitors by bioisosteric replacement: Preparation, activity and binding. Eur J Med Chem. 2017;135:159–173. doi: 10.1016/j.ejmech.2017.04.035. [DOI] [PubMed] [Google Scholar]
- 65.Karsisiotis AI, et al. Solution structures of the Bacillus cereus metallo-beta-lactamase BcII and its complex with the broad spectrum inhibitor R-thiomandelic acid. Biochem J. 2013;456:397–407. doi: 10.1042/BJ20131003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cain R, et al. In silico fragment based design identifies subfamily B1 metallo-beta-lactamase inhibitors. J Med Chem. 2018 doi: 10.1021/acs.jmedchem.7b01728. [DOI] [PubMed] [Google Scholar]
- 67.Zhang E, et al. NOTA analogue: A first dithiocarbamate inhibitor of metallo-beta-lactamases. Bioorg Med Chem Lett. 2018;28:214–221. doi: 10.1016/j.bmcl.2017.10.074. [DOI] [PubMed] [Google Scholar]
- 68.Huang PJ, et al. Inhibiting the VIM-2 Metallo-beta-Lactamase by Graphene Oxide and Carbon Nanotubes. ACS Appl Materials Interfaces. 2015;7:9898–9903. doi: 10.1021/acsami.5b01954. [DOI] [PubMed] [Google Scholar]
- 69.Ouyang X, et al. A DNA nanoribbon as a potent inhibitor of metallo-beta-lactamases. Chemical communications (Cambridge, England) 2017;53:8878–8881. doi: 10.1039/c7cc04483f. [DOI] [PubMed] [Google Scholar]
- 70.Kim SK, et al. Inhibition of Bacillus anthracis metallo-β-lactamase by compounds with hydroxamic acid functionality. J Enzyme Inhibition Med Chem. 2016;31:132–137. doi: 10.1080/14756366.2016.1222580. [DOI] [PubMed] [Google Scholar]
- 71.McGeary RP, et al. Structure-activity relationship study and optimisation of 2-aminopyrrole-1-benzyl-4,5-diphenyl-1H-pyrrole-3-carbonitrile as a broad spectrum metallo-beta-lactamase inhibitor. Eur J Med Chem. 2017;137:351–364. doi: 10.1016/j.ejmech.2017.05.061. [DOI] [PubMed] [Google Scholar]
- 72.Holloway AJ, et al. Cystatin 9 and C: A novel immunotherapy that protects against multi-drug resistant New Delhi metallo-beta-lactamase-1 producing Klebsiella pneumoniae. Antimicrob Agents Chemother. 2018 doi: 10.1128/AAC.01900-17. [DOI] [PMC free article] [PubMed] [Google Scholar]