Abstract
Mitochondria contribute to key processes of cellular function, while mitochondrial dysfunction is implicated in metabolic disorders, neurodegenerative diseases, and cardiovascular diseases, in which angiogenesis- the formation of new blood capillaries- is dysregulated. The Hippo signaling transducer, Yes-associated protein (YAP1) binds to the TEA domain (TEAD1) transcription factor and controls angiogenesis. YAP1 also regulates glucose metabolism through peroxisome proliferator-activated receptor gamma co-activator 1-alpha (PGC1α), a major player controlling mitochondrial biogenesis. However, the role of YAP1-TEAD1-PGC1α signaling in mitochondrial structure, cellular metabolism, and angiogenesis in endothelial cells (ECs) remains unclear. We now find that knockdown of TEAD1 decreases the expression of PGC1α and suppresses mitochondrial biogenesis, glycolysis, and oxygen consumption in ECs. A YAP1 mutant construct, YAP1S127A, which stimulates binding of YAP1 to TEAD1, upregulates the expression of PGC1α, induces mitochondrial biogenesis, and increases oxygen consumption and glycolytic flux in ECs; in contrast, YAP1S94A, which fails to bind to TEAD1, attenuates these effects. PGC1α knockdown inhibits YAP1S127A-induced EC sprouting in vitro and vascular morphogenesis in the fibrin gel subcutaneously implanted on mice, while overexpression of PGC1α reverses vascular morphogenesis suppressed by YAP1S94A. These results suggest that YAP1-TEAD1 signaling induces mitochondrial biogenesis in ECs and stimulates angiogenesis through PGC1α. Modulation of YAP1-TEAD1-PGC1α signaling in ECs may provide a novel intervention for angiogenesis-related diseases.
Keywords: angiogenesis, mitochondrial biogenesis, YAP1, TEAD1, PGC1α
Introduction
Mitochondria are not only a crucial cellular source of ATP but also contribute to key cellular processes such as cell signaling and growth (Osellame et al., 2012; Schinzel and Dillin, 2015). Mitochondrial dysfunction is implicated in diabetes mellitus (Sawada et al., 2014; Silva et al., 2000), aging-related diseases (Mora et al., 2017), and cardiovascular diseases (Diebold et al., 2015; Patten and Arany, 2012; Rowe et al., 2010; Sawada et al., 2014), in which angiogenesis is dysregulated. Peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC1α) is expressed in endothelial cells (ECs) and controls mitochondrial biogenesis (Fan and Evans, 2015; Patten and Arany, 2012) and angiogenesis (Arany et al., 2008; Patten et al., 2012; Sawada et al., 2014). PGC1α also upregulates the expression of angiogenic factor VEGFA in skeletal muscle and retinal müller cells and stimulates angiogenesis (Arany et al., 2008; Chinsomboon et al., 2009; Patten and Arany, 2012; Rowe et al., 2014; Saint-Geniez et al., 2013; Thom et al., 2014). However, the signaling cascade involving PGC1α during angiogenesis is not fully understood.
A Hippo signaling transducer, Yes-associated protein (YAP1) acts as a transcriptional co-activator and controls organ size and regeneration (Panciera et al., 2017; Piccolo et al., 2013). The TEA domain (TEAD) transcription factor (TF) family members play a dominant role as primary mediators of YAP1-dependent gene regulation (Ota and Sasaki, 2008; Stein et al., 2015). YAP1-TEAD1 signaling controls angiogenesis and vascular function in ECs through various signaling pathways, including angiopoietin2 (Ang2), microfibrillar-associated protein 5, matrix metalloproteinase 2, VE-cadherin, and CDC42 (Al-Moujahed et al., 2017; Choi et al., 2015; Kim et al., 2017; Marti et al., 2015; Nakajima et al., 2017; Sakabe et al., 2017; Wei et al., 2017). For example, YAP1 controls retinal angiogenesis through Ang2-Tie2 signaling (Choi et al., 2015). YAP/TAZ also regulates EC morphology and junctional integrity by modulating actin cytoskeleton structures (Kim et al., 2017; Sakabe et al., 2017). YAP1 activity is mechanosensitive and endothelial YAP1 maintains vascular stability to flow in a zebrafish model (Nakajima et al., 2017). YAP signaling also regulates mitochondrial structure and function during development of Drosophila (Nagaraj et al., 2012) and mediates metabolic stress-induced inhibition of angiogenesis (Yuan et al., 2017). In addition, YAP1 controls glucose metabolism through PGC1α in hepatocytes (Hu et al., 2017), suggesting a potential role of YAP1-PGC1α in angiogenesis. However, it remains unknown the role of YAP1-TEAD1 signaling in mitochondrial structure/function in ECs and whether PGC1α is a common link between YAP1-TEAD1 signaling and mitochondrial biogenesis in ECs and angiogenesis.
Here we found that YAP1-TEAD1-PGC1α signaling controls mitochondrial biogenesis in ECs. Endothelial YAP1-TEAD1 signaling regulates angiogenesis through PGC1α in vitro and in a subcutaneously implanted fibrin gel in mice. Modulation of endothelial YAP1-TEAD1-PGC1α signaling could be an efficient therapeutic tool for angiogenesis-related diseases.
Materials and Methods
Materials
Anti-TEAD1 antibody was from Transduction Laboratories (Lexington, KY). Anti-β-actin monoclonal antibody was from Sigma (St. Louis, MO). Anti-PGC1α polyclonal antibody was from Millipore (Billerica, MA). Anti-YAP1 antibody was from Santa Cruze Biotechnology (Dallas, TX). Human umbilical vascular endothelial (HUVE) cells (Lonza, passage 3–5) were cultured in EBM2 medium containing 5% FBS and growth factors (vascular endothelial growth factor (VEGF), basic fibroblast growth factor (bFGF) and platelet-derived growth factor (PDGF)) (Mammoto et al., 2016a). Human adult lung fibroblasts (ATCC, Manassas, VA, passage 3–5) were cultured in Eagle’s Minimum Essential Medium (EMEM, ATCC) (Mammoto et al., 2016a).
Plasmid construction and gene knockdown
The retroviral full-length pQCXIH-myc-YAP1 (human) and pQCXIH-flag-YAP1-S127A (human) were gifts from Kunliang Guan (Addgene plasmid # 33091 and # 33092) (Zhao et al., 2007) and pLX304-YAP1 (S94A) (human) construct was a gift from William Hahn (Addgene plasmid # 59145) (Shao et al., 2014). pLenti-PGC1α (mouse) construct was kindly provided by Dimitri Krainc (Northwestern University) (Cui et al., 2006). Lentiviral construct of pTRIPZ-PGC1α shRNA (human) was produced by the Blood Center of Wisconsin Hybridoma Core Laboratory (Kadlec et al., 2017). The lentiviral pHAGE-GFP construct (Mammoto et al., 2009) was used for labeling HUVE cells. As a control, plasmid with vector only was used. Generation of retroviral/lentiviral vectors was accomplished as reported (Mammoto et al., 2009). Viral supernatants were collected starting 48 h after transfection, for four consecutive times every 12 h, pooled, and filtered through a 0.45 μm filter. Viral supernatants were then concentrated 100-fold by ultracentrifugation in a Beckman centrifuge for 1.5 h at 16,500 rpm. HUVE cells were incubated with viral stocks in the presence of 5 μg/ml polybrene (Sigma) for 3 days in the BSL2 facility (Mammoto et al., 2009). We confirm the transduction efficiency using immunocytochemistry before using them for the assay. If the efficiency is less than 80–90%, second round of transduction is performed, which achieves 80–90% transduction efficiency. Cells were used for the assays 3 days after transduction.
Molecular biological and biochemical methods
RNA was isolated using an RNeasy mini kit (Qiagen, Valencia, CA, USA). Quantitative reverse transcription (qRT)-PCR was performed with the iScript reverse transcription and iTaq SYBR Green qPCR kit (BioRad, Hercules, CA) using the BioRad real time PCR system (BioRad). β2 microglobulin controlled for overall cDNA content. The primers used for human VEGFR2, Tie2, and β2 microglobulin were previously described (Mammoto et al., 2009; Mammoto et al., 2012). The primers used for human TEAD1 forward; ATGGAAAGGATGAGTGACTCTGC and reverse; TCCCACATGGTGGATAGATAGC, human PGC1α forward; AGCTGCTGAAGAGGCAAGAG and reverse; TTCCCCTAAACCAAGCACAC, human YAP1 forward; TAGCCCTGCGTAGCCAGTTA and reverse; TCATGCTTAGTCCACTGTCTGT, human mitofusion (MFN) 1 forward; TGGCTAAGAAGGCGATTACTGC and reverse; TCTCCGAGATAGCACCTCACC, human MFN2 forward; CTCTCGATGCAACTCTATCGTC and reverse; TCCTGTACGTGTCTTCAAGGAA, human dynamin-related protein (DRP1) forward; CTGCCTCAAATCGTCGTAGTG and reverse; GAGGTCTCCGGGTGACAATTC, and human fission mitochondrial 1 (FIS1) forward; GTCCAAGAGCACGCAGTTTG, reverse; ATGCCTTTACGGATGTCATCATT.
Gene knockdown was performed using the RNA interference technique (Mammoto et al., 2009; Mammoto et al., 2013c). siRNA for human TEAD1 (No. 1) was ON-TARGETplus TEAD1siRNA (J-012603-08; Thermo Scientific) and siRNA for human TEAD1 (No. 2) was TEAD1_5FlexiTUbe siRNA (SI04181261; QIAGEN) (Stein et al., 2015). Target sequence of siRNA for human PGC1α was 5′-GACGAAGCAGACAAGACCGGU-3′. HUVE cells were transfected using siLentFect (BioRad, Hercules, CA) (Mammoto et al., 2009; Mammoto et al., 2013c) and used for the assays 3 days after transfection. As a control, siRNA duplex with an irrelevant sequence (QIAGEN) was used.
To measure mitochondrial DNA (mtDNA), HUVE cells were treated with virus or siRNA for 3 days. Genomic DNA was extracted using PureLink® Genomic DNA Kits. mtDNA was quantified by calculating the ratio of a mitochondrion-encoded gene (NADH:ubiquinone oxidoreductase core subunit 1 (ND1)) to a nuclear-encoded gene (solute carrier organic anion transporter family member 2B1 (SLCO2B1)) (TAKARA mtDNA monitoring primer set, Takara, Shiga, Japan) using qPCR, and expressing it as mtDNA copy number per cell (Diebold et al., 2015). Mitochondrial morphometry was assessed by MitoTracker staining 3 days after viral transduction or siRNA transfection (Diebold et al., 2015). The mitochondrial area/cell was determined using imageJ software and the same contrast and brightness were used to compare the images.
Measurement of oxygen consumption rate (OCR) and extracellular acidification rate (ECAR)
OCR and ECAR measurements were performed using the XF96 Extracellular Flux analyzer (Seahorse Bioscience, North Billerica, MA) according to the manufacturer instructions (TeSlaa and Teitell, 2014; Wu et al., 2007). Briefly, HUVE cells treated with siRNA or virus for 3 days were plated into XF96 (V3) polystyrene cell culture plates (Seahorse Bioscience) at 10,000 cells/well and incubated for 24 hours. OCR and ECAR were normalized by total protein amount in each well.
In vitro fibrin gel angiogenesis assay
Fibrin gel angiogenesis assays were performed as previously described (Mammoto et al., 2016a; Nakatsu et al., 2007). Briefly, 1×105 HUVE cells were incubated with 3000 Cytodex 3 microcarrier beads (GE Healthcare Life Sciences, Pittsburgh, PA) in 1 ml EGM2 in a glass tube for 4 hours with gentle agitation. The beads coated with cells were transferred to 25 cm2 tissue culture flask and incubated with or without virus or siRNA treatment. As a control, cells were treated with control virus (vector alone). After 16 h incubation, 250 beads coated with HUVE cells were suspended in 500 μl of 2.5 mg/ml fibrinogen solution (Sigma) and mixed with 500 μl of thrombin solution (0.5 U) in a 24-well plate. After fibrin gels were solidified, 1 ml of EGM2 containing 2×104 human lung fibroblasts was seeded on top of each fibrin gel in a 24-well plate. The medium was changed every other day. After incubation of beads inside the fibrin gels for 5 days, the area of the sprout from the beads and the length of the sprout were quantified using ImageJ software.
Fibrin gel subcutaneous implantation in vivo
The in vivo animal study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was reviewed and approved by the Animal Care and Use Committee of Medical College of Wisconsin. NOD scid gamma (NSG) mice (Jackson Laboratory) were used for the study. To label HUVE cells, cells were treated with viral stock expressing GFP in the presence of 5 μg/ml polybrene (Sigma). We confirm the transduction efficiency using immunocytochemistry before using for the assay. Fibrin gel was fabricated as described (Mammoto et al., 2016b; Mammoto and Mammoto, 2014; Mammoto et al., 2018). Briefly, we added 20 μl of thrombin (2.5 U/ml) to 20 μl of fibrinogen solution (12.5 mg/ml), supplemented gel with GFP-labeled HUVE cells (1×106 cells), in which gene expression was manipulated, and lung fibroblasts (3×105 cells). The drops of the gel were incubated at 37 °C for 30 min until they solidified (Mammoto et al., 2016b; Mammoto and Mammoto, 2014; Mammoto et al., 2018). We then implanted the gel subcutaneously on the back of NSG mice for 7 days as described previously (Mammoto et al., 2009). Supplemented GFP-labeled ECs (Supplementary Fig. S3B) formed vascular lumen structures in the implanted gel 7 days after implantation (Supplementary Fig. S3C). Vascular network formation of GFP-labeled HUVE cells was evaluated by measuring the area of GFP-labeled blood vessels from five different areas of the gel (Mammoto et al., 2009; Mammoto et al., 2012; Mammoto et al., 2016b; Mammoto and Mammoto, 2014; Mammoto et al., 2018). Fluorescent images were taken on a Leica TCS SP5 confocal laser scanning microscope and morphometric analysis was performed using ImageJ software as we reported (Mammoto et al., 2009; Mammoto et al., 2012; Mammoto et al., 2016b; Mammoto and Mammoto, 2014; Mammoto et al., 2018).
Statistical Analysis
All phenotypic analysis was performed by masked observers unaware of the identity of experimental groups. Error bars (SEM) and p values were determined from the results of three or more independent experiments. The F test (for two samples) or the Levene test (for more than two samples) was performed to confirm that the variances are homogeneous. Student’s t-test was used for statistical significance for two groups. For more than two groups, one-way ANOVA with a post-hoc analysis using the Bonferroni test was conducted.
Results
YAP1-TEAD1 signaling is necessary for PGC1α expression in ECs in vitro
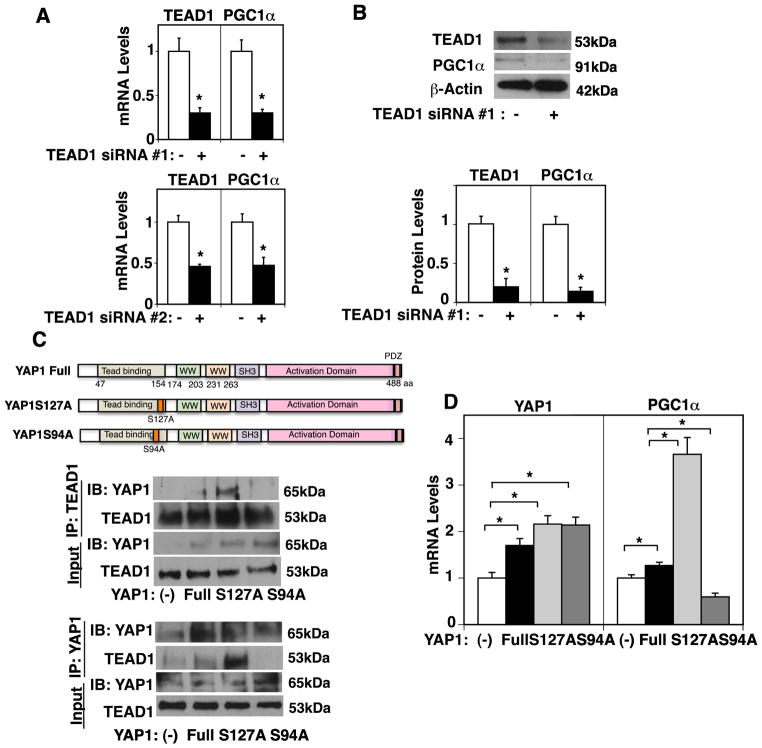
TEAD1 is a primary mediator of YAP1-dependent gene regulation (Ota and Sasaki, 2008; Stein et al., 2015). The mRNA and protein expression levels of PGC1α, a major player controlling mitochondrial biogenesis, were 78% and 92% lower, respectively, in HUVE cells treated with TEAD1 siRNA #1 compared to those treated with control siRNA with scrambled sequences (Fig. 1A, B, Supplementary Fig. S1A). We confirmed the results using a second siRNA targeting TEAD1, which exhibits similar results (Fig. 1A). Therefore, we used TEAD1 siRNA #1 for the rest of the study. It has been reported that a point mutant construct of YAP1 Ser94 (S94), the site essential for the interaction with TEAD1, fails to bind to TEAD1 (Li et al., 2010; Ota and Sasaki, 2008; Shao et al., 2014). Phosphorylation of YAP1 at the serine127 (S127) residue by large tumor suppressor (LATS) sequesters YAP1 to the cytoplasm and has a potent role in suppressing YAP1 activity (Ota and Sasaki, 2008; Piccolo et al., 2014). Therefore, we next examined the effects of YAP1S94, which mediates YAP1-TEAD1 binding, and YAP1S127 phosphorylation, which suppresses YAP1 nuclear localization and inhibits its activity, on YAP1-TEAD1 interaction and PGC1α expression in ECs. Consistent with previous reports in fibroblasts (Li et al., 2010; Ota and Sasaki, 2008), overexpression of YAP1S127A mutant construct, a constitutively active form of YAP1, stimulated, while the YAP1S94A mutant construct abolished the interaction of YAP1 with TEAD1 in HUVE cells when analyzed using a co-immunoprecipitation assay (Fig. 1C). Overexpression of YAP1S127A increased PGC1α mRNA expression by 3.1-fold in HUVE cells, while the levels of PGC1α were lower by 46% in HUVE cells treated with YAP1S94A mutant construct compared to those treated with non-mutated full-length YAP1 (Fig. 1D). Overexpression of YAP1S127A also increased PGC1α protein expression by 2.4-fold, while the protein levels of PGC1α were lower by 48% in HUVE cells treated with YAP1S94A mutant compared to those treated with non-mutated full-length YAP1 (Supplementary Fig. S1B), suggesting that YAP1-TEAD1 signaling is required for PGC1α expression in ECs in vitro.
Fig. 1. YAP1-TEAD1 signaling controls PGC1α expression in HUVE cells.
A) Graphs showing TEAD1 and PGC1α mRNA levels in HUVE cells treated with TEAD1 siRNA#1, #2, or control siRNA with irrelevant sequences (*, p<0.05). Error bars represent s.e.m. of three independent experiments. B) Immunoblots showing TEAD1, PGC1α, and β-actin protein levels in HUVE cells treated with TEAD1 siRNA#1 or control siRNA with irrelevant sequences (top). Graph showing TEAD1 and PGC1α protein levels normalized by β-actin protein levels in HUVE cells treated with TEAD1 siRNA #1 or control siRNA with irrelevant sequences (bottom, *, p<0.05). Error bars represent s.e.m. of three independent experiments. C) Diagram showing the structure of full-length YAP1 and the YAP1S127A and S94A mutant constructs (top). Immunoblots showing YAP1 and TEAD1 immunoprecipitated with TEAD1 antibody in HUVE cells overexpressing full-length YAP1, YAP1S127A, YAP1S94A, or control vector (middle). Immunoblots showing YAP1 and TEAD1 immunoprecipitated with YAP1 antibody in HUVE cells overexpressing full-length YAP1, YAP1S127A, YAP1S94A, or control vector (bottom). D) Graph showing YAP1 and PGC1α mRNA expression levels in HUVE cells overexpressing full-length YAP1, YAP1S127A, YAP1S94A, or control vector alone (*, P<0.05). Error bars represent s.e.m. of three independent experiments.
YAP1-TEAD1 signaling controls mitochondrial structure and function
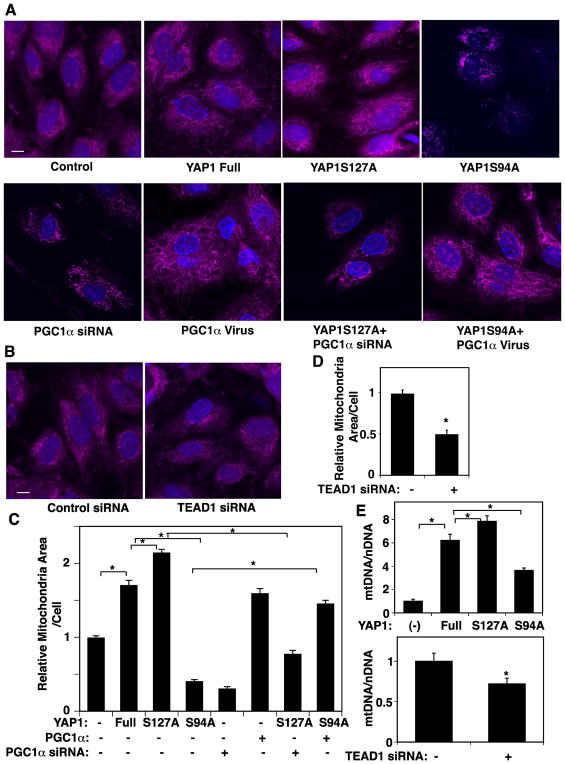
Since PGC1α is a major player in mitochondrial biogenesis (Fan and Evans, 2015; Patten and Arany, 2012), we next examined whether YAP1-TEAD1 signaling modulates mitochondrial biogenesis. First, we analyzed the mitochondrial structure using MitoTracker Orange staining. Consistent with the results of PGC1α expression (Fig. 1), mitochondrial area increased in ECs overexpressing YAP1S127A by 23%, while the mitochondrial area decreased in ECs overexpressing YAP1S94A by 76% compared to those expressing full-length YAP1 (Fig. 2A, C). These effects are mediated by PGC1α, because PGC1α knockdown attenuated the effects of YAP1S127A, while overexpression of PGC1α reversed the effects of YAP1S94A on mitochondrial structure (Fig. 2A, C). TEAD1 knockdown also decreased the mitochondrial area by 51% compared to that in control siRNA-treated ECs (Fig. 2B, D). Mitochondrial biogenesis is controlled by nuclear-encoded proteins that are essential for replication of mtDNA. Thus, we also assessed mtDNA copy numbers in each cell by measuring the copy number of mitochondrially encoded ND1 as a marker of mtDNA and SLCO2B1 as a marker for nuclear DNA and took ratio of mtDNA to nuclear DNA (Diebold et al., 2015; Hiramitsu et al., 2014). Consistent with MitoTracker staining, overexpression of YAP1S127A increased the mtDNA content by 30%, while YAP1S94A decreased the mtDNA quantity by 37% in HUVE cells compared to those treated with non-mutated full-length YAP1 (Fig. 2E). TEAD1 knockdown also decreased the mtDNA content (Fig. 2E), indicating that YAP1-TEAD1 signaling is necessary for mitochondrial biogenesis in ECs.
Fig. 2. YAP1-TEAD1 signaling is required for mitochondrial biogenesis in HUVE cells.
A) Immunofluorescence micrographs showing mitochondrial morphology analyzed by MitoTracker Orange staining in HUVE cells treated with virus overexpressing full-length YAP1, YAP1S127A, YAP1S94A or in combination with PGC1α siRNA or PGC1α virus. Scale bar, 10 μm. B) Immunofluorescence micrographs showing mitochondrial morphology analyzed by MitoTracker Orange staining in HUVE cells treated with TEAD1 siRNA or control siRNA with irrelevant sequences. Scale bar, 10 μm. C) Graph showing quantification of the areas of mitochondria in HUVE cells treated with virus overexpressing full-length YAP1, YAP1S127A, YAP1S94A or in combination with PGC1α siRNA or PGC1α virus (*, p<0.05). D) Graph showing quantification of the areas of mitochondria in HUVE cells treated with TEAD1 siRNA or control siRNA with irrelevant sequences (*, p<0.05). Error bars represent s.e.m. of three independent experiments. E) Graphs showing quantification of mtDNA copy number assessed by the ratio of ND1 to nuclear SLCO2B1 in HUVE cells overexpressing full-length YAP1, YAP1S127A, YAP1S94A, or control vector alone (top, *, p<0.05), or TEAD1 siRNA or control siRNA with irrelevant sequences (bottom, *, p<0.05). Error bars represent s.e.m. of three independent experiments.
The balance between mitochondrial fission and fusion is important for the maintenance of mitochondrial integrity. Therefore, we also investigated whether the expression levels of MFN1 and 2, which control mitochondrial fusion, and DRP1 and FIS1, which control mitochondrial fission, are regulated by YAP1-TEAD1 signaling in HUVE cells. The mRNA levels of MFN1, MFN2, DRP1, and FIS1 were upregulated in ECs expressing YAP1S127A. The levels of MFN1 and MFN2 decreased in ECs overexpressing YAP1S94A compared to those expressing full-length YAP1, while the levels of DRP1 and FIS1 were not changed by YAP1 S94A mutation (Supplementary Fig. S2A, B). TEAD1 knockdown did not change the mRNA levels of MFN1 and 2, but decreased the mRNA levels of DRP1 and FIS1 (Supplementary Fig. S2A, B). These results suggest that YAP1 may control mitochondrial biogenesis and dynamics through separate mechanisms.
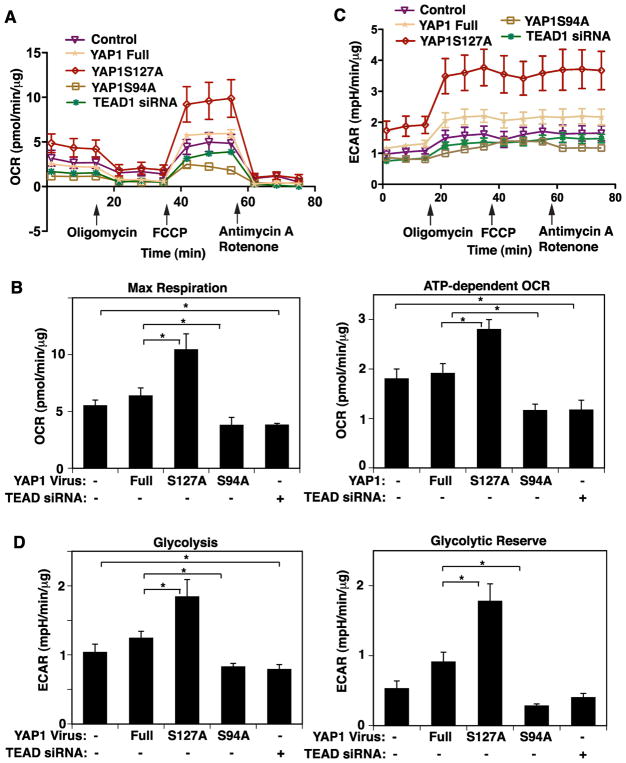
To determine whether YAP1-TEAD1 signaling influences mitochondrial function, cellular respiration was measured using a Seahorse extracellular flux analyzer (Wu et al., 2007). When we treated HUVE cells with retrovirus encoding YAP1S127A, maximum respiration rate and ATP-dependent OCR increased by 1.7-times and 1.4-times, respectively, compared to those treated with non-mutated full-length YAP1, while YAP1S94A mutant construct failed to increase oxygen consumption and ATP-dependent OCR (Fig. 3A, B). TEAD1 knockdown also attenuated maximum respiration rate and ATP-dependent OCR by 34% and 42%, respectively, in HUVE cells (Fig. 3A, B). Since glycolysis is known to regulate EC sprouting (Cruys et al., 2016; De Bock et al., 2013), we also measured ECAR in ECs in the same condition using a Seahorse extracellular flux analyzer (TeSlaa and Teitell, 2014). YAP1S127A increased glycolysis and glycolytic reserve by 1.4- and 1.9-times, respectively, compared to those treated with non-mutated full-length YAP1, while YAP1S94A mutant construct failed to increase glycolysis and glycolytic reserve (Fig. 3C, D). TEAD1 knockdown also attenuated glycolysis by 24% and in HUVE cells (Fig. 3C, D), indicating that YAP1-TEAD1 signaling is necessary for oxygen consumption and glycolysis in ECs.
Fig. 3. YAP1-TEAD1 signaling controls mitochondrial function in HUVE cells.
A) Oxygen consumption rate (OCR) of HUVE cells treated with virus overexpressing full-length YAP1, YAP1S127A or S94A mutant construct, TEAD1 siRNA or control vector alone. Error bars represent s.e.m. of three independent experiments. B) Graphs showing maximal respiration following carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone (FCCP) treatment (left) and ATP-dependent OCR (right, *, p<0.05). Error bars represent s.e.m. of three independent experiments. C) Extracellular acidification rate (ECAR) of HUVE cells treated with virus overexpressing full-length YAP1, YAP1S127A or S94A mutant construct, TEAD1 siRNA or control vector alone. Error bars represent s.e.m. of three independent experiments. D) Graphs showing glycolysis (left) and glycolytic reserve (right, *, p<0.05). Error bars represent s.e.m. of three independent experiments.
YAP1 controls angiogenesis through PGC1α in vitro
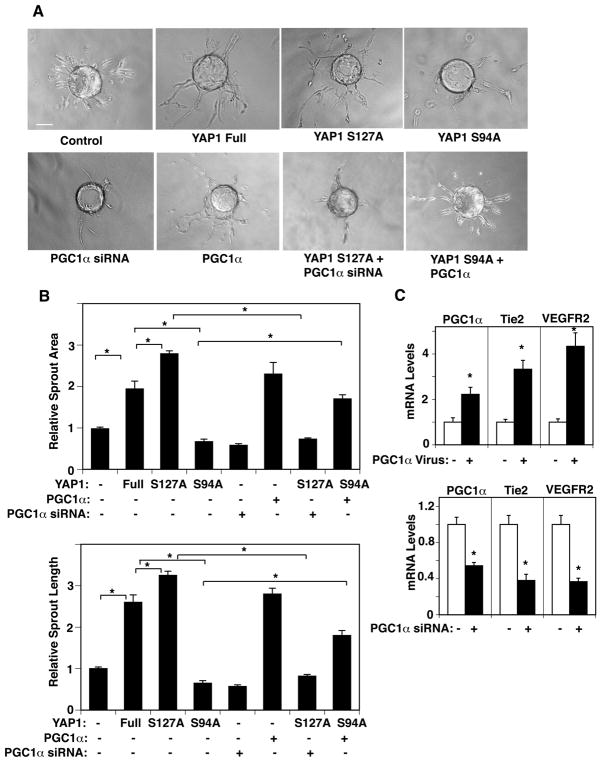
In addition to stimulating mitochondrial biogenesis, PGC1α is known to stimulate the expression of angiogenic factor, such as VEGF in muscle cells and induce angiogenesis (Arany et al., 2008; Chinsomboon et al., 2009; Patten and Arany, 2012; Rowe et al., 2014; Thom et al., 2014). Since YAP1 stimulates angiogenesis and maintains vascular function (Al-Moujahed et al., 2017; Choi et al., 2015; Kim et al., 2017; Marti et al., 2015; Nakajima et al., 2017; Sakabe et al., 2017; Wei et al., 2017), we next examined whether YAP1 controls angiogenesis through PGC1α using an EC sprouting assay in combination with gene manipulation. Overexpression of YAP1S127A induced, while YAP1S94A inhibited EC sprouting; EC sprout area and length were 1.4- and 1.2- times higher in ECs overexpressing YAP1S127A compared to those treated with full-length YAP1, while these parameters were lower by 70 % and 77% in ECs overexpressing YAP1S94A compared to those expressing full-length YAP1 (Fig. 4A, B). When we treated HUVE cells with lentivirus encoding the YAP1S94A mutant construct in combination with PGC1α, PGC1α overexpression, which increases the levels of Tie2 and VEGFR2 (Fig. 4C), restored EC sprouting inhibited by YAP1S94A mutant construct (Fig. 4A, B). In contrast, PGC1α knockdown decreased the levels of Tie2 and VEGFR2 (Fig. 4C) and suppressed EC sprouting induced by the YAP1S127A mutant construct (Fig. 4A, B). These results suggest that YAP1-TEAD1 signaling controls EC sprouting through PGC1α in ECs in vitro.
Fig. 4. YAP1 controls EC sprouting through PGC1α in HUVE cells.
A) Phase contrast images showing EC sprouting from each bead in HUVE cells treated with virus overexpressing full-length YAP1, YAP1S127A, YAP1S94A, or control vector alone, or in combination with PGC1α virus or siRNA (scale bar, 100 μm). B) Graphs showing changes in sprout area (top) and length (bottom) of the sprout in HUVE cells overexpressing full-length YAP1, YAP1S127A, YAP1S94A or control vector alone, or in combination with PGC1α virus or siRNA (*, p<0.05). Error bars represent s.e.m. of three independent experiments. C) Graph showing the mRNA expression levels of PGC1α, Tie2, and VEGFR2 in HUVE cells treated with lentivirus encoding PGC1α or control virus (top, *, P<0.05). Graph showing the mRNA expression levels of PGC1α, Tie2, and VEGFR2 in HUVE cells treated with PGC1α siRNA or control siRNA with irrelevant sequences (bottom, *, P<0.05). Error bars represent s.e.m. of three independent experiments.
YAP1-PGC1α signaling controls angiogenesis in vivo
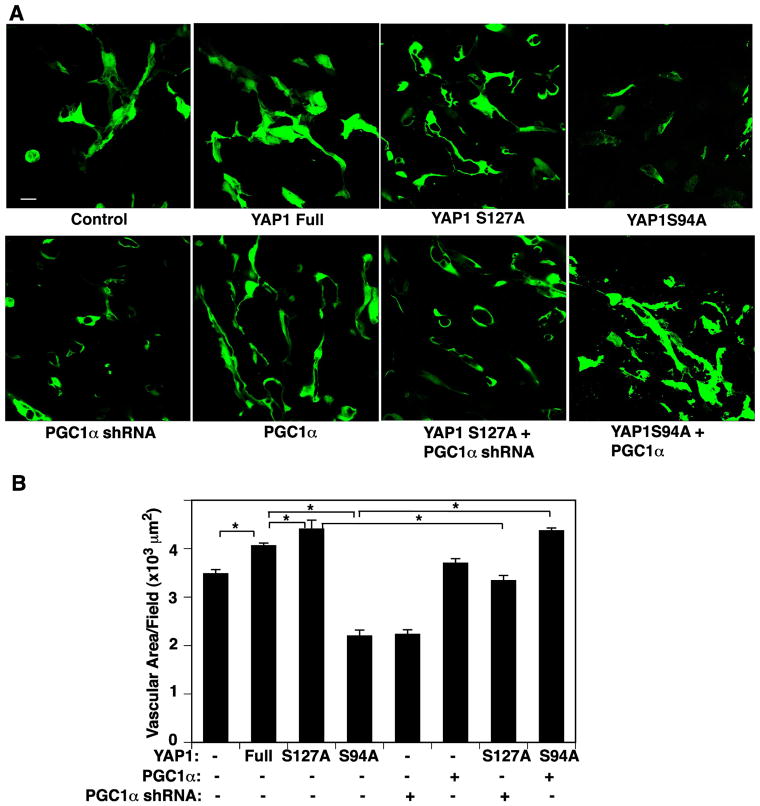
To study whether YAP1 controls angiogenesis through PGC1α signaling in vivo, we subcutaneously implanted fibrin gel supplemented with GFP-labeled HUVE cells overexpressing full-length YAP1, YAP1S127A, YAP1S94A mutant construct, or in combination with PGC1α or PGC1α shRNA and human lung fibroblasts on the back of the mouse. Confocal fluorescence images reveal that GFP-labeled HUVE cells supplemented into the gel (Supplementary Fig. S3B) form a vascular lumen structure in the gel 7 days after implantation (Fig. 5A, Supplementary Fig. S3C). Consistent with an in vitro EC sprouting assay, supplemented fluorescently labeled HUVE cells overexpressing YAP1S127A enhanced vascular network formation compared with those treated with full-length non mutated YAP1, while PGC1α knockdown (Supplementary Fig. S3D) suppressed blood vessel formation induced by YAP1S127A in the implanted gel (Fig. 5A, B). The YAP1S94A mutant construct failed to form a vascular structure in the gel, while PGC1α overexpression restored blood vessel formation in the implanted gel (Fig. 5A, B). These results suggest that YAP1 controls angiogenesis through PGC1α signaling in the gel implanted on mice.
Fig. 5. YAP1 signaling controls vascular formation through PGC1α in a fibrin gel implantation assay.
A) Immunofluorescence micrographs showing GFP-labeled HUVE cells treated with virus overexpressing full-length YAP1, YAP1S127A, YAP1S94A, control vector, or in combination with PGC1α or PGC1α shRNA in the subcutaneously implanted fibrin gel for 7 days. Scale bar, 20 μm. B) Graph showing the changes in vascular area in HUVE cells treated with virus overexpressing full-length YAP1, YAP1S127A, YAP1S94A, control vector, or in combination with PGC1α or PGC1α shRNA in the subcutaneously implanted fibrin gel for 7 days (n=6, *, p<0.05). Error bars represent s.e.m.
Discussion
In this report, we have demonstrated that YAP1-TEAD1 controls the expression of PGC1α and regulates mitochondrial biogenesis/dynamics, oxygen consumption, and glycolytic flux in ECs. YAP1-TEAD1 also controls angiogenesis through PGC1α in vitro and in the fibrin gel subcutaneously implanted on mice. Angiogenesis is a complex process; the establishment of stable and functional blood vessel networks requires cooperation of multiple signaling pathways (Carmeliet and Jain, 2011; Chung and Ferrara, 2011; Herbert and Stainier, 2011). In addition to conventional angiogenic signaling pathways, mitochondria, which are responsible for energy production and contribute to the formation of various organs (e.g., liver, heart, muscle) (Call et al., 2017; Han et al., 2015; Nakada et al., 2017), play important roles in angiogenesis and endothelial function (Arany et al., 2008; Chinsomboon et al., 2009; Kluge et al., 2013; Patten and Arany, 2012; Rowe et al., 2014; Thom et al., 2014). Thus, these findings, which provide a novel signaling axis regulating mitochondrial biogenesis/dynamics and angiogenesis, offer new insights into the regulation of angiogenesis.
Overexpression of PGC1α stimulates expression of angiogenic factor receptors, Tie2 and VEGFR2, in HUVE cells (Fig. 4C). PGC1α also upregulates the expression of angiogenic factor VEGFA in skeletal muscle and retinal müller cells (Arany et al., 2008; Chinsomboon et al., 2009; Patten and Arany, 2012; Rowe et al., 2014; Saint-Geniez et al., 2013; Thom et al., 2014). It has been reported that YAP1-TEAD1 signaling controls angiogenesis and vascular integrity in ECs through various signaling pathways (Al-Moujahed et al., 2017; Choi et al., 2015; Kim et al., 2017; Marti et al., 2015; Nakajima et al., 2017; Sakabe et al., 2017; Wei et al., 2017). We have found that YAP1 also controls Tie2 expression in HUVE cells; YAP1S127A increased, while YAP1S94A decreased Tie2 mRNA levels compared to those in cells overexpressing the non-mutant YAP1 full-length construct (Supplementary Fig. S3A). Thus, manipulation of YAP1-TEAD1- PGC1α signaling, which controls the multiple angiogenic signaling pathways, could be an optimal strategy for modulating angiogenesis. It has been demonstrated that PGC1α maintains the levels of a number of anti-oxidant genes (Patten and Arany, 2012; Valle et al., 2005; Won et al., 2010) and prevents oxidative cell injury in ECs and counteracts endothelial dysfunction (Patten and Arany, 2012). Thus, although PGC1α expression in ECs is relatively low compared to other cell types in organs (Saint-Geniez et al., 2013), manipulation of PGC1α expression in ECs, which affects the expression of multiple downstream genes and influences cellular metabolism (Patten and Arany, 2012), may have a significant impact on angiogenesis and vascular functions. One possible limitation of targeting this pathway may be that modulation of YAP1 activity and/or subsequent changes in the levels of PGC1α may have additive toxicity and detrimental off-target effects resulting from the control of multiple genes (e.g., various types of cancer (Bhalla et al., 2011; Lau et al., 2014), pulmonary fibrosis (Liu et al., 2015; Mora et al., 2017), pulmonary hypertension (Bertero et al., 2015; Diebold et al., 2015)). It has been reported that endothelial PGC1α inhibits endothelial function and angiogenesis in diabetes (Sawada et al., 2014). PGC1α may have divergent roles in angiogenesis and contribute to multiple aspects of vascular morphogenesis and function in a context-dependent manner. Further characterization of the mechanisms by which YAP1-PGC1α signaling controls angiogenesis will maximize its angiogenic ability and minimize the potential toxicity.
As a translational application of YAP-TEAD signaling, Verteporfin (VP), a benzoporphyrin derivative, is clinically used in photodynamic therapy for neovascular macular degeneration (Miller et al., 1999). Recently, it has been reported that VP also inhibits growth of various types of cancer cells including pancreatic cancer (Wei et al., 2017), retinoblastoma (Brodowska et al., 2014), and glioma (Al-Moujahed et al., 2017) through disruption of YAP-TEAD complex and subsequent downregulation of angiogenic factor expression. VP is also known as an autophagy inhibitor to enhance its anti-tumor activity (Donohue et al., 2014). Characterization of the effects of VP on YAP-TEAD signaling and mitochondrial biogenesis/dynamics will lead to the development of additional therapeutic strategy for cancer and other angiogenesis-related diseases.
YAP1 is known as a mechanosensitive gene and its activity is controlled by various mechanical stimuli (e.g., cell density (Choi et al., 2015; Ota and Sasaki, 2008), extracellular matrix (ECM) stiffness (Bertero et al., 2015), mechanical tension (Liu et al., 2016), flow (Nakajima et al., 2017; Wang et al., 2016)) as well (Panciera et al., 2017). We have reported that mechanical forces control vascular morphogenesis and function (Mammoto et al., 2009; Mammoto et al., 2013a; Mammoto et al., 2013b; Mammoto et al., 2013d). For example, mechanosensitive transcription factors (e.g., TFII-I, GATA2, Twist1) control angiogenesis and endothelial cell integrity, and contribute to angiogenesis-related diseases (e.g., pulmonary fibrosis, pulmonary hypertension) (Mammoto et al., 2009; Mammoto et al., 2016b; Mammoto et al., 2013c; Mammoto et al., 2018). Mitochondria are associated with the actin cytoskeleton, which is involved in cellular responses to various mechanical factors (Mammoto and Ingber, 2009). It has been demonstrated that changes in cytoskeletal structures and mechanical forces control mitochondrial network structure and function (Bartolak-Suki et al., 2017) and that perturbation of mitochondrial structures is involved in various diseases in which mechanical forces and YAP1 play important roles (e.g., atherosclerosis (Kadlec et al., 2016; Wang et al., 2016), fibrosis (Liu et al., 2015; Mora et al., 2017), pulmonary hypertension (Bertero et al., 2015; Diebold et al., 2015), and cancer (Andrzejewski et al., 2017; Lau et al., 2014)) (Panciera et al., 2017). PGC1α is also responsive to shear stress in ECs (Chen et al., 2010), further supporting the mechanosensitive mechanism of YAP1-PGC1α signaling in angiogenesis.
We have demonstrated that angiogenesis is stimulated through YAP1- TEAD1- PGC1α signaling. Other Hippo signaling molecules including transcriptional co-activator with PDZ-binding motif (TAZ), which is known to have distinct biological activities from YAP1 (Panciera et al., 2017), are also involved in angiogenesis (Kim et al., 2017). Although YAP1 is phosphorylated at the S127 residue by LATS kinase, other phosphorylation sites (e.g., S381, S112) are also involved in YAP1 activity (Ota and Sasaki, 2008; Piccolo et al., 2014). In addition, YAP1 is serine phosphorylated by AKT and JNK and tyrosine phosphorylated by Yes/Src and Abl (Basu et al., 2003; Danovi et al., 2008; Levy et al., 2008; Piccolo et al., 2014; Zhao et al., 2010). YAP1 exerts its co-transcriptional activity through other TFs (e.g., TEAD-2/-3/-4, Smads, Runx1/-2, p73, ErbB4, Pax3, AP-1, or TBX5) as well (Stein et al., 2015). In fact, although YAP1S127A construct induces, while YAP1S94A suppresses the expression of MFNs, TEAD1 knockdown had little effect on MFN expression (Supplementary Fig. S2A). Other TFs, which bind to YAP1, may contribute to mitochondrial fusion. Since mitochondrial fusion is known to promote energy production (Westermann, 2012), YAP1S127A may increase oxygen consumption by stimulating mitochondrial fusion as well. The YAP1S127A construct also induces the expression of FIS1 and DRP1, which control mitochondrial fission, while TEAD1 knockdown decreases the expression of these fission-related genes (Supplementary Fig. S2B). YAP1S94A did not change the expression of fission genes in ECs (Supplementary Fig. S2B). Thus, YAP1 and TEAD1 may control mitochondrial dynamics in a distinct way. Other genes that regulate mitochondrial biogenesis (e.g., Nuclear respiratory factor (NRF), transcription factor A, mitochondrial (TFAM)), mitochondrial fusion (e.g., optic atrophy 1 (OPA1)) may also mediate YAP1-dependent changes in mitochondrial structure and function. In fact, although mtDNA contents are higher in ECs treated with full-length YAP1 (Fig. 2E), the levels of oxygen consumption are similar to those treated with control virus (Fig. 3). In addition, YAP1S94A mutant decreases mtDNA content compared to YAP1 full-length non-mutated construct, the levels are not lower compared to those in control untreated ECs; however, mitochondrial area and OCR were lower in ECs treated with YAP1S94A mutant compared to those in control untreated ECs (Figs. 2 and 3). YAP1 may control mitochondrial structure and function by modulating mitochondrial dynamics and/or other signaling pathways. Furthermore, the levels of PGC1α in ECs treated with YAP1S94A were lower compared to those in control untreated ECs. Given that YAP1 interacts with multiple signaling pathways (Zhang et al., 2015) (e.g., Wnt/β-catenin (Azzolin et al., 2014; Heallen et al., 2011; Rosenbluh et al., 2012; Varelas et al., 2010), TGFβ/smad (Alarcon et al., 2009) and ERK (Zhang et al., 2014)), these molecules may further inhibit PGC1α expression and subsequently suppress mitochondrial function and angiogenesis (De Luca et al., 2015; Liu et al., 2012). Thus, the mechanism by which YAP1-TEAD1-PGC1α controls mitochondrial structure and angiogenesis seems to be complex, and these complex signaling mechanisms may be necessary for precise spatiotemporal control of mitochondrial structure and angiogenesis.
In summary, we have demonstrated that YAP1-TEAD1 signaling controls angiogenesis and mitochondrial biogenesis through PGC1α. Given that PGC1α drives functional angiogenesis (Rowe et al., 2014), manipulation of YAP1-TEAD1-PGC1α signaling may potentially lead to the development of new therapeutic strategies for angiogenesis-related diseases.
Supplementary Material
Highlights.
YAP1-TEAD1 signaling controls PGC1α expression in endothelial cells.
YAP1-TEAD1 signaling controls mitochondrial biogenesis.
YAP1 controls angiogenesis through PGC1α
Acknowledgments
We thank G. Konduri and R. Teng for helpful discussion. This work was supported by funds from NIH R21AG054830 (to A.M., to T.M.), Medical College of Wisconsin Research Affair Committee New Investigator Award (to A.M.), Medical College of Wisconsin Faculty Start-up funds (to A.M., to T.M.), NIH R01HL13590101 (to D.G.), NIH T32GM080202 (to MCW Medical Scientist Training Program), and the American Heart Association Predoctoral Fellowship Grant 16PRE29130003 (to A.K.). Seahorse XF96 Flux Analysis was performed by S. Komas in the Medical College of Wisconsin Cancer Center Redox and Bioenergetics Shared Resource and funded by Advancing Healthier Wisconsin.
Abbreviations
- YAP
Yes-associated protein
- TEAD
TEA domain transcription factor
- PGC1α
peroxisome proliferator-activated receptor gamma co-activator 1-alpha
- Ecs
endothelial cells
- TF
transcription factor
- Ang2
angiopoietin2
- HUVE
human umbilical vascular endothelial
- VEGF
vascular endothelial growth factor
- bFGF
basic fibroblast growth factor
- PDGF
platelet derived growth factor
- EMEM
Eagle’s Minimum Essential Medium
- MFN
mitofusion
- DRP-1
dynamin-related protein 1
- FIS1
fission, mitochondrial1
- mtDNA
mitochondrial DNA
- ND1
NADH:ubiquinone oxidoreductase core subunit 1
- SLCO2B1
solute carrier organic anion transporter family member 2B1
- OCR
oxygen consumption rate
- ECAR
extracellular acidification rate
- NSG
NOD scid gamma
- LATS
large tumor suppressor
- VP
verteporfin
- ECM
extracellular matrix
- TAZ
transcriptional co-activator with PDZ-binding motif
- NRF
Nuclear respiratory factor
- TFAM
transcription factor A, mitochondrial
- OPA1
optic atrophy 1
Footnotes
Author Contributions
Conceived and designed the experiments: AM, TM. Performed the experiments: TM, MM, AM. Analyzed the data: AM, TM. Contributed reagents/materials/analysis tools: AM, MM, AK, DG, TM. Wrote the paper: AM, AK, DG, TM. All authors approved the final version of the manuscript.
Disclosure Statement
The authors have declared that no conflict of interest exists.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Al-Moujahed A, et al. Verteporfin inhibits growth of human glioma in vitro without light activation. Sci Rep. 2017;7:7602. doi: 10.1038/s41598-017-07632-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alarcon C, et al. Nuclear CDKs drive Smad transcriptional activation and turnover in BMP and TGF-beta pathways. Cell. 2009;139:757–69. doi: 10.1016/j.cell.2009.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrzejewski S, et al. PGC-1alpha Promotes Breast Cancer Metastasis and Confers Bioenergetic Flexibility against Metabolic Drugs. Cell Metab. 2017;26:778–787 e5. doi: 10.1016/j.cmet.2017.09.006. [DOI] [PubMed] [Google Scholar]
- Arany Z, et al. HIF-independent regulation of VEGF and angiogenesis by the transcriptional coactivator PGC-1alpha. Nature. 2008;451:1008–12. doi: 10.1038/nature06613. [DOI] [PubMed] [Google Scholar]
- Azzolin L, et al. YAP/TAZ incorporation in the beta-catenin destruction complex orchestrates the Wnt response. Cell. 2014;158:157–70. doi: 10.1016/j.cell.2014.06.013. [DOI] [PubMed] [Google Scholar]
- Bartolak-Suki E, et al. Regulation of Mitochondrial Structure and Dynamics by the Cytoskeleton and Mechanical Factors. Int J Mol Sci. 2017:18. doi: 10.3390/ijms18081812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu S, et al. Akt phosphorylates the Yes-associated protein, YAP, to induce interaction with 14-3-3 and attenuation of p73-mediated apoptosis. Mol Cell. 2003;11:11–23. doi: 10.1016/s1097-2765(02)00776-1. [DOI] [PubMed] [Google Scholar]
- Bertero T, et al. Matrix Remodeling Promotes Pulmonary Hypertension through Feedback Mechanoactivation of the YAP/TAZ-miR-130/301 Circuit. Cell Rep. 2015;13:1016–32. doi: 10.1016/j.celrep.2015.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalla K, et al. PGC1alpha promotes tumor growth by inducing gene expression programs supporting lipogenesis. Cancer Res. 2011;71:6888–98. doi: 10.1158/0008-5472.CAN-11-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodowska K, et al. The clinically used photosensitizer Verteporfin (VP) inhibits YAP-TEAD and human retinoblastoma cell growth in vitro without light activation. Exp Eye Res. 2014;124:67–73. doi: 10.1016/j.exer.2014.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Call JA, et al. Ulk1-mediated autophagy plays an essential role in mitochondrial remodeling and functional regeneration of skeletal muscle. Am J Physiol Cell Physiol. 2017 doi: 10.1152/ajpcell.00348.2016. ajpcell 00348 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmeliet P, Jain RK. Molecular mechanisms and clinical applications of angiogenesis. Nature. 2011;473:298–307. doi: 10.1038/nature10144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, et al. Shear stress, SIRT1, and vascular homeostasis. Proc Natl Acad Sci U S A. 2010;107:10268–73. doi: 10.1073/pnas.1003833107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinsomboon J, et al. The transcriptional coactivator PGC-1alpha mediates exercise-induced angiogenesis in skeletal muscle. Proc Natl Acad Sci U S A. 2009;106:21401–6. doi: 10.1073/pnas.0909131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HJ, et al. Yes-associated protein regulates endothelial cell contact-mediated expression of angiopoietin-2. Nat Commun. 2015;6:6943. doi: 10.1038/ncomms7943. [DOI] [PubMed] [Google Scholar]
- Chung AS, Ferrara N. Developmental and pathological angiogenesis. Annu Rev Cell Dev Biol. 2011;27:563–84. doi: 10.1146/annurev-cellbio-092910-154002. [DOI] [PubMed] [Google Scholar]
- Cruys B, et al. Glycolytic regulation of cell rearrangement in angiogenesis. Nat Commun. 2016;7:12240. doi: 10.1038/ncomms12240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L, et al. Transcriptional repression of PGC-1alpha by mutant huntingtin leads to mitochondrial dysfunction and neurodegeneration. Cell. 2006;127:59–69. doi: 10.1016/j.cell.2006.09.015. [DOI] [PubMed] [Google Scholar]
- Danovi SA, et al. Yes-associated protein (YAP) is a critical mediator of c-Jun-dependent apoptosis. Cell Death Differ. 2008;15:217–9. doi: 10.1038/sj.cdd.4402226. [DOI] [PubMed] [Google Scholar]
- De Bock K, et al. Role of PFKFB3-driven glycolysis in vessel sprouting. Cell. 2013;154:651–63. doi: 10.1016/j.cell.2013.06.037. [DOI] [PubMed] [Google Scholar]
- De Luca A, et al. Mitochondrial biogenesis is required for the anchorage-independent survival and propagation of stem-like cancer cells. Oncotarget. 2015;6:14777–95. doi: 10.18632/oncotarget.4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diebold I, et al. BMPR2 preserves mitochondrial function and DNA during reoxygenation to promote endothelial cell survival and reverse pulmonary hypertension. Cell Metab. 2015;21:596–608. doi: 10.1016/j.cmet.2015.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donohue E, et al. Induction of Covalently Crosslinked p62 Oligomers with Reduced Binding to Polyubiquitinated Proteins by the Autophagy Inhibitor Verteporfin. PLoS ONE. 2014;9:e114964. doi: 10.1371/journal.pone.0114964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan W, Evans R. PPARs and ERRs: molecular mediators of mitochondrial metabolism. Curr Opin Cell Biol. 2015;33:49–54. doi: 10.1016/j.ceb.2014.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han LH, et al. Deceleration of liver regeneration by knockdown of augmenter of liver regeneration gene is associated with impairment of mitochondrial DNA synthesis in mice. Am J Physiol Gastrointest Liver Physiol. 2015;309:G112–22. doi: 10.1152/ajpgi.00435.2014. [DOI] [PubMed] [Google Scholar]
- Heallen T, et al. Hippo pathway inhibits Wnt signaling to restrain cardiomyocyte proliferation and heart size. Science. 2011;332:458–61. doi: 10.1126/science.1199010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert SP, Stainier DY. Molecular control of endothelial cell behaviour during blood vessel morphogenesis. Nat Rev Mol Cell Biol. 2011;12:551–64. doi: 10.1038/nrm3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiramitsu M, et al. Eriocitrin ameliorates diet-induced hepatic steatosis with activation of mitochondrial biogenesis. Sci Rep. 2014;4:3708. doi: 10.1038/srep03708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, et al. YAP suppresses gluconeogenic gene expression through PGC1alpha. Hepatology. 2017;66:2029–2041. doi: 10.1002/hep.29373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadlec AO, et al. Role of PGC-1alpha in Vascular Regulation: Implications for Atherosclerosis. Arterioscler Thromb Vasc Biol. 2016;36:1467–74. doi: 10.1161/ATVBAHA.116.307123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadlec AO, et al. PGC-1alpha (Peroxisome Proliferator-Activated Receptor gamma Coactivator 1-alpha) Overexpression in Coronary Artery Disease Recruits NO and Hydrogen Peroxide During Flow-Mediated Dilation and Protects Against Increased Intraluminal Pressure. Hypertension. 2017;70:166–173. doi: 10.1161/HYPERTENSIONAHA.117.09289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, et al. YAP/TAZ regulates sprouting angiogenesis and vascular barrier maturation. J Clin Invest. 2017;127:3441–3461. doi: 10.1172/JCI93825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluge MA, et al. Mitochondria and endothelial function. Circ Res. 2013;112:1171–88. doi: 10.1161/CIRCRESAHA.111.300233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau AN, et al. Tumor-propagating cells and Yap/Taz activity contribute to lung tumor progression and metastasis. Embo J. 2014;33:468–81. doi: 10.1002/embj.201386082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy D, et al. Yap1 phosphorylation by c-Abl is a critical step in selective activation of proapoptotic genes in response to DNA damage. Mol Cell. 2008;29:350–61. doi: 10.1016/j.molcel.2007.12.022. [DOI] [PubMed] [Google Scholar]
- Li Z, et al. Structural insights into the YAP and TEAD complex. Genes Dev. 2010;24:235–40. doi: 10.1101/gad.1865810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, et al. Mechanosignaling through YAP and TAZ drives fibroblast activation and fibrosis. Am J Physiol Lung Cell Mol Physiol. 2015;308:L344–57. doi: 10.1152/ajplung.00300.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, et al. Low density lipoprotein (LDL) receptor-related protein 6 (LRP6) regulates body fat and glucose homeostasis by modulating nutrient sensing pathways and mitochondrial energy expenditure. J Biol Chem. 2012;287:7213–23. doi: 10.1074/jbc.M111.286724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, et al. MAPK-Mediated YAP Activation Controls Mechanical-Tension-Induced Pulmonary Alveolar Regeneration. Cell Rep. 2016;16:1810–9. doi: 10.1016/j.celrep.2016.07.020. [DOI] [PubMed] [Google Scholar]
- Mammoto A, et al. A mechanosensitive transcriptional mechanism that controls angiogenesis. Nature. 2009;457:1103–8. doi: 10.1038/nature07765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mammoto A, Ingber DE. Cytoskeletal control of growth and cell fate switching. Curr Opin Cell Biol. 2009;21:864–70. doi: 10.1016/j.ceb.2009.08.001. [DOI] [PubMed] [Google Scholar]
- Mammoto A, et al. Control of lung vascular permeability and endotoxin-induced pulmonary edema by changes in extracellular matrix mechanics. Nature Comm. 2013a;4:1759. doi: 10.1038/ncomms2774. [DOI] [PubMed] [Google Scholar]
- Mammoto T, et al. LRP5 Regulates Development of Lung Microvessels and Alveoli through the Angiopoietin-Tie2 Pathway. PLoS ONE. 2012;7:e41596. doi: 10.1371/journal.pone.0041596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mammoto T, et al. Platelet-rich plasma extract accelerates lung regeneration through the LRP5-Tie2 pathway. Am J Respir Cell Mol Biol. 2016a;54:103–113. doi: 10.1165/rcmb.2015-0045OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mammoto T, et al. The Role of Twist1 Phosphorylation in Angiogenesis and Pulmonary Fibrosis. Am J Respir Cell Mol Biol. 2016b;55:633–644. doi: 10.1165/rcmb.2016-0012OC. [DOI] [PubMed] [Google Scholar]
- Mammoto T, et al. A Role of Collagen Matrix in Tumor Angiogenesis and Glioblastoma Multiforme Progression. American journal of pathology. 2013b;183:1293–1305. doi: 10.1016/j.ajpath.2013.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mammoto T, et al. Twist1 Controls Lung Vascular Permeability and Endotoxin-induced Pulmonary Edema by Altering Tie2 Expression. PLoS ONE. 2013c;8:e73407. doi: 10.1371/journal.pone.0073407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mammoto T, et al. ECM structure and tissue stiffness control postnatal lung development through the LRP5-Tie2 signaling system. American journal of respiratory cell and molecular biology. 2013d;49:1009–1018. doi: 10.1165/rcmb.2013-0147OC. [DOI] [PubMed] [Google Scholar]
- Mammoto T, Mammoto A. Implantation of fibrin gel on mouse lung to study lung-specific angiogenesis. J Vis Exp. 2014 doi: 10.3791/52012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mammoto T, et al. Twist1 in hypoxia-induced pulmonary hypertension through TGFβ-Smad signaling. Am J Respir Cell Mol Biol. 2018;58:194–207. doi: 10.1165/rcmb.2016-0323OC. [DOI] [PubMed] [Google Scholar]
- Marti P, et al. YAP promotes proliferation, chemoresistance, and angiogenesis in human cholangiocarcinoma through TEAD transcription factors. Hepatology. 2015;62:1497–510. doi: 10.1002/hep.27992. [DOI] [PubMed] [Google Scholar]
- Miller JW, et al. Photodynamic therapy with verteporfin for choroidal neovascularization caused by age-related macular degeneration: results of a single treatment in a phase 1 and 2 study. Arch Ophthalmol. 1999;117:1161–73. doi: 10.1001/archopht.117.9.1161. [DOI] [PubMed] [Google Scholar]
- Mora AL, et al. Mitochondria in the spotlight of aging and idiopathic pulmonary fibrosis. J Clin Invest. 2017;127:405–414. doi: 10.1172/JCI87440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaraj R, et al. Control of mitochondrial structure and function by the Yorkie/YAP oncogenic pathway. Genes Dev. 2012;26:2027–37. doi: 10.1101/gad.183061.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakada Y, et al. Hypoxia induces heart regeneration in adult mice. Nature. 2017;541:222–227. doi: 10.1038/nature20173. [DOI] [PubMed] [Google Scholar]
- Nakajima H, et al. Flow-Dependent Endothelial YAP Regulation Contributes to Vessel Maintenance. Dev Cell. 2017;40:523–536 e6. doi: 10.1016/j.devcel.2017.02.019. [DOI] [PubMed] [Google Scholar]
- Nakatsu MN, et al. Optimized fibrin gel bead assay for the study of angiogenesis. J Vis Exp. 2007:186. doi: 10.3791/186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osellame LD, et al. Cellular and molecular mechanisms of mitochondrial function. Best Pract Res Clin Endocrinol Metab. 2012;26:711–23. doi: 10.1016/j.beem.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota M, Sasaki H. Mammalian Tead proteins regulate cell proliferation and contact inhibition as transcriptional mediators of Hippo signaling. Development. 2008;135:4059–69. doi: 10.1242/dev.027151. [DOI] [PubMed] [Google Scholar]
- Panciera T, et al. Mechanobiology of YAP and TAZ in physiology and disease. Nat Rev Mol Cell Biol. 2017 doi: 10.1038/nrm.2017.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patten IS, Arany Z. PGC-1 coactivators in the cardiovascular system. Trends Endocrinol Metab. 2012;23:90–7. doi: 10.1016/j.tem.2011.09.007. [DOI] [PubMed] [Google Scholar]
- Patten IS, et al. Cardiac angiogenic imbalance leads to peripartum cardiomyopathy. Nature. 2012;485:333–8. doi: 10.1038/nature11040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccolo S, et al. Molecular pathways: YAP and TAZ take center stage in organ growth and tumorigenesis. Clin Cancer Res. 2013;19:4925–30. doi: 10.1158/1078-0432.CCR-12-3172. [DOI] [PubMed] [Google Scholar]
- Piccolo S, et al. The biology of YAP/TAZ: hippo signaling and beyond. Physiol Rev. 2014;94:1287–312. doi: 10.1152/physrev.00005.2014. [DOI] [PubMed] [Google Scholar]
- Rosenbluh J, et al. beta-Catenin-driven cancers require a YAP1 transcriptional complex for survival and tumorigenesis. Cell. 2012;151:1457–73. doi: 10.1016/j.cell.2012.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe GC, et al. PGC-1 coactivators in cardiac development and disease. Circ Res. 2010;107:825–38. doi: 10.1161/CIRCRESAHA.110.223818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe GC, et al. PGC-1alpha induces SPP1 to activate macrophages and orchestrate functional angiogenesis in skeletal muscle. Circ Res. 2014;115:504–17. doi: 10.1161/CIRCRESAHA.115.303829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint-Geniez M, et al. PGC-1alpha regulates normal and pathological angiogenesis in the retina. Am J Pathol. 2013;182:255–65. doi: 10.1016/j.ajpath.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakabe M, et al. YAP/TAZ-CDC42 signaling regulates vascular tip cell migration. Proc Natl Acad Sci U S A. 2017;114:10918–10923. doi: 10.1073/pnas.1704030114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada N, et al. Endothelial PGC-1alpha mediates vascular dysfunction in diabetes. Cell Metab. 2014;19:246–58. doi: 10.1016/j.cmet.2013.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schinzel R, Dillin A. Endocrine aspects of organelle stress-cell non-autonomous signaling of mitochondria and the ER. Curr Opin Cell Biol. 2015;33:102–10. doi: 10.1016/j.ceb.2015.01.006. [DOI] [PubMed] [Google Scholar]
- Shao DD, et al. KRAS and YAP1 converge to regulate EMT and tumor survival. Cell. 2014;158:171–84. doi: 10.1016/j.cell.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva JP, et al. Impaired insulin secretion and beta-cell loss in tissue-specific knockout mice with mitochondrial diabetes. Nat Genet. 2000;26:336–40. doi: 10.1038/81649. [DOI] [PubMed] [Google Scholar]
- Stein C, et al. YAP1 Exerts Its Transcriptional Control via TEAD-Mediated Activation of Enhancers. PLoS Genet. 2015;11:e1005465. doi: 10.1371/journal.pgen.1005465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TeSlaa T, Teitell MA. Techniques to monitor glycolysis. Methods Enzymol. 2014;542:91–114. doi: 10.1016/B978-0-12-416618-9.00005-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thom R, et al. Hypoxic induction of vascular endothelial growth factor (VEGF) and angiogenesis in muscle by truncated peroxisome proliferator-activated receptor gamma coactivator (PGC)-1alpha. J Biol Chem. 2014;289:8810–7. doi: 10.1074/jbc.M114.554394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle I, et al. PGC-1alpha regulates the mitochondrial antioxidant defense system in vascular endothelial cells. Cardiovasc Res. 2005;66:562–73. doi: 10.1016/j.cardiores.2005.01.026. [DOI] [PubMed] [Google Scholar]
- Varelas X, et al. The Hippo pathway regulates Wnt/beta-catenin signaling. Dev Cell. 2010;18:579–91. doi: 10.1016/j.devcel.2010.03.007. [DOI] [PubMed] [Google Scholar]
- Wang KC, et al. Flow-dependent YAP/TAZ activities regulate endothelial phenotypes and atherosclerosis. Proc Natl Acad Sci U S A. 2016;113:11525–11530. doi: 10.1073/pnas.1613121113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H, et al. Verteporfin suppresses cell survival, angiogenesis and vasculogenic mimicry of pancreatic ductal adenocarcinoma via disrupting the YAP-TEAD complex. Cancer Sci. 2017;108:478–487. doi: 10.1111/cas.13138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann B. Bioenergetic role of mitochondrial fusion and fission. Biochim Biophys Acta. 2012;1817:1833–8. doi: 10.1016/j.bbabio.2012.02.033. [DOI] [PubMed] [Google Scholar]
- Won JC, et al. Peroxisome proliferator-activated receptor-gamma coactivator 1-alpha overexpression prevents endothelial apoptosis by increasing ATP/ADP translocase activity. Arterioscler Thromb Vasc Biol. 2010;30:290–7. doi: 10.1161/ATVBAHA.109.198721. [DOI] [PubMed] [Google Scholar]
- Wu M, et al. Multiparameter metabolic analysis reveals a close link between attenuated mitochondrial bioenergetic function and enhanced glycolysis dependency in human tumor cells. Am J Physiol Cell Physiol. 2007;292:C125–36. doi: 10.1152/ajpcell.00247.2006. [DOI] [PubMed] [Google Scholar]
- Yuan L, et al. Palmitic acid dysregulates the Hippo-YAP pathway and inhibits angiogenesis by inducing mitochondrial damage and activating the cytosolic DNA sensor cGAS-STING-IRF3 signaling mechanism. J Biol Chem. 2017;292:15002–15015. doi: 10.1074/jbc.M117.804005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, et al. The hippo pathway effector YAP regulates motility, invasion, and castration-resistant growth of prostate cancer cells. Mol Cell Biol. 2015;35:1350–62. doi: 10.1128/MCB.00102-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, et al. Downstream of mutant KRAS, the transcription regulator YAP is essential for neoplastic progression to pancreatic ductal adenocarcinoma. Sci Signal. 2014;7:ra42. doi: 10.1126/scisignal.2005049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, et al. The Hippo-YAP pathway in organ size control and tumorigenesis: an updated version. Genes Dev. 2010;24:862–74. doi: 10.1101/gad.1909210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–61. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.