Abstract
J3-crystallin, one of the three major eye-lens proteins of the cubomedusan jellyfish (Tripedalia cystophora), shows similarity to vertebrate saposins, which are multifunctional proteins that bridge lysosomal hydrolases to lipids and activate enzyme activity. Sequence alignment of deduced J3-crystallin indicates two saposin-like motifs arranged in tandem, each containing six cysteines characteristic of this protein family. The J3-crystallin cDNA encodes a putative precursor analogous to vertebrate prosaposins. The J3-crystallin gene has seven exons, with exons 2–4 encoding the protein. Exon 3 encodes a circularly permutated saposin motif, called a swaposin, found in plant aspartic proteases. J3-crystallin RNA was found in the cubomedusan lens, statocyst, in bands radiating from the pigmented region of the ocellus, in the tentacle tip by in situ hybridization, and in the embryo and larva by reverse transcription–PCR. Our data suggest a crystallin role for the multifunctional saposin protein family in the jellyfish lens. This finding extends the gene sharing evolutionary strategy for lens crystallins to the cnidarians and indicates that the putative primordial saposin/swaposin J3-crystallin reflects both the chaperone and enzyme connections of the vertebrate crystallins.
Complex eyes with cellular lenses are scattered throughout the animal phyla from cubomedusan jellyfish to humans. The optical properties of the transparent lens depend on a diverse group of water-soluble, multifunctional proteins called crystallins, many of which are related or identical to stress proteins or metabolic enzymes (1, 2). The α-crystallins are small heat-shock protein/molecular chaperones (3–5), and the β/γ-crystallins (6) are related to microbial stress-protective proteins (7); the α- and the β/γ-crystallins are present ubiquitously in vertebrate lenses. By contrast, the enzyme-crystallins are expressed selectively in different species. Crystallins are critical for the optical properties of the lens and are essentially defined by their abundance (collectively 80–90% of the water-soluble protein) in the lens. Most, if not all, of the crystallins also are expressed at lower amounts outside of the lens where they have nonoptical roles. This dual function of crystallins, which depends on their tissue location and quantitative levels, has been called gene sharing (8, 9).
Recruitment of enzymes as lens crystallins also has occurred in invertebrates (10). This recruitment was first shown for the glutathione S-transferase-related S-crystallins (11, 12) and subsequently for the aldehyde dehydrogenase-related Ω-crystallin (13, 14) of cephalopods. More recently, aldehyde dehydrogenase/Ω-crystallin was shown to be the only crystallin in the scallop eye lens (15). Except for one minor S-crystallin (16), the multiple S-crystallins (13, 17, 18) and Ω-crystallin (14, 15, 19) seem enzymatically inactive.
Much less is known about lens crystallins of other invertebrates. Drosocrystallin is a secreted protein forming the transparent sheath of ommatidia in the compound eye of Drosophila (20). It is likely that drosocrystallin has an additional function, because it is expressed in the brain as well as the ommatidia.
We have been investigating the crystallins of the cubomedusan jellyfish. These cnidarians have remarkably well developed eyes called ocelli that contain cellular lenses filled with J1-, J2-, and J3-crystallin (21, 22). The three extremely similar J1-crystallins (23) appear homologous to a zebrafish expressed sequence tag (Fb98a02). Here, we show that J3-crystallin appears similar to the ubiquitous saposin family of proteins that are associated with lysosomal storage disorders in humans (24, 25). Saposins show homology to domains of sphingomyelinase (26) and acyloxyacyl hydrolase (27) in humans and aspartate proteases in plants (28). Saposins have multiple functions, including that of enhancing lysosomal enzyme–lipid interactions by solubilizing the lipids and/or activating the enzymes (24, 29–32). Thus, J3-crystallin of this cnidarian reflects the enzyme- and chaperone-related evolutionary pathways of the diverse crystallins of vertebrates.
Materials and Methods
Collection of Jellyfish and Rhopalia.
T. cystophora Conant (phylum Cnidaria, class Cubozoa) cubomedusae were collected in the mangroves of La Parguera, Puerto Rico. The rhopalia were excised and stored at −80°C, as described earlier (22).
Sequencing of J3-Crystallin Peptides.
J3-crystallin was purified from rhopalia by SDS/PAGE; its tryptic peptides were separated by HPLC, and three of the peptides were sequenced by Edman degradation by the Microchemistry Facility, Biological Laboratories at Harvard University, as described earlier (22).
Isolation of RNA.
Total RNA was extracted from 3,600 rhopalia by homogenization in RNAzol (Tel-Test, Friendswood, TX) by using the manufacturer's instructions. An average of 5.3 ng of total RNA was obtained per rhopalium. Poly(A+) RNA was prepared by using the Dynabeads mRNA purification kit (Dynal, Oslo, Norway).
Construction of cDNA Library.
Approximately 380 ng of poly(A+) RNA was used to make a cDNA library by using a λZAP-cDNA synthesis and cloning kit (Stratagene). The library had a titer of 3.5 × 104 pfu/ml and was amplified to 1.4 × 107 pfu/ml; the average insert size was 1.53 kbp with a range from 400 bp to 1.75 kbp.
Screening and Sequencing the cDNA Library.
Oligodeoxynucleotides encoding J3-crystallin tryptic peptides using the most commonly observed codons in humans (33) were made in forward and reverse orientations. The forward primer (5′-gccaccatccagatcgtggactcctccctggccatcttc-3′, derived from peptide JFS3-NT53) and the reverse primer (5′-tctcacgatgttctcacactc-3′, derived from peptide JFS-NT12) produced a 137-bp band by reverse transcription–PCR using rhopalial RNA as template. This product was purified by agarose-gel electrophoresis, electro-eluted, ethanol-precipitated, radiolabeled by random primers, and used as a probe to screen the cDNA library. DNA sequencing was performed with the Sequenase kit (United States Biochemical Corp.) and 27 primers to generate overlapping sequence data on both strands of DNA.
Construction and Screening of Genomic Libraries.
Genomic libraries of gonadal DNAs that were digested either with EcoRI or XbaI were constructed by using the λZAP II cloning kit (Stratagene). The EcoRI library had a titer of 6.5 × 105 pfu/ml, and the XbaI library had a titer of 1.2 × 106 pfu/ml. A 2.2-kbp clone (G1) was obtained from the EcoRI library by using the 1.76-kbp J3 cDNA as a probe. DNA clones of 4.7 kbp (G2) and 7.4 kbp (G3) were obtained from the XbaI library by using G1 and the J3 cDNA as probes, respectively.
5′ End Rapid Amplification of cDNA Ends (RACE).
Total (1 μg) RNA was subjected to 5′ RACE analysis (Life Technologies, Grand Island, NY) by using the following 3′ primers derived from the third exon of the J3-crystallin gene. GSP1 (5′-ccagcttatcgatttcaagc-3′) was used for the reverse transcription step; GSP2 (5′-tgaaaattgccaatgaggaa-3′) and GSP3 (5′-ggatggtagctcggcagata-3′) were used for two successive rounds of nested-PCR amplification. The PCR products were subcloned into SrfI-digested pCR-Script (Stratagene), screened by hybridization, and sequenced. Ten clones were isolated with the same 5′ end, thus extending the original cDNA by 32 bp.
Generation of 5′ End of the J3-Crystallin Gene by PCR.
Genomic DNA was used as a template to produce a clone that contained exon 1. The 5′ primer was derived from the 5′ RACE product (5′-atgcttaactctgagcacttc-3′); the 3′ primer was derived from the 5′ end of G2 in intron 1 (5′-caacgaacgaccactatagtgaa-3′). Each primer contained an EcoRI linker. The 2.75-kbp PCR product was cloned into pBluescript SK− (Stratagene). Partial sequencing verified that the clone contained exon 1, intron 1, and the 3′ end of G2.
Analysis of Protein Sequences.
Multiple sequence alignment was performed with clustralw program (available at http://www2.ebi.ac.uk). The GenBank accession numbers of vertebrate saposins used for alignment are AB003471 (for chicken), AB108655 (for zebrafish), and 1360694 (for human). Secondary structure was predicted by phdsec program; the percentage of similarity/identity between J3-crystallin and saposin A, B, C, and D of the human prosaposin precursor was determined by blast (available at http://www.ncbi.nlm.nih.gov/BLAST/).
J3-Crystallin Model Building.
The N- and C-terminal portions of the J3-crystallin sequence from jellyfish were aligned with the sequence of NK-lysin (GenBank accession no. 2392473). The known structure of NK-lysin (ID code 1NKL) was taken into account, and no insertions or deletions were introduced within the helices of the NK-lysin. With this alignment and the NK-lysin structure as a template, a homology model was built for each half of J3-crystallin by using the SegMod algorithm (34) as implemented in the program genemine.
To demonstrate the plausibility of a disulfide bond between Cys-38 and Cys-118, the model of the N-terminal structure was manipulated within the program charmm (35) to establish the disulfide bond. Simulated annealing was performed during which only residues 36–39 and 114–120 were free to move. Electrostatic interactions were approximated with a distance-dependent dielectric and were shifted to zero at 10 Å. Soft nonbond interactions were used at short range to facilitate conformational sampling (36). After preliminary energy minimization, the model was heated to 900 K during 100 ps of simulated molecular dynamics, during which the force constant of a nuclear Overhauser effect restraint between the sulfur atoms of Cys-38 and Cys-118 was gradually increased to a maximum of 300 kcal/mol/(AA). The model then was cooled to 300 K during 150 ps of dynamics. After 10 ps of dynamics at 300 K and energy minimization, the disulfide bond was introduced into the structure. Additional energy minimization resulted in the final N-terminal model.
The residues in the N- and C-terminal models were then color coded according to hydropathy (37) and rendered by using the programs molscript (38) and RASTER3D (39).
In Situ Hybridization.
Nonradioactive whole-mount and cryosection in situ hybridizations were performed as described (40). Jellyfish were fixed overnight in a sea water:fixative [4% (vol/vol) paraformaldehyde/0.6 M NaCl/150 mM phosphate buffer, pH 7.4] mixture (1:1), transferred into ascending concentrations of methanol, and stored in 100% methanol at −20°C. For cryosectioning, fixed jellyfish were transferred through descending concentrations of methanol into diethyl pyrocarbonate-treated PBS, then into 20% (wt/vol) sucrose in PBS, and, finally, into OCT compound (Fisher Scientific). Digoxigenin probes were generated by creating a PCR amplicon with the T7 RNA polymerase promoter incorporated in the appropriate orientation by using partial J3- and J1A-crystallin cDNAs in pBluescript SK− as templates. A 484-bp J3-crystallin antisense probe was amplified with the following primers: 5′-atgagatccaccgtacatct-3′ and 5′-taatacgactcactatagtaaaactaaaggcagacggaa-3′. The 510-bp J1A-crystallin antisense probe was amplified with the following primers: 5′-atgtcttccgatcaagctaaa-3′ and 5′-taatacgactcactatagcgacttgttcgacgtaggagt-3′.
Results
Isolation and Characterization of J3-Crystallin cDNA.
The 19-kDa polypeptide band (J3-crystallin) derived from jellyfish rhopalia was eluted from an SDS-polyacrylamide gel, and three tryptic peptides (boxed in Fig. 1) separated by HPLC were sequenced. This abundant protein band has been defined as J3-crystallin by reason of its being one of the three major protein bands present in the isolated lenses (22). A λZAP cDNA library made from rhopalial total RNA was screened with double-stranded oligodeoxynucleotide probes encoding these tryptic peptides using the preferred codon usage derived from humans (33). The 5′ end of a positive J3-crystallin cDNA was extended 32 nucleotides by 5′ RACE (see Methods). Radioactively labeled J3-crystallin cDNA hybridized to an ≈1.7-kb RNA from rhopalia in a Northern blot, consistent with the size of the J3-crystallin cDNA (data not shown). The complete J3-crystallin cDNA sequence (Fig. 1) was deposited into GenBank (accession no. AF175577).
Figure 1.
cDNA and deduced protein sequences of J3-crystallin cDNA. The arrowheads signify positions of introns (see Fig. 4). The boxed amino acid sequences match the tryptic peptide sequences obtained from the purified protein (JFS-NT12: ECENIVR; JFS3-NT37: DLSPGDV-AVLGY; JFS3-NT53: ATIQIVDSSLAIF-YTELE-DKLVM). The putative polyadenylation signal is underlined.
The tryptic peptides obtained from the J3-crystallin polypeptide are boxed in the deduced protein sequence in Fig. 1, confirming the identity of the cDNA. Unexpectedly, the J3-crystallin cDNA encodes a 28.6-kDa protein rather than a 19-kDa protein indicated by SDS/PAGE. As discussed below, this discrepancy suggests that the 19-kDa J3-crystallin band is derived by cleavage of a precursor protein. A putative atypical polyadenylation signal is underlined in Fig. 1. The arrowheads mark the positions of the introns in the J3-crystallin gene (see below).
Similarity of Deduced J3-Crystallin with Saposins.
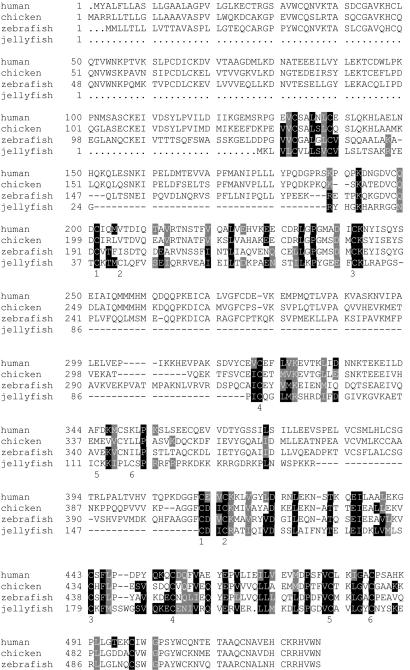
Computer-assisted analysis (http://www.ncbi.nlm.nih.gov/BLAST/) showed that the deduced amino acid sequence of J3-crystallin shows 25–29% identity and 43–50% similarity to saposins A–D (Fig. 2). Saposins are a family of multifunctional glycoproteins that bridge lysosomal enzymes and lipids and activate enzyme activity (24, 29–32). Two tandemly arranged sets of six cysteines in J3-crystallin can be aligned with the six cysteines required for the structural integrity of the vertebrate saposin motifs (41). Similarity also is seen at a number of other amino acids.
Figure 2.
Sequence comparison of J3-crystallin with human, chicken, and zebrafish prosaposin. Prosaposin amino acids that are identical to those of J3-crystallin in the alignment are highlighted in black; prosaposin residues that are similar to those in J3-crystallin are highlighted in gray. The six characteristic cysteines identifying the two putative saposin motifs in J3-crystallin are indicated under the J3-crystallin sequence. The accession numbers of the human, chicken, and zebrafish prosaposins are given in Methods.
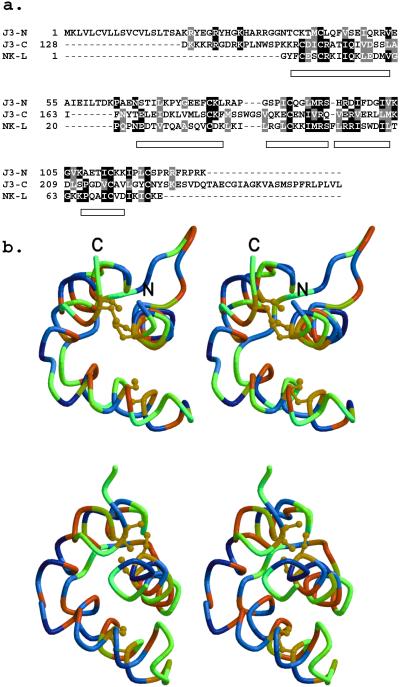
Alignment of the N- and C-terminal regions of J3-crystallin with NK-lysin suggests that there has been an intragenic duplication (Fig. 3a). The open rectangles in Fig. 3a delineate the regions that are α-helical in the NK-lysin structure determined by NMR spectroscopy (41). No insertions or deletions were permitted in these regions. Most importantly, the cysteines in each half can be aligned with those in NK-lysin, which have been shown by NMR spectroscopic analysis to be critical for the saposin structural motif. The one exception is Cys-38, which is one residue away from alignment (Fig. 3a), but is still close enough in the three-dimensional model (Fig. 3b) to form the disulfide bond with Cys-118.
Figure 3.
Evidence for two putative saposin motifs within J3-crystallin and their predicted structure based on comparisons with NK-lysin. (a) Multiple sequence alignment of N- and C-terminal parts of J3-crystallin and NK-lysin. Sequence identities and similarities are shaded black and gray, respectively. Helices in the NK-lysin structure (PDB ID code 1NKL) are represented as rectangles. The alignment introduces no insertions or deletions within these helices. J3-N has conserved five of the six cysteines that are disulfide bonded in NK-lysin; the first one of these in J3-crystallin (Csy-38) precedes that in NK-lysin by one residue. All six cysteines of the putative saposin motif are conserved in J3-C. (b) Wall-eyed view of models of J3-N (Upper) and J3-C (Lower) portions of J3-crystallin based on the 1NKL template structure. All disulfide bonds are conserved. The α-carbon traces are colored according to hydropathy (37) with hydrophobic and hydrophilic residues at the red and blue ends of the spectrum, respectively. The lack of hydrophilic residues in the modeled interiors is evident.
The homology models of the first (N-terminal) and second (C-terminal) saposin motifs of J3-crystallin were based on the NMR-derived structure of NK-lysin. Wall-eyed stereo views of these proposed structures provide further evidence for their relationship to saposins (Fig. 3b). No hydrophilic residues (blue end of the spectrum) are found in the interiors of the models; rather, hydrophobic residues (red end of the spectrum) form the core of the models. These hydropathy patterns support the alignments of J3-crystallin to NK-lysin and suggest that the modeled structures represent a reasonable hypothesis to guide future investigations.
SDS/PAGE of rhopalial proteins heated to 60°C indicated that J2- and J3-crystallin are considerably more heat stable than J1-crystallin (23; data not shown), consistent with the heat stability of saposin motifs (41, 42).
Isolation and Characterization of the J3-Crystallin Gene.
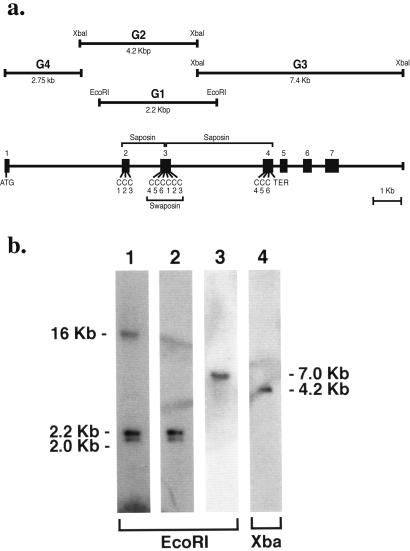
The J3-crystallin gene was cloned and characterized (Fig. 4a). Intron 2 divides Cys 3 and 4 of the first saposin domain, and intron 3 divides Cys 3 and 4 of the second saposin domain. This gene structure places Cys 4–6 of the first saposin domain contiguously with Cys 1–3 of the second saposin domain on exon 3, encoding a circularly permutated swaposin similar to that in plant aspartate proteases (26).
Figure 4.
Gene structure and genomic Southern blot analysis of J3-crystallin. (a) The J3-crystallin gene was cloned as follows. Five identical 2.2-kbp genomic fragments (G1) were obtained from an EcoRI genomic library by using J3-crystallin cDNA as a probe. Next, a 4.7-kbp clone (G2) was obtained from a XbaI genomic library by screening with G1. Partial sequencing and PCR with primers derived from the J3-crystallin cDNA were used to identify part of exon 2 and all of exon 3 in G1 and exons 2 and 3 in G2. Exons 4–7 were identified on a 7.4-kbp clone (G3) that was obtained from the XbaI library by screening with three deoxyoligonucleotides (5′-gagtgcgagaacatagtgcga-3′; 5′-ctgtaattattcgaaggaaag-3′; 5′ctgtgcagtgctgggctact-3′) derived further downstream on the J3-crystallin cDNA. The 5′ end of the J3-crystallin gene was determined by cloning a PCR product using primers with EcoRI linkers on their ends. The 5′ primer was derived from the beginning of the cDNA and the 3′ primer was derived from the 5′ end of G2. The resulting clone (G4) contained exon 1 at its 5′ end. (b) Southern blot of genomic J3-crystallin sequences. Lane 1, J3-crystallin cDNA probe; lanes 2 and 4, G1 probe; lane 3, G4 probe.
A Southern hybridization blot using pooled EcoRI-digested genomic DNAs from jellyfish gonads probed with the J3-crystallin cDNA showed 16-, 2.2-, and 2.0-kbp bands (Fig. 4b, lane 1). The same blot probed with G1 gave only the expected 2.2-kbp band (equivalent to G1) and a 2.0-kbp band (presumably a polymorphic G1 band; Fig. 4b, lane 2). G4 (Fig. 4b, lane 3) and a synthetic exon 1 oligonucleotide probe each gave a single 7.0-kbp EcoRI band. XbaI-digested DNA gave a single 4.2-kbp band (presumably representing G2) when probed with G1, a single 1.45-kbp band when probed with G4 (data not shown), and a single 3.4-kbp band when probed with an exon 1 synthetic oligonucleotide (data not shown). These results suggest one J3-crystallin gene with expected polymorphisms within the natural jellyfish population.
Expression Patterns of the J3- and J1A-Crystallin Genes.
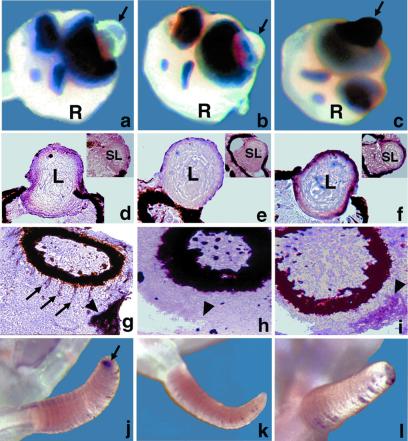
The expression pattern of the J3-crystallin gene was explored by in situ hybridization using an antisense (Fig. 5 a, d, g, and j) and a control sense (Fig. 5 d, e, h, and k) riboprobe. Parallel tests were conducted with antisense riboprobe for J1A-crystallin mRNA (23) for comparison (Fig. 5 c, f, i, and l). On the whole mount rhopalium (R in Fig. 5 a–c), staining was intense for J1A-crystallin mRNA (Fig. 5c), but not for J3-crystallin mRNA (Fig. 5a) on the outer surface of the lens of the large ocellus (arrow). The in situ hybridization tests on cryosections showed that both J3-crystallin (Fig. 5d) and J1A-crystallin (Fig. 5f) mRNAs are expressed in the outer layers of the lens of the large ocellus, with the latter being stained more intensely than the former. A positive signal also was observed for J1A-crystallin mRNA in the outmost layer of the lens of the small ocellus (Fig. 5f, Inset). It is unlikely that the slight darkening of the lens of the small ocellus for J3-crystallin mRNA (Fig. 5d, Inset) is significant because J3-crystallin was absent from purified lenses of the small ocellus in previous electrophoretic tests (22).
Figure 5.
J3 and J1A-crystallin gene expression in adult jellyfish rhopalia and tentacles. In situ hybridization on whole jellyfish (a–c, j–l) and cryosections (d–i). J3-crystallin (a, d, g, j) and J1A-crystallin (c, f, i, l) antisense riboprobes; J3-crystallin sense riboprobe (b, e, h, k). R, rhopalium; L, lens of large ocellus; SL, lens of small ocellus. (a, b, c) Arrows point to the surface of the lens of the large ocellus. (g, h, i) Arrowheads point to the tissue surrounding the lumen of the statocyst. (g) Arrows point to the staining in the outer aspect of the pigmented region of the ocellus. (j) Arrow points to the tip of a tentacle.
The J3- and J1-crystallin genes were differentially expressed outside of the lens. Strong J3-crystallin (Fig. 5g, large arrowhead) and weaker J1A-crystallin (Fig. 5i, large arrowhead) signals were present in the tissue surrounding the lumen of the statocyst. J3-crystallin (Fig. 5g, arrows) but not J1A-crystallin (Fig. 5i) mRNA appeared in equally spaced bands radiating from the outer aspect of the pigmented region of the ocellus. Finally, the tips of the tentacles stained for J3-crystallin (Fig. 5j, arrow) but not J1A-crystallin (Fig. 5l) RNA. The sporadic staining within the creases along the length of the tentacle was inconsistent and presumably artifact. Control tests with the J3-crystallin sense riboprobe were negative (Fig. 5 b, e, h, and k). Reverse transcription–PCR tests indicated that J3- and J1A-crystallin mRNAs are expressed in the embryonic and larval stages of development as well as in the rhopalia (data not shown), consistent with their having a noncrystallin role outside of the lens.
Discussion
The intensely studied vertebrate crystallins are identical or related to stress proteins (small heat-shock proteins or microbial stress-related proteins) or metabolic enzymes (1, 2, 43, 44). Although much less is known about lens crystallins of invertebrates, their recruitment from enzymes has taken place in molluscs (10). S-crystallins are homologous to glutathione S-transferase in cephalopods (squid and octopus; refs. 11–13, and 18), and Ω-crystallin is homologous to aldehyde dehydrogenase class 1 and 2 in cephalopods (13, 14, 19) and scallops (15). In contrast to many enzyme-crystallins of vertebrates, S-crystallins (except for one member; ref. 16) and Ω-crystallin have tested negative for enzyme activity. However, scallop Ω-crystallin seems to be expressed in various nonocular tissues, suggesting that it has a yet-to-be-discovered noncrystallin role (15). The present study shows that J3-crystallin of the jellyfish lens is similar to the saposins, a family of multifunctional proteins containing a structural motif conserved from plants to humans (24, 29–32). This extends the general rule that lens crystallins have been recruited from preexisting proteins with nonrefractive functions to cnidarians.
Our data suggesting a J3-crystallin/saposin homology is supported by the fact that the six cysteines critical for the saposin structural motif and a number of other identical or conserved amino acids of J3-crystallin align with vertebrate saposins. We also show that two physically reasonable homology models can be built by assuming that both the N- and C-terminal portions of J3-crystallin resemble the NMR-derived model of NK-lysin (41), a homologue of the saposins. Another argument favoring a J3-crystallin/saposin homology is that the J3-crystallin cDNA encodes a protein with two tandemly arranged saposin motifs with an SDS/PAGE-deduced Mr that is higher (28 kDa) than that derived for J3-crystallin (19 kDa). This observation suggests that J3-crystallin is cleaved from a precursor protein. In vertebrates, four distinct saposins are cleaved from a prosaposin precursor (45–49). Another saposin homologue, surfactant-associated polypeptide (50), also is cleaved from a precursor protein with three linked saposin motifs (51).
We do not know yet whether one or both of the putative saposin motifs are represented in a J3-crystallin polypeptide, or whether J3-crystallin comprises two saposin-like polypeptides that comigrate on the SDS-polyacrylamide gel. The actual polypeptide molecular weight of J3-crystallin(s) is also uncertain, inasmuch as saposins are glycosylated, increasing their apparent size in SDS-polyacrylamide gels. Finally, we do not know yet whether the saposin motif in J3-crystallin has the same compact three-dimensional structure as that present in the saposins and NK-lysin (41). Although the relative high-heat stability of J3-crystallin is consistent with the thermostability of the saposins (42), we did not see a reduction in heat stability or a change in the electrophoretic mobility of J3-crystallin upon reduction (data not shown). This observation suggests that J3-crystallin may have a more open configuration than do the compact saposins.
The linkage of four saposins (A–D) on the prosaposin precursor in vertebrates indicates that these proteins have evolved by intragenic duplications (52). The deduced saposin motifs encoded in the J3-crystallin cDNA suggest that at least one intragenic duplication took place in invertebrates at the beginning of metazoan evolution. Of particular interest is our finding that exon 3 of the J3-crystallin gene has a contiguous saposin motif whose cysteines are circularly permutated (Cys 4–6 are on the N terminus, and Cys 1–3 are on the C terminus of the peptide encoded by exon 3). This arrangement is precisely the situation in the saposin motif, called a swaposin, of plant aspartic proteases (28, 53). Possibly exon 3 of the jellyfish J3-crystallin gene and the swaposin-coding region of plant aspartic proteases are orthologous. Our data also raise the possibility that the primordial saposin motif was encoded by a single exon with the permutated cysteines (a swaposin), and that introns were added during evolution as the gene underwent duplications.
In addition to plant aspartic proteases (28, 53), saposin-type motifs are present in the N-terminal region of acid sphingomyelinase (26) and in acyloxyacyl hydrolase (27) of humans. Remarkably, the saposin domain of acyloxyacyl hydrolase is cleaved from a precursor protein and subsequently attached covalently as the small subunit of this heterodimeric enzyme (27). The saposin-type domain of these two enzymes probably possesses lipid-binding and/or enzyme-activating properties.
Saposins have been associated with numerous functions. Saposin C contains a neurotrophic peptide (54–56) that is present in mammalian but not in chicken prosaposin (47) or in J3-crystallin (present study), suggesting a later evolutionary development. This active peptide, called prosaptide, acts as a Schwann-cell survival factor via a phosphorylation signal-transduction pathway associated with myelin formation (57–59). Moreover, prosaposin's complex developmental and tissue-specific expression pattern in mice differs from other lysosomal hydrolases, suggesting that it has multiple functions (60). In the present study, J3-crystallin also shows a tissue-specific (especially lens) and complex expression pattern, as does mouse prosaposin.
The recruitment of a lens crystallin from saposins provides a new role for these multifunctional proteins and fits elegantly within the framework of crystallin evolution. There is a conceptual link between the solubilizing and bridging functions of saposins and the vertebrate α-crystallins, which are small heat-shock proteins (3, 4) and molecular chaperones (5). There is also a connection between the functional partnerships of saposins with metabolic enzymes and the enzyme-crystallins of vertebrates (1, 2, 9, 10). The saposin-related J3-crystallin seems to represent another case of the innovation of a new protein function, possibly without loss of the original function(s), being associated with changes in gene regulation by a gene-sharing strategy (43). The extension of this strategy to the lens of the complex eye of the evolutionarily distant cubomedusan jellyfish suggests that gene sharing by multifunctional proteins applies to the evolutionary history of lens crystallins throughout the animal kingdom.
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF175577).
References
- 1.Wistow G, Piatigorsky J. Annu Rev Biochem. 1988;57:479–504. doi: 10.1146/annurev.bi.57.070188.002403. [DOI] [PubMed] [Google Scholar]
- 2.de Jong W W, Hendriks W, Mulders J W, Bloemendal H. Trends Biochem Sci. 1989;14:365–368. doi: 10.1016/0968-0004(89)90009-1. [DOI] [PubMed] [Google Scholar]
- 3.Ingolia T D, Craig E A. Proc Natl Acad Sci USA. 1982;79:2360–2364. doi: 10.1073/pnas.79.7.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klemenz R, Frohli E, Steiger R H, Schafer R, Aoyama A. Proc Natl Acad Sci USA. 1991;88:3652–3656. doi: 10.1073/pnas.88.9.3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Horwitz J. Proc Natl Acad Sci USA. 1992;89:10449–10453. doi: 10.1073/pnas.89.21.10449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lubsen N H, Aarts H J M, Schoenmakers J G G. Prog Biophys Mol Biol. 1988;51:47–76. doi: 10.1016/0079-6107(88)90010-7. [DOI] [PubMed] [Google Scholar]
- 7.Wistow G, Summers L, Blundell T. Nature (London) 1985;316:622–624. doi: 10.1038/315771a0. [DOI] [PubMed] [Google Scholar]
- 8.Piatigorsky J, O'Brien W E, Norman B L, Kalumuck D, Wistow G J, Borras T, Nickerson J M, Wawrousek E F. Proc Natl Acad Sci USA. 1988;85:3479–3483. doi: 10.1073/pnas.85.10.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Piatigorsky J, Wistow G. Cell. 1989;57:197–199. doi: 10.1016/0092-8674(89)90956-2. [DOI] [PubMed] [Google Scholar]
- 10.Tomarev S I, Piatigorsky J. Eur J Biochem. 1996;235:449–465. doi: 10.1111/j.1432-1033.1996.00449.x. [DOI] [PubMed] [Google Scholar]
- 11.Wistow G, Piatigorsky J. Science. 1987;236:1554–1556. doi: 10.1126/science.3589669. [DOI] [PubMed] [Google Scholar]
- 12.Tomarev S I, Zinovieva R D. Nature (London) 1988;336:86–88. doi: 10.1038/336086a0. [DOI] [PubMed] [Google Scholar]
- 13.Tomarev S I, Zinovieva R D, Piatigorsky J. J Biol Chem. 1991;266:24226–24231. [PubMed] [Google Scholar]
- 14.Montgomery M K, McFall-Ngai M J. J Biol Chem. 1992;267:20999–21003. [PubMed] [Google Scholar]
- 15.Piatigorsky J, Kozmik Z, Horwitz J, Ding L, Carosa E, Robison W G, Jr, Steinbach P J, Tamm E R. J Biol Chem. 2000;275:41064–41073. doi: 10.1074/jbc.M005625200. [DOI] [PubMed] [Google Scholar]
- 16.Tomarev S I, Sambath C, Piatigorsky J. J Mol Evol. 1995;41:1048–1056. doi: 10.1007/BF00173186. [DOI] [PubMed] [Google Scholar]
- 17.Tomarev S I, Zinovieva R D, Piatigorsky J. J Biol Chem. 1992;267:8604–8612. [PubMed] [Google Scholar]
- 18.Tomarev S I, Zinovieva R D, Guo K, Piatigorsky J. J Biol Chem. 1993;268:4534–4542. [PubMed] [Google Scholar]
- 19.Zinovieva R D, Tomarev S I, Piatigorsky J. J Biol Chem. 1993;268:11449–11455. [PubMed] [Google Scholar]
- 20.Jansens H, Gehring W J. Dev Biol. 1999;207:204–214. doi: 10.1006/dbio.1998.9170. [DOI] [PubMed] [Google Scholar]
- 21.Laska G, Hunden M. Zool J Anat. 1982;1088:107–123. [Google Scholar]
- 22.Piatigorsky J, Horwitz J, Kuwabara T, Cutress C E. J Comp Physiol A. 1989;164:577–587. doi: 10.1007/BF00614500. [DOI] [PubMed] [Google Scholar]
- 23.Piatigorsky J, Horwitz J, Norman B L. J Biol Chem. 1993;268:11894–11901. [PubMed] [Google Scholar]
- 24.O'Brien J S, Kishimoto Y. FASEB J. 1991;5:301–308. doi: 10.1096/fasebj.5.3.2001789. [DOI] [PubMed] [Google Scholar]
- 25.Chang M H Y, Bindloss C A, Grabowski G A, Qi X, Winchester B, Hopwood J J, Meikle P J. Clin Chem (Washington, DC) 2000;46:167–174. [PubMed] [Google Scholar]
- 26.Ponting C P. Protein Sci. 1994;3:359–361. doi: 10.1002/pro.5560030219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hagen F S, Grant F J, Kuijper J L, Slaughter C A, Moomaw C R, Orth K, O'Hara P J, Munford R S. Biochemistry. 1991;30:8415–8423. doi: 10.1021/bi00098a020. [DOI] [PubMed] [Google Scholar]
- 28.Guruprasad K, Tormakangas K, Kervinen J, Blundell T L. FEBS Lett. 1994;352:131–136. doi: 10.1016/0014-5793(94)00935-x. [DOI] [PubMed] [Google Scholar]
- 29.Kishimoto Y, Hiraiwa M, O'Brien J S. J Lipid Res. 1992;33:1255–1267. [PubMed] [Google Scholar]
- 30.Furst W, Sandhoff K. Biochim Biophys Acta. 1992;1126:1–16. doi: 10.1016/0005-2760(92)90210-m. [DOI] [PubMed] [Google Scholar]
- 31.Munford R S, Sheppard P O, O'Hara P J. J Lipid Res. 1995;36:1653–1663. [PubMed] [Google Scholar]
- 32.Vaccaro A M, Salvioli R, Tatti M, Ciaffoni F. Neurochem Res. 1999;24:307–314. doi: 10.1023/a:1022530508763. [DOI] [PubMed] [Google Scholar]
- 33.Lathe R. J Mol Biol. 1985;183:1–12. doi: 10.1016/0022-2836(85)90276-1. [DOI] [PubMed] [Google Scholar]
- 34.Levitt M. J Mol Biol. 1992;226:507–533. doi: 10.1016/0022-2836(92)90964-l. [DOI] [PubMed] [Google Scholar]
- 35.Brooks B R, Bruccoleri R E, Olafson B D, States D J, Swaminathan S, Karplus M. J Comput Chem. 1983;4:187–217. [Google Scholar]
- 36.Vieth M, Hirst J D, Kolinski A, Brooks C L., III J Comput Chem. 1998;19:1612–1622. [Google Scholar]
- 37.Kyte J, Doolittle R F. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 38.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 39.Merritt E A, Bacon D J. Methods Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- 40.Wilkinson D G, Nieto M A. Methods Enzymol. 1993;225:361–373. doi: 10.1016/0076-6879(93)25025-w. [DOI] [PubMed] [Google Scholar]
- 41.Liepinsh E, Andersson M, Ruysschaert J-M, Otting G. Nat Struct Biol. 1997;4:793–795. doi: 10.1038/nsb1097-793. [DOI] [PubMed] [Google Scholar]
- 42.Waring A J, Chen Y, Faull K F, Stevens R, Sherman M A, Fluharty A L. Mol Genet Metab. 1998;63:14–25. doi: 10.1006/mgme.1997.2646. [DOI] [PubMed] [Google Scholar]
- 43.Piatigorsky J. J Biol Chem. 1992;267:4277–4280. [PubMed] [Google Scholar]
- 44.Sax C M, Piatigorsky J. Adv Enzymol Relat Areas Mol Biol. 1994;69:155–201. doi: 10.1002/9780470123157.ch5. [DOI] [PubMed] [Google Scholar]
- 45.O'Brien J S, Kretz K A, Dewji N, Wenger D A, Esch F, Fluharty A L. Science. 1988;241:1098–1101. doi: 10.1126/science.2842863. [DOI] [PubMed] [Google Scholar]
- 46.Collard M W, Sylvester S R, Tsuruta J K, Griswold M D. Biochemistry. 1988;27:4557–4564. doi: 10.1021/bi00412a050. [DOI] [PubMed] [Google Scholar]
- 47.Azuma N, Seo H-C, Lie O, Fu Q, Gould R M, Hiraiwa M, Burt D W, Paton I R, Morrice D R, O'Brien J S, Kishimoto Y. Biochem J. 1998;330:321–327. doi: 10.1042/bj3300321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Madar-Shapiro L, Pasmanik-Chor M, Vaccaro A M, Dinur T, Dagan A, Gatt S, Horowitz M. Biochem J. 1999;337:433–443. [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao Q, Morales C R. J Biol Chem. 2000;275:24829–24839. doi: 10.1074/jbc.M003497200. [DOI] [PubMed] [Google Scholar]
- 50.Andersson M, Curstedt T, Jornvall H, Johansson J. FEBS Lett. 1995;362:328–332. doi: 10.1016/0014-5793(95)00268-e. [DOI] [PubMed] [Google Scholar]
- 51.Zaltash S, Johansson J. FEBS Lett. 1998;423:1–4. doi: 10.1016/s0014-5793(97)01582-2. [DOI] [PubMed] [Google Scholar]
- 52.Rorman E G, Scheinker V, Grabowski G A. Genomics. 1992;13:312–318. doi: 10.1016/0888-7543(92)90247-p. [DOI] [PubMed] [Google Scholar]
- 53.Ponting C P, Russell R B. Trends Biol Sci. 1995;20:179–180. doi: 10.1016/s0968-0004(00)89003-9. [DOI] [PubMed] [Google Scholar]
- 54.O'Brien J S, Carson G S, Seo H-C, Hiraiwa M, Kishimoto Y. Proc Natl Acad Sci USA. 1994;91:9593–9596. doi: 10.1073/pnas.91.20.9593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.O'Brien J S, Carson G S, Sea H-C, Hiraiwa M, Weiler S, Tomich J M, Barranger J A, Kahn M, Azuma N, Kishimoto Y. FASEB J. 1995;9:681–685. doi: 10.1096/fasebj.9.8.7768361. [DOI] [PubMed] [Google Scholar]
- 56.Kotani Y, Matsuda S, Wen T-C, Sakanaka M, Tanaka J, Maeda N, Kondoh K, Ueno S-I, Sano A. J Neurochem. 1996;66:2197–2200. doi: 10.1046/j.1471-4159.1996.66052197.x. [DOI] [PubMed] [Google Scholar]
- 57.Campana W M, Hiraiwa M, O'Brien J S. FASEB J. 1998;12:307–314. doi: 10.1096/fasebj.12.3.307. [DOI] [PubMed] [Google Scholar]
- 58.Campana W M, Darin S J, O'Brien J S. J Neurosci Res. 1999;57:332–341. [PubMed] [Google Scholar]
- 59.Hiraiwa M, Campana W M, Mizisin A P, Mohiuddin L, O'Brien J S. Glia. 1999;26:353–360. [PubMed] [Google Scholar]
- 60.Sun Y, Witte D P, Grabowski G A. Am J Path. 1994;145:1390–1398. [PMC free article] [PubMed] [Google Scholar]