Abstract
Breast cancer remains the leading cause of mortality worldwide. Human papilloma virus 16 (HPV16) may serve a function in the pathogenesis and development of breast cancer. However, the detection rate of HPV16 in breast carcinoma may vary by region. In the present study, the expression of HPV16 E7 in paraffin-embedded tissues from patients with breast cancer from North China was detected. Additionally, the molecular mechanisms underlying the function of HPV16 E7 in the proliferation of breast cancer cells were examined. The results demonstrated that the DNA of HPV16 E7 was detected in 30.5% of the samples, and that HPV16 E7 promoted the proliferation of breast cancer cells in vitro and in vivo. Additionally, HPV16 E7-mediated proliferation of breast cancer cells was suppressed in response to treatment with cyclooxygenase-2 (COX-2)-specific small interfering RNA and celecoxib. The results of the present study revealed that HPV16 E7 may promote the proliferation of breast cancer cells by upregulating COX-2, suggesting that COX-2 may be a potential therapeutic target for HPV16 E7-mediated progression of breast cancer.
Keywords: breast cancer, human papilloma virus 16, cyclooxygenase-2, celecoxib, proliferation
Introduction
It has been reported that breast cancer is one of the main causes of female mortality from cancer worldwide in 2017 (1). The incidence of human breast cancer is associated with various factors, including lifestyle, medical conditions, oncogenes and virus infection (2–7). It has been reported that human papillomaviruses (HPVs), including high risk-HPV (HR-HPV), may be responsible for the pathogenesis and development of breast cancer. HPV16 is the most prevalent type of HR-HPVs. The integration of the DNA of HPV16 into host cells promotes a constitutive high expression of E7 oncoprotein, leading to carcinogenesis in vivo (8). A number of HPV vaccines have been developed to prevent HPV16-mediated cancer progression (9,10). However, these vaccines have been proven to be inefficient for the treatment of HPV16-mediated cancer. Polymerase chain reaction (PCR) has been employed for the diagnosis of HR-HPVs (11). However, the detection rate of HPV16 in breast carcinoma may vary by region (12,13). As ~80% of all breast cancers are invasive ductal carcinoma, the expression of HPV16 E7 was examined in samples from patients with invasive breast ductal carcinoma from North China in the present study (14). Additionally, novel therapeutic targets for HPV16 E7-mediated cancer were investigated.
Cyclooxygenase-2 (COX-2) is a rate-limiting catalyzer in the conversion of arachidonic acid to prostaglandins (15). Furthermore, COX-2 is highly modulated in various malignancies, including breast carcinoma (16,17). Several reports have demonstrated that the suppression of COX-2 by selective inhibitors has antiviral effects on viral agents, including herpes simplex virus, avian influenza A (H5N1) and hepatitis C virus (18–20). Celecoxib is a selective COX-2 inhibitor and has been reported to be used for the treatment of various types of cancer (21–23). Evidence suggests that COX-2 is overexpressed in various HPV-induced lesions (24,25). However, it remains unknown whether the upregulation of COX-2 results from HPV E7-mediated progression in breast carcinoma.
In the present study, the expression of HPV16 E7 was detected in samples from patients with invasive breast ductal carcinomas from North China. Additionally, the molecular mechanisms underlying the function of HPV16 E7 in the proliferation of breast cancer cells were examined.
Materials and methods
Tissue specimens
A total of 59 cases of invasive breast ductal carcinoma were collected from the patients who underwent resection in the Department of Pathology, Third Affiliated Hospital of Xinxiang Medical University (Henan, China) between January 2010 and March 2012. All patients were females and did not receive chemotherapy, radiotherapy or immunotherapy prior to surgery. All specimens were subjected to haematoxylin and eosin staining and diagnosed as invasive ductal breast carcinoma by two pathologists. All participants provided written informed consent and the tissue acquisition protocol was approved by the Institutional Board of Xinxiang Medical University (Xinxiang, China).
DNA isolation and quantitative (q)PCR
Invasive breast ductal carcinoma tissues were fixed in neutral buffered formalin, embedded in paraffin and stored at room temperature. Paraffin-embedded tissue samples were cut into 4 µm-thick sections. The sections were deparaffinized in xylene and then were rehydrated in a descending ethanol series. The sections were dried at room temperature and digested with Proteinase K (Qiagen GmbH, Hilden, Germany) overnight at 56°C. The following day, the samples were incubated for 1 h at 90°C to inactivate Proteinase K and total DNA was extracted from paraffin-embedded tissues using QIAamp DNA FFPE Tissue kit (Qiagen GmbH), according to the manufacturer's protocol. The primers for HPV16 E7 and COX-2 are presented in the Table I. The thermocycling conditions were as follows: Initial denaturation for 1 min at 95°C followed by 40 cycles of denaturation at 95°C for 30 sec, 50°C for 45 sec, 72°C for 30 sec and a final extension at 72°C for 5 min. DNA content was measured using Applied Biosystems 7500 Sequence Detection system and SYBR Green I (Applied Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA, USA). The data were normalized to the geometric mean of the housekeeping gene GAPDH and quantified using the 2−ΔΔCq method (26).
Table I.
Primer sequences.
| Gene | Forward primer | Reverse primer |
|---|---|---|
| HPV16E7 | 5′-GCATGGAGATACACCTACATTG-3′ | 5′-TGGTTTCTGAGAACAGATGG-3 |
| COX-2 | 5′-CGAGGTGTATGTATGAGTGT-3 | 5′-AGTGGGTAAGTATGTAGTGC-3 |
| GAPDH | 5′-GACTCATGACCACAGTCCATGC-3 | 5′-AGAGGCAGGGATGATGTTCTG-3 |
HPV, human papilloma virus; COX-2, cyclooxygenase-2.
Immunohistochemistry
The expression of HPV16 E7 and COX-2 in invasive breast ductal carcinoma tissues were detected using immunohistochemistry streptavidin peroxidase (SP) method. IHC SP-9000 kit which includes 3% H2O2, goat serum, biotinylated second antibody and avidin-biotin complex reagent. (SP-9000, Ready to use, ZSGB-BIO: OriGene Technologies, Inc., Beijing, China). Paraffin-embedded tissue samples were cut into 4 mm-thick sections and incubated at 60°C for 1 h. The sections were deparaffinized in xylene and then were rehydrated in a descending ethanol series. Endogenous peroxidase activity was quenched by treatment with 3% hydrogen peroxide at room temperature for 30 min. Antigens were retrieved with citrate buffer (0.01 M; pH 6). The tissue sections were blocked for 20 min with normal non-immune serum and then incubated with mouse anti-HPV16 E7 (sc-51951; 1:50; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) or rabbit anti-COX-2 (12375-1-AP; 1:100; ProteinTech Group, Inc., Chicago, IL, USA) primary antibodies overnight at 4°C. Following the primary incubation, the sections were incubated with pre-diluted biotinylated second antibody and avidin-biotin complex reagent according to the manufacturer's instructions at room temperature for 20 min. The staining was visualized using diaminobenzidine. In negative controls, the primary antibody was replaced by PBS. The results of immunohistochemical staining were evaluated by two independent pathologists blind to the clinical findings. The staining intensity of COX2 was assessed with a standard immunoreactive score (27).
Cell culture
MCF-7 cells stably expressing HPV16 E7 (MCF-7/HPV16 E7) or negative control (MCF-7/vector) were established previously (28). Cells were maintained in Dulbecco's modified Eagle's medium (Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum (FBS; Thermo Fisher Scientific, Inc.) and 1% penicillin/streptomycin (Thermo Fisher Scientific, Inc.) with the temperature of 37°C and 5% CO2.
Western blot analysis
Total protein was extracted from cells with 1X sample buffer (3.8 g Tris base, 2.5 ml 10% SDS, 50 ml Glycerin, 75 ml 20% SDS, then ddH2O2 added to a total volume of 500 ml). Protein lysates obtained from the cells and protein determination was measured by BCA protein assay kit (20201ES86; Shanghai Qcbio Science and Technologies Co., Ltd, Shanghai, China). A total of 20 µl protein lysates was loaded per lane and then was separated by SDS-PAGE (10.5% gels) and transferred onto a polyvinylidene difluoride membrane (Nanjing KeyGen Biotech Co., Ltd., Nanjing, China). The membranes were blocked in 5% non-fat milk, which was suspended in tris-buffered saline with Tween 20 in room temperature for 2 h and then were incubated with mouse anti-HPV16 E7 (1:200) or rabbit anti-COX-2 (1:200) primary antibodies overnight at 4°C. The membranes were incubated with the HRP-conjugated secondary antibody (7076s; anti-mouse IgG; 1:2,000; 7074s; anti-rabbit IgG; 1:2,000; CST Biological Reagent Co., Ltd., Shanghai, China) in room temperature for 1 h. The protein bands were visualized by enhanced chemiluminescence (Tanon Science and Technology Co., Ltd., Shanghai, China).
MTT assay
A total of 1×103 cells were seeded onto 96-well plates and cultured for 24 h. Then, 20 µl MTT solution (Merck KGaA, Darmstadt, Germany) was added to each well prior to incubation at 37°C for an additional 4 h. Following careful removal of the medium, 150 µl dimethyl sulfoxide (Merck KGaA) was added to the wells. The absorbance was measured at 450 nm using a microplate autoreader (Bio-Rad Laboratories, Inc., Hercules, CA, USA). Three individual experiments were performed.
Colony formation assay
Cells were trypsinized, plated on 6-well plates (200 cells/well) and cultured for 2 weeks. Subsequently, the cells were fixed with 4% paraformaldehyde for 5 min and stained with hematoxylin for 30 min. Visible colonies consisting of >50 cells were counted. Three independent experiments were performed.
Soft agar assay
A total of 1×103 cells were resuspended in 2 ml 0.3% low melting point agarose in DMEM/20% FBS and plated on top of 1 ml 0.6% agarose in the same medium in 6-well culture plates. The plates were incubated at 37°C in a humidified atmosphere containing 5% CO2 for 2 weeks. The cell colonies were photographed using light microscope (magnification, ×200).
In vivo proliferation assay
Female BALB/c nude mice aged 4–5 weeks (weight, 15–18 g) were obtained from the Center of Laboratory Animal Science of Guangdong (Guangzhou, China). The mice were kept in the plastic cage with sealed air filter at 27°C, in ad libitum feeding and maintained 10 h of light and 14 h of dark daily. All animal experiments were conducted in accordance with the China Guidelines for Animal Care and Ethic for Animal Experiments. The study was approved by the Institutional Animal Care and Use Committee of Xinxiang Medical University (Henan, China). For the in vivo proliferation assay, a total of 2×106 MCF-7/HPV16 E7 or MCF-7/vector cells were injected subcutaneously into the hind limb (n=6/group) of the mice. Tumor size was measured using a slide caliper twice weekly (volume = length × width × height). After 3 weeks, mice were euthanized and tumors were excised, fixed in 10% neutral buffered formalin and embedded in paraffin. Paraffin-embedded tissue samples were cut into 4-µm thick sections. Sections were deparaffinized, transferred to xylene, and rehydrated in descending concentrations of ethanol (100, 100, 95, 95, 85, 70 and 0%). The sections were then stained with hematoxylin and eosin. Additionally, the tissue sections were washed three times in PBS. Citrate buffer (0.01 M, pH 6.0) was applied to the sections for 5 min under 95°C for antigen retrieval. Then IHC SP-9000 kit (SP-9000, Ready to use, ZSGB-BIO: OriGene Technologies, Inc.) was used in the detection of ki-67 expression according to the manufacturer's protocol. Firstly, the sections were incubated in 3% H2O2 and goat serum at room temperature for 30 min respectively and then were incubated in 4°C for 10 h with primary antibody against mouse ki-67 (MX006; 1:100; Fuzhou Maixin Biotech Co., Ltd., Fuzhou, China). Subsequently, the slides were incubated with biotinylated second antibody and avidin-biotin complex reagent (SP-9000 kit) at room temperature for 20 min.
Treatment with COX-2-specific small interfering RNA (siRNA) and celecoxib
COX-2 siRNA (sequence, 5′-GCTCAGCCATACAGCAAAT-3′) was purchased from Guangzhou RiboBio Co., Ltd. (Guangzhou, China). Cells were transfected Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol (siRNA, 100 nmol/l; 37°C for 24 h). The control siRNA was provided by Guangzhou RiboBio Co., Ltd. (Guangzhou, China) which was a random sequence of siRNA and incompatible with the target gene. Cells were treated with celecoxib (Selleck Chemicals, Houston, TX, USA) at the concentration of 1 mol/l (diluted in DMSO) at 37°C for 48 h (DMSO without celecoxib as the control).
Statistical analysis
Data were analyzed using SPSS software (v20.0; IBM Corp., Armonk, NY, USA). Data are expressed as the mean ± standard deviation. Three individual experiments were performed. One-way analysis of variance followed by least significant difference (LSD) test was used to examine differences between more than two group means. Mann-Whitney U-test was used to examine differences between two groups medians. The association between HPV16 E7 and COX-2 expression was analyzed using Chi-square test. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression of HPV16 E7 DNA in invasive ductal breast cancer
The DNA of HPV16 E7 was detected in 18 out of 59 cases of paraffin-embedded invasive ductal breast carcinoma samples by qPCR (Table II). Therefore, HPV16 E7 was positive for 30.5% cases of invasive ductal breast carcinoma from North China.
Table II.
Expression of HPV16 E7 and COX-2 in invasive ductal breast cancer.
| Sample | Sex | Ct GAPDH (mean ± SD) | Ct of HPV16E7 (mean ± SD) | Ct of COX-2 (mean ± SD) |
|---|---|---|---|---|
| 1 | Female | 25.15±0.07 | Undetermined | 27.56±0.43 |
| 2 | Female | 24.27±0.18 | 25.53±0.29 | 23.62±0.23 |
| 3 | Female | 25.51±0.32 | Undetermined | 27.68 ±0.39 |
| 4 | Female | 23.55±0.18 | 24.29±0.09 | 21.38±0.38 |
| 5 | Female | 26.42±0.23 | 27.58±0.31 | 25.36±0.24 |
| 6 | Female | 26.09±0.32 | Undetermined | 28.29±0.10 |
| 7 | Female | 28.07±0.07 | Undetermined | 27.18±0.11 |
| 8 | Female | 24.58±0.20 | 22.22±0.04 | 23.22±0.19 |
| 9 | Female | 26.94±0.05 | Undetermined | 29.52±0.18 |
| 10 | Female | 27.59±0.50 | Undetermined | 25.67±0.34 |
| 11 | Female | 25.07±0.27 | Undetermined | 27.77±0.35 |
| 12 | Female | 29.35±0.16 | Undetermined | 28.77±0.44 |
| 13 | Female | 25.06±0.12 | Undetermined | 27.53±0.41 |
| 14 | Female | 21.30±0.21 | Undetermined | 26.64±0.91 |
| 15 | Female | 25.44±0.26 | Undetermined | 27.56±0.29 |
| 16 | Female | 24.33±0.15 | 32.17±0.03 | 24.21±0.17 |
| 17 | Female | 26.49±0.30 | Undetermined | 28.32±0.21 |
| 18 | Female | 26.28±0.19 | 28.16±0.05 | 24.42±0.15 |
| 19 | Female | 28.28±0.22 | Undetermined | 27.30±0.10 |
| 20 | Female | 27.51±0.40 | Undetermined | 28.52±0.18 |
| 21 | Female | 26.62±0.20 | 33.21±0.05 | 26.44±0.20 |
| 22 | Female | 27.60±0.50 | Undetermined | 26.52±0.23 |
| 23 | Female | 24.57±0.23 | Undetermined | 27.58±0.21 |
| 24 | Female | 29.44±0.23 | Undetermined | 29.13±0.18 |
| 25 | Female | 25.67±0.17 | Undetermined | 24.55±0.37 |
| 26 | Female | 24.67±0.20 | 25.32±0.27 | 22.33±0.17 |
| 27 | Female | 27.36±0.15 | Undetermined | 27.52±0.37 |
| 28 | Female | 25.19±0.13 | 30.27±0.13 | 24.12±0.03 |
| 29 | Female | 29.34±0.27 | Undetermined | 27.19±0.29 |
| 30 | Female | 25.52±0.26 | 28.13±0.10 | 23.56±0.28 |
| 31 | Female | 24.81±0.15 | Undetermined | 24.15±0.09 |
| 32 | Female | 26.41±0.19 | Undetermined | 27.45±0.31 |
| 33 | Female | 23.28±0.10 | Undetermined | 21.67±0.10 |
| 34 | Female | 28.76±0.30 | Undetermined | 28.29±0.15 |
| 35 | Female | 24.54±0.28 | 24.18±0.03 | 22.59±0.13 |
| 36 | Female | 29.60±0.36 | Undetermined | 29.33±0.29 |
| 37 | Female | 27.57±0.31 | Undetermined | 26.47±0.15 |
| 38 | Female | 29.82±0.03 | Undetermined | 29.21±0.04 |
| 39 | Female | 24.70±0.18 | 33.20±0.08 | 22.49±0.35 |
| 40 | Female | 22.80±0.16 | 26.53±0.29 | 21.59±0.11 |
| 41 | Female | 26.29±0.18 | Undetermined | 25.67±0.10 |
| 42 | Female | 27.37±0.48 | Undetermined | 26.29±0.15 |
| 43 | Female | 27.95±0.02 | Undetermined | 26.59±0.13 |
| 44 | Female | 30.49±0.17 | Undetermined | 30.39±0.23 |
| 45 | Female | 26.27±0.42 | Undetermined | 24.74±0.55 |
| 46 | Female | 27.44±0.34 | Undetermined | 27.24±0.22 |
| 47 | Female | 29.74±0.16 | Undetermined | 29.38±0.24 |
| 48 | Female | 22.42±0.25 | 30.58±0.31 | 21.32±0.22 |
| 49 | Female | 23.71±0.20 | 35.29±0.09 | 23.45±0.22 |
| 50 | Female | 23.61±0.44 | 27.18±0.01 | 25.42±0.52 |
| 51 | Female | 25.12±0.05 | Undetermined | 25.39±0.10 |
| 52 | Female | 26.46±0.31 | Undetermined | 26.72±0.33 |
| 53 | Female | 27.41±0.25 | Undetermined | 27.65±0.28 |
| 54 | Female | 29.14±0.12 | Undetermined | 29.22±0.14 |
| 55 | Female | 22.97±0.03 | 23.37±0.23 | 21.61±0.12 |
| 56 | Female | 26.81±0.16 | 29.31±0.15 | 25.27±0.12 |
| 57 | Female | 28.49±0.22 | Undetermined | 30.46±0.46 |
| 58 | Female | 26.10±0.02 | Undetermined | 24.57±0.31 |
| 59 | Female | 25.44±0.43 | Undetermined | 26.15±0.18 |
HPV, human papilloma virus; COX-2, cyclooxygenase-2; SD, standard deviation.
HPV16 E7 promotes the proliferation of breast cancer cells in vitro and in vivo
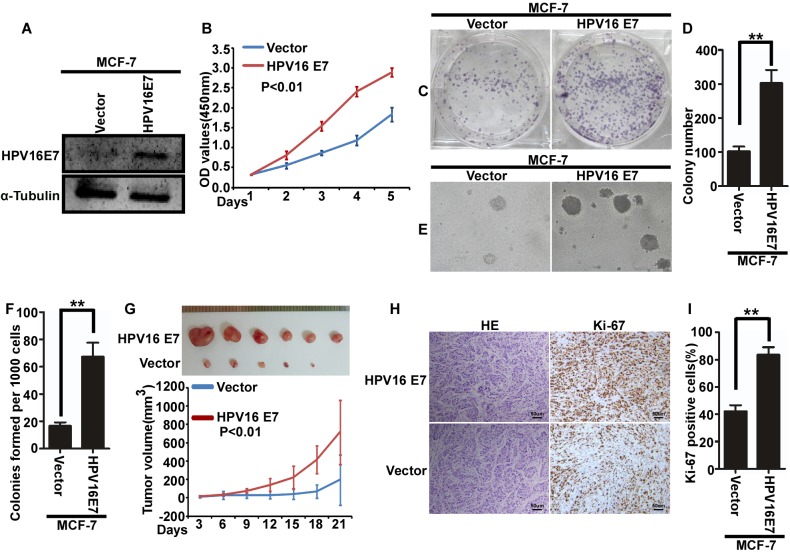
To further explore the function of HPV16 E7 in breast cancer, MCF-7 cells with stable expression of HPV16 E7 were used. MCF-7/vector cells were used as a control (Fig. 1A). Cell proliferation was assessed using a MTT (Fig. 1B), colony formation (Fig. 1C and D) and soft agar (Fig. 1E). The results demonstrated that stable expression of HPV16 E7 in MCF-7 cells promoted the proliferation of MCF-7 cells (Fig. 1B-F). Additionally, female BALB/c nude mice injected with MCF7/HPV16 E7 cells exhibited an increased tumor volume (Fig. 1G) and increased cellular proliferation (Fig. 1H, I) compared with control mice.
Figure 1.
HPV16 E7 promotes the proliferation of breast cancer cells in vitro and in vivo. (A) The expression of HPV16 E7 in MCF-7/HPV16 E7 and MCF-7/vector cells. (B) Cell proliferation as assessed using a MTT assay. (C) Representative images and (D) quantification of colony formation. (E) Representative images and (F) quantification of soft agar colony formation. (G) Tumor volume in mice injected with MCF-7/HPV16 E7 and MCF-7/vector cells (n=6/group). Tumor volume was evaluated at indicated days. (H) Tumor sections were stained with hematoxylin and eosin or with anti-ki-67 to evaluate proliferation, or subjected to IHC staining using an antibody against ki-67. (I) Quantification of ki-67 immunopositive cells in mice injected with MCF-7/HPV16 E7 and MCF-7/vector cells. **P<0.01. HPV, human papilloma virus; OD, optical density; H&E, hematoxylin and eosin.
HPV16 E7 promotes the proliferation of breast cancer cells by upregulating COX-2
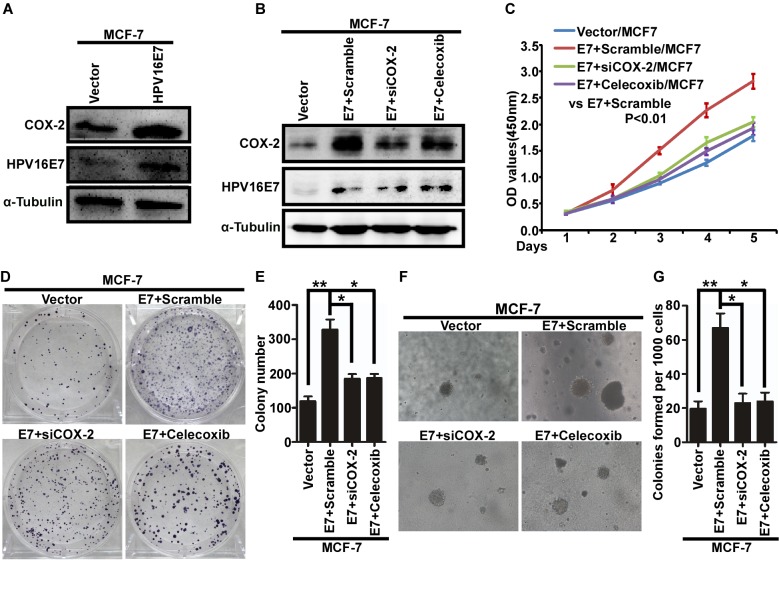
Western blot analysis demonstrated that the expression of COX-2 increased with stable expression of HPV16 E7 in MCF-7 cells (Fig. 2A). Next, MCF-7/HPV16 cells were treated with COX-2-specific siRNA (siCOX-2) and celecoxib to inhibit the expression of COX-2 (Fig. 2B). Proliferation was assessed in response to siCOX-2 and celecoxib treatment using a MTT (Fig. 2C), colony formation (Fig. 2D and E) and soft agar (Fig. 2F, G) and revealed that downregulation of COX-2 significantly inhibited proliferation of MCF-7/HPV16 cells (Fig. 2B-G). Therefore, these results suggest that HPV16 E7 may promote the proliferation of breast cancer cells by upregulating the expression of COX-2.
Figure 2.
HPV16 E7 promotes the proliferation of breast cancer cells by upregulating COX-2. (A) The expression of COX-2 in MCF-7/HPV16 E7 and MCF-7/vector cells using western blot analysis. (B) The expression of HPV16 E7 and COX-2 in response to siCOX-2 or celecoxib treatment using western blot analysis. (C) Analysis of cell proliferation using an MTT assay. (D) Representative images and (E) quantification of colony formation. (F) Representative images and (G) quantification of soft agar colony formation. *P<0.05, **P<0.01. HPV, human papilloma virus; COX-2, cyclooxygenase-2; siCOX-2, COX-2-specific small interfering RNA.
The association between the expression of HPV16 E7 and COX- in invasive breast ductal carcinoma
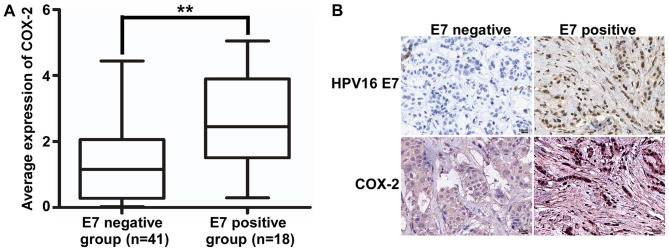
According to the results of IHC, immunopositive cells for HPV16 E7 were detected in the nucleus and the immunopositive cells for COX-2 were detected in the cytoplasm. The expression level of COX-2 DNA in HPV16 E7-positive samples was significantly increased compared with that in HPV16 E7-negative samples (Fig. 3A, Table II). Additionally, the expression of HPV16 E7 and COX-2 was also detected using immunohistochemistry. The results demonstrated that HPV16 E7 protein was expressed in 18 cases of invasive breast ductal carcinoma (Fig. 3B). Additionally, there was an association between HPV16 E7 and COX-2 expression in invasive breast ductal carcinoma (Table III).
Figure 3.
The association between the expression of HPV16 E7 and COX-2 in invasive breast ductal carcinoma. (A) The expression of COX-2 in HPV16 E7-positive and negative samples. **P<0.01. (B) Representative images of the expression of HPV16 E7 and COX-2 in invasive ductal breast carcinoma tissues by immunohistochemistry (magnification, ×400; scale bar, 50 µm). HPV, human papilloma virus; COX-2, cyclooxygenase-2.
Table III.
Association between the expression of HPV16 E7 and COX-2 in invasive ductal breast cancer tissues by immunohistochemistry.
| COX-2 expression | ||||
|---|---|---|---|---|
| HPV16 E7 expression | High | Low | χ2 value | P-value |
| Positive | 17 | 1 | 4.625 | 0.023 |
| Negative | 26 | 15 | ||
HPV, human papilloma virus; COX-2, cyclooxygenase-2.
Discussion
Breast cancer remains the leading cause of mortality in females worldwide. Breast cancer accounts for 1.7 million new cases and 0.5 million cases of mortality worldwide in 2012 (29). Additionally, the incidence and mortality rates of breast cancer have been increasing in the developing countries (1). HPV is a small, double-stranded and circular DNA virus that infects epithelial cells. Accumulating evidence has indicated that various HPVs, including HPV16, may be involved in the pathogenesis and development of breast cancer (12,30). However, the prevalence of HPV16 may vary widely. The detection rate of HPV16 in breast carcinoma varies from 0–86%, indicating an inconsistent association between HPV16 and the progression of breast cancer. A recent meta-analysis suggested an increased risk of breast cancer associated with HPV infection (31). HPV16 encodes a vital oncoprotein E7. HPV16 E7 serves an important function in the viral life cycle by regulating cellular proliferation and differentiation in the epithelium, and thus causing the virus to replicate and be constitutively expressed in differentiating epithelial cells (32). Additionally, HPV16 E7 may be a promising therapeutic target for cervical cancer (33).
In the present study, the expression of HPV16 E7 DNA was detected in patients with invasive breast ductal carcinoma from North China. The results demonstrated that HPV16 E7 DNA was detected in 30.5% cases of invasive breast ductal carcinoma, suggesting that HPV16 E7 may serve an important function in the pathogenesis of breast cancer. Previous studies demonstrated that infection with HPV16 E7 increased the proliferation of breast cancer cells (34,35). The results of the present study demonstrated that the expression of COX-2 in HPV16 E7-positive samples was increased compared with that in HPV16 E7-negative samples, suggesting that the expression of COX-2 may be upregulated by the expression of HPV16 E7.
Recent studies have investigated the function of COX-2 in tumorigenesis (36). COX-2 is an important COX inform, which is usually undetectable in normal tissues. Various factors, including hormones, cytokines and oncogenes may induce the expression of COX-2. It has been reported that COX-2 serve important functions in carcinogenesis and development of various tumors, including breast cancer (37,38). In vivo, it has been demonstrated that increased expression of COX-2 is associated with tumorigenesis (39). Recently, it has been demonstrated that the combination of specific COX-2 inhibitors with conventional chemotherapy may be used as a novel approach for the treatment of breast cancer. Celecoxib is a selective COX-2 inhibitor that has been used for the treatment of osteoarthritis and rheumatoid arthritis. Recently, it has been demonstrated that celecoxib prevented carcinogenesis, delayed cancer progression and enhanced the efficacy of conventional cancer therapies, including chemotherapy and radiation therapy (40–43). Considering the lack of effective therapeutic treatments for HPV-associated breast cancer, the use of celecoxib may be a promising approach. In the present study, the effect of HPV16 E7 on the expression of COX-2 was examined and it was demonstrated that COX-2 may be regulated by HPV16 E7. Additionally, in vitro experiments using COX-2 inhibitors demonstrated that HPV16 E7 promoted the proliferation of breast cancer cells by increasing the expression of COX-2.
The expression of HPV16 E7 and COX-2 was detected in 59 cases of invasive breast ductal carcinoma from North China using immunohistochemistry. The results revealed a positive association between the expression of HPV16 E7 and COX-2. These results suggest that COX-2 may be upregulated by HPV16 E7 and are in accordance with previous studies (24,44). Currently, there is no effective therapeutic approach for HPV-associated cancer (45). Thus, the development of therapeutic strategies for the treatment of HPV-induced cancer is required. It has been reported that celecoxib-treated HPV16+/- animals showed a lower incidence of epidermal dysplasia compared with untreated mice (46). Additionally, celecoxib was reported to significantly decrease breast tumor volume in rats (47). In conclusion, the use of celecoxib may be an effective therapeutic approach for the treatment of HPV-associated breast cancer.
In conclusion, the present study confirmed that HPV16 E7 may be an important factor in the pathogenesis of breast cancer in North China, and HPV16 E7 promoted the proliferation of breast cancer cells by upregulating the expression of COX-2. Additionally, the inhibition of COX-2 by siCOX-2 or celecoxib attenuated the HPV16 E7-mediated proliferation of breast cancer cells. Therefore, the present study may provide a potent therapeutic target for HPV16 E7-associated breast cancer.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Science and Technology Key Project of Henan Province Office Education of China (grant no. 14A310004) and the Scientific Research Fund of Xinxiang Medical University (grant nos. ZD200959, ZD2011-21 and ZD2011-11).
Availability of data and materials
The data generated or analyzed during this study are available from the corresponding author on reasonable request.
Author's contributions
XLQ and JC designed the experiments. YXW and ZYZ conducted experiments and wrote the manuscript. JQW provided research materials and methods and analyzed data.
Ethics approval and consent to participate
All participants provided written informed consent to participate and the tissue acquisition protocol was approved by the Ethic Institutional Board of Xinxiang Medical University.
Consent for publication
The relevant patients were informed and agreed to publication.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Denny L, de Sanjose S, Mutebi M, Anderson BO, Kim J, Jeronimo J, Herrero R, Yeates K, Ginsburg O, Sankaranarayanan R. Interventions to close the divide for women with breast and cervical cancer between low-income and middle-income countries and high-income countries. Lancet. 2017;389:861–870. doi: 10.1016/S0140-6736(16)31795-0. [DOI] [PubMed] [Google Scholar]
- 2.Brody JG, Rudel RA, Michels KB, Moysich KB, Bernstein L, Attfield KR, Gray S. Environmental pollutants, diet, physical activity, body size, and breast cancer: Where do we stand in research to identify opportunities for prevention? Cancer. 2017;109:2627–2634. doi: 10.1002/cncr.22656. [DOI] [PubMed] [Google Scholar]
- 3.Kaiser J. Cholesterol forges link between obesity and breast cancer. Science. 2013;342:1028. doi: 10.1126/science.342.6162.1028. [DOI] [PubMed] [Google Scholar]
- 4.Gage M, Wattendorf D, Henry LR. Translational advances regarding hereditary breast cancer syndromes. J Surg Oncol. 2012;105:444–451. doi: 10.1002/jso.21856. [DOI] [PubMed] [Google Scholar]
- 5.Anothaisintawee T, Wiratkapun C, Lerdsitthichai P, Kasamesup V, Wongwaisayawan S, Srinakarin J, Hirunpat S, Woodtichartpreecha P, Boonlikit S, Teerawattananon Y, Thakkinstian A. Risk factors of breast cancer: A systematic review and meta-analysis. Asia Pac J Public Health. 2013;25:368–387. doi: 10.1177/1010539513488795. [DOI] [PubMed] [Google Scholar]
- 6.Glaser SL, Canchola AJ, Keegan TH, Clarke CA, Longacre TA, Gulley ML. Variation in risk and outcomes of Epstein-Barr virus-associated breast cancer by epidemiologic characteristics and virus detection strategies: An exploratory study. Cancer Causes Control. 2017;28:273–287. doi: 10.1007/s10552-017-0865-3. [DOI] [PubMed] [Google Scholar]
- 7.Wu H, Zhao C, Adhikari VP, Lu L, Huang J, Wei Y, Luo Q, Dai W, Wu Y, Li X, et al. The prevalence and clinicopathological features of breast cancer patients with hepatitis B virusinfection in China. Oncotarget. 2017;8:18185–18190. doi: 10.18632/oncotarget.15305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.White EA, Munger K. Crowd control: E7 conservation is the key to cancer. Cell. 2017;170:1057–1059. doi: 10.1016/j.cell.2017.08.033. [DOI] [PubMed] [Google Scholar]
- 9.Yin W, Duluc D, Joo H, Oh S. Dendritic cell targeting vaccine for HPV-associated cancer. Cancer Cell Microenviron. 2016;3:e1482. [PMC free article] [PubMed] [Google Scholar]
- 10.Torres-Rueda S, Rulisa S, Burchett HE, Mivumbi NV, Mounier-Jack S. HPV vaccine introduction in Rwanda: Impacts on the broader health system. Sex Reprod Healthc. 2016;7:46–51. doi: 10.1016/j.srhc.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 11.Einstein MN, Smith KM, Davis TE, Schmeler KM, Ferris DG, Savage AH, Gray JE, Stoler MH, Wright TC, Jr, Ferenczy A, Castle PE. Clinical evaluation of the cartridge-based GeneXpert human papillomavirus assay in women referred for colposcopy. J Clin Microbiol. 2014;52:2089–2095. doi: 10.1128/JCM.00176-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ilahi NE, Anwar S, Noreen M, Hashmi SN, Murad S. Detection of human papillomavirus-16 DNA in archived clinical samples of breast and lung cancer patients from North Pakistan. J Cancer Res Clin Oncol. 2016;142:2497–2502. doi: 10.1007/s00432-016-2251-z. [DOI] [PubMed] [Google Scholar]
- 13.Omura Y, Jones MK, Nihrane A, Duvvi H, Shimotsuura Y, Ohki M. More than 97% of human papilloma virus type 16 (HPV-16) was found with chrysotile asbestos & relatively smooth round tumor outline, and less than 3% was found with HPV-18 and tremolite asbestos & irregular sawtooth-like zigzag outline in breast cancer tissues in over 500 mammograms of female patients: Their implications in diagnosis, treatment, and prevention of breast cancer. Acupunct Electrother Res. 2013;38:211–230. doi: 10.3727/036012913X13831832269324. [DOI] [PubMed] [Google Scholar]
- 14.Tavassoli FA, Devolee P. Worldhealth organization classification of tumors, pathology and genetics of the breast and female genital organs. IARC press; 2003. [Google Scholar]
- 15.Wu KK. Cyclooxygenase 2 induction: Molecular mechanism and pathophysiologic roles. J Lab Clin Med. 1996;128:242–245. doi: 10.1016/S0022-2143(96)90023-2. [DOI] [PubMed] [Google Scholar]
- 16.Harris RE, Casto BC, Harris ZM. Cyclooxygenase-2 and the inflammogenesis of breast cancer. World J Clin Oncol. 2014;5:677–692. doi: 10.5306/wjco.v5.i4.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu B, Qu L, Yan S. Cyclooxygenase-2 promotes tumor growth and suppresses tumor immunity. Cancer Cell Int. 2015;15:106. doi: 10.1186/s12935-015-0260-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gebhardt BM, Varnell ED, Kaufman HE. Inhibition of cyclooxygenase 2 synthesis suppresses Herpes simplex virus type 1 reactivation. J Ocul Pharmacol Ther. 2005;21:114–120. doi: 10.1089/jop.2005.21.114. [DOI] [PubMed] [Google Scholar]
- 19.Lee SM, Gai WW, Cheung TK, Peiris JS. Antiviral effect of a selective COX-2 inhibitor on H5N1 infection in vitro. Antiviral Res. 2011;91:330–334. doi: 10.1016/j.antiviral.2011.07.011. [DOI] [PubMed] [Google Scholar]
- 20.Lin CK, Tseng CK, Chen KH, Wu SH, Liaw CC, Lee JC. Betulinic acid exerts anti-hepatitis C virus activity via the suppression of NF-κB- and MAPK-ERK1/2-mediated COX-2 expression. Br J Pharmacol. 2015 doi: 10.1111/bph.13233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao Q, Guo J, Wang G, Chu Y, HU X. Suppression of immune regulatory cells with combined therapy of celecoxib and sunitinib in renal cell carcinoma. Oncotarget. 2017;8:1668–1677. doi: 10.18632/oncotarget.13774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tury S, Becette V, Assayag F, Vacher S, Benoist C, Kamal M, Marangoni E, Bièche I, Lerebours F, Callens C. Combination of COX-2 expression and PIK3CA mutation as prognostic and predictive markers forcelecoxib treatment in breast cancer. Oncotarget. 2016;7:85124–85141. doi: 10.18632/oncotarget.13200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Riva B, De Dominici M, Gnemmi I, Mariani SA, Minassi A, Minieri V, Salomoni P, Canonico PL, Genazzani AA, Calabretta B, Condorelli F. Celecoxib inhibits proliferation and survival of chronic myelogeous leukemia (CML) cells via AMPK-dependent regulation of β-catenin and mTORC1/2. Oncotarget. 2016;7:81555–81570. doi: 10.18632/oncotarget.13146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Subbaramaiah K, Dannenberg AJ. Cyclooxygenase-2 transcription is regulated by human papillomavirus 16 E6 and E7 oncoproteins: Evidence of a corepressor/coactivator exchange. Cancer Res. 2007:3976–3985. doi: 10.1158/0008-5472.CAN-06-4273. [DOI] [PubMed] [Google Scholar]
- 25.Zhuang ZH, Tsao SW, Deng W, Wang JD, Xia HH, He H, Feng HC, Wang LD, Gu Q, Lam SK, et al. Early upregulation of cyclooxygenase-2 in human papillomavirus type 16 and telomerase-induced immortalization of human esophageal epithelial cells. J Gastroenterol Hepatol. 2008;23:1613–1620. doi: 10.1111/j.1440-1746.2008.05509.x. [DOI] [PubMed] [Google Scholar]
- 26.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T))method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 27.Friedrichs K, Gluba S, Eidtmann H, Jonat W. Overexpression of p53 and prognosis in breast cancer. Cancer. 1993;72:3641–3647. doi: 10.1002/1097-0142(19931215)72:12<3641::AID-CNCR2820721215>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 28.Li N, Ye YP, Zhu HF, et al. The effect and mechanism of HPV16 E6 and E7 on the invasion of breast cancer. Chongqing Yixue. 2012;41:2572–2574. [Google Scholar]
- 29.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 30.Zhou Y, Li J, Ji Y, Ren M, Pang B, Chu M, Wei L. Inconclusive role of human papillomavirus infection in breast cancer. Infect Agent Cancer. 2016;10:36. doi: 10.1186/s13027-015-0029-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bae JM, Kim EH. Human papillomavirus infection and risk of breast cancer: A meta-analysis of case control studies. Infect Agents Cancer. 2016;11:14. doi: 10.1186/s13027-016-0058-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Münger K, Basile JR, Duensing S, Eichten A, Gonzalez SL, Grace M, Zacny VL. Biological activities and molecular targets of the human papillomavirus E7 oncoprotein. Oncogene. 2001;20:7888–7898. doi: 10.1038/sj.onc.1204860. [DOI] [PubMed] [Google Scholar]
- 33.Grabowska AK, Kaufmann AM, Riemer AB. Identification of promiscuous HPV16-derived T helper cell epitopes for therapeutic HPV vaccine design. Int J Cancer. 2015;136:212–224. doi: 10.1002/ijc.28968. [DOI] [PubMed] [Google Scholar]
- 34.Liu Y, Wang YN, Chen CQ, Zhang J, Qian W, Dong Y, Liu Z, Zhang X, Wang X, Zhang Z, et al. LSD1 binds to HPV16 E7 and promotes the epithelial-mesenchymal transition in cervical cancer by demethylating histones at the Vimentin promoter. Oncotarget. 2017;8:11329–11342. doi: 10.18632/oncotarget.13516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Organista-Nava J, Gómez-Gómez Y, Ocadiz-Delgado R, García-Villa E, Bonilla-Delgado J, Lagunas-Martínez A, Tapia JS, Lambert PF, García-Carrancá A, Gariglio P. The HPV16 E7 oncoprotein increases the expression of Oct3/4 and stemness-related genes and augments cell self-renewal. Virology. 2016;499:230–242. doi: 10.1016/j.virol.2016.09.020. [DOI] [PubMed] [Google Scholar]
- 36.Xu F, Li M, Zhang C, Cui J, Liu J, Li J, Jiang H. Clinicopathological and prognostic significance of COX-2 immunohistochemical expression in breast cancer: A meta-analysis. Oncotarget. 2017;8:6003–6012. doi: 10.18632/oncotarget.13990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Campillo N, Torres M, Vilaseca A, Nonaka PN, Gozal D, Roca-Ferrer J, Picado C, Montserrat JM, Farré R, Navajas D, Almendros I. Role of Cyclooxygenase-2 on intermittent hypoxia-induced lung tumor Malignancy in a mouse model of sleep apnea. Sci Rep. 2017;7:44693. doi: 10.1038/srep44693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hsu HH, Chen MC, Day CH, Lin YM, Li SY, Tu CC, Padma VV, Shih HN, Kuo WW, Huang CY. Thymoquinone suppresses migration of LoVo human colon cancer cells by reducing prostaglandin E2 induced COX-2 activation. World J Gastroenterol. 2017;23:1171–1179. doi: 10.3748/wjg.v23.i7.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Howe LR. Inflammation and breast cancer. Cyclooxygenase/prostaglandin signaling and breast cancer. Breast Cancer Res. 2007;9:210. doi: 10.1186/bcr1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shirali S, Barari A, Hosseini SA, Khodadi E. Effects of six weeks endurance training and aloe vera supplementation on COX-2 and VEGF levels in mice with breast cancer. Asian Pac J Cancer Prev. 2017;18:31–36. doi: 10.22034/APJCP.2017.18.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun J, Liu NB, Zhuang HQ, Zhao LJ, Yuan ZY, Wang P. Celecoxib-erlotinib combination treatment enhances radiosensitivity in A549 human lung cancer cell. Cancer Biomark. 2017;19:45–50. doi: 10.3233/CBM-160323. [DOI] [PubMed] [Google Scholar]
- 42.Pridgen WL, Duffy C, Gendreau JF, Gendreau RM. A famciclovir + celecoxib combination treatment is safe and efficacious in the treatment of fibromyalgia. J Pain Res. 2017;10:451–460. doi: 10.2147/JPR.S127288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu B, Yan S, Qu L, Zhu J. Celecoxib enhances anticancer effect of cisplatin and induces anoikis in osteosarcoma via PI3K/Akt pathway. Cancer Cell Int. 2017;17:1. doi: 10.1186/s12935-016-0378-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu R, Abramson AL, Shikowitz M, Dannenberg AJ, Steinberg BM. Epidermal growth factor-induced cyclooxygenase-2 expression is mediated through phosphatidylinositol-3 kinase, not mitogen-activated protein/extracellular signal-regulated kinase kinase, in recurrent respiratory papillomas. Clin Cancer Res. 2005;11:6155–6161. doi: 10.1158/1078-0432.CCR-04-2664. [DOI] [PubMed] [Google Scholar]
- 45.Yang A, Jeang J, Cheng K, Cheng T, Yang B, Wu TC, Hung CF. Current state in the development of candidate therapeutic HPV vaccines. Expert Rev Vaccines. 2016;15:989–1007. doi: 10.1586/14760584.2016.1157477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.DA Costa RMG, Araújo R, Santos JMO, Fernandes M, Neto T, Sousa H, Ribeiro J, Bastos MMSM, Oliveira PA, Carmo D, et al. Regulation of miRNA-146a and miRNA-150 levels by celecoxib in premalignant lesions of K14-HPV16 Mice. Anticancer Res. 2017;37:2913–2918. doi: 10.21873/anticanres.11644. [DOI] [PubMed] [Google Scholar]
- 47.Alshafie GA, Abou-Issa HM, Seibert K, Harris RE. Chemotherapeutic evaluation of Celecoxib, a cyclooxygenase-2 inhibitor, in a rat mammarytumor model. Oncol Rep. 2000;7:1377–1381. doi: 10.3892/or.7.6.1377. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data generated or analyzed during this study are available from the corresponding author on reasonable request.