Abstract
Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), a well-known lncRNA associated with numerous diseases, particularly cancer, has received increased attention. The expression of MALAT1 was determined to be upregulated in numerous types of tumors and MALAT1 exhibited effects on tumor cell proliferation, migration, invasion and apoptosis. The abnormal expression of MALAT1 was identified in almost in every organ of the digestive system. MALAT1 performed an important role in the pathological alterations of organs that are associated with sex hormones and several reproductive system cancers. MALAT1 participates in molecular pathways. In the clinical application of MALAT1, MALAT1 was considered as a potential biomarker for the diagnosis and prediction of cancers, and may also serve as therapeutic target for treatment of specific tumors. This review summarizes the abnormal expression of MALAT1 in cancer, its significant effect on the primary features of cancer, as well as the underlying molecular mechanisms of MALAT1 in various cancers. According to studies on MALAT1, we introduce the upstream and downstream substances associated with the function of MALAT1. These reviewed studies promote the clinical application of MALAT1 in the aspect of diagnosis and treatment of different cancers, and may help point out new study directions for MALAT1.
Keywords: long non-coding RNA, metastasis-associated lung adenocarcinoma transcript 1, cancer, clinical application, molecular mechanism
1. Introduction
For decades, studies have focused on protein-coding genes, as they are significantly involved in the mechanisms of cancer initiation and progression. Mutation or deletion of the TP53 may lead to development of Li-Fraumeni syndrome, a disease that leads to increased likelihood of developing tumors during early adulthood (1). PTEN may suppress the growth of tumor cells by antagonizing protein tyrosine kinases and may regulate the invasion and metastasis of tumor cells through interaction with focal adhesion (2). Completion of the human genome sequencing project (3), enriched the available information regarding genome organization and content. Along with the identification that non-protein coding RNAs (ncRNAs) account for the majority of human genome-transcripted sequences (4), ncRNAs have attracted markedly growing research interest.
Long non-coding RNAs (lncRNAs) are a type of RNA molecule with a length of >200 nucleotides (nt), which are unable to encode proteins (5). Specifically, lncRNAs have been defined as RNA molecules that may function as either primary or spliced transcripts, and do not match with these known classes of small RNAs or structural RNAs (6). lncRNAs regulate gene expression through various processes, including chromatin modification, transcription and post-transcription (6,7). lncRNAs can regulate the transcription processing by interacting with RNA binding proteins, co-activating transcription factors, or repressing the promoters of target genes (8–10). Although these lncRNAs cannot encode proteins, their roles in cellular functions are indispensable as well as complex.
2. Research background of MALAT1
Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), also termed nuclear enriched abundant transcript 2, is a long intergenic non-coding RNA (lincRNA) with >8,000 nts, located on chromosome 11q13 (11). Sequences of lncRNAs are highly evolutionarily conserved among species, which predicts the potentially important biological functions of MALAT1 (11). In 2003, MALAT1 was firstly identified to be significantly associated with the metastasis of early-stage non-small cell lung cancer (NSCLC), and therefore MALAT1 was proposed to be a prognostic marker for stage I NSCLC (11). Since then, studies about MALAT1 have gradually increased. A search for all the MALAT1-associated publications on PubMed was performed, and the statistical result is shown in Fig. 1. As shown in Fig. 1, between 2003 and December 2014, the amount of relevant studies increased year-by-year, indicating growing interest of researchers in MALAT1. Among all these articles, cancer-associated studies accounted for ~20% of the total. In addition to cancers, several non-cancer diseases, including myocardial infarction and hyperghlycemia, have also been reported to be associated with MALAT1 (12–16). Studies have identified that MALAT1 performs a vital role in regulating the function of endothelial cell and vessel growth (12), and the role was evidenced in the cardiovascular vascular system (13). In addition, the abnormal expression of MALAT1 in patients with myocardial infarction indicated that it performs an important role in cardiovascular diseases (14). Notably, MALAT1 was also involved in hyperglycemia by inducing inflammatory processes (15), and in diabetic retinopathy it was improperly regulated (16). MALAT1 also affected the progression of proliferative vitreoretinopathy (17), hereditary degenerative disease myotonic dystrophy type 1 (18) and keratoacanthoma (19). In addition, the upregulation of MALAT1 was identified in the cerebellum, hippocampus and brain stem of human alcoholics (20) and the progression of microtia (21) were reported to be associated with MALAT1 expression. Therefore, MALAT1 is widely involved in numerous pathological processes.
Figure 1.
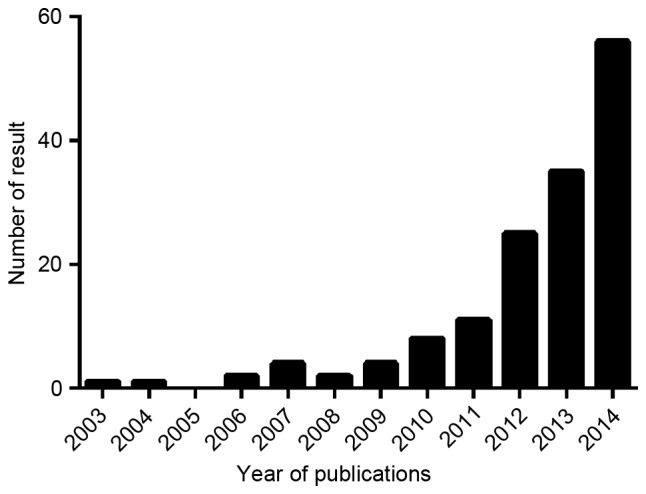
Literature about MALAT1. Articles were searched on PubMed, limited to ‘between 2003 and December 2014’.
3. Association of MALAT1 with cancers
Since the association between MALAT1 and NSCLC was identified, its important roles as an lncRNA in cancer have been considered as a paradigm (22). The expression of MALAT1 was found to be upregulated in numerous types of tumor, and MALAT1 exhibited marked effects on tumor cell proliferation, migration, invasion and apoptosis (22). The advances in clinical study about MALAT1 on cancer during recent years are summarized in Table I.
Table I.
Research advances of MALAT1 in cancers.
| Cancers | Function of MALAT1 | (Refs.) |
|---|---|---|
| NSCLC | Associated with metastasis and survival | (4) |
| A prognostic parameter for stage I and stage II NSCLC | (4,16) | |
| Contributes to brain metastasis | (24) | |
| Esophageal Squamous cell carcinoma | Promotes tumor proliferation and metastasis | (25) |
| Gastric cancer | Promotes the development and peritoneal metastasis of cancer | (26) |
| Promotes cell proliferation | (35) | |
| CRC | A negative prognostic factor to patients with stage II/III CRC | (27) |
| Hepatocellular carcinoma | Predicts a significantly increased risk of tumor recurrence following liver transplantation | (28) |
| Modulating cell proliferation | (59) | |
| Pancreatic cancer | Unfavorable predictor for its clinical progression and prognosis | (29) |
| Promotes cell growth, migration and invasion | (30) | |
| Promotes the stem cell-like phenotypes in cancer cells | (70) | |
| Clear cell renal carcinoma | Associated with proliferation, migration and invasion, indicates tumor progression and poor prognosis | (31) |
| Bladder cancer | Promotes cell migration with the EMT | (32) |
| Mediates the TGFβ-induced EMT and promotes tumor metastasis | (75) | |
| Urothelial carcinoma of the bladder | Associated with proliferation, apoptosis and motility | (33) |
| Castration-resistant prostate cancer | Maintains tumorigenicity | (34) |
| Involved in RNA processing and resulting in incomplete splicing | (35) | |
| Breast cancer | Involved in the inhibitory effect of 17β-Estradiol treatment on breast tumor cell proliferation, migration and invasion | (36) |
| Regulated EMT | (68) | |
| Cervical cancer | Associated with the positive of human papilloma virus in cervical squamous cells | (37) |
| Effected cervical cancer cell proliferation and invasion | (38) | |
| Adrenocortical cancer | The second most upregulated lncRNA | (39) |
| Multiple myeloma | Predicts early progression | (40) |
| Melanoma | Associated with the metastasis | (41) |
| Osteosarcoma | Promotes tumor proliferation and metastasis | (42) |
| Involved in drug action in high-grade osteosarcoma | (51) | |
| Neuroblastoma | Involved in neuroblastoma cells migration and invasion | (48) |
| Glioblastoma | Affects the migration of glioblastoma cells | (81) |
MALAT1, metastasis-associated lung adenocarcinoma transcript 1; EMT, epithelial-mesenchymal transition; lncRNA, long non-coding RNA; TGFβ, transforming growth factor β; CRC, colorectal cancer; NSCLC, non-small cell lung cancer.
Aberrant expression and multiple biological functions of MALAT1 in different cancers
Following the first identification of MALAT1 in NSCLC(11), a subsequent study on NSCLC about MALAT1 hypothesized that the apparent overexpression of MALAT1 in stage I and II NSCLC primary tumors increased the likelihood to metastasis (23). Furthermore, increased MALAT1 expression contributed to brain metastasis by promoting epithelial-mesenchymal transition (EMT) in NSCLC (24).
Undoubtedly, the action of MALAT1 on digestive system cancer deserves wide attention. It was demonstrated that in esophageal squamous cell carcinoma (ESCC), the overexpression of MALAT1 promoted tumor proliferation and metastasis (25). In gastric cancer, the high-level expression of MALAT1 was reported to promote the development and the peritoneal metastasis of cancer (26). A previous clinical study showed that MALAT1 was associated with colorectal cancer (CRC), and its elevated expression may be a negative prognostic factor of patients with stage II/III CRC (27). MALAT1 was also revealed to be upregulated in hepatocellular carcinoma, and its overexpression may indicate a higher risk of tumor recurrence following liver transplantation (28). In a clinical study on pancreatic cancer, the abnormal overexpression of MALAT1 was identified as an unfavorable predictor for its clinical progression and prognosis (29). In accordance with the former research, studies on pancreatic cancer in vitro using seven associated cell lines confirmed the role of MALAT1 in promoting cell growth, migration and invasion (30).
The situation was similar in clear cell renal carcinoma; patients with higher levels of MALAT1 indicated worse tumor progression and poor prognosis, and exploration revealed that knockdown of MALAT1 could inhibit proliferation, migration and invasion in renal cancer cells (31). A previous study on bladder cancer revealed that MALAT1 promoted EMT-associated cell migration and may be activated via Wnt signaling (32). The upregulated MALAT1 was associated with the ability of proliferation, apoptosis and motility in urothelial carcinoma of bladder cancer cells (33). Therefore, its effect on the urinary system was also noteworthy.
It appeared that MALAT1 performed an important role in the pathological changes of organs that are highly associated with sex hormones and several reproductive system cancers (34–37). The upregulation of MALAT1 was involved in the progression of castration-resistant prostate cancer (CRPC) and was associated with the maintenance of tumorigenicity (34). It was also shown to be one of the 10 most highly-expressed transcripts in CRPC, involving RNA processing and resulting in incomplete splicing (35). In addition, the inhibitory effect of 17β-estradiol treatment on breast tumor cell proliferation, migration and invasion was elucidated to be not ERα-dependent but dose-dependent by downregulating the expression level of MALAT1 (36). Furthermore, in cervical cancer, upregulated MALAT1 was associated with positive human papilloma virus (HPV) in cervical squamous cells (37), and MALAT1 was found effect cervical cancer cell proliferation and invasion(38).
MALAT1 was also studied in several other types of cancers (39–42). Among numerous lncRNAs, MALAT1 was revealed to be the second most largely upregulated lncRNA in adrenocortical cancer (39). In patients with multiple myeloma, MALAT1 was reported as overexpressed (40). The markedly high expression of MALAT1 was also shown to be associated with melanoma metastasis (41). In the progression of osteosarcoma, MALAT1 was reported to promote tumor proliferation and metastasis via the phosphoinositide 3-kinase (PI3K)/Akt pathway (42).
In conclusion, these studies demonstrated that MALAT1 serves as an oncogenic gene during the progression of diverse cancers, and that the potential functions of MALAT1 are associated with the basic features of cancer, including proliferation, metastasis, invasion and apoptosis.
Preliminary studies on clinical application of MALAT1 in cancers
As aforementioned, MALAT1 serves a vital role in several cancers, and increasing efforts have been devoted to developing MALAT1-based cancer diagnosis and treatment. The deregulation of MALAT1 in certain types of tumor tissues and association with tumor cell proliferation, migration and invasion make it potential diagnostic biomarkers (4,16,27,29,39). One study has already revealed that urine MALAT1 may act as a promising diagnostic biomarker for prostate cancer based on a multicenter evaluation (43). In addition, MALAT1-derived miniRNA in the plasma may be used to detect and diagnose prostate cancer (44). Other applications about MALAT1 on predicting cancer progression, prognosis, recurrence and metastasis have always been important directions in cancer research. A study about the association between the expression of MALAT1 in the peripheral whole blood of patients and lung cancer progression indicated that its expression level may reflect the host response to lung cancer development (45).
In addition to its advantages as diagnostic or predictive biomarkers, MALAT1 and substances interacting with it may also serve as therapeutic targets in specific tumors (46–48). Several synthetic artificial microRNAs targeting MALAT1 and other genes successfully exhibited anticancer effects on two bladder cancer cell lines (46), which was at the forefront of the application study. The tumor-suppressive ability of Myc-6, a small synthetic chemical pyrrole-imidazole polyamide, on human osteosarcoma MG63 cells was suggested to be partly associated with the specific decrease of MALAT1 (47). In another study about the histone demethylase jumonji domain-containing protein 1A (JMJD1A) and MALAT1, the small molecule JMJD1A inhibitor dimethyloxaloylglycine was demonstrated be able to suppress neuroblastoma cell migration and invasion (48). These studies laid the basis of clinical applications.
Clinical application studies about MALAT1 were not confined to the aforementioned fields. In the study of the genotoxic stress-induced apoptosis, MALAT1 was markedly downregulated in bleomycin-treated HeLa and MCF-7 cells (49). MALAT1 was one of the significant lncRNAs in laryngeal squamous cell carcinoma; the expression of MALAT1 decreased when patients were treated with increasing concentrations of cisplatin and paclitaxel (50). MALAT1 was previously identified to be involved in drug action in high-grade osteosarcoma, suggesting its potential to modulate the drug sensitivity (51). Therefore, the association between MALAT1 and drug action is noteworthy.
4. Molecular mechanisms of MALAT1 in cancers
MALAT1 co-localizes with SC35 splicing domains which were also known as interchromatin granule clusters or nuclear speckles, and the localization of MALAT1 suggests its function in RNA metabolism (52) MALAT1 is a nuclear-retained RNA, and has been shown to be involved in pre-mRNA processing in mammalian cells (53). The 3′ end processing of MALAT1 generates the mature MALAT1 transcript and a tRNA-like RNA cytoplasmic RNA (54). Mature MALAT1 lacking a poly (A) tail was protected by the triple helix structure to maintain the stability of its 3′ ends, and the triple helix could also serve as a translational enhancer (55). MALAT1 was involved in mRNA processing, splicing and exporting (56), and these findings laid the basis of the MALAT1 function.
MALAT1 is regulated by upstream genes or proteins
The expression of MALAT1 was regulated by numerous genes or proteins during transcription and post-transcriptional processing (Fig. 2). Sp1 and JMJD1A were reported to regulate the expression of MALAT1 by binding to the gene promoter, consequently affecting the transcription processing (48,57). A study revealed that Sp1 may bind to the promoter to activate MALAT1, leading to the upregulation of MALAT1 in lung cancer (57). By binding to the promoter and activating the transcription of MALAT1, the histone demethylase JMJD1A upregulated the expression of MALAT1, facilitating neuroblastoma cell migration and invasion (48). In a study on the sex determining region Y-box (Sox) 17 in esophageal cancer, MALAT1 was confirmed as one of the new suppressive downstream genes in its transcriptional network (58). MALAT1 was reported to perform an important role in modulating proliferation of early-stage hematopoietic cells, and p53 regulated the process of hematopoietic differentiation via changing MALAT1 expression (59).
Figure 2.
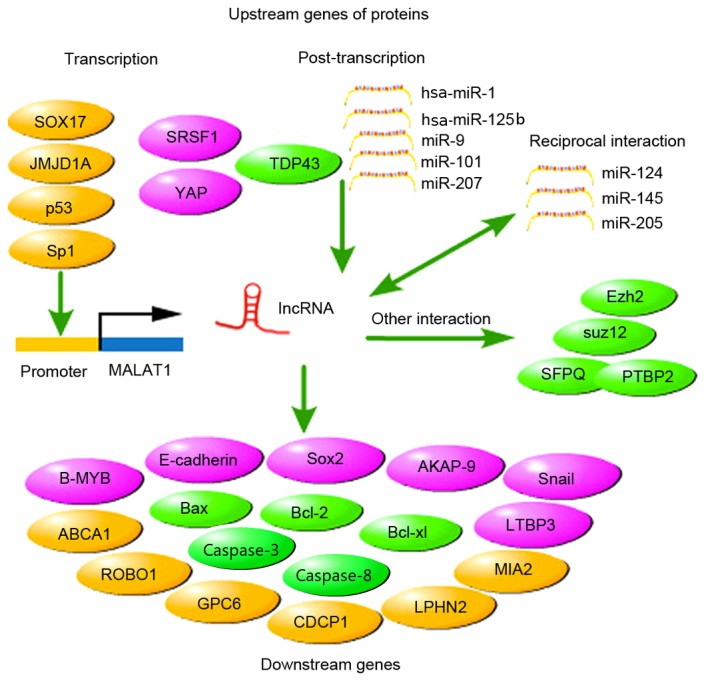
Genes, proteins or miRNAs associated with MALAT1. The upstream regulators during the transcriptional and posttranscriptional procession and the downstream genes modulated by MALAT1 are exhibited. In addition, reciprocal interaction and other associations between MALAT1 and other genes, proteins or miRNAs are shown. MALAT1, metastasis-associated lung adenocarcinoma transcript 1; SOX17, sex-determining region-Y-box 17; SRSF1, serine/arginine-rich splicing factor 1; YAP, yes-associated protein 1; Bcl-2, B-cell lymphoma-2; miR/miRNA, microRNA; ABCA1, ATP-binding cassette transporter member 1; ROBO1, roundabout guidance receptor 1; GPC6, glupican 6; CDCP1, CUB domain-containing protein 1; LPHN2, latrophilin-2, a cell-adhesion G protein-coupled receptor and presumptive α-latrotoxin receptor; MIA2, melanoma inhibitory activity 2; LTBP3, latent transforming growth factor β-binding protein 3; PTBP2, polypyrimidine tract binding protein 2; SFPQ, splicing factor proline-glutamine rich; suz12, suz12 polycomb repressive complex 2 subunit; Ezh2, enhancer of zeste homolog 2; Bax, Bcl-2-associated X protein; Bcl-xl, B-cell lymphoma-extra-large; TDP43, TAR DNA-binding protein 43; lncRNA, long non-coding RNA; AKAP-9, A-kinase anchor protein-9.
The post-transcriptional processing of MALAT1 is complex (60). In NSCLC, TDP-43 may regulate the expression of MALAT1 by directly binding to MALAT1 RNA (61). The interactions with nucleic acids, including miRNAs, during post-transcriptional processing significantly affect the expression and functions of MALAT1 (62–65). By downregulating K-ras and MALAT1, hsa-miR-1 was identified to suppress the development of breast cancer (62). Similarly, MALAT1 was suggested as suppressed by hsa-miR-125b in bladder cancer (63). In the study of the posttranscriptional regulation of MALAT1 in ESCC, studies identified that the posttranscriptional silencing of MALAT1 could suppress tumor proliferation, migration and invasion by miR-101 and miR-217 (64). miR-9 was also reported to modulate the degradation of MALAT1 by targeting AGO2-mediated regulation in the nucleus (65).
As the mechanism underlying the expression and function of MALAT1 is complex, the interactions between upstream regulators and MALAT1 are not simple either. According to a related study, MALAT1 was associated with yes-associated protein (YAP) and serine/arginine-rich splicing factor 1 (SRSF1) at transcriptional and post-transcriptional processing in liver cancer (60), and the associations among MALAT1, YAP and SRSF1 were complicated.
MALAT1 regulates downstream gene expression
Modulation of gene expression is one of the most important functions of lncRNA (6). In lung cancer, the pivotal function of MALAT1 associated with metastasis phenotype was not alternative splicing but regulating the expression of 23 concerning genes, including melanoma inhibitory activity 2, roundabout 1, glypican 6, latrophilin 2, CUB domain containing protein 1 and ATP-binding cassette, subfamily A, member 1 (66). It was previously revealed in NSCLC that the MALAT1 levels affect the expression of B-cell lymphoma (Bcl-2), and Bcl-2 was identified to affect the prognosis (67). Notably, MALAT1 was identified to promote the expression of caspase-3, −8 and Bcl-2-associated X protein, and inhibit the expression of Bcl-2 and Bcl-extra-large, subsequently affecting cervical cancer cells proliferation and invasion (68). PPKA kinase anchor protein 9 was indicated to be the target protein of MALAT1 in colorectal cancer, and the two of them were involved in tumor promotion (69). One study has shown that the promotion of MALAT1 on the stem cell-like phenotypes in pancreatic cancer cells is associated with increased expression of Sox2, a self-renewal associated factor (70). Another study on multiple myeloma revealed that in mesenchymal stem cells, MALAT1 promoted the activation of the promoter of latent transforming growth factor-β binding protein (LTBPS) via regulating recruitment of Sp1 to LTBPS, and the interaction of MALAT1 with Sp1 and LTBP3 promoter increased the expression of LTBPS (71). In terms of its mechanism on regulating the proliferation of cancer cells, MALAT1 modulated the expression level of transcription factor B-MYB, an oncogenic gene involved in cell cycle progression (72). The downstream genes associated with MALAT1 are listed in Fig. 2.
Interaction between MALAT1 and other genes
The underlying mechanism of MALAT1 promoting tumor growth and metastasis in colorectal cancer was revealed to be associated with its competitive binding to the tumor suppressor gene splicing factor proline and glutamine rich (SFPQ) and subsequently releasing SFPQ from the SFPQ/polypyrimidine tract binding protein 2 (PTBP2) complex, which was accompanied by increased SFPQ-detached PTBP2, which served as a proto-oncogene (73). In gastric cancer, MALAT1 promoted cellular proliferation partly by recruiting the splicing factor SF2/alternative splicing factor, a member of the serine/arginine-rich protein family (74). According to a study on bladder cancer, MALAT1 also mediated the TGF-β-induced EMT and was associated with suppressor of zeste12, thus promoting tumor metastasis (75). To investigate the mechanism underlying the promotion of MALAT1, a recent study conducted on renal cell carcinoma showed that the binding of MALAT1 and enhancer of zeste homolog 2 affected the epithelial-mesenchymal transition by affecting the expression of E-cadherin and β-catenin, thus, finally affecting the tumor progression (76). Furthermore, its reciprocal interaction with miR-205 was also disclosed (76). miR-205 was not the only miRNA involved in the reciprocal interaction with MALAT1; it was demonstrated that MALAT1 could regulate the radiosensitivity of HR-HPV+ cervical cancer by the reciprocal repression between MALAT1 and miR-145 (77). The reciprocal interaction between MALAT1 and miR-124 was also involved in the growth and invasion of HR-HPV-positive cervical cancer cells via the MALAT1-miR-124-RBG2 axis (78). In Fig. 2 we show the interaction.
Signaling pathways involving MALAT1
MALAT1 has been found to be associated with a multitude of molecular pathways, which in turn complicated the mechanism of action (25,68,79–83). It modulated nuclear factor-κB/RelA, which is involved in the process of EMT (79). Furthermore, another study on breast cancer revealed that the expression of MALAT1 could regulate EMT through the PI3K-AKT pathway (68), which was also involved in the progression of osteosarcoma (41). In ESCC, the overexpression of MALAT1 was demonstrated to promote tumor proliferation and metastasis by dephosphorylating the ATM-CHK2 pathway (25). In the study of gallbladder carcinoma, researchers found MALAT1 acted as an oncogenic lncRNA by activating the extracellular-signal regulated kinase/mitogen-activated protein kinase pathway (80). Another study on glioblastoma revealed MALAT1 was the top downregulated gene in the WNT inhibitory factor 1-expressing cells, and identified that knockdown of MALAT1 could reduce the migration of glioblastoma cells (81). Furthermore, this study also suggested the possible association between the non-canonical Wnt signaling and MALAT1 (81). Chemokine (C-C motif) ligand 5, derived from tumor-associated dendritic cell, was revealed to be associated with colon cancer progression through the MALAT1/Snail pathway (82). In addition, the PCDH10-Wnt/β-catenin-MALAT1 regulatory axis was established in a study on endometrioid endometrial cancer (83).
5. Perspectives and challenges
Firstly, MALAT1 is an important lncRNA involved in numerous biological processes. The critical roles of MALAT1 in gene regulation and the noticeable effect on the basic function of cells, particularly on tumor cells, have been confirmed by numerous concerning studies (72,84–86). However, certain results of these studies were inconsistent. A study showed that the quantitative loss of MALAT1 had no effect on the phenotypes of human lung or liver cancer cells, including proliferation and cell cycle progression (87), which disagreed with other studies (11,59,88). Similarly, the expression of MALAT1 also differed significantly from another study on hepatocellular carcinoma, according to the expression profiles of lncRNAs (89). Secondly, since numerous molecular mechanisms underlying the action of MALAT1 remain unclear, multiple experiments are required to explore and verify its functions. Furthermore, certain parts of mechanisms underlying the progression of different cancers appear to be the same. For example, the abnormal expression of MALAT1 was found almost in every organ of the digestive system (25–30), MALAT1 and PI3K-AKT pathway are involved in osteosarcoma and breast cancer (42,68). It is then important to confirm whether these shared mechanisms are universal. Furthermore, in terms of the diverse types of diseases associated with MALAT1, research fields should be expanded to its associated functions in addition to cancer. Finally, there is no doubt that the application of MALAT1 was diverse. During the development of MALAT1-based cancer diagnosis and treatment, with the expectation of its advantages as a diagnostic biomarker, predictive biomarker or therapeutic targets in specific tumors, its effect on drug action is also a good study direction on its clinical application.
Acknowledgements
The authors would like to thank Xueqing Hu for writing assistance.
Funding
The National Natural Science Foundation of China (grant no. 81520108031) supported this review.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
SW made substantial contributions to design. QJ and QL were involved in drafting the manuscript or revising it critically for important intellectual content, XL and PG were involved in acquiring, and analyzing related articles and data. MZ was a major contributor in writing the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no conflict of interests.
References
- 1.Hollstein M, Sidransky D, Vogelstein B, Harris CC. p53 mutations in human cancers. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 2.Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang SI, Puc J, Miliaresis C, Rodgers L, McCombie R, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275:1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 3.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 4.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 6.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: Insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 7.Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: Functional surprises from the RNA world. Genes Dev. 2009;23:1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feng J, Bi C, Clark BS, Mady R, Shah P, Kohtz JD. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev. 2006;20:1470–1484. doi: 10.1101/gad.1416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martianov I, Ramadass A, Barros Serra A, Chow N, Akoulitchev A. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature. 2007;445:666–670. doi: 10.1038/nature05519. [DOI] [PubMed] [Google Scholar]
- 10.Wang X, Arai S, Song X, Reichart D, Du K, Pascual G, Tempst P, Rosenfeld MG, Glass CK, Kurokawa R. Induced ncRNAs allosterically modify RNA binding proteins in cis to inhibit transcription. Nature. 2008;454:126–130. doi: 10.1038/nature06992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ji P, Diederichs S, Wang W, Böing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 12.Michalik KM, You X, Manavski Y, Doddaballapur A, Zörnig M, Braun T, John D, Ponomareva Y, Chen W, Uchida S, et al. Long noncoding RNA MALAT1 regulates endothelial cell function and vessel growth. Circ Res. 2014;114:1389–1397. doi: 10.1161/CIRCRESAHA.114.303265. [DOI] [PubMed] [Google Scholar]
- 13.Uchida S, Dimmeler S. Long noncoding RNAs in cardiovascular diseases. Circ Res. 2015;116:737–750. doi: 10.1161/CIRCRESAHA.116.302521. [DOI] [PubMed] [Google Scholar]
- 14.Vausort M, Wagner DR, Devaux Y. Long noncoding RNAs in patients with acute myocardial infarction. Circ Res. 2014;115:668–677. doi: 10.1161/CIRCRESAHA.115.303836. [DOI] [PubMed] [Google Scholar]
- 15.Puthanveetil P, Chen S, Feng B, Gautam A, Chakrabarti S. Long non-coding RNA MALAT1 regulates hyperglycaemia induced inflammatory process in the endothelial cells. J Cell Mol Med. 2015;19:1418–1425. doi: 10.1111/jcmm.12576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu J, Yao J, Li X, Song YC, Wang XQ, Li YJ, Yan B, Jiang Q. Pathogenic role of lncRNA-MALAT1 in endothelial cell dysfunction in diabetes mellitus. Cell Death Dis. 2014;5:e1506. doi: 10.1038/cddis.2014.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou R, Wang X, Yao J, Shen Y, Chen SN, Yang H, Jiang Q, Yan B. Identification and characterization of proliferative retinopathy-related long noncoding RNAs. Biochem Biophys Res Commun. 2015;465:324–330. doi: 10.1016/j.bbrc.2015.07.120. [DOI] [PubMed] [Google Scholar]
- 18.Wheeler TM, Leger AJ, Pandey SK, MacLeod AR, Nakamori M, Cheng SH, Wentworth BM, Bennett CF, Thornton CA. Targeting nuclear RNA for in vivo correction of myotonic dystrophy. Nature. 2012;488:111–115. doi: 10.1038/nature11362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ra SH, Su A, Li X, Zhou J, Cochran AJ, Kulkarni RP, Binder SW. Keratoacanthoma and squamous cell carcinoma are distinct from a molecular perspective. Modern Pathol. 2015;28:799–806. doi: 10.1038/modpathol.2015.5. [DOI] [PubMed] [Google Scholar]
- 20.Kryger R, Fan L, Wilce PA, Jaquet V. MALAT-1, a non protein-coding RNA is upregulated in the cerebellum, hippocampus and brain stem of human alcoholics. Alcohol. 2012;46:629–634. doi: 10.1016/j.alcohol.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 21.Wei G. Bioinformatics analysis of microRNA comprehensive regulatory network in congenital microtia. Int J Pediatr Otorhinolaryngol. 2015;79:1727–1731. doi: 10.1016/j.ijporl.2015.07.036. [DOI] [PubMed] [Google Scholar]
- 22.Gutschner T, Hämmerle M, Diederichs S. MALAT1-a paradigm for long noncoding RNA function in cancer. J Mol Med (Berl) 2013;91:791–801. doi: 10.1007/s00109-013-1028-y. [DOI] [PubMed] [Google Scholar]
- 23.Muller-Tidow C, Diederichs S, Thomas M, Serve H. Genome-wide screening for prognosis-predicting genes in early-stage non-small-cell lung cancer. Lung Cancer. 2004;2(45 Suppl):S145–S150. doi: 10.1016/j.lungcan.2004.07.979. [DOI] [PubMed] [Google Scholar]
- 24.Shen L, Chen L, Wang Y, Jiang X, Xia H, Zhuang Z. Long noncoding RNA MALAT1 promotes brain metastasis by inducing epithelial-mesenchymal transition in lung cancer. J Neurooncol. 2015;121:101–108. doi: 10.1007/s11060-014-1613-0. [DOI] [PubMed] [Google Scholar]
- 25.Hu L, Wu Y, Tan D, Meng H, Wang K, Bai Y, Yang K. Up-regulation of long noncoding RNA MALAT1 contributes to proliferation and metastasis in esophageal squamous cell carcinoma. J Exp Clin Cancer Res. 2015;34:7. doi: 10.1186/s13046-015-0123-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Okugawa Y, Toiyama Y, Hur K, Toden S, Saigusa S, Tanaka K, Inoue Y, Mohri Y, Kusunoki M, Boland CR, Goel A. Metastasis-associated long non-coding RNA drives gastric cancer development and promotes peritoneal metastasis. Carcinogenesis. 2014;35:2731–2739. doi: 10.1093/carcin/bgu200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng HT, Shi DB, Wang YW, Li XX, Xu Y, Tripathi P, Gu WL, Cai GX, Cai SJ. High expression of lncRNA MALAT1 suggests a biomarker of poor prognosis in colorectal cancer. Int J Clin Exp Pathol. 2014;7:3174–3181. [PMC free article] [PubMed] [Google Scholar]
- 28.Lai M, Yang Z, Zhou L, Zhu QQ, Xie HY, Zhang F, Wu LM, Chen LM, Zheng SS. Long non-coding RNA MALAT-1 overexpression predicts tumor recurrence of hepatocellular carcinoma after liver transplantation. Med Oncol. 2012;29:1810–1816. doi: 10.1007/s12032-011-0004-z. [DOI] [PubMed] [Google Scholar]
- 29.Pang EJ, Yang R, Fu XB, Liu YF. Overexpression of long non-coding RNA MALAT1 is correlated with clinical progression and unfavorable prognosis in pancreatic cancer. Tumor Biol. 2015;36:2403–2407. doi: 10.1007/s13277-014-2850-8. [DOI] [PubMed] [Google Scholar]
- 30.Jiao F, Hu H, Yuan C, Wang L, Jiang W, Jin Z, Guo Z, Wang L. Elevated expression level of long noncoding RNA MALAT-1 facilitates cell growth, migration and invasion in pancreatic cancer. Oncol Rep. 2014;32:2485–2492. doi: 10.3892/or.2014.3518. [DOI] [PubMed] [Google Scholar]
- 31.Zhang HM, Yang FQ, Chen SJ, Che J, Zheng JH. Upregulation of long non-coding RNA MALAT1 correlates with tumor progression and poor prognosis in clear cell renal cell carcinoma. Tumor Biol. 2015;36:2947–2955. doi: 10.1007/s13277-014-2925-6. [DOI] [PubMed] [Google Scholar]
- 32.Ying L, Chen Q, Wang Y, Zhou Z, Huang Y, Qiu F. Upregulated MALAT-1 contributes to bladder cancer cell migration by inducing epithelial-to-mesenchymal transition. Mol Biosyst. 2012;8:2289–2294. doi: 10.1039/c2mb25070e. [DOI] [PubMed] [Google Scholar]
- 33.Han Y, Liu Y, Nie L, Gui Y, Cai Z. Inducing cell proliferation inhibition, apoptosis, and motility reduction by silencing long noncoding ribonucleic acid metastasis-associated lung adenocarcinoma transcript 1 in urothelial carcinoma of the bladder. Urology. 2013;81(209):e1–7. doi: 10.1016/j.urology.2012.08.044. [DOI] [PubMed] [Google Scholar]
- 34.Ren S, Liu Y, Xu W, Sun Y, Lu J, Wang F, Wei M, Shen J, Hou J, Gao X, et al. Long noncoding RNA MALAT-1 is a new potential therapeutic target for castration resistant prostate cancer. J Urol. 2013;190:2278–2287. doi: 10.1016/j.juro.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 35.Sowalsky AG, Xia Z, Wang L, Zhao H, Chen S, Bubley GJ, Balk SP, Li W. Whole transcriptome sequencing reveals extensive unspliced mRNA in metastatic castration-resistant prostate cancer. Mol Cancer Res. 2015;13:98–106. doi: 10.1158/1541-7786.MCR-14-0273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao Z, Chen C, Liu Y, Wu C. 17β-estradiol treatment inhibits breast cell proliferation, migration and invasion by decreasing MALAT-1 RNA level. Biochem Biophys Res Commun. 2014;445:388–393. doi: 10.1016/j.bbrc.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 37.Jiang Y, Li Y, Fang S, Jiang B, Qin C, Xie P, Zhou G, Li G. The role of MALAT1 correlates with HPV in cervical cancer. Oncol Lett. 2014;7:2135–2141. doi: 10.3892/ol.2014.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guo F, Li Y, Liu Y, Wang J, Li Y, Li G. Inhibition of metastasis-associated lung adenocarcinoma transcript 1 in CaSki human cervical cancer cells suppresses cell proliferation and invasion. Acta Biochim Biophys Sin (Shanghai) 2010;42:224–229. doi: 10.1093/abbs/gmq008. [DOI] [PubMed] [Google Scholar]
- 39.Glover AR, Zhao JT, Ip JC, Lee JC, Robinson BG, Gill AJ, Soon PS, Sidhu SB. Long noncoding RNA profiles of adrenocortical cancer can be used to predict recurrence. Endocr Relat Cancer. 2015;22:99–109. doi: 10.1530/ERC-14-0457. [DOI] [PubMed] [Google Scholar]
- 40.Cho SF, Chang YC, Chang CS, Lin SF, Liu YC, Hsiao HH, Chang JG, Liu TC. MALAT1 long non-coding RNA is overexpressed in multiple myeloma and may serve as a marker to predict disease progression. BMC Cancer. 2014;14:809. doi: 10.1186/1471-2407-14-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tian Y, Zhang X, Hao Y, Fang Z, He Y. Potential roles of abnormally expressed long noncoding RNA UCA1 and Malat-1 in metastasis of melanoma. Melanoma Res. 2014;24:335–341. doi: 10.1097/CMR.0000000000000080. [DOI] [PubMed] [Google Scholar]
- 42.Dong Y, Liang G, Yuan B, Yang C, Gao R, Zhou X. MALAT1 promotes the proliferation and metastasis of osteosarcoma cells by activating the PI3K/Akt pathway. Tumor Biol. 2015;36:1477–1486. doi: 10.1007/s13277-014-2631-4. [DOI] [PubMed] [Google Scholar]
- 43.Wang F, Ren S, Chen R, Lu J, Shi X, Zhu Y, Zhang W, Jing T, Zhang C, Shen J, et al. Development and prospective multicenter evaluation of the long noncoding RNA MALAT-1 as a diagnostic urinary biomarker for prostate cancer. Oncotarget. 2014;5:11091–11102. doi: 10.18632/oncotarget.2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ren S, Wang F, Shen J, Sun Y, Xu W, Lu J, Wei M, Xu C, Wu C, Zhang Z, et al. Long non-coding RNA metastasis associated in lung adenocarcinoma transcript 1 derived miniRNA as a novel plasma-based biomarker for diagnosing prostate cancer. Eur J Cancer. 2013;49:2949–2959. doi: 10.1016/j.ejca.2013.04.026. [DOI] [PubMed] [Google Scholar]
- 45.Guo F, Yu F, Wang J, Li Y, Li Y, Li Z, Zhou Q. Expression of MALAT1 in the peripheral whole blood of patients with lung cancer. Biomed Rep. 2015;3:309–312. doi: 10.3892/br.2015.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fu X, Liu Y, Zhuang C, Liu L, Cai Z, Huang W. Synthetic artificial microRNAs targeting UCA1-MALAT1 or c-Myc inhibit malignant phenotypes of bladder cancer cells T24 and 5637. Mol Biosyst. 2015;11:1285–1289. doi: 10.1039/C5MB00127G. [DOI] [PubMed] [Google Scholar]
- 47.Taniguchi M, Fujiwara K, Nakai Y, Ozaki T, Koshikawa N, Toshio K, Kataba M, Oguni A, Matsuda H, Yoshida Y, et al. Inhibition of malignant phenotypes of human osteosarcoma cells by a gene silencer, a pyrrole-imidazole polyamide, which targets an E-box motif. FEBS Open Bio. 2014;4:328–334. doi: 10.1016/j.fob.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tee AE, Ling D, Nelson C, Atmadibrata B, Dinger ME, Xu N, Mizukami T, Liu PY, Liu B, Cheung B, et al. The histone demethylase JMJD1A induces cell migration and invasion by up-regulating the expression of the long noncoding RNA MALAT1. Oncotarget. 2014;5:1793–1804. doi: 10.18632/oncotarget.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Özgür E, Mert U, Isin M, Okutan M, Dalay N, Gezer U. Differential expression of long non-coding RNAs during genotoxic stress-induced apoptosis in HeLa and MCF-7 cells. Clin Exp Med. 2013;13:119–126. doi: 10.1007/s10238-012-0181-x. [DOI] [PubMed] [Google Scholar]
- 50.Chen H, Xin Y, Zhou L, Huang JM, Tao L, Cheng L, Tian J. Cisplatin and paclitaxel target significant long noncoding RNAs in laryngeal squamous cell carcinoma. Med Oncol. 2014;31:246. doi: 10.1007/s12032-014-0246-7. [DOI] [PubMed] [Google Scholar]
- 51.Fellenberg J, Bernd L, Delling G, Witte D, Zahlten-Hinguranage A. Prognostic significance of drug-regulated genes in high-grade osteosarcoma. Mod Pathol. 2007;20:1085–1094. doi: 10.1038/modpathol.3800937. [DOI] [PubMed] [Google Scholar]
- 52.Hutchinson JN, Ensminger AW, Clemson CM, Lynch CR, Lawrence JB, Chess A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics. 2007;8:39. doi: 10.1186/1471-2164-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zong X, Tripathi V, Prasanth KV. RNA splicing control: Yet another gene regulatory role for long nuclear noncoding RNAs. RNA Biol. 2011;8:968–977. doi: 10.4161/rna.8.6.17606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wilusz JE, Freier SM, Spector DL. 3′ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell. 2008;135:919–932. doi: 10.1016/j.cell.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wilusz JE, JnBaptiste CK, Lu LY, Kuhn CD, Joshua-Tor L, Sharp PA. A triple helix stabilizes the 3′ ends of long noncoding RNAs that lack poly(A) tails. Gene Dev. 2012;26:2392–2407. doi: 10.1101/gad.204438.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Spector DL, Lamond AI. Nuclear speckles. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a000646. pii: a000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li S, Wang Q, Qiang Q, Shan H, Shi M, Chen B, Zhao S, Yuan L. Sp1-mediated transcriptional regulation of MALAT1 plays a critical role in tumor. J Cancer Res Clin Oncol. 2015;141:1909–1920. doi: 10.1007/s00432-015-1951-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kuo IY, Wu CC, Chang JM, Huang YL, Lin CH, Yan JJ, Sheu BS, Lu PJ, Chang WL, Lai WW, Wang YC. Low SOX17 expression is a prognostic factor and drives transcriptional dysregulation and esophageal cancer progression. Int J Cancer. 2014;135:563–573. doi: 10.1002/ijc.28695. [DOI] [PubMed] [Google Scholar]
- 59.Ma XY, Wang JH, Wang JL, Ma CX, Wang XC, Liu FS. Malat1 as an evolutionarily conserved lncRNA, plays a positive role in regulating proliferation and maintaining undifferentiated status of early-stage hematopoietic cells. BMC Genomics. 2015;16:676. doi: 10.1186/s12864-015-1881-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang J, Wang H, Zhang Y, Zhen N, Zhang L, Qiao Y, Weng W, Liu X, Ma L, Xiao W, et al. Mutual inhibition between YAP and SRSF1 maintains long non-coding RNA, Malat1-induced tumourigenesis in liver cancer. Cell Signal. 2014;26:1048–1059. doi: 10.1016/j.cellsig.2014.01.022. [DOI] [PubMed] [Google Scholar]
- 61.Guo F, Jiao F, Song Z, Li S, Liu B, Yang H, Zhou Q, Li Z. Regulation of MALAT1 expression by TDP43 controls the migration and invasion of non-small cell lung cancer cells in vitro. Biochem Biophys Res Commun. 2015;465:293–298. doi: 10.1016/j.bbrc.2015.08.027. [DOI] [PubMed] [Google Scholar]
- 62.Liu R, Li J, Lai Y, Liao Y, Liu R, Qiu W. Hsa-miR-1 suppresses breast cancer development by down-regulating K-ras and long non-coding RNA MALAT1. Int J Biol Macromol. 2015;81:491–497. doi: 10.1016/j.ijbiomac.2015.08.016. [DOI] [PubMed] [Google Scholar]
- 63.Han Y, Liu Y, Zhang H, Wang T, Diao R, Jiang Z, Gui Y, Cai Z. Hsa-miR-125b suppresses bladder cancer development by down-regulating oncogene SIRT7 and oncogenic long non-coding RNA MALAT1. FEBS Lett. 2013;587:3875–3882. doi: 10.1016/j.febslet.2013.10.023. [DOI] [PubMed] [Google Scholar]
- 64.Wang X, Li M, Wang Z, Han S, Tang X, Ge Y, Zhou L, Zhou C, Yuan Q, Yang M. Silencing of long noncoding RNA MALAT1 by miR-101 and miR-217 inhibits proliferation, migration, and invasion of esophageal squamous cell carcinoma cells. J Biol Chem. 2015;290:3925–3935. doi: 10.1074/jbc.M114.596866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Leucci E, Patella F, Waage J, Holmstrøm K, Lindow M, Porse B, Kauppinen S, Lund AH. microRNA-9 targets the long non-coding RNA MALAT1 for degradation in the nucleus. Sci Rep. 2013;3:2535. doi: 10.1038/srep02535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gutschner T, Hämmerle M, Eissmann M, Hsu J, Kim Y, Hung G, Revenko A, Arun G, Stentrup M, Gross M, et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013;73:1180–1189. doi: 10.1158/0008-5472.CAN-12-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schmidt LH, Görlich D, Spieker T, Rohde C, Schuler M, Mohr M, Humberg J, Sauer T, Thoenissen NH, Huge A, et al. Prognostic impact of Bcl-2 depends on tumor histology and expression of MALAT-1 lncRNA in non-small-cell lung cancer. J Thorac Oncol. 2014;9:1294–1304. doi: 10.1097/JTO.0000000000000243. [DOI] [PubMed] [Google Scholar]
- 68.Xu S, Sui S, Zhang J, Bai N, Shi Q, Zhang G, Gao S, You Z, Zhan C, Liu F, Pang D. Downregulation of long noncoding RNA MALAT1 induces epithelial-to-mesenchymal transition via the PI3K-AKT pathway in breast cancer. Int J Clin Exp Pathol. 2015;8:4881–4891. [PMC free article] [PubMed] [Google Scholar]
- 69.Yang MH, Hu ZY, Xu C, Xie LY, Wang XY, Chen SY, Li ZG. MALAT1 promotes colorectal cancer cell proliferation/migration/invasion via PRKA kinase anchor protein 9. Biochim Biophys Acta. 2015;1852:166–174. doi: 10.1016/j.bbadis.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jiao F, Hu H, Han T, Yuan C, Wang L, Jin Z, Guo Z, Wang L. Long noncoding RNA MALAT-1 enhances stem cell-like phenotypes in pancreatic cancer cells. Int J Mol Sci. 2015;16:6677–6693. doi: 10.3390/ijms16046677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li B, Chen P, Qu J, Shi L, Zhuang W, Fu J, Li J, Zhang X, Sun Y, Zhuang W. Activation of LTBP3 gene by a long noncoding RNA (lncRNA) MALAT1 transcript in mesenchymal stem cells from multiple myeloma. J Biol Chem. 2014;289:29365–29375. doi: 10.1074/jbc.M114.572693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tripathi V, Shen Z, Chakraborty A, Giri S, Freier SM, Wu X, Zhang Y, Gorospe M, Prasanth SG, Lal A, Prasanth KV. Long noncoding RNA MALAT1 controls cell cycle progression by regulating the expression of oncogenic transcription factor B-MYB. PLoS Genet. 2013;9:e1003368. doi: 10.1371/journal.pgen.1003368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ji Q, Zhang L, Liu X, Zhou L, Wang W, Han Z, Sui H, Tang Y, Wang Y, Liu N, et al. Long non-coding RNA MALAT1 promotes tumour growth and metastasis in colorectal cancer through binding to SFPQ and releasing oncogene PTBP2 from SFPQ/PTBP2 complex. Br J Cancer. 2014;111:736–748. doi: 10.1038/bjc.2014.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang J, Su L, Chen X, Li P, Cai Q, Yu B, Liu B, Wu W, Zhu Z. MALAT1 promotes cell proliferation in gastric cancer by recruiting SF2/ASF. Biomed Pharmacother. 2014;68:557–564. doi: 10.1016/j.biopha.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 75.Fan Y, Shen B, Tan M, Mu X, Qin Y, Zhang F, Liu Y. TGF-β-induced upregulation of malat1 promotes bladder cancer metastasis by associating with suz12. Clin Cancer Res. 2014;20:1531–1541. doi: 10.1158/1078-0432.CCR-13-1455. [DOI] [PubMed] [Google Scholar]
- 76.Hirata H, Hinoda Y, Shahryari V, Deng G, Nakajima K, Tabatabai ZL, Ishii N, Dahiya R. Long noncoding RNA MALAT1 promotes aggressive renal cell carcinoma through Ezh2 and interacts with miR-205. Cancer Res. 2015;75:1322–1331. doi: 10.1158/0008-5472.CAN-14-2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lu H, He Y, Lin L, Qi Z, Ma L, Li L, Su Y. Long non-coding RNA MALAT1 modulates radiosensitivity of HR-HPV+ cervical cancer via sponging miR-145. Tumor Biol. 2016;37:1683–1691. doi: 10.1007/s13277-015-3946-5. [DOI] [PubMed] [Google Scholar]
- 78.Liu S, Song L, Zeng S, Zhang L. MALAT1-miR-124-RBG2 axis is involved in growth and invasion of HR-HPV-positive cervical cancer cells. Tumor Biol. 2016;37:633–640. doi: 10.1007/s13277-015-3732-4. [DOI] [PubMed] [Google Scholar]
- 79.Li X, Zhu M, Brasier AR, Kudlicki AS. Inferring genome-wide functional modulatory network: a case study on NF-κB/RelA transcription factor. J Comput Biol. 2015;22:300–312. doi: 10.1089/cmb.2014.0299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wu XS, Wang XA, Wu WG, Hu YP, Li ML, Ding Q, Weng H, Shu YJ, Liu TY, Jiang L, et al. MALAT1 promotes the proliferation and metastasis of gallbladder cancer cells by activating the ERK/MAPK pathway. Cancer Biol Ther. 2014;15:806–814. doi: 10.4161/cbt.28584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vassallo I, Zinn P, Lai M, Rajakannu P, Hamou MF, Hegi ME. WIF1 re-expression in glioblastoma inhibits migration through attenuation of non-canonical WNT signaling by downregulating the lncRNA MALAT1. Oncogene. 2016;35:12–21. doi: 10.1038/onc.2015.61. [DOI] [PubMed] [Google Scholar]
- 82.Kan JY, Wu DC, Yu FJ, Wu CY, Ho YW, Chiu YJ, Jian SF, Hung JY, Wang JY, Kuo PL. Chemokine (C-C Motif) ligand 5 is involved in tumor-associated dendritic cell-mediated colon cancer progression through non-coding RNA MALAT-1. J Cell Physiol. 2015;230:1883–1894. doi: 10.1002/jcp.24918. [DOI] [PubMed] [Google Scholar]
- 83.Zhao Y, Yang Y, Trovik J, Sun K, Zhou L, Jiang P, Lau TS, Hoivik EA, Salvesen HB, Sun H, Wang H. A novel Wnt regulatory axis in endometrioid endometrial cancer. Cancer Res. 2014;74:5103–5117. doi: 10.1158/0008-5472.CAN-14-0427. [DOI] [PubMed] [Google Scholar]
- 84.Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lin R, Roychowdhury-Saha M, Black C, Watt AT, Marcusson EG, Freier SM, Edgington TS. Control of RNA processing by a large non-coding RNA over-expressed in carcinomas. FEBS Lett. 2011;585:671–676. doi: 10.1016/j.febslet.2011.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tano K, Mizuno R, Okada T, Rakwal R, Shibato J, Masuo Y, Ijiri K, Akimitsu N. MALAT-1 enhances cell motility of lung adenocarcinoma cells by influencing the expression of motility-related genes. FEBS lett. 2012;584:4575–4580. doi: 10.1016/j.febslet.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 87.Eißmann M, Gutschner T, Hämmerle M, Günther S, Caudron-Herger M, Groß M, Schirmacher P, Rippe K, Braun T, Zörnig M, Diederichs S. Loss of the abundant nuclear non-coding RNA MALAT1 is compatible with life and development. RNA Biol. 2012;9:1076–1087. doi: 10.4161/rna.21089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang B, Arun G, Mao YS, Lazar Z, Hung G, Bhattacharjee G, Xiao X, Booth CJ, Wu J, Zhang C, Spector DL. The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Rep. 2012;2:111–123. doi: 10.1016/j.celrep.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Liu W, Lu X, Tang GH, Ren JJ, Liao WJ, Ge PL, Huang JF. LncRNAs expression signatures of hepatocellular carcinoma revealed by microarray. World J Gastroenterol. 2014;20:6314–6321. doi: 10.3748/wjg.v20.i20.6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.


