Abstract
Background
Rab proteins form the largest family of the Ras superfamily of small GTP-binding proteins and regulate intracellular trafficking pathways. However, the function of the Rab proteins in woody species is still an open question.
Results
Here, a total of 67 PtRabs were identified in Populus trichocarpa and categorized into eight subfamilies (RabA-RabH). Based on their chromosomal distribution and duplication blocks in the Populus genome, a total of 27 PtRab paralogous pairs were identified and all of them were generated by whole-genome duplication events. Combined the expression correlation and duplication date, the PtRab paralogous pairs that still keeping highly similar expression patterns were generated around the latest large-scale duplication (~ 13 MYA). The cis-elements and co-expression network of unique expanded PtRabs suggest their potential roles in poplar development and environmental responses. Subcellular localization of PtRabs from each subfamily indicates each subfamily shows a localization pattern similar to what is revealed in Arabidopsis but RabC shows a localization different from their counterparts. Furthermore, we characterized PtRabE1b by overexpressing its constitutively active mutant PtRabE1b(Q74L) in poplar and found that PtRabE1b(Q74L) enhanced the salt tolerance.
Conclusions
These findings provide new insights into the functional divergence of PtRabs and resources for genetic engineering resistant breeding in tree species.
Electronic supplementary material
The online version of this article (10.1186/s12870-018-1342-1) contains supplementary material, which is available to authorized users.
Keywords: Populus, Rab GTPase, Phylogenetic analysis, Co-expression network, Subcellular localization, Salt tolerance
Background
Plant cells are divided into several biochemically distinct membrane-bound organelles, which have specific functions permitting the maintenance of specialized environments for various chemical reactions [1]. Communication and transport between these membrane compartments are vital to not only basic cellular activities but also development and environmental responses. This transport is maintained through complex and precise regulation pathways, which includes membrane fusion between transport vesicles and target organelles [2]. The vesicular fusion is mediated by two important group proteins: the soluble N-ethylmaleimide-sensitive factor attachment receptors (SNARE) and the Rab GTPases [1].
Rab GTPase are involved in the entire process of vesicle transport including budding from donor organelle, docking, tethering and fusing with the target membrane [3, 4]. Similar with other small GTPases, Rabs can be recycled between the GTP-bound active form and GDP-bound form; and this recycle mechanism is conserved in all eukaryotes [5]. There are four conserved motifs involved in nucleotide binding and hydrolysis in Rab proteins. Mutating the specific residues can generate dominant negative or constitutively active forms that exhibit altered nucleotide-binding or hydrolysis characteristics, which can be used to investigate Rab functions [1].
The Rab family in plants is divided into eight subfamilies (RabA-RabH) [6]. Each type of Rab has a characteristic distribution on organelle membranes. In other word, each organelle has at least one type of Rab proteins on its cytosolic surface [7]. Arabidopsis RabA1 GTPases are involved in transport between the trans-Golgi network (TGN) and the plasma membrane (PM) [8]. Tobacco NtRab2 (RabB) regulates vesicle trafficking between the endoplasmic reticulum (ER) and the Golgi bodies [9]. RabD is involved in ER–Golgi traffic [10]. RabE has the Golgi apparatus and PM localization [11, 12]. RabF/Rab5 and RabG/Rab7 are major regulators of endosomal/vacuolar trafficking in eukaryotes. RabH is located in Golgi apparatus [13]. The hypervariable C-terminal domain (HVD) of Rab proteins is post-translationally modified by isoprenyl moieties, which is important for protein target to membrane structure. But the complete mechanism is needed to elucidate the localization [14–16].
As the key player in fundamentally cellular activities, plant Rab proteins are involved in various regulatory processes of development and stress response. For example, Rab proteins are implicated in pollen tubes growth [9], leaf morphology [12], xylem development [17], autophagy [18], salinity stress [19] and pathogen defense [12]. Eukaryotic cells recognize their environment mainly through proteins on the PM, including receptors and sensors, which evoke intracellular signal transduction to respond to environmental cues. To date, membrane traffic and associated members are well studied in yeast, mammalian and Arabidopsis cells. However, there were finite studies on Rab GTPase functional characterization in woody plants. Different with the annual plants, perennial woody species not only undergo seasonally environmental changes, but also have specific developmental processes such as strong secondary growth. Therefore, study the Rab gene family is important to understand the regulation of trafficking during these specific environmental response or development in woody plants. As perennial woody model plant, poplars are widespread distributed group of economic species having the rapid growth and high production of plant biomass features [20]. The expansion of gene family along with the whole genome duplication events of Populus genome also provide the possibility for functional divergence of genes.
In this study, we comprehensively analyzed the Populus Rab GTPase gene family. A total of 67 PtRab genes were identified and categorized into eight subfamilies in P. trichocarpa. And 27 paralogous pairs were identified and all of them were generated by whole genome duplication (WGD) events, which implying tandem duplication was missed in PtRab family expansion. The PtRab paralogous pairs with highly similar expression patterns were generated around the latest WGD event (~ 13 MYA). In addition, the different localization differences among PtRab subfamilies provide basis for their functional divergence. Based on the cis-acting elements and the co-expression network, several Rab genes were associated with stress response. Among them, PtRabE1b was significantly induced by salt stress. Finally, we overexpressed constitutively active mutant PtRabE1b(Q74L) in poplar and found PtRabE1b(Q74L) enhanced the salt tolerance in poplar. These findings provide new insights into the functional divergence of PtRab genes and resources for genetic engineering resistant breeding in tree species.
Results
Identification and phylogenetic analysis of Rab gene family in Populus
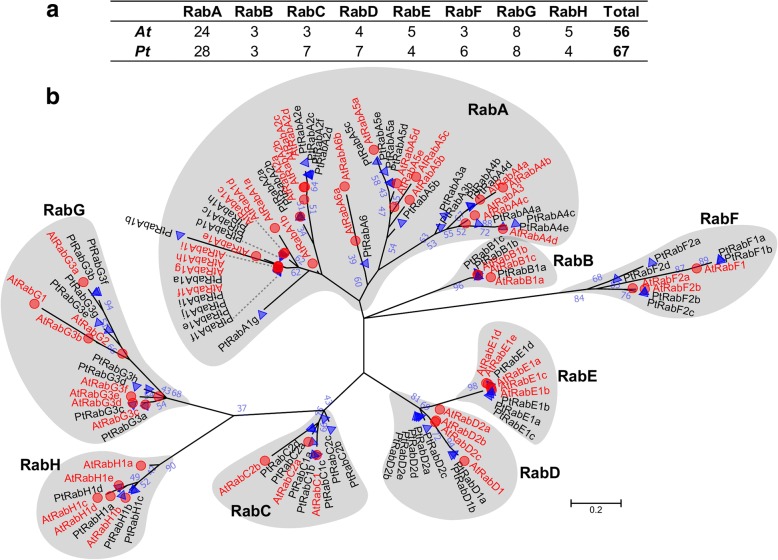
To identify Rab GTPase genes in Populus, the 56 known Arabidopsis Rab sequences [1] were used as queries in BLAST searches against the P. trichocarpa genome (release V3.0). A total of 67 putative Rab GTPase genes were identified in Populus (Additional file 1: Table S1). To examine the phylogenetic relationships of the PtRab gene family, a multiple sequence alignment was performed using full-length protein sequences of PtRabs and AtRabs, and a phylogenetic tree was constructed. As shown in Fig. 1, the PtRab family was grouped into eight clades (RabA-RabH) based on the sequence similarity and PtRabs were named according to their orthologous in Arabidopsis. Compared to Arabidopsis, four (RabA, RabC, RabD and RabF) of eight Rab subfamilies were extended in Populus. The Populus RabB and RabG subfamilies had the same size with Arabidopsis, while Populus RabE and RabH subfamilies were smaller than Arabidopsis.
Fig. 1.
Rab family members and their phylogenetic relationships in A. thaliana and P. trichocarpa. a Eight Rab subfamilies (RabA-H) members in A. thaliana (At) and P. trichocarpa (Pt). b Phylogenetic tree was constructed using full-length protein sequences by the maximum likelihood (ML) method with 1000 bootstrap replicates using MEGA 7.0.26. Numbers above branches of the tree indicate percentage of bootstrap values
Structure of Rab genes and conserved motifs of Rab proteins in Populus
To further investigate the structural features of Populus Rab genes, their exon-intron structures and protein motif composition were analyzed. Structure divergence of exon-intron play a pivotal role during evolution. Generally, paralogous genes are highly conversed in gene structure and this conservation is sufficient to reveal their evolutionary relationships [21]. In Populus, Rabs within the same subfamily shared similar gene structures (intron number and exon length), especially the members in RabA, RabC and RabH subfamilies. Compared to other multi-intron Rab genes, the members in RabA subfamily only have one intron (Additional file 2: Figure S1). Despite the gene length and gene structures were varies among the members in different Rab subfamilies, the protein length of most Rabs was consistently ~ 200 aa in both Populus and Arabidopsis.
Classical Rab proteins including four conserved motifs, which were involved in nucleotide binding and hydrolysis. We then analyzed the conserved motifs in PtRabs. Like Rabs in many other species, almost all the PtRabs including the four conserved motifs (motif 1–4; Additional file 2: Figure S1B). Although a little sequence bias was existed in the four motifs among different Rab subfamilies, the basic framework of the four motifs was relatively conserved in PtRabs. Based on previous studies, several specific amino acids in the conserved motifs of Rabs are involved in nucleotide binding and hydrolysis [1, 11]. In the PtRab family, three residues (S in motif-1, Q in motif-2 and N in motif-3; asterisks in Additional file 3: Figure S2) could be used to generate the dominant negative or constitutively active mutants through altering nucleotide-binding or hydrolysis characteristics for functional investigation.
Restricted by currently available annotation of the Populus genome, the structures of several genes were not well characterized. In the PtRab family, four genes (PtRabA1b, PtRabD1b, PtRabF2a and PtRabG3 g) were not full-length (Additional file 2: Figure S1). PtRabH1a had a complete open reading frame, while it lost ~ 50 aa including the conserved motif-1 in the N-terminus, and the distance between motif-3 and motif-4 was much closer than the other PtRabs. Furthermore, a pseudogene PtRabA1g.ψ were identified which coding protein only retained 92 aa in N-terminus including motif-1.
Chromosomal distribution and duplication among PtRab genes
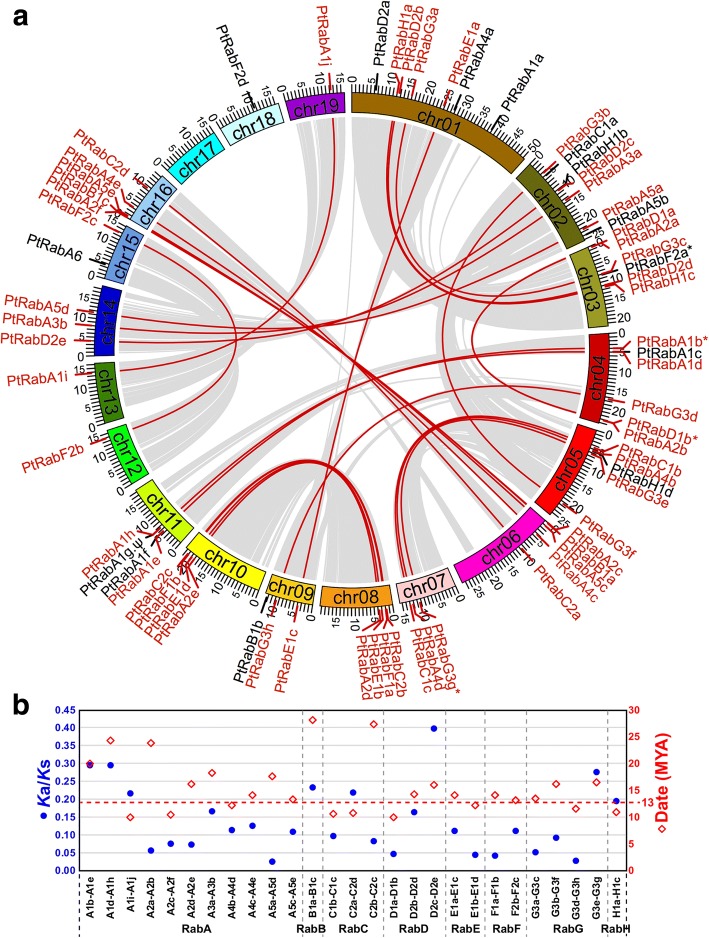
To explore the expansion mechanism of the PtRab family, we then analyzed its duplication patterns. All the 67 PtRabs unevenly distributed on 18 of 19 Populus chromosomes (Chr). Chr1 and Chr2 contain the most Rab genes (seven on each), while Chr17 does not contain any Rab gene (Fig. 2a). the Populus genome experienced at least two whole genome duplication (WGD) events and a series of chromosomal reorganizations [20]. In PtRab gene family, a total of 27 WGD paralogous pairs were identified, while no tandem duplication event was involved (Fig. 2 and Table 1). The nonsynonymous versus synonymous substitution rate ratios (Ka/Ks) were calculated to test whether Darwinian positive selection was involved in the PtRab gene divergence after duplication [22]. All the Ka/Ks ratios were prominent lower than 0.5 suggesting a purifying selection plays the dominate role in duplication of PtRab paralogous pairs (Fig. 2 and Table 1). Based on the divergence rate of λ = 9.1 × 10− 9 for Populus [23], the PtRab paralogous pairs were estimated to have occurred between 9.96 to 28.14 million years ago (MYA; Table 1). Recent large-scale genome duplication event in Populus occurred in ~ 13 MYA [20], all the PtRab paralogous pairs in five subfamilies (RabD-RabH) were generated around this stage, while PtRabB1a/B1c and PtRabC2b/C2c were generated in 28.14 and 27.32 MYA, respectively (Table 1 and Fig. 2b).
Fig. 2.
Genome-wide duplication of PtRabs. a Chromosome locations and duplication of PtRabs. Duplicated PtRabs were labelled as red and linked with red lines. Grey links between chromosomes indicate duplicated blocks generated by whole-genome duplication events. b Ka/Ks (left y-axis, blue dots) and duplication date (right y-axis, red dots) of pairwise PtRabs paralogous pairs
Table 1.
Divergence between PtRab paralogous pairs
| Subfamily | Gene 1 | Gene 2 | Duplication | Ka | Ks | Ka/Ks | Date (MYA) |
|---|---|---|---|---|---|---|---|
| RabA | PtRabA1b | PtRabA1e | W | 0.108 | 0.366 | 0.295 | 20.13 |
| PtRabA1d | PtRabA1h | W | 0.132 | 0.444 | 0.297 | 24.37 | |
| PtRabA1i | PtRabA1j | W | 0.040 | 0.182 | 0.218 | 9.98 | |
| PtRabA2a | PtRabA2b | W | 0.025 | 0.435 | 0.058 | 23.88 | |
| PtRabA2c | PtRabA2f | W | 0.014 | 0.191 | 0.075 | 10.50 | |
| PtRabA2d | PtRabA2e | W | 0.022 | 0.296 | 0.073 | 16.25 | |
| PtRabA3a | PtRabA3b | W | 0.056 | 0.334 | 0.167 | 18.35 | |
| PtRabA4b | PtRabA4d | W | 0.026 | 0.223 | 0.115 | 12.26 | |
| PtRabA4c | PtRabA4e | W | 0.032 | 0.257 | 0.126 | 14.10 | |
| PtRabA5a | PtRabA5d | W | 0.008 | 0.322 | 0.025 | 17.71 | |
| PtRabA5c | PtRabA5e | W | 0.027 | 0.243 | 0.110 | 13.35 | |
| RabB | PtRabB1a | PtRabB1c | W | 0.120 | 0.512 | 0.235 | 28.14 |
| RabC | PtRabC1b | PtRabC1c | W | 0.019 | 0.193 | 0.098 | 10.59 |
| PtRabC2a | PtRabC2d | W | 0.043 | 0.195 | 0.220 | 10.73 | |
| PtRabC2b | PtRabC2c | W | 0.042 | 0.497 | 0.084 | 27.32 | |
| RabD | PtRabD1a | PtRabD1b | W | 0.009 | 0.181 | 0.047 | 9.96 |
| PtRabD2b | PtRabD2d | W | 0.043 | 0.260 | 0.165 | 14.30 | |
| PtRabD2c | PtRabD2e | W | 0.117 | 0.293 | 0.399 | 16.12 | |
| RabE | PtRabE1a | PtRabE1c | W | 0.029 | 0.258 | 0.112 | 14.15 |
| PtRabE1b | PtRabE1d | W | 0.010 | 0.223 | 0.045 | 12.27 | |
| RabF | PtRabF1a | PtRabF1b | W | 0.011 | 0.257 | 0.043 | 14.10 |
| PtRabF2b | PtRabF2c | W | 0.027 | 0.239 | 0.111 | 13.14 | |
| RabG | PtRabG3a | PtRabG3c | W | 0.013 | 0.246 | 0.051 | 13.54 |
| PtRabG3b | PtRabG3f | W | 0.028 | 0.296 | 0.094 | 16.28 | |
| PtRabG3d | PtRabG3h | W | 0.006 | 0.212 | 0.029 | 11.63 | |
| PtRabG3e | PtRabG3g | W | 0.084 | 0.302 | 0.277 | 16.60 | |
| RabH | PtRabH1a | PtRabH1c | W | 0.039 | 0.200 | 0.195 | 10.99 |
Notes: W, whole genome-wide duplication; Ka, non-synonymous substitution rate; Ks, synonymous substitution rate; MYA, million years ago
Variety of cis-acting elements in the promoter regions of PtRabs
As transcription factors (TFs) binding sites, cis-acting elements located in gene promoter region are implicated in control of gene expression [24]. The 1.5 kb sequences upstream of translation start sites (TSS) of the PtRab genes were analyzed and the number of hormone responses and/or stress-related cis-acting elements were identified in the promoter regions of PtRabs (Additional file 4: Figure S3). Among the 67 PtRabs, 46 (68.7%) and 37 (55.2%) contain SA-responsive element (TCA-element) and MeJA-responsive elements (TGACG-motif or CGTCA-motif), respectively; and 24 PtRabs include both of them. Moreover, GA-, ABA-, ethylene- and auxin-responsive elements were found in promoter regions of 36, 30, 28 and 25 PtRabs, respectively. In addition, abundant stress-responsive elements were existed in the promoters of PtRabs. Top three stress-responsive elements were defense-, heat- and anoxia-responsive elements, which were presented in promoters of 54, 53 and 52 PtRabs, respectively (Additional file 4: Figure S3 and Additional file 5: Figure S4). These results indicated that PtRab genes might be involved in multiple hormone and stress responses.
Differential expression profiles of PtRabs across tissues and under various stresses
To explore the potential roles of PtRabs in various tissues or developmental stages, the publicly available microarray data was used to analyze their expression patterns. Across various tissues, most of the PtRab members were highly expressed in phloem and xylem, especially PtRabC subfamily. In contrast, most of PtRabs were relatively low expressed in reproductive organs (Additional file 6: Figure S5A and Additional file 7: Table S2). During the stem development, more than half of PtRabs were highly expressed in the 5th and 9th internodes, which experience transition from primary growth to secondary growth (Additional file 6: Figure S5B). These results suggested PtRab genes might be involved in biological processes related with stem development.
We then investigated the expression profiles of PtRabs under various abiotic stresses, including low nitrogen, wounding, drought, heat, cold, and salinity stresses. Under low nitrogen treatment, only two PtRabs (A3b and G3f) were slightly reduced in expanding leaves under 8-week-treatment. To response mechanical wounding, 11 PtRabs (A3a, A3b, A4b, A4d, A5a, A5b, A5d, C2a, D1a, G3e and G3f) were up-regulated and 7 PtRabs (A4a, A6, B1a, C1a, F1b, F2c and H1d) were down-regulated in expanding leaved under 1-week-treatment. The expression of PtRabs were not sensitive to drought stress in both Soligo and Carpacio genotypes. Total of 14 PtRabs were affected under heat stress, which including 7 PtRabs (A2c, A4a, A6, B1b, C1a, G3f and H1d) were up-regulated and 7 PtRabs (A3a, A5e, C2a, D1a, E1b, E1d and G3e) were down-regulated. Under cold stress, two PtRabs (A6 and H1d) and three PtRabs (A3a, C2a and G3e) were up- and down-regulated significantly (Additional file 6: Figure S5).
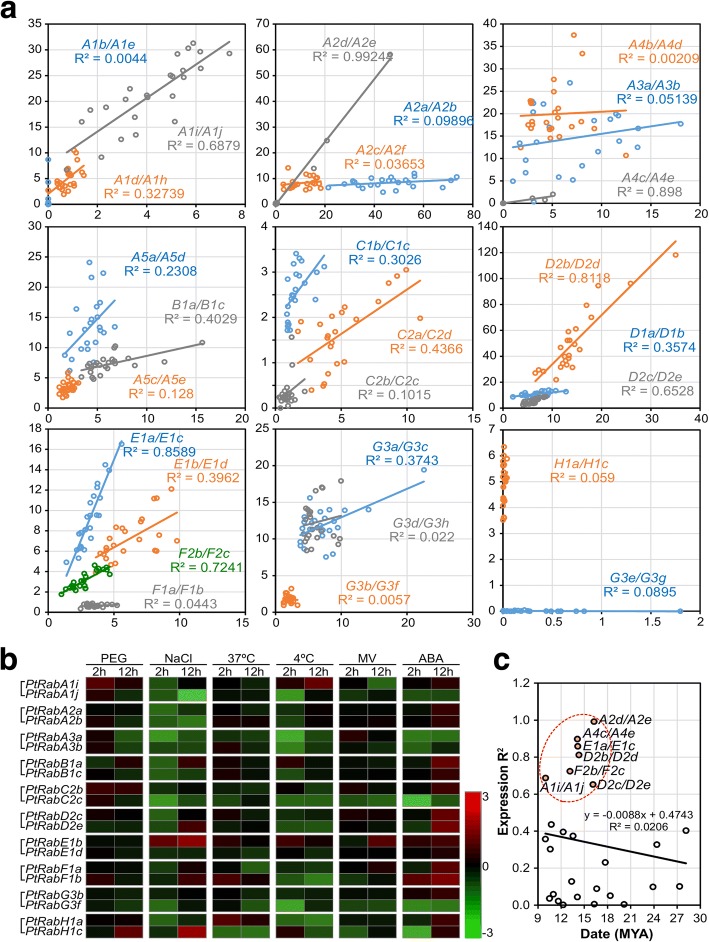
To reveal the divergence between PtRab paralogous pairs, the expression of 27 PtRab paralogous pairs were compared across 24 tissues. Only seven PtRab pairs (A1i/A1j, A2d/A2e, A4c/A4e, D2b/D2d, D2c/D2e, E1a/E1c and F2b/F2c) showed highly similar patterns with R2 > 0.6 across various tissues (Fig. 3a and Additional file 8: Table S3). This indicates most of the PtRab paralogous pairs were divergent during the evolution. Noticeably, four PtRab paralogous pairs (A1b/A1e, F1a/F1b, H1a/H1c and G3e/G3 g) showed significant differences (R2 < 0.1) in expression abundance between two genes in each pair - one has high abundance while another one was extremely low expressed in detected tissues (Fig. 3a). Expression R2 between genes in paralogous pairs was negatively correlated with their duplication date. Noticeably, the seven pairs with high expression R2 were duplicated during 9.98~ 16.25 MYA (red dot circle in Fig. 3c).
Fig. 3.
Expression analysis of 27 PtRab paralogous pairs. a Correlation of expression of 27 PtRab paralogous pairs across various tissues. x- and y-axis indicate FPKM values across 24 samples of two genes in each paralogous pair. For example, x-axis represents PtRabA1b and y-axis represents PtRabA1e in pair PtRabA1b/A1e. Expression values were shown in Table S3. b Expression validation of ten PtRab paralogous pairs response to various abiotic stresses using qRT-PCR. Red and green colors indicate up- and down-regulation of log2 transformed fold changes compared to control plants. c Correlation between duplication date and expression R2 of 27 PtRab paralogous pairs. Red dot circle includes seven pairs with highly similar patterns with R2 > 0.6
We then selected 10 PtRab paralogous pairs from eight subfamilies to validate their responsiveness under various abiotic stresses using qRT-PCR. Similar with the result from microarray data (Additional file 6: Figure S5), most genes in the 10 PtRab paralogous pairs were not sensitive to drought stress (Fig. 3b). Under salt, cold or oxidative stresses, most PtRab genes were down-regulated at 2 and 12 h after stress. While PtRabE1b and PtRabH1c were up-regulated under salt stress, especially PtRabE1b was prompt induced at both 2 and 12 h. This implying PtRabE1b might play a role in salt stress tolerance. Overall, the expression patterns of 10 selected PtRab paralogous pairs were divergent between the pairs in various abiotic stresses.
Co-expression network of PtRabs
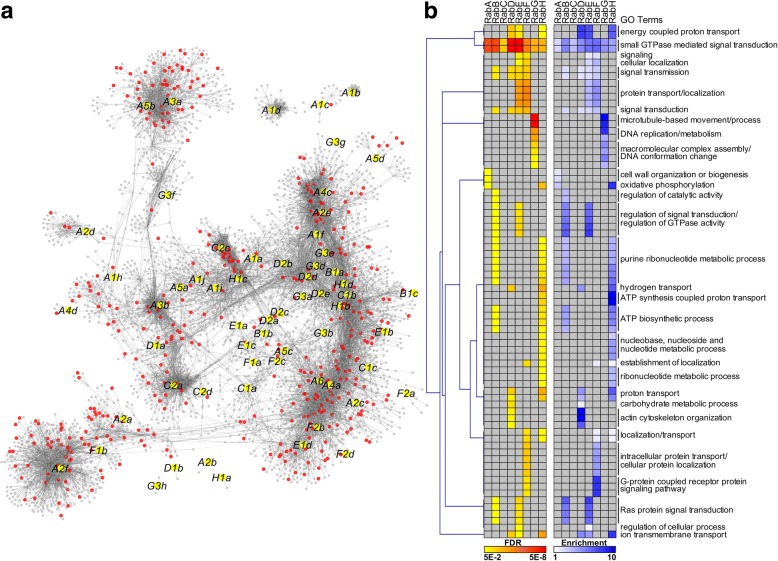
To further reveal the functional differences of PtRabs in different subfamilies, we constructed a co-expression network of PtRabs. Total 61 of 67 PtRabs and 411 TFs were identified in the PtRabs co-expression network (Fig. 4a and Table S4). Among the 6151 genes in PtRabs co-expression network, RabA sub-network represents as the largest sub-network including a total of 4387 genes (71.32%) and 24 RabA members; whereas RabF and RabG sub-networks only including 6.15 and 7.79% in the whole network, respectively (Additional file 9: Figure S6). After screen the TF genes number (Additional file 9: Figure S6) and their enrichment (Additional file 10: Figure S7) in each sub-network, we found four HSFs (HSFA4a, HSFA5a, HSFA5b and HSFB3b) were enriched 7.697-fold in RabE sub-network.
Fig. 4.
Co-expression network of PtRabs. a Co-expression network of PtRabs. Yellow and red nodes indicate PtRabs and transcription factors (TFs) in the co-expression network, respectively. b Hierarchical clustering of enriched GO terms of co-expression sub-networks of eight PtRab subfamilies (RabA-H). Colors represent FDR (yellow to red) or enrichment (white to blue); grey indicates no significant enrichment. Details of genes in PtRabs co-expression network and GO enrichment were shown in Table S4 and S5
Then the genes in sub-networks of different PtRab subfamilies were used for GO enrichment analysis. Noticeably, all the eight (PtRabA-H) sub-networks were enriched in “intracellular signaling cascade” and “small GTPase mediated signal transduction” (Fig. 4b, Additional file 11: Table S4, and Additional file 12: Table S5). PtRabE and PtRabF sub-networks were enriched in “signal process”, while PtRabB and PtRabE sub-networks were enriched in “regulation of GTPase activity”. For sub-network specific enriched GO terms, PtRabA sub-network was enriched in “cell wall organization”, PtRabD sub-network was enriched in “actin cytoskeleton organization”, PtRabF sub-network was enriched in “intracellular transport”, while PtRabG sub-network was enriched in “microtubule-based process” (Fig. 4b). In addition, the genes primarily co-expressed with PtRabE1b were functional classified based on their annotation. Total of 24 transport-, 11 stress-, 10 signal-, 5 protein-, 7 metabolic-, 6 development-, 4 cytoskeleton- and 2 autophagy-related genes were primarily co-expressed with PtRabE1b (Additional file 13: Figure S8).
Subcellular localization of PtRab proteins in different subfamilies
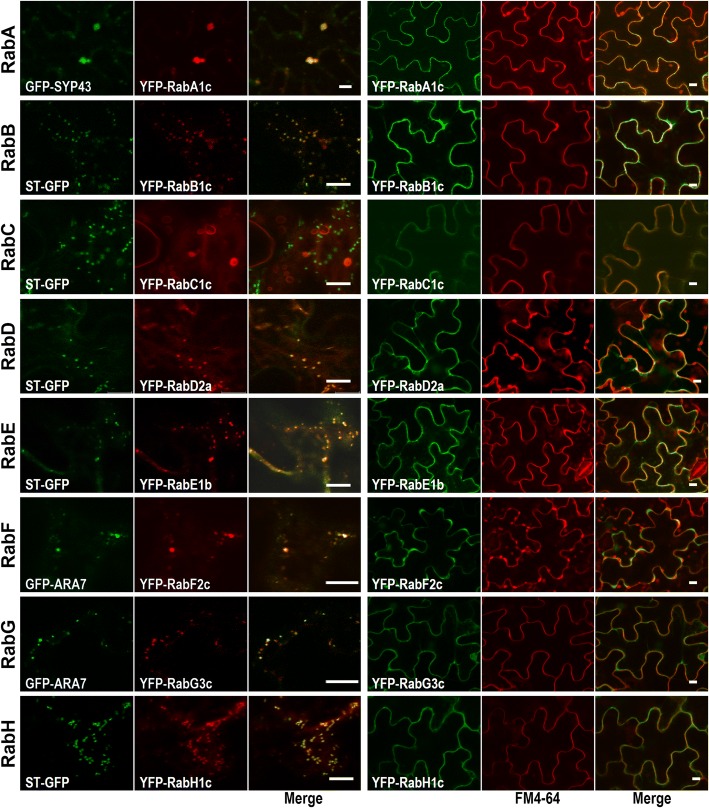
Plant Rab proteins coordinate different steps of endomembrane trafficking and showed specific subcellular localizations [1, 11]. Subsequently, eight PtRabs representing different subfamilies were selected for subcellular localization analysis in Nicotiana benthamiana leaf epidermal cells. The eight fusion proteins (35S::YFP-PtRabs) were detected on the PM as indicated by co-localized with the PM marker FM4–64 (Fig. 5 right panel). In addition, the well-known organelle markers including GFP-SYP43 (TGN), ST-GFP (Golgi), ARA7 (endosome) and GFP-HDEL (ER) were performed to confirm their precise localization in organelle. PtRabA1c was localized in the TGN, four PtRabs (B1c, D2a, E1b and H1c) were localized in the Golgi apparatus, while PtRabF2c and PtRabG3c were localized in the endosome (Fig. 5 left panel). However, PtRabC1c was localized in small vesicles but not co-localized with any organelle marker used in this study, its precise localization needs to be further determined.
Fig. 5.
Subcellular localization of PtRab members in eight subfamilies. Confocal images of tobacco leaf epidermal cells co-expressing YFP-PtRabs (A1c, B1c, C1c, D2a, E1b, F2c, G3c and H1c) and different organelle markers (left panel) or FM4–64 (5 min after staining, right panel). Various organelle markers including GFP-SYP43 (for trans-Golgi network, TGN), ST-GFP (for Golgi) and GFP-ARA7 (for endosome). Bar, 10 μm
Natural variation of PtRabE1b
Based on the expression analysis, PtRabE1b was induced at 2 h after exposure to salt stress, and the induction was enhanced when the exposure was extended to 12 h (Fig. 3b). Furthermore, 11 stress-related genes were primarily co-expressed with PtRabE1b (Additional file 13: Figure S8). We then used PtRabE1b for further functional analysis. The natural variation of single nucleotide polymorphism (SNP) in a gene is known to introduce functional divergence in the gene. To detect if the conserved domains of PtRabE1b were affected in natural condition, the whole genome re-sequencing data of 549 P. trichocarpa natural individuals in North America were analyzed. As shown in Additional file 14: Figure S9, a total of 227 SNPs were identified in the PtRabE1b gene body (including UTR, exon and intron), and four SNPs affect non-synonymous coding (I45N, E100V, E168V and S201P). Noticeably, the four non-synonymous SNPs were not located in the conserved motifs of PtRabE1b (Additional file 14: Figure S9C).
Overexpression of the constitutive activation mutant PtRabE1b(Q74L) confers salt tolerance in poplar
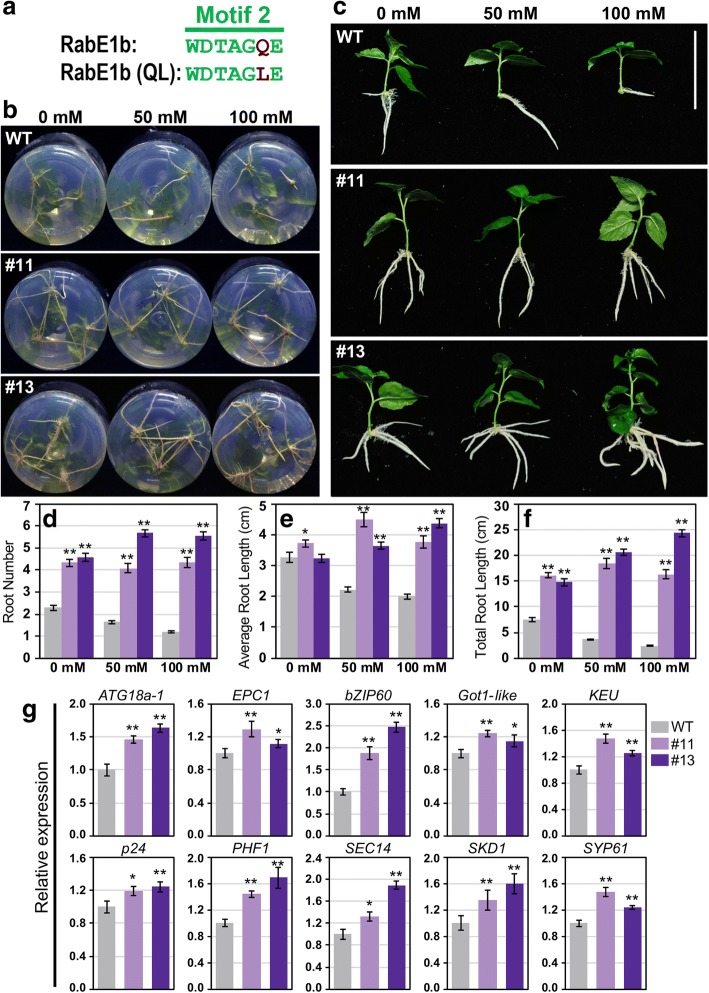
To further investigate the function of PtRabE1b, a constitutively activate mutant PtRabE1b(Q74L) was constructed by point mutation of Q74 to L in motif 2 (Fig. 6a and Additional file 14: Figure S9C). A plant binary construct containing PtRabE1b(Q74L) driven by the CaMV 35S promoter was generated and overexpressed in hybrid poplar (P. alba × P. glandulosa) clone 84 K. A total of 25 independent lines with hygromycin resistance were obtained and the positive transformants were confirmed using qRT-PCR. Two independent transgenic lines (#11 and #13) with the high transcript levels were selected for further analysis. Under normal condition, more adventitious roots (ARs) were differentiated in the two transgenic lines than wild type (WT), but the average root length was not significant differences between the transgenic lines and WT plants. When the seedlings were exposed to salt stresses (50 and 100 mM NaCl), both the root differentiation (root number) and root growth (root length) were inhibited in WT poplar. By contrast, the two transgenic lines maintained great root growth status even under salt stresses (Fig. 6). This indicates that overexpression the constitutive activate RabE1b(Q74L) enhanced salt tolerance in poplar. We then selected 10 different genes from the PtRabE1b co-expression network to validate their expression in PtRabE1b(Q74L) overexpression poplar. Seven trafficking-related genes (Got1-like, KEU, p24, PHF1, SEC14, SKD1 and SYP61), a stress-related TF (bZIP60), an autophagy-related gene (ATG18a-1) and a development-related genes (EPC1) were up-regulated in the two PtRabE1b(Q74L) overexpression lines (Fig. 6g).
Fig. 6.
Overexpression of constitutive activation mutant of PtRabE1b confers salt tolerance in poplar. a Construction of a constitutively activation mutant of PtRabE1b at Q74L. b, c Growth status of wild type and two PtRabE1b(Q74L) overexpression lines (#11 and #13) under salt stress (0 mM, 50 mM and 100 mM NaCl). d-f Root number, average root length and total root length (n = 12). g Expression of selected co-expressed genes of PtRabE1b in wild type and PtRabE1b(Q74L) overexpression lines #11 and #13 under salt stress. Bars represent the mean ± SE of three independent experiments. * and ** represent significant differences at P < 0.05 and P < 0.01 compared with WT plants, respectively
Discussion
Rab GTPase as the important proteins in vesicular transport, are involved in the entire process of vesicle transport including budding from donor organelle, docking, tethering and fusing with the target membrane [3, 4]. Rab family was highly conserved in yeasts, mammalian and higher plants [6, 25], so it provide the consistent foundation for vesicle transport in organism. Here, a total of 67 non-redundant Rab genes were identified in Populus, it’s 1.2-fold than that in Arabidopsis (56 AtRabs) and relative lower than the ratio of 1.4~ 1.6 putative poplar homologs for Arabidopsis gene [20]. It seems that multicellutar organism has larger Rab GTPase family members compared to the 7–11 Rabs are found in the yeasts. It may be that multicellular organism is faced with complicated circumstance and adapt to complex metabolism.
The RabA subfamily is the largest subfamily in poplar and Arabidopsis (Fig. 1). However, yeast and animals only possess a few members corresponding to the RabA subgroup [25]. The Rab2 subgroup is closely related to human Rab11 and Rab25, which are generally thought to be associated with the polarize recycling of proteins at the PM. The studies on pea and tomato indicate RabAs play roles in the targeted secretion of cell-wall components to restricted areas of the cell surface [26]. In poplar, four more RabA members were identified than Arabidopsis, but its proportion among entire Rab family (41.8%; 28 PtRabAs of 67 PtRabs) is relative lower than that in Arabidopsis (42.9%; 24 of 56). Noticeably, the member of RabA6 was lack in poplar than Arabidopsis, which is similar with rice [27]; this implies RabA6 might not play conserved function across different species. During the expansion of the Rab family in the Populus genome, no tandem duplication event was observed. But there are two RabA clusters on Chr4 (PtRabA1b, PtRabA1c and PtRabA1d) and Chr11 (PtRabA1e, PtRabA1f and PtRabA1h), these two clusters might be undergone tandem duplication before the latest WGD. In contrast, the RabC subfamily was expanded to seven in poplar, while there are only three in Arabidopsis with unclear functions. In mammalian cells, its orthologue Rab18 is majorly involved in lipid droplet formation in the ER and possibly for cell metabolism in stressful conditions [28]. The expansion of RabC in poplar might be associated with the specific stress response or development, but it’s function still need further study. Based on the correlation analysis, only seven of 27 PtRabs paralogous pairs kept high correlation with R2 > 0.6, and about half (13 of 27) pairs showed significant divergence with R2 < 0.3 (Fig. 3). In addition, the expression R2 of PtRab paralogous pairs were negatively correlated with their duplication data. The seven PtRab paralogous pairs which still kept high expression similarity were mainly generated around the recent large-scale genome duplication event in Populus (~ 13 MYA; Fig. 3c) [20].
In the investigation of conserved Rab motifs, we found almost all the PtRab members had four conserved motifs, except several PtRabs which were not well annotated in the latest released P. trichocarpa genome. The four conserved motifs play crucial roles for their nucleotide binding and hydrolysis functions [29]. Three residues (S in motif 1, Q in motif 2 and N in motif 3 labelled in Additional file 2: Figure S1) could be used to generate the dominant negative or constitutively active mutants for gene functional investigation. In this study, we constructed an active mutation of PtRabE1b through point mutation of Q74 to L in motif 2 and overexpressed PtRabE1b(Q74L) in poplar. The phenotypes of PtRabE1b(Q74L)-overexpression poplars under normal condition and salt stress proved mutation at Q74 of PtRabE1b can introduce constitutive activity and this mutation can be used for gene functional studies.
The Rab proteins exhibit restricted and specific subcellular localizations. Mammalian Rab11 resides on recycling endosomes and regulates traffic to specific PM domains and to the TGN in diverse cell types [30]. The function of Rab11 orthologous may be conserved either in mammalian or plant. As the orthologous of Rab11 in plants, RabA subfamily members have the same localization. RabA1a, RabA1b and RabA1c co-localized with TGN marker in the division zone of roots [8]. Moreover, members in RabA subfamily had evolved divergent function in distinct steps of subcellular trafficking. In poplar, PtRabA1c was precisely located to TGN and PM, which implied the functional conservation of RabA subfamily. Either transient expression in tobacco epidermal cell or stable expression in Arabidopsis indicate AtRabE1d localized in Golgi apparatus and PM [11, 12], which were consistent with our findings of PtRabE1b. For members in other subfamilies, most of them showed similar localization with the orthologous reported in other plant species [13]. Whereas, the localization evidence of RabC is still scarce. A study using mammalian cells reveals Rab18, an orthologous of RabC, was localized to endosomes through antibody staining. In addition, it was also existed in ER and Golgi apparatus [28]. In our study, PtRabC1c was localized in small vesicles, but it was not overlapped with any marker except PM. Further studies should be conducted to survey the various localization of Rab protein.
Rab GTPases have various roles in development. Arabidopsis RabE down-regulation resulted in drastically altered leaf morphology and reduced plant size [12]. Tobacco NbRabE1 silenced resulted in growth arrest, premature senescence, and abnormal leaf development. The dominant negative mutant of NbRabE1 in Arabidopsis resulted in retardation of shoot and root growth accompanied by defective root hair formation [31]. Except members in RabE subfamily, Rab genes in other subfamilies were also play important roles. Overexpression a constitutively active form of Arabidopsis RabG3b promotes xylem development through the activation of autophagy in the transgenic poplar [17]. Among the processes unique to tree biology, one of the most obvious is the yearly development of secondary xylem from the vascular cambium different from herbaceous plants. From our results, almost all the Rab genes were highly expressed in xylem and phloem. Moreover, the expression level of Rab genes were elevated following the maturity of internode (Additional file 6: Figure S5). It can explain massive materials’ transport within cell during this developmental process. Generally, wood is a product of plant cell wall polymers and wood formation is the consequences of plant cell wall deposition. Various plant cell wall synthesis-related enzymes and metabolites have been shown to be mediated by membrane trafficking. For instance, cellulose synthase, hemicellulose, pectin and other polysaccharides transport from Golgi apparatus to cell membrane or secrete to extracellular space [32–34].
As sessile organisms, plant have acquired different strategies to respond to various biotic and abiotic stresses in their ambient environment [35]. During the stress response, cis-acting elements play pivotal roles in regulating genes expression by controlling TF binding site and promoter efficiency [24]. Our results indicated that the cis-acting elements of PtRab genes were involved in multiple hormone and stress responses. A majority of PtRab promoters contain SA- and MeJA-responsive elements, which implied PtRab genes might be involved in pathogen resistance. In tomato, two RabE members Api2 and Api3 can interact with the Pst DC3000 TTSS effector AvrPto [36]. Overexpression of the constitutively active RabE1d(Q74L) conferring Arabidopsis resistance to P. syringae infection [12]. In addition, Rab genes are involved in abiotic stress tolerance in plants. For instance, both the Arabidopsis RabA1 quadruple mutant (completely knocked out for expression of RabA1a, A1b, A1c and A1d) and expressing the dominant-negative mutant of RabA1b(S27b) exhibited hypersensitivity to salinity stress at 15 mM NaCl concentration [8]. Loss-of-function of ARA6 (also known as RabF1) conferred Arabidopsis salt hypersensitivity; but overexpression of ARA6(Q93L)-GFP conferred tolerance to high-salt stress, which formed discrete speckles on the PM after salt stress with no other PM proteins were affected [37]. Moreover, overexpression of Pennisetum glaucum PgRab7 or Prosopis juliflora PjRab7, which are homologs of Arabidopsis RabG3e, confer salt tolerance to transgenic tobacco [19, 38]. In plants, Rabs are expanded greatly but the function of many of them is unknown. This may reflect the fact that Rabs are expanded to deal with prevailing environmental conditions, which was supported by our cis-acting elements, co-expression network and expression profiles analyses. In our study, overexpression of constitutively active RabE1b(Q74L) confer salt tolerance in transgenic poplar (Fig. 6). From the co-expression network, four HSF including HSFA4a were highly enriched in RabE sub-network (Fig. 4 and Additional file 10: Figure S7). In Arabidopsis, HSFA4a confers salt tolerance and regulated by oxidative stress and MPK3/MPK6 [39]. It’s homologue in Chrysanthemum, CmHSFA4, was also been proved positively regulate salt tolerance through maintain Na+/K+ ion balance with SOS1 and HKT2 and regulate ROS scavenger activities [40]. Among the genes directly co-expressed with PtRabE1b (Additional file 13: Figure S8), SOS2, MPK19 and Ca2+ signaling related genes (CAM7, CKL6 and calcium exchanger) were consistent with these studies on plant salt tolerance. The SOS2 encodes a serine/threonine protein kinase with an amino-terminal catalytic domain and a carboxy-terminal regulatory domain. SOS2 can physically interact with and be activated by SOS3, a myristoylated calcium-binding protein, to form SOS3-SOS2 kinase complex for transcriptional regulation of SOS1. And this pathway was mediated by calcium signal [41]. Mitogen-activated protein kinase (MAPK) cascades are universal signal transduction modules in eukaryotes. Several members in MPK family have been reported were involved in salt stress response, e.g. MPK3, MPK4 and MPK6 are activated by NaCl stress, expression of active MKK9 through activation of endogenous MPK3 and MPK6 enhances salt sensitivity in Arabidopsis [42]. Moreover, MPK6 can phosphorylates the C–terminal fragment of SOS1 to exclude Na+ from cells [43].
Conclusions
In conclusion, we comprehensively analyzed the evolutionary relationship of the Rab gene family in Populus and functionally characterized PtRabE1b, which confers salt tolerance in transgenic poplar. Our findings provide a deeper insight of the structure-localization-function relationships of vesicular transport genes and an abundant resource for genetic engineering to improve stress tolerance in trees.
Methods
Sequence retrieval and phylogenetic reconstruction
Published Arabidopsis Rab sequences [1] were retrieved and were used as queries in BLAST searches against the P. trichocarpa genome database (http://phytozome.jgi.doe.gov/pz/portal.html#!info?alias=Org_Ptrichocarpa) to identify potential PtRab genes. All homologous protein sequences of the predicted Rab family members were accepted if they were satisfied with expectation (E) value < 10− 10. Multiple alignment of Rab proteins from P. trichocarpa and A. thaliana were performed using the Clustal X2.1 [44]. The maximum likelihood (ML) phylogenetic trees were constructed using MEGA 7.0.26 [45] under the Jones-Taylor-Thornton (JTT) amino acid substitution model with 1000 bootstraps.
Bioinformatics analyses of Populus Rab genes
The exon and intron structures were illustrated using Gene Structure Display Server (GSDS) [46]. The chromosomal locations and duplication of the Rab genes were drawn using Circos software [47]. To analyze the putative cis-acting regulatory elements, 1500 bp upstream of translation start site (TSS) were searched using PlantCARE database [24]. The PAL2NAL program (http://www.bork.embl.de/pal2nal/) was used to estimate synonymous (Ks) and nonsynonymous (Ka) substitution rates. The divergence time (T) was calculated according to T = Ks/(2 × 9.1 × 10− 9) million years ago (MYA) for Populus [23]. For co-expression network construction, the expression data was obtained in the Populus Gene Atlas Study from Phytozome (https://phytozome.jgi.doe.gov/pz/portal.html#). Pearson correlations were calculated in parallel between all pairs of gene expression vectors. A threshold greater than or equal to 0.85 was applied to resulting correlations and the remaining correlations were visualized by Cytoscape [48]. GO enrichment of each sub-network was performed using agriGO [49].
Publicly available microarray data analyses
The microarray data for various tissues and developmental stages were obtained from the NCBI Gene Expression Omnibus (GEO) database with the accession numbers GSE21481, GSE21485 and GSE13043. For abiotic stresses, Affymetrix microarray data with the series accession numbers GSE14893 and GSE148515 (nitrogen limitation), GSE16783 and GSE16785 (wounding), GSE17225 (drought), GSE41557 (heat, 42 °C 6 h) and GSE43872 (cold, 4 °C 10 h) were analyzed. Probe sets corresponding to PtRabs genes were identified using the online Probe Match tool POParray (http://aspendb.uga.edu) and were listed in Additional file 3: Table S2. The data was normalized using the Gene Chip Robust Multiarray Analysis (GCRMA) algorithm followed by log transformation and average calculations. For correlation analysis of 27 PtRab paralogous pairs, the expression data of PtRabs across 24 P. trichocarpa tissues were obtained in the Populus Gene Atlas Study from Phytozome (https://phytozome.jgi.doe.gov/pz/portal.html#). Pearson correlations were calculated between paralogous genes in each pair.
Plasmids and constructs
The full-length CDS of PtRabA1c, RabB1c, RabC1c, RabD2a, RabF2c, RabG3c and RabH1c were amplified from the cDNA of P. trichocarpa and was introduced into the pDNOR222.1 (Life technologies, Carlsbad, California, U.S.) to produce pENTR vectors. Subsequently, the genes in the pENTR vectors were validated by sequencing and then subcloned into pEarleyGate104 (ABRC stock DB3–686) to produce 35S::YFP-PtRabs constructs using the Gateway cloning system (Invitrogen). To generate Q74L substitution of PtRabE1b, site-directed mutagenesis was conducted by overlapping PCR. Four primers containing the desired changes (underlined) were used: PtRabE1b-QL-1, 5’-ATGGCTGCACCGCCAG-3′; PtRabE1b-QL-2, 5’-AGTTCGGAAACGTTCCAGGCCTGCTGTATCCA-3′; PtRabE1b-QL-3, 5’-TGGGATACAGCAGGCCTGGAACGTTTCCGAACT-3′; PtRabE1b-QL-4, 5’-TTATGAACCACAGCAAGCTGACT-3′. Through a change at 221 bp of PtRabE1b CDS (A to T), PtRabE1b(Q74L) was obtained. The coding sequences of PtRabE1b(Q74L) was introduced into pDNOR222.1 and subcloned into pMDC32 using the Gateway cloning system (Invitrogen). The expression vectors were transferred into Agrobacterium tumefaciens GV3101 by electroporation.
Plant material, growth conditions and transformation
One-year-old P. trichocarpa grown in a growth chamber under long-day conditions (16/8 h light/dark) at 23–25 °C. Plant materials for qRT-PCR in different tissues (YL, young leaf; ML, mature leaf; PS, primary stem; SS, secondary stem; R, root.) were collected from hybrid poplar (P. alba × P. glandulosa) clone 84 K. Samples were frozen immediately in liquid nitrogen, and stored at − 80 °C for further analysis. Three biological replicates were performed.
For abiotic stress and hormone treatments, P. trichocarpa seedlings were water-cultured in Hoagland’s nutrient solution for 15 days after subculture on 1/2 MS solid medium. Then the seedlings were treated with 10% polyethylene glycol (PEG, for drought stress), 150 mM NaCl (for salt stress), 37 °C (for heat stress), 4 °C (for cold stress), 10 μM methyl viologen (MV, for oxidative stress) or 100 μM abscisic acid (ABA), respectively. The control plants were grown in Hoagland’s nutrient solution under normal condition. During the treatments, three time-points (0, 2 and 12 h) were chosen for samples collection. The above samples were immediately frozen in liquid nitrogen after harvest and stored at − 80 °C for further analysis.
Poplar stable transformation of PtRabE1b(Q74L) was through leaf-disc method as described by Liu et al. [50]. A total of 25 independent transgenic lines carrying the 35S::PtRabE1b(Q74L) construct with hygromycin resistance were obtained. Two lines (#11 and #13) with high transcript levels of PtRabE1b(Q74L) were used for further analysis.
Subcellular localization of PtRabs
Co-expression of 35S::YFP-PtRabs constructs and fluorescent markers in Nicotiana benthamiana leaf lower epidermal cells was performed using Agrobacterium transformation as described by Zhang et al. [51]. The fluorescence of infiltrated tobacco leaves was observed using a LSM 510 confocal laser scanning microscope. For fluorescence imaging of YFP/FM4–64 samples, 488/543-nm excitation and 505–530/585-nm filters were used. For GFP/YFP samples, 458/514-nm excitation and 475–525/520-555 nm filters were used.
RNA isolation and real-time qRT-PCR
Total RNA was extracted using the RNeasy Plant Mini Kit (Qiagen) with on-column treatment with RNase-free DNase I (Qiagen) to remove genomic DNA. First-strand cDNA synthesis was carried out with approximately 1 μg RNA using the SuperScript III reverse transcription kit (Invitrogen) according to the manufacturer’s procedure. Primers with melting temperatures of 58–60 °C and amplicon lengths of 100–250 bp were designed using Primer3 software (http://frodo.wi.mit.edu/primer3/input.htm). All primer sequences are listed in Additional file 15: Table S6. Real-time qRT-PCR was performed in quadruplicate using the SYBR Premix Ex Taq™ II Kit (TaKaRa, Dalian, China) on a Roche LightCycler 480 (Roche Applied Science, Penzberg, Upper bavaria, Germany) according to the manufacturer’s instructions. All experiments were performed in three biological replicates and four technical replicates. The PtActin and PtTubulin genes were used as internal controls.
Statistical analyses
All data are presented as the means ± standard error (SE) of at least three replicates. The Student’s t-test was used to test the significance of differences between the control plants and transgenic lines. Asterisks (* or **) indicate a significant difference between the control and transgenic plants at P < 0.05 or 0.01, respectively.
Additional files
Table S1. Rab gene families in P. trichocarpa. (DOCX 24 kb)
Figure S1. Gene structures and motif composition of PtRabs. (A) The exon-intron structures of PtRab genes. The numbers (0, 1 and 2) indicate the splicing phases (phase 0, 1 and 2, respectively) of the Rab genes. (B) A schematic representation of conserved motifs in Rab proteins. Four conserved motifs (1–4) are represented by different colors. (TIF 3329 kb)
Figure S2. Conserved motifs and their sequences in PtRab proteins. The four motifs are involved in nucleotide binding and hydrolysis. Mutation of the residues (shown in red asterisk) could be used to generate dominant-negative and constitutively active forms that exhibit altered nucleotide-binding or hydrolysis characteristics, which could be used to investigate Rab function. (TIF 1105 kb)
Figure S3. The number of Rab genes containing various cis-acting elements. Auxin-responsive elements: TGA-element, TGA-box and AuxRR-core; gibberellin-responsive elements: TATC-box, P-box and GARE-motif; abscisic acid (ABA)-responsive elements: CE3 and ABRE; salicylic acid (SA)-responsive element: TCA-element; MeJA-responsive elements: TGACG-motif and CGTCA-motif; ethylene-responsive element: ERE; heat-responsive element: HSE; cold-responsive element: LTR and C-repeat/DRE; MYB binding site involved in drought-inducibility: MBS; defense-related elements: Box-W1 and TC-rich repeats; anoxic-related elements: GC-motif and ARE; wound-responsive element: WUN-motif. (TIF 415 kb)
Figure S4. Various cis-acting elements in responsive to hormone and stresses in Rab promoters. (TIF 1113 kb)
Figure S5. Expression profiles of PtRab genes across different tissues and under various abiotic stresses. Heatmap showing expression of PtRab genes across tissues (A), different stem development/growth stages (B) and under various abiotic stresses (C) in poplar. For abiotic stresses, Stress treatments including: low N, nitrogen limitation; wounding, sampled either one week or 90 h after wounding; EAR, early response to water deficit by 36 h; LMI, long-term (10 day) response to mild stress with soil relative extractable water (REW) at 20–35%; LMO, long-term (10 day) response to moderate stress with soil REW at 10–20%; heat, 42 °C for 6 h; and cold, 4 °C for 10 h. Genotypes including P. fremontii × P. angustifolia clones 1979, 3200 and RM5 (low N and wounding), P. deltoids clones Soligo and Carpaccio (drought), and P. simonii (heat and cold). Color scale represents log2 expression values, green represents low level and red indicates high level of transcript abundances. Blank represents a gene has no corresponding probe sets in the microarray data. The microarray data (GSE21481, GSE21485, GSE13043, GSE16786, GSE17230, GSE41557 and GSE43872) were obtained from NCBI Gene Expression Omnibus (GEO) database. (TIF 3420 kb)
Table S2. Probe sets corresponding to PtRab genes. (DOCX 22 kb)
Table S3. Expression data of 27 PtRab paralogous pairs used for correlation analysis. (XLSX 22 kb)
Figure S6. Summary of Rab and TF subfamilies gene numbers in PtRabs co-expression sub- networks. (TIF 945 kb)
Figure S7. TF enrichment of different subfamilies in PtRabs co-expression sub-networks. (TIF 1210 kb)
Table S4. Functional annotation of genes in PtRabs co-expression network. (XLSX 484 kb)
Table S5. Enriched GO terms of genes in PtRabs sub-networks. (XLSX 16 kb)
Figure S8. Functional classification of genes primarily co-expressed with PtRabE1b. (TIF 1141 kb)
Figure S9. Natural variation and protein structure of PtRabE1b. (A) Identification of SNPs in PtRabE1b from 549 P. trichocarpa individuals. (B) Effects of SNPs located in PtRabE1b. (C) Protein structure of PtRabE1b. Four non-synonymous SNPs affected proteins (I45N, E100V, E168V and S201P) were labelled as yellow. Q74L in motif-2 used for constitutive activation was labelled as green. (TIF 631 kb)
Table S6 qRT-PCR Primers used in this study. (DOCX 18 kb)
Acknowledgements
We would like to thank the reviewers and editors for their careful reading and helpful comments on this manuscript.
Funding
This work was supported by the National Natural Science Foundation of China [31270699] to J.C. and a Collaborative Innovation Plan of Jiangsu Higher Education to M.L. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Availability of data and materials
All data generated or analyzed during this study are included in this article (and its supplementary information files) or are available from the corresponding author on reasonable request. Protein sequence and phylogeny data, including alignments, have been deposited in the TreeBASE repository (ID: 22857).
Abbreviations
- ABA
Abscisic acid
- ER
Endoplasmic reticulum
- HVD
Hypervariable C-terminal domain
- Ka
nonsynonymous substitution rate
- Ks
synonymous substitution rate
- MV
Methyl viologen
- MYA
Million years ago
- PEG
Polyethylene glycol
- SNARE
the soluble N-ethylmaleimide-sensitive factor attachment receptors
- SNP
Single nucleotide polymorphism
- TF
Transcription factor
- TGN
Trans-Golgi network
- TSS
Translation start site
- UTR
Untranslated region
- WGD
Whole genome duplication
- YFP
Yellow fluorescent protein
Authors’ contributions
JZ and ML designed the study. JZ, YL, BL and LZ performed the experiments. JZ and YL analyzed the data and wrote the manuscript. LW, JH, JC and HZ contributed to the data collection and data analysis. JZ, HZ and ML revised the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Jin Zhang and Yu Li contributed equally to this work.
Electronic supplementary material
The online version of this article (10.1186/s12870-018-1342-1) contains supplementary material, which is available to authorized users.
Contributor Information
Jin Zhang, Email: zhang007jin@163.com.
Yu Li, Email: linxueliyu@163.com.
Bobin Liu, Email: 163liubobin0377@163.com.
Lijuan Wang, Email: wlj307@caf.ac.cn.
Li Zhang, Email: zhanglisdau86@126.com.
Jianjun Hu, Email: hujj@caf.ac.cn.
Jun Chen, Email: chenjun@caf.ac.cn.
Huanquan Zheng, Email: hugo.zheng@mcgill.ca.
Mengzhu Lu, Email: lumz@caf.ac.cn.
References
- 1.Rutherford S, Moore I. The Arabidopsis Rab GTPase family: another enigma variation. Curr Opin Plant Biol. 2002;5(6):518–528. doi: 10.1016/S1369-5266(02)00307-2. [DOI] [PubMed] [Google Scholar]
- 2.Wickner WSR. Membrane fusion. Nat Struct Mol Biol. 2008;15(7):658–664. doi: 10.1038/nsmb.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonifacino JS, Glick BS. The mechanisms of vesicle budding and fusion. Cell. 2004;116(2):153–166. doi: 10.1016/S0092-8674(03)01079-1. [DOI] [PubMed] [Google Scholar]
- 4.Glick BS, Nakano A. Membrane traffic within the Golgi apparatus. Annu Rev Cell Dev Bi. 2009;25:113. doi: 10.1146/annurev.cellbio.24.110707.175421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zerial M, McBride H. Rab proteins as membrane organizers. Nat Rev Mol Cell Bio. 2001;2(2):107–117. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
- 6.Vernoud V, Horton AC, Yang Z, Nielsen E. Analysis of the small GTPase gene superfamily of Arabidopsis. Plant Physiol. 2003;131(3):1191–1208. doi: 10.1104/pp.013052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saito C, Ueda T. Functions of RAB and SNARE proteins in plant life. Int Rev Cell Mol Biol. 2009;274:183–233. doi: 10.1016/S1937-6448(08)02004-2. [DOI] [PubMed] [Google Scholar]
- 8.Asaoka R, Uemura T, Ito J, Fujimoto M, Ito E, Ueda T, Nakano A. Arabidopsis RABA1 GTPases are involved in transport between the trans-Golgi network and the plasma membrane, and are required for salinity stress tolerance. Plant J. 2013;73(2):240–249. doi: 10.1111/tpj.12023. [DOI] [PubMed] [Google Scholar]
- 9.Cheung AY. Rab2 GTPase regulates vesicle trafficking between the endoplasmic reticulum and the golgi bodies and is important to pollen tube growth. Plant Cell. 2002;14(4):945–962. doi: 10.1105/tpc.000836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Batoko H, Zheng H, Hawes C, Moore I. A Rab1 GTPase is required for transport between the endoplasmic reticulum and Golgi apparatus and for normal Golgi movement in plants. Plant Cell. 2000;12(11):2201–17. doi: 10.1105/tpc.12.11.2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng H, Camacho L, Wee E, Batoko H, Legen J, Leaver CJ, Malho R, Hussey PJ, Moore I. A Rab-E GTPase mutant acts downstream of the Rab-D subclass in biosynthetic membrane traffic to the plasma membrane in tobacco leaf epidermis. Plant Cell. 2005;17(7):2020–2036. doi: 10.1105/tpc.105.031112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Speth EB, Imboden L, Hauck P, He SY. Subcellular localization and functional analysis of the Arabidopsis GTPase RabE. Plant Physiol. 2009;149(4):1824–1837. doi: 10.1104/pp.108.132092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Uemura T, Ueda T. Plant vacuolar trafficking driven by RAB and SNARE proteins. Curr Opin Plant Biol. 2014;22:116–121. doi: 10.1016/j.pbi.2014.10.002. [DOI] [PubMed] [Google Scholar]
- 14.Wu YW, Oesterlin LK, Tan KT, Waldmann H, Alexandrov K, Goody RS. Membrane targeting mechanism of Rab GTPases elucidated by semisynthetic protein probes. Nat Chem Biol. 2010;6(7):534–540. doi: 10.1038/nchembio.386. [DOI] [PubMed] [Google Scholar]
- 15.Li F, Yi L, Zhao L, Itzen A, Goody RS, Wu YW. the role of the hypervariable C-terminal domain in Rab. Proc Natl Acad Sci U S A. 2014;111(7):2572–2577. doi: 10.1073/pnas.1313655111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aivazian D, Serrano RL, Pfeffer S. TIP47 is a key effector for Rab9 localization. J Cell Biol. 2006;173(6):917–926. doi: 10.1083/jcb.200510010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kwon SI, Cho HJ, Lee JS, Jin H, Shin SJ, Kwon M, Noh EW, Park OK. Overexpression of constitutively active Arabidopsis RabG3b promotes xylem development in transgenic poplars. Plant Cell Environ. 2011;34(12):2212–2224. doi: 10.1111/j.1365-3040.2011.02416.x. [DOI] [PubMed] [Google Scholar]
- 18.Kwon SI, Cho HJ, Kim SR, Park OK. The Rab GTPase RabG3b positively regulates autophagy and immunity-associated hypersensitive cell death in Arabidopsis. Plant Physiol. 2013;161(4):1722–1736. doi: 10.1104/pp.112.208108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.George S, Parida A. Over-expression of a Rab family GTPase from phreatophyte Prosopis juliflora confers tolerance to salt stress on transgenic tobacco. Mol Biol Rep. 2011;38(3):1669–1674. doi: 10.1007/s11033-010-0278-9. [DOI] [PubMed] [Google Scholar]
- 20.Tuskan GA, Difazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N, Ralph S, Rombauts S, Salamov A, et al. The genome of black cottonwood, Populus trichocarpa (Torr. & gray) Science. 2006;313(5793):1596–1604. doi: 10.1126/science.1128691. [DOI] [PubMed] [Google Scholar]
- 21.Hardison RC. A brief history of hemoglobins: plant, animal, protist, and bacteria. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(12):5675–5679. doi: 10.1073/pnas.93.12.5675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hurst LD. The Ka/Ks ratio: diagnosing the form of sequence evolution. Trends Genet. 2002;18(9):486–487. doi: 10.1016/S0168-9525(02)02722-1. [DOI] [PubMed] [Google Scholar]
- 23.Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290(5494):1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- 24.Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30(1):325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pereira-Leal JB, Seabra MC. Evolution of the Rab family of small GTP-binding proteins. J Mol Biol. 2001;313(4):889–901. doi: 10.1006/jmbi.2001.5072. [DOI] [PubMed] [Google Scholar]
- 26.Inaba T, Nagano Y, Nagasaki T, Sasaki Y. Distinct localization of two closely related Ypt3/Rab11 proteins on the trafficking pathway in higher plants. J Biol Chem. 2002;277(11):9183–9188. doi: 10.1074/jbc.M111491200. [DOI] [PubMed] [Google Scholar]
- 27.Zhang J, Hill DR, Sylvester AW. Diversification of the RAB guanosine triphosphatase family in dicots and monocots. J Integr Plant Biol. 2007;49(8):1129–1141. doi: 10.1111/j.1672-9072.2007.00520.x. [DOI] [Google Scholar]
- 28.Ozeki S, Cheng J, Tauchi-Sato K, Hatano N, Taniguchi H, Fujimoto T. Rab18 localizes to lipid droplets and induces their close apposition to the endoplasmic reticulum-derived membrane. J Cell Sci. 2005;118(12):2601–2611. doi: 10.1242/jcs.02401. [DOI] [PubMed] [Google Scholar]
- 29.Moore I, Schell J, Palme K. Subclass-specific sequence motifs identified in Rab GTPases. Trends Biochem Sci. 1995;20(1):10–12. doi: 10.1016/S0968-0004(00)88939-2. [DOI] [PubMed] [Google Scholar]
- 30.Ren M, Xu G, Zeng J, De Lemos-Chiarandini C, Adesnik M, Sabatini DD. hydrolysis of GTP on rab11 is required for the direct delivery of transferrin from the pericentriolar recycling compartment to the cell surface but not from sorting endosomes. Proc Natl Acad Sci U S A. 1998;95(11):6187–6192. doi: 10.1073/pnas.95.11.6187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ahn CS, Han JA, Pai HS. Characterization of in vivo functions of Nicotiana benthamiana RabE1. Planta. 2013;237(1):161–172. doi: 10.1007/s00425-012-1760-5. [DOI] [PubMed] [Google Scholar]
- 32.Crowell EF, Bischoff V, Desprez T, Rolland A, Stierhof YD, Schumacher K, Gonneau M, Hofte H, Vernhettes S. Pausing of Golgi bodies on microtubules regulates secretion of cellulose synthase complexes in Arabidopsis. Plant Cell. 2009;21(4):1141–1154. doi: 10.1105/tpc.108.065334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liepman AH, Wightman R, Geshi N, Turner SR, Scheller HV. Arabidopsis - a powerful model system for plant cell wall research. Plant J. 2010;61(6):1107–1121. doi: 10.1111/j.1365-313X.2010.04161.x. [DOI] [PubMed] [Google Scholar]
- 34.Lycett G. The role of Rab GTPases in cell wall metabolism. J Exp Bot. 2008;59(15):4061–4074. doi: 10.1093/jxb/ern255. [DOI] [PubMed] [Google Scholar]
- 35.Ahuja I, de Vos RC, Bones AM, Hall RD. Plant molecular stress responses face climate change. Trends Plant Sci. 2010;15(12):664–674. doi: 10.1016/j.tplants.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 36.Bogdanove AJ, Martin GB. AvrPto-dependent Pto-interacting proteins and AvrPto-interacting proteins in tomato. Proc Natl Acad Sci U S A. 2000;97(16):8836–8840. doi: 10.1073/pnas.97.16.8836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ebine K, Fujimoto M, Okatani Y, Nishiyama T, Goh T, Ito E, Dainobu T, Nishitani A, Uemura T, Sato MH, et al. A membrane trafficking pathway regulated by the plant-specific RAB GTPase ARA6. Nat Cell Biol. 2011;13(7):853–859. doi: 10.1038/ncb2270. [DOI] [PubMed] [Google Scholar]
- 38.Agarwal PK, Agarwal P, Jain P, Jha B, Reddy MK, Sopory SK. Constitutive overexpression of a stress-inducible small GTP-binding protein PgRab7 from Pennisetum glaucum enhances abiotic stress tolerance in transgenic tobacco. Plant Cell Rep. 2008;27(1):105–115. doi: 10.1007/s00299-007-0446-0. [DOI] [PubMed] [Google Scholar]
- 39.Pérez-Salamó I, Papdi C, Rigó G, Zsigmond L, Vilela B, Lumbreras V, Nagy I, Horváth B, Domoki M, Darula Z. The heat shock factor A4A confers salt tolerance and is regulated by oxidative stress and the mitogen-activated protein kinases MPK3 and MPK6. Plant Physiol. 2014;165(1):319–334. doi: 10.1104/pp.114.237891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fei L, Zhang H, Zhao H, Gao T, Song A, Jiang J, Chen F, Chen S. Chrysanthemum CmHSFA4 gene positively regulates salt stress tolerance in transgenic chrysanthemum. Plant Biotechnol J. 2017. 10.1111/pbi.12871. [DOI] [PMC free article] [PubMed]
- 41.Zhu J-K. Cell signaling under salt, water and cold stresses. Curr Opin Plant Biol. 2001;4(5):401–406. doi: 10.1016/S1369-5266(00)00192-8. [DOI] [PubMed] [Google Scholar]
- 42.Xu J, Li Y, Wang Y, Liu H, Lei L, Yang H, Liu G, Ren D. Activation of MAPK kinase 9 induces ethylene and camalexin biosynthesis and enhances sensitivity to salt stress in Arabidopsis. J Biol Chem. 2008;283(40):26996–27006. doi: 10.1074/jbc.M801392200. [DOI] [PubMed] [Google Scholar]
- 43.Yu L, Nie J, Cao C, Jin Y, Yan M, Wang F, Liu J, Xiao Y, Liang Y, Zhang W. Phosphatidic acid mediates salt stress response by regulation of MPK6 in Arabidopsis thaliana. New Phytol. 2010;188(3):762–773. doi: 10.1111/j.1469-8137.2010.03422.x. [DOI] [PubMed] [Google Scholar]
- 44.Larkin MA, Blackshields G, Brown N, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 45.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hu B, Jin J, Guo AY, Zhang H, Luo J, Gao G. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics. 2014;31(8):1296–7. doi: 10.1093/bioinformatics/btu817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19(9):1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smoot ME, Ono K, Ruscheinski J, Wang P-L, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2010;27(3):431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tian T, Liu Y, Yan H, You Q, Yi X, Du Z, Xu W, Su Z. agriGO v2. 0: a GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 2017;45(W1):W122–W129. doi: 10.1093/nar/gkx382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu B, Wang L, Zhang J, Li J, Zheng H, Chen J, Lu M. WUSCHEL-related Homeobox genes in Populus tomentosa: diversified expression patterns and a functional similarity in adventitious root formation. BMC Genomics. 2014;15(1):296. doi: 10.1186/1471-2164-15-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang J, Li JB, Liu BB, Zhang L, Chen J, Lu MZ. Genome-wide analysis of the Populus Hsp90 gene family reveals differential expression patterns, localization, and heat stress responses. BMC Genomics. 2013;14(1):532. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Rab gene families in P. trichocarpa. (DOCX 24 kb)
Figure S1. Gene structures and motif composition of PtRabs. (A) The exon-intron structures of PtRab genes. The numbers (0, 1 and 2) indicate the splicing phases (phase 0, 1 and 2, respectively) of the Rab genes. (B) A schematic representation of conserved motifs in Rab proteins. Four conserved motifs (1–4) are represented by different colors. (TIF 3329 kb)
Figure S2. Conserved motifs and their sequences in PtRab proteins. The four motifs are involved in nucleotide binding and hydrolysis. Mutation of the residues (shown in red asterisk) could be used to generate dominant-negative and constitutively active forms that exhibit altered nucleotide-binding or hydrolysis characteristics, which could be used to investigate Rab function. (TIF 1105 kb)
Figure S3. The number of Rab genes containing various cis-acting elements. Auxin-responsive elements: TGA-element, TGA-box and AuxRR-core; gibberellin-responsive elements: TATC-box, P-box and GARE-motif; abscisic acid (ABA)-responsive elements: CE3 and ABRE; salicylic acid (SA)-responsive element: TCA-element; MeJA-responsive elements: TGACG-motif and CGTCA-motif; ethylene-responsive element: ERE; heat-responsive element: HSE; cold-responsive element: LTR and C-repeat/DRE; MYB binding site involved in drought-inducibility: MBS; defense-related elements: Box-W1 and TC-rich repeats; anoxic-related elements: GC-motif and ARE; wound-responsive element: WUN-motif. (TIF 415 kb)
Figure S4. Various cis-acting elements in responsive to hormone and stresses in Rab promoters. (TIF 1113 kb)
Figure S5. Expression profiles of PtRab genes across different tissues and under various abiotic stresses. Heatmap showing expression of PtRab genes across tissues (A), different stem development/growth stages (B) and under various abiotic stresses (C) in poplar. For abiotic stresses, Stress treatments including: low N, nitrogen limitation; wounding, sampled either one week or 90 h after wounding; EAR, early response to water deficit by 36 h; LMI, long-term (10 day) response to mild stress with soil relative extractable water (REW) at 20–35%; LMO, long-term (10 day) response to moderate stress with soil REW at 10–20%; heat, 42 °C for 6 h; and cold, 4 °C for 10 h. Genotypes including P. fremontii × P. angustifolia clones 1979, 3200 and RM5 (low N and wounding), P. deltoids clones Soligo and Carpaccio (drought), and P. simonii (heat and cold). Color scale represents log2 expression values, green represents low level and red indicates high level of transcript abundances. Blank represents a gene has no corresponding probe sets in the microarray data. The microarray data (GSE21481, GSE21485, GSE13043, GSE16786, GSE17230, GSE41557 and GSE43872) were obtained from NCBI Gene Expression Omnibus (GEO) database. (TIF 3420 kb)
Table S2. Probe sets corresponding to PtRab genes. (DOCX 22 kb)
Table S3. Expression data of 27 PtRab paralogous pairs used for correlation analysis. (XLSX 22 kb)
Figure S6. Summary of Rab and TF subfamilies gene numbers in PtRabs co-expression sub- networks. (TIF 945 kb)
Figure S7. TF enrichment of different subfamilies in PtRabs co-expression sub-networks. (TIF 1210 kb)
Table S4. Functional annotation of genes in PtRabs co-expression network. (XLSX 484 kb)
Table S5. Enriched GO terms of genes in PtRabs sub-networks. (XLSX 16 kb)
Figure S8. Functional classification of genes primarily co-expressed with PtRabE1b. (TIF 1141 kb)
Figure S9. Natural variation and protein structure of PtRabE1b. (A) Identification of SNPs in PtRabE1b from 549 P. trichocarpa individuals. (B) Effects of SNPs located in PtRabE1b. (C) Protein structure of PtRabE1b. Four non-synonymous SNPs affected proteins (I45N, E100V, E168V and S201P) were labelled as yellow. Q74L in motif-2 used for constitutive activation was labelled as green. (TIF 631 kb)
Table S6 qRT-PCR Primers used in this study. (DOCX 18 kb)
Data Availability Statement
All data generated or analyzed during this study are included in this article (and its supplementary information files) or are available from the corresponding author on reasonable request. Protein sequence and phylogeny data, including alignments, have been deposited in the TreeBASE repository (ID: 22857).