Abstract
In the present study, the anti-tumor effects of combination treatment with an siRNA targeting B-Raf proto-oncogene serine/threonine kinase (BRAF)V600E and phosphoinositide 3-kinase (PI3K) signaling pathway inhibitors was investigated in melanoma cell lines harboring BRAFV600E. Human melanoma A375 and WM115 cells were treated with siRNA targeting to BRAF or BRAFV600E, combined with treatment with PI3K signaling pathway inhibitors. CCK-8 and EdU proliferation assays were performed to assess cell viability and proliferation, respectively, following treatment. In addition, flow cytometry analysis was performed to determine cell cycle distribution, and western blot analysis was performed to analyze the activity of the extracellular signal-regulated kinase (ERK) and PI3Ksignaling pathways following treatment. Targeting BRAFV600E using small interfering (si)RNA significantly decreased cell viability and DNA replication in tumor cell lines that harbor oncogenic BRAFV600E. Inhibition of BRAFV600E by siRNA combined with treatment with PI3K or mammalian target of rapamycin signaling pathway inhibitors significantly decreased cell viability and proliferation compared with siRNA or inhibitor treatment alone. Concomitant BRAFV600E and PI3K inhibition led to G1/S phase arrest in melanoma cells. However, melanoma cells in which oncogenic BRAFV600E is not highly expressed (WM115 cells) were not sensitive to BRAFV600E targeted therapy. The PI3K signaling pathway inhibitors were more effective in this cell line. The results from the present study provide an insight into the potential effectiveness of combination therapy and personalized cancer treatments.
Keywords: melanoma, BRAFV600E siRNA, ERK pathway, PI3K pathway, combination therapy
Introduction
Malignant melanoma is the most dangerous form of skin tumor, accounting for the majority of cancer-associated mortalities (1). Due to the high rate of metastasis at an early stage of the disease and resistance to conventional chemotherapy and radiotherapy, the morbidity and mortality rates for malignant melanoma increase faster each year than any other type of cancer (1,2). Previous studies have demonstrated that melanoma is a heterogeneous disease (3,4) and the optimal treatment for patients with melanoma is personalized based on the presence of specific molecular abnormalities. Numerous abnormalities have been identified in melanoma, including mutations in receptor tyrosine kinase (RTK) genes [including KIT proto-oncogene (KIT), EPH receptor A2, erb-b2 receptor tyrosine kinase 4 and platelet derived growth factor receptor (PDGFR)], the RAS family of small G protein genes [including NRAS proto-oncogene GTPase (NRAS) and HRAS proto-oncogene GTPase] and cytoplasm kinase genes [B-Raf proto-oncogene serine/threonine kinase (BRAF), RAF family, mitogen-activated protein kinase kinase (MEK)1/2, AKT serine/threonine kinase 3 and phosphatase and tensin homolog] (5). Mutations in the BRAF, NRAS and KIT genes are more active compared with other genes (6).
BRAF kinases have been demonstrated to be associated with melanoma and other types of cancer (7,8). The substitution of glutamic acid for valine at position 600 (BRAFV600E) accounts for >80% of BRAF mutations and ~66% of melanomas. These mutations lock the BRAF kinase enzyme into a constitutively activated state, which promotes tumorigenesis by hyperactivating the extracellular signal-regulated kinase (ERK) signaling pathway without stimulation by RTKs (9). Therefore, the BRAFV600E mutation has been considered as an attractive therapeutic target for melanoma, leading to the development of specific BRAFV600E inhibitors. In 2011 and 2013, respectively, vemurafenib and dabrafenib were approved by the US Food and Drug Administration (FDA) for single agent or combinational treatment of metastatic and unresectable BRAF-mutated melanomas. Although administration of these drugs leads to significant levels of tumor shrinkage in patients who harbor the BRAFV600E mutation, ≤25% of patients develop cutaneous squamous cell carcinomas (primarily karatoacanthomas) and drug resistance. The underlying molecular mechanisms mediating resistance to BRAF inhibitors in melanoma are complex (10). The paradoxical activation of the mitogen activated protein kinase (MAPK) pathway in BRAF wild-type cells by homo- or hetero-dimerization of RAF isoforms or the upregulation of RTKs, including PDGFβR or insulin-like growth factor 1 receptor, are mechanisms of drug resistance as well as mutations in KRAS, NRAS (Q61K and Q61R) (11), MEK1 (C121S and P124L) or MEK2 (Q60P) (12).
However, previous studies have demonstrated that ATP competitive RAF inhibitors are not only poor inhibitors of wild-type BRAF but also increase Raf-1 proto-oncogene serine/threonine kinase activity to activate the ERK signaling pathway in BRAF wild-type cells (13–16). Compared with small molecular inhibitors, gene therapy targeting of disease-associated genes demonstrates high efficiency, high specificity and relatively low toxicity to healthy tissue. Small interfering (si)RNA targeted to active oncogenic BRAFV600E kinase could provide a promising treatment for BRAF-mutant melanomas (17).
Cancer cells constantly adapt to promote survival and escape the immune system, thus ensuring tumor growth. The phosphoinositide-3-OH kinase (PI3K)/AKT/mammalian target of rapamycin (mTOR) signaling pathway has an important role in cell proliferation, survival and apoptosis. In melanoma, the PI3K/RAC-α serine-threonine-protein kinase (AKT)/mTOR and Raf/MEK/ERK signaling cascades are interconnected with multiple points of convergence, cross-talk, and feedback loops. Inhibition of the two pathways could be more effective than inhibiting either pathway alone (18). At present, multiple clinical trials are underway with combined inhibition of the ERK and PI3K pathways, including Vemurafenib with PX-866 (PI3K inhibitor) (trial no. NCT01616199), or Dabrafenib with GSK2141795 (AKT inhibitor) (trial no. NCT01902173) to determine if this strategy can stop tumors from developing drug resistance in BRAF-mutant melanoma (19).
BRAFV600E siRNA in combination with PI3K signaling pathway inhibition may benefit patients with BRAFV600E-positive tumors and act with high efficiency and low toxicity. The present study aimed to examine the effects of BRAFV600E siRNA combined with different types of PI3K signaling pathway inhibitors on BRAFV600E mutant melanoma cell lines.
Materials and methods
Reagents
Antibodies used in the present study were as follows: Anti-BRAF (catalog no., Ab33899; Abcam, Cambridge, UK); anti-MEK1 (catalog no., 9146), anti-MEK2 (catalog no.,), anti-ERK1/2 (catalog no., 4695), anti-phospho-AKT (Ser473) (catalog no., 4060s), anti-phospho-MEK1/2 (Ser217/211) (catalog no., 9154), anti-phospho-ERK1/2 (Thr202/Tyr204) (catalog no., 4377), anti-phospho-S6 (Ser240/244) (catalog no., 5364), anti-GAPDH (catalog no., 2118) and anti-β-actin (catalog no., 3700) (Cell Signaling Technology, Inc., Danvers, MA, USA); and horseradish peroxidase (HRP)-conjugated anti-rabbit immunoglobulin G (IgG) (catalog no., ZDR-5306) and HRP-conjugated anti-mouse IgG (catalog no., ZDR-5307) (ZSGB-Bio, Beijing, China). The PI3K/AKT/mTOR signaling pathway inhibitors PI-103 (catalog no., S1038), GSK690693 (catalog no., S1113), ZSTK474 (catalog no., S1072) and AZD8055 (catalog no., S1555) were purchased from Selleck Chemicals (Houston, TX, USA). All siRNA sequences were synthesized and purified with high performance liquid chromatography by GenePharma Co., Ltd. (Shanghai, China). All other reagents were of analytical grade and obtained from commercial sources.
Cell culture
Human embryonic kidney HEK293A and melanoma A375 cell lines were obtained from the Cell Resource Center, Institute of Basic Medical Sciences, Chinese Academy of Medical Sciences/Peking Union Medical College (Beijing, China). The melanoma cell line WM115 was obtained from the American Type Culture Collection (Manassas, VA, USA). All cells were cultured in Dulbecco's modified Eagle's medium (Macgene, Beijing, China). All cells were supplemented with 10% fetal bovine serum (FBS; Corning Incorporated, Corning, NY, USA) at 37°C with 5% CO2.
Cell transfection
For siRNA silencing, cells were seeded at 2×105 cells/35-mm2/well one day prior to transfection. Cells were transfected using Lipofectamine® RNAiMAX transfection reagent (Thermo Fisher Scientific, Inc., Waltham, MA, USA) in 0.5 ml of GenOpti (Macgene) with siRNAs as follows: siMB3 (siBraf-mu; targeted to BRAFV600E; antisense, 5′-AUCGAGAUUUCUCUGUAGCdtdt-3′ and sense, 5′-GCUACAGAGAAAUCUCGAUdtdt-3′); siWTM (siBraf-wtm; targeted to wild-type BRAF and BRAFV600E; antisense, 5′-AUGAUCCAGAUCCAAUUCUdtdt-3′ and sense, 5′-AGAAUUGGAUCUGGAUCAUdtdt-3′); siMEK1 (antisense, 5′-AGCAUGAACCAUGAGUUGCdtdt-3′ and sense, 5′-GCAACUCAUGGUUCAUGCUdtdt-3′); siMEK2 (antisense, 5′-TGCTGTGAGGCTCTCCTTCdtdt-3′ and sense, 5′-GAAGGAGAGCCUCACAGCAdtdt-3′) or negative control siRNA (siControl; Guangzhou Ribo Bio, Co., Ltd., Guangzhou, China) at different concentrations, as stated in the appropriate figure legends, into 1.5 ml medium containing 10% FBS, according to the manufacturer's protocol. Cells were treated and harvested following the transfection as described in the figure legends.
Gene expression evaluation by reverse transcription-polymerase chain reaction (RT-PCR)
Total RNA was isolated from cells with TRIzol (Thermo Fisher Scientific, Inc.), then chloroform was added and the sample was mixed and centrifuged (12,000 × g) for 15 min at 4°C. Total RNA was extracted from the supernatant and purified according to the manufacturer's protocol. Total RNA was reverse transcribed using the Reverse Transcription System (Promega Corporation, Madison, WI, USA). Reverse transcription conditions were as follows: 42°C for 15 min, 95°C for 5 min, and 0–5°C for 5 min. The complementary DNA was subjected to PCR with GoTaq® Green Master Mix (Promega Corporation) using the Techne TC-5000 PCR Thermal Cycler (GMI, Ramsey, MN, USA) according to the manufacturer's protocol. The primers used were as follows: β-actin forward, 5′-CCAACCGCGAGAAGATGA-3′ and reverse, 5′-CCAGAGGCGTACAGGGATAG-3′); Total-braf forward, 5′-CTGCCTCATTACCTGGCTCACTA-3′ and reverse, 5′-CACCATGCCACTTTCCCTTGT-3′; and Braf(T1599A) forward, 5′-TGGTGTGAGGGCTCCAGCTTGT-3′ and reverse, 5′-ATGGGACCCACTCCATCGAGATTTCT-3′. PCR was performed as follows: 95°C for 5 min (1 cycle), 95°C for 30 sec, 60°C for 45 sec, 72°C for 45 sec (25 cycles for β-actin and 31 cycles for Total-braf and Braf(T1599A)) and 72°C for 5 min (1 cycle). The reaction solution was analyzed by 1% agarose gel electrophoresis with ethidium bromide staining. β-actin was used as a control. Finally, the products were analyzed by Image Lab™ software 6.0 (ChemiDoct XRS System; Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Western blot analysis
Treated cells were harvested with cell lysis buffer (20 mM TrisCl pH 7.5, 150 mM NaCl, 1% Triton X-100; catalog no., P1003; Beyotime Institute of Biotechnology, Haimen, China) supplemented with a protease inhibitor cocktail (Pierce; Thermo Fisher Scientific, Inc.). Cell extracts were normalized for protein content using a BCA protein assay kit (Pierce; Thermo Fisher Scientific, Inc.). Total protein (8 µg) was separated by 10% SDS-PAGE, transferred to polyvinylidene fluoride membranes (catalog no., IPVH00010; EMD Millipore, Billerica, MA, USA) and blocked with 5% (w/v) milk in TBS-Tween. Western blot analysis was performed following standard protocols using the indicated antibodies [anti-BRAF, anti-MEK1, anti-MEK2, anti-ERK1/2, anti-phospho-AKT (Ser473), anti-phospho-MEK1/2 (Ser217/211), anti-phospho-ERK1/2 (Thr202/Tyr204), anti-phospho-S6 (Ser240/244), anti-GAPDH, anti-β-actin, HRP-conjugated anti-rabbit IgG and HRP-conjugated anti-mouse IgG]. The primary antibodies were incubated at 25°C for 1 h, and the secondary antibodies were incubated at 25°C for 3 h. The membrane was developed using the chemiluminescent HRP substrate kit followed by the procedure provided by the manufacturer (EMD Millipore). The densitometric quantification of the protein bands was determined using the ChemiDoc™ XRS+System (Bio-Rad Laboratories, Inc.).
Cell viability assay
Cells (A375 or WM115) in 96-well plates (4,000 cells/well) were transfected with siRNA or treated with gradient concentrations of the compounds at 37°C for 48 or 72 h. Cell proliferation was determined using a Cell Counting Kit-8 (CCK-8; Dojindo Molecular Technologies, Inc., Kumamoto, Japan) according to the manufacturer's protocol. Briefly, CCK-8 was added 1.5 h prior to detection of the optical density (OD) at 450 nm with a micro-plate reader. Cell viability was calculated as follows: Cell viability=(ODsample-ODblank)/(ODNC-ODblank). ODNC indicated the OD value of culture medium of the cells that were transfected by negative control siRNA, which was used as a control. ODblank indicated the OD value of culture medium at 450 nm. The half-maximal inhibitory concentration (IC50) was calculated using GraphPad Prism software (version 5; GraphPad Software, Inc., La Jolla, CA, USA) and presented as the mean with 95% confidence limits.
5-ethynyl-2′-deoxyuridine (EdU) proliferation assay
Cell proliferation was determined using the Cell-Light™ EdU Apollo®567 In Vitro Imaging kit (Guangzhou RiboBio, Co., Ltd.). A375 cells were seeded in 96-well plates (4,000 cells/cell) one day prior to treatment. Following treatment, cells were incubated with 50 µM EdU at the indicated times as stated in the figure legends for 2 h prior to fixation, permeabilization and staining. Cell nuclei were stained with 1X Hoechst 33342 for 30 min. The images were obtained with a High Content Screening machine Operetta™ (PerkinElmer, Inc., Waltham, MA, USA) and the images were analyzed using Harmony 3.5.1 (PerkinElmer, Inc.). The border cells with irregular nuclei were considered, which were removed with a common filter (using the ‘Select Population’ function, with ‘Nuclei’ as ‘Population’, the ‘Common Filter’ selected as the ‘Method’ and ‘hoechst’ selected as the selective objects), and cells whose intensity in the Cy3 channel was 1.5 times higher than the background were defined as EdU-positive cells. The percentage of EdU-positive cells was calculated from 12 randomly selected fields/well. The data were normalized to the control cells and presented as percentages.
Cell cycle analysis
Treated cells (105-106 cells/plate) were harvested and washed with PBS and then fixed with pre-cooled 70% ethanol at 4°C overnight. The cell pellets were washed and suspended in PBS containing 20 µg/ml RNase A at 37°C for 30 min. DNA was stained with 20 µg/ml propidium iodide (M&C) and 0.1% Triton X-100 (Thermo Fisher Scientific, Inc.). The cells were analyzed using a FACSCalibur™ flowcytometer (BD Biosciences, Franklin Lakes, NJ, USA), and cell cycle analysis was performed using ModFit LT3.2 software (Verity Software House, Topsham, ME, USA).
Statistical analysis
All data are presented as the mean ± standard deviation. Statistical analyses were performed using a two-tailed unpaired t-test with GraphPad Prism software (version 5; GraphPad Software, Inc.). P<0.05 was considered to indicate a statistically significant difference. Unless otherwise specified, all assays were performed in triplicate.
Results
siRNA targeting of mutant BRAFV600E decreased the viability of BRAFV600E mutant melanoma cell lines
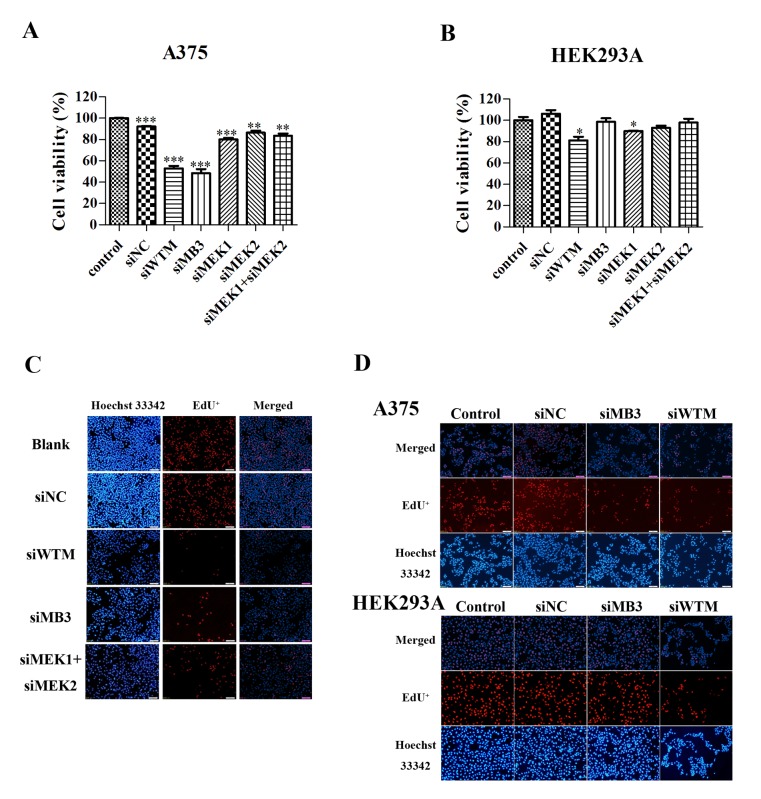
The specificity and efficiency of siWTM and siMB3 on the A375 melanoma cell line, which harbors the Braf(T1599A) mutation, and normal HEK293A cells was investigated. siWTM, which targets wild-type BRAF and mutant BRAFV600E, significantly decreased the viability of A375 and HEK293A cells (P<0.001 and P<0.05, respectively; Fig. 1A and B). The siMB3 siRNA, which specifically targets BRAFV600E, significantly decreased the viability of A375 cells (P<0.001), but did not significantly decrease HEK293A cell viability compared with siWTM. MEK1 silencing significantly decreased the viability of A375 and HEK293A cells (P<0.001 and P<0.05, respectively; Fig. 1A and B); however, MEK2 and MEK1/2 combined silencing significantly decreased the viability of A375 cells (P<0.01; Fig. 1A), but not HEK293A cells.
Figure 1.
BRAF and BRAFV600E-targeted siRNAs significantly decrease A375 cell viability. Viability of (A) A375 and (B) HEK293A cells following treatment with 30 nmol/l of siWTM, siMB3, siMEK1, siMEK2 or siNC siRNA for 48 h. (C) DNA replication was assessed by the EdU method in melanoma A375 cells. Cells were treated with or without 30 nM of siWTM, siMB3, siMEK1 +siMEK2, and siControl for 48 h. EdU was stained cells (red) and Hochest 33342 (blue) was stained the nuclei of total cells. (D) The DNA replication measurement on A375 and HEK293A cells with 30 nM of siWTM or siMB3. EdU was stained (red) following 48 h treatment. Data are represented as the mean ± standard deviation (n=3). *P<0.05, **P<0.01, ***P<0.001, compared with the corresponding control. siRNA, small interfering RNA; EdU, 5-ethynyl-2′-deoxyuridine; BRAF, B-Raf proto-oncogene serine threonine kinase; siWTM, siRNA targeting wild type BRAF and mutant BRAFV600E; siMB3, siRNA targeting mutant BRAFV600E; MEK, mitogen-activated protein kinase kinase; NC, negative control.
To investigate the molecular mechanisms underlying the effects of the siRNA treatments on cell viability, EdU retention assays were performed to examine the regulatory effect of the two BRAF-targeted siRNAs on DNA replication in A375 and HEK293A cell lines. A375 and HEK293Acells treated with siMB3 and siWTM exhibited decreased DNA replication 48 h following treatment (Fig. 1C and D). In addition, A375 cells treated with siMEK1/2 exhibited decreased DNA replication. This data demonstrates that targeting BRAFV600E with siRNA markedly decreases cell viability and inhibits DNA replication.
Activity of the RAF/MEK/ERK and PI3K/AKT/mTOR signaling pathways in melanoma cell lines
Comparison of the expression of BRAFV600E in HEK293A, A375 and WM115 cells lines revealed that BRAFV600E expression in WM115 cells was markedly lower than that in A375 cells (Fig. 2A). HEK293A cells were included as a control, as they did not express the mutant protein BRAFV600E. Western blot analysis revealed that levels of phosphorylated (p)ERK1/2, pAKT (S473) and pS6 ribosomal protein [pS6; a surrogate readout for mammalian target of rapamycin complex 1 (mTORC1) activity] were markedly increased in WM115 cells compared with A375 cells, indicating that the activity of the RAF/MEK/ERK and PI3K/AKT/mTOR signaling pathways was increased in WM115 cells compared with A375 cells (Fig. 2B). In A375 cells, the hyperactivation of the ERK pathway by the BRAFV600E mutation is a major contributor to tumorigenesis (8,9). siMB3 treatment decreased cell viability of the melanoma A375 cell line (almost 60% inhibition) (Fig. 2C). Although WM115 cells exhibited increased expression of pERK, pAKT (S483) and pS6 protein compared with A375 cells (Fig. 2B), they exhibited decreased expressionof mutant BRAF at the mRNA level (Fig. 2A) and exhibited less sensitivity to siRNA targeting BRAFV600E compared with the A375 cells.
Figure 2.
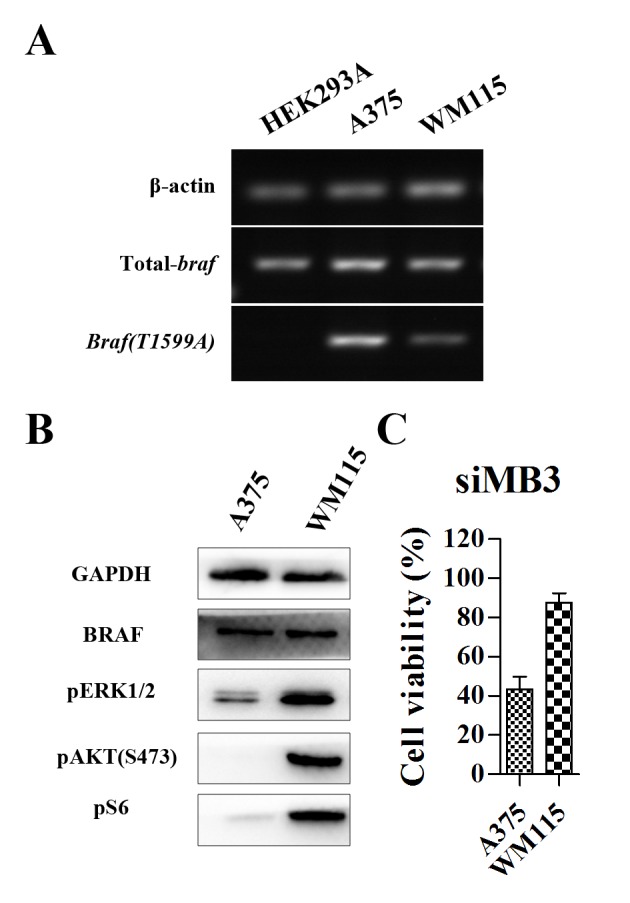
Expression of total and mutant BRAF is increased in A375 compared with HEL293A and WM115 cells. (A) Measurement of mRNA level of mutant BRAFV600E in melanoma A375, WM115 and HEK293A cell lines. (B) Measurement of the protein expression of BRAF, pERK1/2, pAKT(S473), pS6 in A375 and WM115 melanoma cell lines by western blot analysis. (C) Cell viability of A375 and WM115 cells, measured using a CCK-8 assay following treatment with 5 nM of siMB3 for 72 h. p, phosphorylated; ERK, extracellular signal-regulated kinase; AKT, RAC-α serine-threonine-protein kinase; S6, S6 ribosomal protein.
Effects of BRAFV600E siRNA combined with PI3K signaling pathway inhibitors on cell viability
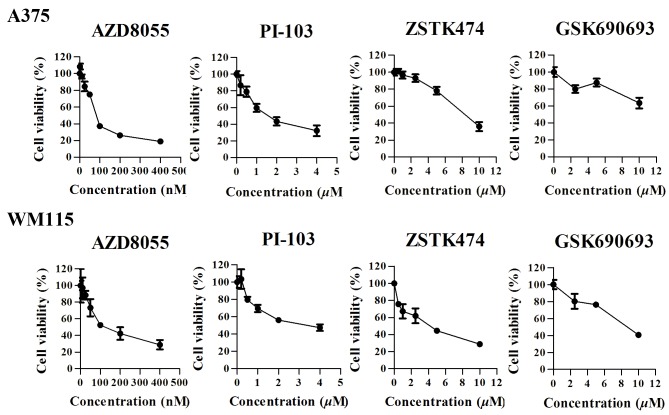
To explore the role of the PI3K signaling pathway in the regulation of cell growthinBRAFV600-positive cell lines, A375 and WM115 cells were treated with four types of kinase inhibitors, including the dual PI3K/mTOR inhibitor PI-103, the mTOR inhibitor AZD8055, the pen-class I PI3K inhibitor ZSTK474 and the AKT inhibitor GSK690693. A375 and WM115 cells were treated with increasing concentrations of the four inhibitors, and cell viability was measured following treatment for 72 h using a CCK-8 assay. The four drugs exhibited different effects on cell viability in the two cell lines (Fig. 3). AZD8055 exhibited a dose-dependent antitumor effect on A375 cells, with an IC50 value of 91.67 nM (95%CI: 85.22 to 98.11 nM). At a concentration of 100 nM the growth inhibition rate was 63.7±2.69%. The IC50 value for PI-103 was 1.67 µM (95% CI: 1.333–2.012 µM), and at a concentration of 2 µM, the growth inhibition rate was 56.7±4.97%. The IC50 value for ZSTK474 was 8.0343 µM (95% CI: 7.472–8.638 µM). A375 cells were not sensitive to GSK690693.
Figure 3.
Cell-growth response curves for melanoma cell lines A375 and WM115. Cells were treated with the indicated concentrations of PI-103, AZD8055, ZSTK474, or GSK6906930 for 72 h and cell viability was measured using a CCK-8 assay. Results represent the mean ± standard deviation of three independent experiments.
The WM115 cell line exhibited decreased expression of BRAFV600E, but increased PI3K signaling pathway activity compared with A375 cells (Fig. 2). All four inhibitors exhibited antitumor effects on WM115 cells, with IC50 values of139.9 nM, 2.943, 3.436, and 7.885 µM (AZD8055 >PI-103 > ZSTK474 > GSK690693), respectively.
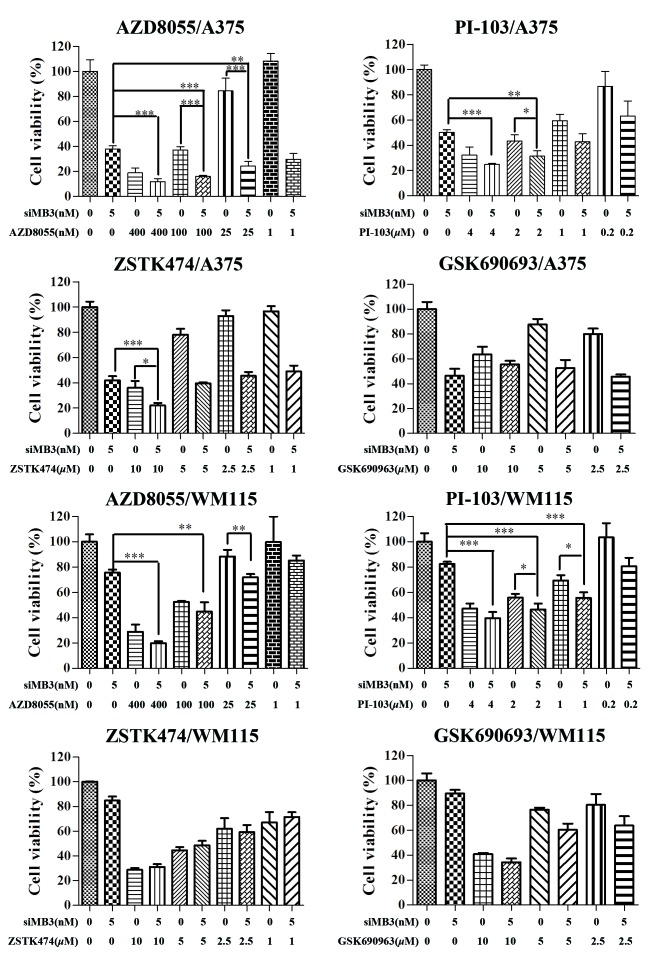
Combination treatment with BRAFV600E siRNA and PI3K signaling pathway inhibitors may be an effective treatment for patients with BRAFV600E-positive tumors. Therefore, in the present study, the combinational effects of four PI3K signaling pathway inhibitors with siMB3 on cell viability were measured (Fig. 4). Combining mTOR inhibition (AZD8055), dual PI3K/mTOR inhibition (PI-103), or PI3K inhibition (ZSTK474) with BRAFV600E siRNA resulted in significantly greater growth inhibition than inhibitor or siMB3 alone in A375 cells. The combination of AZD8055 with siMB3 had the greatest effect on cell viability compared with that of siMB3 with PI-103 or ZSTK474. Treatment of A375 cells with siMB3 and AZD8055 significantly decreased cell viability compared with AZD8055 or siMB3 alone (84.8±0.62 compared with 62.9±2.69 and 62.1±2.54%, respectively; P<0.001). Although no significant effect on cell viability was observed with AZD8055 at a low concentration (1 nM), the synergistic effect with siMB3 was significant compared with AZD8055 treatment alone (P<0.005). When cells were co-treated with 5 nM of siMB3 and 400, 100, 25 or 1 nM AZD8055, the cell inhibition rates were 75.8±4.45, 75.9±3.62, 73.5±7.00, and 70.3±4.6, respectively.
Figure 4.
Combination treatment of siMB3 with phosphoinositide 3-kinase/RAC-α serine-threonine-protein kinase/mammalian target of rapamycin inhibitors significantly decreases cell viability compared with siMB3 or inhibitor alone. Cells were treated with the indicated concentrations of AZD8055, PI-103, ZSTK474, or GSK690693 alone or with 5 nM of siMB3 for 72 h. Cell viability was measured using a CCK-8 assay. Results represent the mean of three replicates ± standard deviation. *P<0.05, **P<0.01, ***P<0.001 compared with the corresponding control.
There was no significant improvement with combination treatment in WM115 cells compared with siMB3 or inhibitor treatment alone. WM115 exhibit low levels of BRAFV600E, therefore the ERK signaling pathway may be activated by the hyperactivation of growth factor signaling or other receptor signaling pathways. This may explain why WM115 cells were not sensitive to BRAFV600E inhibition. Inhibiting the PI3K/AKT/mTOR and MEK1/2 signaling pathways may provide an efficient treatment for WM115 cells.
Effect of siMB3/AZD8055 combination treatment on DNA replication and cell cycle progression
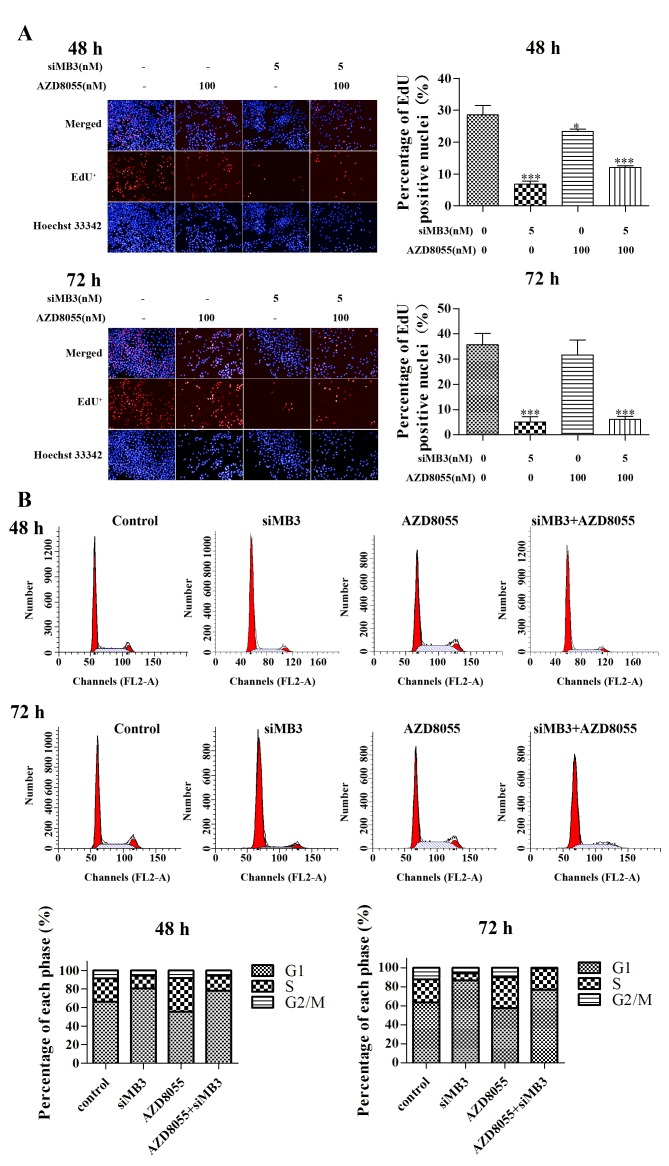
The effect of combined treatment with siMB3 and AZD8055 on DNA replication was measured using the EdU method. siMB3 alone, AZD8055 treatment alone and siMB3/AZD8055 combination treatment significantly inhibited DNA replication compared with control untreated A375 cells (P<0.001, P<0.05 and P<0.001, respectively; Fig. 5A). The siMB3 alone and siMB3/AZD8055 combination treatments exhibited >80% inhibition of DNA replication following treatment for 72 h. AZD8055 arrested A375 cells at S phase, which correlated with the observed inhibition of DNA replication (Fig. 5B). At 72 h following treatment with AZD8055 and siMB3, all cells were arrested in either the G1 or S phase.
Figure 5.
DNA replication assay in A375 cells following treatment with siMB3 in combination with AZD8055 or with siMB3 or inhibitor alone. (A) Representative images and quantification of DNA replication of A375 cells treated with siMB3, AZD8055, and siMB3/AZD8055 combination using the EdU method. A375 cells were incubated with or without 5 nM siMB3, 100 nM AZD8055 or siMB3/AZD8055 combination, and EdU was used to stain cells (red) following 48 or 72 h treatment. Cell nuclei were stained with Hoechst 33342 (blue). Graphs represent the percentage of total cells with EdU-positive nuclei. (B) Cell cycle assay of siMB3, AZD8055, and siMB3/AZD8055 combination at 48 or 72 h. A375 cells were incubated with or without 5 nM of siMB3, 100 nM of AZD8055, and siMB3/AZD8055 combination, stained with propidium iodide and subjected to flow cytometry analysis 48 or 72 h following treatment.
Effects of siMB3/AZD8055 combination treatment on ERK and PI3K signaling pathways
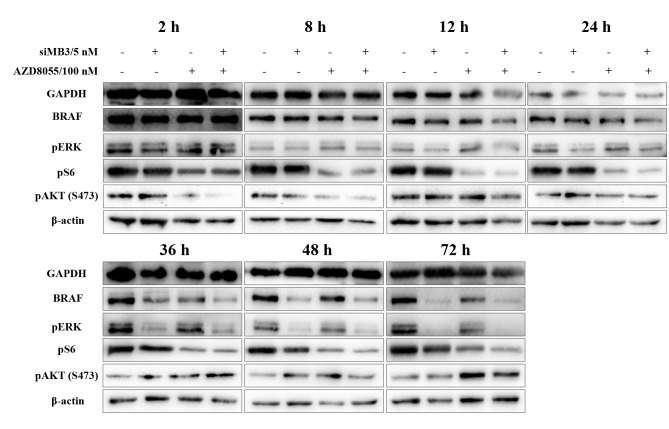
AZD8055 is a potent and specific ATP-competitive inhibitor, which has been demonstrated to inhibit the phosphorylation of the mTORC1 substrates p70S6K and pS6, in addition to inhibiting the phosphorylation of the mTORC2 substrate AKT (S483) and downstream proteins in multiple cancer clinical studies (20). The expression of BRAF in cells treated with siMB3 alone and siMB3/AZD8055 was markedly reduced at 12 h following treatment and was completely knocked down at the 36 h time point (Fig. 6). In addition, ERK signaling pathway activity was markedly inhibited in a time-dependent manner under the same conditions. pS6 expression was reduced from the 2 h time point following AZD8055 alone or siMB3/AZD8055 treatment; however, pS6 expression levels in the combination group were decreased compared with the AZD8055 alone group. Phosphorylation of AKT(S473) in the AZD8055 alone and siMB3/AZD8055 combination groups was markedly decreased at the 2 h time point; however, the expression increased again following the 36 and 72 h time points.
Figure 6.
Representative western blot analysis of expression levels of BRAF, pERK1/2 (240/244), pS6 (202/204) and pAKT(S473) in A375 cells following 2, 8, 12, 24, 48 and 72 h treatment with 5 nM of siMB3, 100 nM of AZD8055 or siMB3/AZD8055 combination. The expression levels of GAPDH and β-actin were included as loading controls. The results are representative of three independent experiments.
Therefore, down regulation of BRAF in BRAFV600E-positive cells with siRNA leads to a decrease in pERK, while treatment with AZD8055 leads to a decrease in pS6. However, continuous administration of AZD8055 would be required to inhibit pAKT levels and further decrease cell viability. Therefore, the combination treatment with siMB3 and AZD8055 leads to a decrease in pERK and pS6 levels, which may contribute to their combined effect on the viability of A375 cells.
siMB3/PI-103 and siMB3/ZSTK474 combination treatments decrease cell viability, and inhibit the PI3K/AKT/mTOR and Ras/MEK/ERK signaling cascades
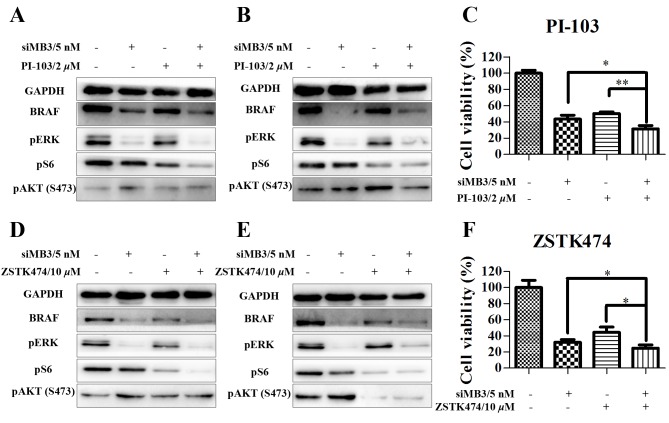
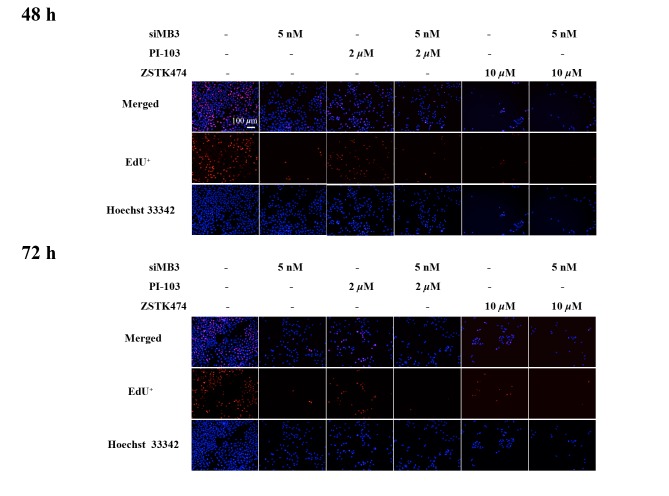
Next, the effects of siMB3/PI-103 combination treatment were investigated in A375 cells. The phosphorylation of ERK and S6 was markedly decreased at 48 and 72 h following combination treatment; however, phosphorylation of AKT (S473) did not recover at a later time point (Fig. 7A and B). In addition, cell growth inhibition significantly increased in the combination treatment (5 nM of siMB3 and 2 µM of PI-103) group compared with the PI-103 treatment alone group (68.4±4.2% compared with 56.7±4.9%, respectively; P<0.01; Fig. 7C). Similar effects were observed with ZSTK474 (Fig. 7D-F). In addition, compared with siMB3 alone or PI-103 alone groups, siMB3/PI-103 or siMB3/ZSTK474 combination treatments markedly inhibited DNA replication and decreased cell number (Fig. 8).
Figure 7.
PI3K/RAC-α serine-threonine-protein kinase/mammalian target of rapamycin signaling pathway modulation in A375 cells following BRAFV600E knockdown and PI3K inhibition. (A and B) Western blot analyses of A375 cells treated with siMB3 and PI-103 alone or in combination after (A) 48 or (B) 72 h treatment. GAPDH was used as a loading control. (C) Viability of A375 cells following treatment with siMB3/PI-103 combination for 72 h using aCCK-8 assay. (D and E) Western blotting analyses of A375 cells treated with siMB3 and ZSTK474 alone or in combination after (D) 48 or (E) 72 h treatment. GAPDH was used as a loading control. (F) Viability of A375 cells following treatment with siMB3/ZSTK474 combination for 72 h using a CCK-8 assay. *P<0.05 **P<0.01, **P<0.001, compared with the corresponding control.
Figure 8.
DNA replication assay in A375 cells treated with siMB3, PI-103, ZSTK474, siMB3/PI-103 or siMB3/ZSTK474 combination using the EdU method at 48 or 72 h following treatment. Cell nuclei were stained with Hoechst 33342 (blue).
Discussion
Melanoma has become one of the most extensively studied cancer types in order to develop effective targeted therapies. Prior to 2011, only two drugs (interferon-α-2b and interleukin-2) had been previously approved for the treatment of melanoma (1). By2015, the FDA had approved 6 first-in-class drugs specific for melanoma (18). Of these, ipilimumab, nivolumab and pembrolizumab are immunotherapies, whereas the other 3 drugs were developed for thespecific treatment of BRAF-mutant melanoma (21–23).
The oncogenic BRAFV600E allele is a common mutation of the BRAF gene and has been identified in ~50% of advanced-stage metastatic melanomas (2). Vemurafenib and dabrafenib/trametinib combination treatments have demonstrated success in patients with melanoma (24); however, small-molecule BRAF inhibitors exhibit side effects such as fast development of drug resistance (19).
Targeting mRNA degradation using siRNA may be a potential strategy for the treatment of a number of diseases (25). Although there remain challenges with siRNA-based treatments in terms of appropriate delivery vehicles, siRNA could be efficient and potent anticancer agents (26). Synthetic siRNA or short hairpin RNAs have been widely used for drug development and a number of phase I and II clinical trials are in progress (27).
siRNA-based treatments that only deplete the oncogenic form of BRAF (BRAFV600E) could be a successful strategy for the treatment for BRAFV600E-positive tumor types. In the present study, siMB3, a 19mer overlapping the T1799A mutation site of BRAFV600E, inhibited ERK signaling pathway activity and decreased the viability of cell lines with this mutation, including A375 and WM115, but not HEK293A cells, as these do not harbor the mutation. Previous studies have demonstrated that a 25mer siRNA (containing the 19mer of siBraf-mu) targeted to BRAFV600E significantly inhibited melanoma tumor growth and reduced lung metastases in UACC 903, 1205 Lu, and C8161 cell lines (17,28,29). In addition, data from the present study demonstrated that A375 cells were more sensitive to the effects of siMB3 on cell viability than WM115 cells due to their increased expression of BRAFV600E. Preliminary data from a current study from our group demonstrated that siMB3 treatment may be effective in vivo; however, the cytotoxic effects of siMB3 on normal cells limit its application in vivo (Xinmeng Fan et al, unpublished data).
The RAF/MEK/ERK and PI3K/AKT/mTOR signaling pathways have been implicated in the tumorigenesis of melanoma (18,30). The combination inhibitory activity of these two pathways can increase antitumor activity and specificity and thus reduces side effects on normal tissues. Previous studies have suggested that combined targeting of the ERK and PI3K pathways increases antitumor activity and may serve as a novel treatment for patients with NRAS mutant-positive melanoma, for which there are currently no effective therapies (30). Sanchez-Hernandez et al (17), demonstrated that combined with BRAFV600E deletion by siRNA, PI3K/AKT/mTOR signaling pathway inhibitors synergized to increase apoptosis levels to a greater extent than that achieved by inhibitor alone in BRAFV600E mutant melanomas, and suggested that mTOR was a convergence point of BRAF and PI3K signals in these cells (17,18).
In the present study, PI3K, mTOR and AKT inhibitors exhibited different antitumor effects on A375 cells. Based on the cell viability assays, the efficacy decreased in the following order: AZD8055 (mTOR inhibitor) > PI-103 (dual PI3K/mTOR inhibitor) > ZSTK474 (pen-class I PI3K inhibitor) >GSK690693 (AKT inhibitor). AZD8055 inhibition of both mTORC1 (IC50=27±3 nM for pAKT473 in MDA-MB-468 cells) and mTORC2 (IC50=24±9 nM for pS235/236) exhibited potent efficacy and selectivity compared with rapamycin in vitro and in vivo (20,31). The inhibition of mTORC1 by rapamycin results in the release of the negative feedback loop between ribosomal protein S6 kinase and insulin receptor substrate 1, leading to hyperactivation of AKT (32).
Silencing of BRAFV600E by siRNA combined with treatment with AZD8055 significantly decreased the viability of A375 cells. Western blot analysis demonstrated that AZD8055 inhibited the phosphorylation of S6 (S240/244) and AKT (S473) at 2 h following treatment, whereas the pAKT (S473) expression level recovered 36 h following treatment.
AKT kinases are important in melanoma tumorigenesis. Selective activation of AKT3 protein promoted cell survival and tumor development in ~70% of melanomas (33). In addition, a previous study on drug resistant cancer types suggested that the hyperactivation of AKT was associated with a shorter tumor progression time (33). In the present study, AZD8055 was unable to simultaneously inhibit both mTORC1 and mTORC2 in A375 melanoma cells 36 h following treatment. The pAKT level may be upregulated by other upstream kinases, including PI3K, or RTKs. This indicated that the enhanced inhibition effect on tumor progression by AZD8055 in combination with siRNA treatment may not last in the absence of AKT inhibition. However, siMB3/PI-103 and siMB3/ZSTK474 combination treatments significantly decreased the pAKT expression level compared to that of control groups.
The results from the present study demonstrate that the efficacy of siRNA treatment on mutant genes is dependent on the percentage of mutation in melanoma cells. siRNA treatment of cells harboring the BRAFV600E mutation may be a potent treatment for melanoma cells that harbor high expression and activating levels of BRAFV600E mutation. However, in those melanoma cells in which the oncoprotein BRAFV600E is not the major carcinogenic factor, patients may not be sensitive to targeted BRAFV600E therapy, including inhibition by treatment with vemurafenib or elimination of BRAFV600E expression with siRNA. Targeting the PI3K/AKT/mTOR signaling pathway, which induces cell apoptosis and decrease cell survival, may represent an improved treatment strategy. A more comprehensive understanding of the molecular mechanisms underlying melanoma will aid with developing, personalized cancer treatments. The data from the present study may also be relevant for other types of cancer that harbor the BRAFV600E mutation, including colorectal carcinomas and non-small cell lung cancer (23).
In conclusion, the present study demonstrated that silencing BRAFV600E by using siRNA in combination with PI3K signaling pathway inhibitors improved the effect of PI3K/mTOR inhibitors on tumor cell viability. Concomitant BRAFV600E and PI3K inhibition resulted in G1 and S phase arrest. Future studies are required in order to develop an efficient and safe delivery system for siRNA-based therapy.
Acknowledgements
Not applicable.
Funding
This study was supported by grants from the National Natural Sciences Foundation of China (grant no. 81302626) and the Specialized Research Fund for the Doctoral Program of Higher Education (grant no. 20120001120023). The funding agencies had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Availability for data and materials
All data generated or analyzed during this study are included in this published article.
Author's contributions
HH, XN and SL performed the experiments, YW contributed to the writing of the manuscript. YW, ZY, LiaZ and LihZ conceived of the study. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Schadendorf D, Fisher DE, Garbe C, Gershenwald JE, Grob JJ, Halpern A, Herlyn M, Marchetti MA, McArthur G, Ribas A, et al. Melanoma. Nat Rev Disease Primers. 2015;1:15003. doi: 10.1038/nrdp.2015.3. [DOI] [PubMed] [Google Scholar]
- 2.Batus M, Waheed S, Ruby C, Petersen L, Bines SD, Kaufman HL. Optimal management of metastatic melanoma: Current strategies and future directions. Am J Clin Dermatol. 2013;14:179–194. doi: 10.1007/s40257-013-0025-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gray-Schopfer V, Wellbrock C, Marais R. Melanoma biology and new targeted therapy. Nature. 2007;445:851–857. doi: 10.1038/nature05661. [DOI] [PubMed] [Google Scholar]
- 4.Smalley KS, Haass NK, Brafford PA, Lioni M, Flaherty KT, Herlyn M. Multiple signaling pathways must be targeted to overcome drug resistance in cell lines derived from melanoma metastases. Mol Cancer Ther. 2006;5:1136–1144. doi: 10.1158/1535-7163.MCT-06-0084. [DOI] [PubMed] [Google Scholar]
- 5.Bis S, Tsao H. Melanoma genetics: The other side. Clin Dermatol. 2013;31:148–155. doi: 10.1016/j.clindermatol.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 6.Rapp UR, Goldsborough MD, Mark GE, Bonner TI, Groffen J, Reynolds FH, Jr, Stephenson JR. Structure and biological-activity of v-raf, a unique oncogene transduced by a retrovirus. Proc Natl Acad Sci USA. 1983;80:4218–4222. doi: 10.1073/pnas.80.14.4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 8.Wellbrock C, Hurlstone A. BRAF as therapeutic target in melanoma. Biochem Pharmacol. 2010;80:561–567. doi: 10.1016/j.bcp.2010.03.019. [DOI] [PubMed] [Google Scholar]
- 9.Bucheit AD, Davies MA. Emerging insights into resistance to BRAF inhibitors in melanoma. Biochem Pharmacol. 2014;87:381–389. doi: 10.1016/j.bcp.2013.11.013. [DOI] [PubMed] [Google Scholar]
- 10.Nazarian R, Shi H, Wang Q, Kong X, Koya RC, Lee H, Chen Z, Lee MK, Attar N, Sazegar H, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–977. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Emery CM, Vijayendran KG, Zipser MC, Sawyer AM, Niu L, Kim JJ, Hatton C, Chopra R, Oberholzer PA, Karpova MB, et al. MEK1 mutations confer resistance to MEK and B-RAF inhibition. Proc Natl Acad Sci USA. 2009;106:20411–20416. doi: 10.1073/pnas.0905833106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poulikakos PI, Zhang C, Bollag G, Shokat KM, Rosen N. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature. 2010;464:427–430. doi: 10.1038/nature08902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hatzivassiliou G, Song K, Yen I, Brandhuber BJ, Anderson DJ, Alvarado R, Ludlam MJ, Stokoe D, Gloor SL, Vigers G, et al. RAF inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth. Nature. 2010;464:431–435. doi: 10.1038/nature08833. [DOI] [PubMed] [Google Scholar]
- 14.Heidorn SJ, Milagre C, Whittaker S, Nourry A, Niculescu-Duvas I, Dhomen N, Hussain J, Reis-Filho JS, Springer CJ, Pritchard C, Marais R. Kinase-dead BRAF and oncogenic RAS cooperate to drive tumor progression through CRAF. Cell. 2010;140:209–221. doi: 10.1016/j.cell.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Corcoran RB, Dias-Santagata D, Bergethon K, Iafrate AJ, Settleman J, Engelman JA. BRAF gene amplification can promote acquired resistance to MEK inhibitors in cancer cells harboring the BRAF V600E mutation. Sci Signal. 2010;3:ra84. doi: 10.1126/scisignal.2001148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sharma A, Trivedi NR, Zimmerman MA, Tuveson DA, Smith CD, Robertson GP. Mutant B-V599E-RAF regulates growth and vascular development of malignant melanoma tumors. Cancer Res. 2005;65:2412–2421. doi: 10.1158/0008-5472.CAN-04-2423. [DOI] [PubMed] [Google Scholar]
- 17.Sánchez-Hernández I, Baquero P, Calleros L, Chiloeches A. Dual inhibition of (V600E)BRAF and the PI3K/AKT/mTOR pathway cooperates to induce apoptosis in melanoma cells through a MEK-independent mechanism. Cancer Lett. 2012;314:244–255. doi: 10.1016/j.canlet.2011.09.037. [DOI] [PubMed] [Google Scholar]
- 18.Fedorenko IV, Gibney GT, Sondak VK, Smalley KS. Beyond BRAF: Where next for melanoma therapy? Br J Cancer. 2015;112:217–226. doi: 10.1038/bjc.2014.476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li S, Li Y, Hu R, Li W, Qiu H, Cai H, Wang S. The mTOR inhibitor AZD8055 inhibits proliferation and glycolysis in cervical cancer cells. Oncol Lett. 2013;5:717–721. doi: 10.3892/ol.2012.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sondak VK, Smalley KS, Kudchadkar R, Grippon S, Kirkpatrick P. Ipilimumab. Nat Rev Drug Discov. 2011;10:411–412. doi: 10.1038/nrd3463. [DOI] [PubMed] [Google Scholar]
- 21.Mullard A. 2014 FDA drug approvals. Nat Rev Drug Discov. 2015;14:77–81. doi: 10.1038/nrd4545. [DOI] [PubMed] [Google Scholar]
- 22.Mullard A. 2013 FDA drug approvals. Nat Rev Drug Discov. 2014;13:85–89. doi: 10.1038/nrd4239. [DOI] [PubMed] [Google Scholar]
- 23.Flaherty KT, Yasothan U, Kirkpatrick P. Vemurafenib. Nat Rev Drug Discov. 2011;10:811–812. doi: 10.1038/nrd3579. [DOI] [PubMed] [Google Scholar]
- 24.Kim DH, Rossi JJ. Strategies for silencing human disease using RNA interference. Nat Rev Genet. 2007;8:173–184. doi: 10.1038/nrg2006. [DOI] [PubMed] [Google Scholar]
- 25.Bouchie A. Markets, venture investors and big pharma interest in RNAi soars. Nat Biotechnol. 2014;32:203–204. doi: 10.1038/nbt0314-203. [DOI] [PubMed] [Google Scholar]
- 26.Deng Y, Wang CC, Choy KW, Du Q, Chen J, Wang Q, Li L, Chung TK, Tang T. Therapeutic potentials of gene silencing by RNA interference: Principles, challenges, and new strategies. Gene. 2014;538:217–227. doi: 10.1016/j.gene.2013.12.019. [DOI] [PubMed] [Google Scholar]
- 27.Hingorani SR, Jacobetz MA, Robertson GP, Herlyn M, Tuveson DA. Suppression of BRAF(V599E) in human melanoma abrogates transformation. Cancer Res. 2003;63:5198–5202. [PubMed] [Google Scholar]
- 28.Sharma A, Tran MA, Liang S, Sharma AK, Amin S, Smith CD, Dong C, Robertson GP. Targeting mitogen-activated protein kinase/extracellular signal-regulated kinase kinase in the mutant (V600E) B-Raf signaling cascade effectively inhibits melanoma lung metastases. Cancer Res. 2006;66:8200–8209. doi: 10.1158/0008-5472.CAN-06-0809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Posch C, Moslehi H, Feeney L, Green GA, Ebaee A, Feichtenschlager V, Chong K, Peng L, Dimon MT, Phillips T, et al. Combined targeting of MEK and PI3K/mTOR effector pathways is necessary to effectively inhibit NRAS mutant melanoma in vitro and in vivo. Proc Natl Acad Sci USA. 2013;110:4015–4020. doi: 10.1073/pnas.1216013110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chresta CM, Davies BR, Hickson I, Harding T, Cosulich S, Critchlow SE, Vincent JP, Ellston R, Jones D, Sini P, et al. AZD8055 is a potent, selective, and orally bioavailable ATP-competitive mammalian target of rapamycin kinase inhibitor with in vitro and in vivo antitumor activity. Cancer Res. 2010;70:288–298. doi: 10.1158/0008-5472.CAN-09-1751. [DOI] [PubMed] [Google Scholar]
- 31.O'Reilly KE, Rojo F, She QB, Solit D, Mills GB, Smith D, Lane H, Hofmann F, Hicklin DJ, Ludwig DL, et al. mTOR inhibition induces upstream receptor tyrosine kinase signaling and activates Akt. Cancer Res. 2006;66:1500–1508. doi: 10.1158/0008-5472.CAN-05-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stahl JM, Sharma A, Cheung M, Zimmerman M, Cheng JQ, Bosenberg MW, Kester M, Sandirasegarane L, Robertson GP. Deregulated Akt3 activity promotes development of malignant melanoma. Cancer Res. 2004;64:7002–7010. doi: 10.1158/0008-5472.CAN-04-1399. [DOI] [PubMed] [Google Scholar]
- 33.Shao Y, Aplin AE. Akt3-mediated resistance to apoptosis in B-RAF-targeted melanoma cells. Cancer Res. 2010;70:6670–6681. doi: 10.1158/0008-5472.CAN-09-4471. [DOI] [PMC free article] [PubMed] [Google Scholar]