Abstract
Bioassay-guided metabolomic analyses led to the characterization of four new 20-membered glycosylated polyketide macrolactams – macrotermycins A-D – from a termite-associated actinomycete, Amycolatopsis sp. M39. M39’s sequenced genome revealed the macrotermycin’s putative biosynthetic gene cluster. Macrotermycins A and C had antibacterial activity against human-pathogenic S. aureus and of greater ecological relevance, they also had selective antifungal activity against a fungal parasite of the termite fungal garden.
Graphical Abstract
Explorations of the bacterial symbionts of phylogenetically diverse insects have repeatedly led to biologically active natural products with interesting chemical scaffolds.1 The bacteria provide small molecule chemical defenses that selectively inhibit the insects’ microbial competitors and pathogens. The small molecule defenses found in current studies reflect the numerous rounds of mutation, selection, and amplification between the insects’ defenders and antagonists.
Our earlier studies investigated symbiotic bacteria from ants, beetles, and termites that lived in large colonies and grew fungi in specialized gardens. The bacteria provided chemical defenses against microbial threats, especially against fungal parasites that specialized in consuming the fungal crop. These studies led to selective antifungal agents such as the depsipeptide dentigerumycin from a fungus-growing ant (Apterostigma dentigerum) symbiont,2 and the polyene peroxide mycangimycin from a Southern pine beetle (Dendroctonus frontalis) symbiont.3 While our initial studies of bacteria associated with a species of fungus-growing termites (Macrotermes natalensis)4 led to the discovery of chemically-intriguing compounds like the microtermolides5 and natalamycin A,6 they did not identify any selective antifungal agents. We focused our efforts to find selective agents from bacteria associated with M. natalensis through a systematic combination of LC-HRMS-based dereplication and bioactivity assays against the basidiomycete cultivar (Termitomyces spp.) and an ascomycete competitor (Pseudoxylaria spp.).7 As a result of these studies, the actinomycete Amycolatopsis sp. M39 was found to exhibit both unique metabolomic profile and promising anti-Pseudoxylaria activity. We now report the isolation and structural characterization, biosynthetic analysis, and biological activity of four new macrolactams isolated from Amycolatopsis sp. M39. Two of the four exhibited selective inhibition of the Pseudoxylaria pathogen over the Termitomyces cultivar.
We started our systematic analysis with 41 termite-associated Actinobacteria isolated from a South African M. natalensis colony.8 Principal component analysis (PCA) on preprocessed LC-HRMS traces identified Amycolatopsis sp. M39 as having a unique metabolomic profile. The putative new natural products responsible for the variance observed in the PCA were quasi-molecular ions at m/z 473.2678 and 489.2622. Analysis of the metabolomic profiles indicated a strong time and medium dependence. Whereas the first group of putative new compounds (m/z 473.2678) was mainly detected after seven days of cultivation, the second group of compounds (m/z 489.2622) increased with longer growth times. We also tested Amycolatopsis sp. M39 in ecologically relevant plate challenge assays and found that it suppressed the growth of Pseudoxylaria sp. (strain X802) and insect entomopathogenic fungi such as Beauveria bassiana and Metarhizium anisopliae. The cultivar’s (Termitomyces sp.) relative resistance to M39 was especially noteworthy (Fig. S3).
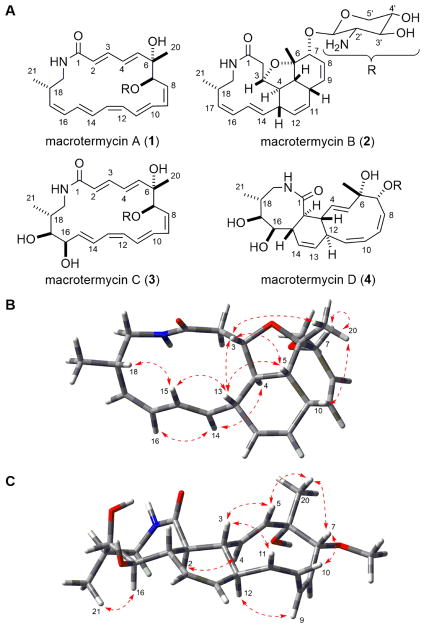
LC-MS analysis extracts from the zone of inhibition (ZOI) revealed a set of metabolites matching the molecular weight of the putative new compounds (m/z 473.3 and 489.3) identified in the metabolomic analysis. To identify these compounds, we performed a preparative scale fermentation of Amycolatopsis sp. M39 on solid ISP-2 agar for 14 d at 30 °C. Agar plates densely covered with Amycolatopsis sp. M39 were extracted using i-PrOH. The crude extract was purified using an activated pre-packed C18 cartridge, followed by preparative and semi-preparative reverse-phase HPLC. LC-MS analysis revealed the HPLC fractions that contained compounds with m/z 473.3 and 489.3. The first isolated compound, macrotermycin A (1), was obtained as a colorless solid with a quasi-molecular ion of 473.2678 [M+H]+ consistent with 1 exhibiting a molecular formula of C26H36N2O6 (Fig. 1A).9 The 1H NMR spectrum in DMSO-d6 indicated a complex overlap of 14 olefinic protons from δH 5.10 to δH 6.60, four oxygenated methine protons between δH 3.00 and δH 4.40, two oxygenated methylene protons at δH 3.01 and 3.68, one amine-bearing methine proton at δH 2.50, an NH signal at δH 7.66, two N-amidated methylene protons at δH 2.72 and 3.10, and nine aliphatic protons that include two methyl groups at δH 0.91 and 1.33. Analysis of the HSQC and HMBC spectra allowed assigning the 14 olefinic carbons between δC 120 and δC 145, one amide carbonyl carbon at δC 165.3, six oxygen-bearing carbons (δC 65–105) that include one quaternary carbon at δC 75.3 and an anomeric carbon at δC 104.8, one amine-bearing carbon at δC 57.3, and four aliphatic carbon signals (δC 17–45). 2D NMR (COSY, TOCSY, HMBC) analysis revealed a macrolactam core structure coupled with a deoxypentopyranose aminosugar group. The long-range correlation of H-1′ (anomeric proton) to C-7 in the HMBC experiment showed the linkage of the aminosugar unit to the macrocyclic lactam. The stereochemistry of the double bonds was determined as 2E, 4E, 8Z, 10E, 12Z, 14E, and 16Z on the basis of the characteristic coupling constants observed in the homo J-resolved 1H NMR spectrum and corresponding NOESY correlations. The stereochemistry of the three chiral centers of the macrotermycin aglycone was established by NOESY correlations and biosynthetic relation. An NOE correlation between H-7/H-10 was observed in a similar fashion to that seen for macrotermycin D (4) (vide infra), which sets the stereoconfiguration of C-7 as R* (Fig. 1 and S6). The configuration of C-6 and C-18 cannot unambiguously be resolved by NOE data alone; instead, C-6 and C-18 were assigned on the basis of the configurational analysis of macrotermycin B (2) and 4. The aminosugar moiety was found to be 2-deoxy-2-amino-β-L-xylopyranose using a combination of 1D and 2D NMR analysis, coupling constant analysis and Snatzke’s method, and was found to be identical for all isolated macrotermycin derivatives.8 In total, the data indicate 1 as the most probable structure for macrotermycin A.
Figure 1.
A) Structures of macrotermycins A-D. B) Computational models of a macrotermycin B derivative, and C) of a macrotermycin D derivative calculated at the DFT B3LYP/6–31+G(d,p) level of theory without geometric constraints and supported by key NOE/ROE correlations (amino sugar moiety was replaced by a methoxy group to simplify calculations).
Macrotermycin B (2), which is less polar than 1, has the same quasi-molecular ion of 473.2678 [M+H]+ that is consistent with a molecular formula of C26H36N2O6. Compound 2 exhibited a very different 1H NMR spectrum with isolated 1H spin systems and only eight distinct olefinic protons. Analysis of gCOSY, TOCSY, and HMBC correlations revealed the presence of a macrolactam as part of a fused polycyclic ring system (Fig. 1A). NOE correlations observed between H-3/H-5, H-3/H-13, and H-5/H-13 in conjunction with observing good agreement between relevant experimental vicinal scalar coupling constants (J3,4 = 9.0, J4,5 = 12.5; J5,10 = 6.0, J4,13 = 10.0 Hz) and estimated values (J3,4 = 10.0, J4,5 = 12.7; J5,10 = 6.7, J4,13 = 11.2 Hz), as determined in the Schrödinger software suite using the coupling constant measuring tool on a DFT optimized analog of 2 (Fig. 1B).8 The values indicated that H-3, H-5, H-10, and H-13 exhibit cis stereoconfiguration. C-6 configuration was assigned as S* through further computational analysis and observing NOE correlations between H-3/H3-20, H-7/H3-20, and H-10/H3-20. The same strong H-7/H3-20 NOE correlation along with the presumed biosynthetic similarity with other macrotermycins facilitated predicting C-7 as R*. An NOE correlation between H-15/H-18 affirmed C-18 as S*.8
We then focused on the second set of putative new compounds with quasi-molecular ion 489.2622 [M-H2O+H]+ corresponding to those compounds exhibiting a molecular formula of C26H38N2O8. A comparative NMR analysis of macrotermycin C (3) vs. 1 revealed that 3 contains 12 olefinic protons and two additional hydroxyl groups (δC 66.0 and δC 79.0), which accounted for the higher polarity of 3, and led to a proposed structure (Fig. 1A). All alkene geometries were confirmed by analyzing J-J resolved 2D proton spectra and ROESY correlations between H-2/H-4 and H-3/H-5 were also identified. The stereochemistry of the aglycone chiral centers was determined using ROE correlations and biosynthetic relation, and is predicted to be homologous to compounds 1, 2 and 4 respectively.8 2D NMR analysis of macrotermycin D (4) revealed eight olefinic protons and bonds between C-2/C-15 and C-3/C-12, where the latter yields a tricyclic ring system (Fig. 1A). Strong ROE correlations between H-2/H-4, H-3/H-11, H-3/H-5, H-7/H-10 and H-7/H-1′ as well as comparing experimental coupling constants (J2,3 = 11.0; J2,15 = 10.0; J3,12 = 10.0 Hz) to those estimated for a DFT optimized analog of 4 (Fig. 1C) (J2,3 = 12.2; J2,15 = 12.1; J3,12 = 10.6 Hz) revealed an all trans substitution pattern for the cyclohexenyl ring and allowed for C-7 stereoconfigurational assignment. A strong ROE correlation between H3-20/H-5 and H3-20/H-7 along with relation to other macrotermycins allows predicting that C-6 exhibits S* stereoconfiguration. Based on comparing experimental coupling constants (J15,16 = 9.0 Hz, J16,17 = 2.0 Hz, J17,18 = 0.5 Hz) with estimated values (J15,16 = 9.9 Hz, J16,17 = 0.9 Hz, J17,18 = 1.7 Hz), the dihedral angle between H-15 and H-16 is calculated to be close to 180°, whereas the dihedral angles between H-16/H-17 and H-17/H-18 are assumed to be close to 90° defining cis diol stereoconfiguration (C-16, C-17) (Fig. 1C). An ROE correlation between H-16/H3-21 allowed defining C-18 as R* (vs. S* in 1 and 2 due to a Cahn-Ingold-Prelog priority change arising from the C-17 hydroxyl).8
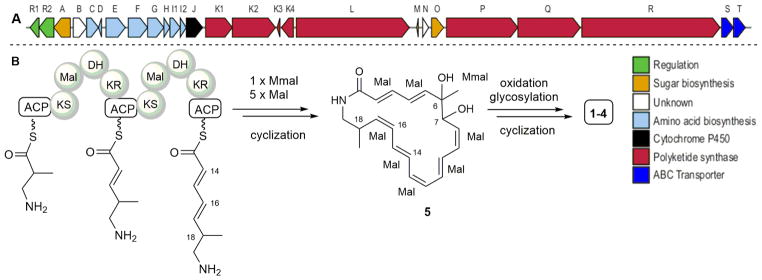
The core structure of the macrotermycins (5) belongs to a family of macrolactam polyketides that include incednine,10 vicenistatin,11 silvalactam,12 heronamides,13 verticilactam,14 ciromicins,15 mirilactams and lobosamides.16 Both 1 and 3 can be viewed as polyene precursors for intramolecular cycloadditions leading to 2 and 4, respectively, and the relevant reaction pathways, whether enzymatically catalyzed or occurring spontaneously either in situ or during purification, are being investigated. Formation of the substituted tetrahydrofuranyl ring in 2 can occur through a 1,4-conjugate addition of a C-6 hydroxyl group to proximal Δ2,3 alkenyl bond in an unisolated precursor. To gain further insights into macrotermycin biogenesis, the Amycolatopsis sp. M39 genome was sequenced and annotated (accession number: LWSF00000000, Fig. 2).8 The likely macrotermycin biosynthetic gene cluster was identified based on significant homology to the known vicenistatin cluster.17 The cluster includes the biosynthesis of the 3-amino-2-methylpropionate starter unit (MetC-MteI). It also harbors four PKS genes (MteL, MteP-R), while a fifth PKS (MteK) is fragmented and a likely pseudogene. Only PKS module MteQ is lacking a dehydratase and terminates with a ketoreductase (KR), which was classified as an ‘A-type’ reductase based on comparative alignment of the amino acid sequence of the ketoreductase catalytic site.18 In this analysis, the polyketide chain is released from the PKS by the termination domains in either MteK or MteL. Oxidation at position C-6, C-16 and C-17 are presumably catalyzed by the encoded cytochrome P-450 (MteJ). O-glycosylation at position C-7 likely involves the glycosyltransferase MteA or MteO. This preliminary biogenetic analysis is also under investigation.
Figure 2.
Organization of the putative macrotermycin biosynthetic gene cluster from Amycolatopsis sp. M39. A) Genetic map of the Mte gene cluster; each arrow represents the direction of transcription of open reading frame (ORF). B) Putative biosynthetic scheme of macrotermycin aglycone (5) formation.
Macrotermycins A-D were assayed for activity against Gram-negative (Escherichia coli ATCC 11775) and Gram-positive (Bacillus subtilis ATCC 6051; Staphylococcus aureus ATCC 25923) bacteria, and yeasts (Candida albicans ATCC 24433, Saccharomyces cerevisiae ATCC 9763). While 1 showed moderate to good antimicrobial activity, 3 was only weakly active (Table 1). Both compounds (1 and 3) modestly inhibited Pseudoxylaria sp. X802 growth. Importantly, growth of the termites’ symbiotic fungus Termitomyces sp. T112 was not significantly affected by 1 or 3. In contrast, compounds 2 and 4 showed no activity against all tested strains in the tested concentration range (MIC > 100 μg/mL).
Table 1.
Minimal inhibitory concentrations (μg/mL) of 1 and 3 (positive controls: ciprofloxacin and amphotericin B).
| strain | 1 | 3 | positive control |
|---|---|---|---|
| MIC (μg/mL) | |||
| ciprofloxacin | |||
|
B. subtilis ATCC 6051 |
1.0 | 15 | 0.3 |
|
S. aureus ATCC 25923 |
1.5 | 10 | 0.5 |
| amphotericin B | |||
|
C. albicans ATCC 10231 |
10 | 25 | 0.3 |
|
S. cerevisiae ATCC9763 |
5.0 | 20 | 0.2 |
|
Termitomyces sp. T112 |
>100 | >100 | ~ 65 |
|
Pseudoxylaria sp. X802 |
~ 50 | ~ 80 | ~ 10 |
In summary, we have isolated and characterized four previously unreported macrolactam polyketides, macrotermycins A-D, from Amycolatopsis sp. M39, proposed their relative structures, and presumptively identified the biosynthetic gene cluster responsible for their production. Macrotermycins A (1) and C (3) were shown to exhibit selective inhibition of the termite fungal garden competitor Pseudoxylaria sp. X802. The identification of these selective antifungal compounds demonstrates the potential of using ecologically relevant assays to identify biologically active compounds.
Supplementary Material
Acknowledgments
We are grateful for financial support through the NIH (R01-GM086258 to J.C., RC4-GM096347 to J.C. and C.R.C., R01-GM104192 to T.S.B., and F32-GM108415 to T.R.R.) and through the German National Academy of Sciences Leopoldina for a postdoctoral fellowship to C.B. (LPDS 2011-2). This research was also supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT, & Future Planning (2015R1C1A1A02037383 to K.H.K.). Quantum-chemical calculations were run on the Odyssey cluster supported by the FAS Division of Science, Research Computing Group at Harvard University. We thank Dr. Duur K. Aanen (Wageningen University) for providing T112, Z. Wilhelm de Beer, Michael J. Wingfield and the staff and students at the Forestry and Agricultural Biotechnology Institute, University of Pretoria, for hosting fieldwork.
Footnotes
Notes
The authors declare no competing financial interest.
Procedures for isolation and characterization of Amycolatopsis sp. M39, dereplication methods, challenge assays, compound purification, HRMS and NMR spectra, detailed NMR and computational analysis, activity assays, computational procedures, and Cartesian coordinates. The Supporting Information is available free of charge on the ACS Publications website.
References
- 1.(a) Ramadhar TR, Beemelmanns C, Currie CR, Clardy J. J Antibiot. 2013;67:53. doi: 10.1038/ja.2013.77. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Seipke RF, Kaltenpoth M, Hutchings MI. FEMS Microbiol Rev. 2012;36:862. doi: 10.1111/j.1574-6976.2011.00313.x. [DOI] [PubMed] [Google Scholar]; (c) Crawford JM, Clardy J. Chem Commun. 2011;47:7559. doi: 10.1039/c1cc11574j. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Bode HB. Angew Chem Int Ed. 2009;48:6394. doi: 10.1002/anie.200902152. [DOI] [PubMed] [Google Scholar]; (e) Kaltenpoth M. Trends Microbiol. 2009;17:529. doi: 10.1016/j.tim.2009.09.006. [DOI] [PubMed] [Google Scholar]; (f) Brownlie JC, Johnson KN. Trends Microbiol. 2009;17:348. doi: 10.1016/j.tim.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 2.Oh DC, Poulsen M, Currie CR, Clardy J. Nat Chem Bio. 2009;5:391. doi: 10.1038/nchembio.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oh DC, Scott JJ, Currie CR, Clardy J. Org Lett. 2009;11:633. doi: 10.1021/ol802709x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Visser AA, Nobre T, Currie CR, Aanen DK, Poulsen M. Microb Ecol. 2012;63:975. doi: 10.1007/s00248-011-9987-4. [DOI] [PubMed] [Google Scholar]
- 5.Carr G, Poulsen M, Klassen JL, Hou Y, Wyche TP, Bugni TS, Currie CR, Clardy J. Org Lett. 2012;14:2822. doi: 10.1021/ol301043p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim KH, Ramadhar TR, Beemelmanns C, Cao S, Poulsen M, Currie CR, Clardy J. Chem Sci. 2014;5:4333. doi: 10.1039/C4SC01136H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hou Y, Braun DR, Michel CR, Klassen JL, Adnani N, Wyche TP, Bugni TS. Anal Chem. 2012;84:4277. doi: 10.1021/ac202623g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.For further details and additional references, see the Supporting Information.
- 9.U.S. Pat. Appl. Publ. 20110136752 A1 20110609 US. 2011
- 10.(a) Futamura Y, Sawa R, Umezawa Y, Igarashi M, Nakamura H, Hasegawa K, Yamasaki M, Tashiro E, Takahashi Y, Akamatsu Y, Imoto M. J Am Chem Soc. 2008;130:1822. doi: 10.1021/ja710124p. [DOI] [PubMed] [Google Scholar]; (b) Takaishi M, Kudo F, Eguchi T. Tetrahedron. 2008;64:6651. [Google Scholar]
- 11.Shindo K, Kamishohara M, Odagawa A, Matsuoka M, Kawai H. J Antibiot. 1993;46:1076. doi: 10.7164/antibiotics.46.1076. [DOI] [PubMed] [Google Scholar]
- 12.Schulz D, Nachtigall J, Geisen U, Kalthoff H, Imhoff JF, Fiedler HP, Süssmuth RD. J Antibiot. 2012;65:369. doi: 10.1038/ja.2012.33. [DOI] [PubMed] [Google Scholar]
- 13.Raju R, Piggott AM, Conte MM, Capon RJ. Org Biomol Chem. 2010;8:4682. doi: 10.1039/c0ob00267d. [DOI] [PubMed] [Google Scholar]
- 14.Nogawa T, Okano A, Takahashi S, Uramoto M, Konno H, Saito T, Osada H. Org Lett. 2010;12:4564. doi: 10.1021/ol1018618. [DOI] [PubMed] [Google Scholar]
- 15.Derewacz DK, Covington BC, McLean JA, Bachmann BO. ACS Chem Biol. 2015;10:1998. doi: 10.1021/acschembio.5b00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schulze CJ, Donia MS, Siqueira-Neto JL, Ray D, Raskatov JA, Green RE, McKerrow JH, Fischbach MA, Linington RG. ACS Chem Biol. 2015;10:2373. doi: 10.1021/acschembio.5b00308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.(a) Ogasawara Y, Katayama K, Minami A, Otsuka M, Eguchi T, Kakinuma K. Chem Biol. 2004;11:79. doi: 10.1016/j.chembiol.2003.12.010. [DOI] [PubMed] [Google Scholar]; (b) Kudo F, Kitayama T, Kakinuma K, Eguchi T. Tetrahedron Lett. 2006;47:1529. [Google Scholar]
- 18.Caffrey P. ChemBioChem. 2003;4:654. doi: 10.1002/cbic.200300581. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.