Abstract
Background
Molecular profiling of non-small cell lung cancer (NSCLC) is essential for therapeutic decision-making. Pleural effusion obtained by a non-invasive, repeatable procedure may provide an opportunity for molecular profiling and thereby possibly provide information enabling targeted therapy. In this study, we aimed to evaluate the diagnostic performance of pleural effusion as a specimen for molecular analysis.
Methods
Thirty patients with paired malignant pleural effusion and thoracic biopsy specimens were included. Clinically actionable mutations were assessed using a validated targeted next generation sequencing assay. EGFR/KRAS/ALK mutation status in thoracic biopsy specimens was tested using ARMS PCR.
Results
The concordance rate between gene status identified by ARMS and next-generation sequencing (NGS) analysis in the thoracic biopsy and pleural effusion samples was 86.7% (26/30). Compared with the thoracic biopsy specimens, the diagnostic performance of pleural effusion showed a sensitivity of 92.3%, a specificity of 50.0%, and a positive predictive value of 92.3%. Therefore, cases with a low percentage of tumor cells (<5%) can successfully be used to detect actionable mutations in pleural effusion specimens.
Conclusions
These results suggest that pleural effusions are suitable specimens for oncogene mutation analysis and enable targeted therapy for patients with advanced NSCLC.
Keywords: Pleural effusion, lung adenocarcinoma, actionable genomic alteration, next generation sequencing
Introduction
Lung cancer is the leading cause of cancer death in China, accounting for 1/4 of all cancer-related mortality in the country (1). The introduction of targeted therapy for EGFR and ALK has dramatically changed the therapeutic strategy for advanced lung adenocarcinoma (2-4). However, the treatment effect of tyrosine kinase inhibitors (TKIs) is largely associated with the mutation status of EGFR and ALK. EGFR-activating mutations are predominantly observed in adenocarcinomas and are more common in tumors from never-smokers, women, and in East Asians (5-7). Thus, molecular profiling of lung adenocarcinoma has become increasingly crucial in predicting the response to EGFR- and ALK TKI-targeted therapy.
Surgical tumor tissue has been considered the preferred sample type for EGFR and ALK mutation screening. However, only early lung adenocarcinoma patients are suitable for surgical resection, contrasting patients with advanced non-small cell lung cancer (NSCLC) (70%) for which only cytology materials were obtained using non-surgical sampling techniques will be available for analysis severely limiting opportunity to perform mutation screening (8). Pleural effusion may be an alternative material for analysis when tumor tissues are not available or adequate for mutation analysis. Pleural effusion is a common complication of lung cancer, especially adenocarcinoma, and indicates an advanced stage of disease (9,10). Given that sampling of pleural effusion fluid is usually easy, non-invasive, and repeatable, and previous studies have demonstrated the benefits of using body fluids for detecting EGFR/KRAS mutations (11-16). Targeted next-generation sequencing (NGS) allows to rapid detection of a variety of cancer-related genes on a single platform (such as Ion Torrent Proton and MiSeq) (17-19). This novel approach may overcome the limitations in the detection of somatic mutations from small amounts of DNA.
In the present study, we attempt to detect actionable mutations in matched tumor tissues and pleural effusion samples, using both ARMS PCR and targeted NGS. To our knowledge, this is the first study to use an ion torrent sequencing platform to detect gene mutations in pleural effusion samples.
Methods
Patients and samples
We selected NSCLC patients with malignant pleural effusion at the time of diagnosis and who underwent biopsy at the First Affiliated Hospital of Guangzhou Medical University between December 2016 and March 2017. The pleural effusion samples were centrifugated, then cell pellets were collected and prepared into cell blocks. The cell block sections were stained with hematoxylin and eosin. Malignant pleural effusion was confirmed by demonstration of malignant cells on cytology examination and tumor cell percentage for each sample were estimated by pathologists. A total of 30 malignant pleural effusion and matched-biopsy specimens with a documented EGFR/KRAS/ALK mutation were available for testing. Relevant clinical and pathological information on the subject, including age, sex, smoking history, histological type, and cancer cell content, were obtained from the electronic health record. All subjects provided written informed consent, and the study protocol was approved by the Institutional Review Board of the First Affiliated Hospital of Guangzhou Medical University. All the molecular tests were conducted in accredited clinical genetics laboratories.
NGS-based genotyping
The pleural effusion specimens were reviewed to assess tissue adequacy before testing. DNA was extracted from unstained FFPE resections, using the QIAamp DNA FFPE Tissue Kit (Qiagen, Hilden, Germany). A well validated targeted NGS assay was used to identify the clinically relevant mutation (20). Briefly, barcoded genomic DNA libraries were constructed for massively parallel sequencing. DNA libraries were pooled by mixing 200 ng DNA from each library. Solution-phase hybrid capture for 145 clinically relevant genes associated with lung cancer was performed. Multiplexed hybrid capture libraries were pooled proportionally and were then sequenced to >200× average unique coverage, using Ion Proton Sequencers (Thermo Fisher, Waltham, MA, USA). A clinically validated bioinformatics pipeline named “Otype” was used to detect clinically relevant genomic alterations in 9 genes (EGFR, KRAS, PIK3CA, BRAF, MET, ERBB2, ALK, ROS1, and RET). This NGS assay showed a concordance rate of 100% between results obtained by NGS and conventional test platforms in a validation study using 61 previously profiled clinical tumor samples (20).
ARMS PCR
Genomic DNA in thoracic biopsy samples were extracted using QIAamp DNA FFPE tissue kits (Qiagen, Hilden, Germany). Mutations of EML4-ALK, EGFR, and KRAS were analyzed using commercially available kits from Amoy Diagnostics (Xiamen, China), based on the ARMS real time PCR technology. The EGFR kit detects 29 mutations in exons 18-21, including T790M, L858R, L861Q, S768I, G719S, G719A, G719C; three insertions in exon 20; and 19 deletions in exon 19. The KRAS kit detects seven mutations, namely, G12D, G12A, G12V, G12S, G12R, G12C, and G13D. The EML4-ALK kit detects nine fusions. The detection limit for mutations in EGFR and KRAS ranges from 1% to 2.5%. The ALK gene fusion detection kit allows detection of 25 copies/µL ALK gene fusions plasmid DNA.
Statistical analysis
Statistical analysis was performed using R studio 19.0 (RStudio, Inc., Boston, MA, USA). Concordance of results between thoracic biopsy and pleural effusion specimens was determined using Cohen’s statistic. A two-tailed P value of less than 0.05 was considered statistically significant. Concordance rate, sensitivity, specificity, and positive predictive value were calculated for mutations in pleural effusion specimens and compared with those in thoracic biopsy specimens (regarded as the reference standard).
Results
Patients and specimens
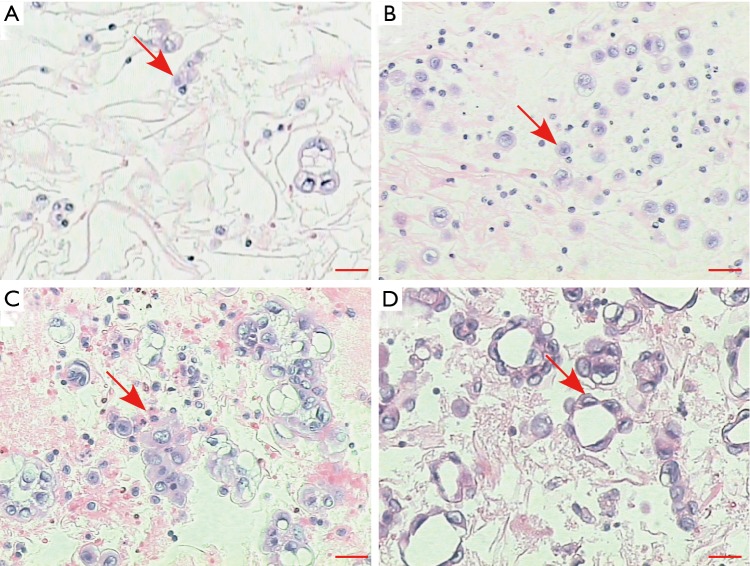
With the aims to evaluate the diagnostic performance of pleural effusion as a specimen for molecular analysis, we selected patients who were highly suspected to have genetic alternations based on the clinical and histological features. Table 1 shows the demographic characteristics of the enrolled patients. The mean age of the patients was 60 years, and 17 patients were male. All 30 patients were diagnosed as having advanced lung adenocarcinoma based on biopsy specimens. For each patient, the EGFR/KRAS/ALK mutation status assessed by ARMS PCR of the thoracic biopsy specimen was considered as a reference to determine the accuracy of molecular tests conducted on pleural effusions. Twenty-two (73.3%) of the 30 thoracic biopsy specimens harbored EGFR mutations. The most frequent mutations were the point mutation (L858R) in exon 21 (11/22, 50.0%) and the deletion mutation in exon19 (9/22, 40.9%). Aside from the EGFR mutation, 2 cases exhibited KRAS mutations (G12V), 1 case harbored ALK fusions, and 5 cases were wild type. Of these paired pleural effusions, neoplastic cell content ranged from 1% to 20%, 17 specimens had less than 10% neoplastic cells (Figure S1). DNA was extracted from all 30 pleural effusions in amounts ranging from 53 ng to 2.9 µg (Table S1).
Table 1. Patient characteristics.
| Characteristic | N=30 | % |
|---|---|---|
| Gender | ||
| Male | 17 | 56.7 |
| Female | 13 | 43.3 |
| Age | ||
| Median | 60 | |
| Range | 37–84 | |
| Smoker | ||
| Never | 18 | 60 |
| Ever | 12 | 40 |
| Histology | ||
| Adenocarcinoma | 30 | 100 |
| Tumor cell % (pleura effusion specimen) | ||
| 1–5 | 7 | 23.3 |
| 6–10 | 10 | 33.3 |
| 11–15 | 5 | 16.7 |
| 16–20 | 8 | 26.7 |
Figure S1.
Representative images of pleural effusion specimens. (A) Tumor cell percentage 1–5%; (B) tumor cell percentage 6–10%; (C) tumor cell percentage 11–15%; (D) tumor cell percentage 16–20%. Hematoxylin and eosin. Scale bars, 20 µm.
Table S1. Tumor cells percentage and DNA/RNA amount extracted from the thoracic biopsy specimens and pleural effusion specimen.
| Sample ID | Thoracic biopsy specimen | Pleural effusion specimen | |||||
|---|---|---|---|---|---|---|---|
| Tumor cell % | DNA amount (ng) | RNA amount (ng) | Tumor cell % | DNA amount (ng) | RNA amount (ng) | ||
| #1 | 25 | 378 | 825 | 11–15 | 540 | 980 | |
| #2 | 20 | 240 | 600 | 1–5 | 56 | 116 | |
| #3 | 20 | 160 | 465 | 6–10 | 65 | 145 | |
| #4 | 30 | 460 | 960 | 11–15 | 78 | 136 | |
| #5 | 25 | 420 | 1,110 | 15–20 | 1,510 | 2,978 | |
| #6 | 25 | 200 | 540 | 6–10 | 75 | 132 | |
| #7 | 25 | 330 | 712 | 1–5 | 81 | 178 | |
| #8 | 20 | 290 | 645 | 6–10 | 68 | 149 | |
| #9 | 50 | 630 | 1,222 | 1–5 | 53 | 140 | |
| #10 | 45 | 540 | 1,177 | 1–5 | 98 | 194 | |
| #11 | 35 | 380 | 900 | 6–10 | 228 | 479 | |
| #12 | 30 | 250 | 585 | 11–15 | 1,830 | 3,240 | |
| #13 | 20 | 230 | 525 | 16–20 | 1,000 | 1,680 | |
| #14 | 30 | 420 | 945 | 6–10 | 160 | 397 | |
| #15 | 25 | 350 | 750 | 16–20 | 720 | 1,875 | |
| #16 | 25 | 270 | 510 | 11–15 | 635 | 1,770 | |
| #17 | 40 | 460 | 930 | 16–20 | 144 | 254 | |
| #18 | 30 | 340 | 705 | 16–20 | 164 | 259 | |
| #19 | 35 | 490 | 1,005 | 6–10 | 2,960 | 4,730 | |
| #20 | 40 | 560 | 1,260 | 16–20 | 725 | 1,828 | |
| #21 | 35 | 390 | 795 | 1–5 | 360 | 950 | |
| #22 | 30 | 270 | 615 | 6–10 | 60 | 240 | |
| #23 | 40 | 460 | 1,035 | 11–15 | 194 | 350 | |
| #24 | 35 | 420 | 795 | 1–5 | 165 | 430 | |
| #25 | 25 | 380 | 780 | 16–20 | 740 | 1,380 | |
| #26 | 35 | 420 | 945 | 1–5 | 143 | 320 | |
| #27 | 30 | 360 | 885 | 6–10 | 115 | 278 | |
| #28 | 20 | 220 | 510 | 6–10 | 62 | 153 | |
| #29 | 25 | 330 | 750 | 16–20 | 1,000 | 2,312 | |
| #30 | 40 | 530 | 1,140 | 6–10 | 248 | 560 | |
Genotyping by NGS in pleural effusions
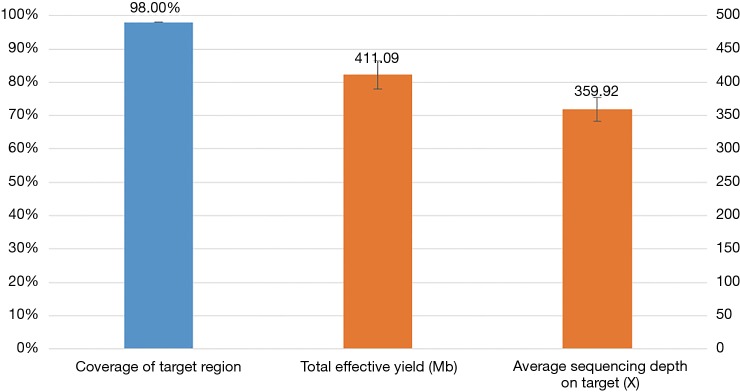
Mutation status was tested in all 30 pleural effusion specimens using targeted NGS. Hybrid captured libraries were sequenced to a mean of 2.4 million sequencing reads per sample with high uniform depth (an average of 359× coverage by unique reads, with >98% of target region at coverage >100×) (Figure S2). Detailed comparisons of gene mutation analyses of thoracic biopsy and pleural effusion specimens by ARMS PCR and targeted NGS are listed in Table 2. Of the 30 paired samples, 20 presented the same EGFR mutations [10 L858R mutations, 8 exon 19 deletions (Figure 1), 1 G719S, and 1 exon 20 insertion] in both the thoracic biopsy specimens and the corresponding pleural effusion sample (Table 2). The same KRAS mutations (2 G12V) were found in both types of specimens in 2 cases. EML4-ALK fusion was identified by both methods in both specimen types of one patient. No mutation was found in the other 6 genes (PIK3CA, BRAF, MET, ERBB2, ROS1, and RET). Two EGFR mutations (L858R mutation and exon 19 deletion) detected by ARMS PCR were identified as wild type by NGS in pleural effusion specimens, and two EGFR mutations (L858R and G719S) detected by NGS were identified as wild type by ARMS PCR.
Figure S2.
Capture and sequencing performance of 30 pleural effusion specimens.
Table 2. Gene mutation analysis of paired thoracic biopsy specimens and pleura effusion specimens using ARMS and NGS.
| Sample ID | Thoracic biopsy specimen (ARMS PCR) | Pleura effusion specimen | ||
|---|---|---|---|---|
| NGS | Allele fraction (%) | ARMS PCR | ||
| #1 | EGFR L858R | EGFR L858R | 10.5 | EGFR L858R |
| #2 | EGFR exon 20 insertion | EGFR exon 20 insertion | 5.5 | EGFR exon 20 insertion |
| #3 | Wild type | EGFR L858R | 6.1 | EGFR L858R |
| #4 | EGFR exon 19 deletion | EGFR exon 19 deletion | 13.2 | EGFR exon 19 deletion |
| #5 | Wild type | Wild type | – | Wild type |
| #6 | EGFR L858R | EGFR L858R | 4.8 | EGFR L858R |
| #7 | EGFR G719S | EGFR G719S | 3.2 | EGFR G719S |
| #8 | EGFR exon 19 deletion | EGFR exon 19 deletion | 5.0 | EGFR exon 19 deletion |
| #9 | EGFR L858R | EGFR L858R | 5.6 | EGFR L858R |
| #10 | EGFR L858R | EGFR L858R | 6.5 | EGFR L858R |
| #11 | Wild type | EGFR G719S | 7.2 | EGFR G719S |
| #12 | EGFR exon 19 deletion | EGFR exon 19 deletion | 11.0 | EGFR exon 19 deletion |
| #13 | EGFR L858R | EGFR L858R | 16.9 | EGFR L858R |
| #14 | EGFR exon 19 deletion | EGFR exon 19 deletion | 13.0 | EGFR exon 19 deletion |
| #15 | EGFR exon 19 deletion | EGFR exon 19 deletion | 15.1 | EGFR exon 19 deletion |
| #16 | EGFR exon 19 deletion | EGFR exon 19 deletion | 11.1 | EGFR exon 19 deletion |
| #17 | EGFR L858R | EGFR L858R | 16.9 | EGFR L858R |
| #18 | EGFR L858R | EGFR L858R | 11.4 | EGFR L858R |
| #19 | KRAS G12V | KRAS G12V | 6.7 | KRAS G12V |
| #20 | Wild type | Wild type | 12.1 | Wild type |
| #21 | EGFR exon 19 deletion | EGFR exon 19 deletion | 4.1 | EGFR exon 19 deletion |
| #22 | EGFR L858R | EGFR L858R | 7.1 | EGFR L858R |
| #23 | ALK-EML4 fusion | ALK-EML4 fusion | – | ALK-EML4 fusion |
| #24 | KRAS G12V | KRAS G12V | 2.0 | KRAS G12V |
| #25 | EGFR L858R | EGFR L858R | 11.1 | EGFR L858R |
| #26 | EGFR L858R | Wild type | – | Wild type |
| #27 | Wild type | Wild type | – | Wild type |
| #28 | EGFR L858R | EGFR L858R | 6.9 | EGFR L858R |
| #29 | EGFR exon 19 deletion | EGFR exon 19 deletion | 11.4 | EGFR exon 19 deletion |
| #30 | EGFR exon 19 deletion | Wild type | – | Wild type |
NGS, next-generation sequencing.
Figure 1.
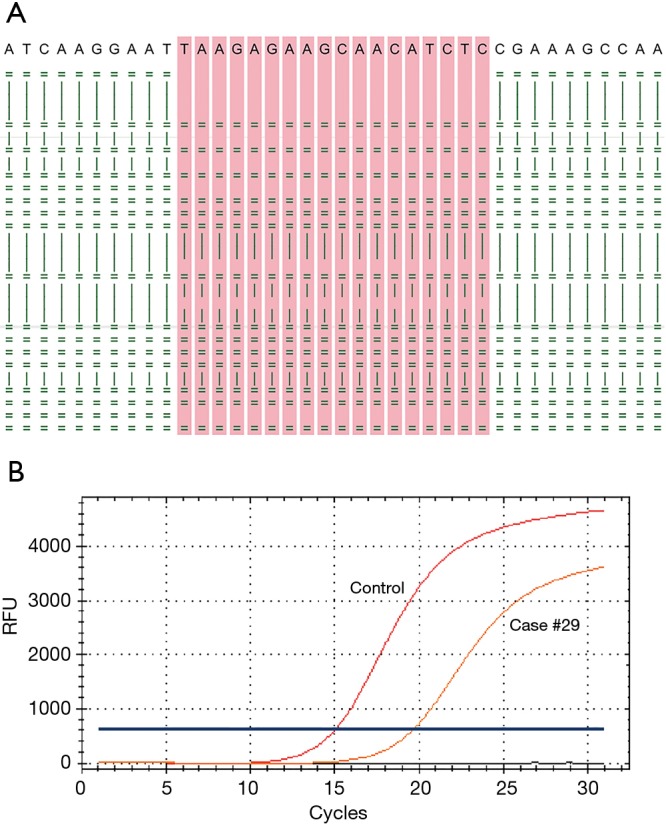
An examples of an EGFR exon 19 deletion detected by both ARMS PCR and targeted NGS in paired thoracic biopsy and pleura effusion samples. (A) The sequence alignment graph depicts an EGFR exon 19 deletion p.L747_P753delinsS (c.2240_2257del) in pleura effusion of case #15, only 26 reads were shown. Reads were aligned to the forward stand (=) or reverse strand (|); (B) the amplification graph corresponds to thoracic biopsy sample of the same case analyzed with the Amoy EGFR mutation test kit.
Diagnostic performance of pleural effusion
The concordance rate between gene status identified by ARMS analysis and NGS in thoracic biopsy and pleural effusion samples was 86.7% (26/30), indicating that pleural effusions could be an alternative material for molecular analyses. Compared with the thoracic biopsy specimens, the diagnostic performance of pleural effusion specimens showed a sensitivity of 92.3%, a specificity of 50.0%, and a positive predictive value of 92.3% (Table 3). The Kappa value indicating the detection efficiency of gene mutation between thoracic biopsy and pleural effusion specimens was 0.42, and the P value was 0.102.
Table 3. Comparison of EGFR/KRAS/ALK mutations between 30 thoracic biopsy specimens detected by ARMS and matched pleural effusion specimens detected by NGS.
| Pleural effusion specimen (NGS) | Thoracic biopsy specimen (ARMS) | ||
|---|---|---|---|
| + | − | Total | |
| + | 24 | 2 | 26 |
| − | 2 | 2 | 4 |
| Total | 26 | 4 | 30 |
+, EGFR/KRAS/ALK positive mutation; −, EGFR/KRAS/ALK negative mutation. NGS, next-generation sequencing.
Discussion
Sampling of pleural effusion is a non-invasive, repeatable procedure that may provide an opportunity for targeted therapy in these patients. The purpose of this study was to evaluate the diagnostic performance of pleural effusion as a specimen for molecular analysis. We compared EGFR, KRAS, and ALK mutation results obtained from 30 thoracic biopsy samples and matched pleural effusion samples, using ARMS PCR and NGS. All of the 30 NSCLC cases had no histological sections available because of the poor physical performance and advanced stage of the disease, and instead, had to undergo another procedure (thoracic biopsy) to obtain materials. We observed high concordance (86.7%) between the pleural effusion and thoracic biopsy specimens. The NGS assay of pleural effusions showed good diagnostic performance. Only four cases of mutation were detected only in pleural effusion or thoracic biopsy specimens. For these four pleural effusion samples, mutations detected by ARMS PCR and NGS were concordant, so the discrepancies might be ascribed to tumor heterogeneity between the materials. Our findings indicate that pleural effusion is a feasible source of tumor DNA for identifying patients who can benefit from the targeted therapy.
So far, only few studies have demonstrated the feasibility of oncogene analysis in pleural effusions from patients with advanced lung cancer (11,13-16). In these studies, allele-specific PCR-based platforms (such as ARMS PCR, Sanger sequencing) were often used for somatic mutation profiling. However, these techniques require an amount of gDNA when testing multiple genes (loci); therefore, only EGFR mutations can be assessed because of the small amount of DNA from pleural effusion samples. Targeted NGS testing, which provides a unifying approach for the detection of multiple categories of genetic alterations, uncovered more actionable options for patients than allele-specific approaches did. The targeted NGS sequencing approach used in this study has been clinically validated in a previous cell line dilution study and reach a mutation detection limit of 5% for base substitutions and 1% for InDels (20). In this study, we used this NGS assay to detect clinically relevant genomic alterations in 9 genes (EGFR, KRAS, PIK3CA, BRAF, MET, ERBB2, ALK, ROS1, and RET). However, no mutation was found in 6 genes (PIK3CA, BRAF, MET, ERBB2, ROS1, and RET). The NGS assay only detected 2 more mutations than ARMS PCR. Beside the mutations in EGFR, one KRAS mutation predicting non-responsiveness to EGFR TKI inhibitors and one ALK rearrangement suggesting eligibility for treatment with the ALK inhibitor crizotinib were detected using pleural effusions. Cases with a low percentage of tumor cells (<5%) can successfully be used to detect actionable mutations in pleural effusion specimens.
The present study has several limitations: (I) it’s a single-center retrospective analysis with a relatively small number of cases; (II) the number of patients whose therapy was changed on the basis of the genomic profiling results is unknown; (III) a prospective study of a large number of patients with pleural effusion is warranted to reveal a correlation between mutation status in pleural effusion fluids and clinical responsiveness to targeted therapies.
In conclusion, we have compared the mutation statuses in pleural effusions and their paired thoracic biopsy specimens from patients with advanced lung adenocarcinoma and demonstrated high concordance rates of EGFR, KRAS, and ALK mutations between the two materials, as detected by ARMS PCR and targeted NGS. Our findings demonstrate that pleural effusions are suitable specimens for oncogene mutation analysis. NGS allows the assessment of more genes than conventional methods with limited amount of DNA from pleural effusions and provides options of personalized therapy for cancer patients at an advanced stage.
Acknowledgements
We appreciate the cooperation of all the patients and their families who contributed to this study.
Funding: This project was supported by the National Key R&D Program of China 2016YFC0905400, Special Foundation for High-level Talents of Guangdong (2016TX03R171), Guangzhou Science and Technology Project (201400000004-5), Pearl River Nova Program of Guangzhou (No. 201506010065) and the Science and Technology Project of Guangdong Province (2015B020232008).
Ethical Statement: The study protocol was reviewed and approved by the Institutional Review Board of the First Affiliated Hospital of Guangzhou Medical University (#2016-07). A written informed consent form, describing the purpose of the study, was signed by all of the participants.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115-32. 10.3322/caac.21338 [DOI] [PubMed] [Google Scholar]
- 2.Maemondo M, Inoue A, Kobayashi K, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med 2010;362:2380-8. 10.1056/NEJMoa0909530 [DOI] [PubMed] [Google Scholar]
- 3.Zhou C, Wu YL, Chen G, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol 2011;12:735-42. 10.1016/S1470-2045(11)70184-X [DOI] [PubMed] [Google Scholar]
- 4.Solomon BJ, Mok T, Kim DW, et al. First-line crizotinib versus chemotherapy in ALK-positive lung cancer. N Engl J Med 2014;371:2167-77. 10.1056/NEJMoa1408440 [DOI] [PubMed] [Google Scholar]
- 5.Rudin CM, Avila-Tang E, Harris CC, et al. Lung cancer in never smokers: molecular profiles and therapeutic implications. Clin Cancer Res 2009;15:5646-61. 10.1158/1078-0432.CCR-09-0377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun Y, Ren Y, Fang Z, et al. Lung adenocarcinoma from East Asian never-smokers is a disease largely defined by targetable oncogenic mutant kinases. J Clin Oncol 2010;28:4616-20. 10.1200/JCO.2010.29.6038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shi Y, Au JS, Thongprasert S, et al. A prospective, molecular epidemiology study of EGFR mutations in Asian patients with advanced non-small-cell lung cancer of adenocarcinoma histology (PIONEER). J Thorac Oncol 2014;9:154-62. 10.1097/JTO.0000000000000033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Little AG, Rusch VW, Bonner JA, et al. Patterns of surgical care of lung cancer patients. Ann Thorac Surg 2005;80:2051-6. 10.1016/j.athoracsur.2005.06.071 [DOI] [PubMed] [Google Scholar]
- 9.Morgensztern D, Waqar S, Subramanian J, et al. Prognostic impact of malignant pleural effusion at presentation in patients with metastatic non-small-cell lung cancer. J Thorac Oncol 2012;7:1485-9. 10.1097/JTO.0b013e318267223a [DOI] [PubMed] [Google Scholar]
- 10.Porcel JM, Gasol A, Bielsa S, et al. Clinical features and survival of lung cancer patients with pleural effusions. Respirology 2015;20:654-9. 10.1111/resp.12496 [DOI] [PubMed] [Google Scholar]
- 11.Wu SG, Gow CH, Yu CJ, et al. Frequent epidermal growth factor receptor gene mutations in malignant pleural effusion of lung adenocarcinoma. Eur Respir J 2008;32:924-30. 10.1183/09031936.00167407 [DOI] [PubMed] [Google Scholar]
- 12.Buttitta F, Felicioni L, Del Grammastro M, et al. Effective assessment of egfr mutation status in bronchoalveolar lavage and pleural fluids by next-generation sequencing. Clin Cancer Res 2013;19:691-8. 10.1158/1078-0432.CCR-12-1958 [DOI] [PubMed] [Google Scholar]
- 13.Yeo CD, Kim JW, Kim KH, et al. Detection and comparison of EGFR mutations in matched tumor tissues, cell blocks, pleural effusions, and sera from patients with NSCLC with malignant pleural effusion, by PNA clamping and direct sequencing. Lung Cancer 2013;81:207-12. 10.1016/j.lungcan.2013.04.023 [DOI] [PubMed] [Google Scholar]
- 14.Liu D, Lu Y, Hu Z, et al. Malignant pleural effusion supernatants are substitutes for metastatic pleural tumor tissues in EGFR mutation test in patients with advanced lung adenocarcinoma. PLoS One 2014;9:e89946. 10.1371/journal.pone.0089946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rodriguez EF, Shabihkhani M, Carter J, Maleki Z, et al. Molecular Alterations in Patients with Pulmonary Adenocarcinoma Presenting with Malignant Pleural Effusion at the First Diagnosis. Acta Cytol 2017:61:214-22. 10.1159/000477148 [DOI] [PubMed] [Google Scholar]
- 16.Carter J, Miller JA, Feller-Kopman D, et al. Molecular Profiling of Malignant Pleural Effusion in Metastatic Non-Small-Cell Lung Carcinoma. The Effect of Preanalytical Factors. Ann Am Thorac Soc 2017:14:1169-76. [DOI] [PubMed] [Google Scholar]
- 17.Bourgon R, Lu S, Yan Y, et al. High-throughput detection of clinically relevant mutations in archived tumor samples by multiplexed PCR and next-generation sequencing. Clin Cancer Res 2014;20:2080-91. 10.1158/1078-0432.CCR-13-3114 [DOI] [PubMed] [Google Scholar]
- 18.Singh RR, Patel KP, Routbort MJ, et al. Clinical massively parallel next-generation sequencing analysis of 409 cancer-related genes for mutations and copy number variations in solid tumours. Br J Cancer 2014;111:2014-23. 10.1038/bjc.2014.518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanagal-Shamanna R, Portier BP, Singh RR, et al. Next-generation sequencing-based multi-gene mutation profiling of solid tumors using fine needle aspiration samples: promises and challenges for routine clinical diagnostics. Mod Pathol 2014;27:314-27. 10.1038/modpathol.2013.122 [DOI] [PubMed] [Google Scholar]
- 20.Shao D, Lin Y, Liu J, et al. A targeted next-generation sequencing method for identifying clinically relevant mutation profiles in lung adenocarcinoma. Sci Rep 2016;6:22338. 10.1038/srep22338 [DOI] [PMC free article] [PubMed] [Google Scholar]