Abstract
Glutathione S-transferase A1 (GSTA1) is a phase II detoxification enzyme and serves a crucial role in anti-cancer drug resistance. In our previous study, GSTA1 was identified to be highly expressed in various subtypes of non-small-cell lung cancer cell lines compared with human embryonic lung fibroblast cell line MRC-5. The aim of the present study was to investigate the effect of GSTA1 expression on the proliferation and apoptosis of A549 cells. GSTA1 expression was knocked down or with overexpressed using lentivirus particles. Western blot analysis and reverse transcription-quantitative polymerase chain reaction (RT-qPCR) were used to assess the protein, and mRNA levels of GSTA1 in A549 cells, respectively. The effect of GSTA1 manipulation on cell proliferation and apoptosis were investigated in vitro using MTT assays, Hoechst 33258 staining and flow cytometry, and in vivo using A549 cell line xenografts in nude mice. The results of the western blot analysis and RT-qPCR revealed that stable cell models of GSTA1 knockdown, and overexpression were established. The data of the MTT assay indicated that the downregulation of GSTA1 significantly inhibited cell proliferation compared with si-control-transfected cells. These si-GSTA1 A549 cells exhibited typical morphological changes of apoptosis, including chromatin condensation and shrunken nuclei compared with the si-control counterparts. An AnnexinV-fluorescein isothiocyanate assay verified that the downregulation of GSTA1 significantly induced cell apoptosis in vitro. In addition, overexpression of GSTA1 significantly promoted tumor growth in vivo. Accordingly, downregulation of GSTA1 suppressed tumor growth. In conclusion, GSTA1 plays an important role in regulation of cell proliferation and cell apoptosis in A549 cell line.
Keywords: glutathione S-transferase A1, A549 cell line, cell apoptosis
Introduction
Lung cancer is one of the most common cancer types worldwide, and is the leading cause of cancer-associated mortality (1). It was reported that ~158,040 Americans were expected to succumb to lung cancer in 2015, accounting for ~27% of all cancer-associated mortalities (2). Non-small-cell lung cancer (NSCLC) accounts for ~85% of all lung cancer cases, including squamous cell carcinoma, adenocarcinoma and large cell carcinoma (3). Treatments include surgery, chemotherapy, radiation therapy and targeted therapy. Recently, targeted therapy has demonstrated to be an effective method to treat with lung cancer (4). To identify a candidate tumor marker, phage display technology was applied to screen specific novel peptides in lung cancer cells (5).
Glutathione S-transferases (GSTs) belong to a super family of phase II detoxification enzymes, which catalyze the conjugation of glutathione (GSH) to variable environmentally and endogenously produced electrophonic substances (6). Human cytosolic GSTs are divided into seven classes abbreviated in Roman capitals A, M, P, S, T, Z and O (7). In addition to their enzymatic roles, GSTs have an important ability in inhibiting cell apoptosis, including in cancer cells.
Evidence has confirmed a high correlation of GSTs with cancer progression and drug resistance (8–10). It has been reported that certain GSTs subtypes are overexpressed in several human tumors and promotes cancer progression (8), while the inhibition of GST activity significantly induces cellular death (11). In addition to the role of regulating cancer cell growth, GSTs serve an essential role in tumor drug resistance. For example, knockdown of GSTO1 expression was demonstrated to abrogate carboplatin-induced breast cancer stem cell enrichment, and decrease tumor recurrence and metastatic capacity (12). In addition, GSTP1 blocks As2O3-induced apoptosis in lymphoma cells by decreasing the intracellular amounts of H2O2. GSTA1 is abundantly expressed in A549 cells, located in the cytoplasm and/or membranes (13). Furthermore, its expression is associated with an increased risk in colorectal, breast and gastric cancer (14–16). The alpha class, GSTA1-A5 are not only expressed in normal human tissues, but also in human cancer (17). GSTA1, GSTA2 and GSTA4 are widely expressed in human tissues. The genetic polymorphism of GSTA1 is characterized by two alleles, GSTA1*A and GSTA1*B (17). However, the roles of GSTA1 in NSCLC cells remain to be elucidated. We hypothesize that downregulation of GSTA1 serves a functional role in inhibiting proliferation and inducing apoptosis in the A549 cell line.
Materials and methods
Chemicals and cell culture
LPP-Z3392-Lv105-400 (lentiviral particles for GSTA1), LPP-e green fluorescent protein (GFP)-Lv105-100 (lentiviral particles for eGFP), LP-HSH008475-5-LvRH1GP-200 [lentiviral particles for small interfering RNA, (si-RNA)] and LP-CSHCTR001-LVRH1GP-050 (lentiviral particles for scramble control) were prepared by GeneCopoeia, Inc. (Rockville, MD, USA). The human lung adenocarcinoma cell line A549 was purchased from Shanghai Institute of Cellular Biology of Chinese Academy of Sciences (Shanghai, China). Cells were maintained in RPMI-1640 medium supplemented with 10% fetal bovine serum and 0.1% penicillin/streptomycin (all from Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) in a humidified atmosphere with 5% CO2 at 37°C.
Animals and xenograft experiments
A total of 24 healthy BALB/c nude mice (half male and female; aged ~3 weeks; weight 17~20 g) were purchased from Guangdong Medical Experimental Center (Guangzhou, China). Mice were maintained under specific pathogen-free conditions at a temperature of 20–25°C with humidity of 40–70%. They were fed in groups, and had free access to water and food. The mice were randomly divided into four groups: GSTA1, vector (lentiviral particles for eGFP), si-control (scramble control) and si-GSTA1 groups with six mice/group. To establish the above four groups of animal A549 tumor models, each nude mouse was injected with 4×105 A549 cells (transfected with GSTA1, vector, si-control and si-GSTA1 respectively) into the right flank. Tumor size was measured using a caliper four times/week. Tumor volumes were calculated by the following formula: 0.5× largest diameter (mm) × smallest diameter2 (mm). Mice were sacrificed and xenograft tumors were removed en bloc following 3 weeks. All xenograft experiments were performed under a protocol approved by the Animal Care and Use Committee. All animal experiments were approved by the Animal Care Committee of Guangdong Pharmaceutical University (Guangzhou, China).
Western blot analysis
Total protein was extracted from the tumor tissues of the four groups or cells cultured in six-well plates. The tumor tissues were ground and lysed in radioimmunoprecipitation assay lysis buffer (#P0013B, Beyotime Institute of Biotechnology, Haimen, China) containing 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 2 mM EDTA, 1% Triton X-100, 1% sodium deoxycholate, 1 µg/ml leupeptin and 0.1% SDS on ice, and then centrifuged at 12,000 × g, 4°C for 20 min to obtain the total protein supernatants. The protein concentration of each extract was measured using a BCA Protein assay kit (Thermo Fisher Scientific, Inc.). A total of 40 µg protein per lane was loaded and separated via 12% SDS-PAGE, and then transferred onto polyvinylidene difluoride membranes (EMD Millipore, Billerica, MA, USA). The membranes were blocked with 5% (w/v) non-fat milk powder in Tris buffered saline with Tween-20 (TBS-T) pH 7.6 solution at room temperature for 1 h. Membranes were then incubated with the primary antibodies goat anti-GSTA1 (#ab53940; 1:3,000 dilution; Abcam, Cambridge, UK) or mouse anti-β-actin (#ab8226; 1:3,000 dilution; Abcam) overnight at 4°C. The membrane was washed with TBS-T buffer three times and incubated with horseradish peroxidase-conjugated polyclonal donkey anti-goat IgG (#ab97110; 1:5,000 dilution; Abcam) or goat anti-mouse IgG (#ab136815; 1:5,000 dilution; Abcam) at room temperature for 1 h. After washing with TBS-T buffer three times, the membranes were visualized after incubation SuperSignal™ West Pico PLUS Chemiluminescent Substrate (#34580, Thermo Fisher Scientific, Inc.). β-actin was used as loading control. The gray value was analyzed by using ImageJ software (version 1.46; National Institutes of Health, Bethesda, MD, USA).
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) assay
Total RNA from the tumor lysates or cells was extracted with TRIzol solution (Life Technologies; Thermo Fisher Scientific, Inc.) following the manufacturer's protocol. cDNA was synthesized using the PrimeScript™ II First Strand cDNA Synthesis kit according to the manufacturer's protocol using 1 µg of total RNA with the Bio-Rad Reverse Transcription system. The temperature protocol was as follows: 42°C for 2 min, then at 37°C for 15 min and 85°C for 5 sec. The RT-qPCR assay was run in a 25 µl reaction system containing 12.5 µl X2 PCR Master mix, 1 µl forward and reverse primer each, 2 µl cDNA and 8.5 µl nuclease-free water on a CFX96™ Real-Time PCR Detection system with SYBR® Premix Ex Taq™ II (Takara Bio, Inc., Otsu, Japan). The primers were synthesized according to the designed sequence by Shanghai Shenggong Biology Engineering Technology Service, Ltd. (Shanghai, China) and were used for qPCR: GSTA1 forward, 5′-GCCTCCATGACTGCGTTATT-3′ and reverse, 5′-CCTGCCCACAGTGAAGAAGT-3′; GAPDH forward, 5′-AACGGATTTGGTCGTATTGGG-3′ and reverse 5′-CCTGGAAGATGGTGATGGGAT-3′. qPCR was performed under the following conditions: 95°C for 30 sec; 40 cycles of 95°C for 5 sec, 60°C for 30 sec; 95°C for 5 sec, 60°C for 1 min; 50°C for 30 sec. GAPDH served as a reference gene. The relative quantity of mRNA expression was calculated by using the 2−ΔΔCt method (18). All experiments were performed in triplicate.
Cell growth assay in vitro
A549 cells were seeded at a density of 5×103 cells/well in 96-well flat bottom plates. A total of 4 µl of 1 ×108 TU/ml lentivirus particles (GSTA1, vector, si-control and si-GSTA1) were serially diluted to 4×106 TU/ml by serum free OptiMEM medium (Gibco; Thermo Fisher Scientific, Inc.) containing 5 µg/ml Polybrene (Santa Cruz Biotechnology, Inc., Dallas, TX, USA). When the cells reached 30–50% confluence, they were transfected with the aforementioned lentivirus particles using 2 µg/ml puromycin (Santa Cruz Biotechnology, Inc., Dallas, TX, USA) for screening stable cell lines. The cells were cultured for 5 days. During this time, 20 µl MTT solution (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) in PBS (5 mg/ml) were added to each well at 1-day interval for 4 h at 37°C, then, 200 µl DMSO was added after removing the supernatants. The absorbance of each well was measured with a microplate reader at 570 nm. All experiments were performed in triplicate.
Lentivirus-meditated GSTA1 overexpression and si-GSTA1 in A549 cell models. The aforementioned lentiviral particles (1×108 TU/ml) were diluted and added (multiplicity of infection: 40, 20, 10, 5) to transfect A549 cells for 24 h. The supernatants containing lentivirus were replaced with OptiMEM medium, and the cells were cultured for an additional 48 h. Successful infection was detected by the presence of green fluorescence in cells due to GFP as observed under an inversed fluorescence microscope (×100).
Cell morphological assay
The si-GSTA1 and si-control A549 cells were seeded in 12-well plates at a density of 1×105 cells/well, and then cultured for up to 5 days. Following treatment, the cells were fixed with 4% formaldehyde for 30 min at room temperature, exposed to Hoechst 33258 (15 µM) for 30 min at room temperature, then observed and imaged using an inverted fluorescence microscope (×100).
Flow cytometric analysis
The si-GSTA1 and si-control A549 cells were cultured in 6-well plates at a density of 5×105 cells/well for 5 days. The cells were collected every day, washed twice with ice-cold PBS and resuspended in binding buffer (cat. no. KGA106; Nanjing KeyGen Biotech, Nanjing, China) containing PBS (pH 7.4), 1% fetal bovine serum and 0.1% NaN3 at a concentration of 1×106 cells/ml. Then, 5 µl of Annexin V-fluorescein isothiocyanate and 5 µl of propidium iodide were added. The cells were cultured for 15 min at room temperature in the dark, after which apoptotic cells were quantified using flow cytometry (FACSCalibur/Calibur; BD Biosciences, Franklin Lakes, NJ, USA). All experiments were performed in triplicate.
Statistical analysis
Data are presented as the mean + standard deviation. Statistical analysis was performed using SPSS software (version 17.0; SPSS, Inc., Chicago, IL, USA). One-way analysis of variance followed by Tukey's post-hoc test was used for multiple comparisons and the paired Student's t-test was applied when only two groups were compared. P<0.05 and P<0.01 were considered to indicate statistically significant difference.
Results
Expression of GSTA1 in each group in vitro
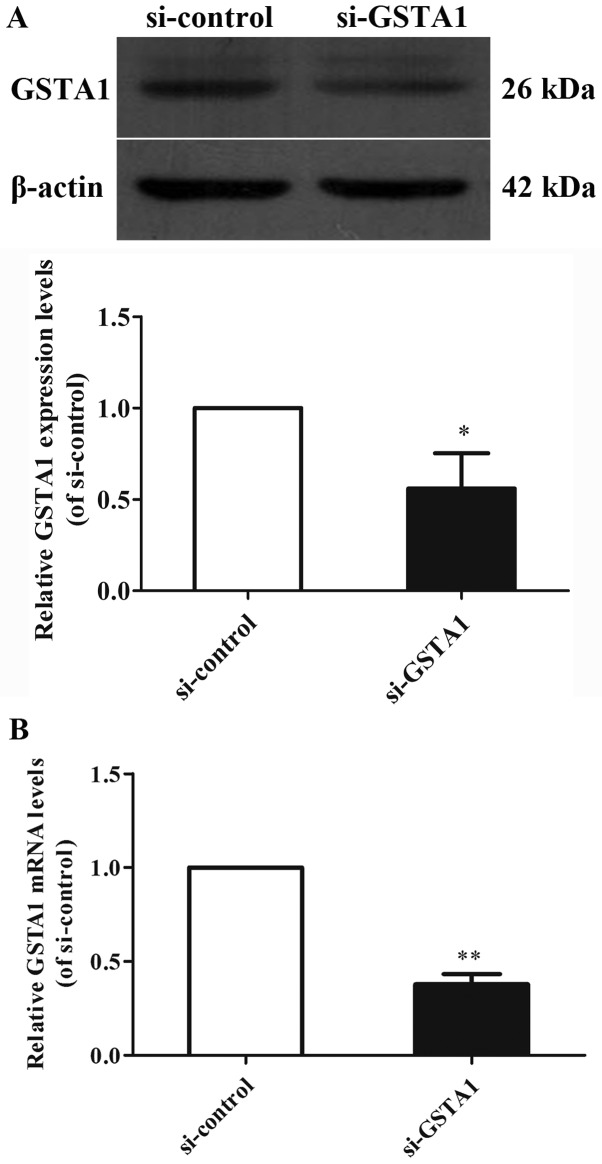
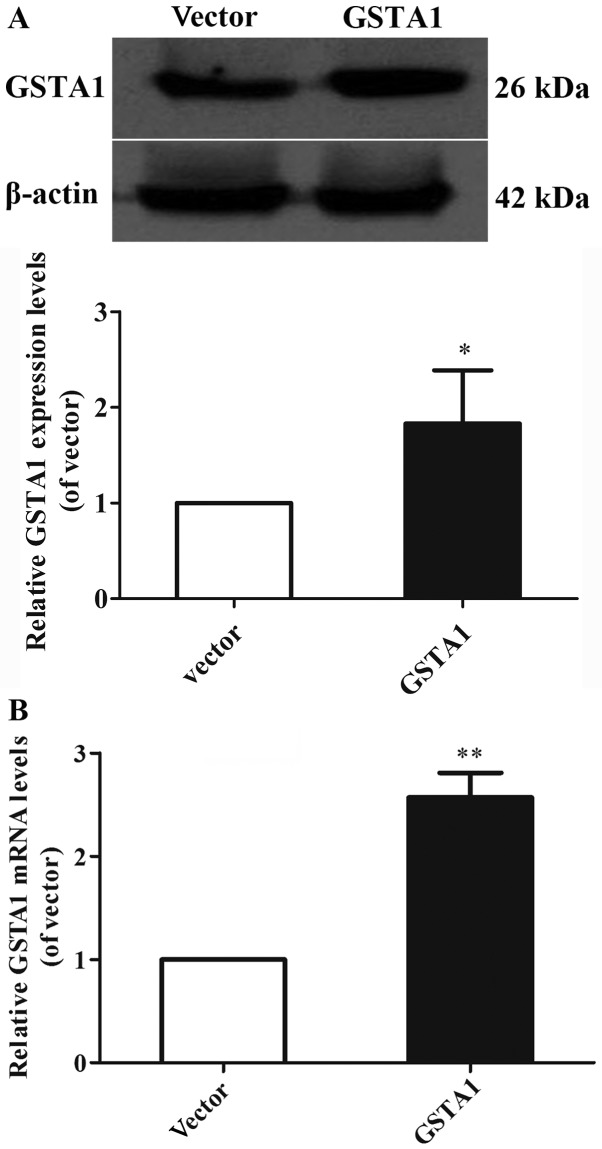
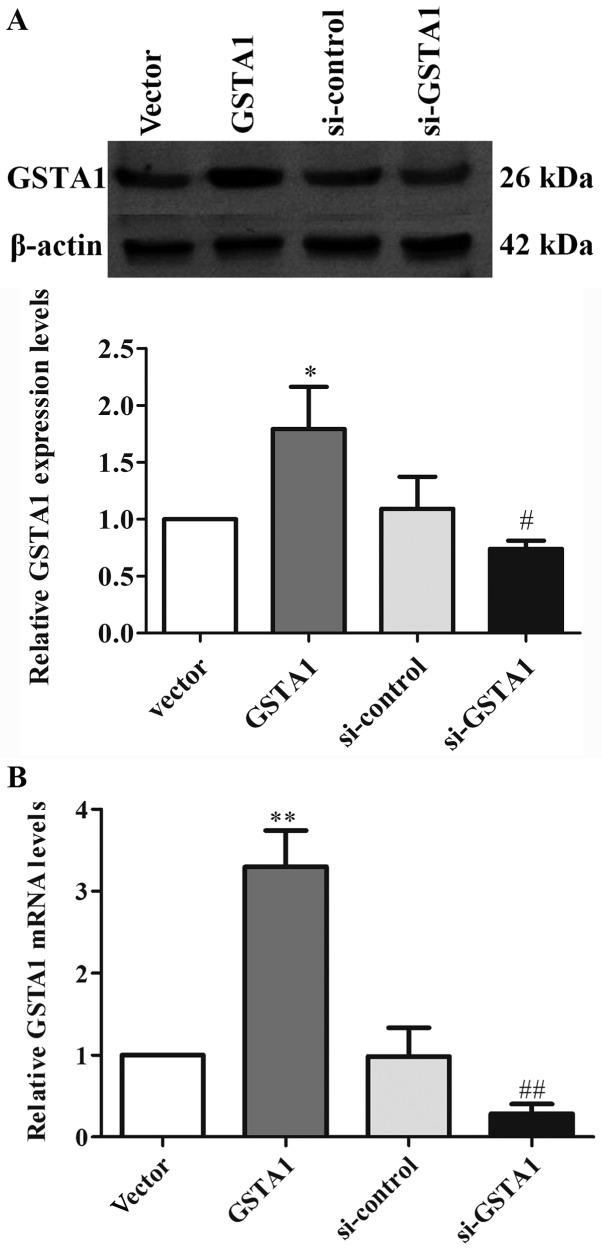
GSTA1 protein and mRNA expression levels were investigated using western blot analysis, and RT-qPCR in cell samples from the GSTA1 and vector groups (Figs. 1 and 2). Si-GSTA1 protein expression (0.56-fold lower) was significantly suppressed compared with the si-control group (Fig. 1A; P<0.01). The data demonstrated that the relative mRNA levels in the si-GSTA1 group were 0.38-fold significantly lower compared with that of the si-control (P<0.01; Fig. 1B). In addition, the GSTA1 protein levels in the GSTA1 overexpression group were 1.83-fold higher compared with that of the vector group (P<0.05; Fig. 2A). Likewise, the GSTA1 mRNA expression in the GSTA1 overexpression group was 2.57-fold higher compared with that of the vector group (P<0.01; Fig. 2B). These results implied that the si-GSTA1 and GSTA1 overexpression cell models were successfully established.
Figure 1.
GSTA1 protein and mRNA expression in si-GSTA1 and si-control A549 cells. (A) The expression of GSTA1 was determined by western blot analysis. β-actin served as a loading control. (B) GSTA1 mRNA levels in si-GSTA1 and si-control A549 cells were detected by a reverse transcription-quantitative polymerase chain reaction assay. GAPDH served as a loading control. Data are presented as the mean + standard deviation. *P<0.05, **P<0.01 vs. si-control. GSTA1, glutathione S-transferase A1; si, small interfering RNA.
Figure 2.
GSTA1 protein, mRNA expression in GSTA1 and vector A549 cells. (A) The expression of GSTA1 was determined by western blot analysis. β-actin served as a loading control. (B) GSTA1 mRNA levels in GSTA1 and vector A549 cells were detected by a reverse transcription-quantitative polymerase chain reaction assay. GAPDH served as a loading control. Data are presented as the mean + standard deviation. *P<0.05, **P<0.01 vs. vector. GSTA1, glutathione S-transferase A1.
Downregulation of GSTA1 inhibits cell proliferation and induces cell apoptosis in vitro
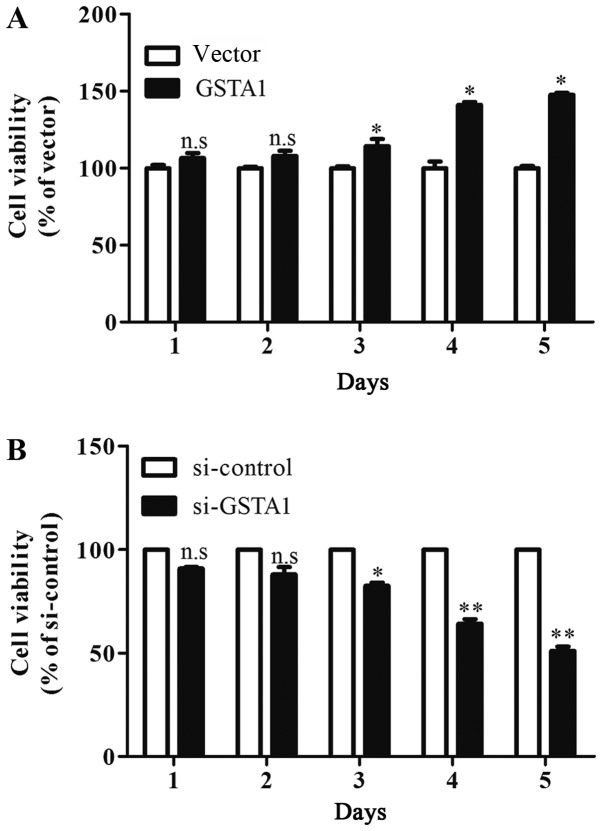
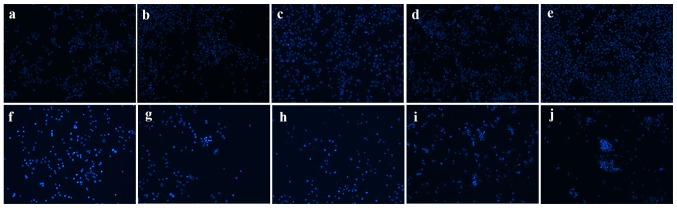
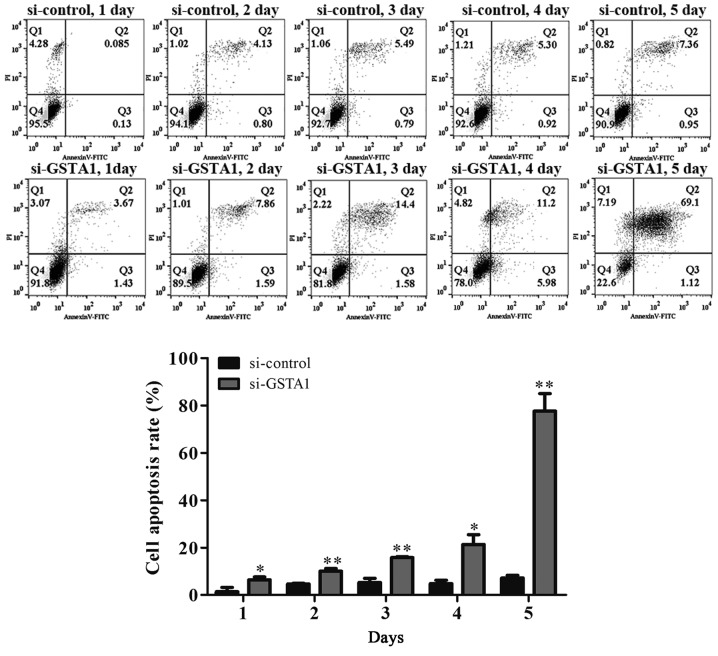
The results of the MTT assay indicated that cell viability in the GSTA1 group after culturing for 3 days were significantly improved when compared with the vector group (P<0.05; Fig. 3A). In addition, si-GSTA1 transfection resulted in significantly decreased cell viability compared with the si-control following 3 days of culture (Fig. 3B; P<0.05). Hoechst 33258 staining was used to observe the morphological changes in the cell nuclei. As shown in Fig. 4, the cell florescence of A549 cells transfected with si-GSTA1 was relatively brighter compared with the si-control groups. When compared with si-control A549 cells, the cell amounts of si-GSTA1 were few. In order to further confirm that the cell death induced by si-GSTA1 was apoptosis, AnnexinV-FITC/PI staining was used to detect early apoptosis cells and middle-late apoptotic cells (19). A time-dependent increase in apoptotic cells was observed from 6.42 on day 1 to 10.06, 15.8, 21.3 and 77.61% in si-GSTA1 A549 cells cultured for 5 days compared with the control counterparts, which saw an increase from 1.38 on day 1 to 4.63, 5.30, 4.84 and 7.22% (Fig. 5). These results demonstrated that the downregulation of GSTA1 in A549 cell lines is able to suppress cell proliferation and induce cell apoptosis in vitro.
Figure 3.
Cell viability of A549 cells in vitro. (A) GSTA1 A549 cells were compared with vector control. *P<0.05 vs. vector. (B) si-GSTA1 A549 cells compared with i-control. *P<0.05, **P<0.01 vs. si-control. Data are presented as the mean + standard deviation. GSTA1, glutathione S-transferase A1; si, small interfering RNA; n.s, no significance.
Figure 4.
Morphological changes of si-GSTA1 and si-control A549 cells. Apoptotic nuclei manifested condensed or fragmented DNA brightly stained by Hoechst 33258 (magnification, ×100). (a-e) Si-control A549 cells cultured for 1, 2, 3, 4 and 5 days, respectively. (f-j) Si-GSTA1 A549 cells cultured for 1, 2, 3, 4 and 5 days, respectively. GSTA1, glutathione S-transferase A1; si, small interfering RNA.
Figure 5.
Cell apoptosis of si-GSTA1 and si-control A549 cells cultured for 5 days were assessed by Annexin V-FITC/PI analysis. Data are presented as the mean + standard deviation. *P<0.05, **P<0.01 vs. si-control. FITC, fluorescein isothiocyanate; GSTA1, glutathione S-transferase A1; si, small interfering RNA; PI, propidium iodide.
GSTA1 induces tumor growth in xenograft models
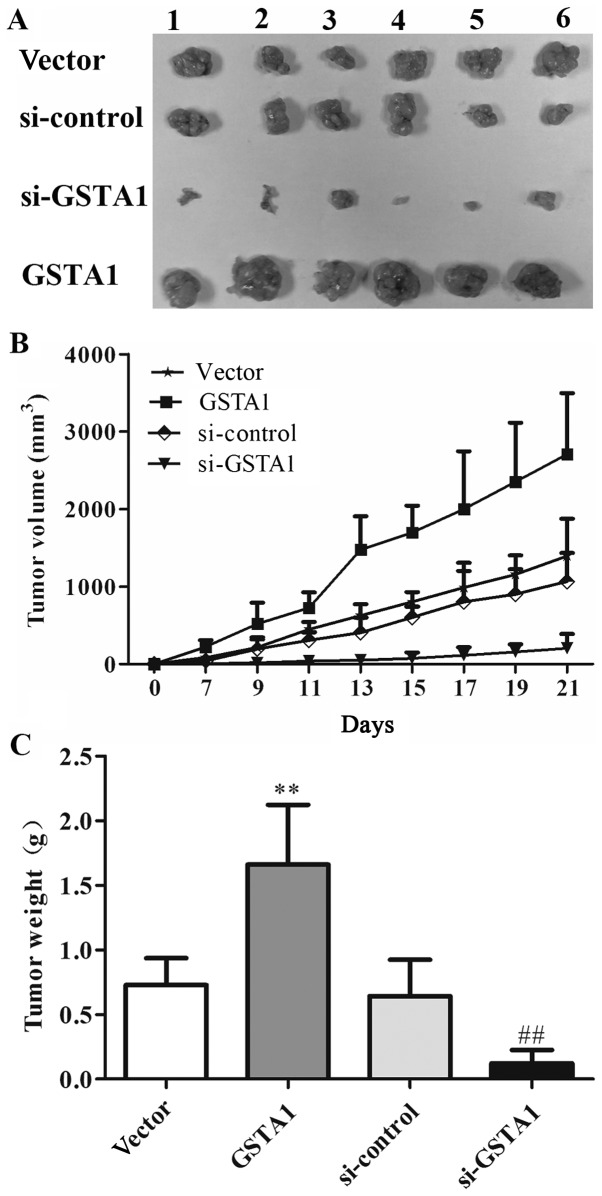
Based on the aforementioned results in vitro, the potential effect of GSTA1 in vivo in A549 cell xenografts was explored (Fig. 6A). As shown in Fig. 6B, the tumor volume in the GSTA1 group was 2,709.27 mm3, whereas that of the vector group was 1,395.43 mm3 after 21 days. The tumor volume in the si-GSTA1 group was 204.12 mm3 compared with that of the si-control group (1,066.07 mm3). Furthermore, as shown in Fig. 6C, the tumor weight in vector, GSTA1, si-control and si-GSTA1 groups were 0.73, 1.66, 0.64, and 0.12 g, respectively. These results indicated that GSTA1 significantly induced tumor growth compared with the vector control, while si-GSTA1 significantly inhibited tumor growth compared with the si-control (Fig. 6).
Figure 6.
Tumor weight and volume of A549 cells xenograft in vivo. (A and B) Tumor volume and (C) tumor weight were measured. Data are presented as the mean + standard deviation. **P<0.01 vs. vector, ##P<0.01 vs. si-control. GSTA1, glutathione S-transferase A1; si, small interfering RNA.
GSTA1 protein and mRNA expression of tumors in each group
In order to further verify the role of GSTA1 in lung cancer cell growth in vivo, western blotting and RT-qPCR were used to analyze the expression of GSTA1. The gray values of GSTA1/β-actin in the vector, GSTA1 overexpression, si-control and si-GSTA1 groups were 0.50, 1.28, 0.52, and 0.38, respectively (Fig. 7A). As shown in Fig. 7B, the relative GSTA1 mRNA levels of the four tumor groups were 1.00, 3.30, 0.98 and 0.28, which demonstrated that tumor growth may have a significant association with the expression of GSTA1.
Figure 7.
GSTA1 protein and mRNA expression levels of tumors in vivo. The four groups included vector, GSTA1, si-control and si-GSTA1. (A) GSTA1 protein expression levels were detected by western blotting. β-actin served as a loading control. (B) Reverse transcription-quantitative polymerase chain reaction was applied to detect the relative GSTA1 mRNA expression in vivo. Data are presented as the mean + standard deviation, *P<0.05, **P<0.01 vs. vector, #P<0.05, ##P<0.01 vs. si-control. GSTA1, glutathione S-transferase A1; si, small interfering RNA.
Discussion
To evaluate the function of GSTA1 in lung cancer cells, GSTA1 knockdown and overexpression was established in the A549 cell line. Western blot analysis and RT-qPCR analysis revealed significantly high expression levels of GSTA1 protein and mRNA in GSTA1 A549 cells compared with that of the vector control. Likewise, the protein and mRNA expression levels of si-GSTA1-transfected A549 cells were significantly lower compared with that of the si-control. According to the cell viability assay, it was evident that GSTA1 overexpression promoted cell proliferation and si-GSTA1 suppressed cell growth in vitro, and in vivo. In addition, the downregulation of GSTA1 in A549 cells significantly induced cell apoptosis in vitro. To the best of our knowledge, little has been reported regarding the biofunctional role of GSTA1 on cell proliferation and apoptosis in lung cancer cells. In the study of Adnan et al (19), siRNA-mediated downregulation of GSTA1 significantly increased cell proliferation and did not alter sodium butyrate-induced apoptosis in Caco-2 cells (19), which are different to the results of the present study. This discrepancy may be due to the heterogeneity of cancer, which may lead to the polymorphisms of GSTA1 in different cancer cell types. It is a normal phenomenon that one gene serves opposite roles in different tumor types. Previously, the polymorphisms of GSTM1 and GSTT1 were identified to be associated with lung cancer risk: Liu et al (20) demonstrated that the GSTM1 null genotype exhibited a significant association between squamous carcinoma (SC), adenocarcinoma (AC) and small cell lung carcinoma, and an association between the GSTT1 null genotype in SC and AC. Several studies have investigated the association between GSTA1 polymorphisms, and colorectal, prostate, breast and bladder cancer (21–24). Deng et al (17) reported that the GSTA1 BB genotype was associated with an increased cancer risk, particularly for colorectal cancer in Caucasians. The present results suggest that GSTA1 is important in tumor growth, and that the downregulation of GSTA1 suppresses proliferation and induces apoptosis in A549 cells. However, the potential underlying mechanisms of GSTA1 in lung cancer require confirmation. It may involve the binding of GSTA1 to certain cellular proteins that serve prominent roles in the regulation of the stress response, cell proliferation and apoptosis.
To date, the best-characterized GST-interacting protein is JUN N-terminal kinase (JNK1), which is a member of a large family of Ser/Thr kinases known as the mitogen-activated protein kinase family (25). JNKs serve an important role in the response to various stress stimuli, and regulate the activities of several target proteins involved in a variety of biological processes, including apoptosis, cell cycle regulation and proliferation (26). Yang et al (27) reported that overexpression of GSTA2 reduced the activation of JNK in K562 cells and attenuated H2O2-induced apoptosis. Furthermore, overexpression of hGSTA1 reduced JNK activation, c-jun phosphorylation and the cytotoxic effects of H2O2, interleukin-1β, and ultraviolet irradiation (28). Taken together, these studies indicate the pivotal role GSTA may act as one of the determinants of the differential activation of JNK. On the basis of the present results, the effects of si-GSTA1 and si-control on JNK phosphorylation, and expression in A549 cells should be investigated.
In conclusion, to the best of our knowledge, the present study demonstrated for the first time that GSTA1 overexpression acts as a tumor promoter in vivo, and si-GSTA1 is able to suppress proliferation and induce apoptosis of A549 cells in vitro. The results suggested that checking the expression of GSTA1 in clinical tests may be useful for diagnosis of lung cancer. Furthermore, GSTA1 may be used as a target molecule for developing anti-NSCLC drugs.
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the National Natural Science Foundation of China (grant no. 81102465).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
XDP and ZPY designed the study. LQZ, GXW and SZ performed experiments. GFJ, HL and ZCY analyzed the data. HL wrote the manuscript.
Ethics approval and consent to participate
All animal experiments were approved by the Animal Care Committee of Guangdong Pharmaceutical University.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 3.Sher T, Dy GK, Adjei AA. Small cell lung cancer. Mayo Clin Proc. 2008;83:355–367. doi: 10.4065/83.3.355. [DOI] [PubMed] [Google Scholar]
- 4.Zappa C, Mousa SA. Non-small cell lung cancer: Current treatment and future advances. Transl Lung Cancer Res. 2016;5:288–300. doi: 10.21037/tlcr.2016.06.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zang L, Shi L, Guo J, Pan Q, Wu W, Pan X, Wang J. Screening and identification of a peptide specifically targeted to NCI-H1299 from a phage display peptide library. Cancer Lett. 2009;281:64–70. doi: 10.1016/j.canlet.2009.02.021. [DOI] [PubMed] [Google Scholar]
- 6.Singh S. Cytoprotective and regulatory functions of glutathione S-transferases in cancer cell proliferation and cell death. Cancer Chemother Pharmacol. 2015;75:1–15. doi: 10.1007/s00280-014-2566-x. [DOI] [PubMed] [Google Scholar]
- 7.Board PG, Menon D. Glutathione transferases, regulators of cellular metabolism and physiology. Biochim Biophys Acta. 2013;1830:3267–3288. doi: 10.1016/j.bbagen.2012.11.019. [DOI] [PubMed] [Google Scholar]
- 8.Townsend DM, Tew KD. The role of glutathione-S-transferase in anti-cancer drug resistance. Oncogene. 2003;22:7369–7375. doi: 10.1038/sj.onc.1206940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Housman G, Byler S, Heerboth S, Lapinska K, Longacre M, Snyder N, Sarkar S. Drug resistance in cancer: An overview. Cancers (Basel) 2014;6:1769–1792. doi: 10.3390/cancers6031769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Searchfield L, Price SA, Betton G, Jasani B, Riccardi D, Griffiths DF. Glutathione S-transferases as molecular markers of tumour progression and prognosis in renal cell carcinoma. Histopathology. 2011;58:180–190. doi: 10.1111/j.1365-2559.2010.03733.x. [DOI] [PubMed] [Google Scholar]
- 11.Zhuo R, Kosak KM, Sankar S, Wiles ET, Sun Y, Zhang J, Ayello J, Prestwich GD, Shami PJ, Cairo MS, et al. Targeting Glutathione S-transferase M4 in Ewing sarcoma. Front Pediatr. 2014;2:83. doi: 10.3389/fped.2014.00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu H, Chen I, Shimoda LA, Park Y, Zhang C, Tran L, Zhang H, Semenza GL. Chemotherapy-Induced Ca2+ release stimulates breast cancer stem cell enrichment. Cell Rep. 2017;18:1946–1957. doi: 10.1016/j.celrep.2017.02.001. [DOI] [PubMed] [Google Scholar]
- 13.Pan XD, Yang ZP, Tang QL, Peng T, Zhang ZB, Zhou SG, Wang GX, He B, Zang LQ. Expression and function of GSTA1 in lung cancer cells. Asian Pac J Cancer Prev. 2014;15:8631–8635. doi: 10.7314/APJCP.2014.15.20.8631. [DOI] [PubMed] [Google Scholar]
- 14.Cordoba EE, Abba MC, Lacunza E, Fernande E, Guerci AM. Polymorphic variants in oxidative stress genes and acute toxicity in breast cancer patients receiving radiotherapy. Cancer Res Treat. 2016;48:948–954. doi: 10.4143/crt.2015.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eichholzer M, Rohrmann S, Barbir A, Hermann S, Teucher B, Kaaks R, Linseisen J. Polymorphisms in heterocyclic aromatic amines metabolism-related genes are associated with colorectal adenoma risk. Int J Mol Epidemiol Genet. 2012;3:96–106. [PMC free article] [PubMed] [Google Scholar]
- 16.Nguyen TV, Janssen MJ, van Oijen MG, Bergevoet SM, te Morsche RH, van Asten H, Laheij RJ, Peters WH, Jansent JB. Genetic polymorphisms in GSTA1, GSTP1, GSTT1, and GSTM1 and gastric cancer risk in a Vietnamese population. Oncol Res. 2010;18:349–355. doi: 10.3727/096504010X12626118080064. [DOI] [PubMed] [Google Scholar]
- 17.Deng Q, He B, Pan Y, Sun H, Liu X, Chen J, Ying H, Lin K, Peng H, Wang S. Polymorphisms of GSTA1 contribute to elevated cancer risk: Evidence from 15 studies. J BUON. 2015;20:287–295. [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 19.Adnan H, Quach H, MacIntosh K, Antenos M, Kirby GM. Low levels of GSTA1 expression are required for Caco-2 cell proliferation. PLoS One. 2012;7:e51739. doi: 10.1371/journal.pone.0051739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu K, Lin X, Zhou Q, Ma T, Han L, Mao G, Chen J, Yue X, Wang H, Zhang L, et al. The associations between two vital GSTs genetic polymorphisms and lung cancer risk in the Chinese population: Evidence from 71 studies. PLoS One. 2014;9:e102372. doi: 10.1371/journal.pone.0102372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ahn J, Gammon MD, Santella RM, Gaudet MM, Britton JA, Teitelbaum SL, Terry MB, Neugut AI, Eng SM, Zhang Y, et al. Effects of glutathione S-transferase A1 (GSTA1) genotype and potential modifiers on breast cancer risk. Carcinogenesis. 2006;27:1876–1882. doi: 10.1093/carcin/bgl038. [DOI] [PubMed] [Google Scholar]
- 22.Komiya Y, Tsukino H, Nakao H, Kuroda Y, Imai H, Katoh T. Human glutathione S-transferase A1, T1, M1, and P1 polymorphisms and susceptibility to prostate cancer in the Japanese population. J Cancer Res Clin Oncol. 2005;131:238–242. doi: 10.1007/s00432-004-0634-z. [DOI] [PubMed] [Google Scholar]
- 23.Martinez C, Martin F, Fernandez JM, García-Martín E, Sastre J, Díaz-Rubio M, Agúndez JA, Ladero JM. Glutathione S-transferases mu 1, theta 1, pi 1, alpha 1 and mu 3 genetic polymorphisms and the risk of colorectal and gastric cancers in humans. Pharmacogenomics. 2006;7:711–718. doi: 10.2217/14622416.7.5.711. [DOI] [PubMed] [Google Scholar]
- 24.Matic M, Pekmezovic T, Djukic T, Mimic-Oka J, Dragicevic D, Krivic B, Suvakov S, Savic-Radojevic A, Pljesa-Ercegovac M, Tulic C, et al. GSTA1, GSTM1, GSTP1, and GSTT1 polymorphisms and susceptibility to smoking-related bladder cancer: A case-control study. Urol Oncol. 2013;31:1184–1192. doi: 10.1016/j.urolonc.2011.08.005. [DOI] [PubMed] [Google Scholar]
- 25.Lo HW, Ali-Osman F. Genetic polymorphism and function of glutathione S-transferases in tumor drug resistance. Curr Opin Pharmacol. 2007;7:367–374. doi: 10.1016/j.coph.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 26.Dhanasekaran DN, Reddy EP. JNK signaling in apoptosis. Oncogene. 2008;27:6245–6251. doi: 10.1038/onc.2008.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang Y, Cheng JZ, Singhal SS, Saini M, Pandya U, Awasthi S, Awasthi YC. Role of glutathione S-transferases in protection against lipid peroxidation. Overexpression of hGSTA2-2 in K562 cells protects against hydrogen peroxide-induced apoptosis and inhibits JNK and caspase 3 activation. J Biol Chem. 2001;276:19220–19230. doi: 10.1074/jbc.M100551200. [DOI] [PubMed] [Google Scholar]
- 28.Romero L, Andrews K, Ng L, O'Rourke K, Maslen A, Kirby G. Human GSTA1-1 reduces c-Jun N-terminal kinase signalling and apoptosis in Caco-2 cells. Biochem J. 2006;400:135–141. doi: 10.1042/BJ20060110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.