Abstract
The glycoprotein dickkopf 1 (DKK1) is highly expressed in lung cancer cell lines and tissues. Our previous study demonstrated that DKK1 promoter activity is low in lung cancer cell lines. This may be because it lacks the necessary transcriptional regulatory elements (TREs) required for higher activity levels. However, it is difficult to computationally predict functionally significant TREs, as TREs from different locations can affect large segments of distant DNA. The Encyclopedia of DNA Elements project features multiple integrated technologies and approaches for the discovery and definition of functional elements, including enhancer elements and enhancer-blocking insulators. In the present study, DNase I hypersensitive sites and histone modifications of DKK1 were investigated in the A549 lung cancer cell line using the UCSC Genome Browser. A set of cis-acting enhancer elements were identified by a dual-luciferase reporter gene assay system to increase activity of the DKK1 promoter with lung cancer specificity. To the best of our knowledge, these data provide the first insight into the role of the DKK1 locus in lung cancer, and confirm the contribution of intronic cis-acting elements to the regulation of DKK1 expression, providing a new insight into gene regulation in lung cancer, which could inform the development of targeted therapy.
Keywords: dickkopf 1, transcriptional regulatory elements, lung cancer, enhancer, dual-luciferase reporter gene assay system
Introduction
Non-small cell lung cancer (NSCLC) is the most common type of lung cancer, and a major public health burden (1). Therefore, the molecular analysis of NSCLC is necessary to identify novel molecular targets for determining the disease prognosis and informing the design of targeted therapies.
There are 4 dickkopf (DKK) proteins: DKK1, 2, 3 and 4. DKK1 antagonizes the canonical Wnt signaling associated with the pathogenesis of malignant tumors by forming a ternary complex with LDL receptor-related proteins (2,3). High levels of DKK1 expression have been observed in multiple lung cancer cell lines and lung cancer tissue samples (4). DKK1 is expressed at very low levels in normal adult human tissues, and higher levels in embryonic tissues (5). The use of DKK1 and other biomarkers may increase the sensitivity of lung cancer detection. Our previous study demonstrated that the activity of the DKK1 promoter was low in lung cancer cells (6). Thus, it is likely that enhancer elements outside the basal promoter region contribute to its wide range of expression patterns (7).
Promoters, located at the 5′ends of genes, surround the transcriptional start site and act to initiate transcription (8,9). However, it is not fully understood how one transcriptional regulatory element (TRE) can precisely define a human genetic locus. Furthermore, not all functional elements have yet been determined for each transcript. In general, both a promoter and its associated transcript are embedded in a collection of positively and negatively regulating elements, which can be positioned anywhere in relation to the transcriptional start site, including at 5′, 3′ and intronic locations, and may act across large segments of DNA up to megabases in length (10). Therefore, the computational prediction of functionally significant TREs is challenging. The Encyclopedia of DNA Elements (ENCODE) project provides systematically mapped regions of transcription, transcription factor association, chromatin structure and histone modification (11,12). It has integrated multiple technologies and approaches to identify and define functional elements, including enhancer elements and enhancer-blocking insulators. This collective effort may be used to screen for potential evolutionarily conserved TREs of the DKK1 promoter.
Regulatory elements have been characterized downstream of a range of human genes, so analyzing multiple intronic and extragenic DNase I hypersensitive sites (DHSs) at +27 kb around a gene in different epithelial cell types can identify regions that are often associated with gene regulatory elements (13,14). This strategy has previously been successfully used to identify enhancers of the c-Myc and cystic fibrosis transmembrane conductance regulator genes (15,16). Not all DHSs contain TREs; they may be associated with structural elements that function in chromatin organization instead, which may have a major effect on the combination of DNA with transcriptional regulation factors (17). Therefore, the extent to which gene-specific combinatorial patterns of histone modifications exist remains to be determined. Numerous covalent histone modifications, including methylation and acetylation, which occur mainly at N-terminal tails, affect gene transcription (18). However, the extent to which combinatorial patterns of histone modifications exist in the DHS remains unknown. Histone modification is one of the most important epigenetic factors in the regulation of gene expression (19,20). Thus, active promoters are often marked by the acetylation of various histone residues, including H3K4ac, H3K27ac and H3K9ac, or methylation, including H3K4me1/2, H3K27me3 and H3K9me3 (21). This modification is largely conserved across species.
With the development of methodology that can evaluate regulatory elements in the whole genome (22), key regulatory elements of the expression and function of one large proximal promoter fragment of the DKK1 locus were investigated. The present study aimed to comprehensively predict potential regulatory elements of the lung cancer-specific DKK1 locus to identify widespread enhancers.
Materials and methods
Cell lines and cell culture
A549, H446 and H460 human lung cancer cell lines, and HEK293(T) cells were obtained from the American Type Culture Collection (Manassas, VA, USA). The squamous cell carcinoma cell line, Eca-109, and human umbilical vein endothelial cells (HUVECs) were supplied by the Cell Bank of Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). The 293 cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.). All other cells were cultured in RPMI 1640 medium (HyClone; GE Healthcare Life Sciences, Logan, UT, USA) containing 10% FBS, and all cells were maintained at 37°C in 5% CO2.
Computational prediction of TREs at the DKK1 locus
To screen for evolutionarily conserved TREs in the genetically identified DKK1 promoter, computational tools were used to download and process information from the UCSC Genome Browser (https://genome-cancer.ucsc.edu/), released by the ENCODE project (23). A human genome sequence of ~27 kb surrounding the lung cancer-specific DKK1 locus was downloaded. Enhancer predictions were performed by separately analyzing histone modification patterns and the DHS of DKK1 in A549 cells, HUVECs and normal human lung fibroblasts (NHLFs). CCCTC binding factor (CTCF) is a multi-zinc finger protein that binds to insulators (24). To investigate whether histone methylation-defined chromatin domains are demarcated by CTCF-bound insulators (25), the genome-wide distribution of CTCF was determined in these cells. The enhancer modules were extended to include the most adjacent blocks with high conservation scores by using PhastCons conservation track and regulatory potential track at the UCSC genome website. The top 12 enhancers located within 3,500 bp upstream and 4,700 bp downstream of the DKK1 transcription start site in each cell were selected. Sequences and detailed alignments are presented in Table I.
Table I.
DKK1 distal promoter fragment primers.
| Promoter fragment | Length (bp) | Primer sequences | Restriction enzymes |
|---|---|---|---|
| DKK1-9 (−938-40) | 978 | F: TCTCCACATTAGCCCACCAC | HindIII/XhoI |
| R: CTGCGGTCCCAGAGTCCT | |||
| DKK1-5 (533–40) | 573 | F: ACTGCGACTCTAAAGGGTTAATG | HindIII/XhoI |
| R: CTGCGGTCCCAGAGTCCT | |||
| DKK1-3 (−350-40) | 390 | F: CCCCTCGGCTCTGTAAAGTAT | |
| R: TGCGGTCCCAGAGTCCT | |||
| DKK1-2 (−282-40) | 320 | F: CAAGTTCCCAGAGTTCCTGCT | HindIII/XhoI |
| R: CTGCGGTCCCAGAGTCCT | |||
| SV40 | 420 | F: CGCAGCACCATGGCCTGA | BgIII/Xho I |
| R: TTGCAAAAGCCTAGGCCTCCA |
DKK1, dickkopf 1; F, forward; R, reverse.
Strongest candidate TREs distally regulate the DKK1 promoter
To interpret these datasets, it was confirmed that the 12 candidate DNA sequences function as enhancers of DKK1. Broad classes of chromatin states were distinguished, referred to as promoters, enhancers, and insulators. To ensure that elements capable of overcoming the effects of strong enhancers were identified, the assay utilized an enhancer from the human β-globin locus control region. Known as DHS II (HS2), this enhancer functions in multiple cell lines, with multiple promoters (8,26). Furthermore, the presence of HS2 provides a large window of expression to reliably measure loss-of-function effects (27). The HS2 enhancer was incorporated into silencer and TRE plasmids to confirm the ability to overcome strong activating events, and as a measure of the interference between promoter and enhancer interactions (28). However, it is not known whether HS2 has an enhancing effect on the DKK1 promoter. The expression clones prepared for transfection are listed in Table II.
Table II.
Primer sequences of the 12 candidate TREs.
| TRE | Locus | Length (bp) | Primer sequences |
|---|---|---|---|
| TRE1 | chr10:53,992,381–53,992,930 | 550 | F: TGGTTCATATTTTGTTTTTCTTGTG |
| R: ACCTAAGTTATTAAGTTTTGTCTCA | |||
| TRE2 | chr10:54,016,954–54,017,517 | 565 | F: TTTATGCTAAGACAAGGAGGTGT |
| R: TTTTGAAGAATAAACATAACATGAGAA | |||
| TRE3 | chr10:54,073,726–54,074,219 | 498 | F: GCAAGGGCACCCAAGTTC |
| R: CCAGAGCCATCATCTCAGAA | |||
| TRE4 | chr10:54,078,638–54,079,344 | 707 | F: AATGTCTGTTGTTGTTGCTGTG |
| R: CCAGGCTCATTCTTATCAGTAG | |||
| TRE5 | chr10:54,086,158–54,086,541 | 384 | F: TTTTCATCCCTTTCCCTCACT |
| R: GGGCAAGGAGAATCAGCTC | |||
| TRE6 | chr10:54,128,561–54,128,810 | 250 | F: CATGCCAGGCTCTCAGTAAG |
| R: CTGTTGAGTCAGGGGTTTGG | |||
| TRE7 | chr10:54,203,100–54,204,074 | 975 | F: TCGCCTAGTGTATCTTTTAG |
| R: TTTGTTGATATTTCATAATCATTGGA | |||
| TRE8 | chr10:54,211,579–54,212,928 | 1,350 | F: TATTCATTTGCATAAAAGATAAAGCC |
| R: ATTTTCTTATTCATTCATTTCCTACCG | |||
| TRE9 | chr10:54,218,904–54,219,195 | 293 | F: GATGCTAAACATAGGTACTTTTGAA |
| R: AATTTGACTATGGGCTTTTAGG |
TRE, transcriptional regulatory element; chr, chromosome.
Construction of reporter plasmids
A total of 4 different fragments of the DKK1 promoter were amplified from A549 cell line genomic DNA, then cloned into HindIII and XhoI sites upstream of the firefly luciferase reporter gene in the pGL3-basic vector (Promega Corporation, Madison, WI, USA) to produce pGL3-DKK1-2, pGL3-DKK1-3, pGL3-DKK1-5 and pGL3-DKK1-9 constructs. The TK promoter-driven Renilla luciferase reporter gene, derived from a commercial vector (pRL-TK; Promega Corporation), was used to normalize for firefly luciferase activity. As a control, the SV40 enhancer/promoter fragment was excised from pRL-TK and inserted in forward and reverse orientations into the pGL3-basic plasmid at the XhoI and BglII sites to produce pGL3-SV40. Genomic DNA extraction, polymerase chain reaction (PCR) amplification and concentration quantification were performed as previously described (6). All primer sequences and cloning sites are listed in Table I. Subcloned fragments were confirmed by sequencing. Predicted TREs were amplified by PCR according to the same protocol (6); Table II presents the primer sequences. The products were ligated into the multiple cloning site of the pGL3-DKK1-5 vector, upstream of a candidate DKK1 promoter-driven luciferase reporter gene.
Plasmid transfection
Plasmids were purified using a Plasmid Midi kit (Qiagen GmbH, Hilden, Germany) according to the manufacturer's protocol. Cells were plated into 96-well plates at 8×103 cells per well, 24 h prior to transfection, and were transfected at 80–90% confluence. The cells were co-transfected with the recombined vectors carrying the firefly (50 ng) and Renilla (1 ng) luciferase reporter genes. X-tremeGENE HP (Roche Diagnostics, Basel, Switzerland) was added at 0.25 µl/well, according to the manufacturer's protocol. The pGL3-basic vector (10 ng) and recombined pGL3-SV40 (10 ng) were used as negative controls.
Firefly/Renilla luciferase quantification
Luciferase activity was measured 48 h post-transfection using Dual-Luciferase® Reporter Assay System (Promega Corporation), according to the manufacturer's protocol. Firefly luciferase reporter gene activity was normalized to Renilla luciferase activity by calculating the Renilla/firefly luciferase activity ratio for each construct. The promoter control, DKK1-5, exhibited a baseline level of luciferase activity, which was normalized to 1. The activity of each test construct was calculated relative to the DKK1 promoter construct. Transfections were performed in triplicate, in 3 independent experiments.
Statistical analysis
The data are presented as the mean ± standard deviation and were analyzed using SPSS 17.0 software (SPSS, Inc., Chicago, IL, USA) by one-way ANOVA with a Student-Newman-Keuls post hoc test. P<0.05 was considered to indicate a statistically significant difference.
Results
Identification of DKK1 DHSs
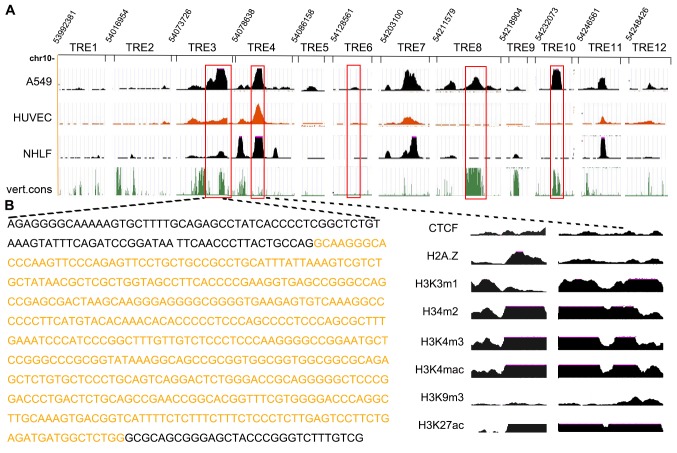
To identify evolutionarily conserved regulatory elements, the genomic DNA sequence of A549 cells was downloaded from the UCSC Genome Browser. DHSs were identified within the 27-kb region of the human genome sequence surrounding DKK1 in the non-cancer human cell lines, HUVECs and NHLFs. A total of 12 potential TREs were identified, the majority of which were located within highly conserved sites of the DKK1 locus (Fig. 1A). The locations of these modules, termed TRE 1–12, are presented in Table II. The strongest signals, representing higher DNase I hypersensitive sites and larger areas of histone interaction, occurred around chr10: 54,073,726–54,074,219 (TRE3). Many of the cancer-associated TREs are located within, or in close proximity to, predicted DHSs, including TRE8, 10 and 12. TRE1, 2, 5 and 6 were associated with weak signaling compared with TRE3 and TRE4.
Figure 1.
Computationally predicted TREs in DKK1, a lung cancer-associated gene. (A) A major DHS was identified at the distal DKK1 promoter in 3 cell lines that express the DKK1 gene, including a 100% vert.cons region. (B) Histone modification patterns at the 2 DHSs of the DKK1 gene in A549; the highlighted sequence is the previously identified TRE3. TRE, transcriptional regulatory element; DKK1, dickkopf 1; DHS, DNase I hypersensitive sites; vert.cons, conserved by vertebrates; chr, chromosome; CTCF, CCCTC binding factor; HUVECs, human umbilical vein endothelial cells; NHLF, normal human lung fibroblasts.
Analysis of DKK1 histone modification patterns
Fig. 1B illustrates the histone modification patterns at the 2 hypersensitive sites of the DKK1 gene in A549 cells. TRE maps of 6 histone acetylation sites (H3K4me1, H3K4me2, H3K4me3, H3K4ac, H3K9ac and H3K27ac), histone variant H2A.Z and a tissue-specific CTCF recognition site in the data from 3 cell lines from ENCODE were also analyzed, with no explicit knowledge of any previous annotation (29). Data analysis revealed H2A.Z, H3K4me1, H3K4me2, H3K4me3, H3K4ac and H3K27ac signals in the intergenic region between TRE3 and 4, and TRE7, 8, 11 and 12 in both A549 and HUVEC cells, but not in NHLF cells. Enrichment patterns of these modifications were mapped, and the chromatin signatures for TRE1, 2, 5, 6, 9 and 10 were described. H3K4me1, H3K4me2 and H3K27ac signals were highly localized to intronic DHSs of TRE7 and 11 and 12 in HUVEC cells, while the other signals were distributed over a broader region. Furthermore, the significantly enriched CTCF signals at TRE6 and 10 suggest that enhancer-blocking insulators flank the DKK1 locus.
DKK1-5 has high functionality as a proximal promoter in different cells
Differences in relative luciferase activity were observed among candidate promoters (Fig. 2). As was hypothesized, compared with an SV40 promoter, the DKK1 promoter greatly increases luciferase activity in NSCLC cell lines (A549 and H460) but not in SCLC cell lines (H446). In contrast, DKK1 activity was lower than that of SV40 in 293(T) cells. There was no significant difference among the 4 segments (pGL3-DKK1-2, pGL3-DKK1-3, pGL3-DKK1-5 and pGL3-DKK1-9) among the different cell types. Once length was taken into consideration, the newly screened DKK1-5 fragment was determined to have the most potential for application as a proximal promoter in subsequent experiments.
Figure 2.
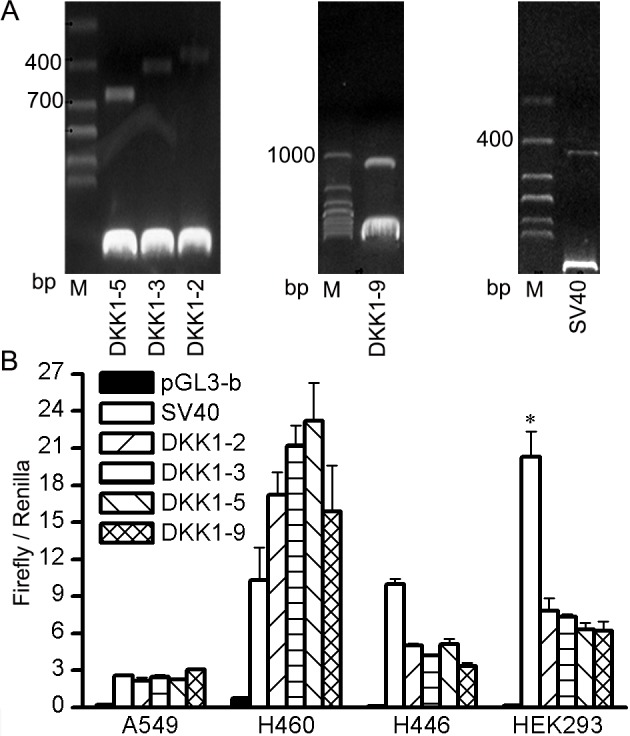
Recombination of pGL3-DKK1 fragments and pGL3-SV40, and measurement of their activity. (A) Verification of reconstructed plasmid by restriction enzyme digestion. (B) Luciferase activity of DKK1-luciferase constructs in A549, H460, H446 and 293 cell lines. *P<0.05 vs. pGL3-b DKK1, dickkopf 1; M, marker.
Construction of TRE-luciferase plasmids
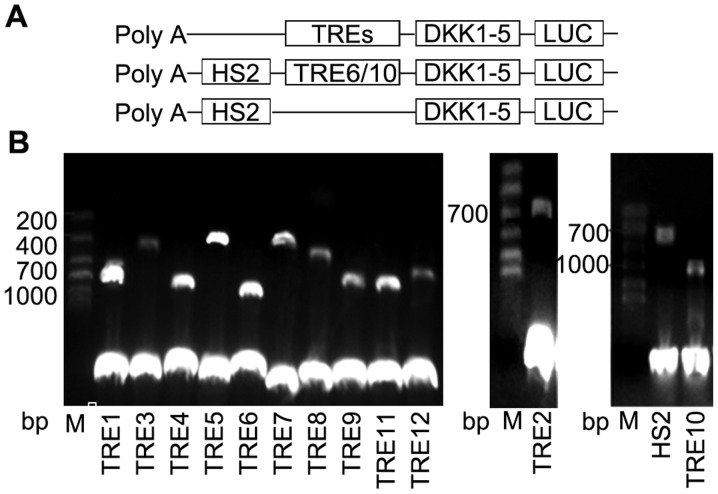
To determine whether the computationally identified TREs possessed biological functionality, they were cloned upstream of either a DKK1-5 promoter-driven luciferase reporter construct or a native HS2 enhancer driven luciferase reporter construct (Fig. 3A). The HS2 enhancer is well characterized and has been used in similar reporter assays (30–32). The candidate enhancers were cloned into the enhancer site of the pGL3 DKK1-5 promoter vector. The observed high CTCF signal predicted 2 insulating fragments, TRE6 and 10. As a silencing test, the insulator was subcloned upstream of the DKK1-5 promoter and downstream of the HS2 enhancer to determine whether the elements had a cooperative effect on luciferase expression (Fig. 3A). The reconstructed TRE reporter plasmids were validated by restriction enzyme digestion (Fig. 3B).
Figure 3.
Recombination of pGL3-DKK1-5-TREs. (A) The location and order of TREs, HS2, DKK1-5 and luciferase in the constructs. The candidate TREs were cloned in front of a DKK1-5 promoter-driven luciferase reporter pGL3-basic vector. (B) Identification of reconstructed TRE reporter plasmids by restriction enzyme digestion. The position on the 1.0% agarose gel corresponds to the specific TRE, as indicated. DKK1, dickkopf 1; TRE, transcriptional regulatory element; HS2, DNase I hypersensitive site II; Poly-A, polyadenylated tail; LUC, luciferase.
Functional validation of transcriptional regulatory elements predicted at the DKK1 locus
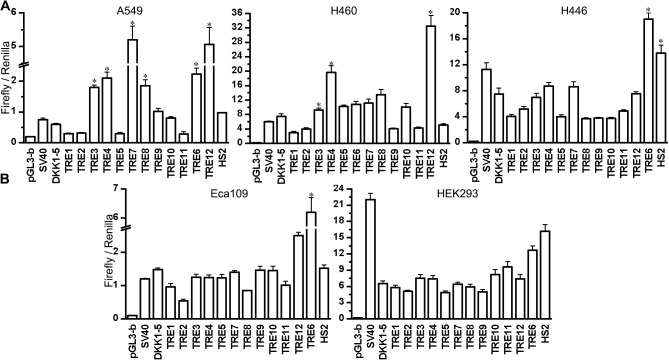
The relative luciferase activity of TRE-DKK1-luciferase constructs was investigated in A549 and H460 NSCLC cells, the small cell lung cancer cell line H446, the esophageal cancer cell line Eca-109, and 293 cells (Fig. 4). The enhancer-control revealed the upper threshold of expression contributed by the HS2 enhancer, and was used as a normalization control. Luciferase reporter assays in A549 cells revealed that TRE7 and 12 act as strong transcriptional enhancers, leading to an 8-fold increase in DKK1 promoter activity. TRE3, 4, 6 and 8 also increased the activity of the DKK1 promoter in A549 cells, reflecting the DHS and histone modification results (Fig. 1). Importantly, HS2 together with TRE6 increased DKK1 promoter activity in A549 and H446 cells, whereas HS2 together with TRE10 and HS2 alone had a minor or no effect. This was also observed in H460 cells, albeit to a lesser extent. This suggests that TRE6 and TRE10 do not act as a silencer in A549, H446 and H460 cells. In H460 cells, it was demonstrated that TRE3, 4 and 12 significantly increased the activity of the DKK1 promoter, while TRE6, 7 and 8 had little effect. Furthermore, TRE3, 4, 7 and 12 had no effect on DKK1 activity in H446 cells. The majority of the putative enhancers did not significantly stimulate the DKK1 promoter in Eca-109 or 293, except for TRE6 which resulted in high DKK1 promoter activity in Eca-109 cell line.
Figure 4.
Enhancer dual luciferase reporter assay analysis of functionality. The relative luciferase activity of the candidate promoters in (A) lung cancer cells A549, H460, H446 and (B) the squamous cell carcinoma cell line Eca-109 and normal cell 293T. *P<0.05 vs DKK1. DKK1, dickkopf 1; TRE, transcriptional regulatory element; HS2, DNase I hypersensitive site II.
Collectively, these results demonstrate that the 6 identified DNA sequences act as efficient TREs. TRE3, 4, 6, 7, 8 and 12 significantly increased the activity of the DKK1 promoter in A549 cells, and TRE7 increased the activity to the highest degree. TRE3 and 4 increased promoter activity in H460 cells, while TRE12 showed a strong effect in A549, H460 and Eca-109 cells. Furthermore, TRE7 and 8 did not increase promoter activity in H460 cells, suggesting that TRE3 and 4 may have NSCLC-specificity.
Discussion
DKK1 has been reported to be a biomarker in lung cancer, with high levels of specificity and expression (4). However, our previous study demonstrated low DKK1 promoter activity in lung cancer cells (6). In the present study, 12 TREs were predicted using DHS and histone modification analysis of the lung cancer-specific DKK1 locus in A549 cells using the UCSC genome browser. Several of these protocols were applied to distinguish between active and poised enhancers. The functionality of the TREs was investigated using a dual-luciferase reporter assay in 5 different cell types. It was demonstrated that several of these DHS regions possess cooperative enhancer activity in vitro.
It has been identified that specific TREs may be associated with low promoter activity (7,33). However, to the best of our knowledge, no TREs were previously defined for the lung cancer-specific expression of DKK1. Methods to detect DNA sequence elements on a genome-wide scale and to examine the role of different one-dimensional regulatory signals remain a challenge to develop (34,35). However, this work uses a computational approach to predict TREs that contain cell line-specific long-range interactions between enhancers and promoters. The present study also presents methods to distinguish between different classes of functional TREs, which may contribute to the elucidation the mechanisms for cell type-specific gene expression.
The majority of predicted enhancers are distal to core promoters, and can be detected by locating tissue-specific DHSs within and around the gene (36). Previous studies have demonstrated that a number of histone modification patterns of promoters, including histone acetyltransferase p300, are enriched at the enhancer site in fetal mouse tissues (37,38). Thus, it is likely that TREs are highly evolutionary conserved and are marked by histone acetylation (39) or the binding of coactivator proteins such as p300 and MED1 (26). The effect of insulators at human enhancers may contribute to the understanding of how enhancers function in tissue-specific gene regulation (40). H3K4me1/2, H3K27ac, H3K9ac and H3K4ac modifications are associated with active enhancers (39,41). The histone variant H2A.Z associates with functional regulatory elements, and was previously demonstrated to be enriched in active promoters and strong enhancers, while the insulator binding protein, CTCF, marks the boundaries of histone methylation domains (42). However, in the present study, H3K9me1/3 and H3K27me3 signals were modestly elevated at silent promoters, and little change was observed in the intergenic regions of the cell lines. By integrating the predictions of DHSs and selected histone modifications in A549, HUVEC and NHLF cell ENCODE datasets, 12 candidate DNA sequences were proposed as potential TREs. A set of cis-acting elements were characterized within the DKK1 locus, and demonstrated that enhancer elements located within cancer-associated regions can regulate DKK1 promoter activity (Fig. 4). As the activity of the DKK1 promoter was only increased in NSCLC cells, it is likely that the most crucial sequences (TRE3, 4, 7 and 8) are NSCLC-specific.
An important aspect of this study was its initial predictive framework for the computational prediction of enhancer-promoter interactions from datasets. The observed changes in reporter activity indicate that these constructive TREs may have potential regulatory ability. However, it should be noted that only the specific TREs of DKK1 expression in cell lines were examined in the present study, and that additional in vivo studies are required to verify the results. Finally, the power of these techniques as a strategy to predict and identify TREs on the basis of their epigenetic characteristics, independent of motifs or other sequence features, was demonstrated in the present study, with particular respect to DHS mapping and histone acetylation patterns. A detailed enhancer map may be used to inform future therapeutic genome editing (43), as demonstrated by a recent report applying the CRISPR-Cas9 system to enhancers in situ, which confirmed that the BCL11A enhancer may be a novel target for intractable diseases (44).
In conclusion, the present study confirms the value of using computational tools to predict TREs, and provides the first insight into the intronic cis-acting elements that control the regulation of DKK1 expression. A systematic strategy to predict cell line-specific enhancer-promoter interactions using a minimal dataset is proposed. Furthermore, the present study suggests that the active global enhancer network offers important insights into cancer development and potentially, targeted therapy.
Acknowledgements
The authors would like to thank Mr Li and Dr. Duan (Department of Nuclear Medicine & Institute of Anesthesiology and Pain, Taihe Hospital, Hubei University of Medicine) for the reagent gifts, and for assistance with cell culture in our laboratory. Their ideas also provided a valuable added dimension to the study.
Funding
The present study was supported by the National Natural Science Foundation of China (grant no., 81401447), the Science and Technology Development Foundation of Shiyan City (grant no., 16Y16) and the Key Discipline Project of Hubei University of Medicine.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
ZP and YG designed the study, interpreted the experimental results and modified the manuscript. XD and JZ constructed plasmids, interpreted results and drafted the manuscript. YG and ZP prepared the figures. RW and YC performed statistical analysis. WL and FL did computational prediction of TREs at the DKK1 locus. HZ and YY cultured cells and measured luciferase activity. All authors have read and approved the manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Hadjihannas MV, Brückner M, Jerchow B, Birchmeier W, Dietmaier W, Behrens J. Aberrant Wnt/beta-catenin signaling can induce chromosomal instability in colon cancer. Proc Natl Acad Sci USA. 2006;103:10747–10752. doi: 10.1073/pnas.0604206103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Niehrs C. Function and biological roles of the Dickkopf family of Wnt modulators. Oncogene. 2006;25:7469–7481. doi: 10.1038/sj.onc.1210054. [DOI] [PubMed] [Google Scholar]
- 4.Yamabuki T, Takano A, Hayama S, Ishikawa N, Kato T, Miyamoto M, Ito T, Ito H, Miyagi Y, Nakayama H, et al. Dikkopf-1 as a novel serologic and prognostic biomarker for lung and esophageal carcinomas. Cancer Res. 2007;67:2517–2525. doi: 10.1158/0008-5472.CAN-06-3369. [DOI] [PubMed] [Google Scholar]
- 5.Forget MA, Turcotte S, Beauseigle D, Godin-Ethier J, Pelletier S, Martin J, Tanguay S, Lapointe R. The Wnt pathway regulator DKK1 is preferentially expressed in hormone-resistant breast tumours and in some common cancer types. Br J Cancer. 2007;96:646–653. doi: 10.1038/sj.bjc.6603579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu R, Guo LJ, Xin J, Li WM, Gao Y, Zheng YX, Guo YH, Lin YJ, Xie YH, Wu YQ, Xu RA. Luciferase assay to screen tumour-specific promoters in lung cancer. Asian Pac J Cancer Prev. 2013;14:6557–6562. doi: 10.7314/APJCP.2013.14.11.6557. [DOI] [PubMed] [Google Scholar]
- 7.Zhang C, Dai H, de Crombrugghe B. Characterization of Dkk1 gene regulation by the osteoblast-specific transcription factor Osx. Biochem Biophys Res Commun. 2012;420:782–786. doi: 10.1016/j.bbrc.2012.03.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chung JH, Bell AC, Felsenfeld G. Characterization of the chicken beta-globin insulator. Proc Natl Acad Sci USA. 1997;94:575–580. doi: 10.1073/pnas.94.2.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Niu J, Wang J, Hu H, Chen Y, An J, Cai J, Sun R, Sheng Z, Liu X, Lin S. Cross-talk between freezing response and signaling for regulatory transcriptions of MIR475b and its targets by miR475b promoter in Populus suaveolens. Sci Rep. 2016;6:20648. doi: 10.1038/srep20648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Naumova N, Smith EM, Zhan Y, Dekker J. Analysis of long-range chromatin interactions using Chromosome Conformation Capture. Methods. 2012;58:192–203. doi: 10.1016/j.ymeth.2012.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sanyal A, Lajoie BR, Jain G, Dekker J. The long-range interaction landscape of gene promoters. Nature. 2012;489:109–113. doi: 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ecker JR, Bickmore WA, Barroso I, Pritchard JK, Gilad Y, Segal E. Genomics: ENCODE explained. Nature. 2012;489:52–55. doi: 10.1038/489052a. [DOI] [PubMed] [Google Scholar]
- 13.Deng T, Zhu ZI, Zhang S, Postnikov Y, Huang D, Horsch M, Furusawa T, Beckers J, Rozman J, Klingenspor M, et al. Functional compensation among HMGN variants modulates the DNase I hypersensitive sites at enhancers. Genome Res. 2015;25:1295–1308. doi: 10.1101/gr.192229.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sherwood RI, Hashimoto T, O'Donnell CW, Lewis S, Barkal AA, van Hoff JP, Karun V, Jaakkola T, Gifford DK. Discovery of directional and nondirectional pioneer transcription factors by modeling DNase profile magnitude and shape. Nat Biotechnol. 2014;32:171–178. doi: 10.1038/nbt.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sotelo J, Esposito D, Duhagon MA, Banfield K, Mehalko J, Liao H, Stephens RM, Harris TJ, Munroe DJ, Wu X. Long-range enhancers on 8q24 regulate c-Myc. Proc Natl Acad Sci USA. 2010;107:3001–3005. doi: 10.1073/pnas.0906067107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Uslu VV, Petretich M, Ruf S, Langenfeld K, Fonseca NA, Marioni JC, Spitz F. Long-range enhancers regulating Myc expression are required for normal facial morphogenesis. Nat Genet. 2014;46:753–758. doi: 10.1038/ng.2971. [DOI] [PubMed] [Google Scholar]
- 17.Khurana E, Fu Y, Chakravarty D, Demichelis F, Rubin MA, Gerstein M. Role of non-coding sequence variants in cancer. Nat Rev Genet. 2016;17:93–108. doi: 10.1038/nrg.2015.17. [DOI] [PubMed] [Google Scholar]
- 18.Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, Zhao K. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schones DE, Zhao K. Genome-wide approaches to studying chromatin modifications. Nat Rev Genet. 2008;9:179–191. doi: 10.1038/nrg2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou VW, Goren A, Bernstein BE. Charting histone modifications and the functional organization of mammalian genomes. Nat Rev Genet. 2011;12:7–18. doi: 10.1038/nrg2905. [DOI] [PubMed] [Google Scholar]
- 21.Ernst J, Kellis M. Large-scale imputation of epigenomic datasets for systematic annotation of diverse human tissues. Nat Biotechnol. 2015;33:364–376. doi: 10.1038/nbt.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gallo SM, Gerrard DT, Miner D, Simich M, Des Soye B, Bergman CM, Halfon MS. REDfly v3.0: Toward a comprehensive database of transcriptional regulatory elements in Drosophila. Nucleic Acids Res. 2011;39:D118–D123. doi: 10.1093/nar/gkq999. (Database Issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.ENCODE Project Consortium: A user's guide to the encyclopedia of DNA elements (ENCODE) PLoS Biol. 2011;9:e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bell AC, West AG, Felsenfeld G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999;98:387–396. doi: 10.1016/S0092-8674(00)81967-4. [DOI] [PubMed] [Google Scholar]
- 25.Cuddapah S, Jothi R, Schones DE, Roh TY, Cui K, Zhao K. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res. 2009;19:24–32. doi: 10.1101/gr.082800.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petrykowska HM, Vockley CM, Elnitski L. Detection and characterization of silencers and enhancer-blockers in the greater CFTR locus. Genome Res. 2008;18:1238–1246. doi: 10.1101/gr.073817.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Talbot D, Grosveld F. The 5′HS2 of the globin locus control region enhances transcription through the interaction of a multimeric complex binding at two functionally distinct NF-E2 binding sites. EMBO J. 1991;10:1391–1398. doi: 10.1002/j.1460-2075.1991.tb07659.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Elnitski L, Miller W, Hardison R. Conserved E boxes function as part of the enhancer in hypersensitive site 2 of the beta-globin locus control region role of basic helix-loop-helix proteins. J Biol Chem. 1997;272:369–378. doi: 10.1074/jbc.272.1.369. [DOI] [PubMed] [Google Scholar]
- 29.Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 30.Wu C, Chen Z, Dardalhon V, Xiao S, Thalhamer T, Liao M, Madi A, Franca RF, Han T, Oukka M, Kuchroo V. The transcription factor musculin promotes the unidirectional development of peripheral Treg cells by suppressing the TH2 transcriptional program. Nat Immunol. 2017;18:344–353. doi: 10.1038/ni.3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fleetwood MR, Ho Y, Cooke NE, Liebhaber SA. DNase I hypersensitive site II of the human growth hormone locus control region mediates an essential and distinct long-range enhancer function. J Biol Chem. 2012;287:25454–25465. doi: 10.1074/jbc.M112.365825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jane SM, Ney PA, Vanin EF, Gumucio DL, Nienhuis AW. Identification of a stage selector element in the human gamma-globin gene promoter that fosters preferential interaction with the 5′HS2 enhancer when in competition with the beta-promoter. EMBO J. 1992;11:2961–2969. doi: 10.1002/j.1460-2075.1992.tb05366.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mina M, Magi S, Jurman G, Itoh M, Kawaji H, Lassmann T, Arner E, Forrest AR, Carninci P, Hayashizaki Y, et al. Promoter-level expression clustering identifies time development of transcriptional regulatory cascades initiated by ErbB receptors in breast cancer cells. Sci Rep. 2015;5:11999. doi: 10.1038/srep11999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schoenfelder S, Furlan-Magaril M, Mifsud B, Tavares-Cadete F, Sugar R, Javierre BM, Nagano T, Katsman Y, Sakthidevi M, Wingett SW, et al. The pluripotent regulatory circuitry connecting promoters to their long-range interacting elements. Genome Res. 2015;25:582–597. doi: 10.1101/gr.185272.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wittkopp PJ, Kalay G. Cis-regulatory elements: Molecular mechanisms and evolutionary processes underlying divergence. Nat Rev Genet. 2011;13:59–69. doi: 10.1038/nrg3095. [DOI] [PubMed] [Google Scholar]
- 36.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 37.Visel A, Prabhakar S, Akiyama JA, Shoukry M, Lewis KD, Holt A, Plajzer-Frick I, Afzal V, Rubin EM, Pennacchio LA. Ultraconservation identifies a small subset of extremely constrained developmental enhancers. Nat Genet. 2008;40:158–160. doi: 10.1038/ng.2007.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blow MJ, McCulley DJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, et al. ChIP-Seq identification of weakly conserved heart enhancers. Nat Genet. 2010;42:806–810. doi: 10.1038/ng.650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, Tsutsumi S, Nagae G, Ishihara K, Mishiro T, et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- 41.Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pougach K, Voet A, Kondrashov FA, Voordeckers K, Christiaens JF, Baying B, Benes V, Sakai R, Aerts J, Zhu B, et al. Duplication of a promiscuous transcription factor drives the emergence of a new regulatory network. Nat Commun. 2014;5:4868. doi: 10.1038/ncomms5868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Canver MC, Smith EC, Sher F, Pinello L, Sanjana NE, Shalem O, Chen DD, Schupp PG, Vinjamur DS, Garcia SP, et al. BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis. Nature. 2015;527:192–197. doi: 10.1038/nature15521. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.