Abstract
Objective
A low prevalence of intestinal parasites has been identified in individuals with irritable bowel syndrome (IBS), but potential associations with alterations in the bacterial microbiome remain largely unexplored. We aimed to investigate the relationship between parasites and bacteria in individuals with IBS in order to identify potential trans-kingdom microbial characteristics.
Design
Stool samples were collected from the Danish background population classified into IBS (n = 119), unspecific gastrointestinal (GI) symptoms (n = 114), and asymptomatic controls (n = 186) based on the Rome III criteria for IBS. Bacterial (16S) and eukaryotic (18S) ribosomal DNA was sequenced, and 18S data were merged with data from conventional parasite laboratory tests. The bacterial microbiome was analyzed according to symptom group and parasite colonization status.
Results
Bacterial richness and diversity were similar for IBS and controls but higher in those with unspecific GI symptoms. A higher abundance of Bacteroides and a lower abundance of Faecalibacterium were detected in individuals with IBS and unspecific GI symptoms compared with controls. Principal component analyses indicated differences in bacterial composition related to parasite colonization rather than symptom group. Parasites were detected at the lowest frequency in the IBS group (39%) and in samples dominated by Bacteroides. Higher bacterial richness and diversity were found in parasite-positive samples from controls and those with unspecific GI symptoms but not in individuals with IBS.
Conclusion
Parasite colonization, rather than bacterial composition, differed between individuals with IBS and healthy controls. Parasite colonization was associated to a rich and diverse bacterial microbiome; however, this association was altered in IBS.
Introduction
The intestinal parasites Dientamoeba fragilis and Blastocystis were previously found to colonize individuals with IBS at a lower frequency compared with asymptomatic controls1. This finding indicates that our understanding of the role of some intestinal parasites in gastrointestinal (GI) health and disease is limited. Several studies have reported a difference in the composition of the bacterial microbiome in relation to IBS2–16, although with inconsistent findings. Organisms from the different kingdoms of the gut microbiome coexist and interact17, and given the fact that some intestinal parasites, such as Blastocystis and D. fragilis, are common colonizers of the human GI tract1,18,19, it is relevant to investigate the lower prevalence of intestinal parasites in individuals with IBS in relation to possible differences in the bacterial microbiome. Limited investigation in the field has focused on the association between Blastocystis and the bacterial microbiome in IBS11,20, identifying differences across some bacterial groups in relation to colonization with Blastocystis.
The association between parasites, including D. fragilis and Blastocystis, and the bacterial microbiome in IBS is still largely unexplored, but could contribute to a trans-kingdom understanding of the possible role of the microbiome in IBS.
The aim of this study was to investigate characteristics of the bacterial microbiome in IBS related to colonization with intestinal parasites, focusing on D. fragilis and Blastocystis, by comparing stool microbiome data from individuals with IBS with asymptomatic controls and with individuals with GI symptoms not fulfilling the criteria for IBS.
Methods
We conducted an internet-based cohort study of the Danish background population in 2010, with a follow-up in 2011; described elsewhere1,21. Briefly, the cohort consisted of 6112 persons (age, 18–50 years [yr]), who completed a questionnaire based on the Rome III criteria for IBS22. Respondents were classified into four symptom groups: asymptomatic controls, individuals with IBS (fulfilling the Rome III criteria for IBS and reporting no organic GI diagnosis) subtyped by the Rome III criteria according to stool type22, individuals with unspecific GI symptoms (reporting GI symptoms within the past 3 months not fulfilling the Rome III criteria for IBS), and individuals with organic GI disease (reporting a GI diagnosis).
The classification into symptom groups by the web-based questionnaire was validated by telephone interviews conducted by a blinded physician on a sample of the population as described elsewhere21.
Consecutive respondents were asked to provide stool samples, until a pre-specified number was reached (see the paragraph on Statistics for details). Sampling kits were sent by postal mail to those agreeing to submit stool samples. The kit included two test tubes without additives. Samples were returned by mail, delivered in the laboratory the following day. Upon receipt, stool samples were tested for ova and parasites by microscopy, short-term in vitro culture for Blastocystis23, and a multiplex quantitative polymerase chain reaction (qPCR) for D. fragilis, Cryptosporidium spp., Entamoeba histolytica, Entamoeba dispar, and Giardia intestinalis24–26. Purified DNA was amplified using four primers targeting nuclear ribosomal genes (16S for prokaryotes and 18S for eukaryotes). For prokaryotes, a modified version of the published universal prokaryotic primers 341F/806R27 was used, while three in-house primers (G3F1/G3R1, G4F3/G4R3, and G6F1/G6R1) were used to amplify eukaryotic DNA (see supplementary material for details). Sequences were mapped using BION, a k-mer-based mapping software described previously28. A unique taxon was defined by denotation to a specific evolutionary taxonomic group.
The data on fungi are still undergoing taxonomic validation and will be presented elsewhere. Parasite sequencing data were merged with data from routine laboratory tests, including microscopy, culture, and qPCR presented earlier1 to ensure the broadest possible and most sensitive approach to parasite detection. Four categories of parasite colonization status were studied in relation to the composition of bacteria: (i) any parasite-positive/-negative, (ii) D. fragilis-positive/-negative, (iii) Blastocystis-positive/-negative, and (iv) positive for multiple parasites (>1)/not positive for multiple parasites (≤1).
Statistics
Sample size was estimated with the aim to detect a statistical difference at a significance level of 5% in the prevalence of intestinal parasites between individuals with IBS (expected prevalence, 30%29) and asymptomatic controls (expected prevalence, 12%30). Seventy-three persons with IBS and 73 controls were required, and we aimed to receive samples from 100 individuals in each group. In 2010, we requested stool samples from 200 individuals with IBS and 300 controls. In 2011, we requested samples from 200 individuals with unspecific GI symptoms.
Differences in the proportion of positive samples according to the four parasite colonization categories between the symptom groups were tested by the χ2-test. BION data were analyzed in R31 with the PhyloSeq32, and Vegan packages33, using ggplot234 and Plotly35 for graphical presentation.
Data were rarefied to 10,000 sequences in order to enable comparison of data from samples with a large variation in number of sequence reads. Richness was defined as a simple count of unique taxa and reported as the mean (standard deviation [SD]) of observed unique taxa. Shannon’s diversity index was used as a measure of diversity, defined by two components, richness and evenness; the latter is a term referring to how equal the abundance of unique taxa is. Differences in bacterial richness and diversity were compared according to symptom group and according to parasite colonization status by two-sided t-tests (when comparing two groups) or one-way ANOVA (when comparing more than two groups). In order to visualize general differences of the bacterial microbiome according to symptom group and parasite colonization status, principal coordinate analyses (PCoA), which reflects inter-sample dissimilarity, were carried out based on Bray–Curtis dissimilarity of the rarefied data. Relative abundance data were based on rarefied sequences and refers to the percentage of total sequences in a sample assigned to a particular taxon. The relative abundance of the two most common phyla and genera was compared between symptom groups by a two-sided t-test, and bar plots were used to display the relative abundance of the ten most common genera for each symptom group.
Based on presence/absence of species, we hypothesized that if any bacterial species should have a significant role in IBS, it should be present at a lower or higher prevalence in the IBS group. We defined that potential species of relevance to IBS should be present with a difference in prevalence of at least 10% between the IBS group and asymptomatic controls based on the number of samples collected. Differences in prevalence of the selected species between the IBS group and asymptomatic controls were tested for statistical significance by the χ2-test.
Species present at a higher or lower prevalence of at least 10% (1) in the group with unspecific GI symptoms compared with the IBS group and/or the control group and (2) between positive and negative samples across the four parasite colonization status categories were also reported.
All authors had access to the study data and reviewed and approved the final manuscript. The study was approved by the ethics committee in Region Zealand, Denmark (SJ-144 and SJ-199) and by the Danish Data Protection Agency. The study was performed according to the Declaration of Helsinki.
Results
Stool samples were obtained from 124 individuals with IBS and 204 asymptomatic controls in 2010 and from 118 individuals with unspecific GI symptoms in 2011. We performed 16S rRNA and 18S rRNA gene sequencing on 119 samples from individuals with IBS (62% women; mean age, 37.2 yr [SD: 7.8]), 186 samples from asymptomatic controls (39% women; mean age, 37.8 yr [SD: 8.1]), and 114 persons with unspecific GI symptoms (68% women; mean age, 36.4 yr [SD: 8.0]). For the remaining samples, stool was not stored for DNA extraction by mistake. IBS subtypes according to stool pattern were distributed as follows: Mixed-IBS, 49 (41%); IBS with diarrhea, 38 (32%); IBS with constipation, 19 (16%); and un-subtyped IBS, 10 (8%) (missing data for three individuals).
Parasite colonization
Of the 419 samples analyzed, 197 (47%) samples were positive for parasites with 67 (16%) samples being positive for multiple parasite species. There was no difference in frequency of colonization with parasites according to sex or age (data not shown). Parasite colonization status for all three symptom groups is presented in Table 1. The prevalence of parasites was highest in asymptomatic individuals and lowest in the IBS group across all four parasite colonization categories.
Table 1.
Prevalence of intestinal parasites in the three symptom groups
| Positive for any parasite, n (%) | Dientamoeba fragilis-positive, n (%) | Blastocystis-positive, n (%) | Positive for multiple parasites species, n (%) | |
|---|---|---|---|---|
| Individuals with IBS, n = 119 | 46 (39) | 29 (24) | 21 (18) | 12 (10) |
| Individuals with unspecific GI symptoms, n = 114 | 54 (48) | 34 (30) | 28 (25) | 17 (15) |
| Asymptomatic controls, n = 186 | 97 (52) | 71 (38) | 51 (27) | 38 (20) |
| P value IBS vs asymptomatic | 0.02 | 0.01 | 0.05 | 0.02 |
Difference in prevalence tested by χ2-test
GI gastrointestinal, IBS irritable bowel syndrome
An overview of parasites detected by sequencing and routine laboratory methods are listed at genus level in Table S1. The primers used to amplify eukaryotic DNA appeared to have low sensitivity with regard to detection of D. fragilis and to some extent for Entamoeba spp.
Bacterial composition
Richness and diversity
BION mapped 38,534,562 sequences to 1536 unique taxa across all 419 samples. Richness and diversity did not differ between the IBS and the control group; however, higher diversity and richness were found in samples from individuals with unspecific GI symptoms (Table 2). Richness and diversity did not differ according to IBS subtype or time since onset of symptoms (data not shown). Richness and diversity were significantly higher in samples positive for any parasite, D. fragilis or Blastocystis, and multiple parasite species (Table 2). For asymptomatic individuals and those with unspecific GI symptoms, bacterial richness and diversity was higher in parasite-positive samples in the four parasite colonization status categories (Table 3). However, in the IBS group, richness did not differ according to parasite status, and diversity was only higher in samples positive for Blastocystis or multiple parasites (Table 3).
Table 2.
Bacterial richness and diversity according to symptom group and parasite colonization status
| Observed unique taxa Mean (SD) [P value] |
Shannon’s diversity index Mean (SD) [P value] |
|
|---|---|---|
| Individuals with IBS, n = 119 | 117.6 (16.6) [0.36]a | 3.00 (0.29) [0.59]a |
| Asymptomatic controls, n = 186 | 115.8 (16.6) | 2.98 (0.32) |
| Individuals with unspecific GI symptoms, n = 114 | 126.0 (21.0) [<0.0001]b | 3.07 (0.31) [0.06]b |
| Positive for any parasite, n = 197 | 124.2 (17.5) [<0.0001]c | 3.09 (0.3) [<0.0001]c |
| Negative for any parasite, n = 222 | 114.5 (17.9) | 2.94 (0.3) |
| Dientamoeba fragilis-positive, n = 134 | 124.0 (17.0) [<0.0001]c | 3.08 (0.3) [0.002]c |
| Dientamoeba fragilis-negative, n = 285 | 116.7 (18.6) | 2.98 (0.3) |
| Blastocystis-positive, n = 100 | 126.0 (18.1) [<0.0001]c | 3.11 (0.2) [<0.0001]c |
| Blastocystis-negative, n = 319 | 116.9 (17.9) | 2.98 (0.3) |
| Positive for multiple parasite species, n = 67 | 126.3 (16.0) [<0.001]c | 3.11 (0.3) [<0.001]c |
| Negative for multiple parasite species, n = 352 | 117.7 (18.5) | 2.99 (0.3) |
GI gastrointestinal, IBS irritable bowel syndrome, SD standard deviation
aDifference between the IBS group and asymptomatic control group tested by two-sided t-test
bDifference between all three symptom groups tested by one-way ANOVA
cDifference between positive and negative samples tested by two-sided t-test
Table 3.
Bacterial diversity and richness according to parasite colonization status, specified for each symptom group
| Mean (SD) | Positive for any parasite | Negative for any parasite | Dientamoeba fragilis-positive | Dientamoeba fragilis-negative | Blastocystis-positive | Blastocystis-negative | Positive for multiple parasite species | Negative for multiple parasite species |
|---|---|---|---|---|---|---|---|---|
| IBS, n = 119 | ||||||||
| Observed unique taxa | 120.6 (14.2) | 115.7 (17.7) | 121.6 (12.5) | 116.3 (17.6) | 121.3 (16.5) | 116.8, 16.6 | 123.2 (13.3) | 117.0 (16.8) |
| Shannon’s diversity index | 3.03 (0.3) | 2.98 (0.3) | 3.00 (0.3) | 3.00 (0.3) | 3.12 (0.2)a | 2.98 (0.3) | 3.19 (0.2)a | 2.98 (0.3) |
| Unspecific GI symptoms, n = 114 | ||||||||
| Observed unique taxa | 132.9 (21.2)b | 119.7 (19.0) | 133.4 (22.3)a | 122.8 (19.8) | 136.4 (19.0)a | 122. 6 (20.6) | 137.5 (14.9)a | 124.0 (21.4) |
| Shannon’s diversity index | 3.18 (0.3)b | 2.97 (0.3) | 3.16 (0.3)a | 3.03 (0.3) | 3.21 (0.1)b | 3.02 (0.3) | 3.22 (0.2)a | 3.04 (0.3) |
| Asymptomatic, n = 186 | ||||||||
| Observed unique taxa | 121.1 (15.1)a | 110. (16.3) | 120.5 (14.0)b | 112.9 (17.5) | 122.2 (16.0)b | 113.4 (16.2) | 122.2 (15.2)a | 114.2 (16.6) |
| Shannon’s diversity index | 3.08 (0.3)b | 2.88 (0.3) | 3.07 (0.3)a | 2.93 (0.3) | 3.06 (0.3)a | 2.96 (0.3) | 3.04 (0.3) | 2.96 (0.3) |
Difference between groups tested by a two-sided t-test
GI gastrointestinal, IBS irritable bowel syndrome, SD standard deviation
aDifference between positive and negative samples significant with a P value ≤0.05
bDifference between positive and negative samples significant with a P value ≤0.0001
PCoA
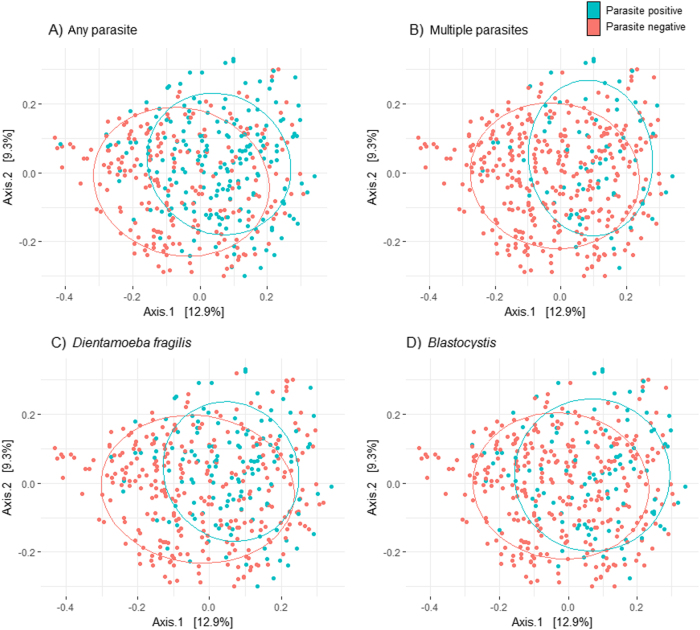
PCoA revealed no separation of samples according to symptom group, indicating no clear differences in the bacterial microbiome composition between individuals with IBS, individuals with unspecific GI symptoms, and asymptomatic controls (Figure S1). According to parasite colonization status, there was no clear separation of samples; nevertheless, a relative clustering of parasite-positive samples was apparent, indicating that the presence of parasites is associated with basic differences in the bacterial microbiome (Fig. 1).
Fig. 1. PCoA of the bacterial microbiome according to parasite colonisation status.
Principal coordinate analysis visualizing differences in the bacterial microbiome between samples positive and negative for any parasite (a), multiple parasites (b), Dientamoeba fragilis (c), and Blastocystis (d). All samples are included in the analysis (n = 419). Blue dots represent samples positive in the given parasite category while negative samples are represented by red dots. The closer the dots are ordinated, the more similar the microbiome. Axes summarize the variability in the data, and the value indicates the variation captured in the axis
Relative abundance
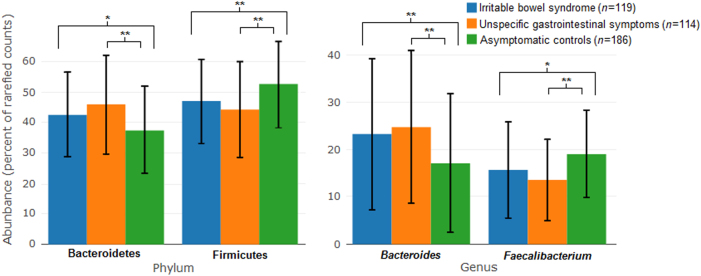
The IBS group had a higher abundance of Bacteroidetes and a lower abundance of Firmicutes compared with asymptomatic controls (Fig. 2). This was reflected in a higher abundance of Bacteroides and a lower abundance of Faecalibacterium at genus level in the IBS group compared with asymptomatic controls (Fig. 2). There was a similar difference in the dominating phyla and genera between the group with unspecific GI symptoms and the asymptomatic controls, and with even more pronounced differences between the groups.
Fig. 2. Mean relative abundance of the two most abundant phyla and genera in the three symptom groups.
Error bars indicate 1 s.d. of the mean. A significant difference between symptom groups according to a two-sided t-test is marked with *p < 0.05 or **p < 0.001
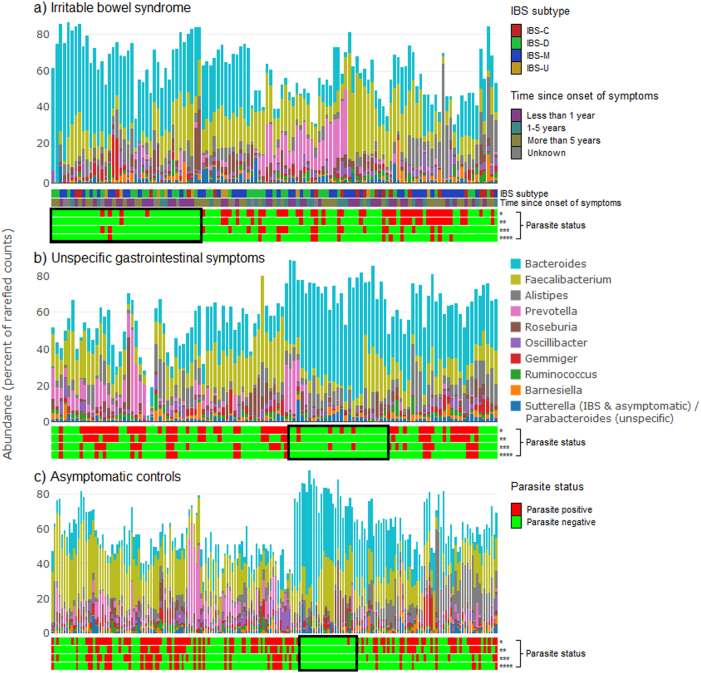
The abundance of the ten most common bacterial genera in combination with parasite colonization status is visualized in Fig. 3. In general, a low frequency of parasites was found in samples with a high proportion of Bacteroides. The samples with a high proportion of Bacteroides had more than 50% of the bacterial abundance accounted for by the ten most common genera. Samples not dominated by Bacteroides consisted to a greater extent of several different, low-abundant genera, reflecting a more diverse microbiome.
Fig. 3. Relative abundance of the ten most abundant bacterial genera found in each of the three symptom groups.
Each bar illustrates the bacterial abundance in one stool sample from an individual. Samples are ordered according to a hierarchical clustering, grouping samples with a similar bacterial composition. Plot a includes samples from individuals with irritable bowel syndrome (IBS), plot b includes samples from individuals with unspecific GI symptoms, and plot c includes samples from asymptomatic controls. The two horizontal bars below plot a show the IBS subtype and time since onset of symptoms of each sample. Additionally, the four horizontal bars below each of the three plots indicate the presence or absence of *, any parasite; **, Dientamoeba fragilis; ***, Blastocystis; and ****, multiple parasites. Notable clusters of samples with low prevalence of parasites are highlighted with a black box
There was no clear clustering of IBS subtype or time since onset of symptoms in relation to the clustering based on abundance of genera (Fig. 3).
Prevalence of bacterial species
Based on our definition of species of particular relevance to IBS, five species were found at a higher prevalence in the IBS group with the difference in prevalence reaching statistical significance for all five (Table 4). All five species, except Blautia faecis, were also found at a higher prevalence in parasite-negative samples compared with parasite-positive samples (Table 4).
Table 4.
Bacterial taxa classified at species level with a difference in prevalence of more than 10% between the IBS group and the group of asymptomatic controls
| IBS n = 119 |
Asymptomatic n = 186 |
Unspecific GI symptoms n = 114 |
Any parasite Positive n = 197 Negative, n = 222 |
D. fragilis
Positive n = 134 Negative n = 285 |
Blastocystis
Positive n = 100 Negative n = 319 |
Multiple parasites Positive n = 67 Negative n = 352 |
|
|---|---|---|---|---|---|---|---|
| Species most prevalent in IBS | |||||||
| Alistipes finegoldii | 70 (59) a | 83 (45) | 58 (51) | Positive: 86 (44) | Positive: 63 (47) | Positive: 36 (36) | Positive: 23 (34) |
| Negative: 125 (56) b | Negative: 148 (52) | Negative: 175 (55) b | Negative: 188 (53) b | ||||
| Bacteroides fragilis | 53 (45) a | 61 (33) | 47 (41) | Positive: 60 (30) | Positive: 43 (32) | Positive: 30 (30) | Positive: 19 (28) |
| Negative: 101 (45) b | Negative: 118 (42) | Negative: 131 (41) b | Negative: 142 (40) | ||||
| Blautia faecis | 102 (86) a | 134 (72) | 88 (77) | Positive: 156 (79) | Positive: 107 (80) | Positive: 77 (77) | Positive: 51 (76) |
| Negative: 168 (76) | Negative: 217 (76) | Negative: 245 (77) | Negative: 273 (76) | ||||
| Flavonifractor plautii | 52 (44) a | 44 (24) | 31 (27) | Positive: 29 (14) | Positive: 107 (8) | Positive: 9 (9) | Positive:11 (14) |
| Negative: 98 (46) b | Negative: 114 (40) b | Negative: 118 (37) b | Negative: 115 (33) b | ||||
| Parabacteroides distasonis | 73 (61) a | 78 (42) | 66 (58) | Positive: 86 (44) | Positive: 51 (38) | Positive: 47 (47) | Positive: 24 (34) |
| Negative: 131 (59) b | Negative: 165 (58) b | Negative: 169 (53) | Negative: 193 (55) b | ||||
| Species most prevalent in asymptomatic | |||||||
| Bifidobacterium adolescentis | 68 (57) | 126 (68) | 80 (70) | Positive: 143 (73) b | Positive: 99 (74) b | Positive: 74 (74) b | Positive: 50 (75) |
| Negative: 131 (59) | Negative: 177 (62) | Negative: 200 (63) | Negative: 224 (64) | ||||
| Clostridium bartlettii | 23 (19) | 56 (30) a | 54 (47) | Positive: 66 (34) | Positive: 50 (37) | Positive: 27 (27) | Positive: 18 (27) |
| Negative: 66 (28) | Negative: 83 (29) | Negative: 105 (33) | Negative: 114 (32) | ||||
| Coprococcus eutactus | 48 (40) | 92 (50) | 59 (52) | Positive: 122 (62) b | Positive: 88 (66) b | Positive: 63 (63) b | Positive: 45 (67) b |
| Negative: 77 (35) | Negative: 111 (39) | Negative: 137 (43) | Negative: 154 (44) | ||||
| Dialister invisus | 28 (24) | 63 (34) a | 41 (36) | Positive: 58 (29) | Positive: 44 (33) | Positive: 22 (22) | Positive: 12 (18) |
| Negative: 74 (33) | Negative: 88 (31) | Negative: 112 (35) b | Negative: 120 (34) b | ||||
| Roseburia faecis | 95 (80) | 167 (90) a | 100 (88) | Positive: 176 (89) | Positive: 121 (90) | Positive: 90 (90) | Positive: 60 (90) |
| Negative: 186 (84) | Negative: 242 (85) | Negative: 274 (86) | Negative: 302 (86) | ||||
Species prevalence is reported according to symptom group and parasite colonization status GI gastrointestinal, IBS irritable bowel syndrome
aSignificant difference in prevalence of species between the IBS group and asymptomatic controls by χ2-test
bSignificant difference in prevalence of species between samples positive and negative for parasites according to parasite colonization status by χ2-test
Five species were found at a higher prevalence in asymptomatic controls compared with the IBS group, reaching statistical significance for three (Table 4).
For the group with unspecific GI symptoms, 21 species were found with a difference in prevalence of at least 10% compared with the IBS group and/or the asymptomatic controls (Table S2). In relation to parasite colonization status, 34 species were found with a difference in prevalence of at least 10% between samples positive and negative for parasites (Table S3).
Discussion
In this study, we investigated the composition of the bacterial microbiome in IBS in relation to colonization by common intestinal parasites. The bacterial microbiome displayed marked differences in relation to parasite colonization status, and we found indications of an altered relationship between parasites and the bacterial microbiome in individuals with IBS.
In general, little difference in the bacterial microbiome between the IBS group and asymptomatic controls was observed. A higher abundance of Bacteroides and a lower abundance of Faecalibacterium were found in individuals with IBS. However, there was no difference in the overall composition of the microbiome between IBS and asymptomatic controls according to richness and diversity or by PCoA.
On the other hand, clear differences in the structure of the bacterial microbiome according to parasite colonization status were observed in several analyses. PCoA plots revealed a more distinct dissimilarity between samples in relation to parasite colonization status than in relation to symptom group. Individuals colonized with parasites exhibited a more diverse and rich bacterial microbiome compared with individuals not colonized by parasites, which confirms earlier findings related to Blastocystis36,37.
High bacterial diversity and richness is considered an attribute of a healthy microbiome38, and a decline in diversity and richness has been linked to several disease states39–41. The concept of parasites as inhabitants of a healthy, diverse microbiome complements associations of parasites to health-promoting features of the microbiome42,43 and the hygiene hypothesis44–46.
Higher richness and diversity in parasite-positive samples was observed in asymptomatic individuals and in individuals with unspecific GI symptoms but could not be generalized to the IBS group, which suggests that the relationship between parasites and the bacterial microbiome is altered in individuals with IBS compared with non-IBS individuals.
The overall association between abundance of bacteria and parasite colonization appeared similar in the three symptom groups (Fig. 3); samples with a high proportion of Bacteroides were dominated by this genus to an extent leading to less diversity, also reflected by a very low frequency of colonization with parasites.
The inverse relationship between colonization with D. fragilis and Blastocystis and a high abundance of Bacteroides confirms earlier findings20,37,47.
Interestingly, even though we did not find any major differences in the composition of the microbiome between IBS and controls, the bacterial microbiome of individuals belonging to the group with unspecific GI symptoms displayed distinct features.
The symptoms represented by the group with unspecific symptoms are very common, as they are reported by 29.5% of the Danish background population48. Most individuals with unspecific symptoms fulfilled ≥two of three IBS criteria regarding the association of abdominal pain to bowel habits as defined by the Rome III criteria, but symptoms were reported to occur at a lower frequency and of shorter duration (<6 months)48. Our data suggest that these unspecific GI symptoms are associated with a more pronounced alteration in the bacterial microbiome compared with the chronic condition of IBS. The group with unspecific GI symptoms reported a high yearly incidence of IBS (18.7%)21, and investigating a possible association between the high incidence of IBS and the detected aberrations of the bacterial microbiome in this group, would be of great interest. Interestingly, the finding of pronounced changes of the bacterial microbiome in the group with unspecific symptoms rather than in the IBS group does not apply to findings related to parasite colonization. Individuals with IBS displayed the greatest deviation from the high prevalence of parasite colonization observed in asymptomatic controls.
Findings of a similar diversity and richness in individuals with IBS and controls confirms a previous study2, while most studies found a lower diversity in the IBS population3–5,7,8,12–14,49,50, and two studies reported a higher diversity in IBS15,51. The finding of Bacteroidetes being more prevalent in IBS is supported by some studies3,5,15 while others find that Bacteroidetes is more abundant in healthy controls8,11–13.
When comparing the ten species with a difference in prevalence between IBS and controls (Table 4) with data from previous studies2–16, we could not find support for any particular clinical relevance to IBS.
The strengths of our study include the fact that stool samples were collected in the background population and that we had large sets of samples representing each symptom group. We compared the structure of the microbiome in individuals with IBS not only with that of asymptomatic controls, but also with individuals with unspecific GI symptoms, as alterations in the microbiome should not only enable discrimination between IBS and asymptomatic individuals, but also between individuals with IBS and those with GI symptoms not categorized as IBS, in order to be of clinical relevance. We established the presence of parasites by both sequencing of the 18s gene and by routine laboratory tests and a low sensitivity for D. fragilis and to some extent Entamoeba spp. was evident by 18S sequencing. This is a relevant finding with regard to interpretation of analyses based on sequencing data as they could underestimate the presence of D. fragilis in samples.
Limitations to the present study include the limited characterization of the study population. Information on demographics and characterization of GI symptoms according to the Rome III criteria were available, but we had no information on possible diagnoses not related to the GI tract, medication use and diet, which are all factors that could influence the composition of the gut microbiome. Stool samples were returned by postal mail in tubes with no additives, which could affect the composition of the microbiome compared with immediate freezing at −80 °C52. However, studies have shown that the effect of interperson variability is larger than effect of storage methods on bacterial composition53,54.
Numerous studies of the bacterial microbiome in IBS have not been able to identify distinct reproducible changes in relation to IBS which questions a possible significant and causal relation. However, many factors have the potential to affect data on the microbiome; both factors unrelated to the study setting and factors embedded in the study design leading to variance in findings between studies. Even so, a distinct feature of the bacterial gut microbiome directly involved in the pathogenesis of IBS should be expected to be evident with some coherence between studies. Two studies have found that only a subgroup of IBS patients differ in composition of the bacterial microbiome compared with healthy controls8,15, which could explain why no distinct changes were found in the overall comparison of IBS and controls. Data from this study did not cluster IBS samples into such subgroups (data not shown).
This study sought to analyze the bacterial microbiome in the context of parasite colonization status and illustrated the importance of integrating the various taxonomic kingdoms when studying the composition of the microbiome. Observations of parasite colonization being associated to healthy features of the gut microbiome should differentiate our view of intestinal parasites beyond the focus on pathogenicity. Further investigation into the altered relationship of parasites and the bacterial microbiome in IBS could contribute to clarifying its potential for distinguishing IBS from both asymptomatic individuals and individuals with GI symptoms not fulfilling the criteria for IBS.
In conclusion, little difference in bacterial composition was observed between individuals with IBS and healthy controls. Parasite colonization was associated to a healthy state, with parasites colonizing 50% of asymptomatic controls, while a significantly lower colonization rate was observed in individuals with IBS. The bacterial microbiome displayed clear differences related to parasite colonization status. Presence of parasites was associated to a rich and diverse microbiome in healthy controls and individuals with unspecific GI symptoms; however, this association was altered in individuals with IBS. The significance of this should be investigated further to understand possible trans-kingdom changes of the microbiome in IBS.
Study Highlights
What is current knowledge?
Alterations of the bacterial microbiome is reported in IBS, although with inconsistent findings.
Individuals with IBS are less frequently colonized with intestinal parasites.
What is new here?
Colonization with parasites is associated with a high diversity and richness of the bacterial microbiome in asymptomatic controls and individuals with non-IBS symptoms.
This association between parasites and bacteria is altered in individuals with IBS.
Electronic supplementary material
Conflict of interest
Guarantor of the article: Laura Rindom Krogsgaard.
Specific authors contributions: L.R.K.: Study concept and design, acquisition of data, analysis and interpretation of data, drafted the manuscript, statistical analyses, and obtained funding. L.O'B.A.: Performed 16s and 18s analysis, expertise in use of BION software, analysis and interpretation of data, critical revision of the manuscript for important intellectual content. T.B.J.: Analysis and interpretation of data, statistical analyses, bio-informatics, critical revision of the manuscript for important intellectual content. A.L.E.: Study concept and design, interpretation of data, critical revision of the manuscript for important intellectual content. C.R.S.: Study concept and design, interpretation of data, critical revision of the manuscript for important intellectual content. H.V.N.: Study concept and design, administrative support. P.B.: Study concept and design, interpretation of data, critical revision of the manuscript for important intellectual content, administrative support, and study supervision.
Financial support: The study was funded by Region Zealand’s Health Sciences Research Foundation, University of Copenhagen, the Aksel Meyer Nielsen and Vetsera Meyer Nielsen trust, the Medical Society of Copenhagen, and the Trust of 1870.
Potential competing interests: The authors declare that they have no conflict of interest.
Footnotes
Supplementary Information
Supplementary Information accompanies this paper at 10.1038/s41424-018-0027-2.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Krogsgaard LR, Engsbro AL, Stensvold CR, Nielsen HV, Bytzer P. The prevalence of intestinal parasites is not greater among individuals with irritable bowel syndrome: a population-based case-control study. Clin. Gastroenterol. Hepatol. 2015;13:507–513. doi: 10.1016/j.cgh.2014.07.065. [DOI] [PubMed] [Google Scholar]
- 2.Tap J, et al. Identification of an intestinal microbiota signature associated with severity of irritable bowel syndrome. Gastroenterology. 2016;152:111–123. doi: 10.1053/j.gastro.2016.09.049. [DOI] [PubMed] [Google Scholar]
- 3.Liu Y, et al. Similar fecal microbiota signatures in patients with diarrhea-predominant irritable bowel syndrome and patients with depression. Clin. Gastroenterol. Hepatol. 2016;14:1602–1611. doi: 10.1016/j.cgh.2016.05.033. [DOI] [PubMed] [Google Scholar]
- 4.Sundin J, et al. Altered faecal and mucosal microbial composition in post-infectious irritable bowel syndrome patients correlates with mucosal lymphocyte phenotypes and psychological distress. Aliment. Pharmacol. Ther. 2015;41:342–351. doi: 10.1111/apt.13055. [DOI] [PubMed] [Google Scholar]
- 5.Pozuelo M, et al. Reduction of butyrate- and methane-producing microorganisms in patients with irritable bowel syndrome. Sci. Rep. 2015;5:12693. doi: 10.1038/srep12693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jalanka-Tuovinen J, et al. Faecal microbiota composition and host-microbe cross-talk following gastroenteritis and in postinfectious irritable bowel syndrome. Gut. 2014;63:1737–1745. doi: 10.1136/gutjnl-2013-305994. [DOI] [PubMed] [Google Scholar]
- 7.Carroll IM, Ringel-Kulka T, Siddle JP, Ringel Y. Alterations in composition and diversity of the intestinal microbiota in patients with diarrhea-predominant irritable bowel syndrome. Neurogastroenterol. Motil. 2012;24:521–530. doi: 10.1111/j.1365-2982.2012.01891.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jeffery IB, et al. An irritable bowel syndrome subtype defined by species-specific alterations in faecal microbiota. Gut. 2012;61:997–1006. doi: 10.1136/gutjnl-2011-301501. [DOI] [PubMed] [Google Scholar]
- 9.Rajilic-Stojanovic M, et al. Global and deep molecular analysis of microbiota signatures in fecal samples from patients with irritable bowel syndrome. Gastroenterology. 2012;141:1792–1801. doi: 10.1053/j.gastro.2011.07.043. [DOI] [PubMed] [Google Scholar]
- 10.Kassinen A, et al. The fecal microbiota of irritable bowel syndrome patients differs significantly from that of healthy subjects. Gastroenterology. 2007;133:24–33. doi: 10.1053/j.gastro.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 11.Nagel R, Traub RJ, Allcock RJN, Kwan MMS, Bielefeldt-Ohmann H. Comparison of faecal microbiota in Blastocystis-positive and Blastocystis-negative irritable bowel syndrome patients. Microbiome. 2016;4:47. doi: 10.1186/s40168-016-0191-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rangel I, et al. The relationship between faecal-associated and mucosal-associated microbiota in irritable bowel syndrome patients and healthy subjects. Aliment. Pharmacol. Ther. 2015;42:1211–1221. doi: 10.1111/apt.13399. [DOI] [PubMed] [Google Scholar]
- 13.Krogius-Kurikka L, et al. Microbial community analysis reveals high level phylogenetic alterations in the overall gastrointestinal microbiota of diarrhoea-predominant irritable bowel syndrome sufferers. BMC Gastroenterol. 2009;9:95. doi: 10.1186/1471-230X-9-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Durbán A, et al. Structural alterations of faecal and mucosa-associated bacterial communities in irritable bowel syndrome. Environ. Microbiol. Rep. 2012;4:242–247. doi: 10.1111/j.1758-2229.2012.00327.x. [DOI] [PubMed] [Google Scholar]
- 15.Labus JS, et al. Differences in gut microbial composition correlate with regional brain volumes in irritable bowel syndrome. Microbiome. 2017;5:49. doi: 10.1186/s40168-017-0260-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Casén C, et al. Deviations in human gut microbiota: a novel diagnostic test for determining dysbiosis in patients with IBS or IBD. Aliment. Pharmacol. Ther. 2015;42:71–83. doi: 10.1111/apt.13236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Norman JM, Handley Sa, Virgin HW. Kingdom-agnostic metagenomics and the importance of complete characterization of enteric microbial communities. Gastroenterology. 2014;146:1459–1469. doi: 10.1053/j.gastro.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jokelainen P, et al. Dientamoeba fragilis, a commensal in Children in Danish Day Care Centers. J. Clin. Microbiol. 2017;55:1707–1713. doi: 10.1128/JCM.00037-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scanlan PD, Marchesi JR. Micro-eukaryotic diversity of the human distal gut microbiota: qualitative assessment using culture-dependent and -independent analysis of faeces. ISME J. 2008;2:1183–1193. doi: 10.1038/ismej.2008.76. [DOI] [PubMed] [Google Scholar]
- 20.Nourrisson C, et al. Blastocystis is associated with decrease of fecal microbiota protective bacteria: comparative analysis between patients with irritable bowel syndrome and control subjects. PLoS ONE. 2014;9:e111868. doi: 10.1371/journal.pone.0111868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krogsgaard LR, Engsbro AL, Jones MP, Bytzer P. The epidemiology of irritable bowel syndrome: symptom development over a 3-year period in Denmark. A prospective, population-based cohort study. Neurogastroenterol. Motil. 2017;29:12986. doi: 10.1111/nmo.12986. [DOI] [PubMed] [Google Scholar]
- 22.Longstreth GF, et al. Functional bowel disorders. Gastroenterology. 2006;130:1480–1491. doi: 10.1053/j.gastro.2005.11.061. [DOI] [PubMed] [Google Scholar]
- 23.Clark CG, Stensvold CR. Blastocystis: isolation, xenic cultivation, and cryopreservation. Curr. Protoc. Microbiol. 2016;43:20A.1.1–20A.1.8. doi: 10.1002/cpmc.18. [DOI] [PubMed] [Google Scholar]
- 24.Verweij J, et al. Real-time PCR for the detection of Dientamoeba fragilis in fecal samples. Mol. Cell. Probes. 2007;21:400–404. doi: 10.1016/j.mcp.2007.05.006. [DOI] [PubMed] [Google Scholar]
- 25.Verweij J, et al. Simultaneous detection of Entamoeba histolytica, Giardia lamblia, and Cryptosporidium parvum in fecal samples by using multiplex real-time PCR. J. Clin. Microbiol. 2004;42:1220–1223. doi: 10.1128/JCM.42.3.1220-1223.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Verweij J, et al. Short communication: prevalence of Entamoeba histolytica and Entamoeba dispar in northern Ghana. Trop. Med. Int. Health. 2003;8:1153–1156. doi: 10.1046/j.1360-2276.2003.01145.x. [DOI] [PubMed] [Google Scholar]
- 27.Yu Y, Lee C, Kim J, Hwang S. Group-specific primer and probe sets to detect methanogenic communities using quantitative real-time polymerase chain reaction. Biotechnol. Bioeng. 2005;89:670–679. doi: 10.1002/bit.20347. [DOI] [PubMed] [Google Scholar]
- 28.Ring HC, et al. The follicular skin microbiome in patients with hidradenitis suppurativa and healthy controls. JAMA Dermatol. 2017;153:897–905. doi: 10.1001/jamadermatol.2017.0904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Engsbro AL, Stensvold CR, Nielsen HV, Bytzer P. Prevalence, incidence, and risk factors of intestinal parasites in Danish primary care patients with irritable bowel syndrome. Scand. J. Infect. Dis. 2014;46:204–209. doi: 10.3109/00365548.2013.861609. [DOI] [PubMed] [Google Scholar]
- 30.Stensvold CR, et al. Blastocystis: unravelling potential risk factors and clinical significance of a common but neglected parasite. Epidemiol. Infect. 2009;137:1655–1663. doi: 10.1017/S0950268809002672. [DOI] [PubMed] [Google Scholar]
- 31.R Development Core Team. R: A language and environment for statistical computing (R Foundation for Statistical Computing, Vienna, Austria, 2008).
- 32.McMurdie PJ, Holmes S. Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE. 2013;8:e21217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jari, O., et al. Vegan: community ecology package. R package version 2.4-2 (2017). https://cran.r-project.org/web/packages/vegan/index.html
- 34.Wickham H. ggplot2: Elegant Graphics for Data Analysis. New York: Springer-Verlag; 2009. [Google Scholar]
- 35.Sievert, C. et al. plotly: create interactive web graphics via “plotly.js”. R package version 4.5.6. 2016. https://cran.r-project.org/web/packages/plotly/index.html
- 36.Audebert C, et al. Colonization with the enteric protozoa Blastocystis is associated with increased diversity of human gut bacterial microbiota. Sci. Rep. 2016;6:25255. doi: 10.1038/srep25255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O’Brien Andersen L, Bonde I, Nielsen HB, Stensvold CR. A retrospective metagenomics approach to studying Blastocystis. FEMS Microbiol. Ecol. 2015;91:fiv072. doi: 10.1093/femsec/fiv072. [DOI] [PubMed] [Google Scholar]
- 38.Bäckhed F, et al. Defining a healthy human gut microbiome: current concepts, future directions, and clinical applications. Cell Host Microbe. 2012;12:611–622. doi: 10.1016/j.chom.2012.10.012. [DOI] [PubMed] [Google Scholar]
- 39.Turnbaugh PJ, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Le Chatelier E, et al. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500:541–546. doi: 10.1038/nature12506. [DOI] [PubMed] [Google Scholar]
- 41.Manichanh C, et al. Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut. 2006;55:205–211. doi: 10.1136/gut.2005.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramanan D, et al. Helminth infection promotes colonization resistance via type 2 immunity. Science. 2016;352:608–612. doi: 10.1126/science.aaf3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lukes J, Stensvold CR, Jirku-Pomajbikova K, Parfrey LW. Are human intestinal eukaryotes beneficial or commensals? PLoS Pathog. 2015;11:7–12. doi: 10.1371/journal.ppat.1005039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Versini M, et al. Unraveling the hygiene hypothesis of helminthes and autoimmunity: origins, pathophysiology, and clinical applications. BMC Med. 2015;13:81. doi: 10.1186/s12916-015-0306-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rook GAW. 99th Dahlem conference on infection, inflammation and chronic inflammatory disorders: darwinian medicine and the “hygiene” or “old friends” hypothesis. Clin. Exp. Immunol. 2010;160:70–79. doi: 10.1111/j.1365-2249.2010.04133.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Okada H, Kuhn C, Feillet H, Bach JF. The “hygiene hypothesis” for autoimmune and allergic diseases: an update. Clin. Exp. Immunol. 2010;160:1–9. doi: 10.1111/j.1365-2249.2010.04139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.O’Brien Andersen L, et al. Associations between common intestinal parasites and bacteria in humans as revealed by qPCR. Eur. J. Clin. Microbiol. Infect. Dis. 2016;35:1427–1431. doi: 10.1007/s10096-016-2680-2. [DOI] [PubMed] [Google Scholar]
- 48.Krogsgaard LR, Engsbro AL, Bytzer P. The epidemiology of irritable bowel syndrome in Denmark. A population-based survey in adults ≤50 years of age. Scand. J. Gastroenterol. 2013;48:523–529. doi: 10.3109/00365521.2013.775328. [DOI] [PubMed] [Google Scholar]
- 49.Carroll IM, et al. Molecular analysis of the luminal- and mucosal-associated intestinal microbiota in diarrhea-predominant irritable bowel syndrome. Am. J. Physiol. Gastrointest. Liver Physiol. 2011;301:G799–G807. doi: 10.1152/ajpgi.00154.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Noor SO, et al. Ulcerative colitis and irritable bowel patients exhibit distinct abnormalities of the gut microbiota. BMC Gastroenterol. 2010;10:134. doi: 10.1186/1471-230X-10-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ponnusamy K, Choi JN, Kim J, Lee SY, Lee CH. Microbial community and metabolomic comparison of irritable bowel syndrome faeces. J. Med. Microbiol. 2011;60:817–827. doi: 10.1099/jmm.0.028126-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Choo JM, Leong LEX, Rogers GB. Sample storage conditions significantly influence faecal microbiome profiles. Sci. Rep. 2015;5:1–10. doi: 10.1038/srep16350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tedjo DI, et al. The effect of sampling and storage on the fecal microbiota composition in healthy and diseased subjects. PLoS ONE. 2015;10:e0126685. doi: 10.1371/journal.pone.0126685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu GD, et al. Sampling and pyrosequencing methods for characterizing bacterial communities in the human gut using 16S sequence tags. BMC Microbiol. 2010;10:206. doi: 10.1186/1471-2180-10-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.