Abstract
Optimization of critical factors affects transduction efficiency and is able to reduce reagent consumption. The present study aimed to determine the optimum transduction conditions of small hairpin (sh)RNA against peroxiredoxin 4 (PRDX4) in the HepG2 cell line. Cell viability assays were conducted based on serum condition, incubation time, polybrene concentration and antibiotic dose selection. Non-targeting control shRNA was transduced into HepG2 cells in a 5-fold serial dilution, and colonies positive for green fluorescent protein were counted using ImageJ software. Reverse transcription-quantitative polymerase chain reaction and western blot analysis were performed to validate PRDX4 expression. The optimum cell density for transduction was 5.0×103 cells/well in 96-well plates to achieve 40 to 50% confluency the following day. The transduction media consisted of 10% fetal bovine serum (FBS) and 12 µg/ml polybrene, and was used to dilute lentiviral particles at a functional titer of 4.9×105 TU/ml for multiplicity of infection (MOI) of 20, 15 and 10, for 24 h of incubation. Selection with 7 µg/ml puromycin was performed in transduced cells. shRNA 3 was revealed to inhibit PRDX4 mRNA and protein expression. In conclusion, PRDX4 was successfully silenced in 5.0×103 HepG2 cells cultured with 10% FBS and 12 µg/ml polybrene, at a 4.9×105 TU/ml functional titer for MOI of 20, 15 and 10.
Keywords: optimization, critical factors, transduction, peroxiredoxin 4, HepG2
Introduction
Transduction is the process of introducing small hairpin RNA (shRNA) sequences that are encoded in viral DNA into cells via viral vector. Upon binding to the cell, the viral genome is delivered into the cytoplasm, and Dicer protein enzymatically cleaves the short form of the shRNA from the viral origin. Then, the shRNA intermediate is imported into the host cell nucleus, where it is stably integrated into the host genome and interacts with several proteins to form an RNA-induced silencing complex (RISC). RISC silences the target gene by recognizing the corresponding mRNA sequences, and causing their degradation (1). With each cellular division, the integrated virus is replicated and passed on to the daughter cells, thus ensuring continued expression of the targeted sequence throughout the population. shRNAs are 19–22 bp double-stranded RNA, and are complementary to specific targets (2).
Cancer cell lines are commonly used to study carcinogenesis, potential biomarkers and alternative interventions, as they are homogenous and their supplies are unlimited. The present study used HepG2, which is a hepatoblastoma cell line (3). The HepG2 cell line has been misidentified as hepatocellular carcinoma cell line in previous scientific reports (4). However, the misidentification will not affect the outcome of the present study because it aims to provide a general guideline to optimize lentiviral transduction in a cell, and is not specific to either hepatocellular carcinoma or hepatoblastoma. In the present study, HepG2 was used as a model to silence a gene using the general guidelines mentioned. Comparison of the outcome in hepatocellular carcinoma and hepatoblastoma is beyond of the scope of the present study.
The development of genetic manipulation in the HepG2 cell line is challenging, with transfection methods resulting in inefficient transduction of the cells. As an alternative, lentiviral vectors and cationic polymers, including polybrene, have been used to increase transduction efficiency in the HepG2 cell line (5,6). HepG2 cells are of interest because previous studies have demonstrated that γ-tocotrienol treatment increases peroxiredoxin 4 (PRDX4) gene expression in HepG2 cells (7) and this gene may act as a sensitive marker of oxidative stress in liver injury (8). These studies suggested that PRDX4 may be involved in mechanisms that protect against oxidative stress, which remain unclear. This gap in knowledge requires elucidation at the molecular level in order to determine the function of PRDX4. Transduction is a useful approach to determine PRDX4 function, allowing modification of its expression in different conditions.
Several critical factors need to be considered in order to achieve specific and efficient transduction (9–11). These include cell density, polybrene concentration, serum condition, incubation time and puromycin antibiotic selection dosage (12–15). Functional titers and multiplicity of infection (MOI) determination further help to optimize shRNA delivery into the cells. In addition, ideal shRNA vector construction and design, which are crucial to control shRNA expression, also influence the transduction efficiency. The cells will suffer cytotoxicity and unwanted side effects if shRNA expression is too high, whereas transfection will be suboptimal if shRNA expression is too low. Significant progress has been made in predicting which shRNA vectors and target sequences are the most effective at reducing gene expression (16,17).
At present, the only way to measure the efficiency of specific shRNA target sequences is by direct experimentation. Variation in cell susceptibility to transduction is one of the major challenges in determining ideal conditions for high efficiency of gene transfer events (18,19). Thus, optimization of each transduction step is recommended to ensure the transduction conditions to fine-tune shRNA expression. The ability to control shRNA expression levels is important as silencing of the targeted gene contributes to cell survival and development (20). Thus, the present study was performed to determine the optimum conditions of PRDX4-shRNA transduction in HepG2 cells, and to determine the critical factors needed to ensure successful silencing.
Materials and methods
Cell line
HepG2 cells (American Type Culture Collection, Manassas, VA, USA) were cultured in complete culture media (CCM): Earle's modified Eagle's medium (Hyclone; GE Healthcare Life Sciences, Logan, UT, USA) containing 4 mM L-glutamine, 0.1 mM minimum essential medium (MEM) and non-essential amino acids, supplemented with 10% fetal bovine serum (FBS; Biowest USA, Riverside, MO, USA) in a humidified 37°C incubator containing 5% CO2. HepG2 cells with <10 passages were used for subsequent experiments.
shRNA sequence and design
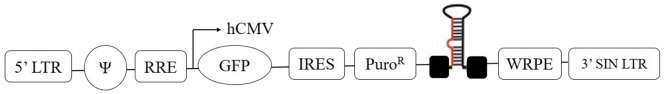
The pGIPZ lentiviral vector of 11.8 kb size (GE Healthcare Dharmacon Inc., Lafayette, CO, USA) harbors a reporter and a puromycin resistance gene. The encoded reporter gene is translated into a green fluorescent protein (GFP). The lentiviral vector is controlled by the immediate-early cytomegalovirus (CMV) promoter (Fig. 1). HepG2 cells were transduced with three individual shRNA clones against PRDX4 (NM_006406.1): sequence 1 (cat. no. 370039), sequence 2 (cat. no. 370042) and sequence 3 (cat. no. 200074) (GE Healthcare Dharmacon Inc.). Non-targeting negative control shRNA (cat. no. RHS4346), consisting of a random sequence and a positive control GAPDH-shRNA lentivirus (cat. no. RHS4372), were used as a control (GE Healthcare Dharmacon Inc.). The sequences are listed in Table I.
Figure 1.
Schematic drawings of the shRNA construct. The PRDX4 shRNA was cloned into the pGIPZ vector under control of hCMV. shRNA, short hairpin RNA; LTR, long terminal repeat; RRE, rev response element; hCMV, human cytomegalovirus promoter; GFP, green fluorescent protein; IRES, internal ribosomal entry sites; PuroR, puromycin; WRPE, woodchuck hepatitis post-transcriptional regulatory element; SIN, self-inactivating.
Table I.
Sequences of PDX4 shRNA and non-targeting negative control shRNA.
| shRNA | Sequence (5′-3′) |
|---|---|
| GAPDH-shRNA positive control | |
| Sense | ATGGTTTACATGTTCCAAT |
| Antisense | ATTGGAACATGTAAACCAT |
| Non-targeting negative control | |
| Sense | ATCTCGCTTGGGCGAGAGTAAG |
| Antisense | CTTACTCTCGCCCAAGCGAGAG |
| shRNA PRDX4 sequence 1 | |
| Sense | GAGCTGAAGTTAACTGATT |
| Antisense | AATCAGTTAACTTCAGCTC |
| shRNA PRDX4 sequence 2 | |
| Sense | AGCTGAAGTATTTCGATAA |
| Antisense | TTATCGAAATACTTCAGCT |
| shRNA PRDX4 sequence 3 | |
| Sense | ACCTGGTAGTGAAACAATA |
| Antisense | TAATTGTTTCACTACCAGGT |
shRNA, small hairpin ribonucleic acid, PRDX4, peroxiredoxin 4.
Optimization of basic transduction conditions
HepG2 cells were seeded at a density of 5×103, 7.5×103 and 10×103 cells/well in a final volume of 100 µl CCM in two 96-well plates, repeated in triplicate for each cell density, and incubated in a humidified 37°C incubator with 5% CO2 overnight. Cell density was observed using an inverted microscope (CK 40; Olympus Corporation, Tokyo, Japan) at 24 h post-transfection. Cell density that reached 40–50%, confluency was selected to be used in subsequent experiments and cell densities that resulted in >50% confluency were excluded. Transduction media (TM) with serum was freshly prepared, and consisted of a 1:1 ratio of Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) to MEM (Hyclone; GE Healthcare Life Sciences), 10% FBS and range of polybrene concentrations (0, 2, 4, 6, 8, 10, 12 and 14 µg/ml; Merck KGaA, Darmstadt, Germany). Culture media was discarded and 50 µl TM with and without serum and the aforementioned range of polybrene concentrations was added to each well. A total of 100 µl CCM with 10% FBS was added to the well with TM and 10% FBS 6 h after the initial addition of TM, whereas 100 µl CCM with 15% FBS was added to each well that contained TM without FBS. The same procedure was repeated for the second plate following incubation for 24 h at 37°C in a humidified atmosphere containing 5% CO2. A cell viability assay was conducted after 24 h of CCM addition.
The viability assay was conducted by adding 120 µl of a Cell Titer 96® aQueous One Solution reagent (Promega Corporation, Madison, WI, USA) that contains a MTS reagent diluted in CCM medium (20 µl MTS/100 µl CCM) in a 96-well for 2 h at 37°C. The purple formazan crystals were dissolved and the viability was analyzed at a wavelength of 490 nm using a microplate reader (EnSpire; PerkinElmer, Inc., Waltham, MA, USA). Three individual experiments were performed.
Determination of cytotoxicity
HepG2 cells were plated at a density of 5×103 cells/well into a 96-well plate in triplicate, and reached 40–50% confluency the following day. A stock solution of 10 mg/ml puromycin (Santa Cruz Biotechnology, Inc., Dallas, TX, USA) was reconstituted with 50 mM dimethyl sulfoxide (Merck KGaA, Darmstadt, Germany). A working concentration range of 1–10 µg/ml was used. Puromycin stock solution was diluted using CCM to concentrations of 1.5, 3.0, 5.0, 7.0 and 10.0 µg/ml. Culture media were discarded, and 100 µl of each puromycin concentration was added to each well. Media containing the selection antibiotic for each concentration was replaced every 2–3 days for up to 7 days. Cell viability assay was performed as aforementioned once per day until day 7 to determine the visual toxicity effect on HepG2 cells. Three independent experiments were performed.
Determination of functional titer and MOI range
To establish the functional titer of lentiviral particles in HepG2 cells, cells were plated at a density of 7×104 cells/well in 24-well plates in duplicate, and were permitted to reach 40 to 50% confluency by culturing overnight in humidified 37°C incubator with 5% CO2. The TM consisted of a 1:1 ratio of DMEM and MEM, 10% FBS and 12 µg/ml polybrene, and was freshly prepared. Culture medium was removed and 225 µl TM was added to each well of the 24-well plate. Non-targeted negative control lentiviral particles were diluted to 5-fold serial dilutions (5, 25, 125, 625, 3,125, 15,625, 78,125 and 390,625 fold) in a flat-bottom 96-well plate with TM as a diluent, as previously described (3). This dilution range was according to the manufacturer's protocol. The diluted viral stock was incubated at room temperature for 10 to 20 min prior to the addition of 25 µl diluted lentiviral particles to the plates. Following incubation for 24 h, 100 µl CCM was added to the wells. GFP expression in the HepG2 cells was observed under a fluorescence microscope (Evos XL Core Imaging System; Thermo Fisher Scientific, Inc.) 48 h after transduction with the pGIPZ lentivirus. To ensure maximum accuracy and to compensate for multiple transduction events per cell, the titers were calculated only from transductions with dilution factors resulting in <25% transduced cells. In addition, the number of GFP-positive colonies were counted in duplicate, for two wells infected with two consecutive dilutions. GFP-positive colonies were counted at ×10 magnification for 3 to 5 random fields using ImageJ software (version 1.41; National Institutes of Health, Bethesda, MD, USA).
Functional titer was determined using the following formula: [493 (number of GFP colonies) × 25 (non-targeting control dilution)]/[0.025 ml (volume of diluted lentiviral particles used)]=4.9×105 TU/ml. The functional titer of the non-targeting control was stated in the certificate of analysis provided by the manufacturer. This functional titer was used in the calculation of the volume of lentiviral particles required to infect HepG2 cells at the desired MOI.
HepG2 cell transduction
Three control groups and three shRNA sequences were established. Each group was set-up in technical triplicate wells. The control group consisted of HepG2 cells with TM, the positive control consisted of HepG2 cells and GAPDH-shRNA lentivirus, and the negative control consisted of HepG2 cells and non-targeting-shRNA lentivirus. HepG2 cells were cultured in a 96-well plate at a density of 5×103 cells/well, reached 40–50% confluency following 24 h incubation at 37°C and underwent transduction using optimized optimal conditions. The lentiviral particles of sequences 1, 2 and 3 were diluted at 4.9×105 TU/ml functional titer for MOI of 20, 15 and 10 with TM consists of 1 MEM: 1 DMEM, 10% FBS and 12 µg/ml polybrene (Merck KGaA, Darmstadt, Germany).
The volume of lentivirus required was calculated using the following formula: i) [5×103 (number of cells seeded) × MOI 20]/[4.9×105 TU/ml (functional titer of HepG2 cells)]=lentiviral volume, and ii) Lentiviral volume (µ1) ×7=final volume of lentiviral stock (µ1).
The lentiviral volume was multiplied to 7 to allow 3 volumes for triplicate wells of MOI 20, 3-fold dilution to achieve MOI 15, and 10, 5 and 1 volume for liquid overage during pipetting. The final volume lentiviral stock was calculated for MOI 20. Serial dilutions were performed for MOI 15, 10 and 5 from the MOI 20 stock. The diluted lentiviral particles were incubated at room temperature between 10 and 20 min. In total, 50 µl diluted virus particles were added at the indicated MOIs (20, 15, 10 and 5) and the cells was incubated at 37°C. After 24 h, 100 µl CCM with no virus particles was added, and the cells were incubated for an additional 48 h at 37°C for GFP expression. The cells were harvested using trypLE (Gibco; Thermo Fisher Scientific, Inc.) and centrifuged at 500 × g for 10 min at room temperature. The supernatant was discarded, the pellet was resuspended with 1 ml selection media (MEM, 10% FBS and 7 µg/ml puromycin) then, cultured in T25 culture flasks (Merck KGaA). Cells were maintained at 37°C in a humidified atmosphere containing 5% CO2. Selection media was replaced every ~2–3 days. The selection media was used for subsequent experiments to ensure that only stably transduced cell were cultured.
Reverse transcription-quantitative polymerase chain reaction (qPCR) analysis
Stably silenced cells from each control and shRNA group were lysed using TRIzol reagent (Gibco: Thermo Fisher Scientific, Inc.) and the RNA was extracted according to the manufacturer's protocol. The extracted RNA was quantified using the NanoDrop 1000 spectrophotometer (NanoDrop Technologies; Thermo Fisher Scientific, Inc.). Subsequently, 1 µg total RNA was used as the template for cDNA synthesis, which was performed using a QuantiNova Reverse Transcription kit (Qiagen, Inc., Valencia, CA, USA) according to the manufacturer's protocol. cDNA was amplified using the QuantiNova SYBR Green real time-PCR kit (Qiagen, Inc.) and the BioRad iQ5 Multicolour Real-time PCR machine (Bio-Rad Laboratories, Inc., Hercules, CA, USA). GAPDH was used as an internal control. Three independent experiments were performed on the same group. The primer sequences for GAPDH (NM_001256799.2) forward, 5′-TCCCTGAGCTGAACGGGAAG-3′, and reverse, 5′-GGAGGAGTGGGTGTCGCTGT-3′, yielded a 245 bp product and the primer sequences for the PRDX4 gene (NM_006406.1) forward, 5′-GCAAAGCGAAGATTTCCAAG-3′ and reverse, 5′-GGCCAAATGGGTAAACTGTG-3′ yielded a 217 bp product.
The PCR conditions were as follows: Initial melting at 95°C for 2 min; followed by 30 cycles at 95°C for 5 sec; 60°C for 10 sec; and 95°C for 1 min. The final extension was performed at 55°C for 10 min. Analysis of the melting curve for the primers was conducted to confirm the specificity of the PCR product, and the threshold cycle (Cq) value for triplicate reactions was averaged. The relative expression of PRDX4 mRNA for each sample was calculated as follows: ΔCq=Cq (sample)-Cq (GAPDH), ΔΔCq (sample)=ΔCq (sample)-ΔCq (calibrator). The fold changes in mRNA were calculated through relative quantification (2−ΔΔCq) (21).
Western blot analysis
Following selection, transduced cell were washed three times with cold 1X phosphate buffered saline and lysed using lysis buffer comprised of protease inhibitors (Promega Corporation, Madison, WI, USA) and RIPA buffer (Merck KGaA). The cell lysates were suspended in 200 µl lysis buffer and were incubated on ice for 30 min. Following this, the cell lysate was centrifuged at 18,506 × g for 30 min at 4°C and the supernatant was collected. The extracted proteins were quantified using the Bradford protein assay (Bio-Rad Laboratories, Inc.). Total protein (40 µg per lane) was treated with SDS sample buffer and heated at 95°C for 2 min. The protein samples were separated on a 12% SDS-polyacrylamide gel and transferred onto polyvinylidene difluoride membranes (GE Healthcare Life Sciences). The membranes were blocked with 5% skimmed milk diluted with Tween-20 phosphate buffered saline (TPBS; 998 ml 1X phosphate buffered saline, 2 ml Tween-20) for 1 h at room temperature. Then, the membranes were incubated overnight with primary mouse monoclonal anti-PRDX4 (1:500; cat no. ab68344; Abcam, Cambridge, UK) and mouse monoclonal anti-β-actin (1:1,000; cat no. ab8224; Abcam) at 4°C. The membranes were then washed with TPBS at intervals of 5 min. Following this, the membranes were incubated with polyclonal secondary horseradish peroxidase-conjugated goat anti-mouse immunoglobulin G (1:2,500; cat no. ab97023; Abcam) for 1 h at room temperature, followed with three washes with TPBS at 5 min intervals. The bands were visualized using enhanced chemiluminescence reagents (Advansta, Inc., Menlo Park, CA, USA) and visualized on a Gel Documentation System (GE Healthcare Life Sciences). Three independent experiments were performed.
Statistical analysis
GraphPad Prism 5 software (GraphPad Software, Inc., La Jolla, CA, USA) was used for data processing. Comparisons between groups were performed using one-way analysis of variance followed by the Tukey's multiple range test. Quantitative data are expressed as the mean ± standard deviation. Figures were generated using GraphPad Prism 5. P<0.05, P<0.01 and P<0.001 were considered to indicate a statistically significant difference.
Results
Optimization of basic transduction conditions
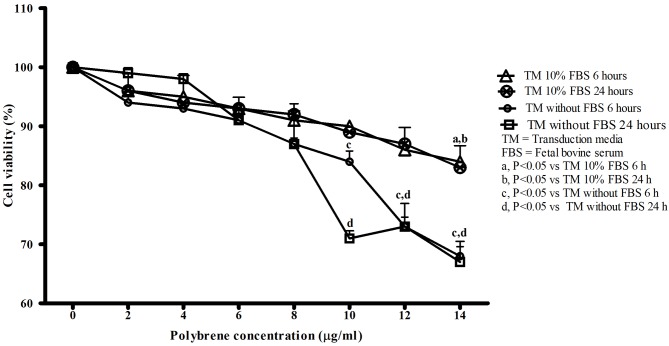
Factors measured included cell density, incubation time, serum conditions and polybrene concentration. The cell densities used were 5.0×103, 7.5×103 and 10.0×103 cells/well, and cell confluence was observed under a light microscope on the following day. The results revealed that densities of 7.5×103 and 10.0×103 cells/well resulted in >50% confluence, while a density of 5.0×103 cells/well did not. Therefore, 5.0×103 cells/well was the selected HepG2 cell density that was used for subsequent experiments. There was no significant difference when comparing 6 and 24 h of incubation for groups with and without FBS in TM. However, as presented in Fig. 2, cells incubated in TM without FBS for 6 and 24 h had decreased cell viability compared with cells incubated in TM with 10% FBS for 6 and 24 h, with increasing polybrene concentration. The absence of serum caused polybrene to significantly reduce cell viability earlier, at 10, 12 and 14 µg/ml (P<0.001) for the two incubation times. In comparison, when cells were incubated with TM with 10% FBS, cell viability was only significantly reduced by 14 µg/ml polybrene (P<0.05). The highest polybrene concentration which did not affect cell viability during incubation in TM with 10% FBS was 12 µg/ml. Incubation times of 6 and 24 h for TM with 10% FBS did not significantly affect cell viability. An increased duration of exposure to lentivirus may increase cell transduction efficiency, so an incubation time of 24 h was selected. Thus, a density of 5×103 cells/well in a 96 well plate, 12 µg/ml polybrene in TM with 10% FBS and a 24 h incubation time were selected as optimal transduction conditions for HepG2 cells.
Figure 2.
Basic transduction condition. HepG2 cell viability in transduction media in the absence and presence of serum at 6 and 24 h, as the polybrene concentration increases. Data are presented as the mean ± standard deviation, and each group consists of a technical triplicate for 3 biological replicates. a, P<0.05 vs. TM 10% FBS 6 h; b, P<0.05 vs. TM 10% FBS 24 h; c, P<0.05 vs. TM without FBS 6 h and d, P<0.05 vs. TM without FBS 24 h. TM, transduction media; FBS, fetal bovine serum.
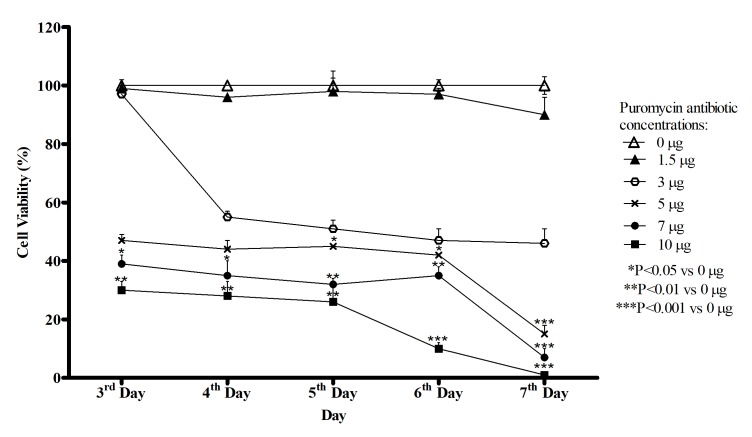
Titration of puromycin
HepG2 cells were subjected to increasing doses of puromycin from day 3 to 7. The optimal dose was selected. As presented in Fig. 3, the viability of HepG2 cells decreased with increasing doses of puromycin. Based on a cell viability assay, HepG2 cells exhibited decreased viability (up to 90% of cell death) at day 7 in response to 7 and 10 µg/ml puromycin. The optimal puromycin concentration is the lowest dose that will kill 90–99% non-selected HepG2 cells within 7 days, therefore a dose of 7 µg/ml puromycin was selected. The transduced cells were selected and cultured in selection media that consisted of base media, 10% FBS and 7 µg/ml puromycin to maintain the stable cell lines generated.
Figure 3.
Kill curve determination. HepG2 kill curves were constructed from day 3–7 using a range of puromycin concentrations. Data are presented as the mean ± standard deviation, and each group consists of technical triplicates for 3 biological replicates. *P<0.05 vs. 0 µg, **P<0.01 vs. 0 µg and ***P<0.001 vs. 0 µg.
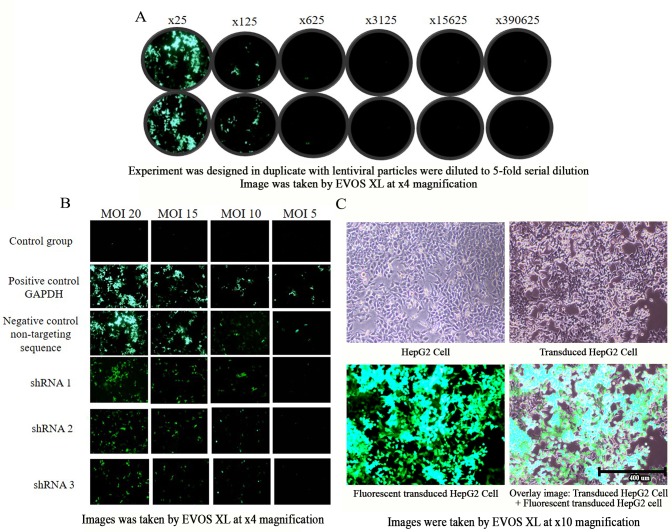
Functional titer and MOI range determination
Serial dilutions (5-fold) of the non-targeting lentiviral control were transduced into HepG2 cells in duplicate, and the number of colonies positive for GFP were used to calculate functional titer. GFP-positive colonies were counted at the low dilution of 25X and 125X, while no GFP colonies were generated for dilutions of 625X to 390,635X (Fig. 4A). A functional titer of 4.9×105 lentiviral particles was obtained. The optimized functional titer and transduction conditions were used to transduce a GAPDH positive control, a non-targeting negative control and three PRDX4-shRNAs with different sequences at MOI 20, 15, 10 and 5. The results revealed that no GFP-positive colonies were generated from the control group for all MOIs, whereas for the GAPDH positive control group and non-targeting negative control group, GFP was generated for all MOIs (Fig. 4B). Transduced colonies decreased as MOI decreased, and only MOI of 20, 15 and 10 were positive for GFP when transduced with shRNA 1, 2 and 3. Therefore, MOI 20, 15 and 10 were used to transduce HepG2 cells for subsequent experiments. An overlay of transduced cells with generated GFP colonies and a comparison of transduced and non-transduced HepG2 cells is presented in Fig. 4C. The transduced cells have clumpy morphology and are slower growing compared with non-transduced cells.
Figure 4.
Green fluorescent protein expression of HepG2 cells under a fluorescence microscope, EVOS XL. (A) Functional titer determination. The experiment was designed in duplicate with lentiviral particle were to 5-fold serial dilutions as follows: ×25, ×125, ×625, ×3125, ×15625 and ×390625. Image was taken by fluorescent microscope, EVOS XL (magnification, ×40). (B) MOI range of HepG2 cells. Image was taken by fluorescent microscope, EVOS XL (magnification, ×40). (C) Overlaid image of transduced and non-transduced HepG2 cells (magnification, ×10). The scale bar represents 400 µm. MOI, multiplicity of infection; shRNA, small hairpin ribonucleic acid.
PRDX4 gene expression
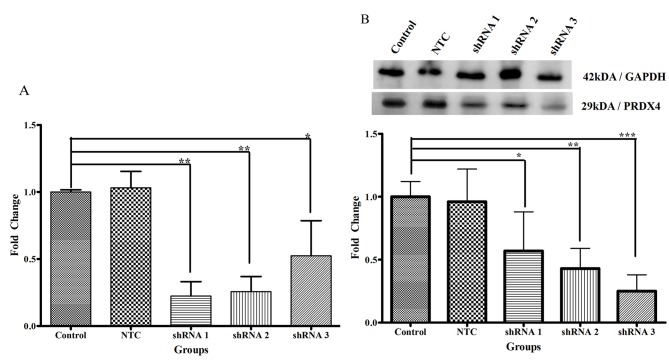
Following transduction for 48 h, shRNA 1 (P<0.01) and 2 (P<0.01) was significantly reduced PRDX4 mRNA expression, up to 8-fold, whereas shRNA 3 resulted in a 5-fold reduction compared with the control group (P<0.05; Fig. 5A). Expression in the non-targeting control transduction and control groups was not significantly different. PRDX4 mRNA expression levels in the negative control, shRNA 1, shRNA 2 and shRNA 3 groups were 1.03±0.12, 0.22±0.11, 0.26±0.11 and 0.52±0.26, respectively. PRDX4 mRNA levels were the lowest in the cells treated with PRDX4-shRNA 1 and shRNA 2 compared with the control group.
Figure 5.
Validation of PRDX4 expression. (A) Fold change of PRDX4 mRNA expression. (B) Fold change of PRDX4 protein expression. Data are presented as the mean ± standard deviation normalized to the control group. NTC refers to a non-targeting control, control refers to non-transduced cells, and each group consists of a technical triplicate for 3 biological replicates. *P<0.05, **P<0.01 and ***P<0.001. PRDX4, peroxiredoxin 4; NTC, non-targeting control; shRNA, small hairpin ribonucleic acid.
PRDX4 protein expression
Transduction with shRNA 1 (P<0.05), 2 (P<0.01) and 3 (P<0.001) for 48 h resulted in a significant reduction of PRDX4 protein expression (5, 6 and 8-fold, respectively) compared with the control group (Fig. 5B). PRDX4 protein expression levels for the non-targeting control, shRNA 1, shRNA 2 and shRNA 3 were 0.96±0.26, 0.57±0.31, 0.43±0.16 and 0.25±0.13. Compared with the control group, PRDX4 protein expression levels were lowest in the cells transduced with PRDX4-shRNA 3.
Discussion
Stable cell lines were generated when GIPZ lentiviral DNA integrated into the host genome and permanently expressed the shRNA as the cells divided. Several factors affected transduction efficiency, including cell density, polybrene concentration, incubation time, serum condition and antibiotic selection dose. The best optimization method should consist of at least two potential shRNA sequences that silence the target gene. This would provide control for off-target knockdowns, since it is unlikely for different sequences to produce the same off-target effects, which increasing the potential for successful gene knockdown (22). Furthermore, optimized shRNA constructs require a relatively low copy number of lentiviral particles, resulting in fewer off-target effects and a reduction in unwanted silencing effects (23). A good experimental design consists of a positive control, negative control and untreated control to further validate the significant and specific silencing effect (24,25). GFP colonies were generated when the GIPZ GAPDH positive control was transduced into HepG2 cells (Fig. 4B). This confirmed that the plasmid construct was successfully delivered into the cells and activated the RNA interference pathway without affecting cell viability or function (26). Well-characterized positive controls target the housekeeping genes that are abundantly expressed in the cells. This demonstrated that expression was neither affected by experimental treatments nor fluctuated with the cell cycle. One of the major problems faced by shRNA delivery in mammalian systems is the probability of triggering off-target effects, inconsistent phenotypes and cellular toxicity due to interactions between sense and antisense shRNA strands (27–29). Thus, GIPZ non-targeting negative controls (NTC) are important to distinguish sequence-specific silencing from off-target effects. The NTC sequence does not match with known mammalian genes, having at least three or more mismatches against any gene as determined via nucleotide alignment. An ideal NTC sequence produces a minimal effect on cell viability, and mRNA and protein expression levels should remain comparable with the control (Fig. 5A and B). When the NTC is comparable with the control group, it is possible to conclude that the gene knockdown is specific and that any downstream effect can be attributed to silencing of the target gene. Furthermore, it is possible to measure transduction efficiency by assessing the number of GFP-positive colonies following transfection with NTC shRNA (Fig. 4A). Untreated control determines the baseline level of cell viability and target gene level compared with NTC control (Fig. 5A and B).
Efficient shRNA delivery into cells is paramount. There should be enough shRNA production to create gene knockdown at a concentration that does not over activate the RISC complex or result in toxic effects to the cells. Vector-based shRNA transduction allows the long-term downregulation of target genes and but may have variable transduction efficiency, since it is cell type-dependent. Lentiviral vectors, enhanced promoters and GIPZ shRNA are elements that serve essential functions in the maximization of efficient shRNA production in HepG2 cells, and may increase the probability of transduction events occurring (30,31). Lentiviral-based vectors with human (h)CMV enhancer have been reported to infect up to 95% of HepG2 cells and stably integrate into the host genome for long-term transgene expression (32–34). In addition, lentiviral vectors result in low toxicity and high stability with no effect on cell phenotype or cell type (35,36). GIPZ shRNA design is based on native microRNA-30 primary transcripts to enable processing by endogenous RNA interference pathways, resulting in specific silencing and minimized cellular toxicity (37,38). In addition, the presence of selectable markers, which are a puromycin resistance gene and a GFP gene reporter, assist in selecting a stable cell line with an incorporated vector and expression of the inserted gene to be cultured. To achieve successful gene silencing, screening >2 shRNA fragments and scaling down to a single fragment which regulates high levels of transcription may increase the silencing effect. Silencing activity does not only depend on the shRNA being 100% complementary to the target gene, but also on the location where shRNA binding occurs. Different shRNA fragments have random integration sites and will generate a variety of dependent expression levels (39). The present study was initiated by the construction of three different shRNA sequences targeting PRDX4, and later transduced into HepG2 cells using polybrene to improve the efficiency. Finally, one shRNA demonstrated the required effect.
Cell density, polybrene concentration, incubation time and transduction media conditions should be systematically examined for every cell type and, once optimized, must be kept constant in all future experiments to ensure reproducible results. Previous studies have stated that HepG2 cells line maintained the same expression of drug metabolizing enzymes compared with that in primary hepatocytes until passage 16 (40,41). Healthy, actively dividing cells take up foreign nucleic acids more efficiently than quiescent cells (42). Thus, it is advisable to select cell lines with <16 passages that are actively dividing for this approach. It is recommended to maintain the cells at least 24 h prior initiating the transduction procedure, to ensure they are in optimum physiological condition for transduction. A cell confluency >50% is considered to be over-confluent and may cause contact inhibition, resulting in poor uptake of nucleic acids, thus decreasing the expression of the transduced gene. However, too few cells in the culture may result in poor growth without cell-to-cell contact (43). In the present study, the transduction process took at least 96 h for the cells to grow, thus, maintaining a standard seeding protocol from each experiment will ensure that optimal confluence is reliably achieved. Generally, a density of 5×103 cells/well seeded in a 96-well plate reached ~40–50% confluence after 24 h, providing optimized results for the HepG2 cell line.
A previous study reported that the addition of polybrene to cells in serum-free media with enhanced lentiviral transduction efficiency by 2–10-fold (11). However, serum free media and high concentration of polybrene may reduce cell viability due to the toxic conditions. Thus, MTS assays were conducted on non-transduced cells to determine the effect of transduction media condition on cell viability. FBS provides a higher transduction efficiency compared with bovine calf serum (9). However, while the presence of serum in culture media boosts cell growth, it also increases the chance of endotoxin contamination. It is important to control for variability among the different brands of serum, as the quality can significantly affect cell growth and transduction results. In contrast, heat inactivation of FBS did not exert a discernible effect on the efficiency of lentiviral transduction (9). The results obtained demonstrated that an absence of serum in the culture media reduced HepG2 cell viability following incubation for 6 and 24 h, compared with cells incubated in the presence of serum (Fig. 2). Thus, TM with 10% FBS is used to transduce HepG2 cells. Cellular and viral lipid membranes possess a net-negative charge. The addition of cationic polymers, including polybrene, to TM has been suggested since these polymers bind to the cell surface and neutralize the surface charge, increasing transduction efficiency by enhancing adsorption of the lentiviral particles. High polybrene concentrations increase the probability of transduction events, however, these may also reduce cell viability. Thus, the optimal concentration of polybrene needs to be determined to avoid toxicity to the cells. The highest polybrene concentration of TM with 10% FBS which did not significantly reduced HepG2 cell viability was 12 µg/ml. If the polybrene concentration failed to induce significant cellular toxicity after 24 h incubation, this incubation time may be used for subsequent transductions (44). Incubation for 6 and 24 h with 10% FBS did not affect cell viability in the present study. In addition, an increased duration of exposure to the lentivirus will maximize lentiviral infection, increasing transduction efficiency (12).
A GIPZ lentiviral construct that encodes for a puromycin resistance gene will only allow the transduced cells to grow in culture with puromycin, which will kill non-transduced cells by interrupting their protein synthesis. Construction of a kill curve is recommended since puromycin concentration is cell type-dependent. The kill curve construction involved a dose-response experiment where non-transduced cells were subjected to increasing concentrations of puromycin to determine the minimum antibiotic concentration needed to kill all cells over the course of 3–7 days. Puromycin reached optimum effectiveness after 48 h of selection and, in general, it took 7 days for a single transduced colony to form a population of transduced HepG2 cells. The selection medium, which was comprised of base medium, 10% FBS and the optimized puromycin concentration (7 µg/ml) was changed daily for a week. This eliminated potentially toxic substances produced by the dying cells and maintained the concentration of the antibiotic at a constant level during the selection process. Antibiotic dosage is considered low when minimal visual toxicity is apparent even after 7 days of antibiotic exposure, while the antibiotic dose is considered high when visual toxicity is evident within the first 2–3 days of antibiotic selection. The optimal dose of puromycin was 7 µg/ml, which was the lowest antibiotic concentration to kill 90–99% of cells (Fig. 3) after 7 days of antibiotic selection.
The most critical factor to determine for successful lentiviral transduction is the functional titer. The functional titer is referred as smallest transduction unit of virus capable of infecting susceptible cells and expressing the transgene per ml solution (TU/ml). Serial dilution of non-targeting lentiviral stock helped to determine the optimal concentration of functional lentiviral required in HepG2 cells. The functional titer was 4.9×105 TU/ml, and was calculated from a number of GFP transduced colonies developed after 48 h transduction. An optimized functional titer only requires a relatively small volume of lentiviral particles to transduce cells at a constant ratio, referred as multiplicity of infection (MOI), and thus increases transduction efficiency. One of the factors that affects the functional titer is shRNA production. A potent promoter and vector are essential to produce sufficient amounts of shRNA in HepG2 cells, in order to maximize transduction efficiency. Low shRNA production will cause transduction to be suboptimal, and higher levels of shRNA production may inhibit transduction. Functional titer is also cell type-dependent. Each cell has different level of susceptibility towards lentiviral infection (45) and an actively dividing cell line would give a higher transduction rate than non-dividing types of cell (46). One of the major advantages of the lentiviral delivery method is the ability to control the number of viral genomes that enter HepG2 cells by adjusting the MOI. It is important to note that different cell types may require different MOIs for knockdown of the target gene. The sensitivity of cells to lentiviral infection is MOI-dependent: The higher the MOI, the higher the transduction efficiency. To obtain optimal expression, a range of MOIs from 20, 15, 10 and 5 were tested on HepG2 cells in the present study. The results obtained revealed that only MOI 20, 15 and 10 result in significant levels of cell transduction (Fig. 4B). Constant MOIs of 20, 15 and 10 were used to transduce the HepG2 cells with a 4.9×105 TU/ml functional titer. Working in the smallest well format and volume helps achieve higher MOI, by using fewer lentiviral particles to increase transduction efficiency.
Although shRNA functions at the mRNA level, ultimately it is the reduction in protein level that causes the observed phenotype. Therefore, validation of gene silencing was assessed by measuring the corresponding protein and gene reduction using western blot analysis and qPCR (47). Gene silencing mediated by mRNA may occur at three stages, including pre-translational, co-translational, and post-translational steps, which exert direct and indirect effects on translation machinery (48). This justifies the non-corresponding effect of PRDX4 silencing on protein reduction at the mRNA level measured following transfection with each shRNA sequence, suggesting that PRDX4 expression was not regulated at the mRNA level. mRNA levels have been demonstrated to not always be associated with protein expression levels, for example those observed in human liver tissue (49,50). These studies emphasized the probabilities of post-transcriptional or post-translational modifications, and additional unknown regulatory mechanisms and signaling. Furthermore, different shRNA integration sites may affect the level of expression. The use of lentiviral vectors provides a novel method by which HepG2 cells may be stably genetically manipulated, with minimal effects on the differentiated hepatic phenotype. Lentiviral infection has previously been demonstrated to not affect hepatic hallmarks, including the expression of the nuclear receptors CXADR, Ig-like cell adhesion molecule, nuclear receptor subfamily 1 group I member 2, retinoid C receptor α, and hepatocyte nuclear factor 4α, or albumin expression. Furthermore, HepG2 cells demonstrated that transgene expression is preserved through cell division and is retained for the duration of the culture period (36).
In conclusion, the optimal transduction conditions, which include cell density, polybrene concentration, transduction media condition, incubation time and selective antibiotic dose, were identified to produce a HepG2 cell line with stably silenced PRDX4. The results demonstrated that shRNA 3 significantly suppressed PRDX4 expression at the mRNA and protein level, and this shRNA was therefore used in subsequent experiments. This model may be used to test the effect of nutritional interventions on hepatoblastoma, and may lead to improved understanding of treatment strategies for hepatoblastoma and also for patients with liver cancer in general.
Acknowledgements
The present study was funded by National University of Malaysia Medical Centre (grant no. FF-2016-063) and the Ministry of Higher Learning under the Fundamental Research Grant Scheme (grant no. UKM-AP-2014-024).
Competing interests
The authors declare that they have no competing interests.
References
- 1.Manjunath N, Haoquan W, Sandesh S, Premlata S. Lentiviral delivery of short hairpin RNAs. Adv Drug Deliv Rev. 2009;61:732–745. doi: 10.1016/j.addr.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aigner A. Gene silencing through RNA interference (RNAi) in vivo: Strategies based on the direct application of siRNAs. J Biotechnol. 2006;124:12–25. doi: 10.1016/j.jbiotec.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 3.López-Terrada D, Cheung SW, Finegold MJ, Knowles BB. HepG2 is a hepatoblastoma-derived cell line. Hum Pathol. 2009;40:1512–1515. doi: 10.1016/j.humpath.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 4.Pang RTK, Poon TCW, Wong N, Lai PBS, Wong NLY, Chan CML, Yu JSW, Chan ATC, Sung JJY. Comparison of protein expression patterns between hepatocellular carcinoma cell lines and a hepatoblastoma cell line. Clin Proteomics. 2004;1:313–331. doi: 10.1385/CP:1:3-4:313. [DOI] [Google Scholar]
- 5.Le Bihan O, Chèvre R, Mornet S, Garnier B, Pitard B, Lambert O. Probing the in vitro mechanism of action of cationic lipid/DNA lipoplexes at a nanometric scale. Nucleic Acids Res. 2011;39:1595–1609. doi: 10.1093/nar/gkq921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cesaratto L, Vascotto C, D'Ambrosio C, Scaloni A, Baccarani U, Paron I, Damante G, Calligaris S, Quadrifoglio F, Tiribelli C, Tell G. Overoxidation of peroxiredoxins as an immediate and sensitive marker of oxidative stress in HepG2 cells and its application to the redox effects induced by ischemia/reperfusion in human liver. Free Radic Res. 2005;39:255–268. doi: 10.1080/10715760400029603. [DOI] [PubMed] [Google Scholar]
- 7.Sazli Abdul Rahman F, Jubri Z, Rahman Abdul M, Karsani SA, Top Md AG, Ngah Wan WZ. Gamma-tocotrienol treatment increased peroxiredoxin-4 expression in HepG2 liver cancer cell line. BMC Complement Altern Med. 2015;15:64. doi: 10.1186/s12906-015-0590-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ito R, Takahashi M, Ihara H, Tsukamoto H, Fujii J, Ikeda Y. Measurement of peroxiredoxin-4 serum levels in rat tissue and its use as a potential marker for hepatic disease. Mol Med Rep. 2012;6:379–384. doi: 10.3892/mmr.2012.935. [DOI] [PubMed] [Google Scholar]
- 9.Denning W, Das S, Guo S, Xu J, Kappes JC, Hel Z. Optimization of the transductional efficiency of lentiviral vectors: Effect of sera and polycations. Mol Biotechnol. 2013;53:308–314. doi: 10.1007/s12033-012-9528-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Logan AC, Nightingale SJ, Haas DL, Cho GJ, Pepper KA, Kohn DB. Factors influencing the titer and infectivity of lentiviral vectors. Hum Gene Ther. 2004;15:976–988. doi: 10.1089/hum.2004.15.976. [DOI] [PubMed] [Google Scholar]
- 11.Davis HE, Rosinski M, Morgan JR, Yarmush ML. Charged polymers modulate retrovirus transduction via membrane charge neutralization and virus aggregation. Biophys J. 2004;86:1234–1242. doi: 10.1016/S0006-3495(04)74197-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morgan JR, LeDoux JM, Snow RG, Tompkins RG, Yarmush ML. Retrovirus infection: Effect of time and target cell number. J Virol. 1995;69:6994–7000. doi: 10.1128/jvi.69.11.6994-7000.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andreadis S, Lavery T, Davis HE, Le Doux JM, Yarmush ML, Morgan JR. Toward a more accurate quantitation of the activity of recombinant retroviruses: Alternatives to titer and multiplicity of infection. J Virol. 2000;74:1258–1266. doi: 10.1128/JVI.74.7.3431-3431.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dokka S, Toledo D, Shi X, Ye J, Rojanasakul Y. High-efficiency gene transfection of macrophages by lipoplexes. Int J Pharm. 2000;206:97–104. doi: 10.1016/S0378-5173(00)00531-7. [DOI] [PubMed] [Google Scholar]
- 15.Zhang B, Metharom P, Jullie H, Ellem KA, Cleghorn G, West MJ, Wei MQ. The significance of controlled conditions in lentiviral vector titration and in the use of multiplicity of infection (MOI) for predicting gene transfer events. Genet Vaccines Ther. 2004;2:6. doi: 10.1186/1479-0556-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taxman DJ, Livingstone LR, Zhang J, Conti BJ, Iocca HA, Williams KL, Lich JD, Ting JP, Reed W. Criteria for effective design, construction, and gene knockdown by shRNA vectors. BMC Biotechnol. 2006;6:7. doi: 10.1186/1472-6750-6-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takasaki S, Kotani S, Konagaya A. An effective method for selecting siRNA target sequences in mammalian cells. Cell Cycle. 2004;3:790–795. doi: 10.4161/cc.3.6.892. [DOI] [PubMed] [Google Scholar]
- 18.Luria SE. Cell susceptibility to viruses. Ann N Y Acad Sci. 1955;61:852–855. doi: 10.1111/j.1749-6632.1955.tb42542.x. [DOI] [PubMed] [Google Scholar]
- 19.Yang L, Bailey L, Baltimore D, Wang P. Targeting lentiviral vectors to specific cell types in vivo. Proc Natl Acad Sci USA. 2006;103:11479–11484. doi: 10.1073/pnas.0604993103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gupta S, Schoer RA, Egan JE, Hannon GJ, Mittal V. Inducible, reversible, and stable RNA interference in mammalian cells. Proc Natl Acad Sci USA. 2004;101:1927–1932. doi: 10.1073/pnas.0306111101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Healthcare G. Technical manual: GIPZ Lentiviral shRNA. Thermo Sci. 2015 [Google Scholar]
- 22.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 23.Moore CB, Guthrie EH, Huang MT, Taxman DJ. Short hairpin RNA (shRNA): Design, delivery, and assessment of gene knockdown. Methods Mol Biol. 2010;629:141–158. doi: 10.1007/978-1-60761-657-3_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Svoboda P. Off-targeting and other non-specific effects of RNAi experiments in mammalian cells. Curr Opin Mol Ther. 2007;9:248–257. [PubMed] [Google Scholar]
- 25.Huppi K, Martin SE, Caplen NJ. Defining and assaying RNAi in mammalian cells. Mol Cell. 2005;17:1–10. doi: 10.1016/j.molcel.2004.12.017. [DOI] [PubMed] [Google Scholar]
- 26.Whither RNAi. Nat Cell Biol. 2003;5:489–490. doi: 10.1038/ncb0603-490. [DOI] [PubMed] [Google Scholar]
- 27.Goncz KK, Gruenert DC, Colosimo A. Expression vector system and a method for optimization and confirmation of DNA delivery and quantification of targeting frequency. Journal. 2005 [Google Scholar]
- 28.Feng Y, Nie L, Thakur MD, Su Q, Chi Z, Zhao Y, Longmore GD. A multifunctional lentiviral-based gene knockdown with concurrent rescue that controls for off-target effects of RNAi. Genomics Proteomics Bioinformatics. 2010;8:238–245. doi: 10.1016/S1672-0229(10)60025-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jackson AL, Linsley PS. Noise amidst the silence: Off-target effects of siRNAs? Trends Genet. 2004;20:521–524. doi: 10.1016/j.tig.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 30.Snøve O, Jr, Holen T. Many commonly used siRNAs risk off-target activity. Biochem Biophys Res Commun. 2004;319:256–263. doi: 10.1016/j.bbrc.2004.04.175. [DOI] [PubMed] [Google Scholar]
- 31.Matveeva O, Nechipurenko Y, Rossi L, Moore B, Saetrom P, Ogurtsov AY, Atkins JF, Shabalina SA. Comparison of approaches for rational siRNA design leading to a new efficient and transparent method. Nucleic Acids Res. 2007;35:e63. doi: 10.1093/nar/gkm088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.ter Brake O, 't Hooft K, Liu YP, Centlivre M, von Eije KJ, Berkhout B. Lentiviral vector design for multiple shRNA expression and durable HIV-1 inhibition. Mol Ther. 2008;16:557–564. doi: 10.1038/sj.mt.6300382. [DOI] [PubMed] [Google Scholar]
- 33.Nasri M, Karimi A, Farsani Allahbakhshian M. Production, purification and titration of a lentivirus-based vector for gene delivery purposes. Cytotechnology. 2014;66:1031–1038. doi: 10.1007/s10616-013-9652-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nash KL, Jamil B, Maguire AJ, Alexander GJ, Lever AM. Hepatocyte-specific gene expression from integrated lentiviral vectors. J Gene Med. 2004;6:974–983. doi: 10.1002/jgm.591. [DOI] [PubMed] [Google Scholar]
- 35.Nishitsuji H, Ikeda T, Miyoshi H, Ohashi T, Kannagi M, Masuda T. Expression of small hairpin RNA by lentivirus-based vector confers efficient and stable gene-suppression of HIV-1 on human cells including primary non-dividing cells. Microbes Infect. 2004;6:76–85. doi: 10.1016/j.micinf.2003.10.009. [DOI] [PubMed] [Google Scholar]
- 36.Rao DD, Vorhies JS, Senzer N, Nemunaitis J. siRNA vs. shRNA: Similarities and differences. Adv Drug Deliv Rev. 2009;61:746–759. doi: 10.1016/j.addr.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 37.Zamule SM, Strom SC, Omiecinski CJ. Preservation of hepatic phenotype in lentiviral-transduced primary human hepatocytes. Chem Biol Interact. 2008;173:179–186. doi: 10.1016/j.cbi.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 39.Li L, Lin X, Khvorova A, Fesik SW, Shen Y. Defining the optimal parameters for hairpin-based knockdown constructs. RNA. 2007;13:1765–1774. doi: 10.1261/rna.599107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McIntyre GJ, Arndt AJ, Gillespie KM, Mak WM, Fanning GC. A comparison of multiple shRNA expression methods for combinatorial RNAi. Genet Vaccines Ther. 2011;9:9. doi: 10.1186/1479-0556-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Westerink WM, Schoonen WG. Phase II enzyme levels in HepG2 cells and cryopreserved primary human hepatocytes and their induction in HepG2 cells. Toxicol In Vitro. 2007;21:1592–1602. doi: 10.1016/j.tiv.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 42.Wilkening S, Bader A. Influence of culture time on the expression of drug-metabolizing enzymes in primary human hepatocytes and hepatoma cell line HepG2. J Biochem Mol Toxicol. 2003;17:207–213. doi: 10.1002/jbt.10085. [DOI] [PubMed] [Google Scholar]
- 43.Prijic S, Sersa G. Magnetic nanoparticles as targeted delivery systems in oncology. Radiol Oncol. 2011;45:1–16. doi: 10.2478/v10019-011-0001-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Song H, Yang PC. Construction of shRNA lentiviral vector. N Am J Med Sci. 2010;2:598–601. doi: 10.4297/najms.2010.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wotherspoon S, Dolnikov A, Symonds G, Nordon R. Susceptibility of cell populations to transduction by retroviral vectors. J Virol. 2004;78:5097–5102. doi: 10.1128/JVI.78.10.5097-5102.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Johnston JC, Gasmi M, Lim LE, Elder JH, Yee JK, Jolly DJ, Campbell KP, Davidson BL, Sauter SL. Minimum requirements for efficient transduction of dividing and nondividing cells by feline immunodeficiency virus vectors. J Virol. 1999;73:4991–5000. doi: 10.1128/jvi.73.6.4991-5000.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Orang Valinezhad A, Safaralizadeh R, Kazemzadeh-Bavili M. Mechanisms of miRNA-mediated gene regulation from common downregulation to mRNA-specific upregulation. Int J Genomics. 2014;2014:970607. doi: 10.1155/2014/970607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zeng Y, Cullen BR. Efficient processing of primary microRNA hairpins by Drosha requires flanking nonstructured RNA sequences. J Biol Chem. 2005;280:27595–27603. doi: 10.1074/jbc.M504714200. [DOI] [PubMed] [Google Scholar]
- 49.Anderson L, Seilhamer J. A comparison of selected mRNA and protein abundances in human liver. Electrophoresis. 1997;18:533–537. doi: 10.1002/elps.1150180333. [DOI] [PubMed] [Google Scholar]
- 50.Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]