Abstract
The incidence of pancreatic cancer is increasing annually in Asia as a whole. Pancreatic cancer ranks sixth in terms of incidence of all malignant tumors. Circular RNA (circRNA) is a type of non-coding RNA which forms a covalently closed continuous loop. CircRNA is extensively expressed in the cytoplasm, and is markedly conservative and stable. MicroRNA (miR)-378a-3p and human (hsa)_circ_0006215 were detected using the reverse transcription-quantitative polymerase chain reaction (RT-qPCR) in tissue and cells. Western blot analysis detected the SERPINA4 and hsa_circ_0006215 expression in tissue. A Cell Counting Kit-8 assay was used to determine cell stability. Flow cytometry was used to determine the cell apoptotic rate. Transwell assays were used to determine cell migration. hsa_circ_0006215 was identified as a significantly upregulated circRNA. RT-qPCR results verified that, in 30 samples of pancreatic cancer tissue and paracancerous tissue, hsa_circ_0006215 expression was increased in pancreatic cancer tissue, miR-378a-3p expression was decreased in pancreatic cancer tissue, and SERPINA4 expression was increased in pancreatic cancer tissue (P<0.05). Using bioinformatics database and bioinformatics analysis, the interaction network of hsa_circ_0006215 indicated that this circRNA was most likely to regulate the expression of miR-378a-3p. Further interaction analysis revealed that the SERPINA4 gene was a regulatory target gene most likely to have an influence. The present study identified the effects of hsa_circ_0006215, miR-378a-3p and SERPINA4 signaling pathways in pancreatic cancer cells.
Keywords: pancreatic cancer, circular RNAs, microRNAs, cell viability, cell apoptosis
Introduction
The incidence of pancreatic cancer is increasing annually in Asia. Pancreatic cancer ranks sixth of all malignant tumors, and is the eighth and ninth leading cause of mortality in men and women, respectively (1). One of the greatest problems in the medical field is obtaining an early diagnosis of pancreatic cancer. Using traditional methods, the majority of patients with pancreatic cancer are diagnosed at an advanced stage. The resection rate is markedly low and the prognosis is markedly poor in patients with advanced pancreatic cancer. Early detection is the only approach to improve the prognosis of pancreatic cancer (2).
Endoscopic ultrasonography-guided fine-needle aspiration (EUS-FNA) is able to directly extract the pancreatic tissue (3). Therefore, as it is difficult to distinguish the nature of pancreatic mass using computer tomography (CT) and EUS (4,5), EUS-FNA technology is a good option (6,7). In recent years, with the rapid development of molecular biology and high-throughput sequencing, it is possible to screen molecular markers for pancreatic cancer at the levels of genome, transcriptome and proteome. EUS-FNA provides reliable samples for genetic testing and gene sequencing. Making full use of molecular biology techniques to detect gene changes has promise as a novel diagnostic method.
Circular RNA (circRNA) is a type of non-coding RNA, which forms a covalently closed continuous loop. These tissue-specific transcripts are produced primarily by the exon or intron sequences of housekeeping genes. CircRNAs are extensively expressed in the cytoplasm, and are markedly conservative and stable. These features afford them special functions: Exerting the function of microRNA (miRNA) sponges, adjusting the selective splicing and encoding the protein as a transcription factor. CircRNAs are likely to be ideal biomarkers for cancer diagnosis. A previous study has confirmed that circRNAs are able to regulate the expression of a variety of cancer-associated miRNAs (8). Certain circRNAs have miRNA-response elements, which are able to interact with miRNAs, thereby regulating the expression of the target gene. In islet β-cells, microRNA (miR)-7 inhibitors are able to modulate the mammalian target of rapamycin signaling pathway in the process of cell proliferation (9). Therefore, circRNAs may be a novel mediator of tumorigenesis and progression. CircRNA-miRNA-mRNA interaction networks serve a regulatory function in a number of cancer-associated signaling pathways, and have inhibitory or promoting effects on the occurrence of cancer. Owing to their potential clinical relevance, circRNAs may be the basis of studies concerning the prevention and treatment of cancer.
The human transcriptome contains mRNA and a large number of non-coding RNAs, including miRNAs and circRNAs, which have a regulatory effect by adsorbing miRNAs (10). High-throughput specific microarrays have been used to screen and verify the candidate circRNA for certain types of cancer, suggesting that specific circRNAs may provide novel molecular markers for tumor diagnosis, prognosis and metastasis. RNA interference-mediated selective circRNA silencing may be used in the treatment of certain types of tumor (11–13). CircRNAs are a class of specific endogenous RNAs, and are characterized by stable structure and increased tissue-specific expression (14). A number of patients with pancreatic cancer are at an advanced stage when diagnosed, so they have missed the optimal time to undergo radical surgery. An ongoing problem of pancreatic cancer is the lack of sensitive indicators and diagnostic methods for early diagnosis. Therefore, it is a requirement to identify an ideal biomarker for increasing the early diagnostic rate of pancreatic cancer.
In the present study, the pancreatic cancer candidate biomarker human (hsa)_circ_0006215 for early diagnosis was screened using high-throughput sequencing of the pancreatic cancer transcriptome. The reverse transcription-quantitative polymerase chain reaction (RT-qPCR) was used to verify that, combined with EUS-FNA that is able to obtain the specimen directly, hsa_circ_0006215 is expected to be a new biomarker for early diagnosis of pancreatic cancer. Transcriptional expression profiles and interaction screening results identified that hsa_circ_0006215 was a significantly upregulated circRNA. In the present study, hsa_circ_0006215 expression was verified in 30 pancreatic cancer tissue and paracancerous tissue samples. RT-qPCR results demonstrated that, compared with paracancerous tissue, hsa_circ_0006215 expression was increased in pancreatic cancer tissue, miR-378a-3p expression was decreased in pancreatic cancer tissue and SERPINA4 expression was increased in pancreatic cancer tissue. Further investigation of circRNA function may improve the understanding of the diseases associated with the underlying molecular mechanism of circRNAs, and improve the prevention and diagnosis of circRNAs-associated diseases. Using bioinformatics database and bioinformatics analysis, it was identified that the interaction network of hsa_circ_0006215 is likely to regulate the expression of miR-378a-3p. Further interaction analysis revealed that the SERPINA4 gene is a regulatory target gene likely to exhibit influence. The present study identified the effects of hsa_circ_0006215, miR-378a-3p and SERPINA4 signaling pathways in pancreatic cancer cells using in vitro assays.
Materials and methods
Tissue collection
Tissue samples were divided into an experimental group (pancreatic cancer tissue; n=30) and a control group (paracancerous tissue; n=30). All samples were obtained from patients with pancreatic cancer at the Department of General Surgery, Shengjing Hospital of China Medical University, Shenyang, China, collected between July 2016 and January 2017. Corresponding paracancerous tissue was obtained 3 cm lateral to the edge of the foci, and did not contain evident tumor cells following evaluation by the two experienced pathologists. Pathological results revealed that it was moderately differentiated and poorly differentiated adenocarcinoma without metastasis. Use of all specimens was approved by the Shengjing Hospital of China Medical University Ethics Committee (approval no. 2016PS277K). All patients provided written informed consent. All tissue samples were immediately placed in a cryopreservation tube containing RNAlater RNA stabilization reagent (Qiagen, Inc., Valencia, CA, USA), labeled and stored at −80°C until further use.
RNA extraction from tissue and blood samples
RNA was extracted from pancreatic cancer tissue and paracancerous tissue using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA). RNA was extracted from blood using the kit. RNA concentration and purity were determined using a microplate reader [optical density (OD)260/OD280 ratio]. The integrity of the extracted RNA was determined using 10% agarose gel electrophoresis.
Sanger sequencing
At the back-splice junction site, primers were designed for RT-qPCR (Table I). PCR products were sequenced to confirm that sequencing results were consistent with back-splice junction sequences. RT-qPCR primer specificity was also confirmed. Design, synthesis and sequencing of the primers were performed by Geneseed Biotech Co., Ltd., Guangzhou, China.
Table I.
Primer sequences of candidate genes detected using the quantitative polymerase chain reaction.
| Target gen | Primer sequence (5′-3′) |
|---|---|
| hsa_circ_0006215 | Forward: TCATAGGCATCGCGGA |
| CAC | |
| Reverse: ATACAGCTCTATGACCT | |
| AGC | |
| miR-378a-3p | Forward: CCTGACTACTGGACTTG |
| GAGTCA | |
| Reverse: ATCCAGTGCAGGGTCCG | |
| AGG | |
| GAPDH | Forward: GCACCGTCAAGGCTGA |
| GAAC | |
| Reverse: TGGTGAAGACGCCAGT | |
| GGA |
qPCR assay
The corresponding primers were added for PCR amplification. Total RNA from tissues were reverse transcribed to cDNA using the FastQuant RT kit (including gDNase; cat. no. KR106; Tiangen Biotech Co., Ltd., Beijing, China) in 20 µl reactions. Triplicate qPCR assays were performed in 20 µl reactions using the FastFire qPCR PreMix (SYBRGreen) kit (cat. no. FP207; Tiangen Biotech Co., Ltd.) according to the manufacturer's protocol. The thermocycling conditions were as follows: Initial denaturation at 95°C for 60 sec, 40 cycles of amplification at 95°C for 20 sec and annealing and extension at 60°C for 30 sec. GAPDH was used as an internal control for PCR amplification. Amplification efficiency was determined using amplification curves and product solubility curves. The data from RT-qPCR were analyzed using the ΔCq method (15). The ΔCq value was determined by subtracting the GAPDH Cq value from the target circRNA Cq value. All results are expressed as the mean ± standard deviation of three independent experiments.
Western blot assay
Protein sample extraction: Cryopreserved tissue blocks were weighed, triturated in a triturator and transferred into Eppendorf tubes. Lysis buffer (cat. no. v900854; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) was added to the tubes, which were placed on ice for 20 min. The lysate was aspirated ~15 times with a 1-ml syringe, prior to an additional 10 min of lysis at 4°C. Subsequently, samples were centrifuged at 1,000 g for 20 min and at 400 g for 10 min. Following removal of the supernatant, 1 ml sample was cooled, resuspended in PBS and placed in a 1.5-ml centrifuge tube for centrifugation at 400 g for 5 min. Following discarding of the supernatant, 100 µl cell lysate containing phenylmethylsulfonyl fluoride was added, mixed and lysed on ice for between 15 and 20 min. During lysis, samples were repeatedly aspirated. After 10 min of centrifugation at 1,000 g, the supernatant was transferred to an additional centrifuge tube. The protein concentration was determined by a bicinchoninic acid protein quantification kit (cat. no. 23229; Thermo Fisher Scientific, Inc.). The protein samples (20 µg) were separated by 12% SDS/PAGE and transferred onto PVDF membranes. The membranes were blocked with 5% BSA for 1 h at room temperature, incubated with anti-SERPINA4 primary antibody (1:1,000; cat. no. CSB-PA021060ESR2HU; Cusabio Technology LLC, College Park, MD, USA) and anti-GAPDH antibody (1:10,000; cat. no. ab181602; Abcam, Cambridge, MA, USA) at 4°C overnight, and washed three or four times for between 5 and 10 min with Tris-buffered saline containing Tween-20 (TBST). Subsequently, the membranes were incubated with horseradish peroxidase-conjugated goat anti-rabbit IgG heavy and light chain secondary antibody (1:10,000; cat. no. ab205718; Abcam) at 4°C for 1 h and washed three or four times with TBST. Protein bands were visualized using an enhanced chemiluminescence kit (Pierce; Thermo Fisher Scientific, Inc.) and quantified using Quantity One software (version 4.62; Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Cell culture
The human pancreatic cancer cell line PANC-1 was cultured in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum, and 100 U/l penicillin and streptomycin in a 37°C incubator containing 5% CO2. Medium was replaced every 24 h. At 80% confluence, adhered cells were observed and digested with 0.25% trypsin for cell passage. Each passage was for between 48 and 72 h.
Cell transfection
Plvx-CMV-minicrRNA6215-EF1-GFP-Puro (containing hsa_circ_0006215 cDNA), hsa_circ_0006215 small interfering RNA (siRNA; sense, 5′-AAGAAACTGCTAGGTCATAGA-3′; antisense, 5′-CTGCTAGGTCATAGAGCTGTA-3′) and controls siRNA (sense, 5′-GCGUUCUGGUCUUACUGUUU-3′; antisense, 5′-AGAGAAUAAACCCGCAGACUU-3′) were used for overexpression and silencing analysis. All transfection reactions were performed using Lipofectamine™ 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). For analysis, the plasmid control and siRNA control were set as 1.
Determination of cell viability using a CCK-8 (Cell Counting Kit-8) assay
After 24 h of culture, 106 cells were centrifuged at 800 g for 5 min and the supernatant was discarded. Each cell treatment group was resuspended in culture medium in a 96-well plate and incubated at 4°C for 24 h. CCK-8 reagent (10 µl) was added to each well. Subsequently, the plate was incubated at 37°C for 2 h. The OD450 values as a measure of cell viability were determined using a microplate reader.
Detection of cell apoptosis using flow cytometry
Suspended cells (105) were placed in a centrifuge tube and centrifuged at 800 g for 5 min. Following removal of the culture medium, samples were washed once with PBS and centrifuged at 800 g for 5 min. Cells were resuspended in binding buffer, incubated at room temperature in the dark for 10 min and centrifuged at 800 g for 5 min. Subsequently, cells were washed once with incubation buffer, incubated with fluorescent solution at 4°C for 20 min, agitated in the dark and analyzed using flow cytometry. Annexin V and propidium iodide (PI) were used using the Annexin V-flurescein isothiocyanate/PI apoptosis detection kits (cat. no. KGA105; Nanjing KeyGen Biotech Co., Ltd., Nanjing, China).
Transwell cell migration assay
Cell suspension (100 µl) was added to Transwell chambers. Culture medium (500 µl) containing fetal bovine serum was added to the 24-well plate prior to incubation at 37°C for between 12 and 48 h. The chambers were removed and washed with PBS. Cells in the upper layer were swabbed with a cotton swab. The samples were fixed with 95% alcohol for 5 min, stained with 4 g/l crystal violet and images were captured using an inverted light microscope (magnification, ×200, 5 fields).
Delineation of circRNA-miRNA interactions
miRanda miRNA target prediction software (version 2.0) was used to predict miRNA targets of circRNAs and circRNA-miRNA interactions. To establish a circRNA-miRNA network, miRNA-response elements to circRNAs were searched for using the software, and the miRNAs were selected according to seed-match sequences. The circRNA-miRNA network was created using Cytoscape (version 3.01; http://www.cytoscape.org/).
Statistical analysis
Experiments were performed at least in triplicate. Results are presented as the mean ± standard deviation. Results were analyzed using SPSS software (version 17.0; SPSS, Inc., Chicago, IL, USA). When only two groups were compared, Student's t-test was used. One-way analysis of variance followed by Bonferroni's post-hoc test was used to compare differences between multiple groups. P<0.05 was considered to indicate a statistically significant difference.
Results
RNA quantity and quality
Total RNA was extracted from pancreatic cancer tissue and paracancerous tissue. The RNA concentration was between 1.0 and 2.4 µg/µl. The OD260/OD280 ratio was between 1.9 and 2.0. RNA integrity was observed using a gel imager.
Primer specificity was determined using Sanger sequencing. Sequencing results were as expected and primer design was successful. The original map of the sequencing curve is presented in Fig. 1.
Figure 1.
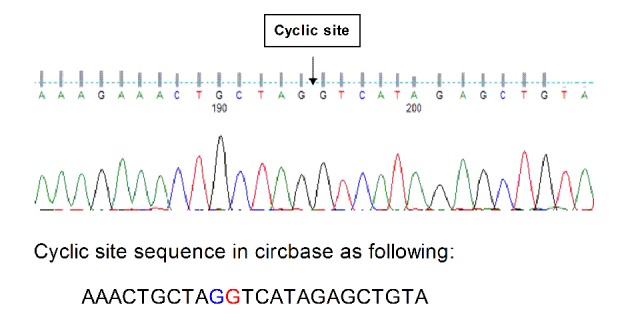
Sequence diagram. The original map of the sequencing curve indicating the cyclic site sequence identified using circbase.
SERPINA4 protein expression in tissues
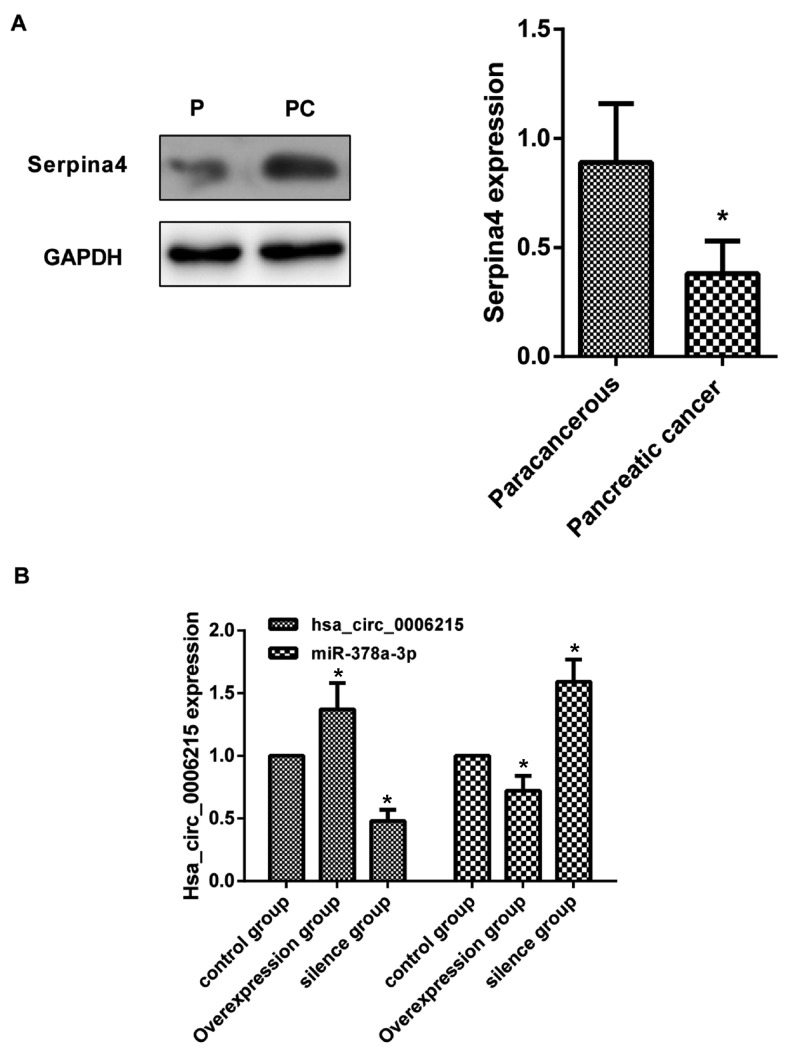
Compared with paracancerous tissue, SERPINA4 expression (OD value of SERPINA4/OD value of GAPDH=0.91±0.17) increased in the pancreatic cancer tissue (P<0.05; Fig. 2A).
Figure 2.
Expression level of SERPINA4, hsa_circ_0006215 and miR-378-3p. (A) Expression level of SERPINA4 quantified by western blot analysis. GAPDH protein was used as an internal control. Results are the mean ± standard error of the mean. *P<0.05. (B) Expression level of hsa_circ_0006215 and miR-378-3p in the overexpression group, silence group and normal group. *P<0.05 vs. control P, paracancerous; PC, pancreatic cancer; hsa, human; circ, circular RNA; miR, microRNA.
Experimental groups and verification
The human pancreatic cancer cell line PANC-1 was divided into three different treatment groups: Overexpression group, silencing group and normal group. Fluorescence microscopy identified that the proportion of cells with green fluorescence to the total number of cells was >90% (data not shown). Following PANC-1 cell transfection, compared with the normal group, hsa_circ_0006215 expression was significantly increased in the overexpression group, but was significantly decreased in the silencing group. Compared with the normal group, miR-378a-3p expression was significantly decreased in the overexpression group, but was significantly increased in the silencing group (P<0.05; Fig. 2B).
Cell viability assay
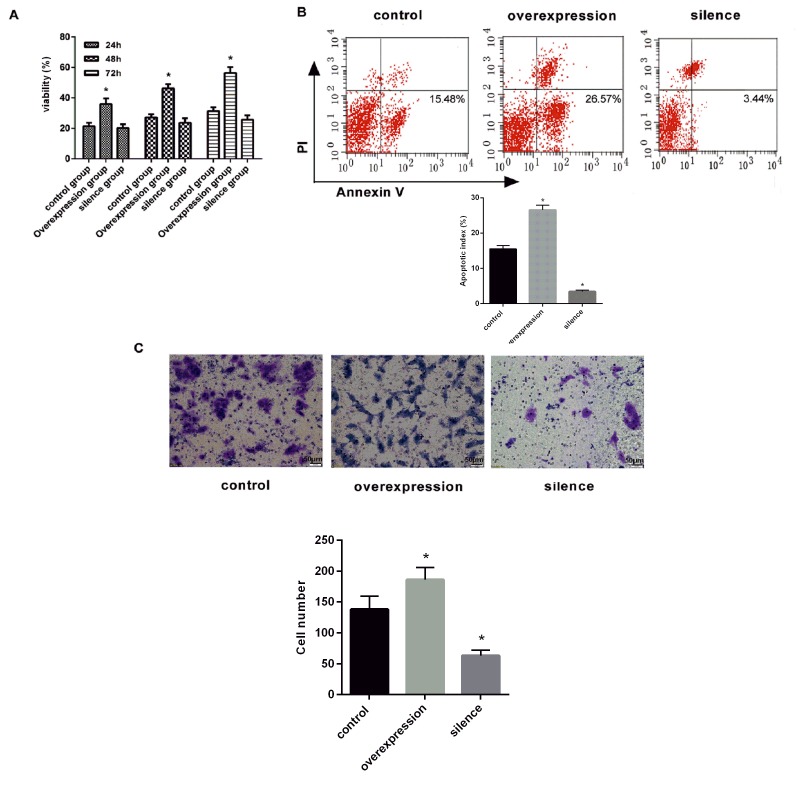
PANC-1 cell viability was determined using a CCK-8 assay at 24, 48 and 72 h. PANC-1 cell viability significantly increased over time in the overexpression group (P<0.05; Fig. 3A). However, no significant increase was observed in the silencing group (Fig. 3A).
Figure 3.
Effect of hsa_circ_0006215 on viability, apoptosis and migration. (A) Cell Counting Kit-8 viability assays were performed on hsa_circ_0006215-overexpressed and -silenced cells, and control cells. Experiments were performed in triplicate. *P<0.05 vs. control. (B) Hsa_circ_0006215 causes apoptosis. Flow cytometry was used to detect the apoptotic rates of cells. Lower-right quadrant, early apoptotic cells; upper-right quadrant, terminal apoptotic cells. Results are the mean ± standard error of the mean. *P<0.05 vs. control. (C) Effect on cell migration by overexpressing or silencing hsa_circ_0006215 cells. Scale bar, 50 µm. *P<0.05 vs. control. hsa, human; circ, circular RNA; PI, propidium iodide.
Detection of cell apoptosis using flow cytometry
Compared with the normal group, cell apoptosis was significantly increased in the overexpression group (apoptotic rate, 26.57±5.51%; P<0.05; Fig. 3B). The apoptotic rate (3.44±3.38%) was significantly decreased compared with the control (P<0.05; Fig. 3B).
Transwell migration assay
Compared with the normal group, migratory ability was increased in the overexpression group, but was decreased in the silencing group (P<0.05 Fig. 3C).
SERPINA4 protein expression in cells
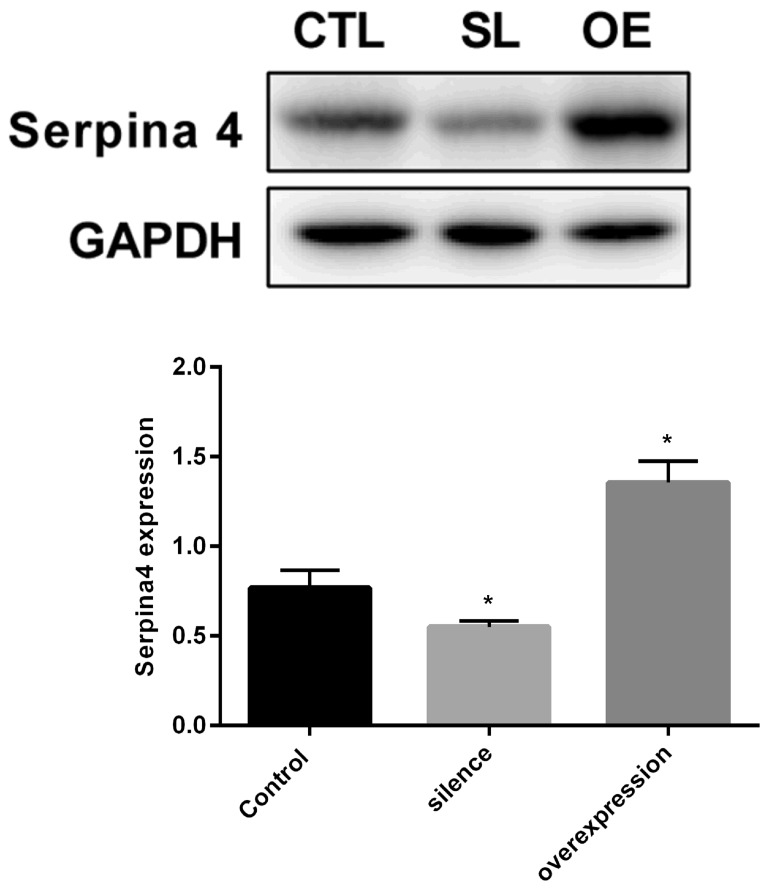
SERPINA4 expression levels increased in the overexpression group compared with the normal group. Expression levels were significantly decreased in the silencing group compared with in the normal group (P<0.05; Fig. 4).
Figure 4.
Western blot analysis of SERPINA4 protein level in hsa_circ_0006215-silenced and -overexpressing cells compared with control cells. GAPDH protein was used as an internal control. Results are the mean ± standard error of the mean. *P<0.05 vs. control. hsa, human; circ, circular RNA; CTL, control group; SL, hsa_circ_0006215-silenced group; OE, hsa_circ_0006215-overexpressing group.
Discussion
CircRNA is a type of non-coding RNA, different from general linear RNA, and forms a covalently closed continuous loop by connecting 3′ and 5′ ends, which makes it more conservative and stable (14). CircRNA exists in a stable form in blood, and previous studies have demonstrated that circRNA was a good biomarker for diagnosis (16,17). Initially, because of their low expression levels, circRNAs are considered to be the products of alternative splicing failure. Nevertheless, with the emergence of novel sequencing and the development of bioinformatics, it is considered that circRNAs have a regulatory function in eukaryotic cells (18–20). Previous studies have suggested that circRNA formation may be regulated by Alu pair inverted repeat sequences, exon skipping, RNA-binding proteins and other factors (21–23). CircRNAs are able to regulate gene expression in a variety of pathological conditions, including the occurrence of cancer (24–26). In addition to its potential function in regulating tumor progression or response to therapy, circRNAs may also be potential clinical biomarkers for cancer and a target for cancer therapy (27).
The reason for the increasing focus on circRNAs is their rich transcription in eukaryotic cells, and also their function. CircRNAs have been identified to have a function in a variety of biological processes, including miRNA binding, protein binding, transcription and post-transcriptional regulation. Previous studies have identified that circRNAs have the following functions: i) CircRNAs may be used as a ‘sponge’ to adsorb miRNAs and inhibit its function; ii) the expression of other RNAs may be directly regulated by complementary base pairing; iii) circRNAs may bind to proteins, inhibit protein activity, recruit components of protein complexes or regulate protein activity; and iv) circRNAs may be used as a template for translation to guide the synthesis of proteins (14,24). Li et al (12) have reported that cir-ITCH competitive adsorption of miR-7, miR-17 and miR-214 results in increased expression of ITCH, which is able to degrade phosphorylated Dvl2, inhibit the Wnt signaling pathway to inhibit the proliferation of esophageal cancer cells. Studies of the application of circRNAs in colorectal cancer, laryngeal cancer, liver cancer and leukemia have also been performed, and results have indicated that associated circRNAs serve an important function in the occurrence or development of these diseases (28). Therefore, studies of circRNAs have been performed, but, to the best of our knowledge, no studies have investigated the molecular mechanisms underlying pancreatic cancer-associated circRNAs. There have been important breakthroughs in the research of miRNAs and early diagnosis of tumors (29). miRNAs are small non-coding RNA molecules that are able to regulate gene expression. miRNAs may be regarded as the main switch of the cell to co-ordinate the expression of a series of genes. Several miRNAs in pancreatic cancer exhibit abnormal expression (13). miRNAs may be used as tumor markers for early diagnosis, and may also provide targets for tumor-targeted therapy (13).
EUS-FNA was accurate in detecting cancerous from non-cancerous tissues (30). Existing imaging methods are sometimes ineffective at distinguishing the nature of pancreatic tumors; however, certainty for pathological diagnosis is required (31–33). Pancreatic pathology assessed using minimally invasive methods, including guided CT, guided ultrasound, guided EUS and guided laparoscopic biopsy by EUS-FNA, has become the primary method to achieve detection of proto-oncogene (K-ras) mutations and of loss of the tumor suppressor gene p16 (34). DPC4 detection may improve the diagnostic accuracy (35). The diagnosis of pancreatic disease by EUS-FNA was first reported by Vilmann et al (36). Subsequently, Chang et al (37,38) identified that a patient with a 1.6-cm pancreatic head tumor underwent the EUS-FNA procedure, which obtained sufficient tissue to confirm the diagnosis of the tumor. Giovanini et al (39) investigated 43 patients with pancreatic cancer, 27 cases of pancreatic cancer, 4 neuroendocrine tumors and 5 cases of cystadenoma using EUS-FNA; the overall sensitivity, specificity and accuracy of EUS-FNA were identified to be 75, 100 and 79%, respectively. Using EUS-FNA, Mallery et al (40) identified no loss in accuracy of EUS-FNA of the tissue samples compared with those obtained using CT or abdominal ultrasound biopsy, and even compared with intraoperative biopsy specimens, although the EUS-guided suction may only rarely be used for histological specimens. The molecular techniques used for the quantitative analysis of K-ras gene mutations in the tissue specimens obtained using EUS-FNA increased the diagnostic information of the specimen (41). In general, EUS-FNA analysis of pancreatic tumors was identified to be safe, and, in a retrospective study of 100 patients, the incidence of acute pancreatitis was only 2% (42).
There are a number of SERPIN family members, which are widely distributed in the body (43). SERPINs have a function in a series of important physiological and pathological processes (44). SERPINs have an important function in blood coagulation, fibrinolysis, angiogenesis, complement activation, immune regulation and inflammatory reaction. Certain SERPINs may act as hormone transport molecules, molecular chaperones and tumor suppressor factors to exert their non-inhibitory function (45). They serve a key function in the regulation of various physiological systems in the body and are important factors to maintain the stability of the environment (46). Analysis of previous genomic database results identified that all multicellular eukaryotes contain multiple SERPIN genes. However, only a few types of SERPIN are present in prokaryotes, which generally contain only one SERPIN gene (47). In total ~36 SERPIN members exist in humans. According to their distribution in vivo, SERPINs may be categorized as either intracellular or extracellular (48). The SERPINA subfamily is predominantly extracellularly distributed and serves an important function in immune regulation (46,49).
Among SERPINA subfamily members, there has been extensive research on the involvement of SERPINA1, SERPINA3 and SERPINA3N in the immune regulatory mechanism (50–52). SERPINA1 is an acute-phase protein, and its level in plasma may increase 3–4-fold following infection (53). SERPINA1 expression is low in high-grade invasive ovarian epithelial cell carcinoma, but high in low malignant potential tumors. However, overexpression of SERPINA1 in vitro is able to promote the invasion and metastasis of tumor cells (54). For certain types of tumor, SERPINA1 is effective for treatment with target enzymes directly. However, for other tumors, SERPIN1 may promote malignant progression. These may be associated with different physiological environments in vivo and in vitro (55). A number of factors are involved in the interaction between SERPINA1 and target enzymes, leading to different results (55). Whether it is the dynamic balance between the SERPIN and its inhibitors that may affect the chronic inflammatory response, or the transfer and invasion of cancer cells, SERPINA3 is the most typical acute-phase protein; SERPINA3 levels in the circulatory system are markedly increased during inflammatory reaction (56). The upregulation of SERPINA3 expression is able to suppress the activity of cathepsin G. A number of diseases are caused by SERPINA3 mutations that result in SERPINA3 deficiency or complete deletion. Thus, the downstream pathway is destroyed and the disordered target enzyme is overproduced, eventually leading to tissue damage (57). The excessive inhibition of cathepsin G is due to the excessive presence of SERPINA3 (57). Cathepsin G serves an important function in antigen presentation, elimination of pathogens and induction of the inflammatory response, thus it may lead to severe diseases (57). SERPINA3N, which is part of the SERPINA clade, is the mouse orthologue of the human anti-chymotrypsin and has been identified as an inhibitor of human and mouse granzyme B in vitro (58–61).
Transcriptome expression profile and interaction screening demonstrated that circRNAs, miRNAs and mRNA are markedly different between the two types of tissue sample. Previous studies have used Cytoscape software to predict the association between circRNA and their target miRNA (62). In the present study, bioinformatics database and bioinformatics analysis revealed that the interaction network of hsa_circ_0006215 is most likely to regulate the expression of miR-378a-3p. Further interaction analysis revealed that the most likely regulatory target is the SERPINA4 gene, whose effect in colorectal cancer has been reported previously (63). A previous study has confirmed that the SERPINA4 gene is present as a cancer-promoting gene in cancer (64). However, to the best of our knowledge, none of these pathways has previously been identified in the study of pancreatic cancer. The results of the present study identified that, following cell transfection, CCK-8 results indicated that cell viability was increased in the overexpression group, but was decreased in the silencing group. Transwell migration assay results identified that cell migration was increased in the overexpression group, but was decreased in the silencing group. Flow cytometric results demonstrated that the apoptotic rate was decreased in the overexpression group, but was increased in the silencing group. The results primarily reveal that hsa_circ_0006215 is able to increase the migration and apoptosis of PANC-1 cells. RT-qPCR results identified that, compared with the normal group, hsa_circ_0006215 expression was increased in the overexpression group, but was decreased in the silencing group. miR-378a-3p expression was decreased in the overexpression group, but was increased in the silencing group. Western blot assay results indicated that, compared with the normal group, SERPINA4 expression was increased in the overexpression group, but was decreased in the silencing group. Therefore, we hypothesize that hsa_circ_0006215 expression is upregulated in the occurrence and development of pancreatic cancer; the upregulated hsa_circ_0006215 downregulated miR-378a-3p and promoted SERPINA4 expression through its sponge function, thereby initiating and promoting the occurrence and development of pancreatic cancer. In the present study, only one cell line was used to analyze the function of circRNA hsa_circ_0006215; the PANC-1 cell line is widely used and it is representative of the molecular trait of pancreatic cancer. In future studies, the circRNA hsa_circ_0006215 will be analyzed further using additional cell lines.
Using RNA sequencing, circRNA microarray, target gene prediction, interaction prediction, structure prediction and bioinformatics analysis, the function of circRNAs has gradually been revealed: CircRNAs are able to act as an miRNA sponge, act as an alternative splicing regulatory factor, transcription factor or even encode a specific protein. By controlling the expression of cancer-associated genes, circRNAs are involved in the regulation of cancer cell viability, proliferation, invasion and metastasis. CircRNAs may act as biomarkers and therapeutic targets for the clinical diagnosis and treatment of cancer. Following the emergence of a variety of novel technologies, EUS has now developed as the core of EUS-FNA technology (65). Elastic imaging, contrast-enhanced ultrasound, biopsy and gene detection are effective and reasonable supplements, and lay the foundation for the early diagnosis of pancreatic cancer. CircRNA research remains in its infancy. The mechanism of production, mode of regulation and biological functions of circRNAs are not fully understood. The progress of studies concerning the association of circRNAs with disease and physiological activity is relatively rapid, but numerous diseases and physical activities have not yet been reported. Cells use a lot of energy and substrates to generate circRNA molecules, therefore these molecules must have further important functions awaiting elucidation.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Liaoning Province Natural Science Foundation (grant no. 20170541024).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
PZ, NG and SS conceived and designed the study, acquired data, interpreted the results and drafted the manuscript. SS also contributed to the acquisition of funding and support. PZ, DL, FY, KZ, JG and XL performed the experiments. SW and GW analyzed the data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Use of all specimens was approved by the Shengjing Hospital of China Medical University Ethics Committee (approval no. 2016PS277K). All patients provided written informed consent.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Wilson LS, Lightwood JM. Pancreatic cancer: Total costs and utilization of health services. J Surg Oncol. 1999;71:171–181. doi: 10.1002/(SICI)1096-9098(199907)71:3<171::AID-JSO7>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 2.Dimastromatteo J, Houghton JL, Lewis JS, Kelly KA. Challenges of pancreatic cancer. Cancer J. 2015;21:188–193. doi: 10.1097/PPO.0000000000000109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lopes CV, Hartmann AA, Almeida RF, Weiss PB. Gastric bulging confirmed as a pancreatic solid pseudopapillary tumor by endoscopic ultrasound-guided fine needle aspiration. Endosc Ultrasound. 2017;6:212–214. doi: 10.4103/2303-9027.208175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gupta R, Mortelé KJ, Tatli S, Girshman J, Glickman JN, Levy AD, Erturk SM, Heffess CS, Banks PA, Silverman SG. Pancreatic intraductal papillary mucinous neoplasms: Role of CT in predicting pathologic subtypes. AJR Am J Roentgenol. 2008;191:1458–1464. doi: 10.2214/AJR.07.3302. [DOI] [PubMed] [Google Scholar]
- 5.Lee TH, Cha SW, Cho YD. EUS elastography: Advances in diagnostic EUS of the pancreas. Korean J Radiol. 2012;(13 Suppl 1):S12–S16. doi: 10.3348/kjr.2012.13.S1.S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khashab MA, Kim K, Lennon AM, Shin EJ, Tignor AS, Amateau SK, Singh VK, Wolfgang CL, Hruban RH, Canto MI. Should we do EUS/FNA on patients with pancreatic cysts? The incremental diagnostic yield of EUS over CT/MRI for prediction of cystic neoplasms. Pancreas. 2013;42:717–721. doi: 10.1097/MPA.0b013e3182883a91. [DOI] [PubMed] [Google Scholar]
- 7.Alizadeh Mohammad AH, Shahrokh S, Hadizadeh M, Padashi M, Zali MR. Diagnostic potency of EUS-guided FNA for the evaluation of pancreatic mass lesions. Endosc Ultrasound. 2016;5:30–34. doi: 10.4103/2303-9027.175879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xin Z, Ma Q, Ren S, Wang G, Li F. The understanding of circular RNAs as special triggers in carcinogenesis. Brief Funct Genomics. 2017;16:80–86. doi: 10.1093/bfgp/elw001. [DOI] [PubMed] [Google Scholar]
- 9.Wang Y, Liu J, Liu C, Naji A, Stoffers DA. MicroRNA-7 regulates the mTOR pathway and proliferation in adult pancreatic β-cells. Diabetes. 2013;62:887–895. doi: 10.2337/db12-0451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kulcheski FR, Christoff AP, Margis R. Circular RNAs are miRNA sponges and can be used as a new class of biomarker. J Biotech. 2016;238:42–51. doi: 10.1016/j.jbiotec.2016.09.011. [DOI] [PubMed] [Google Scholar]
- 11.Li P, Chen S, Chen H, Mo X, Li T, Shao Y, Xiao B, Guo J. Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin Chim Acta. 2015;444:132–136. doi: 10.1016/j.cca.2015.02.018. [DOI] [PubMed] [Google Scholar]
- 12.Li F, Zhang L, Li W, Deng J, Zheng J, An M, Lu J, Zhou Y. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/β-catenin pathway. Oncotarget. 2015;6:6001–6013. doi: 10.18632/oncotarget.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li J, Yang J, Zhou P, Le Y, Zhou C, Wang S, Xu D, Lin HK, Gong Z. Circular RNAs in cancer: Novel insights into origins, properties, functions and implications. Am J Cancer Res. 2015;5:472–480. [PMC free article] [PubMed] [Google Scholar]
- 14.Qu S, Yang X, Li X, Wang J, Gao Y, Shang R, Sun W, Dou K, Li H. Circular RNA: A new star of noncoding RNAs. Cancer Lett. 2015;365:141–148. doi: 10.1016/j.canlet.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 15.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 16.Memczak S, Papavasileiou P, Peters O, Rajewsky N. Identification and characterization of circular RNAs as a new class of putative biomarkers in human blood. PloS One. 2015;10:e0141214. doi: 10.1371/journal.pone.0141214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhuang ZG, Zhang JA, Luo HL, Liu GB, Lu YB, Ge NH, Zheng BY, Li RX, Chen C, Wang X, et al. The circular RNA of peripheral blood mononuclear cells: Hsa_circ_0005836 as a new diagnostic biomarker and therapeutic target of active pulmonary tuberculosis. Mol Immunol. 2017;90:264–272. doi: 10.1016/j.molimm.2017.08.008. [DOI] [PubMed] [Google Scholar]
- 18.Lu Z, Filonov GS, Noto JJ, Schmidt CA, Hatkevich TL, Wen Y, Jaffrey SR, Matera AG. Metazoan tRNA introns generate stable circular RNAs in vivo. RNA. 2015;21:1554–1565. doi: 10.1261/rna.052944.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 20.AbouHaidar MG, Venkataraman S, Golshani A, Liu B, Ahmad T. Novel coding, translation, and gene expression of a replicating covalently closed circular RNA of 220 nt. Proc Natl Acad Sci USA. 2014;111:14542–14547. doi: 10.1073/pnas.1402814111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bartsch D, Zirkel A, Kurian L. Characterization of circular RNAs (circRNA) associated with the translation machinery. Methods Mol Biol. 2018;1724:159–166. doi: 10.1007/978-1-4939-7562-4_13. [DOI] [PubMed] [Google Scholar]
- 22.Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kelly S, Greenman C, Cook PR, Papantonis A. Exon skipping is correlated with exon circularization. J Mol Biol. 2015;427:2414–2417. doi: 10.1016/j.jmb.2015.02.018. [DOI] [PubMed] [Google Scholar]
- 24.Wu Q, Wang Y, Cao M, Pantaleo V, Burgyan J, Li WX, Ding SW. Homology-independent discovery of replicating pathogenic circular RNAs by deep sequencing and a new computational algorithm. Proc Natl Acad Sci USA. 2012;109:3938–3943. doi: 10.1073/pnas.1117815109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilusz J. Circular RNA and splicing: Skip happens. J Mol Biol. 2015;427:2411–2413. doi: 10.1016/j.jmb.2015.05.019. [DOI] [PubMed] [Google Scholar]
- 26.Petkovic S, Muller S. RNA circularization strategies in vivo and in vitro. Nucleic Acids Res. 2015;43:2454–2465. doi: 10.1093/nar/gkv045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sand M, Bechara FG, Sand D, Gambichler T, Hahn SA, Bromba M, Stockfleth E, Hessam S. Circular RNA expression in basal cell carcinoma. Epigenomics. 2016;8:619–632. doi: 10.2217/epi-2015-0019. [DOI] [PubMed] [Google Scholar]
- 28.Vidal AF, Sandoval GT, Magalhaes L, Santos SE, Ribeiro-dos-Santos A. Circular RNAs as a new field in gene regulation and their implications in translational research. Epigenomics. 2016;8:551–562. doi: 10.2217/epi.16.3. [DOI] [PubMed] [Google Scholar]
- 29.Yu X, Koenig MR, Zhu Y. Plasma miRNA, an emerging biomarker for pancreatic cancer. Ann Transl Med. 2015;3:297. doi: 10.3978/j.issn.2305-5839.2015.11.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patil R, Ona MA, Papafragkakis C, Duddempudi S, Anand S, Jamil LH. Endoscopic ultrasound-guided fine-needle aspiration in the diagnosis of adrenal lesions. Ann Gastroenterol. 2016;29:307–311. doi: 10.20524/aog.2016.0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crinò SF, Bellocchi Conti MC, Bernardoni L, Manfrin E, Parisi A, Amodio A, Pretis ND, Frulloni L, Gabbrielli A. Diagnostic yield of EUS-FNA of small (</=15 mm) solid pancreatic lesions using a 25-gauge needle. Hepatob Pancr Dis Int. 2018;17:70–74. doi: 10.1016/j.hbpd.2018.01.010. [DOI] [PubMed] [Google Scholar]
- 32.Yamabe A, Irisawa A, Bhutani MS, Shibukawa G, Fujisawa M, Sato A, Yoshida Y, Arakawa N, Ikeda T, Igarashi R, et al. Efforts to improve the diagnostic accuracy of endoscopic ultrasound-guided fine-needle aspiration for pancreatic tumors. Endosc Ultrasound. 2016;5:225–232. doi: 10.4103/2303-9027.187862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reyes MC, Huang X, Bain A, Ylagan L. Primary pancreatic leiomyosarcoma with metastasis to the liver diagnosed by endoscopic ultrasound-guided fine needle aspiration and fine needle biopsy: A case report and review of literature. Diagn Cytopathol. 2016;44:1070–1073. doi: 10.1002/dc.23540. [DOI] [PubMed] [Google Scholar]
- 34.Chebib I, Albanese E, Scourtas A, Pitman MB. Inspissated cyst fluid in endoscopic ultrasound-guided fine needle aspiration of pancreatic cysts. Diagn Cytopathol. 2018 doi: 10.1002/dc.23912. [DOI] [PubMed] [Google Scholar]
- 35.Salek C, Benesova L, Zavoral M, Nosek V, Kasperova L, Ryska M, Strnad R, Traboulsi E, Minarik M. Evaluation of clinical relevance of examining K-ras, p16 and p53 mutations along with allelic losses at 9p and 18q in EUS-guided fine needle aspiration samples of patients with chronic pancreatitis and pancreatic cancer. World J Gastroenterol. 2007;13:3714–3720. doi: 10.3748/wjg.v13.i27.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vilmann P, Jacobsen GK, Henriksen FW, Hancke S. Endoscopic ultrasonography with guided fine needle aspiration biopsy in pancreatic disease. Gastrointest Endosc. 1992;38:172–173. doi: 10.1016/S0016-5107(92)70385-X. [DOI] [PubMed] [Google Scholar]
- 37.Chang KJ, Nguyen P, Erickson RA, Durbin TE, Katz KD. The clinical utility of endoscopic ultrasound-guided fine-needle aspiration in the diagnosis and staging of pancreatic carcinoma. Gastrointest Endosc. 1997;45:387–393. doi: 10.1016/S0016-5107(97)70149-4. [DOI] [PubMed] [Google Scholar]
- 38.Chang KJ, Albers CG, Erickson RA, Butler JA, Wuerker RB, Lin F. Endoscopic ultrasound-guided fine needle aspiration of pancreatic carcinoma. Am J Gastroenterol. 1994;89:263–266. [PubMed] [Google Scholar]
- 39.Giovannini M, Seitz JF, Monges G, Perrier H, Rabbia I. Fine-needle aspiration cytology guided by endoscopic ultrasonography: Results in 141 patients. Endoscopy. 1995;27:171–177. doi: 10.1055/s-2007-1005657. [DOI] [PubMed] [Google Scholar]
- 40.Mallery JS, Centeno BA, Hahn PF, Chang Y, Warshaw AL, Brugge WR. Pancreatic tissue sampling guided by EUS, CT/US, and surgery: A comparison of sensitivity and specificity. Gastrointest Endosc. 2002;56:218–224. doi: 10.1067/mge.2002.125826. [DOI] [PubMed] [Google Scholar]
- 41.Tada M, Komatsu Y, Kawabe T, Sasahira N, Isayama H, Toda N, Shiratori Y, Omata M. Quantitative analysis of K-ras gene mutation in pancreatic tissue obtained by endoscopic ultrasonography-guided fine needle aspiration: Clinical utility for diagnosis of pancreatic tumor. Am J Gastroenterol. 2002;97:2263–2270. doi: 10.1111/j.1572-0241.2002.05980.x. [DOI] [PubMed] [Google Scholar]
- 42.Gress F, Michael H, Gelrud D, Patel P, Gottlieb K, Singh F, Grendell J. EUS-guided fine-needle aspiration of the pancreas: Evaluation of pancreatitis as a complication. Gastrointest Endosc. 2002;56:864–867. doi: 10.1016/S0016-5107(02)70361-1. [DOI] [PubMed] [Google Scholar]
- 43.Chotwiwatthanakun C, Santimanawong W, Sobhon P, Wongtripop S, Vanichviriyakit R. Inhibitory effect of a reproductive-related serpin on sperm trypsin-like activity implicates its role in sperm maturation of Penaeus monodon. Mol Reprod Dev. 2018;85:205–214. doi: 10.1002/mrd.22954. [DOI] [PubMed] [Google Scholar]
- 44.Gettins PG, Olson ST. Inhibitory serpins. New insights into their folding, polymerization, regulation and clearance. Biochem J. 2016;473:2273–2293. doi: 10.1042/BCJ20160014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Silverman GA, Bird PI, Carrell RW, Church FC, Coughlin PB, Gettins PG, Irving JA, Lomas DA, Luke CJ, Moyer RW, et al. The serpins are an expanding superfamily of structurally similar but functionally diverse proteins. Evolution, mechanism of inhibition, novel functions, and a revised nomenclature. J Biol Chem. 2001;276:33293–33296. doi: 10.1074/jbc.R100016200. [DOI] [PubMed] [Google Scholar]
- 46.Enewold L, Mechanic LE, Bowman ED, Platz EA, Alberg AJ. SERPINA1 and ELA2 polymorphisms are not associated with COPD or lung cancer. Anticancer Res. 2012;32:3923–3928. [PMC free article] [PubMed] [Google Scholar]
- 47.Irving JA, Steenbakkers PJ, Lesk AM, den Camp Op HJ, Pike RN, Whisstock JC. Serpins in prokaryotes. Mol Biol Evol. 2002;19:1881–1890. doi: 10.1093/oxfordjournals.molbev.a004012. [DOI] [PubMed] [Google Scholar]
- 48.Law RH, Zhang Q, McGowan S, Buckle AM, Silverman GA, Wong W, Rosado CJ, Langendorf CG, Pike RN, Bird PI, Whisstock JC. An overview of the serpin superfamily. Genome Biol. 2006;7:216. doi: 10.1186/gb-2006-7-5-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.de Ronde JJ, Lips EH, Mulder L, Vincent AD, Wesseling J, Nieuwland M, Kerkhoven R, Peeters Vrancken MJ, Sonke GS, Rodenhuis S, Wessels LF. SERPINA6, BEX1, AGTR1, SLC26A3, and LAPTM4B are markers of resistance to neoadjuvant chemotherapy in HER2-negative breast cancer. Breast Cancer Res Treat. 2013;137:213–223. doi: 10.1007/s10549-012-2340-x. [DOI] [PubMed] [Google Scholar]
- 50.Hadzik-Blaszczyk M, Zdral A, Zielonka TM, Rozy A, Krupa R, Falkowski A, Wardyn KA, Chorostowska-Wynimko J, Zycinska K. SERPINA1 gene variants in granulomatosis with polyangiitis. Adv Exp Med Biol. 2018 doi: 10.1007/5584_2018_156. [DOI] [PubMed] [Google Scholar]
- 51.Baker C, Belbin O, Kalsheker N, Morgan K. SERPINA3 (aka alpha-1-antichymotrypsin) Front Biosci. 2007;12:2821–2835. doi: 10.2741/2275. [DOI] [PubMed] [Google Scholar]
- 52.Vicuña L, Strochlic DE, Latremoliere A, Bali KK, Simonetti M, Husainie D, Prokosch S, Riva P, Griffin RS, Njoo C, et al. The serine protease inhibitor SerpinA3N attenuates neuropathic pain by inhibiting T cell-derived leukocyte elastase. Nat Med. 2015;21:518–523. doi: 10.1038/nm.3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lim W, Kim HS, Jeong W, Ahn SE, Kim J, Kim YB, Kim MA, Kim MK, Chung HH, Song YS, et al. SERPINB3 in the chicken model of ovarian cancer: A prognostic factor for platinum resistance and survival in patients with epithelial ovarian cancer. PloS One. 2012;7:e49869. doi: 10.1371/journal.pone.0049869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Di Francesco A, Di Germanio C, Panda AC, Huynh P, Peaden R, Navas-Enamorado I, Bastian P, Lehrmann E, Diaz-Ruiz A, Ross D, et al. Novel RNA-binding activity of NQO1 promotes SERPINA1 mRNA translation. Free Radi Biol Med. 2016;99:225–233. doi: 10.1016/j.freeradbiomed.2016.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kloth JN, Gorter A, Fleuren GJ, Oosting J, Uljee S, ter Haar N, Dreef EJ, Kenter GG, Jordanova ES. Elevated expression of SerpinA1 and SerpinA3 in HLA-positive cervical carcinoma. J Pathol. 2008;215:222–230. doi: 10.1002/path.2347. [DOI] [PubMed] [Google Scholar]
- 56.Kwon CH, Park HJ, Choi JH, Lee JR, Kim HK, Jo HJ, Kim HS, Oh N, Song GA, Park DY. Snail and serpinA1 promote tumor progression and predict prognosis in colorectal cancer. Oncotarget. 2015;6:20312–20326. doi: 10.18632/oncotarget.3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Blanchet X, Pere-Brissaud A, Duprat N, Pinault E, Delourme D, Ouali A, Combet C, Maftah A, Pélissier P, Brémaud L. Mutagenesis of the bovSERPINA3-3 demonstrates the requirement of aspartate-371 for intermolecular interaction and formation of dimers. Protein Sci. 2012;21:977–986. doi: 10.1002/pro.2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sipione S, Simmen KC, Lord SJ, Motyka B, Ewen C, Shostak I, Rayat GR, Dufour JM, Korbutt GS, et al. Identification of a novel human granzyme B inhibitor secreted by cultured sertoli cells. J Immunol. 2006;177:5051–5058. doi: 10.4049/jimmunol.177.8.5051. [DOI] [PubMed] [Google Scholar]
- 59.Cullen SP, Martin SJ. Mechanisms of granule-dependent killing. Cell Death Differ. 2008;15:251–262. doi: 10.1038/sj.cdd.4402244. [DOI] [PubMed] [Google Scholar]
- 60.Horvath AJ, Irving JA, Rossjohn J, Law RH, Bottomley SP, Quinsey NS, Pike RN, Coughlin PB, Whisstock JC. The murine orthologue of human antichymotrypsin: A structural paradigm for clade A3 serpins. J Biol Chem. 2005;280:43168–43178. doi: 10.1074/jbc.M505598200. [DOI] [PubMed] [Google Scholar]
- 61.Ang LS, Boivin WA, Williams SJ, Zhao H, Abraham T, Carmine-Simmen K, McManus BM, Bleackley RC, Granville DJ. Serpina3n attenuates granzyme B-mediated decorin cleavage and rupture in a murine model of aortic aneurysm. Cell Death Dis. 2011;2:e209. doi: 10.1038/cddis.2011.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Qian DY, Yan GB, Bai B, Chen Y, Zhang SJ, Yao YC, Xia H. Differential circRNA expression profiles during the BMP2-induced osteogenic differentiation of MC3T3-E1 cells. Biomed Pharmacother. 2017;90:492–499. doi: 10.1016/j.biopha.2017.03.051. [DOI] [PubMed] [Google Scholar]
- 63.Sun HM, Mi YS, Yu FD, Han Y, Liu XS, Lu S, Zhang Y, Zhao SL, Ye L, Liu TT, et al. SERPINA4 is a novel independent prognostic indicator and a potential therapeutic target for colorectal cancer. Am J Cancer Res. 2016;6:1636–1649. [PMC free article] [PubMed] [Google Scholar]
- 64.Péré-Brissaud A, Blanchet X, Delourme D, Pélissier P, Forestier L, Delavaud A, Duprat N, Picard B, Maftah A, Brémaud L. Expression of SERPINA3s in cattle: Focus on bovSERPINA3-7 reveals specific involvement in skeletal muscle. Open Biol. 2015;5:150071. doi: 10.1098/rsob.150071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ge N, Zhang S, Jin Z, Sun S, Yang A, Wang B, Wang G, Xu G, Hao J, Zhong L, et al. Clinical use of endoscopic ultrasound-guided fine-needle aspiration: Guidelines and recommendations from Chinese society of digestive endoscopy. Endosc Ultrasound. 2017;6:75–82. doi: 10.4103/eus.eus_20_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.