Abstract
RNA loops that adopt a characteristic GNRA “tetraloop” fold are common in natural RNAs. Here, we have used in vitro selection by means of mRNA-peptide fusions to select peptides that bind an example of this RNA loop motif. Starting with the RNA recognition domain from the λ N protein, we have constructed libraries containing 150, 1,600, and 9 trillion different peptide sequences as mRNA-peptide fusions and isolated those capable of high-affinity RNA binding. These selections have resulted in more than 80 different peptides that bind the same RNA loop. The highest affinity peptides exhibit low nanomolar dissociation constants as well as the ability to discriminate RNA hairpins differing by a single loop nucleotide. Thus, our work demonstrates that numerous, chemically distinct solutions exist for a particular RNA recognition problem.
The ability to construct high-affinity, high-specificity peptide ligands provides a means to target RNA molecules of interest. Genetic approaches have been developed to isolate novel arginine-rich RNA-binding peptides in vivo (1–3). These systems allow selection in the context of living systems, but limit library sizes to a maximum of ∼105 to 106 sequences, allowing only four residues to be searched exhaustively (204 = 1.6 × 105). In vitro selection experiments to isolate RNA-binding peptides have not been demonstrated. An in vitro approach affords both precise control of the selection environment and the ability to explore much larger libraries.
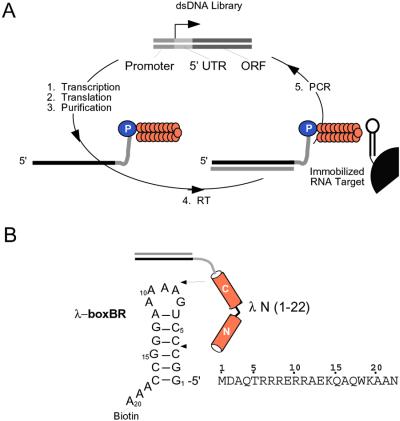
We were interested in using in vitro selection to isolate peptides that bind RNA tetraloops. The tetraloop fold is a common element in many functional RNAs, enhancing duplex stability and participating in tertiary folding interactions (4–6). We had previously developed the mRNA-peptide fusion system to perform in vitro selection on peptides and proteins (7). In the fusion system, cycles are carried out entirely in vitro and libraries of 1013 independent sequences can be constructed (Fig. 1A; refs. 7–10).
Figure 1.
(A) Selection cycle. (B) Constructs used. In the selection cycle, a double-stranded DNA library containing randomized codons is transcribed, generating a pool of mRNA templates. These templates are then ligated to a flexible DNA oligonucleotide containing puromycin at its 3′ end. Translation of these ligated templates in vitro produces peptides covalently attached by means of their C terminus to the 3′ end of their own message by way of a stable amide linkage—a mRNA–peptide fusion. These fusions are then converted into cDNA/mRNA hybrid fusions by using reverse transcriptase and subjected to selection on an affinity matrix.
The arginine-rich peptide corresponding to the RNA-binding domain of the λ N protein served as a starting point for our experiments and libraries. This short peptide (22 residues) recognizes the boxBR RNA hairpin with high affinity and specificity as a bent α-helix (11–14). The hairpin contains a five-base RNA loop that adopts a tetraloop fold with one base extruded (15, 16). Here, we have used in vitro selection experiments to isolate peptides that bind the boxBR RNA motif.
Materials and Methods
Construction of Fusion Template.
Templates and constructs used to assay fusion binding (Fig. 2) correspond to the N-myc construct described previously (17).
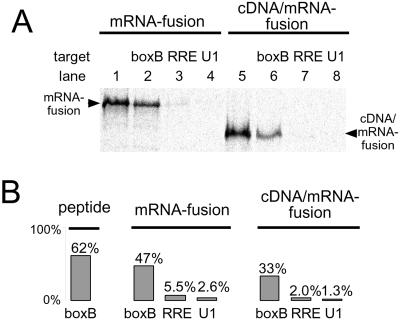
Figure 2.
Binding and specificity of λ N peptide λ N mRNA-peptide fusion, and λ N cDNA/mRNA-peptide fusion constructs. (A) Gel analysis of binding. Binding of 35S-Met-labeled λ N mRNA-peptide fusions and cDNA/mRNA-peptide fusions to immobilized (i) λ boxBR (lanes 2 and 6), (ii) HIV-RRE (lanes 3 and 7) and (iii) U1 hairpin (lanes 4 and 8). Lanes 1 and 5 show the total amount of fusion before binding. Complexes were eluted and resolved on SDS tricine PAGE (21). (B) Scintillation analysis of binding and specificity. 35S-Met-labeled peptide, mRNA, and mRNA/cDNA fusions bound to immobilized boxBR, HIV-RRE, and U1 hairpins. The cDNA/mRNA and mRNA fusions bind the cognate boxBR RNA efficiently, whereas only 1–5% bind the RRE and U1 targets.
Binding Analysis.
Chemically synthesized 3′ biotinylated RNA oligonucleotides of the λ boxBR hairpin (Fig. 1B), HIV Rev RRE binding site (5′-GGUCUGGGCGCAGCGCAAGCUGACGGUACAGGCCAAA-3′), and U1 stem-loop (5′-AAUCCAUUGCACUCCGGAUUAAA-3′) were immobilized on streptavidin agarose beads. Samples were added to 400 μl of N binding buffer (10 mM Hepes pH 7.5/0.5 mM EDTA/100 mM KCl/1 mM MgCl2/1 mM DTT/0.01% Nonidet P-40/10% glycerol) containing 50 μg/ml yeast tRNA and 200 pmol of immobilized RNA. This mixture was incubated at 4°C for 1 h and then washed in a filter centrifuge tube with binding buffer without tRNA. Sample was eluted with either 2× tricine sample buffer (0.1 M Tris⋅Cl pH 6.8/24% glycerol/8% SDS/0.2 M DTT/0.02% Coomassie blue G-250) or 100 mM MgCl2.
Library Construction.
Library 1 was constructed beginning with the N-FLAG-myc synthetic single-stranded DNA (17) containing the sequence 5′-NNCBNG-3′ (n = A,T, G, or C; B = C,G, or T) at codons 6 and 7. This pool encodes the peptide MDAQT X6 X7RERRAEKQAQWKAANDYKDDDDKNSCA. The two random positions, X6 and X7 encode the amino acids FLIVSPTAYHNDCRG at position 6 and LVSPAQEWRG at position 7, respectively. The single sequence that encodes R6R7 encodes an overlapping NgoM IV restriction site. Library 2 was constructed as library 1 and by using a template containing 5′-CNG-3′ at codon 7 and the sequence NNS (S = G, C) at codons 14 and 15. This combination encoded L, P, Q, or R at position 7 and all 20 aa at positions 14 and 15. Library 3 was constructed by inserting NNS codons at residues 13–22 in the N-myc template (17). PCR library construction began with 150 pmols of synthetic DNA which extended 76% yielding a library containing ∼6.9 × 1013 sequences. In round 1, 200 pmols of RNA-puromycin conjugate was translated and ≈15 pmols of fusion were purified and subjected to the selective step resulting in an initial library complexity of ∼9 × 1012 sequences.
Selection Experiments.
The library 1 and library 2 selection rounds were carried out essentially as described (17). In the library 3 selection, selection rounds omitted thiopropyl Sepharose chromatography and preclear. Sequencing of round 11 clones indicated that a number corresponded to the wild-type (wt) λ N control construct of our original experiments, resulting from microscopic contamination of the round 0 pool. This sequence is unlikely to have influenced the pool composition or the selection because of the excess of target RNA in the selective step. The wt sequence was removed during round 12 by subtractive hybridization. A total of 2,000 pmol of round 11 fusion template was incubated in 1 ml of subtractive hybridization buffer (1 M NaCl; 100 mM Tris⋅Cl, pH 8.0/10 mM EDTA) for 1 h at 4°C with 5 μM biotinylated anti-wt oligo (5′-GTTGGCGGCCTTCCACTGGGCCTGCTTCTCAAA-Biotin) immobilized on streptavidin agarose. The eluent was saved and subjected to the procedure two additional times. This process was efficient as none of the round 12 clones contained the wt sequence.
Quantitation by Affinity Precipitation.
Peptides for each clone were produced by 35S-Met-labeled in vitro translation of the mRNA alone under conditions identical to fusion synthesis (17). For library 2 peptides (Fig. 3), 10 μl of crude lysate was mixed with 1 ml 200 nM immobilized boxBR in N binding buffer, washed five times with N-binding buffer and eluted with RNase A (Roche Molecular Biochemicals). For library 3 peptides (Fig. 4), 6–7.5 μl of crude translation reaction was mixed with 160 nM of all three immobilized hairpins in 500 μl of N binding buffer and 50 μg/ml yeast tRNA for 1 h at 4°C and was eluted in tricine loading buffer. In both cases, the eluted peptides were run adjacent to a sample of the translation reaction on a tricine–SDS polyacrylamide gel (21), and the percentage of peptide bound was determined by PhosphorImager counting (Molecular Dynamics) of the respective gel bands.
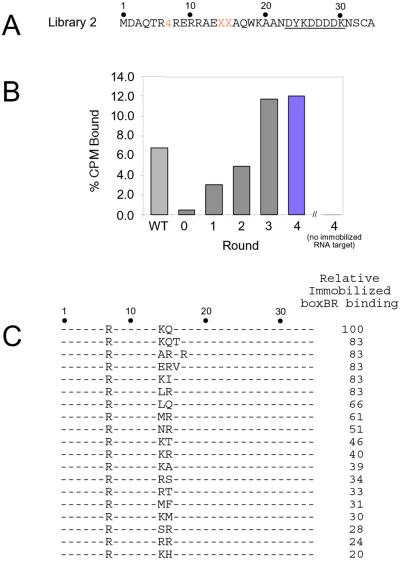
Figure 3.
Selection of λ boxBR binding peptides from library 2. (A) Sequence of library 2. The target is the boxBR RNA (Fig. 1B). Peptide position 7 contains a cassette encoding 4 amino acids, whereas positions 14 and 15 encode all 20 possible amino acids and one stop codon (NNG/C codons). (B) Binding to immobilized boxBR RNA for rounds 0–4 measured by using 35S-Met-labeled fusions. Controls using the wt complex (WT) and agarose with no immobilized RNA (no boxBR) are shown. Material from the fourth round (colored bar) was cloned and sequenced. (C) Peptide sequences and peptide-binding affinities of selected clones. Binding affinities are given relative to the wt sequence in the same construct.
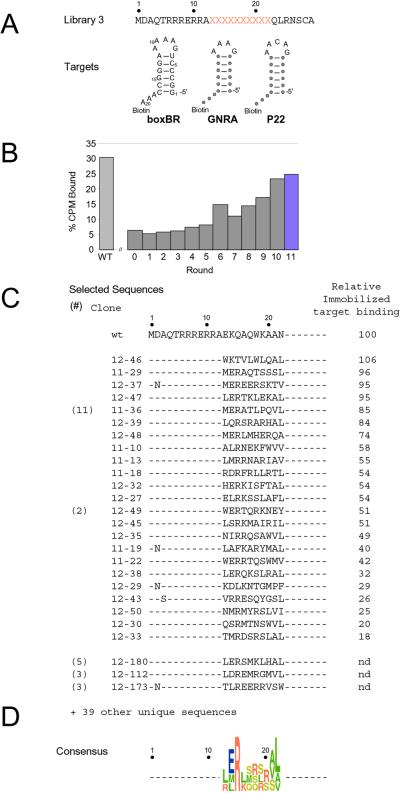
Figure 4.
Selection of RNA-binding peptides from library 3. (A) Sequence of library 3 and the three RNA targets used (λ-boxBR, GNRA tetraloop, and P22 boxBL). Positions of identity are shown as circles. All three stems are identical. The resulting P22 stem differs from the wt sequence by 1 bp. (B) Binding of the rounds 0–11 mRNA-peptide pools (35S-Met-labeled) to beads containing all three immobilized hairpins. (C) Peptide sequences and binding affinities from rounds 11 and 12. The relative binding affinity as compared with the wt boxBR/λ N interaction is given. (D) Consensus sequence of selected clones. The residues are color-coded by amino acid type (green, hydrophobic; red, positively charged; blue, negatively charged; and yellow, polar) and the height indicates the occurrence frequency of each residue.
Fluorescence-Binding Measurements.
Peptides were constructed by means of automated synthesis and fluorenylmethloxycarbonyl or t-butyloxycarbonyl monomers. Crude peptides were purified as single peaks by means of reversed-phase HPLC and the identity was confirmed by mass spectrometry. RNA hairpins containing 2-amino purine at the second loop position (denoted 2AP-2, Table 1; substitution for A8) were constructed by automated RNA synthesis by using 2′-O-methyl 2-aminopurine phosphoramidite (Glen Research, Sterling, VA).
Table 1.
Binding constants and specificity of selected and control peptides
| Name | BoxBR (2AP-2), nM | GNRA (2AP-2), nM | P22 (2AP-2), nM |
|---|---|---|---|
| λ N (1-22) | 1.0 ± 0.2 | 9.0 ± 0.7 | 41 ± 3 |
| Selection 2 | |||
| λ N (1-22; E14R15) | 8.5 ± 2 | 800 ± 150* | 580 ± 20 |
| Selection 3 | |||
| 12-50 (1-22) | 0.4 ± 0.1† | 9 ± 2 | 14 ± 1 |
| 12-39 (1-22) | 0.5 ± 0.1† | 6.5 ± 0.7 | 8.4 ± 0.8 |
| 11-10 (1-22) | 1.9 ± 0.5† | 93 ± 9 | 140 ± 10 |
| 11-36 (1-22) | 3.0 ± 0.2† | 288 ± 7 | 246 ± 7 |
| 12-47 (1-22) | 3.4 ± 0.1 | 81 ± 2 | 60 ± 3 |
| Controls | |||
| λ N (1-11) | 1,290 ± 20 | 19,000 ± 800 | 63,000 ± 7,200 |
| λ N (1-15; E14R15) | 303 ± 8 | 7,500 ± 300 | 12,300 ± 200 |
| 11-36 (1-22) scrambled | 210 ± 25 | 3,400 ± 200 | 10,700 ± 200 |
| 12-47 (1-22) scrambled | 140 ± 12 | 2,000 ± 80 | 5,200 ± 100 |
Binding constants were determined by fluorescence titration at 20°C, 50 mM KOAc/20 mM Tris⋅Oac, pH 7.5. All peptides contain a free amino and carboxyl terminus. λ N (1-11), λ N (1-15, E14R15), 11-36 (1-22) scrambled, and 12-47 (1-22) scrambled contain a C-terminal GY sequence to facilitate quantitation. 2AP-2 denotes a 2′-methoxy 2-aminopurine residue inserted at the second loop position. Error estimates indicate the precision of individual fits. Data for ERV from selection 2 indicates the binding is similar to λ N (1-22, E14R15; unpublished observation).
Contains high uncertainty due to a small fluorescence change upon peptide addition.
Indicates values determined by extrapolation from higher salt.
Fluorescence measurements were made essentially as in Lacourciere et al. (22), with excitation and emission wavelengths of 310 and 370 nm, respectively. Concentrated peptide was added stepwise to a stirred solution of 20 nM to 800 nM 2AP-2 RNA hairpin and the temperature was maintained at 20°C.
CD Spectroscopy.
Spectra were taken on an Aviv 62 DS CD spectrometer at 25°C. The samples contained 5 μM RNA and 6 μM peptide in 10 mM potassium phosphate buffer (pH 7.9). The spectra of the bound peptides were determined by subtracting the free RNA and excess free peptide spectra from the spectra of the complex.
NMR Sample Preparation.
BoxBR RNA 5′-GCCCUGAAAAAGGGC-3′ (15-mer) was synthesized by in vitro transcription by using T7 RNA polymerase. The RNA was purified by 20% urea-PAGE, desalted on a NAP column (Amersham Pharmacia), freeze-dried, and resuspended in NMR buffer (50 mM NaCl/10 mM phosphate, pH 6) in H2O/D2O (90:10, vol/vol). Complexes between the wt λ N (1–22) or 11–36 (1–22) and boxBR RNA were generated by addition of concentrated (≈10 mM) peptide to boxBR RNA (280 μM) with the stoichiometry monitored by inspecting the imino–proton spectra. The final sample concentrations were ≈250 μm for the free RNA and 280 μM for both RNA and peptide in the complexes.
NMR Spectroscopy.
NMR spectra were collected at 25°C on a Varian INOVA 600-MHz spectrometer. A modified double gradient echo Watergate solvent-suppression pulse sequence was used to suppress the solvent peak (23). Assignments were based on reported work (13, 15, 16).
Results and Discussion
It was unclear a priori whether an RNA-binding peptide would be functional as either a mRNA-peptide or cDNA/mRNA-peptide fusion. We synthesized mRNA-peptide fusions containing the λ-N RNA binding domain (17) (Fig. 1B) and tested the ability of these molecules to bind an immobilized target. Fig. 2 shows that both the mRNA-peptide fusion and the cDNA/mRNA-peptide fusion specifically bind the boxBR RNA target. These assays demonstrate that a significant fraction of the desired complex can be isolated (30–60%) whereas little (1–5%) is retained if noncognate biotinylated RNA targets (Rev Response Element, RRE; U1 RNA hairpin loop II, U1) are used instead of the boxBR RNA.
We designed three selections to test our ability to isolate novel tetraloop-binding peptides. In selection 1, we randomized positions 6 and 7 of the wt construct. At these positions, previous mutagenesis and structural work indicated a single sequence, R6R7, should be optimal for binding. The cassette contained 150 different amino acid combinations, only one of which was R6R7. We performed selection 1 (one round), three times in parallel, by using increasing concentrations of competitor tRNA to provide high stringency selection. A majority (85%) of the selected clones contained R6R7 when 5 mg/ml tRNA competitor was used. Thus, increased competitor allowed good enrichment (50- to 150-fold per round) without forcing us to make the target the limiting reagent (18).
In selection 2 (Fig. 3A, library 2) we randomized positions 7, 14, and 15, which total 1,600 possible combinations. In the wt λ N/boxB complexes, residues 14 and 15 lie at the RNA-peptide interface (15, 16) and are important for binding (1, 14). After four rounds of selection, the pool binding was similar to the wt sequence (Fig. 3B). Analysis of 39 round-4 clones yielded 19 different sequences (Fig. 3C). Each peptide was constructed by means of in vitro translation, and the binding was assayed to immobilized boxBR RNA (Fig. 3C). All of the corresponding peptides bind the boxBR RNA and contain arginine at position 7. Aside from the wt (K14Q15), only three of the highest-affinity clones show identity with the original two residues. Indeed, a different sequence containing R15 emerges as a dominant motif, including E14R15, replacing lysine-14 with glutamic acid.
Peptides corresponding to wt λ N (1–22) and λ N (1–22; E14R15) were constructed synthetically, and the binding to the boxBR RNA was measured (Table 1). Both peptides bind with high affinity to the target site (1.9 nM vs. 8.5 nM). The difference is small considering that a single change of K14 to A14 results in loss of more than 2 kcal/mol (13). The high specificity of λ N (1–22; E14R15) may also have played an important role in its selection. The appearance of glutamic acid, though puzzling because of its negative charge, may result from favorable interaction with the helix dipole of residues 12–22 (19, 20).
Selection 3 was designed by randomizing positions 13–22 in the wt λ N peptide, creating a library near the complexity limit of the mRNA-peptide fusion method. The selection was conducted versus three different RNA targets differing only in their loop structures: (i) the boxBR, (ii) a GNRA tetraloop, and (iii) the P22 loop (Fig. 4A). Previous structural work indicated that the C-terminal helix might discriminate between different loops because it contacts the loop in the λ N/boxB structure (15, 16). Additionally, the N-terminal arginine-rich region shows conservation in N proteins from Escherichia coli phages λ and φ21, and Samonella phage p22, despite differences in the RNA targets (11).
Library 3 (Fig. 4A) began with ∼9 trillion individual sequences, capturing the majority of the 10 trillion possible 10-aa cassettes that could exist in the library. We performed 12 rounds of selection against an equimolar mixture of three hairpins (Fig. 4 A and B; rounds 1–11). Binding to any one of the three hairpins was sufficient to allow a peptide to be selected, because each target exceeded the total number of moles of fusion peptide in the selective step. Binding stringency was controlled by addition of yeast tRNA (5 mg/ml) as a nonspecific competitor, rather than limiting target.
The sequences isolated from rounds 11 and 12 are shown in Fig. 4C. We generated 35S-Met-labeled peptides for 23 individual clones by means of in vitro translation, and we measured the fraction of each peptide that bound a mixture of the three hairpins (Fig. 4C). These experiments indicated that each peptide tested could bind at least one of the hairpins in the mixture. The specificity of seven individual clones, the round 11 and round 12 pools were then tested by using each individual hairpin. All showed a marked preference for the boxBR RNA as compared with the GNRA or P22 targets (data not shown).
Peptides corresponding to five of the selected sequences were constructed synthetically, and the binding constants to all three hairpins were measured by fluorescence titration (Table 1, selection 3; Fig. 5A). All show high affinity (Kd ≤ 3.5 nM) for the boxBR target, with two showing equal or better affinity than the wt λ N (1–22) peptide itself. The specificity of the sequences is also quite high, with both 11–36 and 11–10 showing better specificity than wt λ N, particularly against the GNRA hairpin.
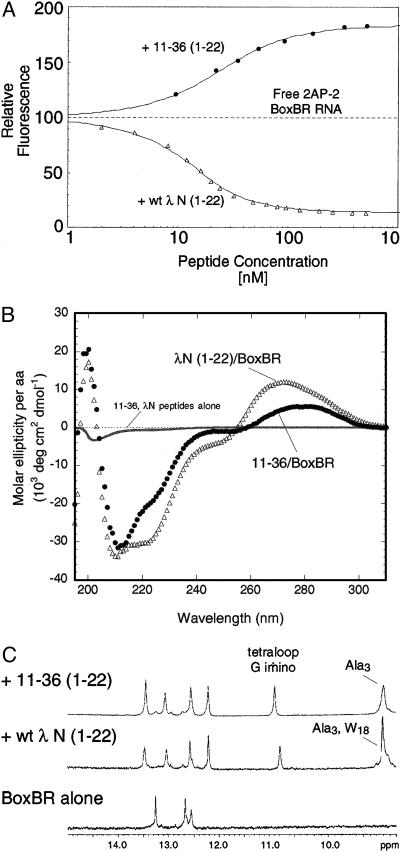
Figure 5.
Properties of 11–36 (1–22) vs. wt λ N (1–22). (A) Binding isotherm for 11–36 (1–22) and wt λ N (1–22) detected by fluorescence of 2AP-2-labeled boxBR in 100 mM KOAc/20 mM Tris⋅OAc. The fluorescence of the RNA is quenched by wt λ N (1–22) and enhanced by 11–36 (1–22). (B) CD spectra of 11–36 (1–22) and wt λ N (1–22). (C) NMR spectra of the BoxBR RNA alone and complex with the wt λ N (1–22) or 11–36 (1–22) peptides. Both peptides protect the G loop nucletide from exchange (10.7 ppm) and give rise to similar amino spectra in the RNA stem.
Given the similar specificity of the peptides, it is surprising how dissimilar the sequences are when compared with wt λ N. On average, there is less than one match per insert. Further, positions known to be important in the binding of λ N (W18, K14, and Q15) show little conservation. There is no similarity with N sequences derived from phages P22 or φ21 (11). Only one sequence, 12–46, contains W18, a residue that cannot be changed in wt λ N without impairing binding (13). Sequence 12–46 is also the only clone that contains K14, revealing that these two positions may be energetically coupled. NMR and fluorescence analysis of a series of mutants at positions 14, 15, and 18 supports this notion (J.R., T.X, T.T.T., and R.W.R. unpublished data).
The selected peptides are somewhat similar to each other, showing an average identity of ∼2.5 residues per cassette when individual clones are compared. The identity results largely from strong conservation of R15 and partial conservation of E14 and L22 (Fig. 4D). Modeling R15 into the λ N structure reveals a possible basis for this conservation—a side chain to main-chain H bond can be formed for both Q15 (wt) and R15. In the R15 model, this H bond would orient the positively charged guanidinium side chain directly toward the A8 and A9 phosphates.
λ N (1–11) and λ N (1–15; E14R15) peptides were synthesized to address the specificity of selected clones and the conserved arginine (Table 1, controls). Surprisingly, λ N (1–11) shows the same selectivity with the full-length recognition domain, λ N (1–22). Binding to the GNRA and P22 hairpins shows a ΔΔG° of 1.3 and 2.2 kcal/mol for λ N (1–22), whereas the binding of λ N (1–11) shows ΔΔG° values of +1.6 and +2.3 kcal/mol. Thus the origin of specificity for the peptide appears to be conferred by the N-terminal 11 residues.
This bias provides an explanation as to why isolation of GNRA and P22 binders proved so challenging—P22- or GNRA-specific clones would require the random region to provide an extra 1.3–2.3 kcal/mol in binding free energy as compared with the boxBR hairpin. In principle, it should be possible to find peptides in the library with specificity for P22 or the GNRA loop. However, further selection for peptides specific to these two hairpins has not resulted in sequences with swapped specificity (J.E.B. and R.W.R., unpublished observations). Unresolved is how the λ N (1–11) peptide discriminates between three hairpins that differ by a single loop nucleotide. The specificity of λ N (1–11) reveals that despite the sequence identity, these three hairpins may have different global structures before peptide binding.
We synthesized a number of peptides to address the role of the residues between positions 13 and 22. The λ N (1–15; E14R15) peptide reveals that addition of R15 cannot be the sole feature that confers low nanomolar binding to the selected peptides. Adding the sequence containing E14R15 to the λ N (1–11) peptide confers only a 3- to 5-fold enhancement in Kd, a ΔΔG° of −0.7 to −1.0 kcal/mol (Table 1, λ N 1–15; E14R15). This result indicates that sequences between positions 16 and 22 contribute ∼3 kcal/mol of binding free energy to the interaction.
We then constructed versions of peptides 12–47 and 11–36 in which the selected insert was scrambled [12–47 (scrambled) = MDAQTRRRERRAELELLTRKKA-GY; 11–36 (scrambled) = MDAQTRRRERRATAPEQVMLR-GY] (Table 1, Controls). If net positive charge and a propensity to form α-helices were sufficient in the random region, then the exact order of the residues should not impair binding. Comparison of the round 11 and round 12 clones with the starting library reveals a net positive charge in the insert and overrepresentation of amino acids that are good helix formers. However, the scrambled sequences show a loss of ∼100-fold in Kd relative to the unscrambled peptides (130 ± 12 nM and 220 nM ± 25, Table 1). Thus, the precise order of residues is necessary to obtain maximal affinity.
Finally, we examined the structure and properties of the 11–36 (1–22) complex as compared with λ N (1–22). The fluorescence titration provides a clear demonstration that the detailed interactions are different in the two complexes (Fig. 5A). Addition of λ N (1–22) produces strong quenching with the 2AP-2 probe, whereas 11–36 enhances the fluorescence at that position. Quenching in the λ N (1–22)/boxBR complex is expected, as W18 is quenched in the complex (13) and should stack directly on top of 2AP-2 (15, 16). In contrast, 11–36 contains no aromatic amino acids. In previous work, 2AP quenching or enhancement has been interpreted as the result of increased or decreased stacking (22). This finding implies that the adenine at loop position 2 is likely less stacked in the 11–36 complex than it is in the free RNA. Profound distortion of the loop is unlikely, though, as both peptides support folding of the tetraloop motif as judged by appearance of the G loop imino at ∼10.8 ppm (Fig. 5C).
In the CD spectra, neither peptide shows any appreciable structure in the absence of RNA (Fig. 5B). The difference spectra of the complex indicates that both peptides fold into α-helices when bound to the RNA (Fig. 5B). Although globally similar, the two complexes display differences in regions indicative of peptide folding (200–225 nm) and RNA folding (260–300 nm). The detailed structure is thus somewhat mercurial and depends on which peptide is bound.
Our results demonstrate that high-affinity RNA ligands may be isolated by using in vitro selection. The large starting library resulted in a great diversity of solutions to this binding/recognition problem. Although general features such as helix propensity and charge may assist recognition, no code for recognition is discernable. Rather, our results are more consistent with a model where RNA-peptide interactions are highly context dependent. In that vein, the combinatorial approach we have used provides a powerful design strategy for interactions where rules may be idiosyncratic at best.
Acknowledgments
We thank Prof. S. L. Mayo, Prof. D. C. Rees, Prof. M. J. Matari′c, R. Austin, S. Li, and B. Ja for comments on the manuscript and T. Snyder for his work defining the round 0 clones. This work was supported by grants from the National Science Foundation (to R.W.R.), National Institutes of Health (to R.W.R.), and Beckman Foundations (to R.W.R.).
Abbreviation
- wt
wild type
References
- 1.Franklin N C. J Mol Biol. 1993;231:343–360. doi: 10.1006/jmbi.1993.1287. [DOI] [PubMed] [Google Scholar]
- 2.Harada K, Martin S S, Frankel A D. Nature (London) 1996;380:175–179. doi: 10.1038/380175a0. [DOI] [PubMed] [Google Scholar]
- 3.Jain C, Belasco J G. Cell. 1996;87:115–125. doi: 10.1016/s0092-8674(00)81328-8. [DOI] [PubMed] [Google Scholar]
- 4.Heus H A, Pardi A. Science. 1991;253:191–194. doi: 10.1126/science.1712983. [DOI] [PubMed] [Google Scholar]
- 5.Cate J H, Gooding A R, Podell E, Zhou K, Golden B L, Szewczak A A, Kundrot C E, Cech T R, Doudna J A. Science. 1996;273:1696–1699. doi: 10.1126/science.273.5282.1696. [DOI] [PubMed] [Google Scholar]
- 6.Cate J H, Gooding A R, Podell E, Zhou K, Golden B L, Kundrot C E, Cech T R, Doudna J A. Science. 1996;273:1678–1685. doi: 10.1126/science.273.5282.1678. [DOI] [PubMed] [Google Scholar]
- 7.Roberts R W, Szostak J W. Proc Natl Acad Sci USA. 1997;94:12297–12302. doi: 10.1073/pnas.94.23.12297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roberts R W. Curr Opin Chem Biol. 1999;3:268–273. doi: 10.1016/S1367-5931(99)80042-8. [DOI] [PubMed] [Google Scholar]
- 9.Liu R, Barrick J, Szostak J W, Roberts R W. Methods Enzymol. 2000;317:268–293. doi: 10.1016/s0076-6879(00)18058-9. [DOI] [PubMed] [Google Scholar]
- 10.Cho G, Keefe A D, Liu R, Wilson D S, Szostak J W. J Mol Biol. 2000;297:309–319. doi: 10.1006/jmbi.2000.3571. [DOI] [PubMed] [Google Scholar]
- 11.Lazinski D, Grzadzielska E, Das A. Cell. 1989;59:207–218. doi: 10.1016/0092-8674(89)90882-9. [DOI] [PubMed] [Google Scholar]
- 12.Tan R, Frankel A D. Proc Natl Acad Sci USA. 1995;92:5282–5286. doi: 10.1073/pnas.92.12.5282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Su L, Radek J T, Hallenga K, Hermanto P, Chan G, Labeots L A, Weiss M A. Biochemistry. 1997;36:12722–12732. doi: 10.1021/bi971408k. [DOI] [PubMed] [Google Scholar]
- 14.Cilley C D, Williamson J R. RNA. 1997;3:57–67. [PMC free article] [PubMed] [Google Scholar]
- 15.Legault P, Li J, Mogridge J, Kay L E, Greenblatt J. Cell. 1998;93:289–299. doi: 10.1016/s0092-8674(00)81579-2. [DOI] [PubMed] [Google Scholar]
- 16.Scharpf M, Sticht H, Schweimer K, Boehm M, Hoffmann S, Rosch P. Eur J Biochem. 2000;267:2397–2408. doi: 10.1046/j.1432-1327.2000.01251.x. [DOI] [PubMed] [Google Scholar]
- 17.Barrick J E, Takahashi T T, Balakin A, Roberts R W. Methods. 2001;23:287–293. doi: 10.1006/meth.2000.1139. [DOI] [PubMed] [Google Scholar]
- 18.Roberts R W, Crothers D M. Proc Natl Acad Sci USA. 1991;88:9397–9401. doi: 10.1073/pnas.88.21.9397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Richardson J S, Richardson D S. Science. 1988;240:1648–1652. doi: 10.1126/science.3381086. [DOI] [PubMed] [Google Scholar]
- 20.Dasgupta S, Bell J A. Int J Peptide Protein Res. 1993;47:499–511. [PubMed] [Google Scholar]
- 21.Schagger H, Jagow G V. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 22.Lacourciere K A, Stivers J T, Marino J P. Biochemistry. 2000;39:5630–5641. doi: 10.1021/bi992932p. [DOI] [PubMed] [Google Scholar]
- 23.Liu M, Mao X, Ye C, Huang H, Nicholson J K, Lindon J. J Magn Res. 1998;132:125–129. [Google Scholar]