Abstract
The aim of the present study was to investigate more colon cancer-related genes in different stages. Gene expression profile E-GEOD-62932 was extracted for differentially expressed gene (DEG) screening. Series test of cluster analysis was used to obtain significant trending models. Based on the Gene Ontology and Kyoto Encyclopedia of Genes and Genomes databases, functional and pathway enrichment analysis were processed and a pathway relation network was constructed. Gene co-expression network and gene signal network were constructed for common DEGs. The DEGs with the same trend were clustered and in total, 16 clusters with statistical significance were obtained. The screened DEGs were enriched into small molecule metabolic process and metabolic pathways. The pathway relation network was constructed with 57 nodes. A total of 328 common DEGs were obtained. Gene signal network was constructed with 71 nodes. Gene co-expression network was constructed with 161 nodes and 211 edges. ABCD3, CPT2, AGL and JAM2 are potential biomarkers for the diagnosis of colon cancer.
Keywords: colon cancer, different stage, biomarker, differently expressed genes
Introduction
Colon cancer together with lung, prostate and breast cancer constitute a threat to human health worldwide (1). Notably, previous findings showed that surgery had minimal adverse reaction in stage 2 patients (2). In addition, some treatment strategies such as adjuvant chemotherapy, were found to be effective, especially for stage 3 colon cancer (3). Thereby, the known critical genes in different stages of colon cancer was necessary for the diagnosis and treatment of this disease.
Various studies have been previously performed to investigate the molecular mechanism of colon cancer. For example, Yu et al found that methylated TBX5 may be a functional tumor suppressor gene for the diagnosis of malignancy of colon cancer (4). Moreover, the metachronous distant metastasis of colon cancer was ensured to be correlated with the downregulation of SASH1 (SAM- and SH3-domain containing 1) expression (5). Schepeler et al showed that the expression level of miR-320 and miR-498 was closely related to the recurrence-free survival of colon cancer (6). miR-21 was also confirmed to be upregulated in stage 2 colon cancer by participating in the immune process (7). In addition, transforming growth factor β1 could prevent an organized hyperplasia in the early transition, and further suppress colon cancer (8). Although these genes, miRNAs and pathways were identified, differentially expressed genes (DEGs) in different staging and their potential mechanism remain to be determined.
In order to investigate more colon cancer-related genes in different stages, DEGs in four stages compared with controls were identified in this study, and series test of clusters were processed. In addition, functional and pathway enrichment analysis was used for these DEGs. Finally, various networks including a pathway relation network, gene signal network and gene co-expression network were constructed.
Materials and methods
Microarray data
The gene expression profile E-GEOD-62932 was extracted from the ArrayExpress Archive, including 64 colon cancer tissues and 4 healthy control samples (9). The 64 colon cancer tissues included 12 samples in stage 1, 17 in stage 2, 20 in stage 3, and 15 in stage 4. Platform information of this profile was GPL570 [HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array.
Data preprocessing and DEGs screening
The obtained probe sets were converted into expression value matrix and background correction, normalization and summarizing were performed using the Robust Multi-chip Average algorithm. Based on the information of Affymetrix (*.transcript.csv), the probe sets were annotated. The annotated data were screened with the threshold of >0.8 median (normalized unscaled standard errors) <1.2 and >0.25 median (relative log expression) <0.25.
DEGs between colon cancer in various stages and normal tissues were identified by significance analysis of microarrays method with the cut-off criteria of P<0.05 (10).
Series test of clusters
The gene expression profiles were always changed with tumor development. In the present study, series test of cluster (STC) analysis was used to obtain the significant trending models related to tumor stage and their involved DEGs (11). Clusters with P<0.05 were regarded as statistically significant.
Functional and pathway enrichment analysis
Gene Ontology (GO) is a cross-species and comprehensive database, which was constructed by the GO Consortium (12). Similarly, the Kyoto Encyclopedia of Genes and Genomes (KEGG) is a powerful tool for analysis of biological metabolism and metabolic network (13). Based on the abovementioned databases, GO functional and KEGG pathway enrichment analysis were processed for identified DEGs with the threshold of P<0.05. The Fisher's exact test and multiple comparisons were used to calculate the significance of GO terms and pathways, and false discovery rate (FDR) was the adjusted P-value.
Construction of pathway relation network
The pathway relation network was constructed based on information from the KEGG database. From this network, a signal transduction relationship was obtained. In addition, upstream regulatory pathways and downstream effective pathways were identified.
Gene signal and gene co-expression network construction for common DEGs
DEGs in GO terms and KEGG pathways were inserted based on the same symbol. The obtained DEGs were regarded as common DEGs. Gene signal information was collected from the KEGG database. The common DEGs were mapped into the whole network and the gene signal network of common DEGs in colon cancer was generated. Additionally, the gene co-expression network was a weighted network that was constructed based on gene expression value and their correlation indexes. The two networks were visualized by Cytoscape (14).
Results
DEG screening and series test of cluster
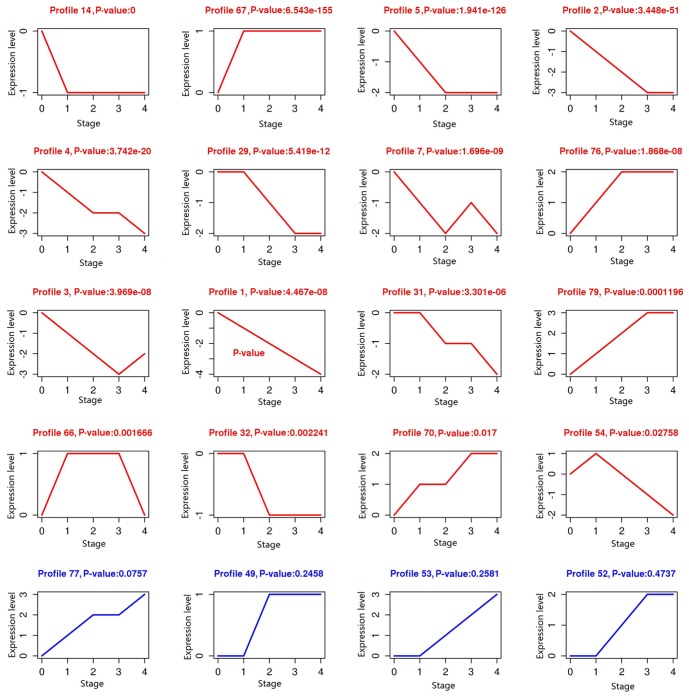
Based on the threshold of DEGs, 1,543 DEGs were screened to be associated with colon tumor development compared with control samples. Then, the DEGs with the same trend were clustered. A total of 16 clusters with statistical significance were obtained, including profile 14, 67, 5 and 2 (Fig. 1).
Figure 1.
Cluster of the screened differentially expressed genes. The red profiles are statistically significant, but the blue profiles have no statistical significance.
Functional and KEGG pathway enrichment
The screened DEGs were enriched into various GO terms, such as small molecule metabolic process (FDR=1.57E-36), cell division (FDR=1.00E-29) and mitotic cell cycle (FDR=9.07E-28) (Table I). In addition, the DEGs were gathered into different pathways including metabolic pathways (FDR=1.57E-21), cell cycle (FDR=8.23E-12) and progesterone-mediated oocyte maturation (FDR=3.56E-09) (Table II).
Table I.
Top 5 GO terms of differentially expressed genes.
| GO ID | GO Name | Diff gene counts in GO | Enrichment Score | P-value | FDR |
|---|---|---|---|---|---|
| GO:0044281 | Small molecule metabolic process | 129 | 3.905874858 | 5.67E-40 | 1.57E-36 |
| GO:0051301 | Cell division | 56 | 7.834124719 | 7.24E-33 | 1.00E-29 |
| GO:0000278 | Mitotic cell cycle | 59 | 6.707641713 | 9.84E-31 | 9.07E-28 |
| GO:0000236 | Mitotic prometaphase | 28 | 11.55533396 | 7.75E-22 | 5.36E-19 |
| GO:0000087 | M phase of mitotic cell cycle | 35 | 7.295034066 | 7.28E-20 | 4.02E-17 |
GO, Gene Ontology; FDR, false discovery rate.
Table II.
The top 5 pathways of differently expressed genes.
| Pathway ID | Pathway name | Diff gene counts in pathway | Enrichment score | P-value | FDR |
|---|---|---|---|---|---|
| 1,100 | Metabolic pathways | 95 | 3.297358904 | 6.37E-24 | 1.57E-21 |
| 4,110 | Cell cycle | 23 | 7.654743119 | 6.66E-14 | 8.23E-12 |
| 4,914 | Progesterone-mediated oocyte maturation | 17 | 8.157835437 | 5.00E-11 | 3.56E-09 |
| 4,114 | Oocyte meiosis | 19 | 7.00099953 | 5.76E-11 | 3.56E-09 |
| 5,200 | Pathways in cancer | 29 | 3.659946318 | 5.29E-09 | 2.61E-07 |
FDR, false discovery rate.
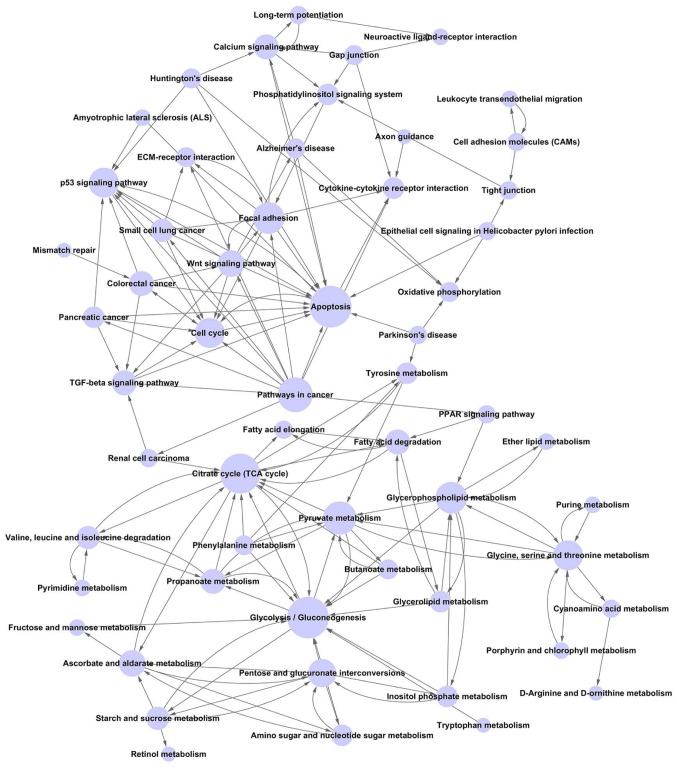
Construction of the pathway relation network
The pathway relation network was constructed with 57 nodes and 158 edges. The hub nodes were glycolysis/gluconeogenesis (Degree=17), apoptosis (Degree=17), citrate cycle (TCA cycle) (Degree=16) and pathways in cancer (Degree=13). Notably, pathways in cancer (Outdegree=13), phenylalanine metabolism (Outdegree=4) and gap junction (Outdegree=4) were upstream pathways, while cytokine-cytokine receptor interaction (Indegree=5) and oxidative phosphorylation (Indegree=4) were downstream pathways (Fig. 2).
Figure 2.
Pathway relation network of pathways enriched by differentially expressed genes. The nodes and edges are pathways and their regulation and relationships, respectively.
Gene signal network and gene co-expression network construction for common DEGs
Based on the same gene symbol, a total of 328 common DEGs were obtained, such as adenosylhomocysteinase-like 2, oxytocin receptor, aquaporin 8 and hedgehog interacting protein. As shown in Fig. 3, the gene signal network was constructed with 71 nodes and 146 edges. The hub nodes were adenylate cyclase 9 (Degree=18), ectonucleoside triphosphate diphosphohydrolase 5 (Degree=18) and protein kinase, cAMP-dependent, catalytic, β (Degree=14).
Figure 3.
Gene signal network of common differentially expressed genes (DEGs). The nodes and edges are common DEGs and their regulation and relationships, respectively.
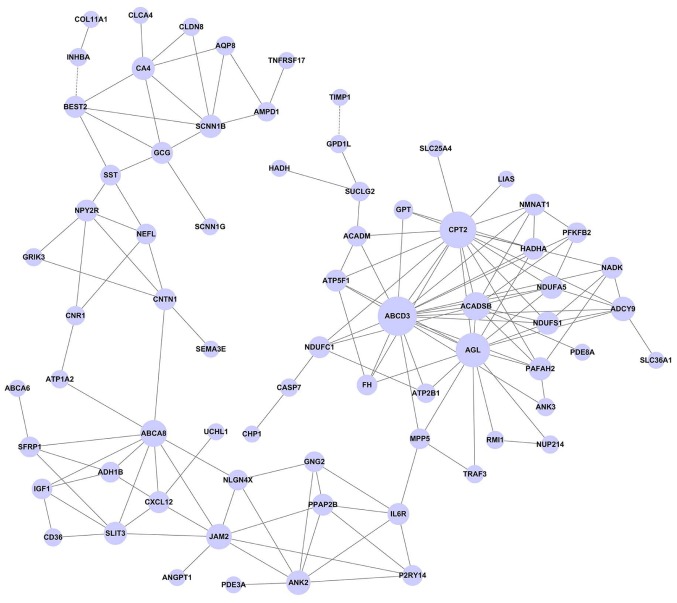
The gene co-expression network was constructed with 161 nodes and 211 edges (Fig. 4). In this network, the hub nodes with a higher degree were ATP-binding cassette, sub-family D, member 3 (ABCD3, Degree=19), carnitine palmitoyltransferase 2 (CPT2, Degree=17) and amylo-α-1, 6-glucosidase, 4-α-glucanotransferase (AGL, Degree=15). Moreover, this network showed that ABCD3 has a positive relationship with various DEGs, including ankyrin 3, adenylate cyclase 9 and acyl-coA dehydrogenase, and short/branched chain.
Figure 4.
Gene co-expression network of common differentially expressed genes (DEGs). The nodes and edges are common DEGs and their regulation and relationships, respectively. The solid lines show the positive correlation while the dashed lines show the negative correlation.
Discussion
Over 75% of colon cancer occurs in individuals with little or no genetic risk (15). However, various gene expressions and their related functions and pathways were altered with the increasing of grade of the tumor. These genes provided the biomarkers for colon cancer of each stage and also suggest effective treatment targets. In the present study, several significant DEGs were screened and resulted in participation in the pathogenesis of colon cancer, including ABCD3, CPT2, AGL and (Junctional adhesion molecule B) JAM2.
ABCD3 encoded a protein of a member of ATP-binding cassette (ABC) transporters, which is involved in peroxisomal import of fatty acids or fatty acyl-CoAs in the organelle (16). Previous findings showed by targeting the expression of the multidrug resistance (MDR)-related ABC transporters, that reversal of MDR could be realized and MDR in cancer could be solved (17). Various ABC transporters such as Ped3p, were confirmed to provide substrates for fatty acid β-oxidation (18). In addition, there was evidence that colorectal cancer cells could be induced to apoptosis by the inhibition of fatty acid oxidation (19,20). Similar to the results in this study, ABCD3 was found to participate in ABC transporters, transmembrane transport, fatty acid β-oxidation and ATP catabolic process. In addition, the results in this study have shown that ABCD3 belonged to profile 5 in series test of cluster. The expression of this gene was significantly decreased in stage 1 and 2 of colon cancer samples. Thus, we inferred that ABCD3 may be used as a biomarker for the early detection of colon cancer.
Moreover, CPT2 was a key DEG in profile 5, which was involved in small molecule metabolic process, cellular lipid metabolic process, carnitine shuttle, fatty acid degradation and peroxisome proliferator-activated receptor (PPAR) signaling pathway in the present study. As known, this gene mainly existed in mitochondrion and nucleus and was closely associated with mitochondrial long-chain fatty-acid oxidation disorders (21). In tumor-bearing rat models, Seelaender et al found that the expression of mitochondrial inner-membrane CPT2 was markedly decreased (22). Additionally, previous bioinformatic findings showed that the pathway of fatty acid metabolism involved various altered expressed genes and was confirmed to have a close link with colorectal carcinogenesis (23). Furthermore, Wang and Dubois found that the PPAR signaling pathway could promote the emergence of chronic colonic inflammation and colitis-related tumors (24). Of note, the results in the present study showed that the PPAR signaling pathway was the upstream pathway of fatty acid degradation. The abovementioned information shows that CPT2 is a potentially critical biomarker for colon cancer by participating in the fatty acid metabolic process and PPAR signaling pathway.
Another critical DEG, AGL was found to have a positive relationship with CPT2 in the present study. AGL encodes the glycogen debrancher enzyme, which is involved in glycogen degradation (25). In bladder tumor, AGL was confirmed to participate in the induction of hyaluronic acid synthesis and further regulate tumor growth (26). To the best of our knowledge, there is currently no correlative literature showing the relevance between AGL and colon. However, AGL was significantly altered and involved in the carbohydrate metabolic process, response to glucocorticoid stimulus and glucose metabolic process. In 1984, Holroyde et al showed that diverse abnormalities of the carbohydrate metabolic process often occurred in cancer cachexia, such as colorectal cancer (27). In addition, aerobic glycolysis was ensured to be a metabolic adaptation that promotes the proliferation of colorectal cancer cells (28). Based on the abovementioned information, we hypothesized that AGL was a potential biomarker for the diagnosis of colon cancer.
JAM2 encodes a type I membrane protein that functions as an adhesive ligand for interacting with the number of immune cell types (29). The findings of a previous study revealed that JAM2 played an important role in motility and its adhesion process of colon cancer cells (30). In addition, Royse et al found that JAM2 altered the expression in early- and late-stage colon tumors, and is also closely associated with PTEN, which is a tumor suppressor (31). In the present study, JAM2 was gathered into tight junction, cell adhesion molecules and in blood coagulation. Consistent with previous studies, the indexes of blood coagulation were closely associated with the progression of colon cancer (32). Moreover, various blood coagulation inhibitors were found to be beneficial for colon cancer patients (33). Thus, JAM2 may be involved in the pathogenesis of colon cancer by participating in cell adhesion molecules and blood coagulation.
In summary, the identified DEGs including ABCD3, CPT2, AGL and JAM2 constitute potential biomarkers for the diagnosis of colon cancer by participating in various functions and pathways. The validation of these biomarkers identified is to be conducted in future studies.
Competing interests
Authors declare they have no competing interests.
References
- 1.Li QC, Liang Y, Tian Y, Hu GR. Arctigenin induces apoptosis in colon cancer cells through ROS/p38MAPK pathway. J BUON. 2016;21:87–94. [PubMed] [Google Scholar]
- 2.Gao XY, Wang XL. An adoptive T cell immunotherapy targeting cancer stem cells in a colon cancer model. J BUON. 2015;20:1456–1463. [PubMed] [Google Scholar]
- 3.Gatzidou E, Mantzourani M, Giaginis C, Giagini A, Patsouris E, Kouraklis G, Theocharis S. Augmenter of liver regeneration gene expression in human colon cancer cell lines and clinical tissue samples. J BUON. 2015;20:84–91. [PubMed] [Google Scholar]
- 4.Yu J, Ma X, Cheung KF, Li X, Tian L, Wang S, Wu CW, Wu WK, He M, Wang M, et al. Epigenetic inactivation of T-box transcription factor 5, a novel tumor suppressor gene, is associated with colon cancer. Oncogene. 2010;29:6464–6474. doi: 10.1038/onc.2010.370. [DOI] [PubMed] [Google Scholar]
- 5.Rimkus C, Martini M, Friederichs J, Rosenberg R, Doll D, Siewert JR, Holzmann B, Janssen KP. Prognostic significance of downregulated expression of the candidate tumour suppressor gene SASH1 in colon cancer. Br J Cancer. 2006;95:1419–1423. doi: 10.1038/sj.bjc.6603452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schepeler T, Reinert JT, Ostenfeld MS, Christensen LL, Silahtaroglu AN, Dyrskjøt L, Wiuf C, Sørensen FJ, Kruhøffer M, Laurberg S, et al. Diagnostic and prognostic microRNAs in stage II colon cancer. Cancer Res. 2008;68:6416–6424. doi: 10.1158/0008-5472.CAN-07-6110. [DOI] [PubMed] [Google Scholar]
- 7.Nielsen BS, Jørgensen S, Fog JU, Søkilde R, Christensen IJ, Hansen U, Brünner N, Baker A, Møller S, Nielsen HJ. High levels of microRNA-21 in the stroma of colorectal cancers predict short disease-free survival in stage II colon cancer patients. Clin Exp Metastasis. 2011;28:27–38. doi: 10.1007/s10585-010-9355-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Engle SJ, Hoying JB, Boivin GP, Ormsby I, Gartside PS, Doetschman T. Transforming growth factor beta1 suppresses nonmetastatic colon cancer at an early stage of tumorigenesis. Cancer Res. 1999;59:3379–3386. [PubMed] [Google Scholar]
- 9.Chen X, Deane NG, Lewis KB, Li J, Zhu J, Washington MK, Beauchamp RD. Comparison of nanostring ncounter® data on FFPE colon cancer samples and affymetrix microarray data on matched frozen tissues. PLoS One. 2016;11:e0153784. doi: 10.1371/journal.pone.0153784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Larsson O, Wahlestedt C, Timmons JA. Considerations when using the significance analysis of microarrays (SAM) algorithm. BMC Bioinformatics. 2005;6:129. doi: 10.1186/1471-2105-6-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang F, Xiao J, Cong W, Li A, Wei F, Xu J, Zhang C, Fan Z, He J, Wang S. Stage-specific differential gene expression profiling and functional network analysis during morphogenesis of diphyodont dentition in miniature pigs, Sus Scrofa. BMC Genomics. 2014;15:103. doi: 10.1186/1471-2164-15-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. The gene ontology consortium: Gene ontology: Tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wixon J, Kell D. The Kyoto Encyclopedia of Genes and Genomes-KEGG. Yeast. 2000;17:48–55. doi: 10.1002/(SICI)1097-0061(200004)17:1<48::AID-YEA2>3.0.CO;2-H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patricia Rios-Ibarra C, Rodriguez-Silva Janeth C, Lopez-Chuken Alonso Y, Ortiz-Lopez R, Fernandez-Castillo E, Del Toro Runzer Beatriz C, Armenta-Perez Paul V, Flores-Gutierrez JP, Quintanilla-Guzman A, Salinas-Santander M, et al. Thymidylate synthase polymorphism in Mexican patients with colon cancer treated with 5-fluorouracil. J BUON. 2016;21:935–940. [PubMed] [Google Scholar]
- 16.Kawaguchi K, Morita M. ABC Transporter Subfamily D: Distinct differences in behavior between ABCD1-3 and ABCD4 in subcellular localization, function, and human disease. Biomed Res Int. 2016;2016:6786245. doi: 10.1155/2016/6786245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stein U, Walther W. Reversal of ABC transporter-dependent multidrug resistance in cancer. Am J Cancer. 2006;5:285–297. doi: 10.2165/00024669-200605050-00001. [DOI] [Google Scholar]
- 18.Hayashi M, Nito K, Takei-Hoshi R, Yagi M, Kondo M, Suenaga A, Yamaya T, Nishimura M. Ped3p is a peroxisomal ATP-binding cassette transporter that might supply substrates for fatty acid beta-oxidation. Plant Cell Physiol. 2002;43:1–11. doi: 10.1093/pcp/pcf023. [DOI] [PubMed] [Google Scholar]
- 19.Hernlund E, Ihrlund LS, Khan O, Ates YO, Linder S, Panaretakis T, Shoshan MC. Potentiation of chemotherapeutic drugs by energy metabolism inhibitors 2-deoxyglucose and etomoxir. Int J Cancer. 2008;123:476–483. doi: 10.1002/ijc.23525. [DOI] [PubMed] [Google Scholar]
- 20.Holla VR, Wu H, Shi Q, Menter DG, DuBois RN. Nuclear orphan receptor NR4A2 modulates fatty acid oxidation pathways in colorectal cancer. J Biol Chem. 2011;286:30003–30009. doi: 10.1074/jbc.M110.184697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ellis JM, Hasek LY, Yurovich EJ, Harris KL, Goergen CJ. Mouse carnitine palmitoyltransferase 2 (CPT2) is required to sustain cardiac function. J FASEB. 2016;30:684–688. [Google Scholar]
- 22.Seelaender MCL, Curi R, Colquhoun A, Williams JF, Zammitt VA. Carnitine palmitoyltransferase II activity is decreased in liver mitochondria of cachectic rats bearing the Walker 256 carcinosarcoma: Effect of indomethacin treatment. Biochem Mol Biol Int. 1998;44:185–193. doi: 10.1080/15216549800201202. [DOI] [PubMed] [Google Scholar]
- 23.Yeh CS, Wang JY, Cheng TL, Juan CH, Wu CH, Lin SR. Fatty acid metabolism pathway play an important role in carcinogenesis of human colorectal cancers by Microarray-Bioinformatics analysis. Cancer Lett. 2006;233:297–308. doi: 10.1016/j.canlet.2005.03.050. [DOI] [PubMed] [Google Scholar]
- 24.Wang D, Dubois RN. PPARδ and PGE2 signaling pathways communicate and connect inflammation to colorectal cancer. Inflamm Cell Signal. 2014;1:10. doi: 10.1186/1478-811X-12-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crushell E, Treacy EP, Dawe J, Durkie M, Beauchamp NJ. Glycogen storage disease type III in the Irish population. J Inherit Metab Dis. 2010;33(Suppl 3):S215–S218. doi: 10.1007/s10545-010-9096-4. [DOI] [PubMed] [Google Scholar]
- 26.Guin S, Ru Y, Agarwal N, Lew CR, Owens C, Comi GP, Theodorescu D. Loss of glycogen debranching enzyme AGL drives bladder tumor growth via induction of hyaluronic acid synthesis. Clin Cancer Res. 2016;1:1274–1283. doi: 10.1158/1078-0432.CCR-15-1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holroyde CP, Skutches CL, Boden G, Reichard GA. Glucose metabolism in cachectic patients with colorectal cancer. Cancer Res. 1984;44:5910–5913. [PubMed] [Google Scholar]
- 28.Straus DS. TNFα and IL-17 cooperatively stimulate glucose metabolism and growth factor production in human colorectal cancer cells. Mol Cancer. 2013;12:78. doi: 10.1186/1476-4598-12-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liang TW, Chiu HH, Gurney A, Sidle A, Tumas DB, Schow P, Foster J, Klassen T, Dennis K, DeMarco RA, et al. Vascular endothelial-junctional adhesion molecule (VE-JAM)/JAM 2 interacts with T, NK, and dendritic cells through JAM 3. J Immunol. 2002;168:1618–1626. doi: 10.4049/jimmunol.168.4.1618. [DOI] [PubMed] [Google Scholar]
- 30.Lian S, Meng L, Xing X, Yang Y, Qu L, Shou C. PRL-3 promotes cell adhesion by interacting with JAM2 in colon cancer. Oncol Lett. 2016;12:1661–1666. doi: 10.3892/ol.2016.4836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Royse KE, Liang C, Uriostegui J, Ittmann M, Graham DY, Hashem ES, Jiao L. Abstract 4591: Differential expression of tight junction polarity genes in human colon cancer. Cancer Res. 2015;75:4591–4591. doi: 10.1158/1538-7445.AM2015-4591. [DOI] [Google Scholar]
- 32.Lee S, Huh SJ, Oh SY, Koh MS, Kim SH, Lee JH, Han JY, Choi HJ, Kim SJ, Kim HJ. Clinical significance of coagulation factors in operable colorectal cancer. Oncol Lett. 2017;13:4669–4674. doi: 10.3892/ol.2017.6058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Im JH, Fu W, Wang H, Bhatia SK, Hammer DA, Kowalska MA, Muschel RJ. Coagulation facilitates tumor cell spreading in the pulmonary vasculature during early metastatic colony formation. Cancer Res. 2004;64:8613–8619. doi: 10.1158/0008-5472.CAN-04-2078. [DOI] [PubMed] [Google Scholar]