Abstract
The present day modern formulation practices for drugs are based on newer tools and techniques toward effective utilization. The methods of antibody formulations are to be revolutionized based on techniques of cell engineering and gene editing. In the present review, we have discussed innovations in cell engineering toward production of novel antibodies for therapeutic applications. Moreover, this review deciphers the use of RNAi, ribozyme engineering, CRISPR-Cas-based techniques for better strategies for antibody production. Overall, this review describes the multidisciplinary aspects of the production of therapeutic proteins that has gained more attention due to its increasing demand.
Keywords: cell line engineering, antibodies, CRISPR-Cas, gene editing, RNAi, ribozymes
Introduction
Antibodies, especially monoclonal antibodies (mAbs) and derived products like antibody-drug conjugates, Fc-fusion proteins and antibody fragments are widely known for their many diagnostic and therapeutic applications (Mahmuda et al., 2017). mAbs constitute the largest group of recombinant proteins that find applications in several diseases such as cancers, autoimmune diseases (rheumatoid arthritis), cardiovascular diseases, etc (Wang C. et al., 2017; Wang Y. et al., 2017; Marston et al., 2018). Thus, it is now a dominant product class within the biopharmaceutical market. As of early 2017, there were about 68 mAbs approved by the US FDA (Cai, 2017). The early approaches for generation of mAbs by hybridoma technology were limited by challenges such as hybridoma instability and the development of human anti-mouse antibodies, which led to their rejection by the patient’s immune system (Kunert and Reinhart, 2016; Wang et al., 2016). Furthermore, during the manufacturing process, several parameters such as reducing time to market, production in large quantities to meet the continuous increasing market demands, cost effectiveness, etc. are major issues which limits its worldwide utility (Li et al., 2010).
So far, the most preferred platforms for expression of any biopharmaceutical, including antibodies has been mammalian cells. This is largely attributed to their ability to produce large volumes of therapeutic antibodies and adaptability in large-scale production systems (Kunert and Reinhart, 2016). Most importantly, mammalian cells are able to carry out desired protein folding and post-translational modifications identical to those in human systems (Wells and Robinson, 2017). Thus, chances of their rejection by patients are low. Other than mammalian cells, alternative expression systems include Pichia pastoris, Aspergillus niger, and Escherichia coli (Shriver-Lake et al., 2017; Magana-Ortiz et al., 2018; Nylen and Chen, 2018. These have been explored for expression of smaller antibody fragments (Li et al., 2010; Kunert and Reinhart, 2016), but these expression platforms have limitations like formation of inclusion bodies, which greatly hamper the final product yield, inability for post-translational modifications and codon bias. Thus, mammalian cells are most preferable (Gupta and Shukla, 2017b).
Further, advances in cell engineering make it more convenient to engineer the mammalian cells (Gupta et al., 2017b; Jazayeri et al., 2018). Engineering is basically used for regulating cellular apoptosis, cell cycle progression to avoid early cell death, control of cellular chaperones and ribozymes for proper post-translational modification of antibodies (Fischer et al., 2014; Vallée et al., 2014; Baek et al., 2017). Further, metabolic engineering can help balancing in-flux to utilized cellular energy toward antibody production. Recent discoveries and advances in gene editing nucleases like zinc finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs) and CRISPR/Cas systems (Figure 1) enable cell engineering more easy and cheaper (Gupta and Shukla, 2017a). These nucleases can alter the original genetic makeup of the cell by editing its genes toward achieving specific goals. Several studies have demonstrated the successful use of these nucleases in cell engineering (Kipniss et al., 2017; Moreno and Mali, 2017; Hashemi, 2018).
FIGURE 1.
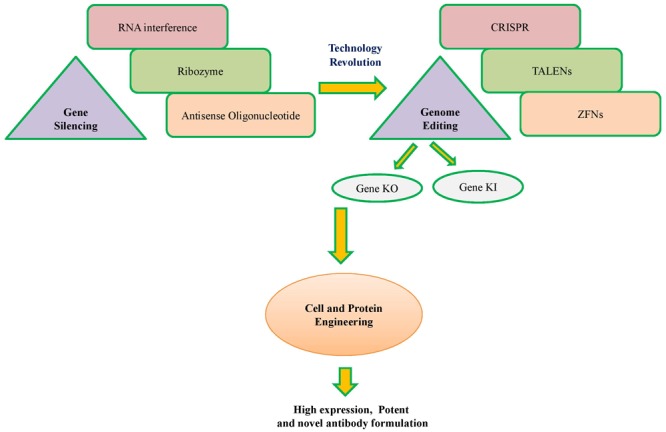
A schematic overview of technology revolution from conventional to modern cell engineering.
This review provides information about conventional and modern cell engineering techniques for more efficient antibody production. Overall, the review emphasizes different RNAi, ribozyme engineering and CRISPR-Cas-based techniques for better mAb production strategies.
Cell Engineering Toward Production of Novel Antibodies for Therapeutic Applications
The most prominent mammalian host cell lines for recombinant mAb expression include the chinese hamster ovary (CHO), mouse myeloma derived NS0 and Sp2/0 cells, human embryonic kidney cells (HEK293), and human embryonic retinoblast-derived PER.C6 cells (Beck et al., 2008). CHO cells are believed to be the most used “workhorse” for today’s industrially produced recombinant products (Bahadori et al., 2017). One of the approaches toward obtaining enhanced production efficiency and improved quality of antibodies from mammalian cell lines is through engineering of the host cell (Wells and Robinson, 2017). Various types of cellular and genetic engineering techniques are therefore directed at modifying features specific to the host cells (Black et al., 2017; Yusufi et al., 2017). These may include approaches to manipulate growth of the cell, prevent death of the cell, promote post-translational modifications etc. This is achieved largely by regulation of apoptosis, metabolic engineering, engineering cells for growth at lower temperature, chaperone engineering and glyco-engineering, which are discussed below.
Regulating Apoptosis in Cells
Apoptosis refers to programmed cell death induced during high stress conditions. Prevention of apoptosis in an antibodies expressing cell line will therefore increase cell viability, suppress cell death, extend cell culture life-span and increase productivity of target antibody product (Fischer et al., 2015a). In view of this, strategies to delay the onset of apoptosis, over-expressanti-apoptotic genes and down-regulate pro-apoptosis genes have been developed (Zustiak et al., 2014; Zhang et al., 2018). Over expression of anti-apoptotic genes such as Mcl-1, 30Kc6, Bcl-2, Bcl-w, Bcl-xL, Aven, E1B-19K and suppression of pro-apoptosis genes such as Bax, Bok, Bak in mammalian host cells have been documented to increase therapeutic protein production, including mAbs (Kim et al., 2012; Baek et al., 2017; Zhang et al., 2018). A recent study reported an 82% increase in production yield of antibody in CHO cells co-transfected with Bcl-x L, and 34% increase in CHO cells co-transfected with Mcl-1 (Zhang et al., 2018). Delaying the onset of apoptosis can be achieved by strategies such as periodic nutrient feeding, use of alternate carbon sources like galactose in place of glucose or use of adenosine (Costa et al., 2010). Inhibition of apoptosis by exosomes in CHO cells has been recently reported (Han and Rhee, 2018). Inhibition of the expression of caspases, which have an important role in the regulation of apoptosis, is another promising strategy (Costa et al., 2010; Zamaraev et al., 2017).
Regulating Cell Cycle Progression
Inhibition of cell cycle progression is another approach which could lead to higher cell viability, density and productivity in mammalian cell cultures. Toward this, approaches such as inducible expression of cell-cycle regulating factors (p27 and p21cip1) and use of rapamycin have been shown to slow down the progression through cell cycle (Costa et al., 2010). Ibarra et al. (2003) showed that arresting the cell cycle in G1 phase of the NS0 6A1/4–9F myeloma cell line through inducible expression of p21cip1 increased IgG4 antibody production. Cell cycle arrest has also been achieved through inhibition of cyclin-dependent kinase (CDK) or over expression of CDK inhibitor. A small molecule, cell cycle inhibitor (CCI) was able to induce complete G0/G1 arrest in CHO cell cultures through selective inhibition of cyclin CDK 4/6 and led to improved specific productivity by twofold to threefold (Du et al., 2015). It has also been predicted that the mammalian target of rapamycin (mTOR) based engineering of mammalian cell lines can significantly influence production levels of therapeutic proteins in the long term (Dreesen and Fussenegger, 2011; Dadehbeigi and Dickson, 2015).
Engineering of Chaperones
Chaperones and foldases have a critical role in mediating the folding of mAbs, produced by recombinant cell lines (Nishimiya et al., 2013). Engineering of chaperones has thus been found to influence production levels of antibodies through alterations in the translational capacity for the recombinant protein product (Pybus et al., 2014). Over expression of protein disulphide isomerase (PID), an enzyme that catalyzes the formation of disulphide bonds, moderately increased expression of mAb from CHO cells (Borth et al., 2005; Mohan et al., 2007). Another binding protein, BIP, associated with the folding pathway of secretory proteins, when over-expressed alone or in combination with PID, was reported to decrease mAb expression (Borth et al., 2005). Over-expression of X-box binding protein 1 (XBP1), another protein linked to the unfolded protein response, could be used as a strategy for enhancing recombinant protein production only when protein accumulation has surpassed the secretory capacity of the host cell (Ku et al., 2008).
Post-translational Modifications
N-glycan structures on antibodies have been demonstrated to have a crucial role in its bioactivity and are linked to improvement in efficacy and safety of the mAb (Pancera et al., 2013). Common mechanisms of action of therapeutic mAbs are through the elicitation of antibody-dependant cell cytotoxicity (ADCC) and complement-dependent cytotoxicity (CDC). Any alterations to the N-glycan structure on antibodies can therefore influence such antibody-specific mechanisms. Antibodies produced in CHO cells are characterized by very low levels of bisecting-N-acetylglucosamine (GlcNAc) and high levels of core fucosylation. N-acetylglucosaminyltransferase III (GnT-III), when over-expressed has been found to increase the bisecting GlcNAc content to eventually improve ADCC (Davies et al., 2001; Popp et al., 2018). Decreasing or eliminating the fucose content on antibodies has also been known to enhance the ADCC activity. Non-fucosylated therapeutic antibodies have also been touted as the next generation of therapeutic antibodies (Mori et al., 2007). Toward this, one of the commonly used methods involves knocking out the fucosyltransferase gene (FUT 8) from CHO cells resulting in expression of non-fucosylated antibody molecules. The ADCC activity markedly improved in case of fucose-deficient mAbs (Shields et al., 2002; Yamane-Ohnuki et al., 2004; Chung et al., 2012).
Metabolic Engineering
Accumulation of ammonia and lactate during recombinant CHO cell culture is common. This usually takes place due to the presence of glutamine and glucose in the culture medium and may cause adverse effects on the growing cells and the recombinant product secreted. Metabolic engineering has therefore been used to inhibit the accumulation of such toxic metabolic by-products. The over-expression of the glutamine synthetase (GS) gene in CHO cells enabled cells to grow in a glutamine-free medium, thus significantly reducing ammonia generation as a of byproduct in the culture (Zhang et al., 2006). Similarly, over-expression of enzymes such as ornithine transcarbamylase or carbamoyl phosphate synthetase I (enzymes associated with the urea cycle) has been found to bring down ammonia production within the culture medium (Park et al., 2000). In order to reduce lactic acid accumulation, various genetic modulation strategies that have been investigated include over-expression of pyruvate carboxylase (Henry and Durocher, 2011; Vallée et al., 2014; Gupta et al., 2017a) or disruption of pyruvate dehydrogenase kinases/lactate dehydrogenase A (LDH-A) (Kim and Lee, 2007; Zhou et al., 2011). LDH-A down regulation along with the GS system in the mAb-producing CHO cell line successfully reduced both ammonia and lactate levels in culture (Noh et al., 2017).
Engineering Cells for Hypothermic Growth
Reducing the cell-culture temperature to improve recombinant protein yields in CHO cells has been well documented. Lowered temperature leads to arrest of growth of cells, prolonging cellular viability and increasing cellular size. Genetic engineering strategies have therefore been developed to improve the volume of recombinant protein production at low temperatures (Mohan et al., 2008; Costa et al., 2010). Expression of cold stress genes, such as cold-inducible RNA-binding protein (CIRP) is altered when mammalian cells are exposed to lower temperatures. Stable over-expression of CIRP at 37°C showed improvement in the productivity and yields of recombinant interferon-γ in the CHO cell line (Tan et al., 2008).
Cell line engineering therefore has great potential for improving CHO cell expression systems, particularly for therapeutic antibody production (Table 1). The benefits of cell line engineering are however not limited to antibody production only, but have also been explored in production of other biopharmaceutical products such as other recombinant proteins, vaccines, fusion proteins, growth factors etc. (Xiao et al., 2014; Genzel, 2015; Johari et al., 2015; Sandig et al., 2017). Cell engineering approaches have also been used to obtain a desired attribute or quality in the target product produced by a cell, reduce those host cell proteins which could act as impurity and adversely impact the final product during downstream processing, or identify of an optimal site in host cell genome in order to target a transgene to that location (Gadgil, 2017).
Table 1.
Cell engineering approaches toward the production of novel antibodies for therapeutic applications.
| Cell engineering approach | Strategies involved | Result | Reference |
|---|---|---|---|
| Regulation of apoptosis in cells | Delay onset of apoptosis | Limit cell-apoptosis | Costa et al., 2010 |
| Over-expression of anti-apoptotic genes | Costa et al., 2010; Baek et al., 2017; Zhang et al., 2018 | ||
| Inhibition or down-regulation of pro-apoptosis genes | Han and Rhee, 2018 | ||
| Regulation of cell cycle progression | Inducible expression of cell-cycle regulating factors | Cell cycle arrest | Ibarra et al., 2003; Costa et al., 2010 |
| Inhibition of cyclin-dependent kinase (CDK) or over expression of CDK inhibitor | Du et al., 2015 | ||
| Use of mTOR- based engineering of mammalian cell lines | Slowed progression through cell cycle | Costa et al., 2010; Dreesen and Fussenegger, 2011; Dadehbeigi and Dickson, 2015 | |
| Engineering of chaperones and foldases | Over expression of protein disulphide isomerase | Increased formation of disulphide bonds in proteins | Borth et al., 2005; Mohan et al., 2007 |
| Post-translational modifications | Knocking out the fucosyltransferase gene (FUT 8) from CHO cells | Enhanced ADCC activity | Shields et al., 2002; Yamane-Ohnuki et al., 2004; Chung et al., 2012 |
| Metabolic engineering | Over-expression of glutamine synthetase gene in CHO cells | Reduction in ammonia generation as by-product within culture | Zhang et al., 2006 |
| Over-expression of ornithine transcarbamylase, carbamoyl phosphate synthetase I | Park et al., 2000 | ||
| Over-expression of pyruvate carboxylase | Reduction in lactic acid accumulation as by-product within culture | Henry and Durocher, 2011; Vallée et al., 2014; Gupta et al., 2017a | |
| Down-regulation of pyruvate dehydrogenase kinases/ lactate dehydrogenase A | Kim and Lee, 2007; Zhou et al., 2011 | ||
| Engineering cells for hypothermic growth | Stable over-expression of cold stress genes, such as cold-inducible RNA-binding protein | Improvement in the productivity and yields of recombinant protein | Tan et al., 2008 |
Ribozyme Engineering in Antibody Formulation
RNAs not only serve to establish linkages between genetic information and proteins, but also play a key role as active regulators of gene expression. MicroRNAs (miRNAs) are one such class of RNA regulatory molecules. These are short non-coding RNAs, which are capable of regulating entire cellular pathways by post-transcriptionally modulating expression of numerous genes. miRNAs are associated with relevant cellular processes such as apoptosis, cell proliferation, or biosynthesis of proteins. Host cell engineering using miRNAs represents an important tool to overcome limitations in industrial cell-line development. miRNAs also provide better advantages over single-gene manipulation due to their ability to affect expression of multiple genes. Moreover, miRNAs, unlike cellular engineering (which relies largely on overexpression of regulatory proteins), are not translated into proteins. Thus, requirement of the translational machinery for miRNA over-expression is not there and there is no translational burden to the production cell line, even as the miRNAs regulate important cellular physiological processes (Druz et al., 2012; Hackl et al., 2012; Jadhav et al., 2013; Patel, 2017; Jazayeri et al., 2018). In recent years, available literature has focused on the identification of miRNAs to improve recombinant protein production (Fischer et al., 2014; Jadhav et al., 2014; Loh et al., 2014; Kelly et al., 2015), or enhanced cell growth (Barron et al., 2011; Druz et al., 2013; Sanchez et al., 2014). Manipulation of miRNA levels in CHO cells has been shown to improve product yield by increasing proliferation and specific productivity, resisting apoptosis and enhancing oxidative metabolism (Figure 2). Some of the recent studies on the effect of miRNA manipulations in CHO cells have been summarized in Table 2 below.
FIGURE 2.
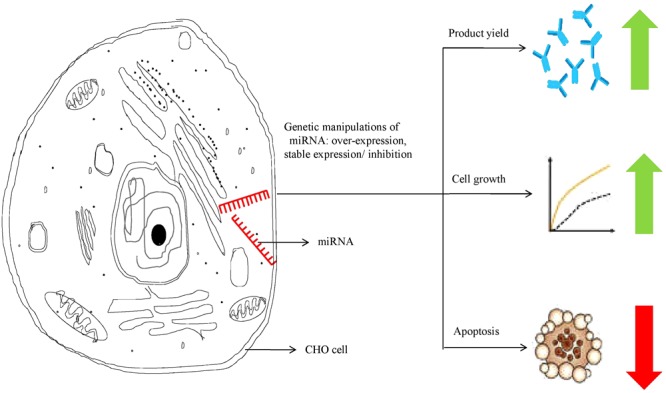
miRNA manipulation in CHO cells leading to improved therapeutic protein productivity.
Table 2.
Recent studies on the effects of miRNA manipulations in CHO cells.
| miRNA | Fate in host cell (CHO cells) | Key observations | Reference |
|---|---|---|---|
| cgr-miR-7 | Over expression | Increase in cell’s specific productivity | Barron et al., 2011 |
| miR-17 | Over expression | Increased cell proliferation; no negative impact on cell’s specific productivity | Jadhav et al., 2012 |
| miR-557, miR-1287 | Stable expression | Enhanced viable cell density and specific productivity of therapeutic IgG1 | Strotbek et al., 2013 |
| Mmu-miR-466h-5p | Stable inhibition | Improved resistance to apoptosis; improved protein production | Druz et al., 2013 |
| miR-30 family | Stable over expression | Improvement in cell’s bioprocess performance | Fischer et al., 2014 |
| miR-17 | Stable over expression | Enhanced cellular growth; cell’s specific productivity increased by two fold | Jadhav et al., 2014 |
| miR-17, miR-1b, miR-92a | Over expression | Enhanced cellular productivity | Loh et al., 2014 |
| miR-2861 | Enhanced cellular productivity | Fischer et al., 2015b | |
| miR-557 | Stable expression | Significant increase in yield of final product | Fischer et al., 2017 |
The Table 2 shows that miRNAs have largely been subjected to stability or over-expression in CHO cell lines, leading to improved protein productivity. For example, CHO cells, which have been engineered to express miR-557, a pro-productive miRNA, have shown an increase in product yield without compromising product quality (Fischer et al., 2017). Expression of miR-30 and miR17 in CHO cells have also led to enhanced specific productivity (Fischer et al., 2014; Jadhav et al., 2014). A study assessing the effect of over-expression of miR-7 on the proteome of CHO cells showed that the cell’s productivity increased with increasing levels of miR-7, albeit at the cost of cell growth (Meleady et al., 2012). Sanchez et al. (2014) have reported an increase in protein production by CHO cells, when miR-7 activity was disrupted using sponge decoy vectors. On the contrary, stable inhibition of mmu-miR-466h-5p in CHO cells has been found to increase resistance to apoptosis and also improve protein production (Druz et al., 2013). Such stable manipulations of miRNA expression can also enable CHO cells to reach a higher cell viability and cell density in comparison to their controls.
miRNA-based cell line engineering provides an opportunity for access to numerous sophisticated therapeutic proteins, including antibodies. While this approach has not yet found a way into routine industrial production processes, there lies a significant potential for their future application in industrial manufacturing cell lines, ultimately contributing to successful biopharmaceutical drug development. While miRNAs are deemed to have multiple applications in medicine and biotechnology, a better understanding of their mechanism at the molecular level is needed to harness their potential toward the development of industrially relevant therapeutic protein-producing cell factories.
Advanced Gene Editing Tools
With the discovery and emergence of modern gene-editing tools, manipulation of production hosts has become relatively easy. It is now possible to maneuver the genome of industrial yeast and mammalian host cells, thereby allowing the development of potential and cost-effective recombinant therapeutic proteins. While previously developed gene editing tools such as TALENs and ZFNs continue to remain useful, the emergence of another new technology, the CRISPR-Cas system is opening new avenues in gene editing opportunities as described below (Sander and Joung, 2014).
Zinc Finger Nucleases (ZFNs)
Most commonly and primitive endonucleases are ZFNs which are artificially designed restriction enzymes (Yang et al., 2017). Zinc finger proteins (ZFPs) derived from eukaryotic transcription factors, which act as a DNA binding domain of ZFN and the nucleotide chopper domain Folk 1 is derived from Flavobacterium okeanokoites (De Souza, 2012). The ZFPs are composed of 30 amino acids which creates two anti-parallel β-sheets opposite an alpha-helix (De Souza, 2012). Each cleavage domain is attached with three to six ZFPs, which are specially designed for a target site. Usually, ZFPs have 18 base pair specificity and DNA-binding domains have 9 base pair specificity, which makes them robust and very precise for target-specific gene editing (Sander et al., 2011b). Till date, the ZFNs have been successfully utilized for HDR-mediated gene knock-in (KI) and non-homologous end joining (NHEJ) based Knock-out (KO) approaches to many prokaryotic and eukaryotic targets (Carroll, 2011). For example CHO cells are modified by KI of GS and dihydrofolate reductase (DHFR) genes, which makes them suitable for consistent protein production in therapeutic biopharmaceutical industries. Even though, after successful experiments, ZFNs are not considered for industrial use now a days due to certain restrictions like: A. All possible nucleotide genomic sequences can’t be targeted, B. Specificity of sequence can be interrupted by neighboring protein domains (Sander et al., 2011b). Apart from these, designing of folks and ZFPs are very complicated and costly which makes it less preferred.
Transcription Activator-Like Effector Nucleases (TALENs)
Transcription activator-like effector nucleases are a moderately innovative gene-editing tool. TAL proteins are the major part of TALENs, which are secreted from the pathogenic bacteria Xanthomonas (Li et al., 2012). TALENs have tandem repeats of 34 amino acids with specific recognition and binding efficiency. The TAL has the capability of recognizing a single nucleotide, which is very specific and not influenced by other domains present around it. The TALENs based gene editing tool is relatively more effective and frequently used for knock-in and knock-out events. Similarly, like ZFNs, TALENs also have two-site two protein domains, which create cuts and second TAL repeats, which helps protein domains to find a specific site for binding and making a notch (Christian et al., 2010). TALENs leave sticky ends to the DNA, which minimize the off-target results (Aryan et al., 2013). Efficiency of targeted KO with TALENs is 30 to 100% while KIs range from 1 to 10%. Many animals like zebrafish, chickens, frogs, rats, and mammalian cells are genetically modified through TALENs (Sander et al., 2011a; Aryan et al., 2013; Kondo et al., 2014).
CRISPR-Cas
CRISPR-Cas is based on the natural defense mechanism in bacteria, which they use for the control of pathogens. Cas9 endonuclease from Streptococcus pyogenes has been one of the most studied with respect to CRISPR-Cas systems (Ran et al., 2015). Cas9 endonuclease binds to a complex of CRISPR RNA (crRNA) and another transactivating crRNA (tracrRNA). The crRNA guides the Cas9 endonuclease activity to cut both strands of target DNA at particular sequences (Briner et al., 2014). The ability of CRISPR-Cas 9 to target the Cas9 endonuclease to specific genomic loci, enabling DNA cleavage at specific sites has allowed it to being considered as an adept tool in gene editing (Doudna and Charpentier, 2014). CRISPR-Cas-mediated gene repair, disruption, insertion or deletion is thus finding applications in several areas of biomedical research, medicine, agriculture and biotechnology (Hwang et al., 2013). Recent studies utilizing CRISPR-Cas-based systems for genetic manipulations in animals, fish and plant models have been summarized in Table 3.
Table 3.
CRIPSR-Cas based gene editing techniques used for manipulation of various organisms.
| Model organism | CRISPR-Cas based gene manipulations | Observations/Remarks | Reference |
|---|---|---|---|
| Zebrafish | Genetic modifications in embryo of zebrafish | Similar efficiency to ZFN and TALENs | Hwang et al., 2013 |
| Zebrafish | Insertion of CreERT2 transgene at otx2 gene locus | otx2:CreER T2 transgenic fish developed; valuable tool for future studies | Kesavan et al., 2018 |
| Mouse | Simultaneous disruption of five genes (Tet1, 2, 3, Sry, Uty - 8 alleles) in embryonic stem cells of the mouse | High efficiency observed | Wang et al., 2013 |
| Mouse | Editing of specific regions of the Duchenne muscular dystrophy gene using AAV vectors | Expression of Cas9 enables direct editing of the mutation, deletion of multiple exons or gene correction | Bengtsson et al., 2017 |
| mdx Mouse | Correction of the dystrophin gene (Dmd) mutation | 2-100% correction of the Dmd gene achieved | Long et al., 2014 |
| Mouse | Deletion of the GAA repeats from frataxin gene | Restoration of frataxin gene activity and protein level | Ouellet et al., 2017 |
| Mouse | Mutation of Pten and p53 in mouse liver | Development of models for liver cancer | Xue et al., 2014 |
| Mouse | Dip2a gene deletion and β-galactosidase (lacz) reporter gene insertion | Development of mouse models and studies on gene function | Zhang L. et al., 2015 |
| Mouse | EGFP transgene or Crygc gene mutation in Spermatogonial Stem Cells (SSCs) | Spermatogenesis observed in mutated SSCs after being transplanted into seminiferous tubules of infertile mouse testes. | Wu et al., 2015 |
| Duchenne muscular dystrophy (DMD) patient-derived induced pluripotent stem cells (iPSCs) | Correction of dystrophin gene | Restoration of the dystrophin protein in patient-derived iPSCs | Li et al., 2015 |
| Human pluripotent stem cells (hPSCs) | Development of an episomal vector-based CRISPR/Cas9 system which enables generation of up to 100% Insertion/Deletion rates | Highly efficient gene knockout in hPSCs | Xie et al., 2017 |
| Indica rice line IR58025B | Deleted DNA fragments in the gene Dense And Erect Panicle1 (DEP1) | Relatively high frequency | Wang Y. et al., 2017 |
| Tomato plants | Somatic mutations in SlIAA9 gene | Morphological changes in leaf shape and seedless fruit observed in mutants | Ueta et al., 2017 |
| Maize plants | Gene sequence ARGOS8 edited | Increased yield of maize grain under stress conditions such as drought | Shi et al., 2017 |
| Oilseed crop Camelina sativa | Mutagenesis of 3 delta 12 desaturase (FAD2) genes | Reduction in polyunsaturated fatty acid levels; increased oleic acid accumulation in oil | Morineau et al., 2017 |
The CRISPR-Cas system has not only enabled gene editing but also has applications across several areas such as gene therapy, development of tissue and animal disease models, drug discovery, deciphering plant disease resistance, transcription regulation, genome imaging, and epigenetic modification (Xiong et al., 2016). CRISPR-Cas-based gene editing has been used for development of animal models, that mimic human diseases. Such models provide an opportunity to predict possible outcomes of clinical trials and simultaneously verify the safety and accuracy of drugs against the particular disease. Recently, CRISPR-Cas9 technology has been used to establish large animal models that can mimic human neurodegenerative diseases for enhanced understanding of pathogenesis of such diseases (Tu et al., 2015). The CRISPR-Cas9 system has also been used for successful correction of diseases or disease-causing mutations in animal models (Wu et al., 2013; Long et al., 2014). In a recent report by Cai et al. (2016) it was summarized how the CRISPR-Cas9-based genome editing technology is opening doors for treatment of diverse human diseases. The technology has recently been applied for the study or treatment of human diseases such as muscular dystrophy, hemophilia, thalassemia, cystic fibrosis etc. The CRISPR-Cas system has been used for correction of the dystrophic gene in Duchenne muscular dystrophy (DMD) patient-derived induced pluripotent stem cells and restoration of the dystrophin protein in the cells (Li et al., 2015). The tool has been used for immunology-based applications, which may lead to designing of treatment regimens of diseases like HIV-AIDs (Hu et al., 2014; Liao et al., 2015). It has also been exploited for its potential to KI or KO specific genes in model organisms for studying genetic diseases (Platt et al., 2014; Kang et al., 2015).
The CRISPR–Cas system also has a big part to play in revolutionizing the role that gene editing plays in drug discovery. However, CRISPR–Cas-facilitated drug discovery is presently largely limited to basic research (Golkar et al., 2016; Luo, 2016). CRISPR’s potential is believed to span each stage of the drug discovery process and will predictably affect the next generation drugs by accelerating drug target identification and validation, discovery of biomarkers, and development of novel therapies (Fellmann et al., 2017; Lu et al., 2017). Alternatively, CRISPR-Cas can also be used in order to generate disease genotypes or phenotypes for use in drug discovery. CRISPR/Cas-based gene editing tools can also play a potential role in antiviral drug development (Chen et al., 2018). Mutations in Hepatitis B virus (HBV) DNA, brought about by the CRISPR-Cas system, inhibit replication and expression of the HBV and may constitute a new therapeutic strategy for HBV infection (Zhen et al., 2015). Similarly, human primary CD4+ T cells have shown HIV-1 resistance owing to disruption of the human CXCR4 gene by CRISPR/Cas9-mediated genome editing (Hou et al., 2015). In addition to the above, CRISPR-Cas system has been implicated in slowing down the spread of antibiotic resistance genes. The system has the ability to limit major routes of horizontal gene transfer and even destroy plasmids thereby restricting the spread of drug resistance (Golkar et al., 2016). This ability of the CRISPR/Cas system also holds potential to be exploited in clinical settings.
In addition to their role in designing disease treatment regimens or drug discovery, advanced gene editing technologies such as CRISPR-Cas9 also hold significant potential for the future in that they will provide new opportunities to stably engineer host cells for antibody production. This has been explored in some of the recently published research. CRISPR/Cas9 based genome editing has been used for knockout of GS-encoding gene resulting in improved recombinant protein production in CHO cells (Grav et al., 2017). The inactivation of GDP-fucose transporter gene in CHO cells by ZFNs, TALENs and CRISPR-Cas9 has been reported to lead to production of fucose-free antibodies (Chan et al., 2016).
However, TALENs and ZFNs are starting to prove their worth in human gene targeting and editing. The biotech company Sangamo Biosciences (California, United States) has stepped forward its ZFN in in vivo gene editing research into the clinic, and the US Food and Drug Administration (US-FDA) has permitted a HIV program in somatic cells (non inherited cells), which has cured more than 80 HIV-infected patients so far (Xu et al., 2017). Meanwhile, TALENs are recently used in the clinic to engineer immune cells to help fight leukemia in a young girl (Le, 2015). Despite these successes, many researchers found CRISPR as a less onerous and more efficient tool as compared to other two gene editing tools described above. The cost savings of the new method is also substantial. Thus, genome editing approaches therefore present a tremendous scope for being exploited for the production of novel antibodies with varying applications.
Conclusion and Future Prospective
Although, mammalian cells are employed for production of more effective mAbs for diagnostics and treatment of many severe and life-threatening diseases, certain limitations prevent their dominancy (Zhu, 2012; Ye et al., 2016). Advances in cell engineering approaches at various stages, therefore enables cells to be used as a sole and improved platform for production of complex natured mAbs. Some of the exiting cell engineering approaches have been discussed in this review. These include by regulation of apoptosis, metabolic engineering, and engineering cells for growth at lower temperature, chaperone engineering and glyco-engineering. Engineering of cells toward regulation of the apoptosis or cell-cycle progression can result in longer cell viability and higher cellular productivity. Specific cellular chaperones and foldases can be engineered for proper folding of nascent polypeptide chains to an antibody (Zhang et al., 2018). Other approaches are targeted for reduction of metabolic by-product accumulation, enhancing cellular productivity at lower cell-culture temperatures or obtaining a desired attribute or quality in the target protein product. Additionally, cell line engineering has also contributed to the production of other biopharmaceutical products, reduction of impure host cell proteins, etc. Host-cell engineering using micro-RNAs also represent a recent and important strategy that can help improve recombinant therapeutic protein production. There lies immense scope for exploring miRNA based cell-line engineering in industrial protein production processes. Discovery of novel gene editing tools including ZFNs, TALENs and CRISPR/Cas have made the cell engineering more convenient to achieve specific goals (Gupta and Shukla, 2017a). Among these three, the CRISPR/Cas9 system is now gaining its own importance in curing human genetic diseases as well as host engineering for potential biopharmaceutical production (Ran et al., 2015). With the advent of the CRISPR/Cas9 tool, it is now possible to target the human genome at specific sites to cure various life threatening genetic diseases such as HIV, Leukemia, Sickle cell anemia etc. This tool has also been potentially used for human gene therapy with success. As CRISPR/Cas9 technology is still in a nascent phase, certain limitations need to be overcome. Off-target effects are the major ones which may cause undesirable editing or mutation in the host cell genome (Zhang X.H. et al., 2015). The designing of sgRNA should be very specific for a particular gene; otherwise, it can edit off-targets (non-specific), which may cause unwanted editing or mutation in the host genome (Doench et al., 2016). Availability of the PAM sequence is also very important because the Cas9 protein recognizes only the PAM (NGG) sequence at the time of binding to insure the spacer’s attachment. If any mutation occurs in the PAM sequence during in vitro processing, the cleavage of the target DNA will be restricted. Therefore, specificity is a must for the locus identification as well as the designing of the sgRNA. It can be predicted that, the CRISPR/Cas9 platform can one day partially replace the drugs, because scientists have developed this tool for treating critical diseases such as cancers, HIV leukemia etc. (Doudna and Charpentier, 2014). However, the CRISPR modifies a gene, the change is permanent. The other limitation is availability of CRISPR, since it comes from bacteria; if it stays in the body for too long, it might trigger a deleterious immune response (Seed et al., 2013). Many ethical issues have already been raised against germ-line cell editing with CRISPR, because it can affect evolution and can raise the ethical and justice issues. It is essential to have strong ethics for the use of CRISPR or any other gene editing tool. These tools have the capability of KI or KO on specific genes at loci for metabolic engineering as well as improved protein production and product quality, which in turn can be beneficial in reducing drug production costs and increased affordability for a large population.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Conflict of Interest Statement
SG and SD were employed by company Ipca Laboratories (India) Mumbai, India. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Authors duly acknowledge M.D. University, Rohtak, India for providing infrastructural facilities. PS acknowledges the infrastructural support from Department of Science and Technology, Government of India through FIST grant (Grant No. 1196 SR/FST/LS-I/2017/4).
References
- Aryan A., Anderson M. A. E., Myles K. M., Adelman Z. N. (2013). TALEN-based gene disruption in the dengue vector Aedes aegypti. PLoS One 8:e60082. 10.1371/journal.pone.0060082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck A., Wagner-Rousset E., Bussat M. C., Lokteff M., Klinguer-Hamour C., Haeuw J. F., et al. (2008). Trends in glycosylation, glycoanalysis and glycoengineering of therapeutic antibodies and Fc-fusion proteins. Curr. Pharm. Biotechnol. 9 482–501. 10.2174/138920108786786411 [DOI] [PubMed] [Google Scholar]
- Baek E., Noh S. M., Lee G. M. (2017). Anti-apoptosis engineering for improved protein production from CHO cells. Methods Mol. Biol. 1603 71–85. 10.1007/978-1-4939-6972-2_5 [DOI] [PubMed] [Google Scholar]
- Bahadori M., Amiri F., Movahed M., Roushandeh A. M., Roudkenar M. H. (2017). Establishment of stable chinese hamster ovary cell line capable of expressing human recombinant hemopexin: a promising therapeutic modality against hemolytic anemia. Gene Cell Tissue 4:e13360 10.5812/gct.13360 [DOI] [Google Scholar]
- Barron N., Kumar N., Sanchez N., Doolan P., Clarke C., Meleady P., et al. (2011). Engineering CHO cell growth and recombinant protein productivity by overexpression of miR-7. J. Biotechnol. 151 204–211. 10.1016/j.jbiotec.2010.12.005 [DOI] [PubMed] [Google Scholar]
- Bengtsson N. E., Hall J. K., Odom G. L., Phelps M. P., Andrus C. R., Hawkins R. D., et al. (2017). Muscle-specific CRISPR/Cas9 dystrophin gene editing ameliorates pathophysiology in a mouse model for Duchenne muscular dystrophy. Nat. Commun. 8:14454. 10.1038/ncomms14454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black J. B., Perez-Pinera P., Gersbach C. A. (2017). Mammalian synthetic biology: engineering biological systems. Annu. Rev. Biomed. Eng. 19 249–277. 10.1146/annurev-bioeng-071516-044649 [DOI] [PubMed] [Google Scholar]
- Borth N., Mattanovich D., Kunert R., Katinger H. (2005). Effect of increased expression of protein disulfide isomerase and heavy chain binding protein on antibody secretion in a recombinant CHO cell line. Biotechnol. Prog. 21 106–111. 10.1021/bp0498241 [DOI] [PubMed] [Google Scholar]
- Briner A. E., Donohoue P. D., Gomaa A. A., Selle K., Slorach E. M., Nye C. H., et al. (2014). Guide RNA functional modules direct Cas9 activity and orthogonality. Mol. Cell 56 333–339. 10.1016/j.molcel.2014.09.019 [DOI] [PubMed] [Google Scholar]
- Cai H. H. (2017). Therapeutic monoclonal antibodies approved by FDA in 2016. MOJ Immunol. 5:00145 10.15406/moji.2017.05.00145 [DOI] [Google Scholar]
- Cai L., Fisher A. L., Huang H., Xie Z. (2016). CRISPR-mediated genome editing and human diseases. Genes Dis. 3 244–251. 10.1016/j.gendis.2016.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll D. (2011). Genome engineering with zinc-finger nucleases. Genetics 188 773–782. 10.1534/genetics.111.131433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K. F., Shahreel W., Wan C., Teo G., Hayati N., Tay S. J., et al. (2016). Inactivation of GDP-fucose transporter gene (Slc35c1) in CHO cells by ZFNs, TALENs and CRISPR-Cas9 for production of fucose-free antibodies. Biotechnol. J. 11 399–414. 10.1002/biot.201500331 [DOI] [PubMed] [Google Scholar]
- Chen S., Yu X., Guo D. (2018). CRISPR-Cas targeting of host genes as an antiviral strategy. Viruses 10:40. 10.3390/v10010040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian M., Cermak T., Doyle E. L., Schmidt C., Zhang F., Hummel A., et al. (2010). Targeting DNA double-strand breaks with TAL effector nucleases. Genetics 186 756–761. 10.1534/genetics.110.120717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S., Quarmby V., Gao X., Ying Y., Lin L., Reed C., et al. (2012). Quantitative evaluation of fucose reducing effects in a humanized antibody on Fcγ receptor binding and antibody-dependent cell-mediated cytotoxicity activities. MAbs 4 326–340. 10.4161/mabs.19941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa A. R., Rodrigues M. E., Henriques M., Azeredo J., Oliveira R. (2010). Guidelines to cell engineering for monoclonal antibody production. Eur. J. Pharm. Biopharm. 74 127–138. 10.1016/j.ejpb.2009.10.002 [DOI] [PubMed] [Google Scholar]
- Dadehbeigi N., Dickson A. J. (2015). Chemical manipulation of the mTORC1 pathway in industrially relevant CHOK1 cells enhances production of therapeutic proteins. Biotechnol. J. 10 1041–1050. 10.1002/biot.201500075 [DOI] [PubMed] [Google Scholar]
- Davies J., Jiang L., Pan L. Z., LaBarre M. J., Anderson D., Reff M. (2001). Expression of GnTIII in a recombinant anti-CD20 CHO production cell line: expression of antibodies with altered glycoforms leads to an increase in ADCC through higher affinity for FCγRIII. Biotechnol. Bioeng. 74 288–294. 10.1002/bit.1119 [DOI] [PubMed] [Google Scholar]
- De Souza N. (2012). Primer: genome editing with engineered nucleases. Nat. Methods 9:27 10.1038/nmeth.1848 [DOI] [PubMed] [Google Scholar]
- Doench J. G., Fusi N., Sullender M., Hegde M., Vaimberg E. W., Donovan K. F., et al. (2016). Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 34 184–191. 10.1038/nbt.3437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna J. A., Charpentier E. (2014). The new frontier of genome engineering with CRISPR-Cas9. Science 346:1258096. 10.1126/science.1258096 [DOI] [PubMed] [Google Scholar]
- Dreesen I. A., Fussenegger M. (2011). Ectopic expression of human mTOR increases viability, robustness, cell size, proliferation, and antibody production of chinese hamster ovary cells. Biotechnol. Bioeng. 108 853–866. 10.1002/bit.22990 [DOI] [PubMed] [Google Scholar]
- Druz A., Betenbaugh M., Shiloach J. (2012). “MicroRNAs as engineering targets,” in MicroRNAs as Tools in Biopharmaceutical Production ed. Barron N. (Berlin: Springer; ) 65–86. [Google Scholar]
- Druz A., Son Y. J., Betenbaugh M., Shiloach J. (2013). Stable inhibition of mmu-miR-466h-5p improves apoptosis resistance and protein production in CHO cells. Metab. Eng. 16 87–94. 10.1016/j.ymben.2012.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z., Treiber D., McCarter J. D., Fomina-Yadlin D., Saleem R. A., McCoy R. E., et al. (2015). Use of a small molecule cell cycle inhibitor to control cell growth and improve specific productivity and product quality of recombinant proteins in CHO cell cultures. Biotechnol. Bioeng. 112 141–155. 10.1002/bit.25332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellmann C., Gowen B. G., Lin P. C., Doudna J. A., Corn J. E. (2017). Cornerstones of CRISPR–Cas in drug discovery and therapy. Nat. Rev. Drug Discov. 16 89–100. 10.1038/nrd.2016.238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer S., Buck T., Wagner A., Ehrhart C., Giancaterino J., Mang S., et al. (2014). A functional high-content miRNA screen identifies miR-30 family to boost recombinant protein production in CHO cells. Biotechnol. J. 9 1279–1292. 10.1002/biot.201400306 [DOI] [PubMed] [Google Scholar]
- Fischer S., Handrick R., Otte K. (2015a). The art of CHO cell engineering: a comprehensive retrospect and future perspectives. Biotechnol. Adv. 33 1878–1896. 10.1016/j.biotechadv.2015.10.015 [DOI] [PubMed] [Google Scholar]
- Fischer S., Paul A. J., Wagner A., Mathias S., Geiss M., Schandock F., et al. (2015b). miR-2861 as novel HDAC5 inhibitor in CHO cells enhances productivity while maintaining product quality. Biotechnol. Bioeng. 112 2142–2153. 10.1002/bit.25626 [DOI] [PubMed] [Google Scholar]
- Fischer S., Marquart K. F., Pieper L. A., Fieder J., Gamer M., Gorr I., et al. (2017). miRNA engineering of CHO cells facilitates production of difficult-to-express proteins and increases success in cell line development. Biotechnol. Bioeng. 114 1495–1510. 10.1002/bit.26280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadgil M. (2017). Cell culture processes for biopharmaceutical manufacturing. Curr. Sci. 112 1478–1488. 10.18520/cs/v112/i07/1478-1488 [DOI] [Google Scholar]
- Genzel Y. (2015). Designing cell lines for viral vaccine production: where do we stand? Biotechnol. J. 10 728–740. 10.1002/biot.201400388 [DOI] [PubMed] [Google Scholar]
- Golkar Z., Rochelle L., Bagasra O. (2016). Crisprs/Cas9 may provide new method for drug discovery and development. J. Mol. Biomark. Diag. 7:280 10.4172/2155-9929.1000280 [DOI] [Google Scholar]
- Grav L. M., la Cour Karottki K. J., Lee J. S., Kildegaard H. F. (2017). Application of CRISPR/Cas9 genome editing to improve recombinant protein production in CHO Cells. Methods Mol. Biol. 1603 101–118. 10.1007/978-1-4939-6972-2_7 [DOI] [PubMed] [Google Scholar]
- Gupta S. K., Sharma A., Kushwaha H., Shukla P. (2017a). Over-expression of a codon optimized yeast cytosolic pyruvate carboxylase (PYC2) in CHO cells for an augmented lactate metabolism. Front. Pharmacol. 8:463. 10.3389/fphar.2017.00463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S. K., Shukla P. (2017a). Gene editing for cell engineering: trends and applications. Crit. Rev. Biotechnol. 37 672–684. 10.1080/07388551.2016.1214557 [DOI] [PubMed] [Google Scholar]
- Gupta S. K., Shukla P. (2017b). Microbial platform technology for recombinant antibody fragment production: a review. Crit. Rev. Microbiol. 43 31–42. 10.3109/1040841X.2016.1150959 [DOI] [PubMed] [Google Scholar]
- Gupta S. K., Srivastava S. K., Sharma A., Nalage V. H., Salvi D., Kushwaha H., et al. (2017b). Metabolic engineering of CHO cells for the development of a robust protein production platform. PLoS One 12:e0181455. 10.1371/journal.pone.0181455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackl M., Borth N., Grillari J. (2012). miRNAs–pathway engineering of CHO cell factories that avoids translational burdening. Trends Biotechnol. 30 405–406. 10.1016/j.tibtech.2012.05.002 [DOI] [PubMed] [Google Scholar]
- Han S., Rhee W. J. (2018). Inhibition of apoptosis using exosomes in Chinese hamster ovary cell culture. Biotechnol. Bioeng. 115 1331–1339. 10.1002/bit.26549 [DOI] [PubMed] [Google Scholar]
- Hashemi A. (2018). CRISPR-Cas system as a genome engineering platform: applications in biomedicine and biotechnology. Curr. Gene Ther. 12 115–124. 10.2174/1566523218666180221110627 [DOI] [PubMed] [Google Scholar]
- Henry O., Durocher Y. (2011). Enhanced glycoprotein production in HEK-293 cells expressing pyruvate carboxylase. Metab. Eng. 13 499–507. 10.1016/j.ymben.2011.05.004 [DOI] [PubMed] [Google Scholar]
- Hou P., Chen S., Wang S., Yu X., Chen Y., Jiang M., et al. (2015). Genome editing of CXCR4 by CRISPR/cas9 confers cells resistant to HIV-1 infection. Sci. Rep. 5:15577. 10.1038/srep1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W., Kaminski R., Yang F., Zhang Y., Cosentino L., Li F., et al. (2014). RNA-directed gene editing specifically eradicates latent and prevents new HIV-1 infection. Proc. Natl. Acad. Sci. 111 11461–11466. 10.1073/pnas.1405186111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang W. Y., Fu Y., Reyon D., Maeder M. L., Tsai S. Q., Sander J. D., et al. (2013). Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 31 227–229. 10.1038/nbt.2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarra N., Watanabe S., Bi J. X., Shuttleworth J., Al-Rubeai M. (2003). Modulation of cell cycle for enhancement of antibody productivity in perfusion culture of NS0 cells. Biotechnol. Prog. 19 224–228. 10.1021/bp025589f [DOI] [PubMed] [Google Scholar]
- Jadhav V., Hackl M., Druz A., Shridhar S., Chung C. Y., Heffner K. M., et al. (2013). CHO microRNA engineering is growing up: recent successes and future challenges. Biotechnol. Adv. 31 1501–1513. 10.1016/j.biotechadv.2013.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jadhav V., Hackl M., Hernandez Bort J. A., Wieser M., Harreither E., Kunert R., et al. (2012). A screening method to assess biological effects of microRNA overexpression in Chinese hamster ovary cells. Biotechnol. Bioeng. 109 1376–1385. 10.1002/bit.24490 [DOI] [PubMed] [Google Scholar]
- Jadhav V., Hackl M., Klanert G., Bort J. A. H., Kunert R., Grillari J., et al. (2014). Stable overexpression of miR-17 enhances recombinant protein production of CHO cells. J. Biotechnol. 175 38–44. 10.1016/j.jbiotec.2014.01.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazayeri S. H., Amiri-Yekta A., Bahrami S., Gourabi H., Sanati M. H., Khorramizadeh M. R. (2018). Vector and cell line engineering technologies toward recombinant protein expression in mammalian cell lines. Appl. Biochem. Biotechnol. [Epub ahead of print]. 10.1007/s12010-017-2689-8 [DOI] [PubMed] [Google Scholar]
- Johari Y. B., Estes S. D., Alves C. S., Sinacore M. S., James D. C. (2015). Integrated cell and process engineering for improved transient production of a ”difficult-to-express fusion protein by CHO cells. Biotechnol. Bioeng. 112 2527–2542. 10.1002/bit.25687 [DOI] [PubMed] [Google Scholar]
- Kang Y., Zheng B., Shen B., Chen Y., Wang L., Wang J., et al. (2015). CRISPR/Cas9-mediated Dax1 knockout in the monkey recapitulates human AHC-HH. ıHum. Mol. Genet. 24 7255–7264. 10.1093/hmg/ddv425 [DOI] [PubMed] [Google Scholar]
- Kelly P. S., Gallagher C., Clynes M., Barron N. (2015). Conserved microRNA function as a basis for Chinese hamster ovary cell engineering. Biotechnol. Lett. 37 787–798. 10.1007/s10529-014-1751-7 [DOI] [PubMed] [Google Scholar]
- Kesavan G., Hammer J., Hans S., Brand M. (2018). Targeted knock-in of CreER T2 in zebrafish using CRISPR/Cas9. Cell Tissue Res. 372 41–50. 10.1007/s00441-018-2798-x [DOI] [PubMed] [Google Scholar]
- Kim J. Y., Kim Y. G., Lee G. M. (2012). CHO cells in biotechnology for production of recombinant proteins: current state and further potential. Appl. Microbiol. Biotechnol. 93 917–930. 10.1007/s00253-011-3758-5 [DOI] [PubMed] [Google Scholar]
- Kim S. H., Lee G. M. (2007). Functional expression of human pyruvate carboxylase for reduced lactic acid formation of Chinese hamster ovary cells (DG44). Appl. Microbiol. Biotechnol. 76 659–665. 10.1007/s00253-007-1041-6 [DOI] [PubMed] [Google Scholar]
- Kipniss N. H., Dingal P. D. P., Abbott T. R., Gao Y., Wang H., Dominguez A. A., et al. (2017). Engineering cell sensing and responses using a GPCR-coupled CRISPR-Cas system. Nat. Communi. 8:2212. 10.1038/s41467-017-02075-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T., Sakuma T., Wada H., Akimoto-Kato A., Yamamoto T., Hayashi S. (2014). TALEN-induced gene knock out in Drosophila. Dev. Growth Differ. 56 86–91. 10.1111/dgd.12097 [DOI] [PubMed] [Google Scholar]
- Ku S. C., Ng D. T., Yap M. G., Chao S. H. (2008). Effects of overexpression of X-box binding protein 1 on recombinant protein production in Chinese hamster ovary and NS0 myeloma cells. Biotechnol. Bioeng. 99 155–164. 10.1002/bit.21562 [DOI] [PubMed] [Google Scholar]
- Kunert R., Reinhart D. (2016). Advances in recombinant antibody manufacturing. Appl. Microbiol. Biotechnol. 100 3451–3461. 10.1007/s00253-016-7388-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le M. (2015). Gene Editing Saves Girl Dying from Leukaemia in World First. London: New Scientist Ltd. [Google Scholar]
- Li F., Vijayasankaran N., Shen A. Y., Kiss R., Amanullah A. (2010). Cell culture processes for monoclonal antibody production. MAbs 2 466–479. 10.4161/mabs.2.5.12720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. L., Fujimoto N., Sasakawa N., Shirai S., Ohkame T., Sakuma T., et al. (2015). Precise correction of the dystrophin gene in Duchenne muscular dystrophy patient induced pluripotent stem cells by TALEN and CRISPR-Cas9. Stem Cell Rep. 4 143–154. 10.1016/j.stemcr.2014.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Liu B., Spalding M. H., Weeks D. P., Yang B. (2012). High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat. Biotechnol. 30 390–392. 10.1038/nbt.2199 [DOI] [PubMed] [Google Scholar]
- Liao H. K., Gu Y., Diaz A., Marlett J., Takahashi Y., Li M., et al. (2015). Use of the CRISPR/Cas9 system as an intracellular defense against HIV-1 infection in human cells. Nat. Commun. 6:6413. 10.1038/ncomms7413 [DOI] [PubMed] [Google Scholar]
- Loh W. P., Loo B., Zhou L., Zhang P., Lee D. Y., Yang Y., et al. (2014). Overexpression of microRNAs enhances recombinant protein production in Chinese hamster ovary cells. Biotechnol. J. 9 1140–1151. 10.1002/biot.201400050 [DOI] [PubMed] [Google Scholar]
- Long C., McAnally J. R., Shelton J. M., Mireault A. A., Bassel-Duby R., Olson E. N. (2014). Prevention of muscular dystrophy in mice by CRISPR/Cas9–mediated editing of germline DNA. Science 345 1184–1188. 10.1126/science.1254445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q., Livi G. P., Modha S., Yusa K., Macarrón R., Dow D. J. (2017). Applications of CRISPR genome editing technology in drug target identification and validation. Expert Opin. Drug Discov. 12 541–552. 10.1080/17460441.2017.1317244 [DOI] [PubMed] [Google Scholar]
- Luo J. (2016). CRISPR/Cas9: from genome engineering to cancer drug discovery. Trends Cancer 2 313–324. 10.1016/j.trecan.2016.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magana-Ortiz D., Fernández F., Loske A. M., Gómez-Lim M. A. (2018). Extracellular expression in Aspergillus niger of an antibody fused to Leishmania sp. antigens. Curr. Microbiol. 75 40–48. 10.1007/s00284-017-1348-1 [DOI] [PubMed] [Google Scholar]
- Mahmuda A., Bande F., Al-Zihiry K. J. K., Abdulhaleem N., Majid R. A., Hamat R. A., et al. (2017). Monoclonal antibodies: a review of therapeutic applications and future prospects. Trop. J. Pharm. Res. 16 713–722. 10.4314/tjpr.v16i3.29 [DOI] [Google Scholar]
- Marston H. D., Paules C. I., Fauci A. S. (2018). Monoclonal antibodies for emerging infectious diseases—borrowing from history. N. Engl. J. Med. 378 1469–1472. 10.1056/NEJMp1802256 [DOI] [PubMed] [Google Scholar]
- Meleady P., Gallagher M., Clarke C., Henry M., Sanchez N., Barron N., et al. (2012). Impact of miR-7 over-expression on the proteome of Chinese hamster ovary cells. J. Biotechnol. 160 251–262. 10.1016/j.jbiotec.2012.03.002 [DOI] [PubMed] [Google Scholar]
- Mohan C., Kim Y. G., Koo J., Lee G. M. (2008). Assessment of cell engineering strategies for improved therapeutic protein production in CHO cells. Biotechnol. J. 3 624–630. 10.1002/biot.200700249 [DOI] [PubMed] [Google Scholar]
- Mohan C., Park S. H., Chung J. Y., Lee G. M. (2007). Effect of doxycycline-regulated protein disulfide isomerase expression on the specific productivity of recombinant CHO cells: thrombopoietin and antibody. Biotechnol. Bioeng. 98 611–615. 10.1002/bit.21453 [DOI] [PubMed] [Google Scholar]
- Moreno A. M., Mali P. (2017). Therapeutic genome engineering via CRISPR-Cas systems. Wiley Interdiscip. Rev. Syst. Biol. Med. 9:e1380. 10.1002/wsbm.1380 [DOI] [PubMed] [Google Scholar]
- Mori K., Iida S., Yamane-Ohnuki N., Kanda Y., Kuni-Kamochi R., Nakano R., et al. (2007). Non-fucosylated therapeutic antibodies: the next generation of therapeutic antibodies. Cytotechnology 55 109–114. 10.1007/s10616-007-9103-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morineau C., Bellec Y., Tellier F., Gissot L., Kelemen Z., Nogué F., et al. (2017). Selective gene dosage by CRISPR-Cas9 genome editing in hexaploid Camelina sativa. Plant Biotechnol. J. 15 729–739. 10.1111/pbi.12671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimiya D., Mano T., Miyadai K., Yoshida H., Takahashi T. (2013). Overexpression of CHOP alone and in combination with chaperones is effective in improving antibody production in mammalian cells. Appl. Microbiol. Biotechnol. 97 2531–2539. 10.1007/s00253-012-4365-9 [DOI] [PubMed] [Google Scholar]
- Noh S. M., Park J. H., Lim M. S., Kim J. W., Lee G. M. (2017). Reduction of ammonia and lactate through the coupling of glutamine synthetase selection and downregulation of lactate dehydrogenase-A in CHO cells. Appl. Microbiol. Biotechnol. 101 1035–1045. 10.1007/s00253-016-7876-y [DOI] [PubMed] [Google Scholar]
- Nylen A., Chen M. T. (2018). “Production of full-length antibody by Pichia pastoris,” in Recombinant Glycoprotein Production. Methods in Molecular Biology Vol. 1674 eds Picanço-Castro V., Swiech K. (New York, NY: Humana Press; ) 37–48. 10.1007/978-1-4939-7312-5_3 [DOI] [PubMed] [Google Scholar]
- Ouellet D. L., Cherif K., Rousseau J., Tremblay J. P. (2017). Deletion of the GAA repeats from the human frataxin gene using the CRISPR-Cas9 system in YG8R-derived cells and mouse models of Friedreich ataxia. Gene Ther. 24 265–274. 10.1038/gt.2016.89 [DOI] [PubMed] [Google Scholar]
- Pancera M., Shahzad-Ul-Hussan S., Doria-Rose N. A., McLellan J. S., Bailer R. T., Dai K., et al. (2013). Structural basis for diverse N-glycan recognition by HIV-1-neutralizing V1-V2-directed antibody PG16. Nat. Struct. Mol. Biol. 20 804–813. 10.1038/nsmb.2600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H. S., Kim I. H., Kim I. Y., Kim K. H., Kim H. J. (2000). Expression of carbamoyl phosphate synthetase I and ornithine transcarbamoylase genes in Chinese hamster ovary dhfr-cells decreases accumulation of ammonium ion in culture media. J. Biotecnol. 81 129–140. 10.1016/S0168-1656(00)00282-0 [DOI] [PubMed] [Google Scholar]
- Patel T. (2017). Manipulation and Exploitation of MicroRNAs for Enhanced Recombinant Protein Production in Chinese Hamster Ovary Cells. Ph.D. thesis, University of Kent; Canterbury. [Google Scholar]
- Platt R. J., Chen S., Zhou Y., Yim M. J., Swiech L., Kempton H. R., et al. (2014). CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell 159 440–455. 10.1016/j.cell.2014.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popp O., Moser S., Zielonka J., Rüger P., Hansen S., Plöttner O. (2018). Development of a pre-glycoengineered CHO-K1 host cell line for the expression of antibodies with enhanced Fc mediated effector function. MAbs 10 290–303. 10.1080/19420862.2017.1405203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pybus L. P., Dean G., West N. R., Smith A., Daramola O., Field R., et al. (2014). Model-directed engineering of “difficult-to-express” monoclonal antibody production by Chinese hamster ovary cells. Biotechnol. Bioeng. 111 372–385. 10.1002/bit.25116 [DOI] [PubMed] [Google Scholar]
- Ran F. A., Cong L., Yan W. X., Scott D. A., Gootenberg J. S., Kriz A. J., et al. (2015). In vivo genome editing using Staphylococcus aureus Cas9. Nature 520 186–191. 10.1038/nature14299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez N., Kelly P., Gallagher C., Lao N. T., Clarke C., Clynes M., et al. (2014). CHO cell culture longevity and recombinant protein yield are enhanced by depletion of miR-7 activity via sponge decoy vectors. Biotechnol. J. 9 396–404. 10.1002/biot.201300325 [DOI] [PubMed] [Google Scholar]
- Sander J. D., Cade L., Khayter C., Reyon D., Peterson R. T., Joung J. K., et al. (2011a). Targeted gene disruption in somatic zebrafish cells using engineered TALENs. Nat. Biotechnol. 29 697–698. 10.1038/nbt.1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander J. D., Dahlborg E. J., Goodwin M. J., Cade L., Zhang F., Cifuentes D., et al. (2011b). Selection-free zinc-finger-nuclease engineering by context-dependent assembly (CoDA). Nat. Methods 8 67–69. 10.1038/nmeth.1542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander J. D., Joung J. K. (2014). CRISPR-Cas systems for editing, regulating and targeting genomes. Nat. Biotechnol. 32 347–355. 10.1038/nbt.2842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandig G., von Horsten H. H., Radke L., Blanchard V., Frohme M., Giese C., et al. (2017). Engineering of CHO cells for the production of recombinant glycoprotein vaccines with xylosylated N-glycans. Bioengineering 4:e38. 10.3390/bioengineering4020038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seed K. D., Lazinski D. W., Calderwood S. B., Camilli A. (2013). A bacteriophage encodes its own CRISPR/Cas adaptive response to evade host innate immunity. Nature 494 489–491. 10.1038/nature11927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., Gao H., Wang H., Lafitte H. R., Archibald R. L., Yang M., et al. (2017). ARGOS8 variants generated by CRISPR-Cas9 improve maize grain yield under field drought stress conditions. Plant Biotechnol. J. 15 207–216. 10.1111/pbi.12603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields R. L., Lai J., Keck R., O’Connell L. Y., Hong K., Meng Y. G., et al. (2002). Lack of fucose on human IgG1 N-linked oligosaccharide improves binding to human FcγRIII and antibody-dependent cellular toxicity. J. Biol. Chem. 277 26733–26740. 10.1074/jbc.M202069200 [DOI] [PubMed] [Google Scholar]
- Shriver-Lake L. C., Goldman E. R., Zabetakis D., Anderson G. P. (2017). Improved production of single domain antibodies with two disulfide bonds by co-expression of chaperone proteins in the Escherichia coli periplasm. J. Immunol. Methods 443 64–67. 10.1016/j.jim.2017.01.007 [DOI] [PubMed] [Google Scholar]
- Strotbek M., Florin L., Koenitzer J., Tolstrup A., Kaufmann H., Hausser A., et al. (2013). Stable microRNA expression enhances therapeutic antibody productivity of Chinese hamster ovary cells. Metab. Eng. 20 157–166. 10.1016/j.ymben.2013.10.005 [DOI] [PubMed] [Google Scholar]
- Tan H. K., Lee M. M., Yap M. G., Wang D. I. (2008). Overexpression of cold-inducible RNA-binding protein increases interferon-γ production in Chinese-hamster ovary cells. Biotechnol. Appl. Biochem. 49 247–257. 10.1042/BA20070032 [DOI] [PubMed] [Google Scholar]
- Tu Z., Yang W., Yan S., Guo X., Li X. J. (2015). CRISPR/Cas9: a powerful genetic engineering tool for establishing large animal models of neurodegenerative diseases. ıMol. Neurodegener. 10:35. 10.1186/s13024-015-0031-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueta R., Abe C., Watanabe T., Sugano S. S., Ishihara R., Ezura H., et al. (2017). Rapid breeding of parthenocarpic tomato plants using CRISPR/Cas9. Sci. Rep. 7:507. 10.1038/s41598-017-00501-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallée C., Durocher Y., Henry O. (2014). Exploiting the metabolism of PYC expressing HEK293 cells in fed-batch cultures. J. Biotechnol. 169 63–70. 10.1016/j.jbiotec.2013.11.002 [DOI] [PubMed] [Google Scholar]
- Wang C., Sun W., Ye Y., Hu Q., Bomba H. N., Gu Z. (2017). In situ activation of platelets with checkpoint inhibitors for post-surgical cancer immunotherapy. Nat. Biomed. Eng. 1:0011 10.1038/s41551-016-0011 [DOI] [Google Scholar]
- Wang C., Ye Y., Hochu G. M., Sadeghifar H., Gu Z. (2016). Enhanced cancer immunotherapy by microneedle patch-assisted delivery of anti-PD1 antibody. Nano Lett. 16 2334–2340. 10.1021/acs.nanolett.5b05030 [DOI] [PubMed] [Google Scholar]
- Wang H., Yang H., Shivalila C. S., Dawlaty M. M., Cheng A. W., Zhang F., et al. (2013). One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153 910–918. 10.1016/j.cell.2013.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Geng L., Yuan M., Wei J., Jin C., Li M., et al. (2017). Deletion of a target gene in Indica rice via CRISPR/Cas9. Plant Cell Rep. 36 1333–1343. 10.1007/s00299-017-2158-4 [DOI] [PubMed] [Google Scholar]
- Wells E., Robinson A. S. (2017). Cellular engineering for therapeutic protein production: product quality, host modification, and process improvement. Biotechnol. J. 12:1600105. 10.1002/biot.201600105 [DOI] [PubMed] [Google Scholar]
- Wu Y., Liang D., Wang Y., Bai M., Tang W., Bao S., et al. (2013). Correction of a genetic disease in mouse via use of CRISPR-Cas9. Cell Stem Cell. 13 659–662. 10.1016/j.stem.2013.10.016 [DOI] [PubMed] [Google Scholar]
- Wu Y., Zhou H., Fan X., Zhang Y., Zhang M., Wang Y., et al. (2015). Correction of a genetic disease by CRISPR-Cas9-mediated gene editing in mouse spermatogonial stem cells. Cell Res. 25 67–79. 10.1038/cr.2014.160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S., Shiloach J., Betenbaugh M. J. (2014). Engineering cells to improve protein expression. Curr. Opin. Struct. Biol. 26 32–38. 10.1016/j.sbi.2014.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y., Wang D., Lan F., Wei G., Ni T., Chai R., et al. (2017). An episomal vector-based CRISPR/Cas9 system for highly efficient gene knockout in human pluripotent stem cells. Sci. Rep. 7:2320. 10.1038/s41598-017-02456-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong X., Chen M., Lim W. A., Zhao D., Qi L. S. (2016). CRISPR/Cas9 for human genome engineering and disease research. Annu. Rev. Genomics Hum. Genet. 17 131–154. 10.1146/annurev-genom-083115-022258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W., Li H., Wang Q., Hua C., Zhang H., Li W., et al. (2017). Advancements in developing strategies for sterilizing and functional HIV cures. Biomed. Res. Int. 2017:6096134. 10.1155/2017/6096134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue W., Chen S., Yin H., Tammela T., Papagiannakopoulos T., Joshi N. S., et al. (2014). CRISPR-mediated direct mutation of cancer genes in the mouse liver. Nature 514 380–384. 10.1038/nature13589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane-Ohnuki N., Kinoshita S., Inoue-Urakubo M., Kusunoki M., Iida S., Nakano R., et al. (2004). Establishment of FUT8 knockout Chinese hamster ovary cells: an ideal host cell line for producing completely defucosylated antibodies with enhanced antibody-dependent cellular cytotoxicity. Biotechnol. Bioeng. 87 614–622. 10.1002/bit.20151 [DOI] [PubMed] [Google Scholar]
- Yang S., Ding S., Xu Q., Li X., Xiong Q. (2017). Genetic manipulation by zinc-finger nucleases in rat-induced pluripotent stem cells. Cell. Reprogram. 19 180–188. 10.1089/cell.2016.0028 [DOI] [PubMed] [Google Scholar]
- Ye Y., Wang J., Hu Q., Hochu G. M., Xin H., Wang C., et al. (2016). Synergistic transcutaneous immunotherapy enhances antitumor immune responses through delivery of checkpoint inhibitors. ACS Nano 10 8956–8963. 10.1021/acsnano.6b04989 [DOI] [PubMed] [Google Scholar]
- Yusufi F. N. K., Lakshmanan M., Ho Y. S., Loo B. L. W., Ariyaratne P., Yang Y., et al. (2017). Mammalian systems biotechnology reveals global cellular adaptations in a recombinant CHO cell line. Cell Syst. 4 530–542.e6. 10.1016/j.cels.2017.04.009 [DOI] [PubMed] [Google Scholar]
- Zamaraev A. V., Kopeina G. S., Prokhorova E. A., Zhivotovsky B., Lavrik I. N. (2017). Post-translational modification of caspases: the other side of apoptosis regulation. Trends Cell Biol. 27 322–339. 10.1016/j.tcb.2017.01.003 [DOI] [PubMed] [Google Scholar]
- Zhang F., Sun X., Yi X., Zhang Y. (2006). Metabolic characteristics of recombinant Chinese hamster ovary cells expressing glutamine synthetase in presence and absence of glutamine. Cytotechnology 51 21–28. 10.1007/s10616-006-9010-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Jia R., Palange N. J., Satheka A. C., Togo J., An Y., et al. (2015). Large genomic fragment deletions and insertions in mouse using CRISPR/Cas9. PLoS One 10:e0120396. 10.1371/journal.pone.0120396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Han L., Zong H., Ding K., Yuan Y., Bai J., et al. (2018). Enhanced production of anti-PD1 antibody in CHO cells through transient co-transfection with anti-apoptotic genes Bcl-x L and Mcl-1. Bioprocess Biosyst. Eng. 41 633–640. 10.1007/s00449-018-1898-z [DOI] [PubMed] [Google Scholar]
- Zhang X. H., Tee L. Y., Wang X. G., Huang Q. S., Yang S. H. (2015). Off-target effects in CRISPR/Cas9-mediated genome engineering. Mol. Ther. Nucleic Acids 4:e264. 10.1038/mtna.2015.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhen S., Hua L., Liu Y. H., Gao L. C., Fu J., Wan D. Y., et al. (2015). Harnessing the clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated Cas9 system to disrupt the hepatitis B virus. Gene Ther. 22 404–412. 10.1038/gt.2015.2 [DOI] [PubMed] [Google Scholar]
- Zhou M., Crawford Y., Ng D., Tung J., Pynn A. F., Meier A., et al. (2011). Decreasing lactate level and increasing antibody production in Chinese Hamster Ovary cells (CHO) by reducing the expression of lactate dehydrogenase and pyruvate dehydrogenase kinases. J. Biotechnol. 153 27–34. 10.1016/j.jbiotec.2011.03.003 [DOI] [PubMed] [Google Scholar]
- Zhu J. (2012). Mammalian cell protein expression for biopharmaceutical production. Biotechnol. Adv. 30 1158–1170. 10.1016/j.biotechadv.2011.08.022 [DOI] [PubMed] [Google Scholar]
- Zustiak M. P., Jose L., Xie Y., Zhu J., Betenbaugh M. J. (2014). Enhanced transient recombinant protein production in CHO cells through the co-transfection of the product gene with Bcl-xL. Biotechnol. J. 9 1164–1174. 10.1002/biot.201300468 [DOI] [PMC free article] [PubMed] [Google Scholar]


