Abstract
CXXC5 is a newly identified CXXC-type zinc finger family protein, which is encoded by the CXXC5 gene localised to the 5q31.3 chromosomal region. Previous studies revealed that CXXC5 is associated with various malignant tumours. The aim of the present study was to investigate the prognosis prediction of CXXC5 in different breast cancer subtypes via the Gene Expression Omnibus database and bc-GenExMiner. CXXC5 overexpression was observed as associated with a poor prognosis for oestrogen receptor positive (ER+) breast cancer. Basal-like breast cancer and triple-negative breast cancer also suggest a poor prognosis, however their CXXC5 expression was low and could not be used as a prognostic factor. The CXXC5 correlated genes and their enriched Gene Ontology (GO) terms were obtained. Among those enriched GO terms, GO:0070062 (extracellular exosome) had the greatest number of associated genes and the associated genes of GO:0000122 (negative regulation of transcription from RNA polymerase II promoter) and GO:0008134 (transcription factor binding) contained CXXC5. These results suggest that overexpression of CXXC5 is a strongly poor prognostic factor in ER+ breast cancer. However, the role of CXXC5 in breast cancer requires further investigation.
Keywords: CXXC5, breast cancer, prognosis, biomarker, bc-GenExMiner
Introduction
Breast cancer is one of the most common malignant tumours among women (1). There were precise data from 2012 related to breast cancer incidence and mortality. Across 5 years, from 2008 to 2012, the average incidence rates for white women were the highest, followed by those of black women (2,3). The current 5-year survival rate of primary breast cancer is relatively high, ranging from 80 to 92% in different populations (4). However, it decreases to <25% when the disease becomes metastatic (4,5). The most important factor to improve the survival rate of patients is to find the most effective treatment, which is guided by tumour cell characteristics (6,7). Once a metastatic lesion is found, accurate characterisation of the tumour cells must be obtained at the start of treatment (8); a possible way to do this is the use of biomarkers (9). Currently, a series of different biomarkers, such as tissue markers, genetic markers, serum markers and non-coding RNA (1,10,11), have been found, but it is much more difficult to assess the effectiveness of the targeted treatment or prognosis of the disease. Therefore, we need to find more biomarkers and determine their clinical utility in future research (9).
CXXC finger protein 5 (CXXC5) is a protein encoded by the CXXC5 gene localised to the 5q31.3 chromosomal region, which is often deleted in myeloid leukaemia (12). Kühnl et al (13) reported that CXXC5 could suppress progression of acute myeloid leukaemia (AML) via inhibiting the Wnt pathway and that downregulation of CXXC5 could predict a better prognosis in AML. The study of Bruserud et al (14) showed that high CXXC5 expression was related to the stem cell signature of AML that has a bad prognostic impact. We know that 17β-oestradiol (E2) plays an important role in the homeodynamic regulation of breast tissue functions, and the oestrogen receptor (ERα) is the primary transcript expressed in breast tissue. Yasar et al (15) reported that E2-ERα could regulate the expression of CXXC5. Therefore, we knew that there was a certain relationship between CXXC5 and breast cancer. Knappskog et al (16) reported that the overexpression of CXXC5 was significantly associated with a bad prognosis in breast cancer. However, the prognostic implications of CXXC5 expression in breast cancers of different molecular types remain unclear. In our study, we used Breast Cancer Gene-Expression Miner v4.0 (bc-GenExMiner v4.0, bcgenex.centregauducheau.fr/BC-GEM/GEM-Accueil.php?js=1) (17), a database that includes a total of 5,861 patients, as the main tool to analyse the role of CXXC5 expression in different breast cancer subtypes. We aimed to show that CXXC5 expression predicts the prognosis of different breast cancer subtypes.
Materials and methods
GEO data analysis
We obtained the dataset of GDS5666 (18) from Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) (19) and analysed it using the Data Analysis Tools of DATASET BROWSER in GEO. Probes of A_51_P234788 (ID_REF) and A_52_P633393 (ID_REF) represented the CXXC5 gene in the platform of GPL7202. We obtained 2 sets of CXXC5 mRNA expression values from these probes. We used the average value of each sample's CXXC5 mRNA expression as the expression value for that sample.
Bioinformatics analysis by bc-GenExMiner v4.0
Using bc-GenExMiner v4.0, we conducted CXXC5 expression analysis, prognostic analysis for CXXC5 through univariate Cox analysis and Kaplan-Meier curve analysis, and gene correlation analysis for CXXC5. Then, we obtained the gene ontology (GO) term results through gene correlation exhaustive analysis. The database of bc-GenExMiner v4.0 had 36 datasets, including a total of 5,861 patients. There were 21 datasets including 3,524 patients for CXXC5 expression analysis and gene correlation analysis among a total of 36 datasets. A total of 3,472 patients from 21 datasets were used for prognostic analysis for CXXC5 with any nodal status, any ER status and any event (AE).
Statistical analysis
In the comparison of CXXC5 expression in primary and metastatic tumours, we used SPSS version 19.0 (IBM SPSS, Armonk, NY, USA) as the software for statistical analysis. Two-tailed unpaired t-tests were used for statistical comparisons. Data are represented as the means ± standard error of the mean. P<0.05 was considered significant. In other research, the statistical analysis for comparison of CXXC5 expression according to ER and Kaplan-Meier survival curves and univariate Cox analysis was performed by bc-GenExMiner v4.0. Box and whiskers plots are displayed, along with Dunett-Tukey-Kramer's test and Welch's t-test for every possible clinical criteria for CXXC5 gene.
Results
CXXC5 expression is increased in 4T1-derived metastatic cancer compared to primary cancer
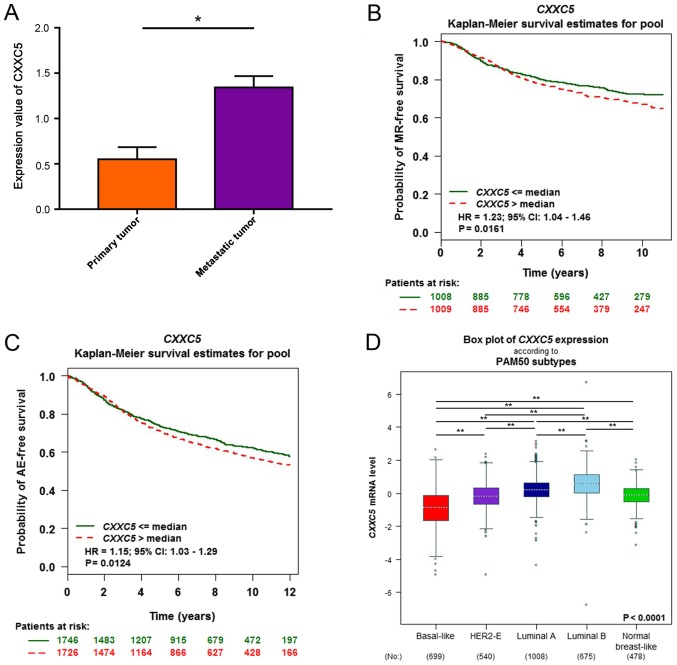
We observed that the expression values of CXXC5 were higher in 4T1-derived metastatic populations than in primary cancers (Fig. 1A). Then, we used bc-GenExMiner v4.0 to determine that CXXC5 upregulation with metastatic relapse (MR) or AE was associated with a poor prognosis of breast cancer (Fig. 1B and C). In the PAM50 breast cancer subtypes, the basal-like subtype had the lowest CXXC5 expression, and CXXC5 expression of luminal tumours was higher than in other types (Fig. 1D).
Figure 1.
CXXC5 expression is increased in 4T1-derived metastatic cancer compared to primary cancer. (A) Expression value of CXXC5 in 4T1-derived metastatic cancer, compared with primary breast cancer, through data analysis of GDS5666. Student's t-test; *P<0.05. The Kaplan-Meier curve of breast cancer with (B) MR-free (C) and AE-free CXXC5 expression. Data were obtained and analysed by using bc-GenExMiner v4.0. (D) Box plot of CXXC5 mRNA expression according to PAM50 subtype in bc-GenExMiner v4.0. Dunnett-Tukey-Kramer's test; **P<0.0001. CXXC5, CXXC finger protein 5; MR, metastatic relapse; AE, any event.
High level of CXXC5 is a poor prognostic factor in oestrogen receptor positive (ER+) breast cancer
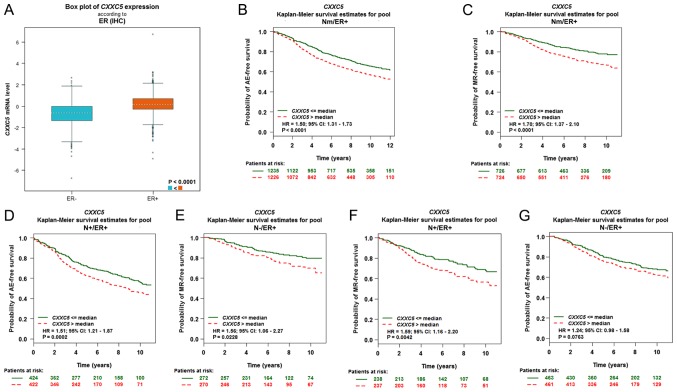
The impact of CXXC5 in breast cancer was considered robust because there were 10 significant results (P<0.05) among the 18 given results (Table I). We determined that the high level of CXXC5 expression is associated with poor prognosis of breast cancer with Nm/ER+/AE, Nm/ER+/MR, N+/ER+/AE, N-/ER+/MR, N+/ER+/MR, Nm/ERm/MR, Nm/ERm/AE, N+/ERm/AE, N+/ER-/AE and N-/ER+/AE through CXXC5 univariate Cox analysis (Table I). In particular, all breast cancer patients with ER+ status had a poor prognosis. CXXC5 expression was significantly higher in ER+ breast cancer than in ER-breast cancer (Fig. 2A). Using the Kaplan-Meier curve, we ascertained that high CXXC5 expression predicted significantly poor AE-free survival in Nm/ER+ status (HR=1.50; 95% CI, 1.31–1.73; P<0.0001) (Fig. 2B), MR-free survival in Nm/ER+ status (HR=1.70; 95% CI, 1.37–2.10; P<0.0001) (Fig. 2C), AE-free survival in N+/ER+ status (HR=1.51; 95% CI, 1.21–1.87; P=0.0002) (Fig. 2D), MR-free survival in N-/ER+ status (HR=1.56; 95% CI, 1.06–2.27; P=0.0228) (Fig. 2E) and MR-free survival in N+/ER+ status (HR=1.59; 95% CI, 1.16–2.20; P=0.0042) (Fig. 2F). However, CXXC5 expression could not predict AE-free survival in N-/ER+ status (P=0.0763) (Fig. 2G).
Table I.
CXXC5 univariate Cox analysis.
| No. | Nodal status | ER status | Event status | P-value | Hazard ratio | 95% CI | No. patients | No. events |
|---|---|---|---|---|---|---|---|---|
| 1 | Nm | ER+ | AE | <0.0001 | 1.31 | 1.22–1.42 | 2,461 | 845 |
| 2 | Nm | ER+ | MR | <0.0001 | 1.53 | 1.34–1.74 | 1,450 | 355 |
| 3 | N+ | ER+ | AE | 0.0001 | 1.30 | 1.14–1.48 | 846 | 348 |
| 4 | N- | ER+ | MR | 0.0002 | 1.61 | 1.25–2.08 | 542 | 113 |
| 5 | N+ | ER+ | MR | 0.0009 | 1.35 | 1.13–1.61 | 475 | 156 |
| 6 | Nm | ERm | MR | 0.0027 | 1.15 | 1.05–1.26 | 2,017 | 539 |
| 7 | Nm | ERm | AE | 0.0057 | 1.09 | 1.02–1.15 | 3,472 | 1,260 |
| 8 | N+ | ERm | AE | 0.0162 | 1.13 | 1.02–1.25 | 1,127 | 503 |
| 9 | N+ | ER- | AE | 0.0201 | 1.26 | 1.04–1.53 | 278 | 155 |
| 10 | N- | ER+ | AE | 0.0347 | 1.15 | 1.01–1.32 | 924 | 277 |
| 11 | N+ | ERm | MR | 0.0540 | 1.15 | 1.00–1.32 | 612 | 224 |
| 12 | N+ | ER- | MR | 0.0540 | 1.32 | 1.00–1.75 | 135 | 68 |
| 13 | Nm | ER- | AE | 0.0784 | 1.10 | 0.99–1.22 | 972 | 406 |
| 14 | Nm | ER- | MR | 0.1103 | 1.13 | 0.97–1.32 | 547 | 181 |
| 15 | N- | ERm | MR | 0.1563 | 1.12 | 0.96–1.32 | 762 | 167 |
| 16 | N- | ER- | MR | 0.5774 | 1.08 | 0.82–1.42 | 205 | 53 |
| 17 | N- | ER- | AE | 0.7933 | 0.98 | 0.81–1.17 | 361 | 118 |
| 18 | N- | ERm | AE | 0.8053 | 0.99 | 0.89–1.09 | 1,306 | 399 |
CXXC5, CXXC finger protein 5; ER, estrogen receptor; CI, confidence interval; m, mixed; +, positive; -, negative; AE, any event; MR, metastatic relapse. Bold text indicates P<0.05.
Figure 2.
High levels of CXXC5 is a poor prognostic factor in ER+ breast cancer. (A) Box plot of CXXC5 mRNA expression according to ER. The Kaplan-Meier curves of breast cancer with (B) Nm/ER+/AE, (C) Nm/ER+/MR, (D) N+/ER+/AE, (E) N-/ER+/MR, (F) N+/ER+/MR and (G) N-/ER+/AE. All data were obtained and analysed by using bc-GenExMiner v4.0. N, nodal status; ER, oestrogen receptor status; +, positive; -, negative; m, mixed.
Basal-like breast cancer and/or TNBC prognostic analysis for CXXC5
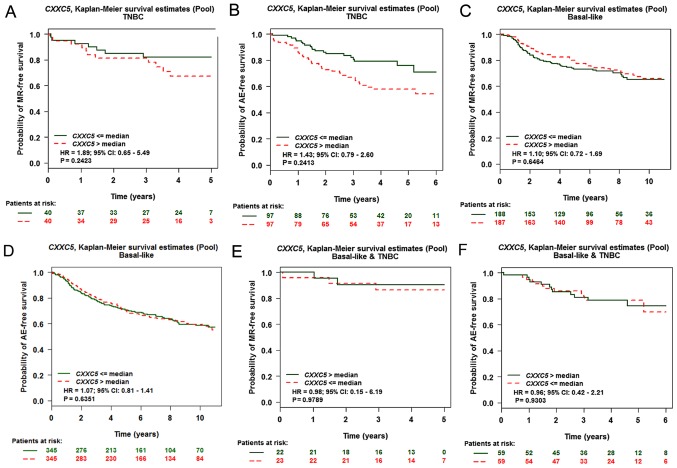
The basal-like breast cancer had lower CXXC5 expression than other subtypes (Fig. 1D). However, basal-like breast cancer could predict bad prognosis, as 75–80% of the triple-negative breast cancers (TNBC) belonged to the group of basal-like breast cancer (20). High levels of CXXC5 expression could predict a bad prognosis in TNBC with MR via CXXC5 univariate Cox analysis (basal-like and/or TNBC) but not in the other group (Tables II and III). However, CXXC5 expression was not associated with prognosis of TNBC with MR through the Kaplan-Meier curve analysis (Fig. 3A). Consistent with the results of CXXC5 univariate Cox analysis (basal-like and/or TNBC), CXXC5 expression was not associated with the prognosis of breast cancer in the other groups (Fig. 3B-F).
Table II.
Univariate Cox analysis (basal-like and/or TNBC) for CXXC5 with MR.
| Population | P-value | HR | 95% CI | No. patients | No. MR |
|---|---|---|---|---|---|
| Basal-like | 0.8288 | 1.02 | 0.84–1.24 | 375 | 109 |
| TNBC | 0.0369 | 1.66 | 1.03–2.67 | 80 | 18 |
| Basal-like + TNBC | 0.7465 | 1.24 | 0.34–4.54 | 45 | 5 |
CXXC5, CXXC finger protein 5; TNBC, triple-negative breast cancer; CI, confidence interval; MR, metastatic relapse. Bold text indicates P<0.05.
Table III.
Univariate Cox analysis (basal-like and/or TNBC) for CXXC5 with AE.
| Population | P-value | HR | 95% CI | No. patients | No. AE |
|---|---|---|---|---|---|
| Basal-like | 0.8185 | 0.99 | 0.87–1.12 | 690 | 251 |
| TNBC | 0.4321 | 1.12 | 0.84–1.50 | 194 | 58 |
| Basal-like + TNBC | 0.7343 | 0.92 | 0.55–1.52 | 118 | 24 |
CXXC5, CXXC finger protein 5; CI, confidence interval; m, mixed; AE, any event; TNBC, triple-negative breast cancer.
Figure 3.
Expression of CXXC5 could not predict the prognosis of basal-like breast cancer and/or TNBC. The Kaplan-Meier curves of TNBC with (A) MR and (B) AE, basal-like breast cancer with (C) MR and (D) AE and basal-like and TNBC with (E) MR and (F) AE. All data were obtained and analysed by using bc-GenExMiner v4.0. TNBC, triple-negative breast cancer; MR, metastatic relapse; AE, any event.
Correlated genes with CXXC5
We obtained the correlated genes with CXXC5 in breast cancer through gene correlation exhaustive analysis. Table IV shows the top 10 best positive/negative correlations with CXXC5. Then, we obtained the GO enrichments of the correlated genes with CXXC5 via GO analysis of bc-GenExMiner v4.0 (Table V). Among them, GO:0070062 (extracellular exosome) had the most associated genes, and the associated genes of both GO:0000122 (negative regulation of transcription from RNA polymerase II promoter) and GO:0008134 (transcription factor binding) contained CXXC5.
Table IV.
Top 10 best positive/negative correlated genes with CXXC5.
| Gene symbol | Pearson's correlation coefficient | P-value | No. of patients |
|---|---|---|---|
| Positive correlation | |||
| FKBP9P1 | 0.5858 | <0.0001 | 214 |
| LOC149401 | 0.5849 | <0.0001 | 155 |
| LOC100288069 | 0.5806 | <0.0001 | 214 |
| ACTG1P20 | 0.5784 | <0.0001 | 326 |
| CA12 | 0.5474 | <0.0001 | 3,524 |
| FOXA1 | 0.5377 | <0.0001 | 3,524 |
| GATA3 | 0.5338 | <0.0001 | 3,524 |
| AGR2 | 0.5287 | <0.0001 | 3,524 |
| PRINS | 0.5245 | <0.0001 | 155 |
| AGR3 | 0.5235 | <0.0001 | 3,023 |
| Negative correlation | |||
| FLJ44715 | −0.8160 | <0.0001 | 155 |
| LOC100507412 | −0.8069 | <0.0001 | 155 |
| LOC100133683 | −0.7873 | <0.0001 | 155 |
| LOC729461 | −0.7741 | <0.0001 | 155 |
| LOC728543 | −0.7698 | <0.0001 | 155 |
| CEP170P1 | −0.6993 | <0.0001 | 155 |
| LOC729324 | −0.6529 | <0.0001 | 155 |
| CEP295NL | −0.6524 | <0.0001 | 155 |
| LOC653739 | −0.5980 | <0.0001 | 155 |
| LOC100507637 | −0.5718 | <0.0001 | 155 |
CXXC5, CXXC finger protein 5.
Table V.
GO enrichments of correlated genes with CXXC5.
| Significant terms | Description | P-value | Associated genes |
|---|---|---|---|
| Biological process | |||
| GO:1902236 | Negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway | 4.53×10−5 | WFS1, TMBIM6, XBP1 |
| GO:0000122 | Negative regulation of transcription from RNA polymerase II promoter | 7.14×10−4 | CXXC5, FOXA1, GATA3, WFS1, CCND1, SPDEF, DACH1, XBP1, BCL11A, LPIN1 |
| GO:0009653 | Anatomical structure morphogenesis | 2.25×10−3 | FOXA1, GATA3, KRT18, SOX10 |
| GO:0043627 | Response to estrogen | 2.40×10−3 | GATA3, ESR1, CCND1 |
| GO:0043433 | Negative regulation of sequence-specific DNA binding transcription factor activity | 2.64×10−3 | WFS1, ESR1, SIGIRR |
| Cellular component | 9.57×10−4 | ||
| GO:0005902 | Microvillus | 4.59×10−3 | FOXA1, STARD10, SLC9A3R1 |
| GO:0030176 | Integral component of endoplasmic reticulum membrane | 4.53×10−5 | WFS1, PIGT, XBP1 |
| GO:0070062 | Extracellular exosome | 6.21×10−3 | ANXA9, SLC9A3R1, KRT18, TFF3, WWP1, GFRA1, FBP1, MLPH, NME3, CMBL, H2AFJ, HAGH, PVRL2, HSPB1, SERPINA5, TSPAN1, GAMT, PSAT1, PM20D2, FAM171A1, SFT2D2 |
| GO:0071944 | Cell periphery | 7.21×10−3 | SLC9A3R1, KRT18 |
| Molecular function | 1.76×10−4 | ||
| GO:0008134 | Transcription factor binding | 4.10×10−3 | CXXC5, FOXA1, GATA3, ESR1, CCND1, FOXC1, SOX10 |
| GO:0000981 | Sequence-specific DNA binding RNA polymerase II transcription factor activity | 7.43×10−3 | FOXA1, SPDEF, XBP1, FOXC1 |
| GO:0001078 | RNA polymerase II core promoter proximal region sequence-specific DNA binding transcription factor activity involved in negative regulation of transcription | 8.49×10−3 | GATA3, DACH1, BCL11A |
| GO:0044212 | Transcription regulatory region DNA binding | 8.49E-03 | FOXA1, GATA3, XBP1, FOXC1 |
GO, gene ontology; CXXC5, CXXC finger protein 5; WFS1, wolframin ER transmembrane glycoprotein; TMBIM6, transmembrane BAX inhibitor motif containing 6; XBP1, X-Box binding protein 1; FOXA1, forkhead Box A1; GATA3, GATA binding protein 3; CCND1, Cyclin D1; SPDEF, SAM pointed domain containing ETS transcription factor; DACH1, dachshund family transcription factor 1; BCL11A, B-cell CLL/lymphoma 11A; LPIN1, Lipin 1; KRT18, keratin 18; SOX10, SRY-box 10; ESR1, estrogen receptor 1; SIGIRR, single Ig and TIR domain containing; STARD10, StAR related lLipid transfer domain containing 10; SLC9A3R1, SLC9A3 regulator 1; PIGT, phosphatidylinositol glycan anchor biosynthesis class T; ANXA9, Annexin A9; TFF3, trefoil factor 3; WWP1, WW domain containing E3 ubiquitin protein ligase 1; GFRA1, GDNF family receptor α1; FBP1, fructose-bisphosphatase 1; MLPH, melanophilin; NME3, NME/NM23 nucleoside diphosphate kinase 3; CMBL, carboxymethylenebutenolidase homolog; H2AFJ, H2A histone family member J; HAGH, hydroxyacylglutathione hydrolase; PVRL2, nectin cell adhesion molecule 2; HSPB1, heat shock protein family B (small) member 1; SERPINA5, serpin family A member 5; TSPAN1, tetraspanin 1; GAMT, guanidinoacetate N-methyltransferase; PSAT1, phosphoserine aminotransferase 1; PM20D2, peptidase M20 domain containing 2; FAM171A1, family with sequence similarity 171 member A1; SFT2D2, SFT2 domain containing 2; FOXC1, forkhead Box C1.
Discussion
CXXC5 is a newly identified CXXC-type zinc finger family protein (21), which is encoded by the CXXC5 gene localised to the 5q31.3 chromosomal region (12). Previous studies showed that CXXC5 was related to AML, myelodysplastic syndromes, human malignant peripheral nerve sheath tumours, prostate cancer, breast cancer, thyroid cancers and metastatic melanomas (16,22–25). Knappskog et al (16) used three independent public microarray datasets, including 599 patients from GEO, to find that CXXC5 was a bad prognostic factor in breast cancer. However, they did not study the effects of CXXC5 on various subtypes in breast cancer.
In our study, using the dataset of GDS5666 from GEO, we found that the expression of CXXC5 was higher in 4T1-derived metastatic populations than in primary breast cancers. Therefore, CXXC5 might be associated with metastasis. Then, we used expression analysis of bc-GenExMiner v4.0 to find that the expression of CXXC5 was significantly different in PAM subtypes. We established that the high level of CXXC5 expression is associated with poor prognosis of ER+ breast cancer through CXXC5 univariate Cox analysis and Kaplan-Meier curve analysis of bc-GenExMiner v4.0. These results propose CXXC5 as a biomarker and potential therapeutic target in ER+ breast cancer. Although basal-like breast cancer and TNBC could predict bad prognosis, their CXXC5 expression was low. In addition, CXXC5 could not predict their prognosis. Finally, we obtained the CXXC5 correlated genes and enriched GO terms of those genes through gene correlation exhaustive analysis of bc-GenExMiner v4.0. Among those enriched GO terms, GO:0070062 (extracellular exosome) had the most associated genes, and the associated genes of both GO:0000122 (negative regulation of transcription from RNA polymerase II promoter) and GO:0008134 (transcription factor binding) contained CXXC5. These GO terms can guide new investigations into understanding the mechanisms of CXXC5 in breast cancer and propose new treatments for ER+ breast cancer.
There is a limitation to the present study. The mechanism of CXXC5 in breast cancer requires further investigation via in vitro and in vivo experiments.
In conclusion, we determined that overexpression of CXXC5 was a strongly poor prognostic factor in ER+ breast cancer through the tools of bc-GenExMiner V4.0 based on a database including a total of 5,861 patients. This means that regardless of the clinical stage of breast cancer, high expression of CXXC5 in patients predicts that the disease is more significantly invasive. As is known, gene expression can be measured in many ways. We hope that measuring the expression of CXXC5 may become a routine inspection to assess the prognosis of breast cancer in different patients. In this way, early intervention and treatment could be used, and the survival rate of patients could improve. However, the pathways of CXXC5 in breast cancer require further investigation. If in-depth research is conducted, we may find the pathways of CXXC5 in breast cancer, and then CXXC5 can be utilized as a potential therapeutic target.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The data that support the findings of this study are available from GEO database (GDS5666; www.ncbi.nlm.nih.gov/geo/) and Breast Cancer Gene-Expression Miner (bcgenex.centregauducheau.fr/BC-GEM/GEM-Accueil.php?js=1).
Authors' contributions
LF, YW and XC conceived and designed the study. LF and YG analyzed and interpreted the data. LF and YW were the contributors in writing the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Donepudi MS, Kondapalli K, Amos SJ, Venkanteshan P. Breast cancer statistics and markers. J Cancer Res Ther. 2014;10:506–511. doi: 10.4103/0973-1482.137927. [DOI] [PubMed] [Google Scholar]
- 2.DeSantis CE, Fedewa SA, Sauer Goding A, Kramer JL, Smith RA, Jemal A. Breast cancer statistics, 2015: Convergence of incidence rates between black and white women. CA Cancer J Clin. 2016;66:31–42. doi: 10.3322/caac.21320. [DOI] [PubMed] [Google Scholar]
- 3.DeSantis CE, Siegel RL, Sauer AG, Miller KD, Fedewa SA, Alcaraz KI, Jemal A. Cancer statistics for African Americans, 2016: Progress and opportunities in reducing racial disparities. CA Cancer J Clin. 2016;66:290–308. doi: 10.3322/caac.21340. [DOI] [PubMed] [Google Scholar]
- 4.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 5.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 6.Ufen MP, Köhne CH, Wischneswky M, Wolters R, Novopashenny I, Fischer J, Constantinidou M, Possinger K, Regierer AC. Metastatic breast cancer: Are we treating the same patients as in the past? Ann Oncol. 2014;25:95–100. doi: 10.1093/annonc/mdt429. [DOI] [PubMed] [Google Scholar]
- 7.Zervoudis S, Iatrakis G, Tomara E, Bothou A, Papadopoulos G, Tsakiris G. Main controversies in breast cancer. World J Clin Oncol. 2014;5:359–373. doi: 10.5306/wjco.v5.i3.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marino N, Woditschka S, Reed LT, Nakayama J, Mayer M, Wetzel M, Steeg PS. Breast cancer metastasis: Issues for the personalization of its prevention and treatment. Am J Pathol. 2013;183:1084–1095. doi: 10.1016/j.ajpath.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nalejska E, Mączyńska E, Lewandowska MA. Prognostic and predictive biomarkers: Tools in personalized oncology. Mol Diagn Ther. 2014;18:273–284. doi: 10.1007/s40291-013-0077-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gasparri ML, Casorelli A, Bardhi E, Besharat AR, Savone D, Ruscito I, Farooqi AA, Papadia A, Mueller MD, Ferretti E, Panici Benedetti P. Beyond circulating microRNA biomarkers: Urinary microRNAs in ovarian and breast cancer. Tumor Biol. 2017;39:1010428317695525. doi: 10.1177/1010428317695525. [DOI] [PubMed] [Google Scholar]
- 11.Ye N, Wang B, Quan ZF, Cao SJ, Wen XT, Huang Y, Huang XB, Wu R, Ma XP, Yan QG. Functional roles of long non-coding RNA in human breast cancer. Asian Pac J Cancer Prev. 2014;15:5993–5997. doi: 10.7314/APJCP.2014.15.15.5993. [DOI] [PubMed] [Google Scholar]
- 12.Pendino F, Nguyen E, Jonassen I, Dysvik B, Azouz A, Lanotte M, Ségal-Bendirdjian E, Lillehaug JR. Functional involvement of RINF, retinoid-inducible nuclear factor (CXXC5), in normal and tumoral human myelopoiesis. Blood. 2009;113:3172–3181. doi: 10.1182/blood-2008-07-170035. [DOI] [PubMed] [Google Scholar]
- 13.Kühnl A, Valk PJ, Sanders MA, Ivey A, Hills RK, Mills KI, Gale RE, Kaiser MF, Dillon R, Joannides M, et al. Downregulation of the Wnt inhibitor CXXC5 predicts a better prognosis in acute myeloid leukemia. Blood. 2015;125:2985–2994. doi: 10.1182/blood-2014-12-613703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bruserud Ø, Reikvam H, Fredly H, Skavland J, Hagen KM, van Hoang TT, Brenner AK, Kadi A, Astori A, Gjertsen BT, Pendino F. Expression of the potential therapeutic target CXXC5 in primary acute myeloid leukemia cells-high expression is associated with adverse prognosis as well as altered intracellular signaling and transcriptional regulation. Oncotarget. 2015;6:2794–2811. doi: 10.18632/oncotarget.3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yasar P, Ayaz G, Muyan M. Estradiol-estrogen receptor α mediates the expression of the CXXC5 gene through the estrogen response element-dependent signaling pathway. Sci Rep. 2016;6:37808. doi: 10.1038/srep37808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Knappskog S, Myklebust LM, Busch C, Aloysius T, Varhaug JE, Lønning PE, Lillehaug JR, Pendino F. RINF (CXXC5) is overexpressed in solid tumors and is an unfavorable prognostic factor in breast cancer. Ann Oncol. 2011;22:2208–2215. doi: 10.1093/annonc/mdq737. [DOI] [PubMed] [Google Scholar]
- 17.Jézéquel P, Campone M, Gouraud W, Guérin-Charbonnel C, Leux C, Ricolleau G, Campion L. bc-GenExMiner: An easy-to-use online platform for gene prognostic analyses in breast cancer. Breast Cancer Res Treat. 2012;131:765–775. doi: 10.1007/s10549-011-1457-7. [DOI] [PubMed] [Google Scholar]
- 18.Tabariès S, Ouellet V, Hsu BE, Annis MG, Rose AA, Meunier L, Carmona E, Tam CE, Mes-Masson AM, Siegel PM. Granulocytic immune infiltrates are essential for the efficient formation of breast cancer liver metastases. Breast Cancer Res. 2015;17:45. doi: 10.1186/s13058-015-0558-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, et al. NCBI GEO: Archive for functional genomics data sets-update. Nucleic Acids Res. 2013;41:D991–D995. doi: 10.1093/nar/gks1193. (Database Issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Badowska-Kozakiewicz AM, Budzik MP. Immunohistochemical characteristics of basal-like breast cancer. Contemp Oncol (Pozn) 2016;20:436–443. doi: 10.5114/wo.2016.56938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang M, Wang R, Wang Y, Diao F, Lu F, Gao D, Chen D, Zhai Z, Shu H. The CXXC finger 5 protein is required for DNA damage-induced p53 activation. Sci China C Life Sci. 2009;52:528–538. doi: 10.1007/s11427-009-0083-7. [DOI] [PubMed] [Google Scholar]
- 22.Astori A, Fredly H, Aloysius TA, Bullinger L, Mansat-De Mas V, de la Grange P, Delhommeau F, Hagen KM, Récher C, Dusanter-Fourt I, et al. CXXC5 (retinoid-inducible nuclear factor, RINF) is a potential therapeutic target in high-risk human acute myeloid leukemia. Oncotarget. 2013;4:1438–1448. doi: 10.18632/oncotarget.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stoddart A, Qian Z, Fernald AA, Bergerson RJ, Wang J, Karrison T, Anastasi J, Bartom ET, Sarver AL, McNerney ME, et al. Retroviral insertional mutagenesis identifies the del(5q) genes, CXXC5, TIFAB and ETF1, as well as the Wnt pathway, as potential targets in del(5q) myeloid neoplasms. Haematologica. 2016;101:e232–e236. doi: 10.3324/haematol.2015.139527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cui Z, Shen Y, Chen KH, Mittal SK, Yang JY, Zhang G. KANK1 inhibits cell growth by inducing apoptosis though regulating CXXC5 in human malignant peripheral nerve sheath tumors. Sci Rep. 2017;7:40325. doi: 10.1038/srep40325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bettin A, Reyes I, Reyes N. Gene expression profiling of prostate cancer-associated genes identifies fibromodulin as potential novel biomarker for prostate cancer. Int J Biol Markers. 2016;31:e153–e162. doi: 10.5301/jbm.5000184. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from GEO database (GDS5666; www.ncbi.nlm.nih.gov/geo/) and Breast Cancer Gene-Expression Miner (bcgenex.centregauducheau.fr/BC-GEM/GEM-Accueil.php?js=1).