Abstract
The breeding of new soft-seeded pomegranate cultivars provides new products for the market and increases farmers’ incomes, yet the genetic architecture mediating seed hardness is largely unknown. Here, the seed hardness and hundred-seed weights of 26 cultivars were determined in 2 successive years. We conducted miRNA and mRNA sequencing to analyse the seeds of two varieties of Punica granatum: soft-seeded Tunisia and hard-seeded Sanbai, at 60 and 120 d after flowering. Seed hardness was strongly positively correlated with hundred-seed weight. We detected 25 and 12 differentially expressed miRNA–mRNA pairs with negative regulatory relationships between the two genotypes at 60 and 120 d after flowering, respectively. These miRNA–mRNA pairs mainly regulated seed hardness by altering cell wall structure. Transcription factors including NAC1, WRKY and MYC, which are involved in seed hardness, were targeted by differentially expressed mdm-miR164e and mdm-miR172b. Thus, seed hardness is the result of a complex biological process regulated by a miRNA–mRNA network in pomegranate. These results will help us understand the complexity of seed hardness and help to elucidate the miRNA-mediated molecular mechanisms that contribute to seed hardness in pomegranate.
Introduction
Pomegranate is an edible fruit that is native to central Asia1. It has gained attention because of its antioxidant properties that have health benefits for humans and protect against several diseases such as hypertension cardiovascular and cancer2. Pomegranate seeds contain phytosterols and have a special fatty acid profile that includes punicic acid, which contributes to their health benefits3. Pomegranates with seeds that are easy to swallow command higher market prices than traditional varieties. In China, the best-known commercial soft-seeded pomegranate variety is Tunisia. This variety has been cultivated in China for more than 30 years4, resulting in cultivar depression. Thus, breeding new soft-seeded cultivars is imperative to meet market demands. Fully characterising the genetic mechanism of seed hardness may be useful to breed new commercially viable pomegranate varieties.
A few recent studies have focused on the seed hardness of pomegranate. Our previous study showed that the seed hardness increased from 60 to 120 d after flowering (DAF) in hard-seeded varieties, but did not change during this period in soft-seeded varieties. Additionally, the latter had lower lignin contents than the former4. These results were consistent with those of Zarein et al.5, in which soft-seeded pomegranate was found to have a higher cellulose content than that of hard-seeded pomegranate at 60 and 120 DAF. Correspondingly, lignin and cellulose biosynthetic genes such as CCR, CAD, CelSy, SuSy, CCoA-OMT, MYB, WRKY and MYC, showed differences in their seed expression levels between soft- and hard-seeded pomegranate genotypes4,5. In another study, four quantitative trait loci (QTLs) associated with seed hardness could explain 15% to 30% of the phenotypic variation6. These results indicated that seed hardness involves complicated physiological processes and is controlled by multiple genetic factors. However, none of the reported genes or QTLs could completely explain the genetic basis of seed hardness in pomegranate.
Genes that determine important traits are usually negatively regulated by microRNAs (miRNAs), small noncoding RNAs of 18–25 nucleotides (nt), through either posttranscriptional degradation or translational repression7. The advent of next-generation sequencing has revealed many miRNAs in plants and highlighted their differential expression levels in different plant species with phenotypic variations. miRNAs are known to participate in numerous biological processes related to human diseases, such as cancer and metabolic diseases8. They also participate in plant growth, development, and stress responses7,9. A recent study identified miRNAs from 10-d-old seedlings, leaves, flowers and arils of pomegranate at different developmental stages10. Both conserved and pomegranate-specific miRNAs were identified, and of the former, the most abundant miRNA family was miR157. Bioinformatics analysis revealed that the miRNAs were mainly involved in regulating linolenic and ascorbate acid metabolism, sugar metabolism, RNA transport, and plant hormone signalling. However, the roles of miRNAs or miRNA targets during seed development, especially related to the development of seed hardness, are yet to be elucidated in pomegranate.
In the study, we conducted deep-sequencing and bioinformatics analysis of seeds at 60 and 120 DAF to identify pomegranate-specific miRNAs, and to determine their expression patterns, in two varieties of P. granatum: soft-seeded Tunisia and hard-seeded Sanbai. The identification of these differentially expressed miRNAs–mRNAs provides new insights into the genetic mechanism of seed hardness in pomegranate.
Results
Phenotypic variations and correlation analysis
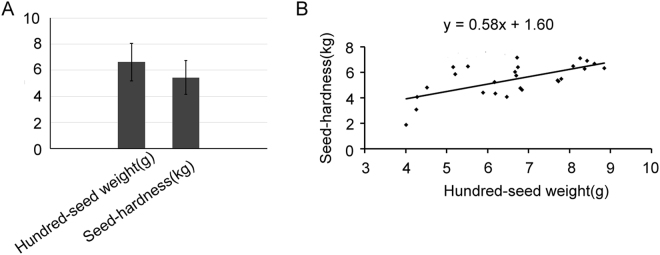
Seed weight is a key factor that controls seed size, and ‘Tunisia’ seeds are smaller and lighter in weight than those of ‘Sanbai’4. To explore the relationship between seed weight and seed hardness, phenotypic variations in hundred-seed weight and seed hardness were analysed for 26 pomegranate cultivars (Supplementary Table S1). The average hundred-seed weight and seed hardness of the cultivars were 6.62 and 5.44, respectively (Fig. 1A). Seed hardness was positively correlated with hundred-seed weight (correlation coefficient, 0.58; P < 0.01) (Fig. 1B). A linear regression analysis of the correlated traits indicated that hundred-seed weight could significantly explain 40.24% of the seed hardness (P < 0.01).
Figure 1.
Phenotypic variation (A) and correlation (B) analyses of seed hardness and hundred-seed weight in pomegranate.
Construction and deep sequencing of small RNAs
The total RNAs were isolated from the seeds of ‘Tunisia’ and ‘Sanbai’ at 60 and 120 DAF. These RNAs were used to construct four small RNA libraries; SS1, SS2, TS1, and TS2 (Supplementary Table S2), and small RNA sequencing generated 24,593,968, 24,362,778, 24,221,594, and 24,222,409 raw reads from these respective libraries. The 5′ and 3′ adaptors and low-quality reads were removed from the raw reads, yielding 23,025,209, 22,362,305, 22,879,827, and 22,523,261 reads from SS1, SS2, TS1, and TS2, respectively. Approximately 93.99%, 95.51%, 91.35%, and 90.68% of the clean reads from SS1, SS2, TS1, and TS2, respectively, were successfully mapped to the reference genome. Among them, reads with sequence lengths of 18–30 nt were filtered to enrich the sample with reads corresponding to the size of typical small RNAs. The distribution of small RNA lengths among the different size categories is shown in Supplementary Fig. S1. In SS1 and TS1, 23- and 24-nt were the most abundant sizes, while in SS2 and TS2, the most abundant small RNAs were 21 and 24 nt in length. Thus, the 24-nt small RNAs dominated the small RNA transcriptome of all of the libraries.
To identify miRNAs from the four small RNA libraries, we used sequence tags of 18–30 nt in length as queries in BLAST searches against the miRBase11 and Rfam12 databases. We identified 207 known miRNAs belonging to 40 miRNAs families (Supplementary Table S3). Among the conserved miRNAs detected, the MIR156 family was the largest, with 31 members (14.98% of the total). The MIR171_1 and MIR172 families contained 15 members each. MIR399 and MIR167_1 both included 10 members, and MIR166 and MIR395 had nine members each. The MIR1511, MIR477, MIR7125, and MIR827_4 families each had a single member. Five known miRNAs (mdm-miR391, mdm-miR477a, mdm-miR7126, mdm-miR7128, and mdm-miR858), could not be assigned to existing miRNA families, indicating that they may be species-specific or present in only some plant species. We used the miRNA prediction software Mireap13 to obtain putative novel miRNAs with their predicted hairpin precursors. By exploring the secondary structure, dicer cleavage sites, and the minimum free energy of the un-annotated small RNA tags that could be mapped to the reference genome, 761 potential novel miRNA candidates were identified from all four libraries (Supplementary Table S4). The putative precursor sequences of these predicted miRNAs were further analysed using RNAfold software to confirm their stem-loop structures. All the precursor sequences folded into hairpin-like structures that were similar to those of other known miRNAs (Supplementary Fig. S2). In total, 830 (137 known and 693 novel), 745 (135 known and 610 novel), 850 (145 known and 705 novel) and 795 (136 known and 659 novel) miRNAs were detected in the SS1, SS2, TS1, and TS2 libraries, respectively (Supplementary Table S2).
Differential expression analysis of miRNAs
miRNAs with least 10 raw read counts from the four equivalent libraries were selected for further analysis. We identified differentially expressed miRNAs between SS1_TS1 and SS2_TS2. Compared with the SS group, the TS group had 94 (31 known and 63 novel) and 120 (26 known and 94 novel) significantly differentially expressed miRNAs at 60 and 120 DAF, respectively, based on the criteria |log 2 (TS/SS)| ≥ 1 and P ≤ 0.01 (Supplementary Table S5).
In total, 62 of the 92 differentially expressed miRNAs in SS1_TS1 and 89 of the 120 differentially expressed miRNAs in SS2_TS2 were up-regulated, while all of the others were down-regulated (Fig. 2A). A Venn diagram (Fig. 2B) showed that the relative expression levels of 14 miRNAs changed in both SS1_TS1 and SS2_TS2. In SS1_TS1, eight out of 14 miRNAs were down-regulated, while in SS2_TS2 10 out of 14 miRNAs were down-regulated (Fig. 2C). Of the 14 common differentially expressed miRNAs, three novel miRNAs were up-regulated in both SS1_TS1 and SS2_TS2. Eighty (56 up-regulated and 24 down-regulated) miRNAs were specifically expressed in SS1_TS1, while 106 (85 up-regulated and 21 down-regulated) miRNAs were specifically expressed in SS2_TS2. The large number of differentially expressed miRNAs may contribute to differences in seed development between the two genotypes. The stage-specific differentially expressed miRNAs exhibited time–space specificity in pomegranate.
Figure 2.
Differentially expressed miRNAs in pomegranate seed. (A) Numbers of miRNAs up- or down-regulated in SS1_TS1 and SS2_TS2. (B) Venn diagram showing unique and shared regulatory miRNAs in SS1_TS1 and SS2_TS2. (C) Hierarchical cluster analysis of 14 regulated miRNAs in SS1_TS1 and SS2_TS2. Fold-change ratios of genes are indicated by different colours. SS1_TS1: comparison between seeds of ‘Sanbai’ and ‘Tunisia’ at 60 d after flowering (DAF). SS2_TS2: comparison between seeds of ‘Sanbai’ and ‘Tunisia’ at 120 DAF.
Correlation analysis of miRNAs and their target mRNAs
We used psRobot14 and TargetFinder15 with position-dependent scoring systems to predict the miRNA targets. These analyses identified 2,646 putative targets; 2,016 miRNA–mRNA pairs identified by psRobot and 1,795 pairs identified by TargetFinder, with 1,165 being common to both (Supplementary Fig. S3). Differences in the scoring matrices of the software explained the discrepancy in the number of targets predicted between the two programs. In total, 408 and 335 miRNA–mRNA pairs were identified in SS1_TS1 and SS2_TS2, respectively (Supplementary Table S6).
Previously, we performed de novo assembly of the seed transcriptome4, and sequenced and assembled the pomegranate genome (unpublished data). Thus, the sequence tags were re-annotated based on our reference genome. Differentially expressed genes between the two varieties were identified by Cuffdiff based on the criteria P ≤ 0.01 and |log2 (TS/SS)| ≥ 1. In this way, we identified 1,713 up-regulated and 1,804 down-regulated genes in SS1_TS1, and 1,400 up-regulated and 1,349 down-regulated genes in SS2_TS2 (Fig. 3). Combined with the differentially expressed miRNA targets, 53 and 38 differentially expressed miRNA–mRNA pairs were independently acquired from SS1_TS1 and SS2_TS2, respectively (Supplementary Table S7).
Figure 3.
Identification of differentially expressed genes in (A) SS1_TS1 and (B) SS2_TS2.
Usually, miRNAs regulate their targets by inducing mRNA degradation16,17. Microarray analyses have shown that miRNA expression decreases the abundance of transcripts with potential miRNA target sites18–20. Thus, to explore the roles of miRNA and mRNA interactions in seed hardness, we studied the negative regulatory relationships between miRNAs and their targets (Table 1). In total, 25 and 12 miRNA–mRNA pairs were identified in SS1_TS1 and SS2_TS2, respectively. The relationships between differentially expressed miRNAs and mRNAs were grouped into three major classes based on their regulatory modes (Fig. 4): (1) one miRNA versus one mRNA; (2) one miRNA or mRNA versus more than one mRNA or miRNA; and (3) one miRNA mediating the expression of another miRNA. An example of type 2 was mdm-miR172b, which repressed Gglean031260.1, Gglean008425.1, Gglean027146.1, Gglean026849.1, Gglean026000.1, and Gglean000051.1. Gglean013488.1 was simultaneously negatively regulated by mdm-miR166h, mdm-miR166e, mdm-miR166f, and mdm-miR166a in the MIR166 family. An example of type 3 was novel_mir671, which negatively regulated Gglean004793.1. Intriguingly, the expression of novel_mir671 and its precursors may be directly regulated by Gglean026964.1, the target gene of mdm-miR164e. This indicated that novel_mir671 and mdm-miR164e together target Gglean026964.1 to contribute to the seed hardness of pomegranate. Only one of the miRNA–mRNA interaction pairs (novel_mir468–Gglean014421.1) was acquired from both SS1_TS1 and SS2_TS2. Together, these results highlighted the complicated interactions between miRNAs and mRNAs during seed development in pomegranate.
Table 1.
Predicted mRNA targets of differentially expressed miRNAs in SS1_TS1 and SS2_TS2.
| miRNA id | P value | log2(TS/SS) | Regulation | Target gene id | P value | log2(TS/SS) | Regulation |
|---|---|---|---|---|---|---|---|
| SS1_TS1 | |||||||
| mdm-miR171i | 0.000 | 4.291 | Up | Gglean015277.1 | 0.001 | −2.792 | Down |
| novel_mir222 | 0.001 | 1.691 | Up | Gglean014013.1 | 0.000 | −1.164 | Down |
| mdm-miR166h | 0.000 | −8.224 | Down | Gglean013488.1 | 0.000 | 1.564 | Up |
| mdm-miR166e | 0.000 | −3.634 | Down | Gglean013488.1 | 0.000 | 1.564 | Up |
| mdm-miR166f | 0.000 | −2.458 | Down | Gglean013488.1 | 0.000 | 1.564 | Up |
| mdm-miR166a | 0.000 | −6.320 | Down | Gglean013488.1 | 0.000 | 1.564 | Up |
| mdm-miR167e | 0.001 | −3.653 | Down | Gglean029426.1 | 0.000 | 7.351 | Up |
| mdm-miR167f | 0.001 | −2.773 | Down | Gglean029426.1 | 0.000 | 7.351 | Up |
| mdm-miR172b | 0.002 | −4.193 | Down | Gglean031260.1 | 0.000 | 1.263 | Up |
| mdm-miR172b | 0.002 | −4.193 | Down | Gglean008425.1 | 0.000 | 5.103 | Up |
| mdm-miR172b | 0.002 | −4.193 | Down | Gglean021449.1 | 0.001 | 2.996 | Up |
| mdm-miR172b | 0.002 | −4.193 | Down | Gglean027146.1 | 0.000 | 2.122 | Up |
| mdm-miR172b | 0.002 | −4.193 | Down | Gglean026849.1 | 0.000 | 1.604 | Up |
| mdm-miR172b | 0.002 | −4.193 | Down | Gglean026000.1 | 0.000 | 1.544 | Up |
| mdm-miR172b | 0.002 | −4.193 | Down | Gglean000051.1 | 0.000 | 9.279 | Up |
| mdm-miR398b | 0.007 | 3.374 | Up | Gglean005184.1 | 0.000 | −1.389 | Down |
| novel_mir671 | 0.000 | 3.853 | Up | Gglean004793.1 | 0.000 | −1.151 | Down |
| novel_mir671 | 0.000 | 3.853 | Up | Gglean026964.1 | 0.000 | −2.145 | Down |
| mdm-miR164e | 0.007 | 3.247 | Up | Gglean026964.1 | 0.000 | −2.145 | Down |
| novel_mir2 | 0.001 | 2.774 | Up | Gglean029881.1 | 0.000 | −2.219 | Down |
| novel_mir367 | 0.000 | −2.738 | Down | Gglean009129.1 | 0.000 | 1.045 | Up |
| novel_mir367 | 0.000 | −2.738 | Down | Gglean023388.1 | 0.000 | 1.194 | Up |
| novel_mir468 | 0.002 | 4.128 | Up | Gglean014421.1 | 0.000 | −2.026 | Down |
| mdm-miR164e | 0.007 | 3.247 | Up | Gglean003381.1 | 0.000 | −4.952 | Down |
| mdm-miR396c | 0.005 | 1.832 | Up | Gglean029554.1 | 0.001 | −1.996 | Down |
| SS2_TS2 | |||||||
| novel_mir608 | 0.000 | 5.483 | Up | Gglean000622.1 | 0.000 | −7.608 | Down |
| novel_mir349 | 0.001 | 3.834 | Up | Gglean028955.1 | 0.000 | −2.365 | Down |
| novel_mir468 | 0.002 | 3.810 | Up | Gglean014421.1 | 0.000 | −1.571 | Down |
| novel_mir725 | 0.000 | 4.828 | Up | Gglean024157.1 | 0.000 | −1.443 | Down |
| mdm-miR164d | 0.000 | 3.948 | Up | Gglean005008.1 | 0.000 | −1.411 | Down |
| novel_mir349 | 0.001 | 3.834 | Up | Gglean021878.1 | 0.000 | −1.242 | Down |
| mdm-miR164d | 0.000 | 3.948 | Up | Gglean026964.1 | 0.000 | −1.100 | Down |
| mdm-miR171h | 0.000 | −2.538 | Down | Gglean025172.1 | 0.000 | 1.128 | Up |
| mdm-miR166e | 0.000 | −6.325 | Down | Gglean012177.1 | 0.000 | 1.775 | Up |
| mdm-miR166c | 0.002 | −3.015 | Down | Gglean012177.1 | 0.000 | 1.775 | Up |
| mdm-miR166i | 0.000 | −5.615 | Down | Gglean012177.1 | 0.000 | 1.775 | Up |
| mdm-miR164f | 0.001 | −3.826 | Down | Gglean016084.1 | 0.000 | 2.355 | Up |
Figure 4.
Combined analysis of negative regulatory miRNA and mRNA expression networks in (A) SS1_TS1 and (B) SS2_TS2. Rectangles represent mRNAs; circles represent miRNAs; blue and red represent up- and down-regulated, respectively.
Validation of miRNA and mRNA expression
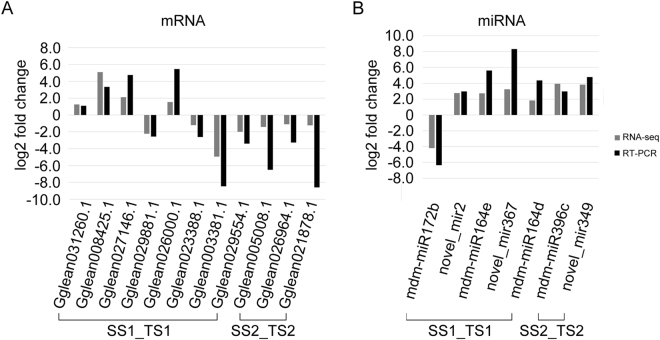
To further confirm the miRNA and mRNA sequencing results, a qRT-PCR analysis was used to validate the expression patterns of the differentially expressed miRNAs and their targets. Seven miRNAs (mdm-miR172b, novel_mir2, novel_mir367, mdm-miR164e, mdm-miR396c, mdm-miR164d, and novel_mir349) and their targets were selected for these analyses. The results of the qRT-PCR analyses were very similar to those obtained from the high-throughput sequencing data (Fig. 5).
Figure 5.
qRT-PCR analysis of selected (A) mRNAs and (B) miRNAs.
GO analysis of targets of differentially expressed miRNAs
To determine the functions of the targets of the differentially expressed miRNAs, we conducted GO analyses of the predicted targets using a false discovery rate correction value at P ≤ 0.05. These analyses showed that ‘cellular component’, ‘molecular function’ and ‘biological process’ were the three main categories enriched with targets of the differentially expressed miRNAs between TS and SS pomegranate (Fig. 6). In the ‘cellular components’ category, genes related to cell, cell part, and organelle were highly enriched as targets of the differentially expressed miRNAs. In the ‘molecular functions’ category, genes related to binding, catalytic activity, and transporter activity were highly enriched as targets of the differentially expressed miRNAs. In the ‘biological processes’ category, genes targeted by differentially expressed miRNAs were involved in biological regulation, cellular component organisation, cellular process, establishment of localisation, and localisation and metabolic process. Cellular process- and metabolic process-related genes were significantly overrepresented in the ‘biological processes’ category, indicating that these two processes were greatly enhanced.
Figure 6.

GO analysis of differentially expressed miRNA targets.
KEGG pathway analysis of differentially expressed miRNA targets
To further explore the functions of differentially expressed miRNAs targets, we conducted KEGG pathway and comparative analyses (Table 2). A total of 14 and eight differentially expressed miRNAs targets were assigned to 18 and eight pathways in SS1_TS1 and SS2_TS2, respectively. Twenty and 10 differentially expressed miRNA–mRNA pairs were detected as acting in these pathways in SS1_TS1 and SS2_TS2, respectively (Fig. 7A and Table 2). Of those, only one pair (novel_mir468-Gglean014421.1) was co-expressed in SS1_TS1 and SS2_TS2 (Fig. 7A and Table 2). We hypothesise that these miRNA–mRNA pairs play important roles in the development of seed hardness.
Table 2.
KEGG pathway analysis of differentially expressed miRNA-mRNAs in SS1_TS1 and SS2_TS2.
| Pomegranate | Arabidopsis thaliana | |||||
|---|---|---|---|---|---|---|
| DE-miRNAs | Regulation | DEGs | Entry | Annotation | Gene | Description |
| SS1-TS1 | ||||||
| mdm-miR171i | Up | Gglean015277.1 | K08193 | solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter) | — | |
| novel_mir222 | Up | Gglean014013.1 | K00517 | indol-3-yl-methylglucosinolate hydroxylase [EC:1.14.-.-] | AT2G46660 (EOD3) | Encodes a member of CYP78A cytochrome P450 monooxygenase |
| mdm-miR166h | Down | Gglean013488.1 | K00820 | glucosamine–fructose-6-phosphate aminotransferase (isomerizing) [EC:2.6.1.16] | — | |
| mdm-miR166e | ||||||
| mdm-miR166f | ||||||
| mdm-miR166a | ||||||
| mdm-miR167e | Down | Gglean029426.1 | K05681 | ATP-binding cassette subfamily G (WHITE) member 2 | — | |
| mdm-miR167f | ||||||
| mdm-miR172b | Down | Gglean031260.1 | K09284 | AP2-like transcription factor | AT2G28550 | related to AP2.7 |
| Gglean008425.1 | K13424 | WRKY transcription factor 33 | — | |||
| K18835 | WRKY transcription factor 2 | — | ||||
| Gglean027146.1 | K13422 | transcription factor MYC2 | — | |||
| Gglean026849.1 | K16225 | WRKY transcription factor 52 | — | |||
| K13425 | WRKY transcription factor 22 | — | ||||
| Gglean026000.1 | K13418 | somatic embryogenesis receptor kinase 1, [EC:2.7.10.1 2.7.11.1] | AT3G25560 | NSP-interacting kinase 2 | ||
| Gglean000051.1 | K01510 | apyrase [EC:3.6.1.5] | — | |||
| novel_mir671 | Up | Gglean026964.1 | K08678 | UDP-glucuronate decarboxylase [EC:4.1.1.35] | AT3G46440 | encodes a protein similar to UDP-glucuronic acid decarboxylase |
| novel_mir2 | Up | Gglean029881.1 | K01115 | phospholipase D1/2 [EC:3.1.4.4] | — | |
| novel_mir367 | Down | Gglean009129.1 | K12355 | coniferyl-aldehyde dehydrogenase [EC:1.2.1.68] | AT3G24503 | aldehyde dehydrogenase AtALDH1a |
| Gglean023388.1 | K01728 | pectate lyase [EC:4.2.2.2] | AT1G04680 | pectin lyase-like superfamily protein | ||
| mdm-miR164e | Up | Gglean003381.1 | K13126 | NAC1 transcription factor | AT1G56010 (NAC1) | encodes a NAC |
| mdm-miR396c | Up | Gglean029554.1 | K05275 | pyridoxine 4-dehydrogenase [EC:1.1.1.65] | — | |
| SS2-TS2 | ||||||
| mdm-miR166e | Down | Gglean012177.1 | K09338 | homeobox-leucine zipper protein | — | |
| mdm-miR166c | Down | |||||
| mdm-miR166i | Down | |||||
| novel_mir608 | Up | Gglean000622.1 | K16578 | CLIP-associating protein 1/2 | AT2G20190 | encodes a microtubule-associated protein |
| mdm-miR171h | Down | Gglean025172.1 | K14494 | DELLA protein | — | — |
| mdm-miR164f | Down | Gglean016084.1 | K08176 | inorganic phosphate transporter | AT5G12460 | fringe-like protein (DUF604) |
| mdm-miR164d | Up | Gglean005008.1 | K13416 | brassinosteroid insensitive 1-associated receptor kinase 1 [EC:2.7.10.1 2.7.11.1] | AT5G16000 | NSP-interacting kinase (NIK1), receptor-like kinase |
| Gglean026964.1 | K08678 | UDP-glucuronate decarboxylase [EC:4.1.1.35] | AT3G46440 | encodes a protein similar to UDP-glucuronic acid decarboxylase | ||
| novel_mir349 | Up | Gglean021878.1 | K05658 | ATP-binding cassette | — | |
| Gglean028955.1 | K14794 | ribosomal RNA-processing protein 12 | — | |||
Figure 7.
KEGG pathway analysis of differentially expressed miRNA–mRNAs. (A) Venn diagram showing unique and shared differentially expressed miRNA–mRNAs acting in pathways in SS1_TS1 and SS2_TS2. (B) Distributions of pathways related to enzymes, transcription factors, and proteins in SS1_TS1 and SS2_TS2.
Eleven miRNA targets (Gglean014013.1, Gglean013488.1, Gglean026000.1, Gglean000051.1, Gglean026964.1, Gglean029881.1, Gglean009129.1, Gglean023388.1, Gglean029554.1, Gglean005008.1, Gglean026964.1, and Gglean021878.1) were annotated as encoding key enzymes that regulate seed hardness in pomegranate (Fig. 7B and Table 2). For example, Gglean026964.1 encodes UDP-glucuronate decarboxylase [EC: 4.1.1.35], which catalyses the production of UDP-xylose. Its ortholog in Arabidopsis thaliana (AT3G46440) encodes a similar protein that produces UDP-xylose, a substrate for many cell-wall carbohydrates, including hemicellulose and pectin. UDP-xylose feedback regulates several cell-wall biosynthetic enzymes.
Five miRNA targets (Gglean031260.1, Gglean008425.1, Gglean027146.1, Gglean026849.1, and Gglean003381.1) were predicted to regulate seven transcription factors (TFs) that affect the formation of seed hardness in pomegranate (Fig. 7B and Table 2). For example, Gglean008425.1 and Gglean027146.1 encode proteins in the WRKY family, which includes WRKY2, WRKY33, WRKY22, and WRKY52. Gglean026849.1 was annotated as MYC2. Gglean031260.1 and its ortholog in A. thaliana (AT2G2855) likely have the same function, i.e., to regulate AP2 TFs.
Eight miRNA targets (Gglean015277.1, Gglean029426.1, Gglean003381.1, Gglean012177.1, Gglean000622.1, Gglean025172.1, Gglean016084.1 and Gglean028955.1) were annotated as encoding regulatory proteins (Fig. 7B and Table 2), including a phosphate transporter, ATP-binding domain, polyadenylate-binding, zipper, CLIP-associating, DELLA, and ribosomal RNA-processing protein. These differentially expressed miRNAs may target their corresponding genes to control their expression levels, thereby influencing the development of pomegranate seed hardness.
Discussion
Phenotypic analysis
After a double-fertilisation event, seed development begins with embryogenesis (cell division), followed by seed maturation (seed filling via accumulation of storage macromolecules) and then desiccation21. The size, weight, and hardness of seeds differ among species and among lines of the same species. All three seed characteristics are inter-related. Here, seed hardness was positively correlated with hundred-seed weight, indicating that decreasing the hundred-seed weight in pomegranate will decrease seed hardness.
Identification of conserved and novel miRNAs in pomegranate seeds
The combination of deep sequencing with a miRNA microarray analysis has revealed that various miRNAs play vital roles throughout the whole seed cycle, including during dormancy modulation22, germination23, development24,25 and maturation26. The results of the present study provide information about the regulatory networks of miRNAs involved in pomegranate seed hardness. Forty known miRNA families were identified, compared with the 30 previously identified known miRNA families10. Most are conserved in plants, such as A. thaliana27 and pomegranate10. The most abundant family was found to be miR156, like in pear28 and apple29,30. Previously, it was reported that miR157 is the most abundant family in pomegranate10. This difference from our result might be because different tissue types were analysed in that study. We identified 92 (31 known and 63 novel) and 120 (26 known and 94 novel) miRNAs that were differentially expressed in SS1_TS1 and SS2_TS2, respectively, consistent with the finding that miRNAs are more abundant in mature seeds than in developing seeds26. Of the differentially expressed miRNAs, 68.49% and 76.67% were newly identified in SS1_TS1 and SS2_TS2, respectively, indicating their potentially important roles in seed development.
Comparative analysis between miRNAs and mRNAs
A single miRNA generally targets a broad range of mRNAs with nearly complementary sequences, resulting in the regulation of a wide range of genes31. Consequently, miRNAs affect a wide range of physiological and developmental processes, including seed development, in plants. Some pre-miRNA and miRNA targets showing differences in abundance were found to be important in Jatropha seed development32. Similarly, the putative targets of 21 novel and 87 known miRNAs were predicted to be involved in various metabolic and biological processes in developing cotton seeds33. In the dry wheat seeds, the target genes of differentially expressed miRNAs between genetically modified and non-genetically modified lines were found to be associated with abiotic stress34.
In the present study, miRNA and mRNA sequencing identified 25 and 12 differentially expressed miRNA–mRNA pairs with negative regulatory relationships from SS1_TS1 and SS2_TS2, respectively. Only one of the miRNA–mRNA interaction pairs (novel_mir468–Gglean014421.1) was co-expressed in both of SS1_TS1 and SS2_TS2. Thus, the development of seed hardness mainly relies on the stage-specific expression of miRNA–mRNA pairs in pomegranate. A KEGG pathway analysis indicated that the functions of the targets of the differentially expressed miRNAs were mainly related to regulation of TFs and metabolic enzymes in SS1_TS1, and to structure-related proteins in SS2_TS2 (Table 2). These results were consistent with seed physiology and development. That is, intense metabolic activity depends on enzymatic activity and TFs that act during early stages, when metabolism is mainly involved in carbohydrate partitioning to supply photoassimilates and accumulate storage compounds21. However, seed maturation requires the deposition of storage macromolecules, including carbohydrates, lipids, and storage proteins35. Thus, the mdm-miRNAs may interact with their corresponding targets to control the production of a diverse range of proteins, metabolic enzymes, and TFs, thus modulating seed hardness in pomegranate.
miRNAs regulate enzymes involved in seed hardness
Enzymes are critical in regulating seed development because they catalyse metabolic processes such as cell-wall formation36, and carbohydrate37, hormone38 and nitrogen metabolism39. Here, a predicted novel_mir367 was found to regulate Gglean009129.1 and Gglean023388.1, encoding coniferyl-aldehyde dehydrogenase [EC: 1.2.1.68] and pectate lyase [EC: 4.2.2.2], respectively, which soften seeds. The two genes showed significantly higher expression levels in soft-seeded cultivars than in hard-seeded cultivars. Coniferyl-aldehyde dehydrogenase participates in the formation of ferulate and sinapate, which impede guaiacyl lignin and syringyl lignin biosynthesis (Supplementary Fig. S4). These pathways are similar to those reported in Arabidopsis and rapeseed40,41. This result may partly explain why soft-seeded pomegranates produces less lignin than hard-seeded ones4,5. Pectate lyase is a depolymerising enzyme that degrades plant cell walls42. Decreasing the abundance of pectate lyase transcripts can severely inhibit cell wall loosening during early fibre development43. In strawberry and tomato, the suppression of the pectate lyase mRNA during ripening resulted in significantly firmer fruit44,45. Thus, we hypothesised that the novel_mir367 controls the expression of genes related to lignin content and loosens the cell wall, resulting in softer pomegranate seeds.
In soft-seeded pomegranate, mdm-miR164e, novel_mir671, and mdm-miR164d down-regulated Gglean026964.1, encoding UDP-glucuronate decarboxylase [EC: 4.1.1.35], thereby reducing the seed hardness. UDP-glucuronate decarboxylase is a key enzyme involved in UDP-xylose biosynthesis, which is required for xylan formation during cell-wall biosynthesis46. The overexpression of antisense UDP-glucuronate decarboxylase in tobacco enhanced cellulose biosynthesis in secondary cell walls47. Thus, these miRNAs may alter cell-wall structure by increasing the cellulose content, which reduces seed hardness. This partly explains the higher proportion of cellulose in soft-seeded pomegranate than in hard-seeded pomegranate5.
The novel_mir222 and mdm-miR164d may affect seed weight, which influences seed hardness. The former regulates indol-3-yl-methylglucosinolate hydroxylase [EC: 1.14.-.-] by suppressing Gglean014013.1. In A. thaliana, its ortholog encodes a member of CYP78A that controls seed size48. We hypothesised that the novel_mir222 controls seed size/weight in pomegranate. The seed weight was significantly correlated with seed hardness in pomegranate (Fig. 1). Seed size appears to be regulated primarily by phytohormones, especially brassinosteroids49. Regulators can stimulate cell division and cell elongation under the influence of brassinosteroid insensitive 1-associated receptor kinase 1 [EC: 2.7.10.1 2.7.11.1] (Supplementary Fig. S5). In rice, brassinosteroids were shown to cause grain expansion50,51. Here, brassinosteroid insensitive 1-associated receptor kinase 1 was found to be regulated by mdm-miR164d through the negative targeting of Gglean005008.1.
miRNAs regulating TFs involved in seed hardness
We found that mdm-miR164e directly down-regulated NAC1 to block seed hardening, while mdm-miR172b upregulated WRKY, MYC and AP2 TFs to achieve the same result. According to our previous transcriptomic analysis, the differential expression of NAC, WRKY, and MYC TFs is related to hardness in pomegranate4, and the trends in their regulation detected in the present study corroborated those results. mdm-miR172b repressed the expression of the AP2-like TF, as previously observed in other studies29,52. AP2-like TFs control seed size and seed mass in Arabdopisis53–55. Thus, mdm-miR172b may regulate the AP2-like TF to control seed size in pomegranate. Similarly, osa-miR172 was shown to regulate grain size in rice56,57. It may, therefore, be inferred that mdm-miR172b simultaneously regulates multiple TFs that regulate seed hardness in pomegranate.
miRNAs regulating proteins involved in seed hardness
The predicted miRNA targets encode a broad range of proteins that participate in seed development26. In soft-seeded pomegranate cultivars, mdm-miR164f was found to increase the activity of the inorganic phosphate transporter by up-regulating Gglean016084.1 (Tables 1 and 2). In a previous study, its increased expression level resulted in an increase in phosphate storage58, which should contribute to greater seed weight. However, the seed weights of soft-seeded pomegranate were lower than those of hard-seeded pomegranate. The contradiction can be explained by the fact that phosphate transport in and out of the vacuolar membrane requires biochemical energy59. A deficiency in energy may limit the phosphate deposition in seed vacuoles, resulting in soft seeds. These results imply that a moderate lack of phosphate could lead to soft seeds. This would be favourable because it would reduce fertiliser consumption and provide a simple way to produce soft-seeded pomegranates58.
In pomegranate, the mdm-miR166e, mdm-miR166c, and mdm-miR166i were found to soften seeds by down-regulating the homeobox-leucine zipper proteins. These proteins are a subset of unique proteins that contain leucine zipper motif-linked homeodomains60. Reducing the abundance of homeobox-leucine zipper proteins is likely to inhibit cell elongation61 and cell division62, which may limit seed hardness.
The novel_mir608 is likely to mediate cell-wall biosynthesis, which affects seed hardness in pomegranate. novel_mir608 was found to negatively regulate Gglean000622.1, which encodes a CLIP-associating protein that prevents xyloglucan formation. Consequently, it disrupts the stability of the microtubule cytoskeleton and the cellulose pattern in primary cell walls63.
Conclusions
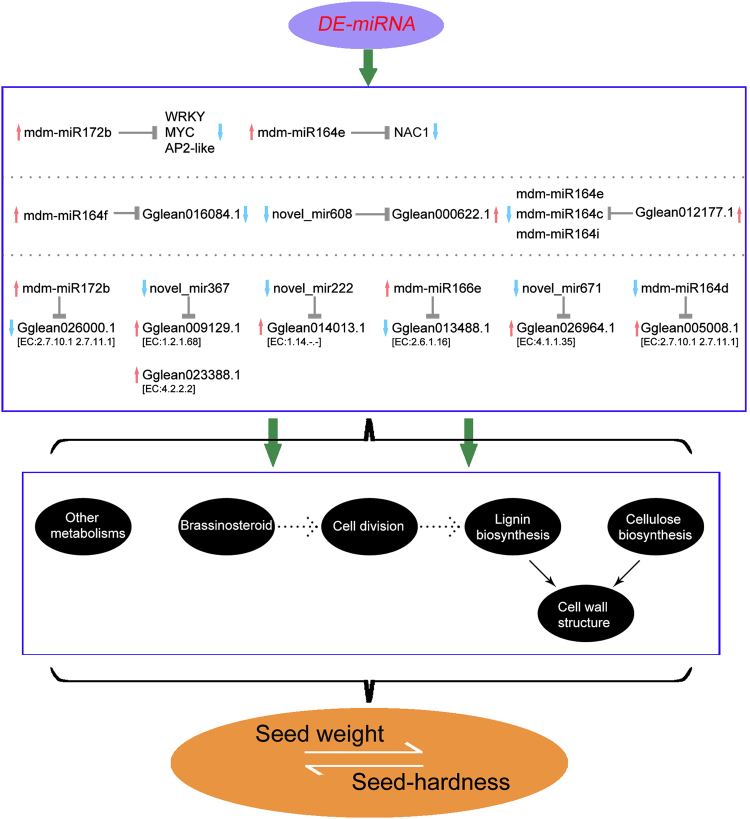
We conducted integrated transcriptomics and miRNA analyses to generate a comprehensive resource focused on identifying key regulatory miRNAs associated with the development of seed hardness in pomegranate. We found that these miRNAs suppressed vital targets, which included genes encoding TFs and enzymes involved in the early stages of seed development, as well as proteins involved in storage compound synthesis and transport in the mature seeds. The miRNA-targets included transcripts encoding proteins involved in brassinosteroid biosynthesis, cell elongation and division, lignin biosynthesis, cellulose biosynthesis, cell-wall biosynthesis and degradation, and other metabolic signalling pathways (Fig. 8). The cell wall is mainly composed of lignin and cellulose64, and their biosynthesis and degradation affects cell-wall structure. From our results, we concluded that the miRNA targets may regulate seed hardness by altering the cell-wall structure in pomegranate. These findings indicate that seed hardness involves a complex biological process regulated by miRNA–mRNA networks in pomegranate. Other enriched miRNAs were also found during different seed developmental stages and their putative corresponding target genes were investigated. These miRNAs may contribute to seed hardness formation by regulating important metabolic processes, as indicated by the highly represented GO terms (Fig. 6). Our understanding of the genetic relationships among seed weight, seed size, and seed hardness remains limited. Further experiments to confirm the targets of miRNAs and the miRNA–mRNA interaction networks in pomegranate are necessary to test our hypotheses. Collectively, these results will contribute to a more comprehensive understanding of seed hardness in pomegranate and help to elucidate miRNA-mediated molecular mechanisms underlying the formation of seed hardness.
Figure 8.
Hypothetical model for regulation of seed hardness in pomegranate through interactions between miRNAs and mRNAs.
Methods
Plant materials and phenotypic analysis
Two pomegranate cultivars, Tunisia (soft-seeded genotype) and Sanbai (hard-seeded genotype), were grown under the same conditions, and were managed in accordance with local standard production practices in Xingyang, China. The fruit developmental periods of the two cultivars were similar. Nine morphologically normal fruits were sampled and pooled in groups of three to obtain three biological replicates from 60 DAF to maturity (120 DAF). All samples were immediately snap frozen in liquid nitrogen and stored at −80 °C. For convenience, the seeds of ‘Tunisia’ and ‘Sanbai’ are abbreviated as TS and SS, respectively. The single seed weights were determined from the average hundred-seed weights. Seed hardness was measured as described by Xue et al. (2017). These two traits were studied in 26 cultivars at maturity in 2016 and 2017, in Zhengzhou, China. Both varieties were obtained from the Zhengzhou Fruit Research Institute (Chinese Academy of Agricultural Sciences, Zhengzhou, China).
Phenotypic variation, correlation, and linear regression analyses were performed using SPSS version 19.0 (IBM Corp., Armonk, NY, USA).
RNA extraction, library construction and deep sequencing
Total RNA was prepared from samples of ‘Tunisia’ and ‘Sanbai’ (control) using TRIzol reagent (Invitrogen, Carlsbad, CA, USA), and the RNA samples were subsequently purified by chloroform extraction. The RNA integrity was checked by electrophoresis on a 0.8% denaturing formaldehyde gel. Only high-quality RNA was used for further analyses. Total RNA (3 mg) was reverse-transcribed to cDNA with RevertAid Reverse Transcriptase (Thermo Fisher Scientific, Waltham, MA, USA) and a random hexamer primer at 42 °C for 60 min. The cDNA was used immediately or stored at −20 °C.
The small RNA libraries were constructed and sequenced using an Illumina Genome Analyzer (Illumina, San Diego, CA, USA) by the BGI Genomics Corporation (Shenzhen, China). Briefly, RNA with integrity >7 was assessed using a Bioanalyzer 2100 (Agilent Technologies, Palo Alto, CA, USA) on an RNA 6000 Nanochip. For small RNA library construction, ~1 μg total RNA was collected into RNA pools according to the Illumina TruSeq Small RNA library preparation protocol. Then, ~16–30 nt gel fragments were selected and ligated to a pair of adapters at the 5′- and 3′-ends using T4 RNA ligase. These small RNAs with adapters were transcribed into cDNA using Super-Script II Reverse Transcriptase (Invitrogen). These cDNAs were subjected to PCR amplification, and then the purified PCR products were sequenced.
Prediction and identification of known and novel miRNAs
To identify known miRNAs in pomegranate, the miRNA categories were mapped to the reference genome (unpublished data) using AASRA65. Alignments with less than two mismatches and more than 16 matches without gaps between the query sequences and known miRNAs were considered. The identified miRNAs were classified into families based on their sequence similarities. The unmatched reads were further processed to predict novel miRNAs using Mireap software13.
The characteristic structures of miRNA precursors, including hairpins, secondary structures, and dicer cleavage sites, and the minimum free energy were used to predict novel miRNAs with the Mireap pipeline. The criteria included the ability of miRNAs to fold into the correct secondary structure and the presence of mature miRNAs on one arm of the hairpin precursor. Additionally, the free energy of hybridisation had to be lower than or equal to −18 kcal/mol, and the mature miRNA strand and its complementary strand had to contain 2-nt 3′ overhangs.
mRNA sequencing data analyses and mRNA–miRNA pair predictions
We previously detected differentially expressed genes between soft-seeded and hard-seeded pomegranate by de novo transcriptome sequencing4. The sequence data were trimmed by removing adaptor sequences, empty reads, reads with more than 5% unknown nucleotides, low-quality sequences (base quality ≤20), and high Ns (ratio >10%) with Trimmomatic66. Clean reads were mapped to the reference genome sequence (unpublished data) using TopHat67 with default parameters. The reads were then assembled into transcripts and compared with reference gene models using Cufflinks68. Gene expression was quantified using the RNA-Seq by Expectation Maximization software69. The data were normalised as fragments per kilobase of transcript per million fragments mapped (FPKM) values70. The differences in transcript abundance between the two genotypes were calculated based on the ratio of FPKM values. The false discovery rate control method was used to identify the threshold of the P-value using Cuffdiff in the Cufflinks software package. Only transcripts with P ≤ 0.001 and |log2(TS/SS)| ≥ 1 were used for further analysis. The relative abundance of a gene or miRNA was calculated as follows: normalised expression = (actual miRNA or gene count/total count of clean reads) × 106. After normalisation, differentially expressed miRNAs between the two pomegranate cultivars were identified based on the criteria P ≤ 0.01 and |log2(TS/SS)| ≥ 1. We used PairFinder software (BGI Genomics Corporation) to predict potential mRNA–miRNA pairs.
qRT-PCR validation of miRNAs and their targets
We used stem-loop qRT-PCR to confirm the miRNA expression levels71. For selected miRNAs, ~1 μg DNA-free total RNA was hybridised with miRNA-specific stem-loop RT primers. The hybridised molecules were reverse transcribed into cDNAs using a Superscript III kit (Thermo Fisher Scientific). We designed forward miRNA-specific primers for the mature miRNA sequences and used a universal reverse primer for the stem-loop sequences. Reactions were repeated three times for each sample set. Each 20-μL reaction mixture contained 1 μL cDNA, 10 μL 2 × FastStart SYBR Green (Roche) and 0.8 μL forward and reverse primers (TaKaRa, Ohtsu, Japan). The PCR amplification conditions were as follows: 95 °C for 10 s and 60 °C for 30 s. The PCRs were conducted using the StepOnePlus Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). Data were analysed using the 2−ΔΔCt method to calculate relative gene expression72. Supplementary Table S8 lists all primers used in the qRT-PCR experiments, including those for miRNAs and their targets.
GO enrichment and pathway enrichment analyses of miRNA targets
The GO enrichment analysis for differentially expressed miRNA targets was conducted using tools at the Blast2GO website (http://www.blast2go.com). Significantly enriched GO terms (P < 0.05) were displayed using the online tool WEGO website (http://wego.genomics.org.cn). The targets of differentially expressed miRNAs were subjected to a KEGG pathway enrichment analysis using tools at the KOBAS2.0 website (http://kobas.cbi.pku.edu.cn/). To analyse the metabolic pathways and functional classifications of the targets of differentially expressed miRNAs, expression data were mapped to metabolic pathways using MapMan software73,74.
Electronic supplementary material
Acknowledgements
This work was funded by grants from the Investigation and Collection of Indigenous Varieties of Deciduous Fruit Trees in the Predominance Region (2012 FY110100) and the Agricultural Science and Technology Innovation Program of the Chinese Academy of Agricultural Sciences (CAAS-ASTIP-2015-ZFRI). We thank Lesley Benyon, PhD, and Jennifer Smith, PhD, from Liwen Bianji, Edanz Group China (www.liwenbianji.cn/ac), for editing the English text of a draft of this manuscript.
Author Contributions
X.L. and S.C. designed the experiment. X.L., X.X., H.L., D.Z. and L.N.C. collected the data. X.L. and Y.L. analysed the data, interpreted the results, and wrote the manuscript. D.C., J.Z., L.C. and H.X. revised the manuscript. All authors have read and approved the current version of the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-27664-y.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Holland D, Hatib K, Barya’Akov I. Pomegranate: botany, horticulture, breeding. Horticultural Reviews. 2009;35:127–191. [Google Scholar]
- 2.Patel C, Dadhaniya PL. Safety assessment of pomegranate fruit extract: acute and subchronic toxicity studies. Food & Chemical Toxicology. 2008;46:2728–2735. doi: 10.1016/j.fct.2008.04.035. [DOI] [PubMed] [Google Scholar]
- 3.Johanningsmeier SD, Harris GK. Pomegranate as a functional food and nutraceutical source. Annual review of food science and technology. 2011;2:181–201. doi: 10.1146/annurev-food-030810-153709. [DOI] [PubMed] [Google Scholar]
- 4.Xue H, et al. De novo transcriptome assembly and quantification reveal differentially expressed genes between soft-seed and hard-seed pomegranate (Punica granatum L.) PloS one. 2017;12:e0178809. doi: 10.1371/journal.pone.0178809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zarei A, et al. Differential expression of cell wall related genes in the seeds of soft- and hard-seeded pomegranate genotypes. Scientia Horticulturae. 2016;205:7–16. doi: 10.1016/j.scienta.2016.03.043. [DOI] [Google Scholar]
- 6.Harel-Beja R, et al. A novel genetic map of pomegranate based on transcript markers enriched with QTLs for fruit quality traits. Tree Genetics & Genomes. 2015;11:109. doi: 10.1007/s11295-015-0936-0. [DOI] [Google Scholar]
- 7.Tang J, Chu C. MicroRNAs in crop improvement: fine-tuners for complex traits. Nature plants. 2017;3:17077. doi: 10.1038/nplants.2017.77. [DOI] [PubMed] [Google Scholar]
- 8.Matsuda A, Yan I, Foye C, Parasramka M, Patel T. MicroRNAs as paracrine signalling mediators in cancers and metabolic diseases. Best Pract. Res. Clin. Endocrinol. Metab. 2016;30:577–590. doi: 10.1016/j.beem.2016.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu J, et al. ROS accumulation and antiviral defence control by microRNA528 in rice. Nature plants. 2017;3:16203. doi: 10.1038/nplants.2016.203. [DOI] [PubMed] [Google Scholar]
- 10.Saminathan T, et al. Genome-wide identification of microRNAs in pomegranate (Punica granatum L.) by high-throughput sequencing. BMC plant biology. 2016;16:122. doi: 10.1186/s12870-016-0807-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kozomara A, Griffithsjones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Research. 2014;42:D68. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nawrocki EP, et al. Rfam 12.0: updates to the RNA families database. Nucleic Acids Research. 2015;43:D130. doi: 10.1093/nar/gku1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Y, et al. Performance comparison and evaluation of software tools for microRNA deep-sequencing data analysis. Nucleic Acids Research. 2012;40:4298. doi: 10.1093/nar/gks043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang P, et al. Identification of a major QTL for silique length and seed weight in oilseed rape (Brassica napus L.). TAG. Theoretical and applied genetics. Theoretische und angewandte Genetik. 2012;125:285–296. doi: 10.1007/s00122-012-1833-7. [DOI] [PubMed] [Google Scholar]
- 15.Fahlgren N, Carrington JC. miRNA Target Prediction in Plants. Methods in Molecular Biology. 2010;592:51. doi: 10.1007/978-1-60327-005-2_4. [DOI] [PubMed] [Google Scholar]
- 16.Bagga S, et al. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 17.Wu L, Belasco JG. Micro-RNA Regulation of the Mammalian lin-28 Gene during Neuronal Differentiation of Embryonal Carcinoma Cells. Molecular & Cellular Biology. 2005;25:9198. doi: 10.1128/MCB.25.21.9198-9208.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li KB, Wang YP. Correlation of expression profiles between microRNAs and mRNA targets using NCI-60 data. Bmc Genomics. 2009;10:218. doi: 10.1186/1471-2164-10-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lim LP, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 20.Jones-Rhoades MW, Bartel DP, Bartel B. MicroRNAS and their regulatory roles in plants. Annual Review of Plant Biology. 2006;57:19. doi: 10.1146/annurev.arplant.57.032905.105218. [DOI] [PubMed] [Google Scholar]
- 21.Garg R, Singh VK, Rajkumar MS, Kumar V, Jain M. Global transcriptome and coexpression network analyses reveal cultivar-specific molecular signatures associated with seed development and seed size/weight determination in chickpea. The Plant journal: for cell and molecular biology. 2017;91:1088–1107. doi: 10.1111/tpj.13621. [DOI] [PubMed] [Google Scholar]
- 22.Liu Y, El-Kassaby YA. Regulatory crosstalk between microRNAs and hormone signalling cascades controls the variation on seed dormancy phenotype at Arabidopsis thaliana seed set. Plant cell reports. 2017;36:705–717. doi: 10.1007/s00299-017-2111-6. [DOI] [PubMed] [Google Scholar]
- 23.Huang D, Koh C, Feurtado J, Tsang E, Cutler A. MicroRNAs and their putative targets in Brassica napus seed maturation. Bmc Genomics. 2013;14:140. doi: 10.1186/1471-2164-14-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kang M, Zhao Q, Zhu D, Yu J. Characterization of microRNAs expression during maize seed development. Bmc Genomics. 2012;13:360. doi: 10.1186/1471-2164-13-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu H, Xie W, Li J, Zhou F, Zhang Q. A whole-genome SNP array (RICE6K) for genomic breeding in rice. Plant biotechnology journal. 2014;12:28–37. doi: 10.1111/pbi.12113. [DOI] [PubMed] [Google Scholar]
- 26.Korbes AP, et al. Identifying conserved and novel microRNAs in developing seeds of Brassica napus using deep sequencing. PloS one. 2012;7:e50663. doi: 10.1371/journal.pone.0050663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meyers BC, et al. Criteria for Annotation of Plant MicroRNAs. The Plant cell. 2008;20:3186. doi: 10.1105/tpc.108.064311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu J, et al. Identification of miRNAs involved in pear fruit development and quality. BMC genomics. 2014;15:1–19. doi: 10.1186/1471-2164-15-953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xing L, et al. Shoot bending promotes flower bud formation by miRNA-mediated regulation in apple (Malus domesticaBorkh.) Plant biotechnology journal. 2016;14:749–770. doi: 10.1111/pbi.12425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xing L, et al. Genome-wide identification of vegetative phase transition-associated microRNAs and target predictions using degradome sequencing in Malus hupehensis. Bmc Genomics. 2014;15:1125. doi: 10.1186/1471-2164-15-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bartel D. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 32.Galli V, et al. Identifying microRNAs and transcript targets in Jatropha seeds. PloS one. 2014;9:e83727. doi: 10.1371/journal.pone.0083727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y, Ding Y, Yu D, Xue W, Liu J. High-throughput sequencing-based genome-wide identification of microRNAs expressed in developing cotton seeds. Science China. Life Sciences. 2015;58:778–786. doi: 10.1007/s11427-015-4877-5. [DOI] [PubMed] [Google Scholar]
- 34.Jiang Q, et al. GmDREB1 overexpression affects the expression of microRNAs in GM wheat seeds. PloS one. 2017;12:e0175924. doi: 10.1371/journal.pone.0175924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Verdier J, Thompson RD. Transcriptional regulation of storage protein synthesis during dicotyledon seed filling. Plant & cell physiology. 2008;49:1263–1271. doi: 10.1093/pcp/pcn116. [DOI] [PubMed] [Google Scholar]
- 36.Anne-Laure CB, et al. Endomembrane proteomics reveals putative enzymes involved in cell wall metabolism in wheat grain outer layers. Journal of Experimental Botany. 2015;66:2649–2658. doi: 10.1093/jxb/erv075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bonome LTDS, Moreira SAF, Oliveira LEMD, Sotero ADJ. Metabolism of carbohydrates during the development of seeds of the brazilian rubber tree [Hevea brasiliensis (Willd. Ex Adr. de Juss) Muell.-Arg.] Acta Physiologiae Plantarum. 2011;33:211–219. doi: 10.1007/s11738-010-0540-8. [DOI] [Google Scholar]
- 38.Song J, et al. Genome-wide identification of gibberellins metabolic enzyme genes and expression profiling analysis during seed germination in maize. Gene. 2011;482:34–42. doi: 10.1016/j.gene.2011.05.008. [DOI] [PubMed] [Google Scholar]
- 39.Yang YZ, Sheng-Ping LI, Qing-Xia WU, Peng FR. The Dynamic Changes of Proteins and Activities of Nitrogen Metabolism Enzymes in Ginkgo biloba Seeds During Germination. Journal of Nanjing Forestry University. 2006;30:119–122. [Google Scholar]
- 40.Mittasch J, Bottcher C, Frolov A, Strack D, Milkowski C. Reprogramming the phenylpropanoid metabolism in seeds of oilseed rape by suppressing the orthologs of reduced epidermal fluorescence1. Plant physiology. 2013;161:1656–1669. doi: 10.1104/pp.113.215491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nair RB, Bastress KL, Ruegger MO, Denault JW, Chapple C. The Arabidopsis thaliana reduced epidermal fluorescence1 gene encodes an aldehyde dehydrogenase involved in ferulic acid and sinapic acid biosynthesis. The Plant cell. 2004;16:544–554. doi: 10.1105/tpc.017509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scavetta RD, et al. Structure of a Plant Cell Wall Fragment Complexed to Pectate Lyase C. The Plant cell. 1999;11:1081–1092. doi: 10.1105/tpc.11.6.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang H, et al. The essential role of GhPEL gene, encoding a pectate lyase, in cell wall loosening by depolymerization of the de-esterified pectin during fibre elongation in cotton. Plant molecular biology. 2009;72:397–406. doi: 10.1007/s11103-009-9578-7. [DOI] [PubMed] [Google Scholar]
- 44.Jimenez-Bermudez S, Redondo-Nevado J-BJ, Caballero JL. Manipulation of strawberry fruit softening by antisense expression of a pectate lyase gene. Plant physiology. 2002;128:751–759. doi: 10.1104/pp.010671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marín-Rodríguez MC, Orchard J, Seymour GB. Pectate lyases, cell wall degradation and fruit softening. Journal of Experimental Botany. 2002;53:2115. doi: 10.1093/jxb/erf089. [DOI] [PubMed] [Google Scholar]
- 46.Du Q, Pan W, Tian J, Li B, Zhang D. The UDP-glucuronate decarboxylase gene family in Populus: structure, expression, and association genetics. PloS one. 2013;8:e60880. doi: 10.1371/journal.pone.0060880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bindschedler LV, et al. Modification of hemicellulose content by antisense down-regulation of UDP-glucuronate decarboxylase in tobacco and its consequences for cellulose extractability. Phytochemistry. 2007;68:2635. doi: 10.1016/j.phytochem.2007.08.029. [DOI] [PubMed] [Google Scholar]
- 48.Fang W, Wang Z, Cui R, Li J, Li Y. Maternal control of seed size by EOD3/CYP78A6 in Arabidopsis thaliana. The Plant journal: for cell and molecular biology. 2012;70:929–939. doi: 10.1111/j.1365-313X.2012.04907.x. [DOI] [PubMed] [Google Scholar]
- 49.Li N, Li Y. Signaling pathways of seed size control in plants. Current opinion in plant biology. 2016;33:23–32. doi: 10.1016/j.pbi.2016.05.008. [DOI] [PubMed] [Google Scholar]
- 50.Tong H, et al. Brassinosteroid regulates cell elongation by modulating gibberellin metabolism in rice. The Plant cell. 2014;26:4376–4393. doi: 10.1105/tpc.114.132092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Che R, et al. Control of grain size and rice yield by GL2-mediated brassinosteroid responses. Nat Plants. 2015;2:15195. doi: 10.1038/nplants.2015.195. [DOI] [PubMed] [Google Scholar]
- 52.Wang L, et al. Coordinated regulation of vegetative and reproductive branching in rice. Proc. Natl. Acad. Sci. USA. 2015;112:15504–15509. doi: 10.1073/pnas.1521949112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ohto MA, Floyd SK, Fischer RL, Goldberg RB, Harada JJ. Effects of APETALA2 on embryo, endosperm, and seed coat development determine seed size in Arabidopsis. Sexual plant reproduction. 2009;22:277–289. doi: 10.1007/s00497-009-0116-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ohto MA, Fischer RL, Goldberg RB, Nakamura K, Harada JJ. Control of seed mass by APETALA2. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:3123–3128. doi: 10.1073/pnas.0409858102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jofuku KD, Omidyar PK, Gee Z, Okamuro JK. Control of seed mass and seed yield by the floral homeotic gene APETALA2. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:3117–3122. doi: 10.1073/pnas.0409893102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Si L, et al. OsSPL13 controls grain size in cultivated rice. Nat. Genet. 2016;48:447–456. doi: 10.1038/ng.3518. [DOI] [PubMed] [Google Scholar]
- 57.Wang S, et al. Control of grain size, shape and quality by OsSPL16 in rice. Nat. Genet. 2012;44:950–954. doi: 10.1038/ng.2327. [DOI] [PubMed] [Google Scholar]
- 58.Liu TY, et al. Identification of plant vacuolar transporters mediating phosphate storage. Nature Communications. 2016;7:11095. doi: 10.1038/ncomms11095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Moradi M, Enkavi G, Tajkhorshid E. Atomic-level characterization of transport cycle thermodynamics in the glycerol-3-phosphate:phosphate antiporter. Nature Communications. 2015;6:8393. doi: 10.1038/ncomms9393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schena M, Davis RW. Structure of Homeobox-Leucine Zipper Genes Suggests a Model for the Evolution of Gene Families. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:8393. doi: 10.1073/pnas.91.18.8393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Son O, et al. ATHB12, an ABA-inducible homeodomain-leucine zipper (HD-Zip) protein of Arabidopsis, negatively regulates the growth of the inflorescence stem by decreasing the expression of a gibberellin 20-oxidase gene. Plant & cell physiology. 2010;51:1537–1547. doi: 10.1093/pcp/pcq108. [DOI] [PubMed] [Google Scholar]
- 62.Hur YS, et al. Arabidopsis thaliana homeobox 12 (ATHB12), a homeodomain-leucine zipper protein, regulates leaf growth by promoting cell expansion and endoreduplication. New Phytologist. 2015;205:316. doi: 10.1111/nph.12998. [DOI] [PubMed] [Google Scholar]
- 63.Xiao C, Zhang T, Zheng Y, Cosgrove DJ, Anderson CT. Xyloglucan Deficiency Disrupts Microtubule Stability and Cellulose Biosynthesis in Arabidopsis, Altering Cell Growth and Morphogenesis. Plant physiology. 2016;170:234–249. doi: 10.1104/pp.15.01395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hambardzumyan A, et al. Structure and optical properties of plant cell wall bio-inspired materials: cellulose-lignin multilayer nanocomposites. Comptes rendus - Biologies. 2011;334:839–850. doi: 10.1016/j.crvi.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 65.Tang, C., Xie, Y. & Yan, W. AASRA: An Anchor Alignment-Based Small RNA Annotation Pipeline (2017). [DOI] [PMC free article] [PubMed]
- 66.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Grabherr MG, et al. Trinity: reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nature Biotechnology. 2011;29:644. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature Protocols. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li H, et al. Investigation of the microRNAs in safflower seed, leaf, and petal by high-throughput sequencing. Planta. 2011;233:611–619. doi: 10.1007/s00425-010-1327-2. [DOI] [PubMed] [Google Scholar]
- 70.Mortazavi A, Williams BA, Mccue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature Methods. 2008;5:621. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 71.Chen C, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Research. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 73.Thimm O, et al. MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant Journal. 2004;37:914–939. doi: 10.1111/j.1365-313X.2004.02016.x. [DOI] [PubMed] [Google Scholar]
- 74.Kanehisa, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45:D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.