Abstract
Metastasis is the most important feature of gastric cancer (GC) and the most widely recognized reason for GC-related deaths. Unfortunately, the underlying mechanism behind this metastasis remains unknown. Mounting evidence suggests the dynamic regulatory role of sirtuin2 (SIRT2), a histone deacetylase (HDAC), in cell migration and invasion. The present study aims to evaluate the biological function of SIRT2 in GC and identify the target of SIRT2 as well as evaluate its therapeutic efficacy. We found that SIRT2 was upregulated in GC tissues compared to adjacent normal tissues, and this was correlated with reduced patient survival. Although CCK8 and colony-formation assays showed that SIRT2 overexpression marginally promoted proliferation in GC cell lines, SIRT2 knockdown or treatment with SirReal2 decreased the migration and invasion of GC cells. We demonstrated both in vitro and in vivo that SirReal2 could inhibit the deacetylation activity of SIRT2 and its downstream target PEPCK1, which is related to mitochondrial metabolism and RAS/ERK/JNK/MMP-9 pathway. Taken together, these results demonstrate for the first time that SirReal2 selectively targets SIRT2 and decreases migration as well as invasion in human GC cells. SirReal2 therefore shows promise as a new drug candidate for GC therapy.
Introduction
Gastric cancer (GC) is the fifth most common cancer in incidence and the third leading cause of cancer mortality [1]. Globally, approximately 841,000 people died due to GC in 2013 [2]. Metastasis is the most important feature of gastric cancer and the most widely recognized reason for GC-related deaths. Unfortunately, the underlying mechanism of metastasis remains unknown [3]. This is not only due to the fact that resistance to conventional anticancer drugs is becoming increasingly commonplace but also a result of the lack of effective biomarkers. Oncogenic pathways identified by genetic studies have likewise proven difficult to target therapeutically [4]. Understanding how the metastasis of GC is dynamically regulated is thus of utmost importance.
In this regard, posttranslational modifications (PTMs) in metabolism regulation have received close attention given their regulation by upstream signaling pathways and ability to respond to changes in cellular metabolic status [5], [6]. Mounting evidence implies the dynamic role of SIRT2, a histone deacetylase (HDAC), in regulating tumorigenesis. The expression of SIRT2 is significantly reduced in basal cell carcinoma [7] as well as gliomas [8], while high SIRT2 levels in melanomas [9] and hepatocellular carcinoma [10] are associated with tumorigenesis. Phosphoenolpyruvate carboxykinase (PEPCK) is the classic downstream target of SIRT2, as well as the rate-limiting enzyme of gluconeogenesis, and catalyzes the conversion of oxaloacetate (OAA) into phosphoenolpyruvate (PEP). PEPCK is found in two forms: cytosolic (PEPCK1) and mitochondrial (PEPCK2) [11]. A recent study confirmed that PEPCK1 could contribute to cancer anabolic metabolism by increasing glucose and glutamine utilization [12]. However, the role of PEPCK1-related metabolism in tumor metastasis still remains unclear.
The tricarboxylic acid (TCA) cycle represents an important aspect of mitochondrial metabolism. It connects cellular carbohydrates, amino acids, and bioenergetics with anabolic and catabolic pathways. As a key enzyme, PEPCK1 controls TCA cycle flux by promoting anabolic metabolism and increasing the utilization of glucose as well as glutamine [12]. The fueled TCA cycle then leads to enhanced mitochondrial metabolism by increasing the generation of ATP, ROS, NADPH, amino acids, nucleotides, and lipids. In addition, it has been associated with tumor metastasis [13] and RAS-mediated tumorigenicity [14]. RAS can activate major downstream mitogen activated protein kinases (MAPKs), as well as extracellular signal regulated kinase (ERK) and c-Jun N-terminal kinase (JNK). As a downstream target of ERK/JNK pathway, matrix metalloproteinase-9 (MMP-9) may be critical for cancer cell metastasis [15].
In the present study, we discovered that SIRT2 plays a critical role in promoting GC migration and invasion. SIRT2-mediated deacetylation in protein posttranslational modification stabilizes PEPCK1 and promotes the mitochondrial metabolism of GC cells. At the molecular level, the RAS/ERK/JNK/MMP-9 pathway was identified as a downstream target of PEPCK1 in promoting GC cell migration and invasion.
Results
SIRT2 Expression Increased in Gastric Cancer and Resulted in Reduced Patient Survival
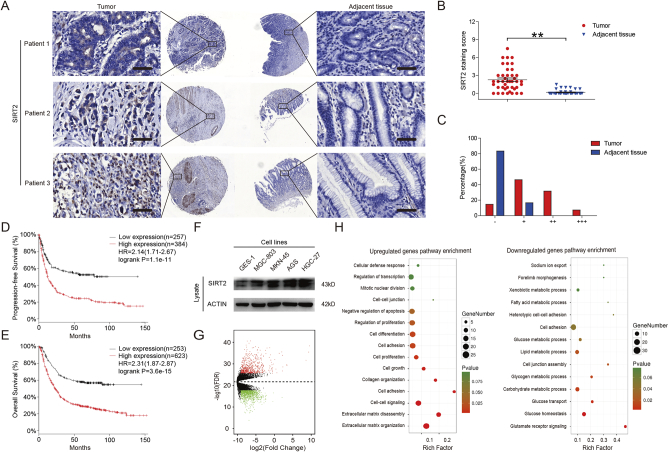
SIRT2 plays a dual role in tumorigenesis. The expression of SIRT2 is significantly reduced in many cancers [7], [8] yet increased in others [9], [10], resulting in an aberrant metabolic status. Though some databases suggest that SIRT2 may be highly expressed in gastric cancer [16], the role of SIRT2 in gastric cancer has not yet been elucidated. Accordingly, we queried tissue microarrays from SHANGHAI OUTDO BIOTECH, which contains 84 gastric cancer samples, and found that SIRT2 was significantly increased in tumor tissues compared to adjacent tissues (Figure 1, A-C). We queried the Kaplan-Meier plotter (http://kmplot.com/analysis) database [17], [18], which contains clinically annotated genomic data from gastric cancer samples, and found that high Sirt2 mRNA expression was associated with poor progression-free survival and overall survival (log-rank test) (Figure 1, D & E). We further assessed the expression of SIRT2 in GC cell lines (AGS, HGC-27, MGC-803 and MKN-45) and normal gastric epithelial cells (GES-1) and found that the expression level of SIRT2 in GC cells was significantly higher than that of GES-1 cells (Figure 1F). These results suggest that SIRT2 expression was increased in GC and associated with worse prognosis. Tumors can exhibit different malignant biological behavior through differential gene expression [19]. Consequently, we analyzed the differentially expressed genes (DEGs) in GC from The Cancer Genome Atlas (TCGA) data. We identified the differentially expressed genes (Figure 1G) and performed Gene Ontology (GO) pathway enrichment analyses to detect the potential biological functions and pathways of the high and low expression genes in GC (Figure 1H). We found the most relevant include metabolic and metastatic pathways in GC and MMP-9 (matrix metallopeptidase 9), a target protein related to tumor migration and invasion, which serves as an important target of malignant biological behavior for GC cells.
Figure 1.
SIRT2 expression was increased in gastric cancer and associated with reduced patient survival. (A) SIRT2 protein levels in tumor and adjacent normal tissues from tissue microarrays were detected and (B) quantified. The magnification is ×400. Scale bars, 50 μm. (C) After scoring, the different proportions of tumor and adjacent normal tissues were shown. (D) Kaplan-Meier curves for GC patients' progression-free survival in the patients stratified by high or low SIRT2 mRNA expression. Median progression-free survival was 5.1 months in the high SIRT2 expression group and 9.8 months in the low SIRT2 expression group (n = 646, HR = 2.14, 95% CI 1.71-2.67, log-rank P < .001). (E) Kaplan-Meier curves for GC patients' overall survival in the patients stratified by high or low SIRT2 mRNA expression. Median overall survival was 9.6 months in the high SIRT2 expression group and 20.1 months in the low SIRT2 expression group (n = 876, HR = 2.31, 95% CI 1.87-2.87, log-rank P < .001). (F) The expression of SIRT2 in GC cell lines and GES-1 cells was determined by Western blot. Data represent the mean ± SEM, n ≥ 3. * P < .05; ** P < .01. A score of “−”=no staining or cytoplasmic staining in less than 10% of the tumor cells; a score of “+”=a faint and partial cytoplasmic staining in greater than 10% of the tumor cells; scores of “++” and “+++”=a weak to moderate and a strong complete cytoplasmic staining in greater than 10% of the tumor cells, respectively. (G) Heat map of DEGs in GC and paired normal gastric samples. Heat map was drawn using the gplots package in Bioconductor. DEGs with fold change (FC) > 2 were shown in red; DEGs with FC < −2 were in green (P < .01) and FDR < 0.01. (H) GO annotation pathways of high and low expression genes in GC. GO annotation was generated using the ggplot2 package in R language. The size of the dots represents the number of genes. Dot color represented the P value. Red: high degree of enrichment, green: low degree of enrichment.
SIRT2 Supports GC Cell Migration and Invasion via RAS/ERK/JNK/MMP-9 Pathway
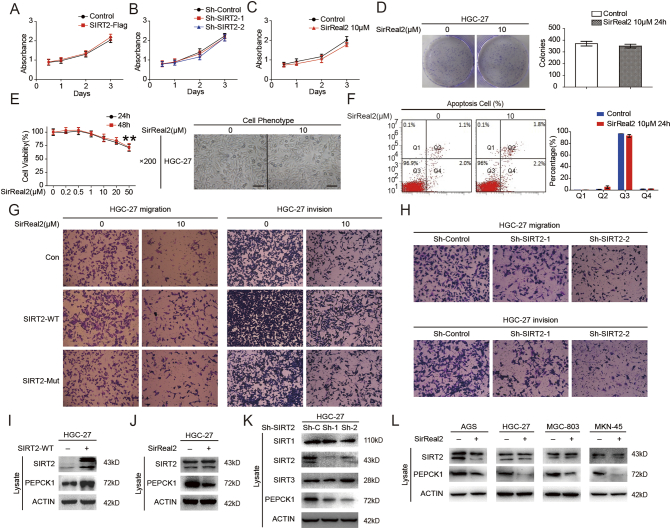
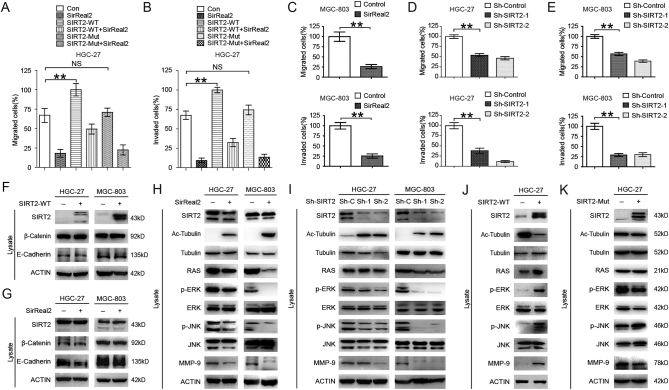
Recent studies have shown that the levels of SIRT2 expression and activity play a critical role in cell epigenetic alterations associated with malignant behaviors and cell metabolism [20]. Since most of the protein targets played a role in promoting tumor proliferation, we evaluated the relationship between cell proliferation and SIRT2 expression in HGC-27 cells. The effectiveness of the SIRT2 overexpression and knockdown was validated through Western blotting, and SIRT2 was found to only marginally affect GC cell proliferation (Supplementary Figure 1, A & B). SirReal2, a selective SIRT2 inhibitor, also demonstrated a marginal effect on GC cell proliferation (Supplementary Figure 1, C-E). Similarly, after inhibiting SIRT2 by SirReal2, no significant difference in the amount of apoptotic cells was observed (Supplementary Figure 1F). The bioinformatics results suggested that metabolic disorders of GC could be related to its metastasis. We next examined the effect of inhibiting SIRT2 on cell invasion, an important event in metastasis. Transwell assay was performed after treatment of GC cells with SirReal2. Consistent with the bioinformatics analysis results, a significantly inhibitory effect on cell migration and invasion was observed when SIRT2 was inhibited (Figure 2, A-C & Supplementary Figure 1G). These results were confirmed by Transwell assays with SIRT2 knockdown (Figure 2, D and E & Supplementary Figure 1H). To further explore the mechanism of SIRT2 induced cell migration and invasion, we investigated the effects of SIRT2 on downstream targets after inhibition of SIRT2 by either SirReal2 treatment or shSIRT2. Several studies demonstrated that the activation of WNT/β-catenin and mitogen activated protein kinase (MAPK) signaling pathways and the initiation of epithelial mesenchymal transition (EMT) were associated with tumor metastasis [21], [22], [23].However, in the present study, no significant difference was observed in the expression levels of β-Catenin and E-cadherin at different SIRT2 expression levels (Figure 2, F & G). Interestingly, both RAS expression and the downstream targets ERK/JNK/MMP-9 were decreased when SIRT2 was inhibited, indicating that SIRT2 induced the migration and invasion of GC cells through RAS/ERK/JNK/MMP-9 pathway (Figure 2, H & I). Furthermore, wild-type SIRT2 overexpression reversed the RAS/ERK/JNK/MMP-9 pathway, and the mutant SIRT2 overexpression failed to show a similar effect (Figure 2, J & K).
Supplementary Figure 1.
(A) HGC-27 cell proliferation was analyzed via CCK8 assay following transfection with SIRT2 overexpression vector. (B) HGC-27 cell proliferation was analyzed via CCK8 assay following infection with lentiviruses expressing shRNA. (C) HGC-27 cells were treated with 10 μM SirReal2 and quantified via CCK-8 assay. (D) HGC-27 cells were treated with 10 μM SirReal2 for 24 hours; cell colonies were stained with crystal violet (left) and quantified (right) after 14 days. (E) HGC-27 cells were treated with indicated SirReal2; the cell viability (left) and morphological changes (right) were observed. Scale bars, 100 μm. (F) HGC-27 cells were treated with 10 μM SirReal2 for 24 hours; apoptotic cells were measured by flow cytometry (left) and quantified (right). (G) HGC-27 cells were treated with 10 μM SirReal2 for 24 hours; cell migration was photographed (left), and cell invasion was photographed (right). (H) SIRT2 was knocked down in HGC-27 cells, cell migration was photographed (up), and cell invasion was photographed (down). (I) HGC-27 cells were transfected with flag tagged SIRT2 plasmid. Cells were harvested and visualized by Western blotting. (J) HGC-27 cells were treated with SirReal2. Cells were harvested and visualized by Western blotting. (K) HGC-27 cells were infected with lentiviruses expressing shRNA. Cells were harvested and visualized by Western blotting. (L) GC cell lines were treated with SirReal2. Cells were harvested and visualized by Western blotting.
Figure 2.
SIRT2 supports GC cell migration and invasion via RAS/ERK/JNK/MMP-9 pathway. (A) HGC-27 cells were treated with 10 μM SirReal2 for 24 hours; cell migration was quantified. (B) HGC-27 cells were treated with 10 μM SirReal2 for 24 hours; cell invasion was quantified. (C) MGC-803 cells were treated with 10 μM SirReal2 for 24 hours; cell migration (up) and invasion (down) were quantified, respectively. (D) SIRT2 was knocked down in HGC-27 cells; cell migration (up) and invasion (down) were quantified, respectively. (E) SIRT2 was knocked down in MGC-803 cells; cell migration (up) and invasion (down) were quantified, respectively. (F) Key proteins associated with EMT and WNT/β-Catenin pathway in HGC-27 and MGC-803 cells were assessed following transfection with wild-type SIRT2 overexpression vector. (G) Key proteins associated with EMT and WNT/β-Catenin pathway in HGC-27 and MGC-803 cells were assessed following 10 μM SirReal2 treatment for 24 hours. (H) The downstream targets of SIRT2 in HGC-27 and MGC-803 cells were visualized following 10 μM SirReal2 treatment for 24 hours. (I) The downstream targets of SIRT2 in HGC-27 and MGC-803 cells were visualized following infection with SIRT2 shRNA. (J-K) The downstream targets of SIRT2 in HGC-27 cells were visualized following transfection with wild-type and mutant SIRT2 overexpression vector. Data represent the mean ± SEM, n ≥ 3, * P < .05, ** P < .01, NS P > .05.
SIRT2 Targets PEPCK1 to Promote GC Cell Migration and Invasion
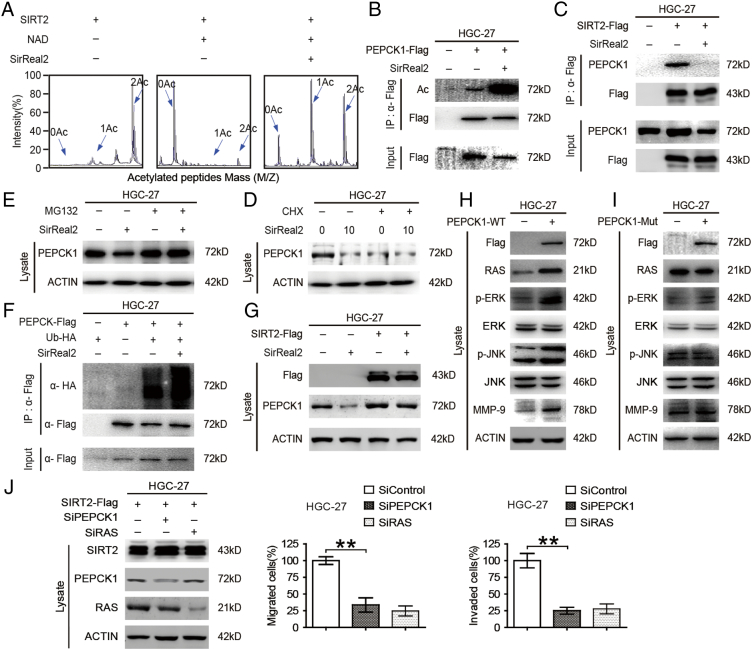
SIRT2 is a key target in promoting mitochondrial metabolism. As such, it seemed reasonable to identify the metabolic downstream target of SIRT2 (PEPCK1, GLS1, PKM2, and LDH) in GC cells [12], [20], [24]. Since Lys70, Lys71, and Lys594 were identified as key acetylated sites of human PEPCK1 and these are deacetylated by SIRT2 [6], [25], we first synthesized a peptide containing acetylated sites of PEPCK1 and incubated it with purified SIRT2 protein. We then assessed peptides by mass spectrometry (MS) (Figure 3A). Combined with the results of immunoprecipitation (Figure 3B), we found that the deacetylation effect of SIRT2 was abolished by SirReal2. Employing immunoprecipitation, we found that SIRT2 could interact with target proteins. Moreover, SirReal2 treatment reduced the binding of SIRT2 to its targets (Figure 3C). Next, we inhibited PEPCK1 synthesis by using CHX (Cycloheximide) and inhibited PEPCK1 proteasomal degradation by using MG132. We found that PEPCK1 degraded faster in presence of SirReal2 and that SirReal2-induced PEPCK1 degradation was blocked by using MG132 (Figure 3, D & E). Moreover, SirReal2 induced PEPCK1 ubiquitination in HGC-27 cells (Figure 3F). These results suggest that the ubiquitin-proteasome pathway mediates the SirReal2-promoted decrease of PEPCK1. We transfected SIRT2-Flag into GC cells and found that endogenous PEPCK1 was significantly increased (Figure 3G & Supplementary Figure 1I). Conversely, inhibition of SIRT2 resulted in a decreased level of PEPCK1 in different gastric cancer cell lines (Supplementary Figure 1, J-L). It is worth noting that only the wild-type PEPCK1 transfection activated the RAS/ERK/JNK/MMP-9 pathway (Figure 3, H-J). Overall, these results support the idea that SIRT2 facilitates the migration and invasion of GC by stabilizing the downstream metabolic target PEPCK1.
Figure 3.
PEPCK1 is the downstream target of SIRT2 in promoting GC cell migration and invasion. (A) Purified SIRT2 protein was incubated with acetylated peptides with or without 10 μM SirReal2 for 4 hours, and the rate of deacetylation was determined using mass spectrometry (MS). (B) Cells were transfected with flag tagged PEPCK1 plasmid and then treated with 10 μM SirReal2 for 24 hours. Cells were harvested and incubated with flag-beads. The immunoprecipitated protein from cell lysates was analyzed via Western blotting. (C) HGC-27 cells were transfected with Flag-SIRT2 and then treated with or without 10 μM SirReal2 for 24 hours. Cells were harvested and incubated with flag-beads. The immunoprecipitated proteins from cell lysates were analyzed by Western blotting. (D) HGC-27 cells treated with indicated CHX, with or without indicated 10 μM SirReal2, for 24 hours. Cells were harvested and lysated, and endogenous PEPCK1 was visualized by Western blotting. (E) HGC-27 cells were treated with 10 μM SirReal2 for 24 hours with or without 10 μM MG132 for 4 hours. Cells were harvested and lysated and visualized by Western blotting. (F) Flag-tagged PEPCK1 and HA-tagged ubiquitin were coexpressed in cells and then treated with 10 μM SirReal2 for 24 hours and 10 μM MG132 for 4 hours. Ubiquitination levels of affinity purified Flag-PEPCK1 proteins were detected and visualized by Western blotting. (G) Flag-tagged SIRT2 was overexpressed in HGC-27 cells and then treated with 10 μM SirReal2 for 24 hours. Flag and PEPCK1 proteins were detected and visualized by Western blotting. (H-I) Flag-tagged wild-type and mutant PEPCK1 was overexpressed in HGC-27 cells. Cells were harvested, lysated, and visualized by Western blotting. (J) PEPCK1 or RAS was knocked down in SIRT2 overexpressed gastric cancer cells. Cells were harvested, the downstream targets of SIRT2 were visualized (left), and cell migration (middle) and invasion (right) were quantified.
SIRT2 Supports GC Cell Migration and Invasion by Promoting Mitochondrial Metabolism
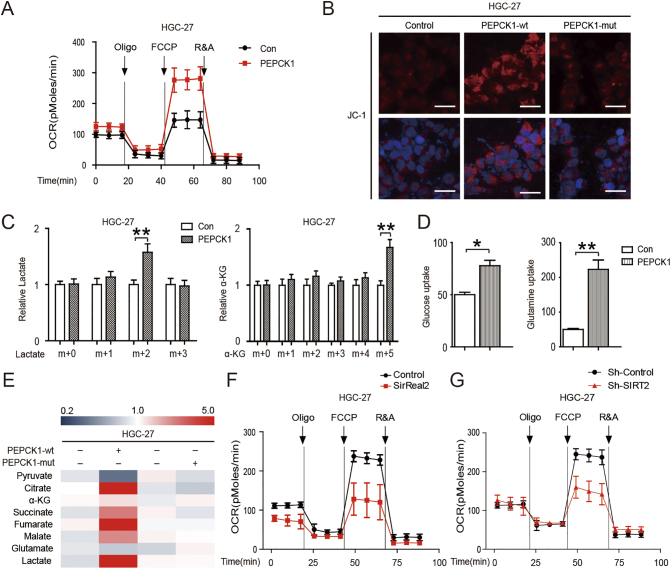
SIRT2 matches cellular energy requirements and availability with direct metabolic pathways to ensure that energy production corresponds to consumption. In addition, SIRT2 promotes the Warburg effect of tumor cells [12], [20], [24], [26], [27]. We investigated the effects of SIRT2 and its target PEPCK1 on cell metabolic changes. Employing the Seahorse bio-energy analyzer, we found that oxygen consumption rates (OCRs) were significantly increased after PEPCK1 overexpression (Figure 4A). These results were confirmed by mitochondrial activity staining with JC-1 (Figure 4B). To further identify the effects of PEPCK1 in GC metabolism, we utilized 13C6 glucose and 13C5 glutamine tracing analysis and found that PEPCK1 overexpressed cells had larger glucose and glutamine uptake, as well as converted them to lactate and α-KG, respectively (Figure 4, C & D). We further measured concentrations of the metabolites in GC cells by mass spectrometry (MS). PEPCK1 increased the mitochondrial metabolism as well as the formation of lactate (Figure 4E). Conversely, SIRT2 inhibition and knockdown decreased mitochondrial metabolism (Figure 4, F & G). This suggests that SIRT2/PEPCK1 is the key target in promoting mitochondrial metabolism as well as in the migration and invasion of GC cells.
Figure 4.
SIRT2 supports the migration and invasion of GC cells by promoting mitochondrial metabolism. (A) The OCRs of PEPCK1-overexpressed HGC-27 cells and control cells were detected at different time points. OCR under oligomycin, carbonyl cyanide-m-chlorophenylhydrazone (FCCP), and antimycin A/rotenone treatments, respectively. (B) HGC-27 cells were overexpressed with different type of PEPCK1 plasmids and stained with JC-1. Red color indicated the mitochondrial membrane potential. DAPI was used to stain nuclei (blue). Greater red fluorescence intensity indicated higher mitochondrial membrane potential in cells. Scale bars, 50 μm. (C) 13C6 glucose (left) and 13C5 glutamine (right) labeling experiments were performed as described in the Methods. PEPCK1-overexpressed and control HGC-27 cells were treated with 13C6 glucose (left) and 13C5 glutamine (right) for 1 hour, respectively, and then relative abundance of 13C3 lactate and 13C5 α-KG was determined by gas chromatography–mass spectrometer (GC-MS). The incorporation of 13C atoms are denoted as m + n, where n is the number of 13C atoms. (D) PEPCK1-overexpressed and control HGC-27 cells were cultured in medium for 3 days, and then supernatant was removed for glucose and glutamine detection. (E) HGC-27 cells were overexpressed with different types of PEPCK1 plasmids, then cells were collected and lysed, and the concentrations of the metabolites in GC cells were measured by MS. (F) OCRs of HGC-27 cells were detected at different time points in the presence or absence of 10 μM SirReal2 for 24 hours. (G) OCRs of HGC-27 cells were detected at different time points following infection with SIRT2 shRNA.
Expression of SIRT2 and PEPCK1 in Human Gastric Cancer Samples and Xenograft Tumors
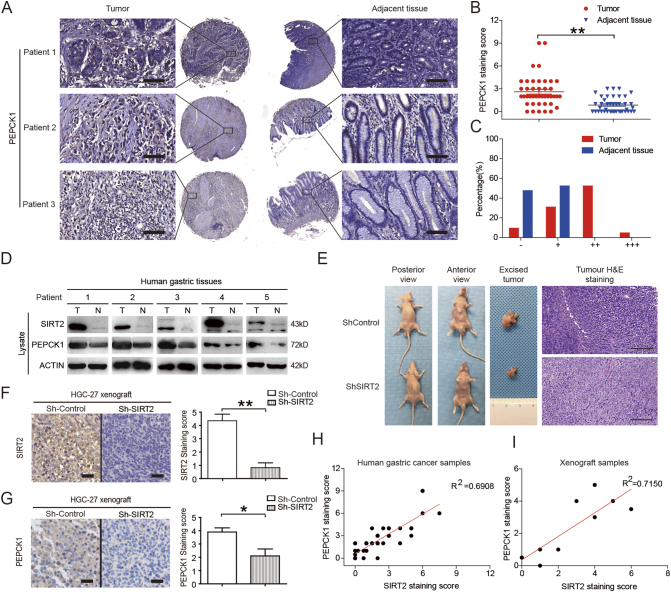
We validated the relationship between SIRT2 and PEPCK1 in clinical specimens. Immunohistochemistry staining demonstrated that SIRT2 and PEPCK1 were colocalized in gastric cancer tissues (Figures 5A & 1A). The level of PEPCK1 was significantly increased in tumor tissues compared to adjacent tissues (Figure 5, B & C). Furthermore, levels of SIRT2 and PEPCK1 had a positive correlation with each other (Figure 5D). We employed the intraperitoneal metastases model by intraperitoneally injecting SIRT2 knockdown GC cells to evaluate the in vivo anticancer effects of SIRT2 inhibition and found that SIRT2 downregulation significantly inhibited the formation of metastatic tumors (Figure 5E). Similar to the results in human gastric cancer samples, the expression of SIRT2 and PEPCK1 in SIRT2 knockdown xenograft tumors was significantly less than that of their control counterparts (Figure 5, F & G). Linear regression analysis showed that the expression of SIRT2 and PEPCK1 was highly correlated in both human gastric cancer samples and xenograft tumors (Figure 5, H & I). These results from human gastric cancer samples and xenograft tumors support the idea that PEPCK1 is the downstream target of SIRT2 in gastric cancer.
Figure 5.
The expression of SIRT2 and PEPCK1 was correlated in gastric cancer samples. (A) PEPCK1 protein levels in tumor and adjacent normal tissues from tissue microarrays were detected and (B) quantified. The magnification is ×400. Scale bars, 50 μm. (C) After scoring, the different proportions of tumor and adjacent normal tissues were shown. (D) Tissues form GC patients were harvested, lysated, and visualized by Western blotting. (E) SIRT2 knockdown suppresses GC cell xenograft tumor growth in nude mice. Tumors were photographed after all animals were sacrificed (n = 10). Xenograft samples were stained with H&E. Scale bars, 100 μm. (F-G) The SIRT2 knockdown GC cell xenograft tumor growth in nude mice. Xenograft samples were stained by SIRT2 and PEPCK1 immunohistochemistry as indicated. Scale bars, 50 μm. (H-I) Correlation analysis of SIRT2 and PEPCK1 protein level in tissue microarrays (n = 42) and xenograft tumors (n = 10). Data represent the mean ± SEM, n ≥ 3. * P < .05; ** P < .01. A score of “−”=no staining or cytoplasmic staining in less than 10% of the tumor cells; a score of “+”=a faint and partial cytoplasmic staining in greater than 10% of the tumor cells; scores of “++” and “+++”=a weak to moderate and a strong complete cytoplasmic staining in greater than 10% of the tumor cells, respectively.
Effects of SIRT2 Knockdown on Tumor Xenografts
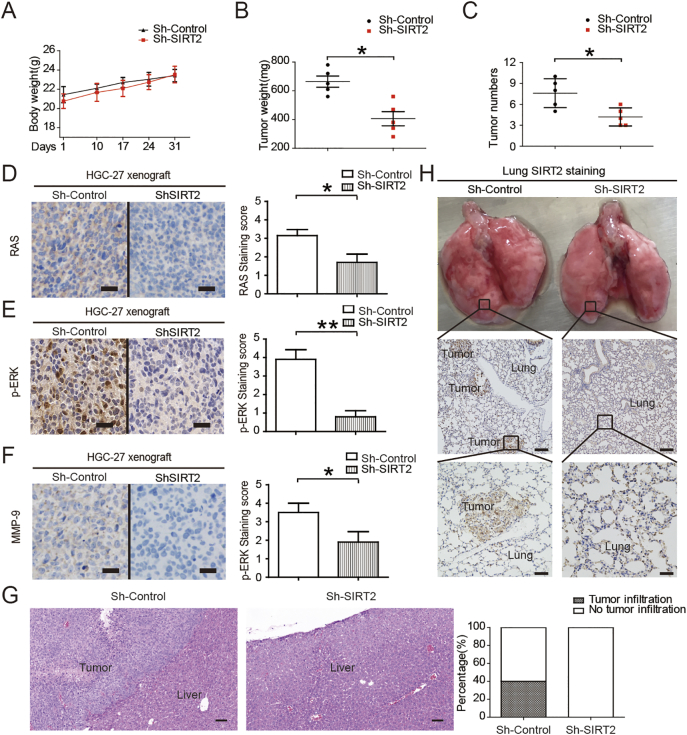
The body weights of treated mice were used as indicators of health [28]. Intraperitoneal injection of SIRT2 knockdown GC cells did not affect mouse body weight, suggesting that the mice did not experience evident toxicity in vivo (Figure 6A). Consistent with these results, we also found that the intraperitoneal metastases from SIRT2 knockdown cells were significantly less than those of their counterparts (Figure 6, B & C). Consistent with in vitro results, SIRT2 inhibition blocked the related downstream pathway and liver metastasis (Figure 6, D-G). Additionally, we evaluated the lung metastases by the tail vein injection model of SIRT2 knockdown GC cells. Multiple metastases were observed in the lung of the control group, but almost no metastases were found in the lung of the nude mice with intravenously administrated SIRT2 knockdown GC cells (Figure 6H). Altogether, these data demonstrate that SIRT2 is a key molecule in the metastasis of gastric cancer and that SIRT2 inhibition can attenuate the tumor metastases of gastric cancer in vivo.
Figure 6.
Effects of SIRT2 inhibition on tumor xenografts. (A) SIRT2 knockdown suppresses GC cell xenograft tumor growth in nude mice. The body weights of tumor-burdened mice (n = 10). (B) The xenograft tumor weights (n = 10). (C) The number of xenograft tumors (n = 10). (D-F) Xenograft samples were stained by RAS, p-ERK, and MMP-9 immunohistochemistry as indicated. Scale bars, 50 μm. (G) The SIRT2 knockdown GC cell xenograft tumor growth in the liver of nude mice (n = 10). The metastases of gastric cancer were stained with H&E. Scale bars, 100 μm. (H) The SIRT2 knockdown GC cell xenograft tumor growth in the lung of nude mice (n = 10). The metastases of gastric cancer were stained with SIRT2 immunohistochemistry. Scale bars were 200 μm (middle) and 50 μm (down), respectively. Data represent the mean ± SEM, n = 5. *P < .05, **P < .01.
Discussion
Metastasis represents a serious worldwide problem threatening the health of millions of gastric cancer patients. Consequently, there is an urgent need to understand the mechanism of GC metastasis and to develop corresponding new agents for its therapy.
Acetylation has emerged as a key posttranslational modification and has likewise been identified as a key metabolic enzyme in cellular regulation [5], [6]. Deacetylases are designated to four classes (I-IV) depending on their amino acid sequence structure. Sirtuins (also known as SIRTs) are NAD+-dependent class III histone deacetylases (HDACs). Seven SIRT homologues have been identified as SIRT1-7 in mammals [29], [30], [31]. Among these sirtuin family members, SIRT2 catalyzes a wide range of biological processes including gene expression, development, and metabolism. Its enzymatic reaction removes the acetyl group from lysine residues and is accompanied with hydrolysis of NAD to generate nicotinamide (NAM), lysine, and O-acetyl-ADP-ribose. Hence, NAM can inhibit this kind of enzymatic reaction [30], [31]. In fact, we evaluated the migration and invasion of GC cells after NAM treatment (data not shown) and found no significant changes. This is probably because NAM is a pan-deacetylation inhibitor, not a SIRT2-specific inhibitor. It is reported that some HDACs share 82% of their identity with each other [32], [33], [34]. Due to the high level of homology among HDACs, the inhibitory effects of the drug on other HDACs besides SIRT2 cannot be ignored. Therefore, selective SIRT2 inhibitors are critical to determine whether SIRT2 is indeed an anticancer target. SirReal2, a new selective SIRT2 inhibitor [35], is considered an isotype-selective drug-like inhibitor with optimized potency and physicochemical properties compared with EX-527, CHIC-35, and other SIRT2 inhibitors [36]. We evaluated the inhibitory effect of SirReal2 on SIRT2 by employing mass spectrometry. At the molecular level, SirReal2 not only partially reversed cell migration and invasion but also reversed the PEPCK1 protein levels and the downstream RAS/ERK/JNK/MMP-9 pathway. This suggests that SirReal2 inhibited GC cell migration and invasion by targeting SIRT2.
SIRT2 is primarily a cytoplasmic protein, and both tubulin [37] and phosphoenolpyruvate carboxykinase (PEPCK) [6] are substrates of this deacetylase. Though studies have demonstrated that SIRT2 inhibitors may have antitumor [29], [38], [39], [40], anti-inflammatory [41], and antidiabetic properties, the impact of SIRT2 on metabolism in GC cells as well as possible molecular mechanisms has not yet been reported. Due to the important role of PEPCK1, its regulation has been extensively studied. Both yeast and human PEPCK1 have been found to have acetylation, with catalytic activity being inactivated following this acetylation [6], [42]. Acetylation of Lys70, Lys71, and Lys594 of human PEPCK1 led to decreased protein stability, reduced protein levels, and decreased gluconeogenesis without affecting mRNA levels [6]. PEPCK1 is an important marker in the evaluation of type II diabetes [43] and can promote glucose and glutamine utilization for anabolic metabolism [12]. However, little is known about the role of PEPCK1 in GC metastasis. Consistent with previous studies, our data demonstrate that SIRT2 is the major enzyme responsible for PEPCK1 acetylation and is the downstream target of SirReal2. Thus, PEPCK1 stability is controlled by the balance between acetylation and deacetylation in response to SirReal2.
Numerous studies have evaluated the role of SIRT2 in cancer, and demonstrated that SIRT2-specific inhibitors with broad anticancer effect in various human cancer cells are promising anticancer agents [44], but few data exist in GC and showed opposite results [16], [45], [46], [47]. Furthermore, SIRT2 has been reported as a tumor suppressor gene in a knockout mouse model and other cancer tissues [7], [8]. It is possible that SIRT2 may promote tumor progression under one circumstance, such as in human pancreatic cancer and GC, as well as suppress tumor progression in other circumstances. Distinct molecular features in different regions of the stomach and various GC cell lines might be another reason for this inconsistency [48], [49]. Our results demonstrated that SIRT2 significantly promotes GC cell migration and invasion, suggesting that SIRT2 may have oncogenic function in gastric cancer. This is consistent with our previous studies, which suggested the idea that SIRT2 is a key promoter of cell invasion in pancreatic cancer [20]. Interestingly, we found that SIRT2 marginally promoted cell proliferation. This may be due to the various downstream targets of SIRT2; as in pancreatic cancer, LDH-A is the downstream target of SIRT2 and led to proliferation [20], [50]. However, we found no difference between LDH-A levels in GC cells and tissues (data not shown), indicating that different substrates of SIRT2 contributed to different malignant biological behaviors of the tumor. In gastric cancer, we found that SIRT2 and its downstream target PEPCK1 promoted the uptake of glucose and glutamine, as well as TCA activity. It has been found that activated mitochondrial metabolism, especially the glutamine-fueled TCA cycle, is critical for oncogene-induced tumorigenicity by RAS activation [14]. It is further confirmed by our results that RAS and its downstream ERK/JNK/MMP-9 are critical for oncogene-induced migration and invasion in gastric cancer. In fact, using the database, we found that Pepck1 mRNA had no correlation with worse overall survival in 876 cases of gastric cancer specimens. We think the reason for this result is that SIRT2 is a regulatory protein; thus, its gene level and protein level are equivalent and always stay at a stable level. However, PEPCK1 is a metabolic protein, which is modified by acetylation and ubiquitination, so its gene level and protein level are thus not quite as stable [6], [25].
Previous studies have shown that mitochondrial metabolism is associated with tumor metastasis [13] and RAS-mediated tumorigenicity [14]. RAS can activate two major downstream MAPKs: ERK and JNK. Activation of ERK/JNK pathway contributes to the upregulation of MMP-9. Moreover, MMP-9 may be one of the most critical molecules in cancer cell metastasis [15], [51]. We found that SIRT2-related PEPCK1 degradation positively correlates with the RAS/ERK/JNK/MMP-9 pathway in GC, suggesting that this pathway is important in SIRT2-mediated gastric cancer.
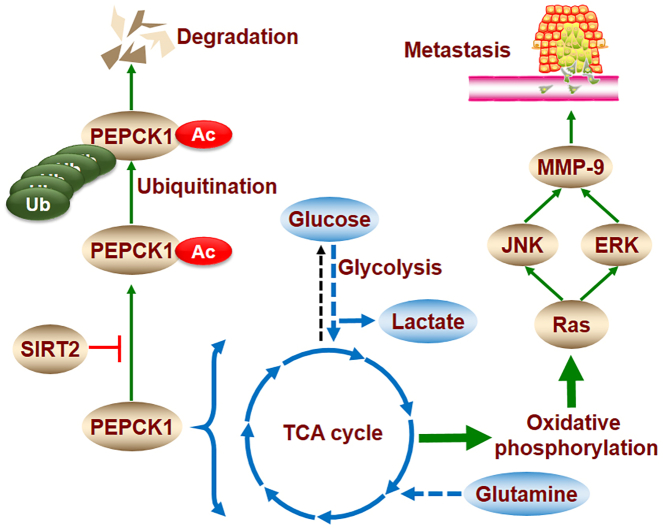
In the present study, we provide insight into the regulation of SIRT2 on gastric cancer metabolism and metastasis. SIRT2 increased PEPCK1 protein levels and mitochondrial activity, as well as induced cell migration and invasion by activating the RAS/ERK/JNK/MMP-9 pathway (Supplementary Figure 2). Based on our results, PEPCK1 is the downstream target of SIRT2, and the inhibition of SIRT2 activity plays an important role in GC cell migration and invasion. Thus, SIRT2 may be a novel molecular target for GC therapy and may shed light on underlying GC metabolic and metastatic mechanisms.
Supplementary Figure 2.
SIRT2 promotes metastasis of gastric cancer through RAS/ERK/JNK/MMP-9 pathway by increasing PEPCK1-related metabolism.
SIRT2 stabilizes the PEPCK1 protein by inhibiting its acetylation associated with ubiquitination. PEPCK1 promotes mitochondrial activity by increasing cellular uptake of glucose and glutamine. Metabolic abnormalities increase migration and invasion by activating the RAS/ERK/JNK/MMP-9 pathway.
Materials and Methods
Ethics, Consent, and Permissions
All experiments utilizing animal and human samples were approved by the Ethical Committee of Medical Research, Nanjing Drum Tower Hospital, Affiliated Hospital of Nanjing University Medical School.
DEGs of Paired-GC from TCGA Data
The GC RNA-Seq data were downloaded from the TCGA database using The GDC Data Portal (https://gdc-portal.nci.nih.gov/). The number of coding gene mRNA expression values was 20,502. The mRNA expression data included a total of 64 samples consisting of 32 normal sample and 32 paired-GC samples. The sequencing data were all publicly available; therefore, no ethical issues were involved. The edgeR package in Bioconductor was used to screen the DEGs in GC and normal gastric tissue samples. The edgeR package is based on the negative binomial (NB) distribution, which can correct the overdispersion problem in RNA-seq data by using a Poisson model and a Bayes procedure. Data with expression values of zero were removed. Genes were deemed to be DEGs if fold change >2, respectively, with both P value < .01 and false discovery rate (FDR) < 0.01.
Functional Annotation
The Database for Annotation Visualization and Integrated Discovery online tool (https://david.ncifcrf.gov/) was used to conduct the functional and pathway enrichment analyses in our study. We performed GO pathway enrichment analyses to detect the potential biological functions and pathways of the high and low expression genes in GC.
Immunohistochemistry
The tissue microarray slides were obtained from SHANGHAI OUTDO BIOTECH (Shanghai, China). Staining intensity was graded as follows: absent staining = 0, weak = 1, moderate = 2, and strong = 3. The percentage of staining was graded as follows: 0 (no positive cells), 1 (<25% positive cells), 2 (25%-50% positive cells), 3 (50%-75% positive cells), and 4 (>75% positive cells). The score for each tissue was calculated by multiplication; the range of this calculation was therefore 0 to 12 [52].
Cell Culture and Reagents
Human GC cell lines (AGS, HGC-27, MGC-803, and MKN-45) and normal gastric epithelial cells (GES-1) were gifts from the Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences (Shanghai, China), and the Zhao lab of Fudan University (Shanghai, China). Cells were maintained in Dulbecco's modified Eagle's medium (Invitrogen, Carlsbad, CA) containing 10% fetal bovine serum (Invitrogen), penicillin (Invitrogen) (100 U/ml) and streptomycin (Invitrogen) (100 U/ml). Full-length PEPCK1 (wild-type, 3K/R, and 3K/Q) and SIRT2 (wild-type and H187Y) plasmids were also gifts from the Zhao lab of Fudan University (Shanghai, China). Antibodies against Flag (Sigma, St. Louis, MO), PEPCK1 (Santa Cruz), SIRT2 (Sigma), and β-actin (Sigma) were all purchased. MG132 (Sigma), SirReal2 (Adooq Bioscience, Irvine, CA), and CHX (Sigma) were all purchased. Control and shSIRT2 lentivirus were purchased (Genechem, Shanghai, China); the sequence of shSIRT2 is 5′-GCTAAGCTGGATGAAAGAGAA-3′ and 5′-GCCAACCATCTGTCACTACTT-3′, respectively. The sequence of control shRNA is 5′-TTCTCCGAACGTGTCACGT-3′. The sequence of siPEPCK1 and siRAS was reported as indicated [53], [54].
Western Blot
Cultured cells were lysed with ice-cold lysis buffer (Biosharp). Extracted proteins were mixed with loading buffer containing 5% 2-mercaptoethanol and then denatured at 100°C for 10 minutes. Samples were separated on 8% to 12% sodium dodecyl sulfate polyacrylamide gels and transferred to PVDF membranes (Millipore). Afterwards, membranes were incubated in TBST containing 5% nonfat milk for 2 hours at room temperature. Next, membranes were incubated according to instructions with the respective primary antibodies overnight at 4°C and then treated with the appropriate horseradish peroxidase (HRP)–conjugated secondary antibodies (1:3000 dilutions). The blots were developed with ECL Western blotting reagents (Millipore). HRP-conjugated anti-mouse and anti-rabbit antibodies were purchased from Cell Signaling Technology (Danvers, MA). The following antibodies were used: SIRT1 (sc15404, Santa Cruz), SIRT2 (s8447, Sigma), SIRT3 (s4072, Sigma), β-Catenin (ab32572, Abcam), E-Cadherin (3195s, CST), PEPCK1 (sc32879, Santa Cruz), JNK (9258, CST), p-JNK (4668, CST), ERK (4695, CST), p-ERK (4370, CST), RAS (3965, CST), MMP-9 (ab137867, Abcam), actin (A5441, Sigma), Tubulin (2144, CST), Ac-Tubulin (5335, CST), anti-mouse IgG (7076, CST), and anti-rabbit IgG (7074, CST).
Apoptosis Assay
Cell apoptosis was measured by flow cytometry using the Annexin V-fluorescein isothiocyanate (FITC) Apoptosis Detection Kit (556,547, BD Biosciences). All cells were treated similarly as in the cell cycle assay. Cultured cells were collected and washed with cold PBS twice. Afterward, cells were resuspended in 100 μl of Annexin V binding buffer and incubated with 5 μl of FITC-conjugated Annexin V and 5 μl of propidium iodide for 15 minutes in the dark. Two hundred microliters of Annexin V binding buffer was then added to each tube. Next, samples were examined using a BD FACSCanto II flow cytometer (BD Biosciences, CA).
Colony Formation Assay
HGC-27 cells were counted and seeded in triplicate in a six-well plate at the concentration of 500 cells/well in normal medium and treated with 10 μM SirReal2. After 24 hours, SirReal2 was removed and normal medium was added. After 14 days, cells were fixed in methanol and stained with 0.1% crystal violet in 25% methanol for 20 minutes. Finally, colonies were counted. Results are presented as the average of the number of colonies counted in each well for each condition.
Migration and Invasion Assays
Cell motility and invasive abilities were assessed by way of Transwell (Corning Life Sciences, Bedford, MA) and Matrigel invasion (BD Biosciences), respectively. For the Transwell migration assay, 5 × 104 cells were seeded; 2 × 105 cells were seeded for the invasion assay. Cells that migrated to the underside of the membrane were fixed and stained with 0.1% crystal violet and were enumerated for 10 microscope fields. Mean values of migrating or invading cells were expressed as percentages. Each experiment was performed in replicate inserts, and the mean value was calculated from three independent experiments.
Deacetylation Assay
Cells were lysed in NP-40 buffer containing 50 mM Tris-HCl (Sigma), 150 mM NaCl (Sangon), 0.5% Nonidet P-40 (Sigma), 1 μg/ml aprotinin (Sigma), 1 μg/ml leupeptin (Sigma), 1 μg/ml pepstatin (Sigma), 1 mM Na3VO4 (Sigma), and 1 mM PMSF (Sigma), pH = 7.5. For immunoprecipitation, 500 μl of cell lysate was incubated with Flag-beads (Sigma) for 12 hours at 4°C with rotation, and the beads were washed three times with lysis buffer before proteins were dissolved in loading buffer. The SIRT2 assay was done using bacterial expression and purification (Biovision, Milpitas, CA). Deacetylation assays were carried out in the presence of 5-μg enzyme and 0.3-μg peptide in 30-μl reaction buffer (30 mM HEPES (Sigma), 0.6 mM MgCl2 (Sangon), 1 mM DTT (Sigma), 1 mM NAD+ (Sigma), and 10 mM PMSF (Sigma)). The deacetylation reaction was incubated for 3 to 5 hours at 37°C before the mixture was desalted by passing it through a C18 ZipTip (Millipore). The desalted samples were analyzed using a MALDI-TOF/TOF mass spectrometer (Applied Biosystems, Grand Island, NY). The acetylated peptide used in the assay was GILRRLKAcKAcYDNCWL (Glssale, Shanghai, China).
Ubiquitination Assay
Thirty-six hours following transfection, cells were lysed in 1% SDS buffer (Tris (Sigma), 0.5 mM EDTA (Sigma), and 1 mM DTT (Sigma), pH = 7.5, as well as boiled for 10 minutes. For immunoprecipitation, the lysates were diluted 10-fold in Tris-HCl buffer. Analyses of ubiquitination were performed using anti-HA blotting.
Cell Viability Assay
Cells viability was determined using the CCK-8 colorimetric assay in 96-well plates (2 × 103 cells/well) (Dojindo, Minato-ku, Tokyo, Japan) and cultured for 24, 48, or 72 hours at 37°C with 5% CO2. After treatment or the indicated times, 10 μl of CCK-8 solutions was added to each well. Then, cells were incubated for one and a half hours. The absorbance of the samples at 450 nm was recorded using a scanning multiwell spectrophotometer. Relative cell viability (%) = (absorbance 450 nm of treated group − absorbance450 nm of blank)/(absorbance 450 nm of control group − absorbance 450 nm of blank) × 100.
Mitochondrial Oxidative Phosphorylation Analysis
XF96 Extracellular Flux Analyzer from Seahorse Bioscience, Inc. (North Billerica, MA) was utilized to detect OCR, representing oxidative phosphorylation (OXPHOS). HGC-27 cells were seeded in 96-well XF cell culture microplates at 1.0 × 104 cells/well. Cells were incubated for 24 hours, and then the medium was replaced with 175 μl/well of XF-96 running media (supplemented with RPMI-1640 without serum). The plates were preincubated at 37°C for 20 minutes in the XF Prep Station incubator (Seahorse Bioscience, Billerica, MA) in the absence of CO2 and then run on the XF96 analyzer to obtain OCR. For each analysis, different compounds that modulate mitochondrial respiration were injected sequentially in each well: for OCR, oligomycin (0.5 μM), carbonylcyanide p-trifluoromethoxy-phenylhydrazone (1 μM), rotenone (1 μM), and antimycin A (1 μM). OCR was recorded during specified programmed time periods (three readings each) as the average numbers between the injections of inhibitors mentioned above. The final data calculation was performed after the readings had been normalized with counting the cell number in each well. OCR is expressed as pmol/min.
Mitochondrial Membrane Potential Assay
Mitochondrial membrane potential was assessed by JC-1 staining (Beyotime Institute of Biotechnology, China). JC-1, a cationic, fluorescent, carbocyanine dye, can be used as a ratiometric indicator of mitochondrial membrane potential (ΔΨm) in cells. HGC-27 cells were seeded at 10× 104 cells per well in six-well plates and incubated for 48 hours at 37°C. Cells were washed with PBS and incubated with medium containing 20 μg/mL JC-1 at 37°C for 20 minutes and observed using a confocal fluorescent microscope. The data are representative of at least three independent experiments.
13C Metabolite Labeling Analysis
13C metabolism labeling experiments were performed as described previously [55]. Medium for 13C labeling experiments contained 10% labeled glucose (2.5 mM) and glutamine (0.4 mM), respectively. Cells were washed in PBS and replenished with the labeling medium for indicated periods of time. After treatment, cells were collected and lysed with 80% pre-cold methanol. Then metabolite extractions were lyophilized and oximated with 20 mg/ml methoxyamine hydrochloride in pyridine at 70°C for 1 hour. Then samples were derivatized with 20 μl N-methyl-N-(tert-butyldimethylsilyl) trifluoroacetamide in 80 μl pyridine at 30°C for 1 hour. After filtration, 2 μl of samples was injected for the GC-MS analysis using an Agilent 6890-5973 GC-MS system with an HP-5MS column (30 m × 0.25 × mm × 0.25 μm). GC oven temperature was programmed from 60°C to 180°C at 5°C/min and from 180°C to 260°C at 10 °C/min. The flow rate of carrier gas was set at 1 ml/min. The mass spectrometer was operated in the electron impact (EI) mode at 70 eV. Relative metabolite abundances were determined by normalizing abundances of each metabolite to the internal standard and to cell number.
Gastric Cancer Xenograft Model
A total of 20 nude mice were purchased from the Department of Laboratory Animal Science, Nanjing Drum Tower Hospital. In intraperitoneal model (n = 10), control HGC-27 cells (5 × 106 cells per mouse, n = 5) and SIRT2 knockdown HGC-27 cells (5 × 106 cells per mouse, n = 5) in FBS-free RPMI-1640 were intraperitoneally injected into the abdomen of mice, respectively. In tail vein injection model (n = 10), control HGC-27 cells (2 × 106 cells per mouse, n = 5) and SIRT2 knockdown HGC-27 cells (5 × 106 cells per mouse, n = 5) in FBS-free RPMI-1640 were injected into the tail vein of mice. Once xenograft tumors were palpable, mice were weighed twice a week for 4 weeks. Mice were sacrificed after 4 weeks, and lungs, livers, and xenograft tumors were harvested. The Animal Welfare Committee of Nanjing Drum Tower Hospital approved all procedures involving animals.
Statistics
Data was expressed as means ± standard error of the mean (SE). The data were analyzed through one-way ANOVAs followed by post hoc Duncan tests (SPSS 17.0). P < .05 was considered significant.
The following are the supplementary data related to this article.
Author Contributions
Mingming Zhang designed the study; Dehua Tang, Lei Xu, Qian Zhou, Yang Li, and Yida Pan did the cell experiments; Yang Li, Yuyao Yin, Yuming Wang, Wenjia Liu, and Zhenguo Zhao collected the tissue samples; Qian Zhou, Shan Huang, and Yang Li performed the protein analysis; Lixing Zhou, Mingming Zhang, Robert G. Dorfman, and Shan Huang drafted the manuscript and performed the immunohistochemistry experiment; Robert G. Dorfman did the language editing. Shan Huang and Mingming Zhang supported the study. All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
We thank the Zhao lab for offering their help.
Footnotes
Grant Support: This work was supported by grants from the National Natural Science Foundation of China (No. 81472756, 81672935 and 81602076), Outstanding Youth Project of Nanjing City (No. JQX17002), the Jiangsu Clinical Medical Center of Digestive Disease (BL2012001), the Natural Science Foundation from the Department of Science & Technology of Jiangsu Province (BK20160113), the Fundamental Research Funds for the Central Universities (No. 021414380244), and the Foundation of Jiangsu Provincial Commission of Health and Family Planning (Q201611).
Conflicts of interest: The authors declare no conflicts of interest.
Contributor Information
Lei Wang, Email: 867152094@qq.com.
Xiaoping Zou, Email: 13770771661@163.com.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Mortality GBD, Causes of Death C Global, regional, and national age-sex specific all-cause and cause-specific mortality for 240 causes of death, 1990-2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet. 2015;385:117–171. doi: 10.1016/S0140-6736(14)61682-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nguyen DX, Bos PD, Massague J. Metastasis: from dissemination to organ-specific colonization. Nat Rev Cancer. 2009;9:274–284. doi: 10.1038/nrc2622. [DOI] [PubMed] [Google Scholar]
- 4.Bruix J. Hepatocellular carcinoma: clinical frontiers and perspectives. Gut. 2014;63:844–855. doi: 10.1136/gutjnl-2013-306627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Q, Zhang Y, Yang C, Xiong H, Lin Y, Yao J, Li H, Xie L, Zhao W, Yao Y. Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science. 2010;327:1004–1007. doi: 10.1126/science.1179687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, Yao J, Zhou L, Zeng Y, Li H. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327:1000–1004. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Temel M, Koc MN, Ulutas S, Gogebakan B. The expression levels of the sirtuins in patients with BCC. Tumour Biol. 2016;37:6429–6435. doi: 10.1007/s13277-015-4522-8. [DOI] [PubMed] [Google Scholar]
- 8.Harting K, Knoll B. SIRT2-mediated protein deacetylation: an emerging key regulator in brain physiology and pathology. Eur J Cell Biol. 2010;89:262–269. doi: 10.1016/j.ejcb.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 9.Wilking-Busch MJ, Ndiaye MA, Liu X, Ahmad N. RNA interference-mediated knockdown of SIRT1 and/or SIRT2 in melanoma: identification of downstream targets by large-scale proteomics analysis. J Proteomics. 2018;170:99–109. doi: 10.1016/j.jprot.2017.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen J, Chan AW, To KF, Chen W, Zhang Z, Ren J, Song C, Cheung YS, Lai PB, Cheng SH. SIRT2 overexpression in hepatocellular carcinoma mediates epithelial to mesenchymal transition by protein kinase B/glycogen synthase kinase-3beta/beta-catenin signaling. Hepatology. 2013;57:2287–2298. doi: 10.1002/hep.26278. [DOI] [PubMed] [Google Scholar]
- 11.Seenappa V, Das B, Joshi MB, Satyamoorthy K. Context dependent regulation of human phosphoenolpyruvate carboxykinase isoforms by DNA promoter methylation and RNA stability. J Cell Biochem. 2016;117:2506–2520. doi: 10.1002/jcb.25543. [DOI] [PubMed] [Google Scholar]
- 12.Montal ED, Dewi R, Bhalla K, Ou L, Hwang BJ, Ropell AE, Gordon C, Liu WJ, DeBerardinis RJ, Sudderth J. PEPCK coordinates the regulation of central carbon metabolism to promote cancer cell growth. Mol Cell. 2015;60:571–583. doi: 10.1016/j.molcel.2015.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.LeBleu VS, O'Connell JT, Gonzalez Herrera KN, Wikman H, Pantel K, Haigis MC, de Carvalho FM, Damascena A, Domingos Chinen LT, Rocha RM. PGC-1alpha mediates mitochondrial biogenesis and oxidative phosphorylation in cancer cells to promote metastasis. Nat Cell Biol. 2014;16:992–1003. doi: 10.1038/ncb3039. [1001–1015] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weinberg F, Hamanaka R, Wheaton WW, Weinberg S, Joseph J, Lopez M, Kalyanaraman B, Mutlu GM, Budinger GR, Chandel NS. Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proc Natl Acad Sci U S A. 2010;107:8788–8793. doi: 10.1073/pnas.1003428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang CQ, Li W, Li SQ, Li J, Li YW, Kong SX, Liu RM, Wang SM, Lv WM. MCP-1 stimulates MMP-9 expression via ERK 1/2 and p38 MAPK signaling pathways in human aortic smooth muscle cells. Cell Physiol Biochem. 2014;34:266–276. doi: 10.1159/000362997. [DOI] [PubMed] [Google Scholar]
- 16.Zhang S, Huang S, Deng C, Cao Y, Yang J, Chen G, Zhang B, Duan C, Shi J, Kong B. Co-ordinated overexpression of SIRT1 and STAT3 is associated with poor survival outcome in gastric cancer patients. Oncotarget. 2017;8:18848–18860. doi: 10.18632/oncotarget.14473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hong SY, Shih YP, Sun P, Hsieh WJ, Lin WC, Lo SH. Down-regulation of tensin2 enhances tumorigenicity and is associated with a variety of cancers. Oncotarget. 2016;7:38143–38153. doi: 10.18632/oncotarget.9411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tian J, Hachim MY, Hachim IY, Dai M, Lo C, Raffa FA, Ali S, Lebrun JJ. Cyclooxygenase-2 regulates TGFbeta-induced cancer stemness in triple-negative breast cancer. Sci Rep. 2017;7 doi: 10.1038/srep40258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shibata T, Aburatani H. Exploration of liver cancer genomes. Nat Rev Gastroenterol Hepatol. 2014;11:340–349. doi: 10.1038/nrgastro.2014.6. [DOI] [PubMed] [Google Scholar]
- 20.Zhao D, Zou SW, Liu Y, Zhou X, Mo Y, Wang P, Xu YH, Dong B, Xiong Y, Lei QY. Lysine-5 acetylation negatively regulates lactate dehydrogenase A and is decreased in pancreatic cancer. Cancer Cell. 2013;23:464–476. doi: 10.1016/j.ccr.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qi J, Yu Y, Akilli Ozturk O, Holland JD, Besser D, Fritzmann J, Wulf-Goldenberg A, Eckert K, Fichtner I, Birchmeier W. New Wnt/beta-catenin target genes promote experimental metastasis and migration of colorectal cancer cells through different signals. Gut. 2016;65:1690–1701. doi: 10.1136/gutjnl-2014-307900. [DOI] [PubMed] [Google Scholar]
- 22.He M, Qin H, Poon TC, Sze SC, Ding X, Co NN, Ngai SM, Chan TF, Wong N. Hepatocellular carcinoma-derived exosomes promote motility of immortalized hepatocyte through transfer of oncogenic proteins and RNAs. Carcinogenesis. 2015;36:1008–1018. doi: 10.1093/carcin/bgv081. [DOI] [PubMed] [Google Scholar]
- 23.Yang S, Liu Y, Li MY, Ng CSH, Yang SL, Wang S, Zou C, Dong Y, Du J, Long X. FOXP3 promotes tumor growth and metastasis by activating Wnt/beta-catenin signaling pathway and EMT in non-small cell lung cancer. Mol Cancer. 2017;16:124. doi: 10.1186/s12943-017-0700-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lv L, Li D, Zhao D, Lin R, Chu Y, Zhang H, Zha Z, Liu Y, Li Z, Xu Y. Acetylation targets the M2 isoform of pyruvate kinase for degradation through chaperone-mediated autophagy and promotes tumor growth. Mol Cell. 2011;42:719–730. doi: 10.1016/j.molcel.2011.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang WQ, Wang SW, Xiao MT, Lin Y, Zhou LS, Lei QY, Xiong Y, Guan KL, Zhao SM. Acetylation regulates gluconeogenesis by promoting PEPCK1 degradation via recruiting the UBR5 ubiquitin ligase. Mol Cell. 2011;43:33–44. doi: 10.1016/j.molcel.2011.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park SH, Ozden O, Liu G, Song HY, Zhu Y, Yan Y, Zou X, Kang HJ, Jiang H, Principe DR. SIRT2-mediated deacetylation and tetramerization of pyruvate kinase directs glycolysis and tumor growth. Cancer Res. 2016;76:3802–3812. doi: 10.1158/0008-5472.CAN-15-2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu Y, Li F, Lv L, Li T, Zhou X, Deng CX, Guan KL, Lei QY, Xiong Y. Oxidative stress activates SIRT2 to deacetylate and stimulate phosphoglycerate mutase. Cancer Res. 2014;74:3630–3642. doi: 10.1158/0008-5472.CAN-13-3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang LT, Liou JP, Li YH, Liu YM, Pan SL, Teng CM. A novel class I HDAC inhibitor, MPT0G030, induces cell apoptosis and differentiation in human colorectal cancer cells via HDAC1/PKCdelta and E-cadherin. Oncotarget. 2014;5:5651–5662. doi: 10.18632/oncotarget.2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kozako T, Suzuki T, Yoshimitsu M, Arima N, Honda S, Soeda S. Anticancer agents targeted to sirtuins. Molecules. 2014;19:20295–20313. doi: 10.3390/molecules191220295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rauh D, Fischer F, Gertz M, Lakshminarasimhan M, Bergbrede T, Aladini F, Kambach C, Becker CF, Zerweck J, Schutkowski M. An acetylome peptide microarray reveals specificities and deacetylation substrates for all human sirtuin isoforms. Nat Commun. 2013;4:1–10. doi: 10.1038/ncomms3327. [DOI] [PubMed] [Google Scholar]
- 31.Balaiya S, Abu-Amero KK, Kondkar AA, Chalam KV. Sirtuins expression and their role in retinal diseases. Oxidative Med Cell Longev. 2017;2017:1–11. doi: 10.1155/2017/3187594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hagelkruys A, Sawicka A, Rennmayr M, Seiser C. The biology of HDAC in cancer: the nuclear and epigenetic components. Handb Exp Pharmacol. 2011;206:13–37. doi: 10.1007/978-3-642-21631-2_2. [DOI] [PubMed] [Google Scholar]
- 33.Yang WM, Tsai SC, Wen YD, Fejer G, Seto E. Functional domains of histone deacetylase-3. J Biol Chem. 2002;277:9447–9454. doi: 10.1074/jbc.M105993200. [DOI] [PubMed] [Google Scholar]
- 34.Yang WM, Yao YL, Sun JM, Davie JR, Seto E. Isolation and characterization of cDNAs corresponding to an additional member of the human histone deacetylase gene family. J Biol Chem. 1997;272:28001–28007. doi: 10.1074/jbc.272.44.28001. [DOI] [PubMed] [Google Scholar]
- 35.Galleano I, Schiedel M, Jung M, Madsen AS, Olsen CA. A continuous, fluorogenic sirtuin 2 deacylase assay: substrate screening and inhibitor evaluation. J Med Chem. 2016;59:1021–1031. doi: 10.1021/acs.jmedchem.5b01532. [DOI] [PubMed] [Google Scholar]
- 36.Rumpf T, Schiedel M, Karaman B, Roessler C, North BJ, Lehotzky A, Olah J, Ladwein KI, Schmidtkunz K, Gajer M. Selective Sirt2 inhibition by ligand-induced rearrangement of the active site. Nat Commun. 2015;6:6263. doi: 10.1038/ncomms7263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee JK, Lee J, Go H, Lee CG, Kim S, Kim HS, Cho H, Choi KS, Ha GH, Lee CW. Oncogenic microtubule hyperacetylation through BEX4-mediated sirtuin 2 inhibition. Cell Death Dis. 2016;7 doi: 10.1038/cddis.2016.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luo J, Bao YC, Ji XX, Chen B, Deng QF, Zhou SW. SPOP promotes SIRT2 degradation and suppresses non–small cell lung cancer cell growth. Biochem Biophys Res Commun. 2017;483:880–884. doi: 10.1016/j.bbrc.2017.01.027. [DOI] [PubMed] [Google Scholar]
- 39.Zhang LL, Zhan L, Jin YD, Min ZL, Wei C, Wang Q, Chen YJ, Wu QM, Hu XM, Yuan Q. SIRT2 mediated antitumor effects of shikonin on metastatic colorectal cancer. Eur J Pharmacol. 2017;797:1–8. doi: 10.1016/j.ejphar.2017.01.008. [DOI] [PubMed] [Google Scholar]
- 40.Jing H, Hu J, He B, Negron Abril YL, Stupinski J, Weiser K, Carbonaro M, Chiang YL, Southard T, Giannakakou P. A SIRT2-selective inhibitor promotes c-Myc oncoprotein degradation and exhibits broad anticancer activity. Cancer Cell. 2016;29:767–768. doi: 10.1016/j.ccell.2016.04.005. [DOI] [PubMed] [Google Scholar]
- 41.Kanda T, Sasaki R, Nakamoto S, Haga Y, Nakamura M, Shirasawa H, Okamoto H, Yokosuka O. The sirtuin inhibitor sirtinol inhibits hepatitis A virus (HAV) replication by inhibiting HAV internal ribosomal entry site activity. Biochem Biophys Res Commun. 2015;466:567–571. doi: 10.1016/j.bbrc.2015.09.083. [DOI] [PubMed] [Google Scholar]
- 42.Lin YY, Lu JY, Zhang JM, Walter W, Dang WW, Wan J, Tao SC, Qian J, Zhao YM, Boeke JD. Protein acetylation microarray reveals that NuA4 controls key metabolic target regulating gluconeogenesis. Cell. 2009;136:1073–1084. doi: 10.1016/j.cell.2009.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matsuoka H, Shima A, Kuramoto D, Kikumoto D, Matsui T, Michihara A. Phosphoenolpyruvate carboxykinase, a key enzyme that controls blood glucose, is a target of retinoic acid receptor-related orphan receptor alpha. PLoS One. 2015;10 doi: 10.1371/journal.pone.0137955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jing H, Hu J, He B, Negron Abril YL, Stupinski J, Weiser K, Carbonaro M, Chiang YL, Southard T, Giannakakou P. A SIRT2-selective inhibitor promotes c-Myc oncoprotein degradation and exhibits broad anticancer activity. Cancer Cell. 2016;29:297–310. doi: 10.1016/j.ccell.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peters CJ, Rees JR, Hardwick RH, Hardwick JS, Vowler SL, Ong CA, Zhang C, Save V, O'Donovan M, Rassl D. A 4-gene signature predicts survival of patients with resected adenocarcinoma of the esophagus, junction, and gastric cardia. Gastroenterology. 2010;139:1995–2004. doi: 10.1053/j.gastro.2010.05.080. [e1915] [DOI] [PubMed] [Google Scholar]
- 46.Hirai S, Endo S, Saito R, Hirose M, Ueno T, Suzuki H, Yamato K, Abei M, Hyodo I. Antitumor effects of a sirtuin inhibitor, tenovin-6, against gastric cancer cells via death receptor 5 up-regulation. PLoS One. 2014;9 doi: 10.1371/journal.pone.0102831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shen X, Li P, Xu Y, Chen X, Sun H, Zhao Y, Liu M, Zhang W. Association of sirtuins with clinicopathological parameters and overall survival in gastric cancer. Oncotarget. 2017;8:74359–74370. doi: 10.18632/oncotarget.20799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cancer Genome Atlas Research N Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–209. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cancer Genome Atlas Research N, Analysis Working Group: Asan U, Agency BCC, Brigham, Women's H, Broad I, Brown U, Case Western Reserve U, Dana-Farber Cancer I, Duke U Integrated genomic characterization of oesophageal carcinoma. Nature. 2017;541:169–175. doi: 10.1038/nature20805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Le A, Cooper CR, Gouw AM, Dinavahi R, Maitra A, Deck LM, Royer RE, Vander Jagt DL, Semenza GL, Dang CV. Inhibition of lactate dehydrogenase A induces oxidative stress and inhibits tumor progression. Proc Natl Acad Sci U S A. 2010;107:2037–2042. doi: 10.1073/pnas.0914433107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xiao LJ, Lin P, Lin F, Liu X, Qin W, Zou HF, Guo L, Liu W, Wang SJ, Yu XG. ADAM17 targets MMP-2 and MMP-9 via EGFR-MEK-ERK pathway activation to promote prostate cancer cell invasion. Int J Oncol. 2012;40:1714–1724. doi: 10.3892/ijo.2011.1320. [DOI] [PubMed] [Google Scholar]
- 52.Xiao Y, Wang J, Qin Y, Xuan Y, Jia Y, Hu W, Yu W, Dai M, Li Z, Yi C. Ku80 cooperates with CBP to promote COX-2 expression and tumor growth. Oncotarget. 2015;6:8046–8061. doi: 10.18632/oncotarget.3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li Y, Luo S, Ma R, Liu J, Xu P, Zhang H, Tang K, Ma J, Zhang Y, Liang X. Upregulation of cytosolic phosphoenolpyruvate carboxykinase is a critical metabolic event in melanoma cells that repopulate tumors. Cancer Res. 2015;75:1191–1196. doi: 10.1158/0008-5472.CAN-14-2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Choi C, Helfman DM. The Ras-ERK pathway modulates cytoskeleton organization, cell motility and lung metastasis signature genes in MDA-MB-231 LM2. Oncogene. 2014;33:3668–3676. doi: 10.1038/onc.2013.341. [DOI] [PubMed] [Google Scholar]
- 55.Frezza C, Zheng L, Folger O, Rajagopalan KN, MacKenzie ED, Jerby L, Micaroni M, Chaneton B, Adam J, Hedley A. Haem oxygenase is synthetically lethal with the tumour suppressor fumarate hydratase. Nature. 2011;477:225–228. doi: 10.1038/nature10363. [DOI] [PubMed] [Google Scholar]