Abstract
Water constitutes and sustains life; however, its pollution afflicts its necessity, further worsening its scarcity. Coliform is one of the largest groups of bacteria evident in fecally polluted water, a major public health concern. Coliform thrive as commensals in the gut of warm-blooded animals, and are indefinitely passed through their feces into the environment. They are also called as model organisms as their presence is indicative of the prevalence of other potential pathogens, thus coliform are and unanimously employed as adept indicators of fecal pollution. As only a limited accessible source of fresh water is available on the planet, its contamination severely affects its usability. Coliform densities vary geographically and seasonally which leads to the lack of universally uniform regulatory guidelines regarding water potability often leads to ineffective detection of these model organisms and the misinterpretation of water quality status. Remedial measures such as disinfection, reducing the nutrient concentration or re-population doesn’t hold context in huge lotic ecosystems such as freshwater rivers. There is also an escalating concern regarding the prevalence of multi-drug resistance in coliforms which renders antibiotic therapy incompetent. Antimicrobials are increasingly used in household, clinical, veterinary, animal husbandry and agricultural settings. Sub-optimal concentrations of these antimicrobials are unintentionally but regularly dispensed into the environment through seepages, sewages or runoffs from clinical or agricultural settings substantially adding to the ever-increasing pool of antibiotic resistance genes. When present below their minimum inhibitory concentration (MIC), these antimicrobials trigger the transfer of antibiotic-resistant genes that the coliform readily assimilate and further propagate to pathogens, the severity of which is evidenced by the high Multiple Antibiotic Resistance (MAR) index shown by the bacterial isolates procured from the environmental. This review attempts to assiduously anthologize the use of coliforms as water quality standards, their existent methods of detection and the issue of arising multi-drug resistance in them.
Keywords: coliform, E. coli, fecal pollution, multi-drug resistance, indicator bacteria, gut microflora, MAR index
Introduction
Water, the elixir of life is indispensable in every facet of existence. Apart from quotidian uses, water is also important in many industries for the production of chemicals, cosmetics, and beverages (Lamikanra, 1999; Mishra et al., 2012). The importance of rivers, the major source of freshwater, is well evidenced by the historic fact that major civilizations were settled along the banks of a river. However, water’s indispensability is handicapped by its pollution which renders it unusable. Water polluted by influx of sewage especially from hospitals, public defecation, slaughter houses, clinics, and animal husbandry contains huge amount of antibiotic resistant bacteria which can ultimately be transferred to humans or livestock (Ahmed et al., 2010; Crimmins and Beltrán-Sánchez, 2011; Carlet et al., 2012). One of the major causes of child mortality under five is waterborne illnesses (WHO and UNICEF, 2000; Howard and Bartram, 2003). The group of coliform bacteria from the family Enterobacteriaceae was chosen as water potability indicators as they inherently populate the gut of warm blooded mammals; end up in their feces and indicate the presence of other pathogens. (Berg and Metcalf, 1978; Mishra et al., 2012; Mishra et al., 2013). These bacteria are very receptive to drug resistance genes and also readily propagate it to the other pathogens prevalent in the vicinity, therefore pose as potent health hazards. Hence, regular monitoring of coliform levels in the environment provides insight into the status of water potability, warns prior incidence of various public health concerns and paves way for designing remedial measures.
The present review focuses on the concern of fecal pollution in surface waters, the use of coliform as water quality indicators, and the alarming incidence of multi-drug resistance in coliforms posing as health risk.
Pollution of River Water and Coliform as Sanitary Standards
The United Nations confirms that almost 2 million tons of waste finds its way to the water bodies globally (UNWAP, 2003; Ross, 2010). About 1.2 billion people defecate along the banks of the rivers in developing and under-developed nations (WHO and UNICEF, 2008). Seventy percentage of the industrial wastes have been estimated to be disposed untreated into the water supplies (UN-WATER, 2009). Although the treated sewage allowed to be discharged into the rivers should not exceed the Biochemical Oxygen Demand (BOD) limit of 30 mg/L, Total Suspended Solids (TSS) limit of 50 mg/L (Tyagi et al., 2013), a mere 18% of the 33,000 mld sewage spawned daily is treated. Several investigations have been undertaken over time to monitor water quality and more precisely its potability; among all the parameters available, the microbiological ones largely influence water usability, as has been established by many studies reporting high levels of contamination in the freshwater bodies (WHO, 2006; Mishra et al., 2012, 2013; Harnisz et al., 2015; Parvez et al., 2016; Gomes Freitas et al., 2017).
Fecal and Non-fecal Coliform
The group of coliform is further subdivided into total and fecal coliforms while total includes both soil intermediates and fecal forms, fecal coliforms confines to those from fecal origin, used as standard microbial indicators of water quality since 1920 (Ashbolt et al., 2001). The fecal coliform group, i.e., Escherichia coli (E. coli), Klebsiella, and Enterobacter inhabit the intestinal tract of warm blooded animals (Dufour, 1977). The use of coliform bacteriological analysis for water potability testing dates back to 1880 when Von Fritsch found Klebsiella pneumoniae and K. rhinoscleromatis in human feces (Geldreich, 1978). Even the USEPA recommends the use of E. coli as efficient indicator for evaluating freshwater quality (United States Environmental Protection Agency [USEPA], 1986).
Methods of Detection- Past and Present
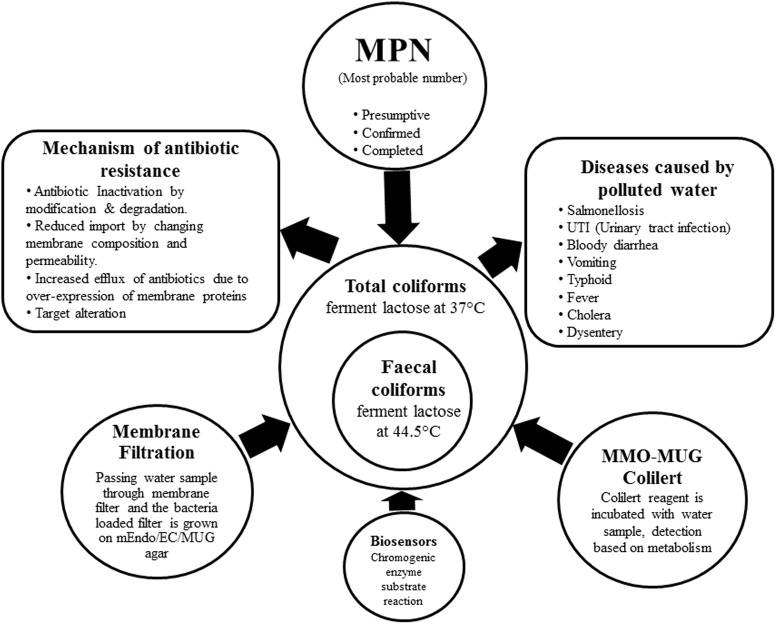
Percy and Grace counted bacteria from water on Robert Koch’s solid gelatin media (Hutchinson and Ridgway, 1977), then Theodor Escherich discovered Bacillus coli (Escherich, 1885); later renamed as E. coli by Castellani and Chambers (1919). By 1893, the ‘Wurtz method’ of enumerating B. coli by directly plating water samples on litmus lactose agar employing the idea of acid formation from lactose fermentation was used by sanitary bacteriologists (Ashbolt et al., 2001). In 1893 Durham’s tubes were used for detecting gas formation from fermentation (Durham, 1898). In 1905 MacConkey’s broth was used as a diagnostic for bile tolerant lactose-fermenting bacteria (MacConkey, 1905). Till date the most preferred and cost-effective method for elaborate detection of coliforms is the Most Probable Number (MPN) comprising of the presumptive, confirmed and completed test. A set of double and single strength lactose broth with inverted durham tubes are inoculated with measured amounts of water to be tested, coliform ferment the lactose present in the broth producing acid and gas. The MPN of coliforms present in 100 ml of water is estimated by the number of positive tubes against the MPN table. Samples from the positive tubes are streaked on EMB agar plates in the confirmed test, discrete black nucleated colonies with green metallic sheen are E. coli. Single colonies are re-inoculated into the Brilliant Green Bile Lactose Broth, while bile inhibits the growth of gram-positive microbes; brilliant green bars the propagation of certain gram-negative bacilli allowing the growth of lactose-fermenting coliforms (Mishra et al., 2012, 2013). To test for thermo-tolerant coliform modified Eijkman test has been proposed by the WHO based on the principle that only fecal coliforms are able to ferment lactose at 44.5°C, hence MacConkey broth with inverted durham’s tubes are inoculated with measured amounts of water samples and are incubated at 44.5°C for the occurrence of acid and/or gas (WHO, 2011). Other existent methods of detection are the membrane filter and MMO-MUG (Colilert®). The different methods of detection have been illustrated in the Figure 1 along with categorization of coliform, their health hazards and mechanism of drug resistance.
FIGURE 1.
The figure lucidly depicts the categorization of coliforms into total coliforms including both soil intermediates and from faecal origin and strictly faecal coliforms. The different methods of detection of coliform from water sample are enumerated. Mechanism of antibiotic resistance developed by coliform and diseases caused by the use of polluted water have been illustrated in the diagram.
An efficient sensor based presence/absence method has also been established for confirming the presence of E. coli. The chromogenic substrate β-galactosidase enzyme assay detects the color change through a live webcam with Wi-Fi connected sensor node, in the water sample when mixed with X-gal containing reagent (Kim and Myung, 2015). Biosensors have also been developed to check the presence of waterborne pathogens in food (Thakur and Ragavan, 2013). Although several other methods such as ELISA and PCR are also existent, however, they are laboratory based methods which necessitate insolation, pure culture of the organism, also require expert personnel, expensive instruments, multiple steps and do not offer real-time data (Yoon and Kim, 2012). With relatively fast generation times but inability to survive longer in limited water samples the duration from field-to-lab massively affects the coliform numbers often leading to inaccurate quantification. Paper based microfluidic diagnostic devices though relatively inexpensive are largely single test based (Yetisen et al., 2013). Nonetheless, when it comes to large water bodies being polluted mere detection of coliform presence is not enough, it needs complete quantitation of the degree of pollution for remedial steps to be devised; hence inexpensive traditional counting methods which give real-time numbers are preferred. So far only one study has professed the use of sensor for detecting and enumerating E. coli in water (Hesari et al., 2016). All the methods available have been enlisted in Table 1.
Table 1.
The various methods developed to detect the presence of coliforms.
| Method (Use of specific media/method/chemical) | Principle | References |
|---|---|---|
| Use of Robert Koch’s solid gelatin media | Counting bacteria based on colony forming unit | Hutchinson and Ridgway, 1977 |
| Wurtz method | Enumerating B. Coli based on lactose fermentation | Ashbolt et al., 2001 |
| Use of durham’s tubes | Positive identification of coliform based on acid and gas production from lactose fermentation | Durham, 1898 |
| Use of MacConkey’s broth | Coliform identification based on being bile tolerant lactose fermenters | MacConkey, 1905 |
| Most Probable Number | Quantitative test recommended by the World Health Organization. Based on lactose fermentation ability of coliforms forming acid and gas, capability of E. coli to reduce methylene blue to exhibit green metallic sheen on Eosin Methylene Blue agar, competence of coliforms to ferment lactose producing gas in the presence of bile | American Public Health Association (APHA), 1992; Clesceri et al., 1996 Mishra et al., 2012; Mishra et al., 2013 |
| Modified Eijkman test | Quantitative test recommended by the World Health Organization. Based on lactose fermentation knack of strictly fecal coliforms forming acid and gas | WHO, 2011;Mishra et al., 2012, 2013 |
| Membrane filtration | Usage of filters would trap the coliforms which could be counted in terms of CFU on media | World Health Organization [WHO] (1996) |
| MMO-MUG (Colilert®, Coliscan®, and Colitag®) | Enzyme based detection of E. coli | United States Environmental Protection Agency [USEPA], 2002 |
| ELISA and PCR | Antibody specificity | Yoon and Kim, 2012 |
| Paper based microfluidic diagnostics | Based on catalase test | Yetisen et al., 2013 |
| Biosensors | Chromogenic substrate- enzyme interaction based | Thakur and Ragavan, 2013; Kim and Myung, 2015; Hesari et al., 2016 |
Need for Universally Uniform Regulatory Guidelines, Continued Surveillance and Detailed Studies
Monitoring fecal pollution of waters used for recreational drinking, and industrial purposes is imperative for public health and economic reasons. Pathogens introduced from fecal contamination leads to diseases in humans (typhoid, Salmonellosis, cholera), livestock and economic losses to industries (Ballester and Sunyer, 2000; Bernhard and Field, 2000).
Coliform densities are influenced by seasonal changes. While in the tropical climate coliform levels are higher during monsoon, owing to sewage seepage and runoffs, in the comparatively cooler regions they are higher during summer due to ambient growth temperature (Kar et al., 2010; Galadima et al., 2011; Panigrahi and Patra, 2013). This geographically inconsistent season based variation in coliform numbers had led to ambiguous standards which don’t hold context in every reference. The stringency in the guidelines was finalized owing to global health concern reports. As per the WHO, there should not be any E. coli or thermo-tolerant coliform detectable in 100 ml of water sample (WHO, 1998, 2011).
Most studies conducted on coliforms only report the MPN level indicating the water potability and no detailed work to further identify and type the procured coliform is done. Routine cultural characterization, biochemical fingerprinting, antibiogram analysis and molecular typing are absolutely indispensable for their genotypic and phenotypic characterization. Even within the same species there occur differences because of variations in the source of bacterial origin, their present habitat and mutations which are unraveled in taxonomic studies.
While coliforms are established as sanitary standards, the startling issue of multi-drug resistance in them is also was on the rise. Coliforms are extremely receptive and altruistic to horizontal gene transfer in nature (Mishra et al., 2013; von Wintersdorff et al., 2016). Hence, many aberrations to the stipulated traits of coliforms have been reported. Unusual Sucrose fermentation was reported by Palchaudhuri et al. (1977) the multi-drug resistant (MDR) strain of E. coli also carried genes for resistance against Streptomycin, Ampicillin, Tetracycline, Chloramphenicol, and sulphonamides. Atypical raffinose fermentation linked with Verocytotoxigenic E. coli (de Lopez et al., 1982; Saif et al., 2008) is plasmid mediated with genes for antibiotic resistance (Orskov and Orskov, 1973; Cornelis et al., 1978). Non-fermentation of rhamnose is a decisive factor of VT+E. coli 0157 (Chapman et al., 1991). The biotype (Rha- and Suc-)” is a quintessential marker for the preliminary detection of SLT-I+ E. coli in SLTEC associated diarrhea (Wieler et al., 1995). The occurrence of certain atypical carbohydrate fermentation genes is largely coupled with multi-drug resistance. These genes are transferred both intra and inter-specifically through horizontal gene transfer mechanism bestowing multi-drug resistance to the isolates which changes their biochemical characteristics. Numerous investigations employ biochemical fingerprinting for identifying coliform (Gililland and Vaughn, 1943; Kühn et al., 1991; Guentzel, 1996; Betsy and Keogh, 2005). While results obtained by most of the biochemical tests are qualitative and used mostly for the identification of the bacterial isolate into genus and species but carbohydrate fermentation, antimicrobial susceptibility, genotypic profiling characterize the bacterial isolate down to the strain level. When coupled together these analyses well substantiate the identification of an individual strain with detailed characterization of its standalone traits. Carson et al. (2001) have used 16S rDNA analysis to discriminate fecal E. coli of human source from collective fecal E. coli isolates of non-human origins and have obtained accuracy up to three host sources. 16S rDNA analysis has been used to differentiate isolates within the same species based on taxonomic classification (Laurent et al., 1996), epidemiological tracking (Bernhard and Field, 2000; Zhao et al., 2005), geographical distribution (Akhtar et al., 2000), population biology and phylogeny (Paradis et al., 2005; Xia et al., 2013).
Antibiotic Resistance in Gut Coliform
Though tagged as “miracle drugs,” antibiotics are increasingly being misused. Their unsystematic use and dumping has led to multi-drug resistance among the indigenous microbiota, in case of a re-infection these antibiotics become futile for therapy. The increasing use of antibiotics in clinical, agricultural and veterinary sectors is analogous with the escalated resistance of bacteria to these frequently used antibiotics (Dhanorkar and Tambekar, 2004; Ahmed et al., 2010). This has exaggerated the expenditure of therapy, the peril of spreading MDR, mortality from diseases through the MDR strains (Martínez and Baquero, 2002; White, 2006; Hawkey and Jones, 2009; Kolar et al., 2010). Antibiotic resistance has been established to be broadcasted in the gut of the warm-blooded animals (Chopra and Roberts, 2001; Launay et al., 2004; Fukao and Yajima, 2012). Studies by Larsson and Fick (2009) and Laxminarayan and Heymann (2012) have reported seepage of about 45 kg of Ciprofloxacin quotidian from factories into the nearby water bodies. Spread of environmental pollution has led to the unearthing of antibiotic-resistant bacteria in uninhabited lands like Antarctica (Sjölund et al., 2008; Hernández et al., 2012). Prolongation of disease either due to scarcity of antibiotics or superfluous use of antibiotics in circumstances where their use is not obligatory is common. Antibiotics usage has seen a precipitous boost over the past few years. Sale of cephalosporins amplified by 60% over 2005–2009, sales of other last line antibiotics escalated six-fold from 2005 to 2010 (Ganguly et al., 2011; Westly, 2012). Drug resistant coliform are the most commonly infecting bacteria, infection is contracted by exposure to unhygienic food, water, to the bacteria or its carriers and unwise use of antibiotics. Hannah et al. (2005) have investigated the association of drug resistant E. coli infections with epidemiological factors: demographic variables, diet, healthcare and antibiotic usage and have found positive correlation between infection with MDR E. coli and contact with infected persons, contaminated food, ambulatory visit, antimicrobial misuse, prolonged medication and frequent meat consumption with self-prescription of drugs. Over-consumption of antibiotics kills the beneficial bacteria in our system enhancing the growth of pathogens which may be insensitive to the antibiotics used thus eventually becoming resistant. Consuming antibiotics lesser than the prescribed amount does not kill the pathogen rather confers resistance upon it. In both the cases treating the MDR bacteria becomes arduous with often, fatal consequences. Antimicrobial susceptibility with the production of Extended Spectrum Beta-Lactamases (ESBL) among the coliform isolates causes concern as they render therapy with beta-lactum antibiotics inefficient (Pitout and Laupland, 2008; Peirano and Pitout, 2010; Rogers et al., 2011; Shibl et al., 2012).
Many investigations have stressed on the concern of multi-drug resistant coliforms in water (Manji et al., 2012; Mishra et al., 2013; Al-Agamy et al., 2014; El-Zanfaly, 2015; Madec et al., 2016; Azzam et al., 2017; Jutkina et al., 2018). Manji et al. (2012) have emphasized concern over the prevalence of multi-drug resistant coliforms and Staphylococcus aureus, in rural water supplies of Nigeria. Over the period of several months their analysis involved bacteria counting, antibiogram and MIC analysis. The presence of pathogens as S. aureus, Bacillus sp., Pseudomonas aeruginosa, with umpteen antibiotics resistant E. coli strains warrant immediate treatment as they pose momentous public health repercussions.
Mishra et al. (2013) have worked on the prevalence of antibiotic resistant E. coli in the River Mahanadi. The water there unintentionally experiences influx of miscellaneous composition. They have employed MPN, Modified Eijkman test, biochemical fingerprinting, antimicrobial susceptibility testing and 16S rDNA ribotyping methods to isolate, enumerate and identify the coliform isolates to be E. coli with established resistance to beta-lactum antibiotics, carboxypenicillin coupled with β-lactamase inhibitor, glycopeptides, carbapenems, macrolides, till fourth generation of fluoroquinolones cephalosporins. They have also reported appalling resistance rates of the isolates against 42 indigenously used antibiotics with MAR indices ranging from 0.51 to 0.90 which indicate severe pollution and risk to the public health.
Transfer of antibiotic resistance cannot be ascribed to horizontal gene transfer mechanisms alone rather integrons too play an important role in certain cases. Xia et al. (2013) used 16S rDNA analysis to accurately isolate coliforms, harboring class 2 integrons and investigated their molecular architecture.
Al-Agamy et al. (2014) have characterized the prevalence of ESBL Escherichia coli (ESBL-EC) in Riyadh. They used antimicrobial susceptibility and pulsed field gel electrophoresis (PFGE) to type and investigate the epidemiology of ESBL-EC and the prevalence of ST131 in them. They have also expressed concern over the dearth of local reports on the prevalence of ESBL-EC despite its universal predominance. Such state of affairs demands the routine and vigilant inspection of the environment.
El-Zanfaly (2015), apprehends that resistance to antibiotics may actually escalate during the sewage treatment processes his study has employed enumeration, antimicrobial susceptibility, granular activated carbon test and transferability of antibiotic resistant character tests to characterize ampicillin, sulfaguanidine penicillin, 2-sulfanilamide pyrimidine, tetracycline, chloramphenicol, neomycin and streptomycin resistant coliforms. Activated carbon application in pilot water treatment plant experiment showed their prevalence with easy transferability of resistance. The work insists on including antibiotic resistance in coliforms as a deciding criterion for judging water quality standards.
Madec et al. (2016) have stressed on the implication of antimicrobial resistance in drinking water as a potent risk indicator for humans in low-income nations. A particular E. coli isolate ST48, procured from drinking water in France, resistant to ceftiofur, tetracyclines and sulfonamides was found to harbor the blaCTX-M-1 IncI1/ST3 plasmid. The plasmid is analogous to the other blaCTX-M-1 IncI1/ST3 plasmids that have been reported in other animals and humans. Based upon their findings they suggest the possibility of human transfer of ESBL genes through drinking water in developing and under-developed nations.
Jutkina et al. (2018) conducted a series of conjugation and MIC assays of both donor and recipient strains to investigate the latent of cefotaxime, ciprofloxacin, gentamicin, erythromycin, sulfamethoxazole, trimethoprim, and chlorhexidine digluconate, hexadecyl trimethyl ammonium chloride and triclosan to stimulate horizontal gene transfer of antibiotic resistance genes. They found that 24.4 μg/L Chlorhexidine (200 times <MIC of the recipient) 0.1 mg/L triclosan (1/20 < MIC), 0.1 mg/L gentamicin (1/16 < MIC) and 1 mg/L sulfamethoxazole (1/16 < MIC) potently escalated the frequencies of antibiotic resistance gene transfer. The study clearly establishes that the presence of certain antimicrobials below their MICs in nature can induce the horizontal transfer and spread of antibiotic resistance.
Conclusion
It is indispensable to study the bacterial isolates endemic to an ecological area in order to devise preventive and remedial mechanisms in the event of nuisance created by it. Owing to the exorbitant gene transfer mechanisms bacteria have been found to vary epidemiologically even within the same species. Knowledge of clinically imperative attributes as virulence and antimicrobial susceptibility is absolutely indispensable both for designing remedial measures and preventive therapy. Therefore, it is essential to thoroughly characterize and report different isolates that vary and adapt according to the changing surroundings. The work holds utmost importance in developing and under-developed countries where parameters for regular monitoring of water quality is limited and advanced molecular biology tools for immediate identification are not ubiquitous, field-to-lab distance is more, availability of skilled personnel is scarce, seasonal abundance of coliform is atypical and quotidian use of natural water body is prevalent. Thus, isolation and complete characterization of water-borne bacteria are quintessential to answer clinically relevant problems.
Author Contributions
MM designed, conceived, and wrote the manuscript. AA and AP helped in writing. NB, TM, and DY critically reviewed, edited, and finalized the manuscript for submission.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are thankful to Dr. Nanda Kishore Mishra (Department of English, Gangadhar Meher University, Odisha) for improving the English language quality.
References
- Ahmed M. O., Clegg P. D., Williams N. J., Baptiste K. E., Bennett M. (2010). Antimicrobial resistance in equine faecal Escherichia coli isolates from North West England. Ann. Clin. Microbiol. Antimicrob. 9:12. 10.1186/1476-0711-9-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhtar M. K., Kaderbhai N., Kaderbhai M. A. (2000). Growth of Escherichia coli on medium containing glycine increases transformation efficiency. Anal. Biochem. 277 273–276. 10.1006/abio.1999.4400 [DOI] [PubMed] [Google Scholar]
- Al-Agamy M. H., Shibl A. M., Hafez M. M., Al-Ahdal M. N., Memish Z. A., Khubnani H. (2014). Molecular characteristics of extended-spectrum β-lactamase-producing Escherichia coli in Riyadh: emergence of CTX-M-15-producing E. coli ST131. Ann. Clin. Microbiol. Antimicrob. 13:4. 10.1186/1476-0711-13-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Public Health Association (APHA) American Water Works Association (AWWA) and Water Pollution Control Federation (WPCF) (1992). Standard Methods for the Examination of Water and Wastewater 18th Edn. Washington, DC: APHA Publication Office. [Google Scholar]
- Ashbolt N. J., Grabow W. O., Snozzi M. (2001). “Indicators of microbial quality,” in Water Quality: Guidelines, Standards and Health, eds Fewtrell F., Batrem J. (Geneva: World Health Organization; ) 289–316. [Google Scholar]
- Azzam M. I., Ezzat S. M., Othman B. A., El-Dougdoug K. A. (2017). Antibiotics resistance phenomenon and virulence ability in bacteria from water environment. Water Sci. 31 109–121. 10.1016/j.wsj.2017.10.001 [DOI] [Google Scholar]
- Ballester F., Sunyer J. (2000). Drinking water and gastrointestinal disease: need of better understanding and an improvement in public health surveillance. J. Epidemiol. Commun Health 54 3–5. 10.1136/jech.54.1.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg G., Metcalf T. G. (1978). “Indicators of viruses in waters,” in Indicators of Viruses in Water and Food ed. Berg G. (Ann Arbor, MI: Ann Arbor Science; ) 267–296. [Google Scholar]
- Bernhard A. E., Field K. G. (2000). Identification of nonpoint sources of fecal pollution in coastal waters by using host-specific 16S ribosomal DNA genetic markers from fecal anaerobes. Appl. Environ. Microbiol. 66 1587–1594. 10.1128/AEM.66.4.1587-1594.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betsy T., Keogh J. E. (2005). Microbiology Demystified. New York, NY: McGraw-Hill. [Google Scholar]
- Carlet J., Jarlier V., Harbarth S., Voss A., Goossens H., Pittet D. (2012). Ready for a world without antibiotics? The pensières antibiotic resistance call to action. Antimicrob. Resist. Infect. Control 1:11. 10.1186/2047-2994-1-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson C. A., Shear B. L., Ellersieck M. R., Asfaw A. (2001). Identification of Fecal Escherichia coli from Humans and Animals by Ribotyping. Appl. Environ. Microbiol. 67 1503–1507. 10.1128/AEM.67.4.1503-1507.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellani A., Chambers A. J. (1919). Manual of Tropical Medicine 3rd Edn. New York, NY: William Wood & Company. 10.5962/bhl.title.84653 [DOI] [Google Scholar]
- Chapman P., Siddons C. A., Zadik P., Jewes L. (1991). An improved selective medium for the isolation of Escherichia coli O 157. J. Med. Microbiol. 35 107–110. 10.1099/00222615-35-2-107 [DOI] [PubMed] [Google Scholar]
- Chopra I., Roberts M. (2001). Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 65 232–260. 10.1128/MMBR.65.2.232-260.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clesceri L. S., Greenberg A. E., Eaton A. D. (1996). Standard Methods for the Examination of Water and Wastewater. Washington DC: APHA. [Google Scholar]
- Cornelis G., Luke R., Richmond M. (1978). Fermentation of raffinose by lactose-fermenting strains of Yersinia enterocolitica and by sucrose-fermenting strains of Escherichia coli. J. Clin. Microbiol. 7 180–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crimmins E. M., Beltrán-Sánchez H. (2011). Mortality and morbidity trends: is there compression of morbidity? J. Gerontol. Series B 66 75–86. 10.1093/geronb/gbq088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lopez A. G., Kadis S., Shotts E. B. (1982). Transfer of drug resistance and enterotoxin production in porcine Escherichia coli strains and relationship between K88 antigen and raffinose (melitose) fermentation. Am. J. Vet. Res. 43 499–501. [PubMed] [Google Scholar]
- Dhanorkar D., Tambekar D. (2004). “Studies on multidrug resistance pattern of clinical isolates,” in Proceedings of the 45th Annual Conference of Association of Microbiologist of India, NDRL Karnal. [Google Scholar]
- Dufour A. (1977). “Escherichia coli: the fecal coliform,” in Bacterial Indicators/Health Hazards Associated with Water Hoadley A. A., Dutka B. J. (West Conshohocken, PA: ASTM International; ). [Google Scholar]
- Durham H. E. (1898). A simple method for demonstrating the production of gas by bacteria. Br. Med. J. 1:1387 10.1136/bmj.1.1952.1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Zanfaly H. T. (2015). Antibiotic resistant bacteria: a factor to be considered in safe drinking water. Environ. Prot. Sustain. Dev. 1 134–143. [Google Scholar]
- Escherich T. (1885). Die Darmbacterien des Neugeborenen und Säuglings I, II. Fortschr. Med. 3 547–554. [Google Scholar]
- Fukao M., Yajima N. (2012). “Assessment of antibiotic resistance in probiotic lactobacilli,” in Antibiotic Resistant Bacteria—A Continuous Challenge in the New Millennium ed. Pana M. (Rijeka: InTech; ), 503–512. [Google Scholar]
- Galadima A., Garba Z., Leke L., Almustapha M., Adam I. (2011). Domestic water pollution among local communities in Nigeria-causes and consequences. Eur. J. Sci. Res. 52 592–603. 12171368 [Google Scholar]
- Ganguly N. K., Arora N. K., Chandy S. J., Fairoze M. N., Gill J. P., Gupta U., et al. (2011). Rationalizing antibiotic use to limit antibiotic resistance in India. Indian J. Med. Res. 134 281–294. [PMC free article] [PubMed] [Google Scholar]
- Geldreich E. E. (1978). “Bacterial populations and indicator concepts in feces, sewage, stormwater and solid wastes,” in Indicators of Viruses in Water and Food ed. Berg G. (Ann Arbor, MI: Ann Arbor Science Publishers Inc; ) 51–96. [Google Scholar]
- Gililland J. R., Vaughn R. H. (1943). Biochemical characteristics of pigmented coliform bacteria. J. Bacteriol. 45 499–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes Freitas D., Silva R. D. R., Bataus L. A. M., Barbosa M. S., Da Silva Bitencourt Braga C. A., Carneiro L. C. (2017). Bacteriological water quality in school’s drinking fountains and detection antibiotic resistance genes. Ann. Clin. Microbiol. Antimicrob. 16:5. 10.1186/s12941-016-0176-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guentzel M. N. (1996). “Escherichia, Klebsiella, Enterobacter, Serratia, Citrobacter, and Proteus,” in Medical Microbiology 4th Edn ed. Baron S. (Galveston, TX: University of Texas, Medical Branch; ). [PubMed] [Google Scholar]
- Hannah E. L., Angulo F. J., Johnson J. R., Haddadin B., Williamson J., Samore M. H. (2005). Drug-resistant Escherichia coli, rural Idaho. Emerg. Infect. Dis. 11 1614–1617. 10.3201/eid1110.050140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harnisz M., Korzeniewska E., Gołaś I. (2015). The impact of a freshwater fish farm on the community of tetracycline-resistant bacteria and the structure of tetracycline resistance genes in river water. Chemosphere 128 134–141. 10.1016/j.chemosphere.2015.01.035 [DOI] [PubMed] [Google Scholar]
- Hawkey P. M., Jones A. M. (2009). The changing epidemiology of resistance. J. Antimicrob. Chemother. 64 i3–i10. 10.1093/jac/dkp256 [DOI] [PubMed] [Google Scholar]
- Hernández J., Stedt J., Bonnedahl J., Molin Y., Drobni M., Calisto-Ulloa N., et al. (2012). Human-associated extended-spectrum β-lactamase in the Antarctic. Appl. Environ. Microbiol. 78 2056–2058. 10.1128/AEM.07320-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesari N., Alum A., Elzein M., Abbaszadegan M. (2016). A biosensor platform for rapid detection of Escherichia coli in drinking water. Enzyme Microb. Technol. 83 22–28. 10.1016/j.enzmictec.2015.11.007 [DOI] [PubMed] [Google Scholar]
- Howard G., Bartram J. (2003). Domestic Water Quantity: Service Level and Health. Geneva: World Health Organization. [Google Scholar]
- Hutchinson M., Ridgway J. (1977). Microbiological Aspects of Drinking Water Supplies. London: Academic Press. [Google Scholar]
- Jutkina J., Marathe N., Flach C.-F., Larsson D. (2018). Antibiotics and common antibacterial biocides stimulate horizontal transfer of resistance at low concentrations. Sci. Total Environ. 616 172–178. 10.1016/j.scitotenv.2017.10.312 [DOI] [PubMed] [Google Scholar]
- Kar P., Pani K., Pattanayak S., Sahu S. (2010). Seasonal variation in physico-chemical and microbiological parameters of mahanadiriver water in and around Hirakud, Orissa (India). Ecoscan 4 263–271. [Google Scholar]
- Kim K., Myung H. (2015). Sensor node for remote monitoring of waterborne disease-causing bacteria. Sensors 15 10569–10579. 10.3390/s150510569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolar M., Bardon J., Chroma M., Hricova K., Stosova T., Sauer P., et al. (2010). ESBL and AmpC beta-lactamase-producing Enterobacteriaceae in poultry in the Czech Republic. Vet. Med. 55 119–124. 10.17221/165/2009-VETMED [DOI] [Google Scholar]
- Kühn I., Allestam G., Stenström T., Möllby R. (1991). Biochemical fingerprinting of water coliform bacteria, a new method for measuring phenotypic diversity and for comparing different bacterial populations. Appl. Environ. Microbiol. 57 3171–3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamikanra A. (1999). Essential Microbiology for Students and Practitioner of Pharmacy, Medicine and Microbiology 2nd Edn. Lagos: Amkra books; 406. [Google Scholar]
- Larsson D. J., Fick J. (2009). Transparency throughout the production chain—a way to reduce pollution from the manufacturing of pharmaceuticals? Regul. Toxicol. Pharmacol. 53 161–163. 10.1016/j.yrtph.2009.01.008 [DOI] [PubMed] [Google Scholar]
- Launay F., Young P., Sterk S., Blokland M., Kennedy D. (2004). Confirmatory assay for zeranol, taleranol and the Fusarium spp. toxins in bovine urine using liquid chromatography-tandem mass spectrometry. Food Addit. Contam. 21 52–62. 10.1080/02652030310001636930 [DOI] [PubMed] [Google Scholar]
- Laurent F., Carlotti A., Boiron P., Villard J., Freney J. (1996). Ribotyping: a tool for taxonomy and identification of the Nocardia asteroides complex species. J. Clin. Microbiol. 34 1079–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laxminarayan R., Heymann D. L. (2012). Challenges of drug resistance in the developing world. BMJ 344:e1567. 10.1136/bmj.e1567 [DOI] [PubMed] [Google Scholar]
- MacConkey A. (1905). Lactose-fermenting bacteria in faeces. Epidemiol. Infect. 5 333–379. 10.1017/S002217240000259X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madec J.-Y., Haenni M., Ponsin C., Kieffer N., Rion E., Gassilloud B. (2016). Sequence Type 48 Escherichia coli carrying the blaCTX-M-1 IncI1/ST3 plasmid in drinking water in france. Antimicrob. Agents Chemother. 60 6430–6432. 10.1128/AAC.01135-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manji P., Antai S., Jacob I. (2012). Incidence of Staphylococcus aureus, coliforms and antibiotic resistant strains of Escherichia coli in rural water supplies in Calabar South Local Government Area. J. Public Health Epidemiol. 4 230–237. 10.5897/JPHE12.055 [DOI] [Google Scholar]
- Martínez J. L., Baquero F. (2002). Interactions among strategies associated with bacterial infection: pathogenicity, epidemicity, and antibiotic resistance. Clin. Microbiol. Rev. 15 647–679. 10.1128/CMR.15.4.647-679.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra M., Patel A., Behera N. (2013). Prevalence of multidrug resistant Escherichia coli in the River Mahanadi of Sambalpur. Curr. Res. Microbiol. Biotechnol. 1 239–244. [Google Scholar]
- Mishra M., Patel A. K., Behera N. (2012). An assessment of coliform bacteria in the river Mahanadi system of Sambalpur. Bioscan 7 463–467. [Google Scholar]
- Orskov I., Orskov F. (1973). Plasmid-determined H2S character in Escherichia coli and its relation to plasmid-carried raffinose fermentation and tetracycline resistance characters. Microbiology 77 487–499. [DOI] [PubMed] [Google Scholar]
- Palchaudhuri S., Rahn S., Santos D., Maas W. (1977). Characterization of plasmids in a sucrose-fermenting strain of Escherichia coli. J. Bacteriol. 130 1402–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigrahi S., Patra A. (2013). Studies on seasonal variations in phytoplankton diversity of river Mahanadi, Cuttack city, Odisha, India. Indian J. Sci. Resour. 4 211–217. [Google Scholar]
- Paradis S., Boissinot M., Paquette N., Bélanger S. D., Martel E. A., Boudreau D. K., et al. (2005). Phylogeny of the Enterobacteriaceae based on genes encoding elongation factor Tu and F-ATPase β-subunit. Int. J. Syst. Evol. Microbiol. 55 2013–2025. 10.1099/ijs.0.63539-0 [DOI] [PubMed] [Google Scholar]
- Parvez A. K., Liza S. M., Marzan M., Ahmed A., Hahedur Rahman M. (2016). Bacteriological quality of drinking water samples across Bangladesh. Arch. Clin. Microbiol. 7:1. 25724966 [Google Scholar]
- Peirano G., Pitout J. D. (2010). Molecular epidemiology of Escherichia coli producing CTX-M β-lactamases: the worldwide emergence of clone ST131 O25: H4. Int. J. Antimicrob. Agents 35 316–321. 10.1016/j.ijantimicag.2009.11.003 [DOI] [PubMed] [Google Scholar]
- Pitout J. D., Laupland K. B. (2008). Extended-spectrum β-lactamase-producing Enterobacteriaceae: an emerging public-health concern. Lancet Infect. Dis. 8 159–166. 10.1016/S1473-3099(08)70041-0 [DOI] [PubMed] [Google Scholar]
- Rogers B. A., Aminzadeh Z., Hayashi Y., Paterson D. L. (2011). Country-to-country transfer of patients and the risk of multi-resistant bacterial infection. Clin. Infect. Dis. 53 49–56. 10.1093/cid/cir273 [DOI] [PubMed] [Google Scholar]
- Ross N. (2010). World Water Quality Facts and Statistics. Paris: UNESCO. [Google Scholar]
- Saif Y., Fadly A., Glisson J., Mcdougald L. (2008). “Newcastle disease, other avian paramyxoviruses, and pneumovirus infections,”in Diseases of Poultry 12th Edn eds Saif Y. M., Fadly A. M., Glisson J. R., McDougald L. R., Nolan L. K., Swayne D. E. (Ames, IA: Blackwell Publishing Professional; ) 75–93. [Google Scholar]
- Shibl A. M., Al-Agamy M. H., Khubnani H., Senok A. C., Tawfik A. F., Livermore D. M. (2012). High prevalence of acquired quinolone-resistance genes among Enterobacteriaceae from Saudi Arabia with CTX-M-15 β-lactamase. Diagn. Microbiol. Infect. Dis. 73 350–353. 10.1016/j.diagmicrobio.2012.04.005 [DOI] [PubMed] [Google Scholar]
- Sjölund M., Bonnedahl J., Hernandez J., Bengtsson S., Cederbrant G., Pinhassi J., et al. (2008). Dissemination of multidrug-resistant bacteria into the Arctic. Emerg. Infect. Dis. 14 70–72. 10.3201/eid1401.070704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur M., Ragavan K. (2013). Biosensors in food processing. J. Food Sci. Technol. 50 625–641. 10.1007/s13197-012-0783-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyagi S., Sharma B., Singh P., Dobhal R. (2013). Water quality assessment in terms of water quality index. Am. J. Water Resour. 1 34–38. [Google Scholar]
- United States. Environmental Protection Agency [USEPA] (1986). Office of Water Regulations, and Standards. Quality Criteria for Water, 1986. Washington, DC: U.S. Environmental Protection Agency. [Google Scholar]
- United States Environmental Protection Agency [USEPA] (2002). Method 1604: Total Coliforms and Escherichia coli in Water by Membrane Filtration Using a Simultaneous Detection Technique (MI Medium), EPA 821-R-02-024. Washington, D.C: U.S. Environmental Protection Agency. [Google Scholar]
- UNWAP (2003). United Nations World Water Assessment Programme. The World Water Development Report 1: Water for People, Water for Life. Paris: UNESCO. [Google Scholar]
- UN-WATER (2009). The United Nations World Water Development Report 3–Water in a Changing World. Paris: United Nations Educational Scientific and Cultural Organization. [Google Scholar]
- von Wintersdorff C. J., Penders J., Van Niekerk J. M., Mills N. D., Majumder S., Van Alphen L. B., et al. (2016). Dissemination of antimicrobial resistance in microbial ecosystems through horizontal gene transfer. Front. Microbiol. 7:173 10.3389/fmicb.2016.00173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westly E. (2012). India moves to tackle antibiotic resistance: drug regulator aims to restrict over-the-counter sales. Nature 489 192–193. 10.1038/489192a [DOI] [PubMed] [Google Scholar]
- White D. G. (2006). “Antimicrobial resistance in pathogenic Escherichia coli from animals,” in Antimicrobial Resistance in Bacteria of Animal Origin ed. Aarestrup F. M. (Washington, DC: ASM Press; ) 145–166. [Google Scholar]
- WHO (1998). Guidelines for Drinking-Water Quality Vol. 2 Health Criteria and Other Supporting Information: Addendum. Geneva: World Health Organization. [Google Scholar]
- WHO (2006). Guidelines for Drinking Water Quality: First Addendum to Third Edition. Geneva: World Health Organization. [PubMed] [Google Scholar]
- WHO (2011). Guidelines for Drinking-Water Quality. Geneva: World Health Organization; 303–304. [Google Scholar]
- WHO and UNICEF (2000). Global Water Supply and Sanitation Assessment 2000 Report. Geneva: World Health Organization. [Google Scholar]
- WHO and UNICEF (2008). Progress on Drinking Water and Sanitation: Special Focus on Sanitation. Geneva: World Health Organization. [Google Scholar]
- Wieler L., Bauerfeind R., Weiss R., Pirro F., Baljer G. (1995). Association of enterohemolysin and non-fermentation of rhamnose and sucrose with Shiga-like toxin genes in Escherichia coli from calves. Zentralbl. Bakteriol. 282 265–274. [PubMed] [Google Scholar]
- World Health Organization [WHO] (1996). WHO Acceptance of APHA Standard Methods. WHO Guidelines For Drinking Water Quality 2nd Edn, Vol. 1. (Geneva: World Health Organization; ) 14–22. [Google Scholar]
- Xia R., Ren Y., Guo X., Xu H. (2013). Molecular diversity of class 2 integrons in antibiotic-resistant gram-negative bacteria found in wastewater environments in China. Ecotoxicology 22 402–414. 10.1007/s10646-012-1034-9 [DOI] [PubMed] [Google Scholar]
- Yetisen A. K., Akram M. S., Lowe C. R. (2013). Paper based microfluidic point-of-care diagnostic devices. Lab Chip 13 2210–2251. 10.1039/c3lc50169h [DOI] [PubMed] [Google Scholar]
- Yoon J.-Y., Kim B. (2012). Lab-on-a-chip pathogen sensors for food safety. Sensors 12 10713–10741. 10.3390/s120810713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S., Maurer J. J., Hubert S., De Villena J. F., Mcdermott P. F., Meng J., et al. (2005). Antimicrobial susceptibility and molecular characterization of avian pathogenic Escherichia coli isolates. Vet. Microbiol. 107 215–224. 10.1016/j.vetmic.2005.01.021 [DOI] [PubMed] [Google Scholar]