Abstract
C-C chemokine receptor type 5 (CCR5) is the main co-receptor for HIV entry into the target CD4+ cells, and homozygous CCR5Δ32/Δ32 cells are resistant to CCR5-tropic HIV infection. However, the CCR5Δ32/Δ32 homozygous donors in populations are rare. Here we developed a simple approach to induce CCR5Δ32/Δ32 homozygotes through CRISPR-Cas9 genome-editing technology. Designing a pair of single-guide RNA targeting the flank region of the CCR5Δ32 mutation locus, we applied the CRISPR-Cas9 and lentiviral packaging system to successfully convert wild-type CCR5 into CCR5Δ32/Δ32 homozygotes in the human Jurkat CD4+ cell line and primary CD4+ cells, exactly the same as the naturally occurring CCR5Δ32/Δ32 mutation. The successful rate is up to 20% in Jurkat cells but less in primary CD4+ cells. The modified CCR5Δ32/Δ32 CD4+ cells are resistant to CCR5-tropic HIV infection. Whole-genome sequencing revealed no apparent off-target sites. This approach has the promise to promote HIV/AIDS therapy from the only cured unique Berlin patient to a routine autologous cell-based therapy.
Keywords: HIV, AIDS, CCR5Δ, 32, CRISPR-Cas9, CD4+ cells
Introduction
In 2009, Dr. Hutter reported that a Berlin patient, Mr. Timothy Brown, who suffered both HIV infection and acute myeloid leukemia, received allogeneic CCR5Δ32/Δ32 bone marrow transplantation.1 The individuals with CCR5Δ32/Δ32 homozygous deletion are resistant to CCR5-tropic HIV infection. The Berlin patient showed no HIV rebound 20 months after the transplantation and the ceasing of antiretroviral therapy (ART).1, 2 Most importantly, he is still healthy and no HIV rebound has occurred after 9 years of stopping the ART.2 Namely, he is cured, and the only cured case until now globally.
Under the encouragement of success in this Berlin patient, CCR5, the main co-receptor of HIV entry into CD4+ cells, has become an important target for gene editing anti-HIV therapy.3, 4, 5, 6, 7, 8 On the other hand, as a retrovirus, HIV reversely transcribes its single-stranded RNA genome into a double-stranded DNA, which integrates into the human genome.9, 10 The integrated viral genome can either actively promote the production of new virions or remain inactive within a CD4+ population of cells.10 Cells harboring inactive viral genomes, known as latent viruses, are not sensitive to ART, and they become a viral reservoir capable of producing infectious virus under altered conditions. Another type of viral reservoir results from the persistent HIV replication in anatomical sites hard to reach with drugs, such as lymphoid tissue, brain, or gut.10, 11 HIV viruses still spread through cell-to-cell despite the ART.10 All of these viral reservoirs contribute to the viral rebound after ART stop, and these reservoirs are the major barriers to an HIV/AIDS cure.10 The HIV reservoirs are established during primary infection and matured in early latent infection.10, 12 CCR5-tropic viruses predominate globally, and they remain dominant throughout the asymptomatic phase of HIV infection.13, 14 Thus, CCR5-tropic viruses are a key target for downsizing the viral reservoirs and preventing further HIV replication.
The Berlin patient is a unique case and dependent on the histocompatibility-matched CCR5Δ32/Δ32 homozygous donors. These donors occur at a little higher rate in the European Caucasian population, but they are very rare in Asians and Africans.15, 16, 17 The recently developed genome-editing technology, such as zinc-finger nuclease (ZFN), transcription activator-like effector nucleases (TALENs), and CRISPR and CRISPR-Cas9, provides powerful approaches for CCR5 artificial modification.8, 18, 19 These approaches are making it possible to modify self-cells to resist HIV infection, and they represent promising approaches for autologous cell-based therapy. Positive results have been obtained in both preclinical and clinical trials in which HIV-infected patients were transplanted with autologous ZFN-disrupted CCR5 CD4+ cells.20, 21, 22 Investigators have also ablated the entire CCR5 gene, but the long-term adverse effects of CCR5-disrupted cell transplantation are unknown. In contrast, naturally occurring CCR5Δ32/Δ32 homozygotes are healthy in the population. Thus, CCR5Δ32/Δ32 induction may be a much better approach than CCR5 disruption. Ye et al.23 successfully created mutant CCR5Δ32/Δ32 homozygotes in induced pluripotent stem cells using the combination of piggyBac transposon technology and TALENs or CRISPR-Cas9 technology. While exciting, their technological approach was complicated. In this paper, we have developed a more simple approach to induce CCR5Δ32/Δ32 homozygotes in human cells using only the CRISPR-Cas9 technology and a pair of single-guide RNAs.
Results
CRISPR-Cas9 Induced CCR5Δ32/Δ32 Mutation in the Jurkat Cell Line
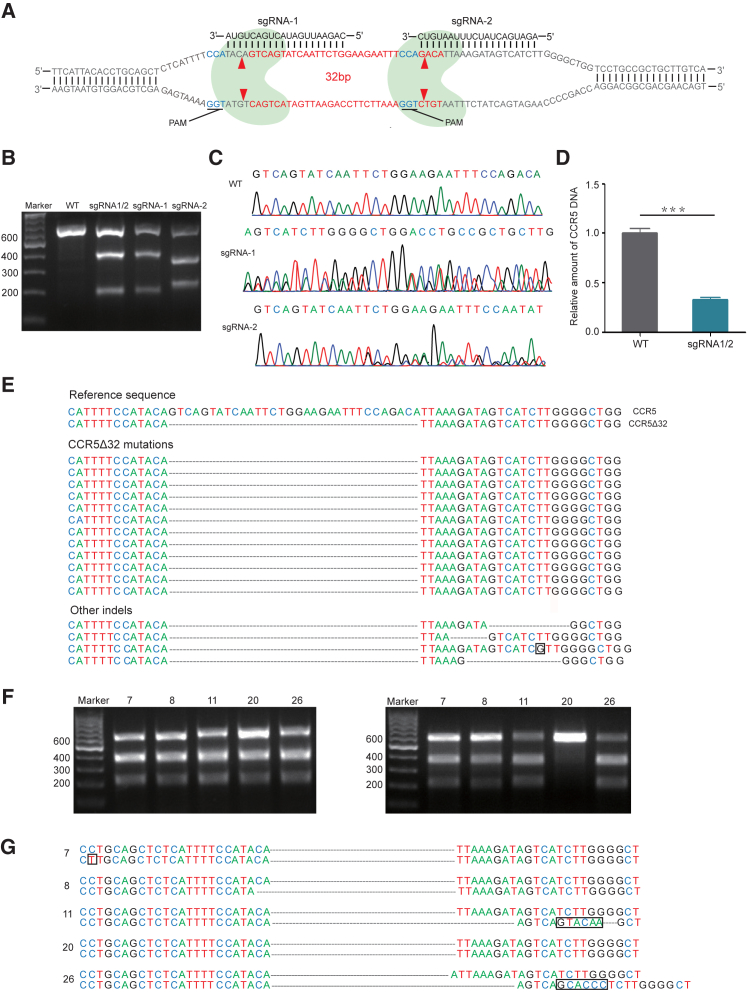
The CRISPR-Cas9 technology is a single RNA guided and limited by the protospacer adjacent motif (PAM) recognition, containing 5′NGG3′. Fortunately, we found a PAM in the flank region of antisense DNA of the CCR5 gene and another PAM in the internal antisense DNA of the CCR5Δ32 mutation (Figure 1A). We then designed a pair of single-guide RNAs (sgRNAs) targeting the CCR5Δ32 locus, sgRNA1 and sgRNA2. Cas9 nuclease cut between the third and fourth nucleotides upstream of the PAM site, as the red arrows indicate (Figure 1A; Table S1). Although the cleavage of the pair of sgRNA1 and sgRNA2 was not exactly as CCR5Δ32 mutation, sgRNA1 cleavage cut 1 less nucleotide T, and this cut was supplemented with 1 more nucleotide T by sgRNA2 cleavage. Thus, the cleavage of the pair sgRNA1 and sgRNA2 resulted in an exact CCR5Δ32 mutation (Figure 1A) (China patent application 201610028603; US patent application PCT/CN2016/079007). The corresponding DNAs of sgRNA1 and sgRNA2 were inserted into the lentiviral vector LentiCRISPRv2 (from Dr. Feng Zhang, McGovern Institute for Brain Research at MIT, Addgene Catalog 52961). As a nuclease, Cas9 causes DNA double-strand breaks (DSBs) under the guide of sgRNAs. DSB recovery is dependent either on homologous recombination (HR) or non-homologous end joining (NHEJ). NHEJ results in nucleotide insertions or deletions (indels) and induces deleterious reading frameshifts.
Figure 1.
CRISPR-Cas9-Mediated CCR5Δ32/Δ32 Homozygotes in the CD4+ Jurkat Cell Line
(A) CCR5 gene and schematic diagram of CRISPR-Cas9-mediated CCR5Δ32 induction. The CCR5Δ32 mutation is in red color. PAM is in dark blue color. Nuclease Cas9 is in light blue color. sgRNA1- and sgRNA2-mediated Cas9 cleavage sites are labeled with red arrows. (B) T7E1 digestion of 609-bp CCR5 PCR products of wild-type (WT) and each sgRNA-treated sample, sgRNA1 (sgRNA-1), or sgRNA2 (sgRNA-2), or both (sgRNA1/2). (C) Sequencing chromatograph of the CCR5 gene, untreated (WT) or treated with sgRNA-1 or sgRNA-2. (D) Real-time qPCR analysis of the CCR5 gene. Error bars represent the average value of two independent experiments, triplicate in each experiment and triplicate in each measurement. ***p = 0.0001. (E) DNA sequence alignment of the CCR5 gene from the sample treated by sgRNA1 and sgRNA2. (F) T7E1 digestion of PCR products from CRISPR-Cas9 sgRNA1/2-treated monoclonal Jurkat cells. PCR product mixture of wild-type with each sample number 7, 8, 11, 20, and 26 (left panel) and PCR products of each sample alone (right panel) are shown. (G) DNA sequence alignment of the CCR5 gene from CRISPR-Cas9 sgRNA1/2-treated monoclonal Jurkat cells. Cell clone number is as indicated. Compared to the wild-type CCR5 gene, some mutations caused by CRISPR-Cas9 are in the black box.
Next, we investigated whether the sgRNA1 and sgRNA2 work alone or in combination to induce CCR5 mutation. First, the human CD4+ Jurkat cell line was used to test sgRNA1, sgRNA2, and the combination. T7 endonuclease 1 (T7E1) can recognize the mismatched double-stranded DNA. After infection of LentiCRISPRv2 containing sgRNA1 and/or sgRNA2, we checked the effects of genome editing. The 609-bp CCR5 gene was amplified from wild-type CCR5 and each sgRNA-treated sample. The PCR products from sgRNA1-, sgRNA2-, and combined sgRNA1 and 2-treated samples could be digested by T7E1, but wild-type CCR5 could not (Figure 1B). These data indicate that sgRNA1, sgRNA2, or the combination can cause genome editing and work well.
PCR products of the CCR5 gene were cloned into the TA vector and sequenced (Table S2). DNA sequencing also confirmed genome editing of the CCR5 gene and the proportion of CCR5Δ32 mutation (Figures 1C and 1E). TA cloning and sequencing showed that 75% bacterial clones from the combined sgRNA1 and sgRNA2-treated DNA sample contained CCR5 mutations, and among them 60% was CCR5Δ32 mutation, which could not be found in sgRNA1 or sgRNA2 singly treated sample (Figures 1C–1E). The other 15% bacterial clones contained non-specific indels (Figure 1E). Real-time qPCR, using sense primer complementary to the CCR5Δ32 deletion and antisense primer complementary to the region downstream of the CCR5Δ32 deletion, further confirmed the significant reduction of the wild-type CCR5 gene (Figure 1D).
We then isolated monoclonal mutated cell clones through serial dilution in a 96-well plate, ensuring roughly one cell/well. Because the vector LentiCRISPRv2 contains a puromycin-screening marker, the LentiCRISPRv2-uninfected Jurkat cells would be killed by puromycin. In 29 randomly selected puromycin-resistant cell clones, all of them were digested by T7E1 (Figure S1), indicating that all of them had the CCR5 mutation. Among 29 cell clones, we randomly selected 5 clones (numbers 7, 8, 11, 20, and 26; Figure 1F) for further analysis. Interestingly, the 609-bp CCR5 PCR product mixtures of wild-type CCR5 and each cell clone could be digested by T7E1 (Figure 1F, left panel). However, the CCR5 PCR products of each cell clone alone could be digested by T7E1 except cell clone 20 (Figure 1F, right panel). DNA sequencing confirmed that the number 20 cell clone was CCR5Δ32/Δ32 homozygotes. The other cell clones were other types of CCR5Δ32 mutations (Figure 1G). Therefore, we successfully obtained the CCR5Δ32/Δ32 homozygous Jurkat cells.
CRISPR-Cas9 Induced CCR5Δ32/Δ32 Mutation in Primary CD4+ Cells
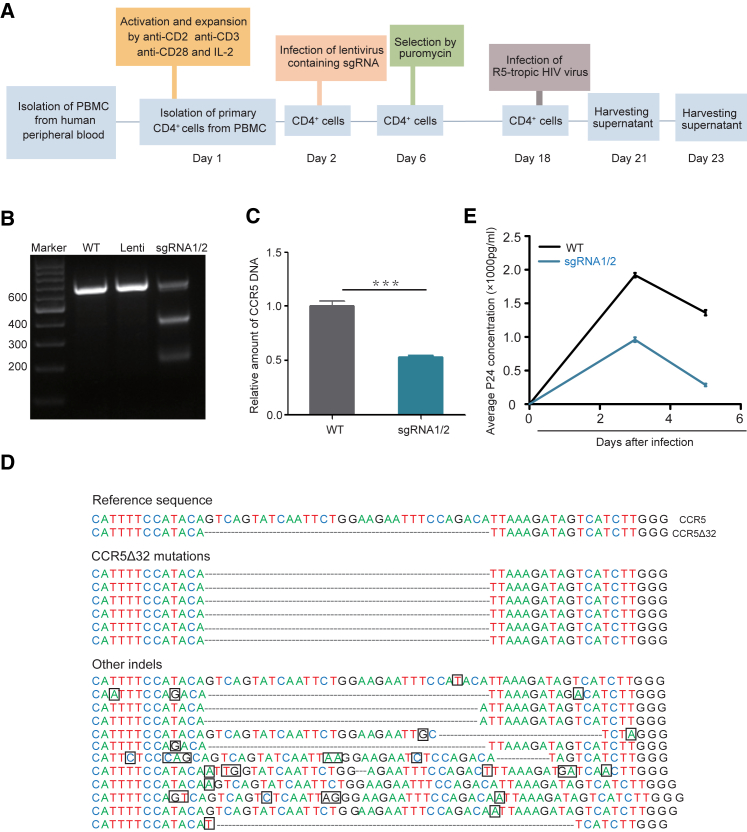
Since CD4+ cells are the target cells of HIV, we then tried this approach in primary CD4+ cells. Primary CD4+ cells from peripheral blood mononuclear cells (PBMCs) of a healthy donor were isolated. The experimental procedure of CRISPR-Cas9 application in primary CD4+ cells is illustrated in Figure 2A. After sgRNA1- and 2-containing LentiCRISPEv2 infection and 12 days of puromycin screening, DNA was extracted from the puromycin-resistant cells and the CCR5 gene was amplified by PCR again. T7E1 digestion assay also illustrated the genome-editing effect of this sgRNA1- and sgRNA2-specific CRISPR-Cas9 technology again (Figure 2B). Real-time qPCR also confirmed the significant reduction of the wild-type CCR5 gene, and the mutation efficiency was 45%–55% (Figure 2C). PCR products were cloned into the TA vector for sequencing.
Figure 2.
CRISPR-Cas9-Mediated CCR5Δ32 Mutation in Primary CD4+ Cells
(A) The experimental procedures for the isolation of primary CD4+ cells from peripheral blood mononuclear cells (PBMCs) and CRISPR-Cas9-mediated CCR5Δ32 induction. (B) T7E1 digestion of samples as indicated. WT, CCR5 wild-type; Lenti, negative control of empty vector LentiCRISPRv2. (C) Real-time qPCR analysis. Error bars represent the average value of two independent experiments, triplicate in each experiment and triplicate in each measurement. ***p = 0.0001. (D) DNA sequence alignment of the CCR5 gene from CRISPR-Cas9 sgRNA1/2-treated primary CD4+ cells. Some mutations caused by CRISPR-Cas9 are in the black box. (E) CCR5-tropic HIV-1397PXJ virus challenge test. Error bars represent the average value of one independent experiment with triplicate measurement. Another independent experiment has a similar trend.
In the selected 30 bacterial monoclonal colonies, only 18 clones carried the CCR5 gene mutations (18/30, 60%), and among them 6 clones were the CCR5Δ32 deletion (6/30, 20%) and the other 12 clones (12/30, 40%) were an unwanted non-CCR5Δ32 deletion (Figure 2D). The CCR5Δ32 induction frequency was obviously lower in primary CD4+ cells (20%) than in Jurkat cells (60%) (Figures 1E and 2D). Like Jurkat cells, we tried to isolate monoclonal cells through limiting dilution method. Unfortunately, for some unknown reasons, we were unable to isolate the monoclonal primary CD4+ cells. Next, sgRNA1 and sgRNA2 CRISPR-Cas9-induced primary CD4+ cells were challenged with HIV. As expected, these induced primary CD4+ CCR5Δ32 cells were resistant to CCR5-tropic virus infection compared to wild-type primary CD4+ cells, when 4 ng CCR5-tropic HIV-1397PXJ virus was used (Figure 2E).
Off-Target Analysis in the Primary CD4+ Cells
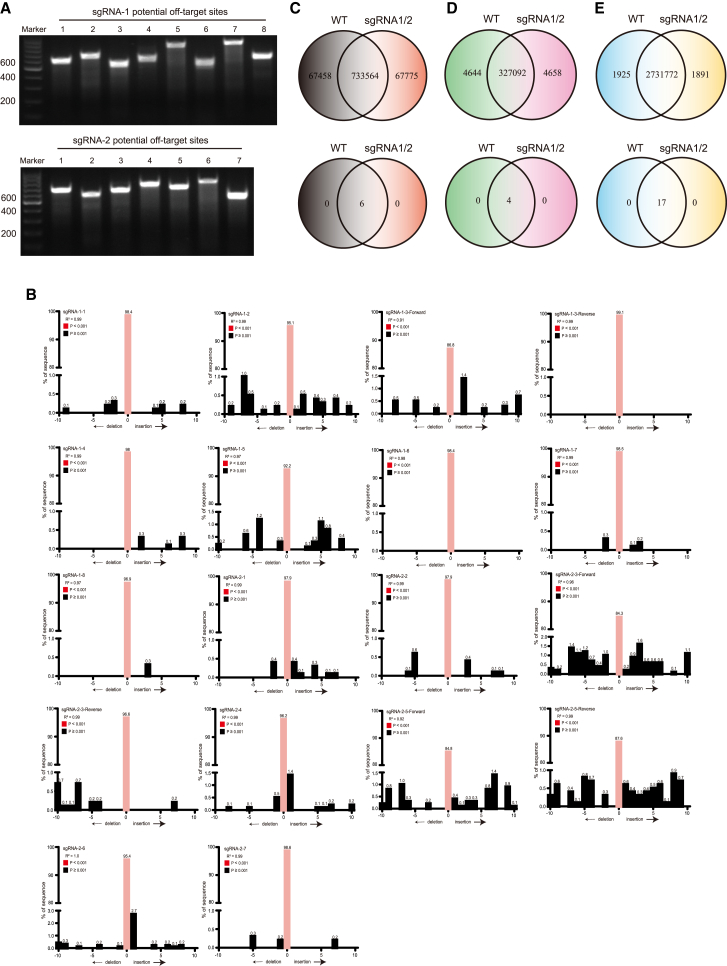
Because CRISPR-Cas9 technology could cause an off-target effect, we analyzed the most possible sites in human genome. Computer analysis predicted the most possible off-target sites in human chromosomes, 8 sites for sgRNA1 and 7 sites for sgRNA2 (Table S3). T7E1 digestion clearly showed that all of the 8 sites for sgRNA1 and 7 sites for sgRNA2 had no off-target effects in the primary CD4+ cells (Figure 3A).
Figure 3.
Off-Target Analysis in the Primary CD4+ Cells
(A) T7E1 digestion of sgRNA1- (upper panel) and sgRNA2-treated primary CD4+ cells (lower panel) in the most possible off-target sites. (B) TIDE analysis of 15 potential off-target sites. (C) Whole-genome sequencing (WGS) shows the joint and respective indels of wild-type or sgRNA1/2-induced CCR5 gene (upper panel) and 49 most possible indels (lower panel). (D) WGS indel data were reanalyzed when the sites were covered more than 20 times by sequencing. (E) WGS shows the joint and respective SNVs of wild-type or sgRNA1/2-induced CCR5 gene (upper panel) and 49 most possible SNVs (lower panel), when the sites were covered more than 20 times by sequencing.
Except T7E1 digestion assay, another simple and quantitative assessment of genome editing is TIDE (Tracking of Indels by Decomposition) analysis.24 TIDE required two parallel PCR reactions for the cell pool and sequencing, and the sequencing data were then analyzed on the R-coded online software (https://tide.nki.nl). We performed TIDE analysis on the 15 potential off-target sites (Figure 3B; Table S4). TIDE showed that, on the cell pools, the on-target editing (indel equals 0) was significantly highly efficient with the rate >90% (Figure 3B). For the sites of on-target editing lower than 90%, we performed two PCRs, one was forward and the other was reverse. TIDE analysis still showed the high on-target efficiency >84% (Figure 3B). Most off-target indels were lower than 1% (Figure 3B).
To further confirm the off-target effect, whole-genome sequencing (WGS) was performed (Figures 3C–3E). Compared to human genome GRCh37/HG19, 733,564 sites were joint indels in wild-type CD4+ cells and sgRNA1/2-induced CD4+ cells, while there were 67,458 indels in wild-type CD4+ cells and 67,775 indels in sgRNA1/2-induced CD4+ cells (Figure 3C), respectively. In contrast to the 49 most possible off-target sites (above 15 sites plus 34 sites in the Materials and Methods; Table S5), 6 indels were all joint sites, which showed no off-target effect (Table S6).
Because of the sequencing error or heterogenicity of cells, the above analysis may be misleading. To ensure the data reliability, more than 20 times of repeat coverage for each site was calculated. Compared to human genome GRCh37/HG19, 327,092 sites were joint indels in wild-type CD4+ cells and sgRNA1/2-induced CD4+ cells, while there were 4,644 indels in wild-type CD4+ cells and 4,658 indels in sgRNA1/2-induced CD4+ cells, respectively (Figure 3D, upper panel). In the 49 most possible off-target sites, 4 indels were all joint sites, and there were no off-target sites in sgRNA1/2-induced CD4+ cells (Figure 3D, lower panel; Table S6). We also analyzed single-nucleotide variants (SNVs). Compared to human genome GRCh37/HG19, 2,731,772 sites were common SNVs in wild-type CD4+ cells and sgRNA1/2-induced CD4+ cells, while there are 1,925 SNVs in wild-type CD4+ cells and 1,891 SNVs in sgRNA1/2-induced CD4+ cells, respectively (Figure 3E, upper panel). In the 49 most possible off-target sites, 17 SNVs were all joint sites, and there were no off-target SNVs in sgRNA1/2-induced CD4+ cells (Figure 3E; Table S7).
CRISPR-Cas9 with One-Cassette sgRNA
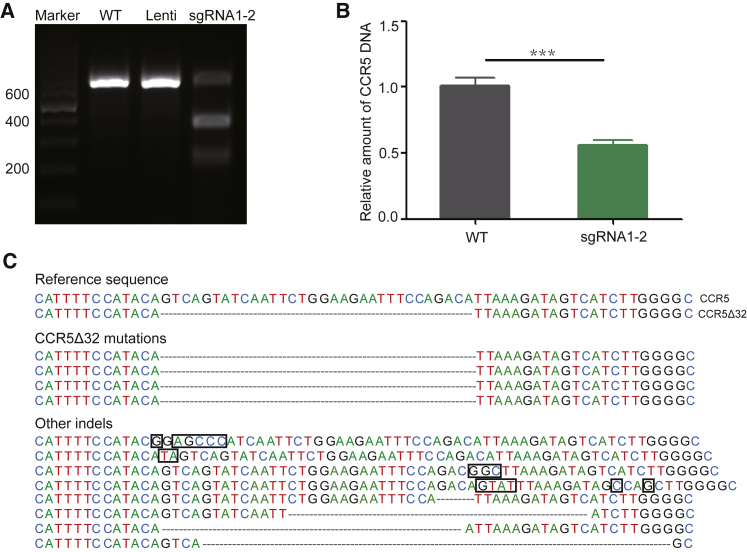
To simplify the process, we tried to put two sgRNAs, sgRNA1 and sgRNA2, into a cassette in one plasmid. Figure 4A shows the T7E1 digestion result of this strategy, indicating this strategy works well. Real-time qPCR and sequencing illustrated the efficacy of this CCR5-targeting CRISPR-Cas9 strategy (Figures 4B and 4C).
Figure 4.
CRISPR-Cas9-Mediated CCR5Δ32 Mutation with One-Cassette sgRNAs
(A) T7E1 digestion of CCR5 PCR products from Jurkat cell samples, respectively, untreated wild-type (WT), or treated with LentiCRISPRv2 vector (Lenti), or with sgRNA1-2 in one cassette in one plasmid (sgRNA1-2). (B) Real-time qPCR analysis. Error bars represent the average value of two independent experiments, triplicate in each experiment and triplicate in each measurement. ***p = 0.0003. (C) DNA sequence alignment of the CCR5 gene from CRISPR-Cas9 sgRNA1-2-treated primary CD4+ cells. Some mutations caused by CRISPR-Cas9 are in the black box.
Discussion
In this study, we successfully converted wild-type CCR5 genes into CCR5Δ32/Δ32 homozygotes in a CD4+ Jurkat cell line and in primary human CD4+ cells by using CRISPR-Cas9 technology. This approach is simple and effective, better than the previous combination use of piggyBac transposon technology and TALENs or CRISPR-Cas9 technology.23 This approach should be easily transferred to hematopoietic stem and progenitor cells (HSPCs), which are longer-lasting than primary CD4+ cells. Ye et al.23 employed the induced pluripotent stem cells (iPSCs). However, the safety of iPSCs is still under study. CRISPR-Cas9 safety is another concern before applying the technique for clinical use. Dual safety problems are enough to prohibit the clinical use of this therapy. Thus, we did not try it in iPSCs. In other studies, adenovirus delivery system or mRNA electroporation was applied, in contrast to the lentiviral delivery system in this study. This CRISPR-Cas9 approach in this study is easily transferred to mRNA electroporation. The lentiviral delivery system can be used ex vivo, but not in vivo.
Following analysis of the Berlin patient, Dr. Hutter reported another six HIV-infected patients received bone marrow transplantation from donors of CCR5Δ32/Δ32 homozygotes. But none of these transplantations succeeded. All patients died either from infections or from the relapse of lymphoma within 1 year.25 Kordelas and colleagues26 also reported a failed case, where the patient died from the CXCR4-tropic variant rapid rebound after the transplantation. Therefore, CXCR4-tropic or dual-tropic HIV viruses are enough to ruin the goal of an HIV/AIDS cure based on CCR5-targeting therapy. The Berlin patient benefited from the primary presence of the CCR5-tropic viruses. Disrupting the CXCR4 gene may not be a practical approach, because CXCR4 plays an important role in hematopoietic stem cell homing and retention.9 However, we do find CCR5-only-tropic virus infections. Circulating Recombinant Form 07_BC (CRF07_BC) is the most predominant epidemic HIV-1 viruses in China. Until now, all of isolated CRF07-BC viruses are CCR5 tropic, and CXCR4-tropic (X4) viruses have thus far not been found in CRF07_BC.27 Moreover, Zhang et al.27 tried to artificially mutate CRF07-BC Env V3 loop into CXCR4-like amino acids, such as two amino acid insertions between positions 13 and 14, as well as an arginine substitution at position 11 or 16 (IG insertion and P16R mutation or MG insertion and S11R mutation). These CXCR4-like mutations in CRF07-BC completely abrogated virus infectivity.27 Therefore, natural or artificial mutated CRF07-BC viruses are all CCR5 tropic. This is good news for this study. At least we can apply a CCR5-targeting strategy in CCR5-tropic virus, such as HIV-1 CRF07-BC.
Toward the HIV/AIDS cure, this study offers a simple approach to induce HIV-1 resistance in cells homozygous for the CCR5Δ32/Δ32 mutant genes. We were surprised to find that, in 29 puromycin-resistant sgRNA1/2-specfic CRISPR-Cas9-induced Jurkat cell clones, all of them harbored CCR5 mutations (100%). Among 29 cell clones, we randomly selected 5 clones, and only one of these (20%, 1/5) was a CCR5Δ32/Δ32 homozygote. However, the TA cloning and sequencing showed that ∼75% mutation is CCR5Δ32 monoallelic or biallelic mutation. Thus, we estimate that the CCR5Δ32/Δ32 homozygote induction rate is 15%–20% in Jurkat cells. This efficiency is lower in primary CD4+ cells, which is estimated around <11% (1/5 of 45%–55%). In this study, we did not get monoclonal primary CD4+ cells. This could be a lack of some cytokines or enough cell density. In the clinical use of this approach, we also should develop a simple way to select the CCR5Δ32/Δ32 homozygotes. Flow cytometry does not seem to work, because there is no specific antibody distinguishing between CCR5Δ32/Δ32 homozygotes from other types of CCR5 mutations.
The top concern for the clinical use of CRISPR-Cas9 technology is safety. Off-target alterations must be avoided to ensure genome integrity and proper cellular function. WGS found no off-target effects among the top 49 predicted off-target sites. WGS did find 4,658 indels and 1,891 SNVs in sgRNA1/2-induced CD4+ cells (Figures 3D and 3E), but these were absent in wild-type CD4+ cells. This finding is similar to the Schaefer report,28 which showed the unexpected mutations in CRISPR-Cas9-edited mice. Schaefer concluded that these mutations were caused by CRISPR-Cas9 editing.28 However, we also found 4,644 indels and 1,925 SNVs in wild-type CD4+ only, and these were absent in sgRNA1/2-induced CD4+ cells. These two numbers are comparable. Therefore, indels and SNVs showed by WGS could not be concluded to be caused by CRISPR-Cas9 editing. Cellular heterogenicity may be a possible explanation.
Although TIDE analysis and WGS showed no apparent off-target effects, we did find unexpected on-target CCR5 mutations (Figures 1E, 1G, 2D, and 4C). These mutations would sometimes be like CCR5Δ32/Δ32 homozygotes, but with unexpected indels or point mutations. The only way to distinguish these mutations from the expected CCR5Δ32/Δ32 mutation is through sequencing.
This study is relevant to the HIV field, but the concept of specifically deleting a region of genomic DNA is also important to geneticists and molecular biologists who are interested in generating knockout cell lines or animals.
In summary, this study successfully induced the CCR5Δ32/Δ32 homozygotes. This approach and other studies are pushing forward the unique Berlin patient to a cell-based HIV/AIDS cure.
Materials and Methods
The Research Ethics Community of School of Medicine, Nankai University reviewed and approved this study.
Plasmids and Cell Culture
The lentiviral vector LentiCRISPRv2 (from Dr. Feng Zhang, McGovern Institute for Brain Research at MIT, Addgene Catalog 52961), lentiviral packaging plasmid psPAX2, and VSVG envelope-expressing plasmid PMD2.G were used in this study. The sequences of sgRNAs and primers are listed in Tables S1 and S2. HEK293T (CRL-11268) cells and human acute T cell leukemia cell line Jurkat E6-1 (TIB-152) were obtained from American Type Culture Collection (ATCC).
T7E1 Analysis, TA Cloning, and Sequencing
The genomic DNA from wild-type cells and lentivirus-infected cells were extracted using the DNA extraction kit (Axygen). DNA fragments across the target sites were amplified by PCR, and analyzed with T7E1 (Polymath Technology). PCR products of interest were cloned into a T vector using a TA cloning kit (Thermo Fisher Scientific). The genotypes of 20–30 bacterial clones were analyzed by DNA sequencing.
HIV-1 Challenge
The CCR5-tropic HIV-1397PXJ virus for challenging test in this study was obtained from the laboratory of Professor Yiming Shao (Chinese Centers for Disease Control and Prevention).
Off-Target Analysis
All potential off-target sites of sgRNA1 and sgRNA2 were predicted using Dr. Feng Zhang’s CRISPR design website (http://crispr.mit.edu:8079/). 15 potential sites were selected, of which 8 were potential off-target sites for sgRNA1 and 7 for sgRNA2 (Table S3). The PCR primers for off-target analysis are listed in the supporting Table S4, and the PCR products alone were analyzed by T7E1 assay.
sgRNA needs to recognize the target sequence by base complementary pairing, and the 8–12 bp nearest the PAM region is the most critical. According to this principle, 34 potential off-target sites with the same 8 bp nearest the PAM side were picked, of which 11 were potential off-target sites for sgRNA1 and 23 were potential off-target sites for sgRNA2 (Table S5). These sites were analyzed in the results of WGS according to their locations.
TIDE Analysis
TIDE analysis was performed according to Brinkman et al.24 Briefly, for 15 potential off-target sites, PCR was conducted and followed by sequencing. The sequencing data were analyzed using the online software (https://tide.nki.nl).
WGS
WGS was performed on the Illumina Hiseq platform at The Beijing Genomics Institute (BGI). The sequencing depth was 30 times. The raw data then were filtered and mapped to the human reference genome GRCh37/HG19. The genomic variations, including indels and SNVs, were detected by the Haplotype Caller of GATK (v.3.3.0) (Tables S6 and S7).
Statistical Analysis
All statistical analyses were performed using statistical software, and the difference between the two groups was determined using the two-tailed upaired t test with Welch’s correction when the confidence intervals were 95%.
Author Contributions
M.W. and C.Q. conceived the project. C.Q. performed major experiments and analyzed the data. D.L., X. Jiang, X. Jia, L.L., Y.W., and J.S. performed the part experiments. Y.S. and M.W. analyzed the data. M.W. and C.Q. wrote the paper. All authors approved the manuscript.
Conflicts of Interest
The authors declare no competing financial interests.
Acknowledgments
We thank Dr. Lawrence Kleiman for English proofreading and suggestions. This work was supported by the National Natural Science Foundation of China (81571991 to M.W.) and Nankai University starting fund (ZB15006101 to M.W.).
Footnotes
Supplemental Information includes one figure and seven tables and can be found with this article online at https://doi.org/10.1016/j.omtn.2018.05.012.
Supplemental Information
References
- 1.Hütter G., Nowak D., Mossner M., Ganepola S., Müssig A., Allers K., Schneider T., Hofmann J., Kücherer C., Blau O. Long-term control of HIV by CCR5 Delta32/Delta32 stem-cell transplantation. N. Engl. J. Med. 2009;360:692–698. doi: 10.1056/NEJMoa0802905. [DOI] [PubMed] [Google Scholar]
- 2.Hütter G. Stem cell transplantation in strategies for curing HIV/AIDS. AIDS Res. Ther. 2016;13:31. doi: 10.1186/s12981-016-0114-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu S., Yao Y., Xiao H., Li J., Liu Q., Yang Y., Adah D., Lu J., Zhao S., Qin L., Chen X. Simultaneous Knockout of CXCR4 and CCR5 Genes in CD4+ T Cells via CRISPR/Cas9 Confers Resistance to Both X4- and R5-Tropic Human Immunodeficiency Virus Type 1 Infection. Hum. Gene Ther. 2018;29:51–67. doi: 10.1089/hum.2017.032. [DOI] [PubMed] [Google Scholar]
- 4.Xu L., Yang H., Gao Y., Chen Z., Xie L., Liu Y., Liu Y., Wang X., Li H., Lai W. CRISPR/Cas9-Mediated CCR5 Ablation in Human Hematopoietic Stem/Progenitor Cells Confers HIV-1 Resistance In Vivo. Mol. Ther. 2017;25:1782–1789. doi: 10.1016/j.ymthe.2017.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holt N., Wang J., Kim K., Friedman G., Wang X., Taupin V., Crooks G.M., Kohn D.B., Gregory P.D., Holmes M.C., Cannon P.M. Human hematopoietic stem/progenitor cells modified by zinc-finger nucleases targeted to CCR5 control HIV-1 in vivo. Nat. Biotechnol. 2010;28:839–847. doi: 10.1038/nbt.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perez E.E., Wang J., Miller J.C., Jouvenot Y., Kim K.A., Liu O., Wang N., Lee G., Bartsevich V.V., Lee Y.L. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat. Biotechnol. 2008;26:808–816. doi: 10.1038/nbt1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang J., Exline C.M., DeClercq J.J., Llewellyn G.N., Hayward S.B., Li P.W., Shivak D.A., Surosky R.T., Gregory P.D., Holmes M.C., Cannon P.M. Homology-driven genome editing in hematopoietic stem and progenitor cells using ZFN mRNA and AAV6 donors. Nat. Biotechnol. 2015;33:1256–1263. doi: 10.1038/nbt.3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi B., Li J., Shi X., Jia W., Wen Y., Hu X., Zhuang F., Xi J., Zhang L. TALEN-Mediated Knockout of CCR5 Confers Protection Against Infection of Human Immunodeficiency Virus. J. Acquir. Immune Defic. Syndr. 2017;74:229–241. doi: 10.1097/QAI.0000000000001190. [DOI] [PubMed] [Google Scholar]
- 9.Wang C.X., Cannon P.M. The clinical applications of genome editing in HIV. Blood. 2016;127:2546–2552. doi: 10.1182/blood-2016-01-678144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sigal A., Baltimore D. As good as it gets? The problem of HIV persistence despite antiretroviral drugs. Cell Host Microbe. 2012;12:132–138. doi: 10.1016/j.chom.2012.07.005. [DOI] [PubMed] [Google Scholar]
- 11.Lorenzo-Redondo R., Fryer H.R., Bedford T., Kim E.Y., Archer J., Pond S.L.K., Chung Y.S., Penugonda S., Chipman J., Fletcher C.V. Persistent HIV-1 replication maintains the tissue reservoir during therapy. Nature. 2016;530:51–56. doi: 10.1038/nature16933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Katlama C., Deeks S.G., Autran B., Martinez-Picado J., van Lunzen J., Rouzioux C., Miller M., Vella S., Schmitz J.E., Ahlers J. Barriers to a cure for HIV: new ways to target and eradicate HIV-1 reservoirs. Lancet. 2013;381:2109–2117. doi: 10.1016/S0140-6736(13)60104-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berger E.A., Murphy P.M., Farber J.M. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu. Rev. Immunol. 1999;17:657–700. doi: 10.1146/annurev.immunol.17.1.657. [DOI] [PubMed] [Google Scholar]
- 14.Keele B.F., Giorgi E.E., Salazar-Gonzalez J.F., Decker J.M., Pham K.T., Salazar M.G., Sun C., Grayson T., Wang S., Li H. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc. Natl. Acad. Sci. USA. 2008;105:7552–7557. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Biloglav Z., Zgaga L., Smoljanović M., Hayward C., Polasek O., Kolcić I., Vitart V., Zemunik T., Boraska V., Torlak V. Historic, demographic, and genetic evidence for increased population frequencies of CCR5Delta32 mutation in Croatian Island isolates after lethal 15th century epidemics. Croat. Med. J. 2009;50:34–42. doi: 10.3325/cmj.2009.50.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zawicki P., Witas H.W. HIV-1 protecting CCR5-Delta32 allele in medieval Poland. Infect. Genet. Evol. 2008;8:146–151. doi: 10.1016/j.meegid.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 17.Zhou C., Sun H., Yin J.X., Zhang H.Y., Lin K.Q., Tao Y.F., Yang Z.Q., Chu J.Y., Huang X.Q. [Detection and preliminary study of a family carrying a CCR5Δ32 deletional mutation] Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2012;29:485–489. doi: 10.3760/cma.j.issn.1003-9406.2012.04.024. [DOI] [PubMed] [Google Scholar]
- 18.Wu X., Scott D.A., Kriz A.J., Chiu A.C., Hsu P.D., Dadon D.B., Cheng A.W., Trevino A.E., Konermann S., Chen S. Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells. Nat. Biotechnol. 2014;32:670–676. doi: 10.1038/nbt.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tebas P., Stein D., Tang W.W., Frank I., Wang S.Q., Lee G., Spratt S.K., Surosky R.T., Giedlin M.A., Nichol G. Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. N. Engl. J. Med. 2014;370:901–910. doi: 10.1056/NEJMoa1300662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DiGiusto D.L., Cannon P.M., Holmes M.C., Li L., Rao A., Wang J., Lee G., Gregory P.D., Kim K.A., Hayward S.B. Preclinical development and qualification of ZFN-mediated CCR5 disruption in human hematopoietic stem/progenitor cells. Mol. Ther. Methods Clin. Dev. 2016;3:16067. doi: 10.1038/mtm.2016.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li L., Krymskaya L., Wang J., Henley J., Rao A., Cao L.F., Tran C.A., Torres-Coronado M., Gardner A., Gonzalez N. Genomic editing of the HIV-1 coreceptor CCR5 in adult hematopoietic stem and progenitor cells using zinc finger nucleases. Mol. Ther. 2013;21:1259–1269. doi: 10.1038/mt.2013.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ye L., Wang J., Beyer A.I., Teque F., Cradick T.J., Qi Z., Chang J.C., Bao G., Muench M.O., Yu J. Seamless modification of wild-type induced pluripotent stem cells to the natural CCR5Δ32 mutation confers resistance to HIV infection. Proc. Natl. Acad. Sci. USA. 2014;111:9591–9596. doi: 10.1073/pnas.1407473111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brinkman E.K., Chen T., Amendola M., van Steensel B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 2014;42:e168. doi: 10.1093/nar/gku936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hütter G. More on shift of HIV tropism in stem-cell transplantation with CCR5 delta32/delta32 mutation. N. Engl. J. Med. 2014;371:2437–2438. doi: 10.1056/NEJMc1412279. [DOI] [PubMed] [Google Scholar]
- 26.Verheyen J., Esser S., Kordelas L. More on shift of HIV tropism in stem-cell transplantation with CCR5 delta32/delta32 mutation. N. Engl. J. Med. 2014;371:2438. doi: 10.1056/NEJMc1412279. [DOI] [PubMed] [Google Scholar]
- 27.Zhang L., Ma L., Wang Z., Wang Y., Zhang J., Wang H., Shao Y. Alterations in HIV-1 gp120 V3 region are necessary but not sufficient for coreceptor switching in CRF07_BC in China. PLoS ONE. 2014;9:e93426. doi: 10.1371/journal.pone.0093426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schaefer K.A., Wu W.H., Colgan D.F., Tsang S.H., Bassuk A.G., Mahajan V.B. Unexpected mutations after CRISPR-Cas9 editing in vivo. Nat. Methods. 2017;14:547–548. doi: 10.1038/nmeth.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.