Abstract
Accumulation of senescent cells over time contributes to aging and age-related diseases. However, what drives senescence in vivo is not clear. Here we used a genetic approach to determine if spontaneous nuclear DNA damage is sufficient to initiate senescence in mammals. Ercc1-/∆ mice with reduced expression of ERCC1-XPF endonuclease have impaired capacity to repair the nuclear genome. Ercc1-/∆ mice accumulated spontaneous, oxidative DNA damage more rapidly than wild-type (WT) mice. As a consequence, senescent cells accumulated more rapidly in Ercc1-/∆ mice compared to repair-competent animals. However, the levels of DNA damage and senescent cells in Ercc1-/∆ mice never exceeded that observed in old WT mice. Surprisingly, levels of reactive oxygen species (ROS) were increased in tissues of Ercc1-/∆ mice to an extent identical to naturally-aged WT mice. Increased enzymatic production of ROS and decreased antioxidants contributed to the elevation in oxidative stress in both Ercc1-/∆ and aged WT mice. Chronic treatment of Ercc1-/∆ mice with the mitochondrial-targeted radical scavenger XJB-5–131 attenuated oxidative DNA damage, senescence and age-related pathology. Our findings indicate that nuclear genotoxic stress arises, at least in part, due to mitochondrial-derived ROS, and this spontaneous DNA damage is sufficient to drive increased levels of ROS, cellular senescence, and the consequent age-related physiological decline.
List of abbreviations: cPus, Cyclopurine DNA lesions; CuZnSOD, Cu/Zn superoxide dismutase; DMPO, 5,5-dimethyl-1-pyrroline-N-oxide; ERCC1, Excision repair cross-complementing group 1; GFAP, Glial fibrillary acidic protein; GSH, Glutathione; GSSG, Glutathione disulfide; IST, Immuno-spin trapping; MnSOD, Manganese superoxide dismutase; NOX, NADPH oxidase; qPCR, Quantitative real-time polymerase chain reaction; ROS, Reactive oxygen species; SA-, Senescence-associated β-galactosidase; SAHFs, Senescence-associated heterochromatic foci; TEMPO, 2,2,6,6-tetramethylpiperdine-1-oxyl; WT, Wild-type; XO, Xanthine oxidase; XPF, Xeroderma pigmentosum complementation group F protein
Keywords: Reactive oxygen species, Free radicals, Genotoxic stress, Oxidative lesions, Endogenous DNA damage, Cellular senescence, Aging
Graphical abstract
Highlights
-
•
Spontaneous damage to the nuclear genome is sufficient to drive cellular senescence.
-
•
Physiological levels of DNA damage mediate increases in ROS and senescence.
-
•
Increased ROS in progeroid and aged mice is multi-factorial and pathologic.
-
•
A mitochondrial radical scavenger reduces nuclear DNA damage and senescence.
1. Introduction
Aging is the primary risk factor for the majority of chronic diseases; hence, aging is now being considered as a therapeutic target [1]. However, this remains a challenge as the precise molecular mechanisms underpinning aging are not well defined. Cellular senescence was recently established to play a causal role in aging [2] and many age-related diseases [3], [4], [5], [6], [7], [8]. Senescence is a programmed cell fate characterized by growth arrest, a metabolic shift, resistance to apoptosis and often a secretory phenotype [9]. The senescent cell burden increases with age in virtually all vertebrates [10], [11], [12]. In replicating human cells, shortened telomeres drive senescence [13]. It has become increasingly clear that non-replicating cells also undergo senescence [14]. However, in non-dividing cells, which are the majority of cells in mammalian organisms, the cause of senescence is not clear.
A variety of cellular stressors including genotoxic, proteotoxic, inflammatory and oxidative have been implicated in driving senescence [9], [15]. However, senescence itself is associated with many of these cellular stressors [16], making it very difficult to decipher cause and effect. For example, DNA damaging agents definitively cause increased senescence (e.g. in cancer patients) [17]. Yet senescent cells are defined by persistent activation of the DNA damage response [18], increased expression of surrogate markers of DNA damage [19] and are able to trigger genotoxic stress in neighboring cells [16]. Therefore, in vivo, the importance of DNA damage as a driver of senescence and aging is debated [20].
Even less is known about endogenous DNA damage as a potential driver of senescence and aging. The vast majority of evidence implicating DNA damage in senescence comes from experiments implementing very high doses of environmental genotoxins such as ionizing radiation, doxorubicin, etoposide or cisplatin [19], [21], [22]. Also of note, all genotoxins damage not only DNA, but also all cellular nucleophiles including phospholipids, proteins and RNA. Thus, it remains unknown whether physiological levels of spontaneous DNA damage is sufficient to drive cellular senescence.
A major source of endogenous DNA damage is reactive oxygen species (ROS) produced during mitochondrial-based aerobic metabolism (e.g. the superoxide anion (O2•-) and the hydroxyl radical (•OH) produced from O2•-or H2O2 via the Fe2+-dependent Fenton or Haber- Weiss reaction) [23]. The DNA lesions caused by ROS include oxidized bases, abasic sites, single-strand breaks and lipid peroxidation-induced adducts such as interstrand crosslinks [24]. Some mitochondrial-derived ROS, such as H2O2, can diffuse throughout the cell, resulting in oxidative damage to lipids, proteins, RNA and DNA [25]. Thus, mitochondrial dysfunction, which leads to an increase in ROS production, was proposed to be central to the aging process [26], [27]. However, this too remains controversial [28].
To address these gaps in knowledge, we utilized a genetic approach to increase endogenous nuclear DNA damage in mice. ERCC1-XPF is an endonuclease complex required for nucleotide excision repair, interstrand crosslink repair and the repair of a subset of DNA double-strand breaks [29]. Mutations that mediate reduced expression of this enzyme cause accelerated aging in humans and mice [29]. ERCC1 is required to stabilize XPF in vivo [30]. Therefore, Ercc1-/Δ mice, with one knock-out and one hypomorphic allele of Ercc1 have 5–10% of the normal complement of ERCC1-XPF [31]. Genetic depletion of DNA repair mechanisms does not increase the amount of damage incurred, it simply accelerates the pace at which damage triggers a demonstrable physiological impact, affording an opportunity to investigate the role of endogenous nuclear DNA damage in driving senescence.
Here, we demonstrate that Ercc1−/Δ mice accumulate oxidative DNA damage and senescent cells more rapidly than age-matched wild-type (WT) controls, yet comparable to WT mice over two years of age. Surprisingly, we found that Ercc1−/Δ mice are also under increased oxidative stress. Increased ROS production and decreased antioxidant buffering capacity contributed to the oxidative stress, which was also observed in aged WT mice. Treatment of Ercc1-/Δ mice with a mitochondrial-targeted radical scavenger (XJB-5–131) was sufficient to suppress oxidative DNA damage, senescence and age-related pathologies. These data demonstrate that damage of the nuclear genome arising spontaneously in vivo is sufficient to drive cellular senescence. Our data also demonstrate that endogenous DNA damage, as a primary insult, is able to trigger increased reactive oxygen species (ROS) and further oxidative damage in vivo.
2. Methods
2.1. Chemicals and reagents
All chemicals were purchased from Sigma-Aldrich (St. Louis, MO) unless otherwise specified. Primary antibodies used for immunoblotting were purchased from Abcam (Cambridge, MA) unless indicated.
2.2. Animal care and experimentation
All animal studies were conducted in compliance with the U.S. Department of Health and Human Services Guide for the Care and Use of Laboratory Animals, and were approved by the Scripps Florida or University of Pittsburgh Institutional Animal Care and Use Committee. Ercc1-/∆ mice were bred and genotyped as previously described [32]. P16-luciferase reporter mice were obtained from Ohio State University [10] and bred to create Albino C57BL/6 p16luc/+;Ercc1+/- and FVB/n p16luc/+;Ercc1+/∆ mice. These mice were further crossed to create f1 p16luc/luc;Ercc1-/Δ mice with white fur for imaging. All animals were genotyped from an ear punch by TransnetYX (Cordova, TN).
2.3. DNA extraction and measurement of cyclopurine DNA lesions
DNA was isolated using a high-salt extraction method [33] from cultured MEFs or liver tissue, which was pulverized with a mortar and pestle under liquid nitrogen. Cyclopurine lesions were measured by LC-MS/MS/MS using an LTQ linear ion trap mass spectrometer using our recently described conditions with some modifications [34]. Nuclease P1 (0.1 U/μg DNA), phosphodiesterase 2 (0.000125 U/μg DNA), 20 nmol of erythro-9-(2-hydroxy-3-nonyl) adenine EHNA and a 20-μL solution containing 300 mM sodium acetate (pH 5.6) and 10 mM zinc chloride were added to isolated nuclear DNA. In this context, EHNA served as an inhibitor for deamination of 2’-deoxyadenosine to 2'-deoxyinosine induced by adenine deaminase [34]. The above digestion mixture was incubated at 37˚C for 48 h. To this mixture were then added alkaline phosphatase (0.05 U/μg DNA), phosphodiesterase 1 (0.00025 U/μg DNA) and 40 μL of 0.5 M Tris-HCl buffer (pH 8.9). The digestion was continued at 37˚C for 2 h and subsequently neutralized by addition of formic acid. To the mixture were then added appropriate amounts of uniformly 15N-labeled standard lesions, which included R-cdG, S-cdG, R-cdA and S-cdA. The digestion mixture was subsequently extracted twice with chloroform. The resulting aqueous layer was subjected to off-line high performance liquid chromatography (HPLC) separation for the enrichment of the lesions under study, following our previously described procedures [34]. The LC-MS/MS/MS experiments were conducted using an LTQ linear ion trap mass spectrometer using our recently described conditions with some modifications [34]. Briefly, a 0.5 × 150 mm Zorbax SB-C18 column (particle size, 5 µm, Agilent) was used for the separation of the above-enriched lesion fractions, and the flow rate was 4.0 μL/min. A solution of 0.1% (v/v) formic acid in water (solution A) and a solution of 0.1% (v/v) formic acid in methanol (solution B) were used as mobile phases for the analyses of all four cyclopurine lesions, i.e. the (5′R) and (5′S) diastereomers of cdA and cdG, after HPLC enrichment, and a gradient of 5 min 0–20% B, 30 min 20–80% B, and 5 min 80% B was employed for the separation.
2.4. Fluorescence in situ hybridization for telomere-specific γH2AX foci
Primary murine embryonic fibroblasts (MEFs) were fixed with 2% paraformaldehyde for 15 min followed by permeabilization (0.2% Triton X-100in PBS) for 15 min. Cells were then blocked (2% BSA, 20% goat serum in PBS) for 2 h. Cells were immuno-stained with mouse anti-γH2AX monoclonal antibody (1:500; Upstate, Billerica, MA) overnight and goat anti-mouse 594 secondary antibody (1:1000) for 1 h. Cells were then fixed in 2% paraformaldehyde for 5 min. Samples were dehydrated in 70%, 95%, 100% ethanol (5 min each) and then denatured for 10 min at 80 °C in hybridization solution (70% deionized formamide, 10% NEN blocking reagent [Roche], 0.1 M Tris-HCl [pH 7.4], MgCl2 buffer [82 mM NaH2PO4, 9 mM citric acid, 20 mM MgCl2], and 0.5 µg/mL Cy3-OO-(CCCTAA)3 PNA probe (Panagene, South Korea). After 2 h hybridization at room temperature, the samples were washed twice with 70% deionized formamide in 10 mM Tris-HCl, pH 7.2. Samples were counterstained with DAPI, mounted onto slides with Gelvatol and images were acquired with a Nikon A1 confocal microscope (Nikon Instruments, Inc.).
2.5. Senescence-associated β-galactosidase (SA-β-gal) staining of tissue
Fresh tissues were fixed in 10% neutral buffered formalin (NBF) for 3–4 h and then transfered to 30% sucrose overnight. Tissues were then embedded in cryo-embedding media (OCT) and cryosectioned at 6 µm for SA-β-gal staining (pH 5.8) at 37oC for 16–24 h in SA-β-gal staining solution (pH 6.0; 40 mM citric acid in sodium phosphate buffer, 5 mM K4[Fe(CN)6] 3H2O, 5 mM K3[Fe(CN)6], 150 mM sodium chloride, 2 mM magnesium chloride and 1 mg/mL X-gal dissolved in N,N-dimethylformamide).
2.6. IVIS in vivo imaging detection of luciferase activity
Isoflurane-anesthetized mice were injected intraperitoneally with D-luciferin substrate (Caliper Life Sciences, Hopkinton, MA; 15 mg/mL in PBS) and were imaged by using an IVIS Lumina (Caliper Life Sciences) as previously described [10].
2.7. RNA isolation and qPCR
Tissues were harvested from euthanized animals and snap frozen in liquid nitrogen. Tissues were homogenized using FastPrep-24 homogenizer (MP Biomedicals, Solon, OH) and total RNA was isolated using Trizol, according to manufacturer’s specifications (Thermo Fisher, Waltham, MA). Total RNA was quantified using a Nanodrop spectrophotometer (Thermo Fisher) and 1 μg of total RNA was used to generate cDNA with the Transcriptor First Strand cDNA synthesis kit (Roche, Basel Switzerland) according to the manufacturer’s specification. Gene expression changes in p16 was quantified by qPCR reactions using 20 μL reaction volumes using a StepOne thermocycler (Thermo Fisher) with input of 100 ng cDNA per reaction. Reactions were performed in duplicate (n = 4–12 mice per group). Data was analyzed by ΔΔCt method and expression was normalized to Gapdh. Primer sequences are as follows: Cdkn2a (p16) Fwd 5’- CCCAACGCCCCGAACT-3’, Cdkn2a (p16) Rev 5’- GCAGAAGAGCTGCTACGTGAA-3’; Gapdh Fwd 5’-AAGGTCATCCCAGAGCTGAA-3’, Gapdh Rev 5’-CTGCTTCACCACCTTCTTGA-3’.
2.8. Biochemical detection of superoxide
Fresh murine tissue slices were incubated in a 30 µM solution of hydroethidine (HE) in PBS for 45 min at 37oC in the dark. The slices were washed with iced cold PBS and placed into a 1.5 mL Eppendorf tube and immediately frozen by immersion in liquid nitrogen. Superoxide levels were measured by the presence of 2-hydroxyethidium (2-OH-E+) using a HPLC system equipped with electrochemical detector as previously reported [35], [36]. Briefly, the separation of the oxidized products of hydroethidine (HE) was performed using an ether-linked phenyl column (Phenomenex, 100 X 4.6 mm, 2.6 µm) and a gradient elution method using two mobile phases with an increasing fraction of acetonitrile (from 25% to 60% over 10 min). The presence of superoxide was also confirmed by electron paramagnetic resonance (EPR) spectroscopy spin-trapping of a 1-hydroxy-3-methoxy-carbonyl-2,2,5,5-tetramethylpyrrolidine (CMH) (Noxygen Science Transfer and Diagnostics, Elzach, Germany) superoxide-sensitive probe and analyzed using a temperature- and O2-controlled Bruker EPR (Millerica, MA) at 37oC as described [37]. n = 3–9 mice per genotype.
2.9. Immuno-spin trapping of biomolecular free radicals
Briefly, mice were injected with 500 mg/kg 5,5-dimethyl-1-pyrroline N-oxide (DMPO, Dojindo, Japan) at 24, 12 and 6 h prior to euthanasia. Tissues were fixed in 2% PFA in PBS for 1 h then submerged in 30% sucrose for 24 h, with several solution exchanges. Tissues were cryopreserved in 2-methylbutane then sectioned on a cryostat (Leica Biosystems, Richmond, IL). Sections were stained with polyclonal anti-DMPO (ALX-210–530-R050; Enzo Life Sciences) followed by secondary antibody (Alexa Fluor 488 anti-rabbit IgG; Life Technologies). Tissues were counterstained with DAPI to detect nuclei and for actin (fluor-conjugated phalloidin) to reveal tissue architecture. For liver, 9 x 9 image sections were stitched together from multiple images with 10% stitching overlap using the Nikon NIS-Elements software. 3–5 mice were used per group.
2.10. Lipid peroxidation
4-Hydroxynonenal-protein adducts, which are by-products of lipid peroxidation, were measured in murine liver using the OxiSelect HNE Adduct Competitive ELISA kit (Cell Biolabs, San Diego, CA). Livers lysates were prepared in RIPA buffer and normalized based on protein concentration. µg of total protein was used for each assay. Four liver samples were measured in duplicate for each group except old WT mice (n = 3–4). Measurements were taken using an EnVision plate reader (Perkin Elmer, Waltham, MA).
2.11. Xanthine oxidase activity
Xanthine oxidase activity was measured as previously described [38]. Briefly, liver samples (50 mg) from 7 to 9 mice per group were homogenized in ice-cold potassium phosphate buffer (50 mM, pH 7.4) and incubated in the presence of 200 µM xanthine and 100 µM oxonic acid for 60 min at 37oC (with and without 200 µM allopurinol). Accumulation of uric acid over this time (above that observed in the presence of allopurinol) frame was assessed via reverse phase HPLC coupled to an electrochemical detector (ESA CoulArray, Chelmsford, MA), (1 Unit = 1 µmole urate/min). Similarly, XO activity was measured from the serum of mice (n = 3–9).
2.12. NADPH oxidase activity
Tissue O2•- production was calculated from the initial linear rate of SOD-sensitive cytochrome c reduction quantified at λ = 550 nm. Briefly, homogenates of frozen liver samples were resuspended in Oxidase Assay Buffer (65 mM sodium phosphate buffer (pH 7.0), 1 mM EGTA, 10 μM FAD, 1 mM MgCl2, 2 mM NaN3, 300 U/mL catalase, and 0.2 mM cytochrome c), in the presence or absence of superoxide dismutase (150 U/mL). After 5 min baseline measurement, NADPH (180 μM) was added and O2•- production was measured at 550 nm using a Biotek Synergy 4 hybrid multimode microplate reader. 6–13 mice per group were used. Data are expressed as fold change from WT.
2.13. Mitochondrial respiration
Mitochondrial respiration in isolated liver mitochondria was measured by the oxygen consumption rate (OCR) using a Seahorse XF96 Extracellular Flux Analyzer (Agilent Seahorse, Santa Clara, CA). Liver mitochondria were isolated as described [39]. 150 μL suspension of liver mitochondria (6 µg protein/well) was plated on a pre-chilled Seahorse PS 96-well microplate reader. The plate was centrifuged at 3220×g for 50 min at 4 °C, subsequently incubated in 37 °C (without CO2) for 15 min then transferred to the XF flux analyzer for respiration measurement. The measurement cycle consisted of a 3 min mixing time and a 4 min time point. After three basal measurements in the presence of complex I substrate pyruvate (5 mM), 150 μM ADP, 2 µg/mL oligomycin (inhibitor of ATP synthase), 4 µM carbonylcyanide p-trifluoromethoxyphenylhydrazone (FCCP; an optimized concentration to give maximum respiratory capacity), 2 µM rotenone and 2 µg/mL antimycin A were auto-injected into the experimental wells, and another three measurement cycles were performed. Each experimental point is an average of a minimum of three replicate wells on four mice per group. State III and maximal respiration were calculated as described [40].
2.14. Metabolite extraction
Liver samples were weighed (10 mg) from 12-week-old WT (n = 7), old WT (120–136-week-old) (n = 7) and 12-week-old Ercc1-/Δ (n = 6) mice. Samples were homogenized in 400 μL methanol/water (80:20 v/v) with 1 mm glass beads (Biospec, Bartlesville, OK, USA) in 1.5 mL glass vials. A Minilyse homogenizer (Bertin Technologies, Montigny le Bretonneux, France) was used for 30 s at 3000 rpm. The samples were sonicated for 15 min and stored overnight at -20oC. The samples were centrifuged at 15,000×g for 15 min at 4oC. The supernatant was transferred to 1.5 mL glass vials and stored at -20oC until later use. The pellet was resuspended in 600 μL acetone and homogenized again for 10 s, and stored at -20oC overnight. The samples were centrifuged at 15,000 ×g for 15 min at 4oC and the supernatant pooled with previously retained supernatant. The samples were dried down in a speedvac and resuspended in 100 μL acetonitrile/water (50/50 v/v), sonicated for 5 min, centrifuged for 15 min at 15,000 ×g, 4oC and transferred to autosampler vials for storage at -80oC until use.
2.15. Global metabolomic analysis
Analyses were performed using a high-performance liquid chromatography (HPLC) system (1200 series, Agilent Technologies) coupled to a 6550 ifunnel quadrupole time-of-flight (Q-TOF) mass spectrometer (Agilent Technologies). Samples were injected (8 μL) onto a Luna Aminopropyl, 3 µm, 150 mm × 1.0 mm I.D. column (Phenomenex, Torrance, CA) for hydrophilic interaction liquid chromatography (HILIC) analysis. Pooled samples were injected every three samples and a blank after every samples for quality control. The standard mobile phase was A = 20 mM ammonium acetate and 40 mM ammonium hydroxide in 95% water and B = 95% acetonitrile in ESI negative mode. The linear gradient elution from 100% B (0–5 min) to 100% A (50–55 min) was applied at a flow rate of 50 μL/min with a 10 min post-run. ESI source conditions were set as following: gas temperature 200˚C, drying gas 11 L/min, nebulizer 15 psig, sheath gas temperature 300˚C, sheath gas flow 9 l/min, fragmentor 360 V, nozzle voltage 500 V, and capillary voltage 2500 V. The instrument was set to acquire over the m/z range 60–1200, with the MS acquisition rate of 2 spectra/s. For the MS/MS of selected precursors the default isolation width was set as medium (~4 m/z), with a MS acquisition rate at 3 spectra/s and MS/MS acquisition at 3 spectra/s. The collision energy was fixed at 20 eV. LC/MS data were processed using XCMS Online [41]. Unpaired parametric tests were carried out. Features were listed in a feature list table and as an interactive cloud plot, containing their integrated intensities (extracted ion chromatographic peak areas) observed fold changes across the two sample groups, and p-values for each sample [42]. Integration of METLIN to XCMS Online allowed for putative identification of metabolites. Identifications were then made by comparing retention times and tandem MS fragmentation patterns to the sample and standard compounds (purchased from Sigma Aldrich, St. Louis, MO).
2.16. Liver proteomic analysis
Male CB6f1, C57BL/6 and female C57BL/6: FVB f1 WT livers were harvested at 5–8 months and 30–32 months, as well as male and female C57BL/6: FVB F1 Ercc1−/Δ livers at 4 months (n = 4–8). Livers were homogenized in 125 mM Tris-HCl, pH 7.6, using a MP Biomedicals Fast Prep 24, Lysing Matrix D. Lysates were brought to 100 mM Tris-HCl, pH 7.6, 4% sodium dodecyl sulfate, and 100 mM dithiothreitol and heated to 99˚C for 5 min. Cooled samples were protein assayed using 660 nm Protein Assay with Ionic Detergent Compatibility Reagent (Pierce, Rockford, IL). Equal protein amounts were dialyzed, alkylated, and digested using the FASP methodology [43], [44]. Briefly, samples were buffer exchanged into 8 M Urea, alkylated with 20 mM iodoacetamide, buffer exchanged into ammonium bicarbonate, and digested with mass spec sequencing grade trypsin (Promega, Madison, WI) in a Millipore Microcon Ultracel YM-30 microcentrifuge filter [45]. Collected peptides were desalted using Discovery DSC-18 vacuum manifold columns with a 50 mg bed weight (Supelco, Bellefonte, PA) [46]. Desalted peptides were dried down in a centrifugal concentrator with inline cold trap (Labconco, Kansas City, MO) [47]. Desalted peptides were resuspended at 1 mg/mL in 0.1% formic acid in mass spectrometry grade water (Burdick & Jackson, Muskegon, MI).
Desalted peptides were separated across a hydrophobicity gradient of 3–32% acetonitrile over 60 min at 300 nL/min using a Waters NanoACQUITY (ultra-high pressure liquid chromatography) UPLC on a 25 cm, 75 µm ID, 5 µM reversed phase C18 heated PicoChip column (New Objective) in line with a high resolution Fourier transform Orbitrap XL mass spectrometer (Thermo Fisher Scientific). A top 4 data dependent acquisition was employed with a 60,000 resolution full scan and four subsequent low resolution MS/MS identification scans performed in the ion trap.
Mass spectrometric raw files were translated and analyzed using the CHORUS cloud computing label free quantitation analysis suite (chorusproject.org). Briefly, chromatographic peaks, features, are separated from noise and placed into appropriate isotope groups before alignment across all samples and quantification using label free differential mass spectrometry [48], [49]. Identification is performed using the Comet and Percolator MS/MS identification engines compared to the Uniprot reference data set for Mus musculus, generating identification and quantification data for all features [50], [51].
Statistical analysis was performed on all identified features by rejecting any feature not found in at least 75% of samples following outlier removal. Feature level data was brought to protein level by taking the median level of all unmodified, unique peptides of a given protein per sample, with a minimum of two unique peptides per protein. A two-tailed Student’s t-test was performed to establish statistical significance for all proteins identified.
2.17. Catalase activity
Catalase activity in liver tissue was determined as previously reported [52]. 50 μg of liver lysate from each mouse were used and analyzed in duplicate. Detection of peroxide (Fisher Scientific, Pittsburgh, PA) at 240 nm was performed using a Cary 300 BIO UV–VIS spectrophotometer (Varian, Palo Alto, CA) at 30 s intervals for a total of 1 min. Catalase activity per milligram of protein (k/mg) was quantified using the following formula: k/mg = [3 ln (Absinitial/Absfinal)] /[milligrams of protein * time] with 3–5 mice were used per group.
2.18. Superoxide dismutase activity
SOD activity (mitochondrial and cytoplasmic) was quantified using the Superoxide Dismutase Assay Kit (Cayman) per the manufacturer’s instructions. All liver samples were normalized based on protein concentration with n = 3 per group.
2.19. Glutathione analysis
Livers were harvested from euthanized mice, fixed in 5% sulfosalicylic acid and extracts were prepared by homogenization in MES buffer (0.2 M 2-(N-morpholino) ethanesulphonic acid, 0.05 M phosphate and 1 mM EDTA, pH 6.0) to prevent post-mortem oxidation of glutathione [53]). Samples were normalized by protein concentration and analyzed for concentration of total GSH and GSSG using a Glutathione Assay Kit (Cayman Chemicals, Ann Arbor, MI) per the manufacturer’s specifications. Sample absorbance was measured at 405 nm using a plate reader. Equation to determine reduced GSH was [Reduced GSH] = [Total GSH]-[GSSG], and ratio was reported as [Reduced GSH]/[GSSG] [54]. n = 3–14 mice per age/genotype.
2.20. Immunoblotting
Liver and kidney samples from 18-week-old Ercc1−/Δ and WT mice (n = 5) were homogenized in RIPA buffer (Pierce, Rockford, IL) with protease inhibitor cocktail (Roche, Indianapolis, IN). Mitochondrial extracts were prepared using Mitochondria Isolation Kit (Pierce) per the manufacturer’s specifications. Samples were separated on 4–20% polyacrylamide gel (Bio-Rad, Hercules, CA), transferred to nitrocellulose membrane, blocked and blotted with anti-PCNA (PC10, Santa Cruz Biotechnology, Santa Cruz, CA), anti-ERCC1 (D-10, Santa Cruz Biotechnology), anti-COXIV (Abcam, Cambridge, MA), anti-XPF (SPM228, Novus Biologicals, Littleton, CO) or anti-GAPDH, anti-MnSOD, anti-CuZnSOD, anti-catalase (3H3L29), anti-XO, and anti-rabbit secondary (all from Life Technologies, Carlsbad, CA) then visualized with ECL reagent (Pierce). Films exposed to membrane were imaged with ImageJ (NIH, Bethesda, MD). GAPDH was used as a loading control.
2.21. Chronic treatment of mice with XJB-5–131
The Ercc1-/Δ mice were given intraperitoneal injections of 2 mg/kg XJB dissolved in sunflower oil (S5007 Sigma-Aldrich, St. Louis, MO) or an equal volume of vehicle only (sunflower oil) three times per week, beginning at five weeks of age, by an investigator blinded to the treatment group. Whenever possible, littermate pairs of Ercc1-/Δ mice were used, with one mouse in each treatment group, to minimize variability. The mice were weighed twice a week and monitored for the onset of age-related symptoms, including dystonia, trembling, ataxia, priapism and urinary incontinence (neurodegenerative symptoms), hind-limb muscle wasting, lethargy (reduced spontaneous activity) and kyphosis (hunched posture). Data from littermate pairs were evaluated to determine the fraction of symptoms delayed in the mouse treated with XJB vs. its sibling treated with vehicle only using a paired Student’s t-test. All mice were euthanized at 20 weeks of age and their tissues were isolated for pathological analysis.
2.22. Micro-computed tomography measurement of bone density
μCT of spines was acquired as previously described [55] using a VivaCT 40 (Scanco USA Inc.) with 15-μm isotropic voxel size resolution, 55 kVp of energy, and 145 μA of current. After the acquisition of transverse 2-dimensional image slices, 3-dimensional reconstruction of the lumbar vertebrae was performed using a constant threshold value of 235, which was selected manually for the bone voxels by visually matching the threshold areas to the gray-scale images.
2.23. Statistics
The mean and standard deviation or standard error of the mean were calculated for all experimental groups and analyzed using unpaired two-tailed Student’s t-tests, or one-way or two-way ANOVA or Tukey's test for multiple comparisons using GraphPad Prism 6.
3. Results
3.1. Ercc1−/Δ mice have accelerated accumulation of spontaneous oxidative DNA damage
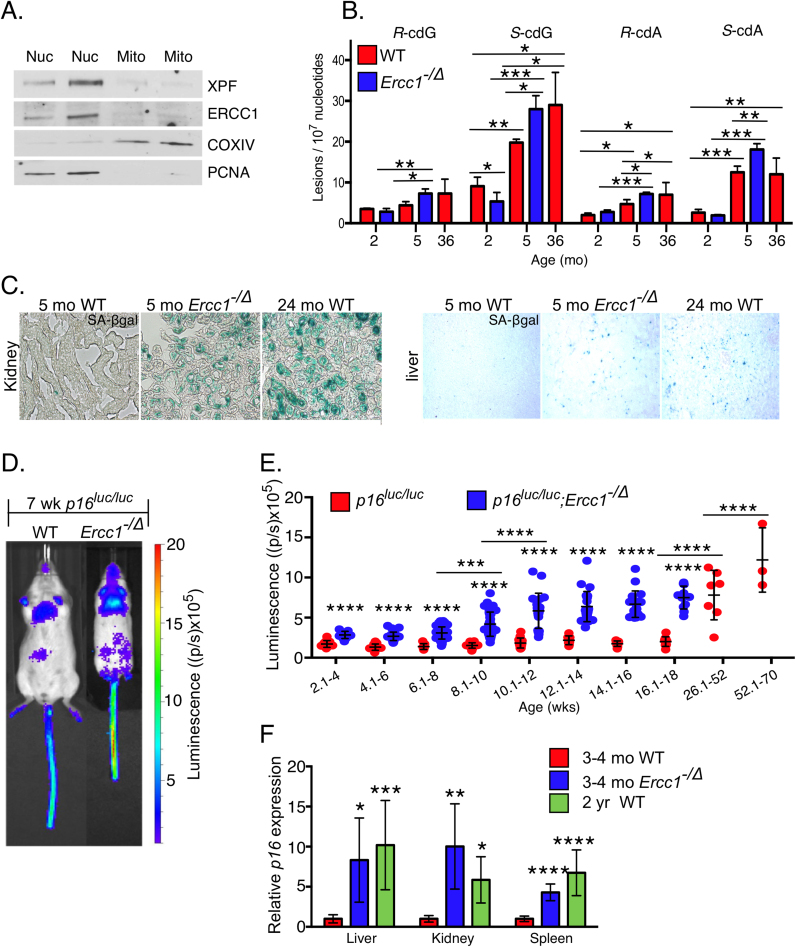
To test conventional wisdom that nucleotide excision repair (NER) is exclusively nuclear and not a mitochondrial DNA repair mechanism [56], ERCC1 and XPF protein levels were measured in fractionated murine liver lysates (Fig. 1A). Both proteins were detected in the nuclear but not mitochondrial fractions, establishing their role in protecting the nuclear genome, exclusively. Cyclopurines (cPus) are DNA lesions generated by endogenous reactive oxygen species [57], which are repaired by NER [58]. Thus, cPus are expected to be increased in Ercc1−/Δ mice compared to age-matched WT animals. LC-MS/MS/MS was used to measure the four cPus lesions (R-cdG, S-cdG, R-cdA and S-cdA) in kidney tissue of mice (Supplemental Fig. 1) [59]. At two months of age, the levels of cPu were not elevated in Ercc1−/Δ mice (Fig. 1B). By five months of age, however, all four lesions were significantly increased in Ercc1−/Δ compared to WT mice. Notably, S-cdG, R-cdA and S-cdA also were significantly increased in old WT mice compared to young animals. Furthermore, adduct levels were equivalent in 5-month-old Ercc1−/Δ and 3-year-old WT mice. This indicates that Ercc1−/Δ mice have an increased burden of endogenous DNA damage than age-matched repair-proficient animals and that they accumulate spontaneous DNA damage faster than WT animals.
Fig. 1.
DNA repair deficient Ercc1-/Δ mice accumulate oxidative damage and senescent cells faster than WT mice. (a) Immunoblot detection of ERCC1 and XPF in fractionated liver lysates from two WT mice. COXIV was used as a loading control for the mitochondria (Mito) and PCNA was used for the nuclear (Nuc) fraction. (b) Levels of 8,5’-cyclopurine-2’-deoxynucleosides in DNA isolated from murine kidney. Graphed are the mean and SD from n = 3 mice per group. *p < 0.05, **p < 0.01, ***p < 0.001 calculated by two-way ANOVA. Data are derived from Wang et al. [59]. (c) Staining for SA-β-gal activity on kidney and liver from Ercc1-/Δ mice and aged WT mice compared to adult WT mice. Images were captured at 20X magnification. (d) Representative images of p16-luciferase signal in age-matched WT and a DNA repair-deficient mouse. (e) Total body luciferase activity in p16luc/luc;Ercc1-/Δ (blue) and p16luc/luc (red) mice with increasing age. Dots represent individual animals. Black bars indicate the mean ± standard deviation. p values were calculated using a two-way ANOVA. ***p < 0.001, ****p < 0.0001. * over the blue dots indicate significant differences between the WT and Ercc1-/Δ mice. * over the black bars indicate a significant difference between Ercc1-/Δ of different age groups. (f) qPCR detection of p16Ink4a expression in liver (n = 6–12), kidney (n = 4–6) and spleen (n = 7–10) of Ercc1-/∆ mice (blue), age-matched WT mice (red) and old WT mice (green). Values represent the mean ± SD, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 determined by one-way ANOVA with Tukey's test.
3.2. Ercc1−/Δ mice have accelerated accumulation of senescent cells
To determine if endogenous DNA damage is sufficient to drive cellular senescence in vivo, multiple markers of senescence were measured in tissues of Ercc1-/∆ and WT mice of various ages. Senescence-associated β − galactosidase (SA-β-gal) activity was increased in 5-month-old Ercc1-/Δ mouse kidney and liver compared to WT littermates (Fig. 1C). Two-year-old WT mice also had increased SA-β-gal activity in these tissues relative to young animals. The level of p16Ink4a expression was measured using a p16Ink4a-luciferase transgenic reporter (Fig. 1D) [10]. Total p16Ink4a-luciferase expression was modestly but significantly increased in mutant animals at weaning (Fig. 1E). The signal increased steadily as the mutant animals aged, in particular, after 8 weeks of age. Notably, the signal level seen in the DNA repair-deficient mice did not exceed that of older WT mice. As both WT and Ercc1-/∆ mice aged, the heterogeneity in signal between animals increased dramatically, as previously reported for WT mice in a different genetic background [10].
Increased p16Ink4a expression was validated by qPCR (Fig. 1F). p16Ink4a mRNA was significantly greater in the liver, kidney and spleen of Ercc1-/∆ mice compared to WT age-matched controls. p16Ink4a expression in 3–4 month-old Ercc1-/∆ mice was comparable to that of 2-year-old WT mice. Taken together, these data document the premature accumulation of senescent cells in DNA repair-deficient Ercc1-/∆ mice. Importantly, ERCC1-XPF-deficient human and murine cells do not show accelerated telomere attrition [60]. To confirm the absence of telomere dysfunction, we measured telomere damage-induced foci [61] in Ercc1-/- and WT mouse embryonic fibroblasts (Supplemental Fig. 2). Notably, the number of γH2AX foci was significantly increased in Ercc1-/- cells compared to WT, as expected for DNA repair-deficient cells. However, γH2AX foci at telomeric DNA was significantly lower in Ercc1-/- cells, confirming prior studies [60]. These data rule-out telomere dysfunction as the driver of senescence in the absence of ERCC1-XPF. This suggests that it is unrepaired, endogenous DNA damage that drives cellular senescence in mammalian tissues.
3.3. Ercc1-/∆ mice demonstrate elevated ROS abundance
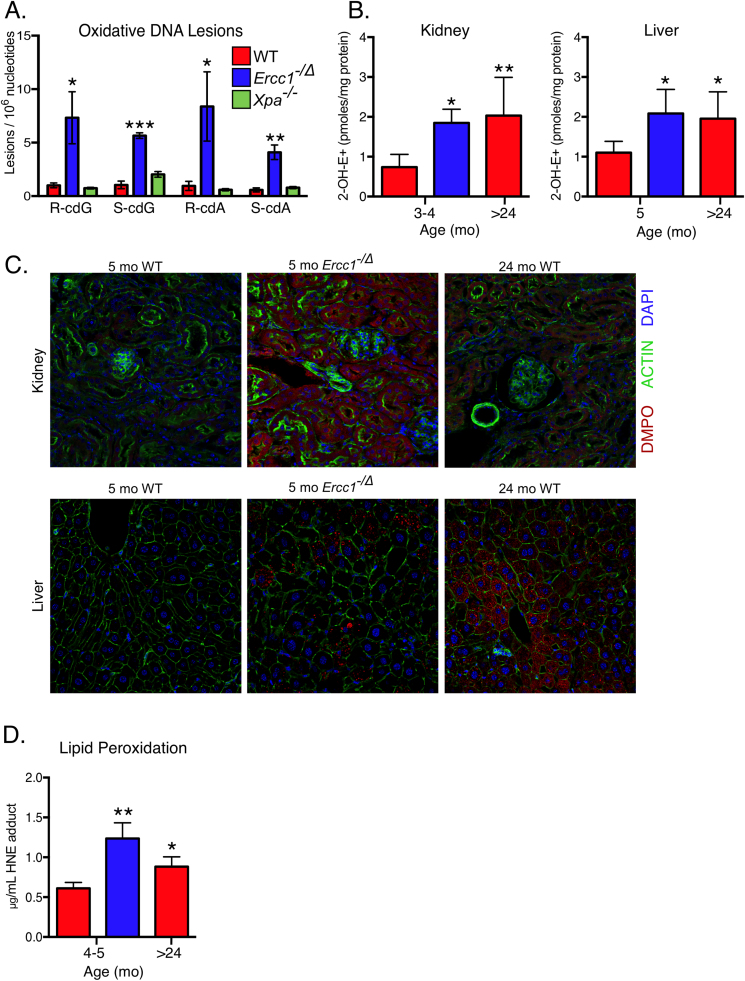
The presumption is that Ercc1-/∆ mice have increased oxidative DNA damage because of their defect in NER. However, it is also possible that Ercc1-/∆ mice are under increased oxidative stress. To test this, we measured cPus in liver tissue of age-matched Ercc1-/∆, WT and Xpa-/- mice. The latter are completely deficient in nucleotide excision repair of cyclopurine (cPu) lesions. Xpa-/- mice also show no signs of accelerated aging [62]. Notably, all four cPus were significantly elevated in liver of Ercc1-/∆ mice compared to WT, but cPus were not elevated in Xpa-/- mouse liver (Fig. 2A). This indicates that lack of DNA repair does not adequately explain the increased oxidative DNA damage in Ercc1-/∆ mice.
Fig. 2.
Increased oxidative stress in tissues of progeroid Ercc1-/Δ mice and old WT mice. (a) Levels of four cyclopurine lesions (R-cdG, S-cdG, R-cdA, S-cdA) measured in liver tissues of 4-month-old WT, Xpa-/- and Ercc1-/Δ mice (n = 3 per genotype) by LC-MS/MS/MS. (b) Detection of endogenous superoxide production by quantifying 2-OH-E+ by HPLC/electrochemical analysis in DHE-treated kidney (n = 3–7) and liver (n = 6–9 animals per genotype/age). (c) Representative images from immuno-spin trapping of endogenous, biomolecular free radicals with 5,5-dimethyl-1-pyrroline-N-oxide (DMPO). The level of oxidant stress was determined by immunodetection of DMPO-adducted biomolecules in renal and liver sections. DMPO staining is illustrated in red, actin in green to illustrate tissue architecture and DAPI in blue to highlight cell nuclei. (d) Lipid peroxidation as measured by quantitation of 4-hydroxynonenal protein adducts via ELISA (n = 3–4 mice per group). For all panels, values represent the mean ± SD, *p < 0.05, **p < 0.01, ***p < 0.001 determined by one-way ANOVA with Tukey's test.
To determine if the Ercc1-/∆ mice are under increased oxidative stress, superoxide anion (O2•-) production was measured in fresh renal and liver tissue by quantification of 2-OH-E+. Oxidation of hydroethidine to its O2•--specific product, 2-hydroxyethidium (2-OH-E+) was assessed by HPLC coupled to electrochemical detection and validated by electron paramagnetic resonance (EPR) spin trapping [36]. Superoxide levels were indeed significantly greater in Ercc1−/Δ mouse tissue compared to age-matched controls (Fig. 2B). Interestingly, O2•- levels were equivalent in 4–5-month-old Ercc1−/Δ and 24–30-month-old WT mice, extending the parallels between normal and accelerated aging.
The presence of elevated endogenous ROS levels was confirmed using a second, in vivo method whereby free radicals are detected by immuno-spin trapping with the nitrone EPR spin trap 5,5-dimethyl-1-pyrroline-N-oxide (DMPO). Renal and liver tissue of 5-month-old Ercc1-/Δ mice had increased DMPO adducts compared to age-matched WT mice (Fig. 2C and Supplemental Fig. 3). Similarly, DMPO signal intensity was elevated in naturally aged WT mouse liver and kidney compared to 5-month-old WT mice.
As a product of lipid peroxidation, 4-hydroxy-2-nonenal (HNE) chemically modifies proteins and DNA [63] and thus is frequently used as a measure of oxidative stress [64]. HNE-protein adducts were significantly elevated in liver lysates of Ercc1−/Δ and old WT mice compared to young adult WT mice (Fig. 2D). Cumulatively, these data provide multiple lines of evidence that Ercc1−/Δ mice are under increased oxidative stress, analogous to what occurs with normal aging.
3.4. Increased ROS production as a source of oxidative stress
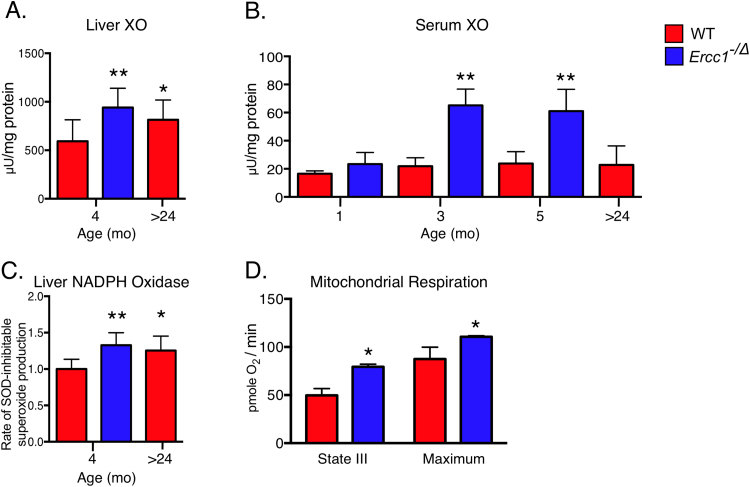
We next examined potential source(s) of increased ROS abundance. The enzymatic activity of xanthine oxidase (XO), a key endogenous enzymatic a source of O2•- and H2O2 [65], was significantly increased in the liver of 4-month-old Ercc1−/Δ mice compared to age-matched WT animals (Fig. 3A). Aged WT mice also demonstrated elevated liver XO activity as previously reported [66]. Notably, XO activity was similar in the progeroid and aged WT mice. Ercc1−/Δ mice also demonstrated elevated serum XO activity compared to WT littermate controls (Fig. 3B). The activity of circulating XO was not elevated at 1 month of age but was increased by 3 months when the mice display aging phenotypes [55]. XO activity was not significantly increased in serum of aged WT mice, in contrast to a previous report [66]. NADPH oxidase (NOX) activity, another key endogenous source of O2•- and H2O2, was significantly elevated in the liver tissue of Ercc1−/Δ and old WT mice compared to young WT animals (Fig. 3C). These data indicate that the enhanced O2•- levels detected in liver and kidney of the progeroid and aged WT mice is likely caused, at least in part, by increased enzymatic production.
Fig. 3.
Increased production of ROS in progeroid Ercc1-/Δ mice and old WT mice. Xanthine oxidase (XO) activity was measured in the (a) livers (n = 7–9 per group) and (b) serum of Ercc1−/Δ and WT mice (n = 3–4 per group) at multiple ages. (c) NADPH oxidase activity was measured in the livers of 4-month-old Ercc1−/Δ and WT mice as well as 24-month-old aged WT mice (n = 6–13 per group). (d) Measurement of mitochondrial respiration using a Seahorse Bioscience XF Analyzer on mitochondria isolated from liver tissues of 2 month-old Ercc1−/Δ and WT mice (n = 4 per group). Values represent the mean ± SD, *p < 0.05, **p < 0.01 as determined by one-way ANOVA with Tukey's test or unpaired two-tailed Student’s t-test.
In addition, a shift towards increased oxidative phosphorylation and oxygen consumption has been reported with aging [67], which is another potential source of increase O2•- production [68]. To determine if this was also the case in the progeroid Ercc1-/Δ mice, we measured mitochondrial respiration in organelles isolated from the liver of adult mutant mice and their littermate controls (Fig. 3D). Indeed, ADP-stimulated respiration and maximum respiration was significantly higher in the Ercc1−/Δ mice compared to age-matched controls; consistent with previous findings in cells from DNA repair-deficient mice [69]. This metabolic shift could serve as another source of increased O2•- production by mitochondria [70].
3.5. Decreased antioxidant activity and levels contribute to excess ROS
Several approaches were used to measure antioxidant status of progeroid and aged mice. Untargeted mass spectrometry (MS)-based metabolomics on liver extracts comparing 3-month-old WT and Ercc1−/Δ mice revealed 7935 aligned features, of which 118 were significantly changed between the two groups. Comparison of liver tissues from 3-month-old and 2.5-year-old WT mice revealed 6812 aligned features, of which 69 were significantly changed between the two groups. Metabolites were identified from both analyses using a combination of MS/MS with spectrum matching on the METLIN database, and confirmed using authentic standards. One of the key nodes identified by metabolomics as significantly altered with accelerated and normal aging was glutathione metabolism (Fig. 4A), a key antioxidant and index of oxidative stress [71].
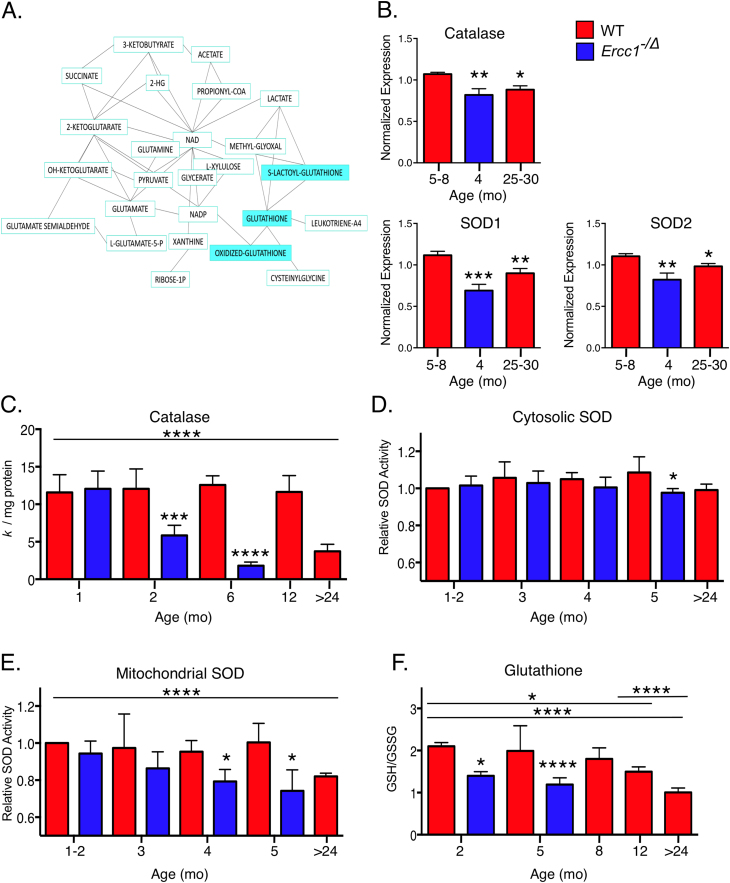
Fig. 4.
Reduced antioxidant capacity in progeroid Ercc1-/Δ mice and old WT mice. (a) Liver metabolite string analysis of 3-month-old WT (n = 6) versus Ercc1−/Δ mice (n = 7). Metabolites shaded in blue are significantly more abundant in the mutant animals. (b) Unbiased differential proteomic analysis of liver from 5 to 8-month-old adult WT, 25–30-month-old aged WT and 4-month-old progeroid Ercc1−/Δ mice (n = 4–8) revealed a significant decrease in the abundance of antioxidant proteins. (c) Catalase activity in the liver of Ercc1−/Δ and WT mice at various ages (n = 3–5 per group). (d) Cytosolic and (e) mitochondrial superoxide dismutase (SOD) activity in the liver of Ercc1−/Δ and WT mice at various ages (n = 3 per group). (f) The ratio of oxidized glutathione to reduced glutathione (GSH/GSSG) in livers from Ercc1−/Δ and WT mice of various ages (n = 3–14 per group). Values represent the mean ± SD, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 determined by one-way ANOVA with Tukey’s test.
Differential MS was used for proteomics analysis to identify redox-related proteins significantly altered in the livers of 3–4 month-old progeroid Ercc1−/Δ mice and old WT mice (>2 years-old) vs. adult WT mice. Expression of catalase, SOD1 (CuZnSOD) and SOD2 (MnSOD) were significantly reduced in Ercc1−/Δ and old WT mice compared to young adult WT mice (Fig. 4B). In fact, numerous proteins affecting redox status were identified as altered in mutant mice, including aconitase 1, cytochrome c oxidase, ATP citrate lyase and microsomal glutathione s-transferase 1 (Supplemental Fig. 4). A very similar pattern of expression changes occurred in old WT mice, relative to younger animals. The MS data were validated by immunodetection of several antioxidants by immunoblot (Supplemental Fig. 5).
To validate the predictions arising from the omics studies, activity of key antioxidants was measured. In liver tissue, catalase activity was significantly decreased in Ercc1−/Δ mice compared to age-matched controls (Fig. 4C). Interestingly, in young mutant animals (1 month-old), catalase activity was normal, but then declined progressively over the rest of their lifespan, reaching a level in the 4–6 month-old Ercc1−/Δ mice comparable to that of 2.5-year-old WT mice. SOD1 (CuZnSOD-cytosolic) and SOD2 (MnSOD-mitochondrial) activity in Ercc1−/Δ was similar to WT mice until they reached 4–5 months of age whereby the enzymatic activity of CuSOD and MnSOD were significantly lower than that of age-matched controls (Fig. 4D-E). Similarly, MnSOD activity was lower in 2.5-year-old WT animals (Fig. 4E).
The reduced form of glutathione (GSH) is the active antioxidant and becomes oxidized to glutathione disulfide (GSSG). A decreased ratio of GSH/GSSG is indicative of a state of oxidative stress as well as antioxidant depletion. The GSH/GSSG was significantly reduced in 2 month-old Ercc1−/Δ mice compared to age-matched controls and increased further by 5 months of age (Fig. 4F). In WT mice, the GSH/GSSG ratio was significantly decreased at one year of age compared to younger animals, then diminished further by 2 years of age, as previously reported in numerous tissues of rodents [72]. These data provide multiple lines of evidence indicating that, in addition to increased ROS production, there is a significant decline in antioxidant buffering capacity in Ercc1−/Δ mice, likely contributing to the enhanced levels of O2•- detected in liver and kidney of the progeroid mice. The parallels between the Ercc1−/Δ mice and aged WT mice suggest a role for spontaneous DNA damage and ROS in normal aging as well.
3.6. A mitochondrial-targeted radical scavenger suppresses senescence
To determine if the increased oxidative stress plays a causal role in driving senescence, Ercc1-/∆ mice were treated with the mitochondrial-targeted free radical scavenger XJB-5–131. XJB-5–131 is a conjugate between the nitroxide TEMPO (2,2,6,6-tetramethylpiperidine-1-oxyl) and the mitochondrial-targeting moiety gramicidin S (Supplemental Fig. 6A) [73]. TEMPO, a stable free radical, is a potent antioxidant due to its proclivity for mimicking SOD in vitro. XJB-5–131 has several advantages over other classes of antioxidants, including its capacity to recycle (Supplemental Fig. 6B), direct acceptance of electrons from the mitochondria respiratory complexes to prevent production of ROS, plus SOD-, catalase- and peroxidase-mimetic activities to neutralize existing ROS [73], [74], [75]. XJB-5–131 localizes to mitochondria within 1 h in primary cell cultures [76] and is enriched 600-fold in the mitochondria fraction of cells over the cytosol [77], [78].
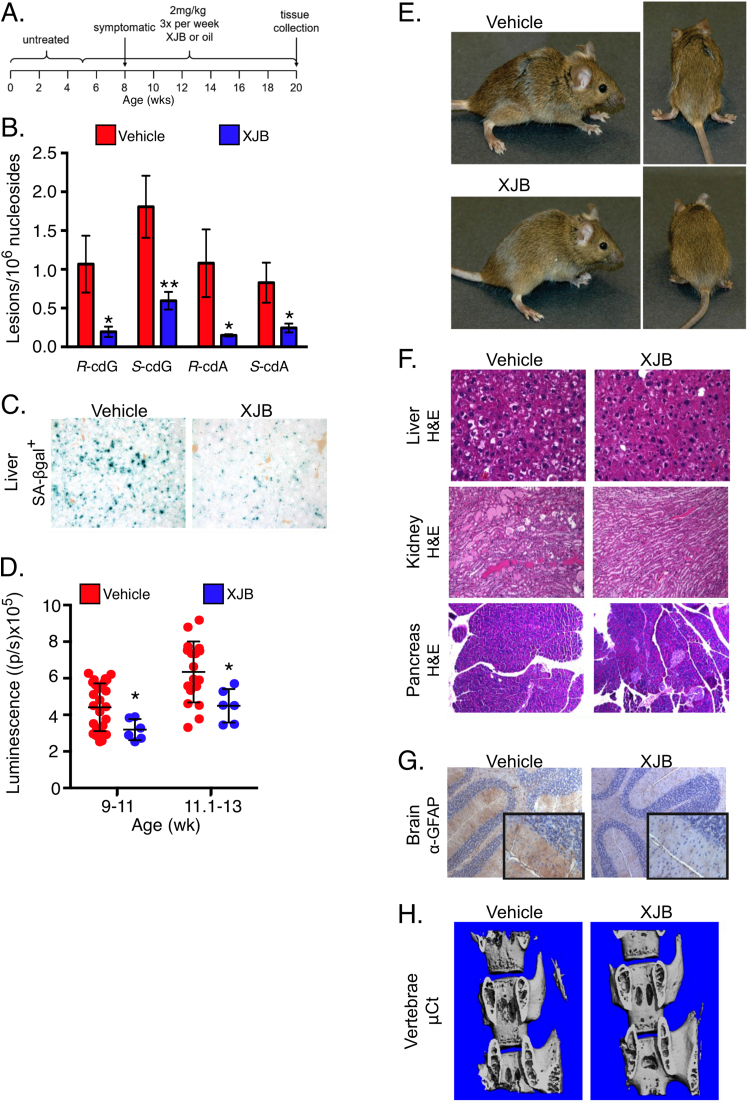
Ercc1-/Δ mice were treated with 2 mg/kg XJB-5–131 or vehicle control, delivered by intraperitoneal injection beginning at 5 weeks of age (Fig. 5A). Chronic exposure of Ercc1-/Δ mice to XJB-5–131 significantly reduced the levels of cPus oxidative DNA lesions in liver tissue (Fig. 5B). XJB-5–131 also suppressed the accumulation of senescent cells. SA-β-gal staining was reduced in the liver of Ercc1-/Δ mice treated with XJB-5–131 (Fig. 5C). Similarly, XJB-5–131 significantly reduced the luciferase signal in Ercc1-/∆;p16Ink4a-luciferase reporter mice (Fig. 5D). Taken together, these results implicate mitochondria-derived ROS as driving endogenous DNA damage and senescence in vivo.
Fig. 5.
A mitochondrial-targeted radical scavenger suppresses endogenous DNA damage, senescence and aging. (a) Schematic diagram of the treatment regimen with XJB-5–131. Littermate pairs of mutant mice were administered either vehicle (sunflower seed oil) or 2 mg/kg XJB-5–131 three times per week for 15 weeks, i.p., starting at five weeks of age. (b) Oxidative DNA damage in the liver of Ercc1-/Δ mice treated with XJB-5–131 or vehicle only (n = 3 per group) was measured by LC-MS/MS/MS detection of cyclopurine adducts (R-cdG, S-cdG, R-cdA, S-cdA) in genomic DNA. Tissues were collected from 20-week-old animals at the end of the study. (c) Representative images of SA-β-gal staining of liver sections from vehicle- or XJB-treated mice. (d) Total body luciferase activity was measured in p16luc/+;Ercc1-/Δ mice treated with 8 mg/kg XJB-5–131, i.p., 3X per week for 4.5 weeks and plotted relative to the signal in Ercc1-/Δ mice treated with vehicle only. Dots represent individual animals. Graphed is the mean ± SD. *p < 0.05 determined by an unpaired two-tailed Student’s t-test. (e) Representative images of 20-week-old Ercc1-/Δ mice (siblings) treated with XJB-5–131 or vehicle only. The vehicle treated mouse shows greater ataxia (splayed-foot gait) and hind-limb wasting than the treated animal. (f) Representative images of H&E stained sections of liver, kidney and pancreas. XJB-5–131 had less necrosis and ballooning degeneration of hepatocytes in liver, fewer hyaline casts in the renal tubules and more islets in the pancreas, compared to mice administered vehicle only. (g) Immuno-stain detection of glial fibrillary acidic protein (GFAP, a marker of neurodegeneration) in cerebellar sections of XJB-treated mice compared to vehicle only. Nuclei were counter-stained with hematoxylin. (h) MicroCT analysis of the vertebral column to detect osteoporotic changes in bone.
3.7. A mitochondrial-targeted radical scavenger suppresses aging symptoms and pathology
XJB-5–131-treated mice were monitored daily for the onset of progeroid symptoms by an investigator blinded to the treatment groups. Mice treated with XJB-5–131 exhibited a significant delay in the onset of dystonia and ataxia, as well as kyphosis, reduced spontaneous activity and hind-limb muscle wasting (Table 1 and Fig. 5E). Seventy percent of the age-related symptoms measured were significantly delayed in the Ercc1-/Δ mice treated with XJB-5–131 compared to their vehicle-treated controls. A sixth symptom (urinary incontinence) was not observed in the 6 male mice in the treatment group, but was seen in 2 of 5 males in the vehicle-only group. XJB-5–131 treatment delayed the onset of symptoms by 1–2 weeks in Ercc1-/Δ mice which is equivalent to 5–8 years in humans, based on a median lifespan of 84 years.
Table 1.
Chronic administration of XJB-5–131 delays the onset of age-related functional decline in Ercc1-/Δ mice.
|
Age at onset (weeks) |
||||
|---|---|---|---|---|
| Symptom | Ercc1-/Δ+ vehicle | na | Ercc1-/Δ+ XJB-5–131 | nb |
| Dystonia | 7.4 | 8 | 8.6* | 10 |
| Trembling | 7.7 | 8 | 8.9 | 10 |
| Kyphosis | 11.7 | 8 | 13.2* | 10 |
| Ataxia | 14.5 | 8 | 16.3* | 10 |
| Hind-limb wasting | 14.0 | 8 | 16.0* | 10 |
| Reduced spontaneous activity | 17.5 | 5 | 20.9* | 5 |
| Urinary incontinence | 12.4 | 2 | n/a | 0 |
| Fraction of symptoms delayed | 10% | 8 | 66% | 8 |
Ercc1-/Δ mice + vehicle; n = 8 mice, 5 ♂; 2 ♀; n indicates the number of mice showing that symptom.
Ercc1-/Δ mice + XJB-5–131; n = 10 mice, 6 ♂; 4 ♀.
Individual symptoms that were significantly delayed in mice treated with XJB-5–131; p < 0.05 one-tailed Student’s t-test.
Mice from both treatment groups were euthanized at 20 wks of age for histopathological analyses. At this age, Ercc1-/Δ mice display significant levels of age-related hepatic lesions including necrosis and ballooning degeneration [79]. Both lesions were reduced in Ercc1-/Δ mice treated with XJB-5–131 compared to those treated with vehicle only (Fig. 5F). Age-related changes in the kidney, including hyaline casts, glomerular and tubule-interstitial injury and inflammation were attenuated by XJB-5–131. The drug also delayed the loss of pancreatic islets in Ercc1-/Δ mice. The brains of XJB-5–131 treated mice showed reduced staining for glial fibrillary acidic protein (GFAP), a marker of neurodegeneration (Fig. 5G). Finally, microcomputed tomography of the spine revealed a significant reduction in osteoporotic changes in Ercc1-/Δ mice treated with XJB-5–131 (Fig. 5H and Supplemental Fig. 7). This demonstrates that a mitochondrial-targeted radical scavenger is sufficient to attenuate endogenous oxidative DNA damage, cellular senescence and aging.
4. Discussion
Although cellular senescence has been demonstrated to drive aging [2], it is not known what endogenous processes are primarily responsible for causing cellular senescence in mammals, particularly in post-mitotic tissues. Here, we used mice in which DNA repair was attenuated genetically. By definition, the primary insult in untreated Ercc1-/Δ mice is unrepaired endogenous DNA damage to the nuclear genome. Not surprisingly, the Ercc1-/Δ mice accumulate senescent cells more rapidly than WT mice. This formally demonstrates that physiologically-relevant types and levels of endogenous DNA damage are able to trigger the time-dependent accumulation of senescent cells.
The surprising discovery is that there is increased ROS in tissues of the Ercc1-/Δ mice. This reveals that spontaneous, endogenous nuclear DNA damage can instigate oxidative stress. We found that elevated ROS is likely due, at least in part, to increased enzymatic production by xanthine oxidase (XO) and NADPH oxidase (NOX), altered mitochondrial metabolism, as well as an attrition of the expression and activity of several key antioxidants, catalase, MnSOD and glutathione. Similar events were found in aged WT mice, consistent with prior studies [80], [81], [82], [83]. The dramatic parallels between the progeroid and naturally aged mice suggest that oxidative stress is an important common denominator in aging.
To determine if this oxidative stress is pathological, we suppressed it pharmacologically in Ercc1-/Δ mice with the mitochondrial-targeted radical scavenger XJB-5–131. Chronic administration XJB-5–131 significantly reduced both oxidative DNA damage and senescence (Fig. 5). The reduced level of senescent cells corresponded to a reduction in age-related morbidity. This is consistent with numerous recent studies demonstrating that genetic or pharmacologic elimination of senescent cells slows age-related decline [2], [4], [7], [8], [84], [85], [86]. The observation that suppressing oxidant production is sufficient to decreases senescence indicates that reactive species are required to ultimately cause or maintain senescence in response to genotoxic stress.
Analogous to our work, recent studies demonstrated increased NOX activity in cells from patients with genome instability disorders such as ataxia telangiectasia (AT) and Nijmegen breakage syndrome [87], as well as Atm-/- mice [88] that model AT. Interestingly, in worms, NOX triggers transcriptional activation of stress response mechanisms [89]. Indeed, increased ROS in Atm-/- mice appears to be pathological [88], [90] as well as a critical signaling mechanism both up and downstream of ATM [91], [92], [93]. Increased ROS has also been reported in cells from xeroderma pigmentosum and Cockayne syndrome patients [94], [95], two diseases caused by a defect in nucleotide excision repair, but the source of ROS is unclear.
Our results are consistent with the oxidative stress theory of aging originally proposed by Denham Harman [26], and the notion that a vicious cycle of ROS generation and oxidative damage is the ultimate driver of aging [27]. Our data also indicate that endogenous nuclear DNA damage is able to trigger this cycle of escalating ROS abundance, oxidative damage, senescent cell accumulation and age-related pathology.
Numerous studies counter the oxidative stress theory of aging. Notably, overexpression of MnSOD, which detoxifies O2•-, does not extend the lifespan of mice [96], [97]. This appears at odds with our data indicating that XJB-5–131 improves the health of Ercc1-/Δ mice. However, XJB-5–131 is able to both prevent ROS production and neutralize existing ROS. The nitroxide radical form of XJB-5–131 can be reduced to a hydroxylamine by accepting an electron from the electron transport chain and subsequent protonation (Supplemental Fig. 6B), thereby preventing electron transfer to O2 and resultant ROS production [98]. Hydroxylamines act as robust reducing agents by hydrogen atom transfer to free radicals and non-radicals such as O=NOO-, resulting in a significant diminution of oxidative/nitrosative stress. While admittedly, reaction of hydroxylamine with O2•- will result in H2O2 generation, it will neutralize O=NOO- and by default, this process regenerates the nitroxide form of XJB-5–131, thus recycling the radical scavenger [75]. In contrast, SOD solely reduces O2•- levels, yet generates H2O2 in the process (2O2•- → H2O2 + O2) which is likely not well-neutralized in older organisms due to the significant reduction in catalase expression and activity (Fig. 4B-C and Supplemental Fig. 5). Thus, in the presence of elevated levels of mitochondrial O2•-, overexpression of MnSOD may yield increased formation of H2O2; an oxidant that is freely diffusible and thus capable of mediating oxidative damage distant from the mitochondrion. This phenomenon has indeed been reported [99] and the combination of SOD with CAT has been shown to afford greater protection against Fe2+-induced radical formation than SOD alone [100].
Another argument against the oxidative stress theory of aging is based on the PolγD257A mice, which age rapidly due to increased mutations in the mitochondrial genome caused by inactivation of the exonuclease domain of the mitochondrial polymerase γ [101]. Unlike the progeroid Ercc1−/Δ mice, ROS is not increased in tissues of PolγD257A mice, countering the notion that oxidative stress drives aging. However, mutagenesis in the PolγD257A mice is stochastic and there are thousands of mitochondrial genomes per cell, making it plausible that phenotypes are less consistent in PolγD257A mice than in Ercc1−/Δ mice where every cell is affected. Therefore, the lack of increased ROS in PolγD257A mice was not considered to counter the mitochondrial theory of aging [101].
In conclusion, we demonstrate that spontaneous, endogenous, nuclear DNA damage leads to an accelerated accumulation of senescent cells in vivo. In addition, we provide novel evidence supporting the oxidative stress theory of aging. In a mammalian system where spontaneous endogenous DNA damage is the primary insult, cellular senescence and ROS abundance is increased, leading to further damage and senescence. Attenuating mitochondrial-ROS defuses this cycle and suppresses age-related decline, implicating it as causative. Taken together, this supports the potential of radical scavengers as a treatment for age-related co-morbidities.
Acknowledgements
This work was supported by the National Institutes of Health [grant numbers P01-AG043376, ES016114, P20 GM109098, K99-AG049126, R00AG036817, CA076541, CA101864, AG044376, AI068021, P30AG024827, 5P20GM109098 and P30CA047904]. LJN had additional funding from the Ellison Medical Foundation (AG-NS-0303-05).
Acknowledgments
Conflict of interest
The authors declare no conflicts of interest.
Author contributions
SQG, AUG, HFS and SM contributed data that led to this manuscript. ARR, CHF, JST, MJY and SJM conducted the in vivo experiments. JW and YW measured cyclopurine adducts. EEK, MJY, TAR, NCM, MAR, SCW, ECP, PJP and CMS measured endogenous ROS. EMS, MCF and PW synthesized, purified and characterized XJB-5-131. CEB developed the p16-luciferase mouse strain and helped with imaging analysis. CET, MJY and NFL contributed to measuring senescence in tissues. LHR did the histopathological analysis. LAN and NVV measured bone density. HBT and NAY did proteomic analysis. CHJ and GS performed metabolomic analysis. JC, BT, RASR and MJY measured oxidative damage. XL measured mitochondrial respiration. HPV and PLO measured TIFs. NVV, CMS, DBS, CEB, PW, YW, PDR and LJN oversaw various aspects of the research. ARR, SQG, AUG, HFS, MJY, PDR, EEK and LJN contributed to the manuscript preparation.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.redox.2018.04.007.
Contributor Information
Eric E. Kelley, Email: eric.kelley@hsc.wvu.edu.
Laura J. Niedernhofer, Email: lniedern@scripps.edu.
Appendix A. Supplementary material
Supplementary material
References
- 1.Burd C.E., Gill M.S., Niedernhofer L.J., Robbins P.D., Austad S.N., Barzilai N., Kirkland J.L. Barriers to the Preclinical Development of Therapeutics that Target Aging Mechanisms. J. Gerontol. A Biol. Sci. Med Sci. 2016;71(11):1388–1394. doi: 10.1093/gerona/glw112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker D.J., Childs B.G., Durik M., Wijers M.E., Sieben C.J., Zhong J., Saltness R.A., Jeganathan K.B., Verzosa G.C., Pezeshki A., Khazaie K., Miller J.D., van Deursen J.M. Naturally occurringp16(Ink4a)-positive cells shorten healthy lifespan. Nature. 2016;530(7589):184–189. doi: 10.1038/nature16932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Childs B.G., Baker D.J., Wijshake T., Conover C.A., Campisi J., van Deursen J.M. Senescent intimal foam cells are deleterious at all stages of atherosclerosis. Science. 2016;354(6311):472–477. doi: 10.1126/science.aaf6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Farr J.N., Xu M., Weivoda M.M., Monroe D.G., Fraser D.G., Onken J.L., Negley B.A., Sfeir J.G., Ogrodnik M.B., Hachfeld C.M., LeBrasseur N.K., Drake M.T., Pignolo R.J., Pirtskhalava T., Tchkonia T., Oursler M.J., Kirkland J.L., Khosla S. Targeting cellular senescence prevents age-related bone loss in mice. Nat. Med. 2017;23(9):1072–1079. doi: 10.1038/nm.4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jeon O.H., Kim C., Laberge R.M., Demaria M., Rathod S., Vasserot A.P., Chung J.W., Kim D.H., Poon Y., David N., Baker D.J., van Deursen J.M., Campisi J., Elisseeff J.H. Local clearance of senescent cells attenuates the development of post-traumatic osteoarthritis and creates a pro-regenerative environment. Nat. Med. 2017;23(6):775–781. doi: 10.1038/nm.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ogrodnik M., Miwa S., Tchkonia T., Tiniakos D., Wilson C.L., Lahat A., Day C.P., Burt A., Palmer A., Anstee Q.M., Grellscheid S.N., Hoeijmakers J.H.J., Barnhoorn S., Mann D.A., Bird T.G., Vermeij W.P., Kirkland J.L., Passos J.F., von Zglinicki T., Jurk D. Cellular senescence drives age-dependent hepatic steatosis. Nat. Commun. 2017;8:15691. doi: 10.1038/ncomms15691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roos C.M., Zhang B., Palmer A.K., Ogrodnik M.B., Pirtskhalava T., Thalji N.M., Hagler M., Jurk D., Smith L.A., Casaclang-Verzosa G., Zhu Y., Schafer M.J., Tchkonia T., Kirkland J.L., Miller J.D. Chronic senolytic treatment alleviates established vasomotor dysfunction in aged or atherosclerotic mice. Aging Cell. 2016;15(5):973–977. doi: 10.1111/acel.12458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schafer M.J., White T.A., Iijima K., Haak A.J., Ligresti G., Atkinson E.J., Oberg A.L., Birch J., Salmonowicz H., Zhu Y., Mazula D.L., Brooks R.W., Fuhrmann-Stroissnigg H., Pirtskhalava T., Prakash Y.S., Tchkonia T., Robbins P.D., Aubry M.C., Passos J.F., Kirkland J.L., Tschumperlin D.J., Kita H., LeBrasseur N.K. Cellular senescence mediates fibrotic pulmonary disease. Nat. Commun. 2017;8:14532. doi: 10.1038/ncomms14532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kirkland J.L., Tchkonia T. Cellular senescence: a translational perspective. EBioMedicine. 2017;21:21–28. doi: 10.1016/j.ebiom.2017.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burd C.E., Sorrentino J.A., Clark K.S., Darr D.B., Krishnamurthy J., Deal A.M., Bardeesy N., Castrillon D.H., Beach D.H., Sharpless N.E. Monitoring tumorigenesis and senescence in vivo with a p16(INK4a)-luciferase model. Cell. 2013;152(1–2):340–351. doi: 10.1016/j.cell.2012.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herbig U., Ferreira M., Condel L., Carey D., Sedivy J.M. Cellular senescence in aging primates. Science. 2006;311(5765):1257. doi: 10.1126/science.1122446. [DOI] [PubMed] [Google Scholar]
- 12.Liu Y., Sanoff H.K., Cho H., Burd C.E., Torrice C., Ibrahim J.G., Thomas N.E., Sharpless N.E. Expression ofp16(INK4a) in peripheral blood T-cells is a biomarker of human aging. Aging Cell. 2009;8(4):439–448. doi: 10.1111/j.1474-9726.2009.00489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.d'Adda di Fagagna F., Reaper P.M., Clay-Farrace L., Fiegler H., Carr P., Zglinicki T. Von, Saretzki G., Carter N.P., Jackson S.P. A DNA damage checkpoint response in telomere-initiated senescence. Nature. 2003;426(6963):194–198. doi: 10.1038/nature02118. [DOI] [PubMed] [Google Scholar]
- 14.van Deursen J.M. The role of senescent cells in ageing. Nature. 2014;509(7501):439–446. doi: 10.1038/nature13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Childs B.G., Durik M., Baker D.J., van Deursen J.M. Cellular senescence in aging and age-related disease: from mechanisms to therapy. Nat. Med. 2015;21(12):1424–1435. doi: 10.1038/nm.4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nelson G., Kucheryavenko O., Wordsworth J., von Zglinicki T. The senescent bystander effect is caused by ROS-activated NF-kappaB signalling. Mech. Ageing Dev. 2018;170:30–36. doi: 10.1016/j.mad.2017.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cupit-Link M.C., Kirkland J.L., Ness K.K., Armstrong G.T., Tchkonia T., LeBrasseur N.K., Armenian S.H., Ruddy K.J., Hashmi S.K. Biology of premature ageing in survivors of cancer. ESMO Open. 2017;2(5):e000250. doi: 10.1136/esmoopen-2017-000250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fumagalli M., Rossiello F., Mondello C., d'Adda di Fagagna F. Stable cellular senescence is associated with persistent DDR activation. PloS One. 2014;9(10):e110969. doi: 10.1371/journal.pone.0110969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rodier F., Munoz D.P., Teachenor R., Chu V., Le O., Bhaumik D., Coppe J.P., Campeau E., Beausejour C.M., Kim S.H., Davalos A.R., Campisi J. DNA-SCARS: distinct nuclear structures that sustain damage-induced senescence growth arrest and inflammatory cytokine secretion. J. Cell Sci. 2011;124(Pt 1):68–81. doi: 10.1242/jcs.071340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sierra F. Is (your cellular response to) stress killing you? J. Gerontol. A Biol. Sci. Med Sci. 2006;61(6):557–561. doi: 10.1093/gerona/61.6.557. [DOI] [PubMed] [Google Scholar]
- 21.Papadopoulou A., Kletsas D. Human lung fibroblasts prematurely senescent after exposure to ionizing radiation enhance the growth of malignant lung epithelial cells in vitro and in vivo. Int J. Oncol. 2011;39(4):989–999. doi: 10.3892/ijo.2011.1132. [DOI] [PubMed] [Google Scholar]
- 22.Chang B.D., Broude E.V., Dokmanovic M., Zhu H., Ruth A., Xuan Y., Kandel E.S., Lausch E., Christov K., Roninson I.B. A senescence-like phenotype distinguishes tumor cells that undergo terminal proliferation arrest after exposure to anticancer agents. Cancer Res. 1999;59(15):3761–3767. [PubMed] [Google Scholar]
- 23.Barnes D.E., Lindahl T. Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annu Rev. Genet. 2004;38:445–476. doi: 10.1146/annurev.genet.38.072902.092448. [DOI] [PubMed] [Google Scholar]
- 24.Yu Y., Cui Y., Niedernhofer L.J., Wang Y. Occurrence, biological consequences, and human health relevance of oxidative stress-induced DNA damage. Chem. Res Toxicol. 2016;29(12):2008–2039. doi: 10.1021/acs.chemrestox.6b00265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cadenas E., Davies K.J. Mitochondrial free radical generation, oxidative stress, and aging. Free Radic. Biol. Med. 2000;29(3–4):222–230. doi: 10.1016/s0891-5849(00)00317-8. [DOI] [PubMed] [Google Scholar]
- 26.Harman D. Origin and evolution of the free radical theory of aging: a brief personal history, 1954–2009. Biogerontology. 2009;10(6):773–781. doi: 10.1007/s10522-009-9234-2. [DOI] [PubMed] [Google Scholar]
- 27.Jang Y.C., Van Remmen H. The mitochondrial theory of aging: insight from transgenic and knockout mouse models. Exp. Gerontol. 2009;44(4):256–260. doi: 10.1016/j.exger.2008.12.006. [DOI] [PubMed] [Google Scholar]
- 28.Dai D.F., Chiao Y.A., Marcinek D.J., Szeto H.H., Rabinovitch P.S. Mitochondrial oxidative stress in aging and healthspan. Longev. Health. 2014;3:6. doi: 10.1186/2046-2395-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gregg S.Q., Robinson A.R., Niedernhofer L.J. Physiological consequences of defects in ERCC1-XPF DNA repair endonuclease. DNA Repair (Amst.) 2011;10(7):781–791. doi: 10.1016/j.dnarep.2011.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Niedernhofer L.J., Garinis G.A., Raams A., Lalai A.S., Robinson A.R., Appeldoorn E., Odijk H., Oostendorp R., Ahmad A., van Leeuwen W., Theil A.F., Vermeulen W., van der Horst G.T., Meinecke P., Kleijer W.J., Vijg J., Jaspers N.G., Hoeijmakers J.H. A new progeroid syndrome reveals that genotoxic stress suppresses the somatotroph axis. Nature. 2006;444(7122):1038–1043. doi: 10.1038/nature05456. [DOI] [PubMed] [Google Scholar]
- 31.Gurkar A.U., Niedernhofer L.J. Comparison of mice with accelerated aging caused by distinct mechanisms. Exp. Gerontol. 2015;68:43–50. doi: 10.1016/j.exger.2015.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ahmad A., Robinson A.R., Duensing A., van Drunen E., Beverloo H.B., Weisberg D.B., Hasty P., Hoeijmakers J.H., Niedernhofer L.J. ERCC1-XPF endonuclease facilitates DNA double-strand break repair. Mol. Cell Biol. 2008;28(16):5082–5092. doi: 10.1128/MCB.00293-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller S.A., Dykes D.D., Polesky H.F. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16(3):1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang J., Yuan B., Guerrero C., Bahde R., Gupta S., Wang Y. Quantification of oxidative DNA lesions in tissues of Long-Evans Cinnamon rats by capillary high-performance liquid chromatography-tandem mass spectrometry coupled with stable isotope-dilution method. Anal. Chem. 2011;83(6):2201–2209. doi: 10.1021/ac103099s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morales R.C., Bahnson E.S., Havelka G.E., Cantu-Medellin N., Kelley E.E., Kibbe M.R. Sex-based differential regulation of oxidative stress in the vasculature by nitric oxide. Redox Biol. 2015;4:226–233. doi: 10.1016/j.redox.2015.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zielonka J., Vasquez-Vivar J., Kalyanaraman B. Detection of 2-hydroxyethidium in cellular systems: a unique marker product of superoxide and hydroethidine. Nat. Protoc. 2008;3(1):8–21. doi: 10.1038/nprot.2007.473. [DOI] [PubMed] [Google Scholar]
- 37.Dikalov S.I., Li W., Mehranpour P., Wang S.S., Zafari A.M. Production of extracellular superoxide by human lymphoblast cell lines: comparison of electron spin resonance techniques and cytochrome C reduction assay. Biochem Pharmacol. 2007;73(7):972–980. doi: 10.1016/j.bcp.2006.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kelley E.E., Trostchansky A., Rubbo H., Freeman B.A., Radi R., Tarpey M.M. Binding of xanthine oxidase to glycosaminoglycans limits inhibition by oxypurinol. J. Biol. Chem. 2004;279(36):37231–37234. doi: 10.1074/jbc.M402077200. [DOI] [PubMed] [Google Scholar]
- 39.Frezza C., Cipolat S., Scorrano L. Organelle isolation: functional mitochondria from mouse liver, muscle and cultured fibroblasts. Nat. Protoc. 2007;2(2):287–295. doi: 10.1038/nprot.2006.478. [DOI] [PubMed] [Google Scholar]
- 40.Brand M.D., Nicholls D.G. Assessing mitochondrial dysfunction in cells. Biochem J. 2011;435(2):297–312. doi: 10.1042/BJ20110162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tautenhahn R., Patti G.J., Rinehart D., Siuzdak G. XCMS Online: a web-based platform to process untargeted metabolomic data. Anal. Chem. 2012;84(11):5035–5039. doi: 10.1021/ac300698c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Patti G.J., Tautenhahn R., Rinehart D., Cho K., Shriver L.P., Manchester M., Nikolskiy I., Johnson C.H., Mahieu N.G., Siuzdak G. A view from above: cloud plots to visualize global metabolomic data. Anal. Chem. 2013;85(2):798–804. doi: 10.1021/ac3029745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wisniewski J.R., Zougman A., Nagaraj N., Mann M. Universal sample preparation method for proteome analysis. Nat. Methods. 2009;6(5):359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 44.Manza L.L., Stamer S.L., Ham A.J., Codreanu S.G., Liebler D.C. Sample preparation and digestion for proteomic analyses using spin filters. Proteomics. 2005;5(7):1742–1745. doi: 10.1002/pmic.200401063. [DOI] [PubMed] [Google Scholar]
- 45.Bell-Temin H., Barber D.S., Zhang P., Liu B., Stevens S.M., Jr. Proteomic analysis of rat microglia establishes a high-confidence reference data set of over 3000 proteins. Proteomics. 2012;12(2):246–250. doi: 10.1002/pmic.201100398. [DOI] [PubMed] [Google Scholar]
- 46.Bell-Temin H., Zhang P., Chaput D., King M.A., You M., Liu B., Stevens S.M., Jr. Quantitative proteomic characterization of ethanol-responsive pathways in rat microglial cells. J. Proteome Res. 2013;12(5):2067–2077. doi: 10.1021/pr301038f. [DOI] [PubMed] [Google Scholar]
- 47.Bell-Temin H., Culver-Cochran A.E., Chaput D., Carlson C.M., Kuehl M., Burkhardt B.R., Bickford P.C., Liu B., Stevens S.M., Jr. Novel molecular insights into classical and alternative activation states of microglia as revealed by stable isotope labeling by amino acids in cell culture (SILAC)-based Proteomics. Mol. Cell Proteom. 2015;14(12):3173–3184. doi: 10.1074/mcp.M115.053926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meng F., Wiener M.C., Sachs J.R., Burns C., Verma P., Paweletz C.P., Mazur M.T., Deyanova E.G., Yates N.A., Hendrickson R.C. Quantitative analysis of complex peptide mixtures using FTMS and differential mass spectrometry. J. Am. Soc. Mass Spectrom. 2007;18(2):226–233. doi: 10.1016/j.jasms.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 49.Wiener M.C., Sachs J.R., Deyanova E.G., Yates N.A. Differential mass spectrometry: a label-free LC-MS method for finding significant differences in complex peptide and protein mixtures. Anal. Chem. 2004;76(20):6085–6096. doi: 10.1021/ac0493875. [DOI] [PubMed] [Google Scholar]
- 50.Eng J.K., Jahan T.A., Hoopmann M.R. Comet: an open-source MS/MS sequence database search tool. Proteomics. 2013;13(1):22–24. doi: 10.1002/pmic.201200439. [DOI] [PubMed] [Google Scholar]
- 51.Kall L., Canterbury J.D., Weston J., Noble W.S., MacCoss M.J. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat. Methods. 2007;4(11):923–925. doi: 10.1038/nmeth1113. [DOI] [PubMed] [Google Scholar]
- 52.Weydert C.J., Cullen J.J. Measurement of superoxide dismutase, catalase and glutathione peroxidase in cultured cells and tissue. Nat. Protoc. 2010;5(1):51–66. doi: 10.1038/nprot.2009.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Anderson M.E. Determination of glutathione and glutathione disulfide in biological samples. Methods Enzymol. 1985;113:548–555. doi: 10.1016/s0076-6879(85)13073-9. [DOI] [PubMed] [Google Scholar]
- 54.Baker M.A., Cerniglia G.J., Zaman A. Microtiter plate assay for the measurement of glutathione and glutathione disulfide in large numbers of biological samples. Anal. Biochem. 1990;190(2):360–365. doi: 10.1016/0003-2697(90)90208-q. [DOI] [PubMed] [Google Scholar]
- 55.Tilstra J.S., Robinson A.R., Wang J., Gregg S.Q., Clauson C.L., Reay D.P., Nasto L.A., Croix C.M., St, Usas A., Vo N., Huard J., Clemens P.R., Stolz D.B., Guttridge D.C., Watkins S.C., Garinis G.A., Wang Y., Niedernhofer L.J., Robbins P.D. NF-kappaB inhibition delays DNA damage-induced senescence and aging in mice. J. Clin. Invest. 2012;122(7):2601–2612. doi: 10.1172/JCI45785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kazak L., Reyes A., Holt I.J. Minimizing the damage: repair pathways keep mitochondrial DNA intact. Nat. Rev. Mol. Cell Biol. 2012;13(10):659–671. doi: 10.1038/nrm3439. [DOI] [PubMed] [Google Scholar]
- 57.Jaruga P., Dizdaroglu M. 8,5'-Cyclopurine-2'-deoxynucleosides in DNA: mechanisms of formation, measurement, repair and biological effects. DNA Repair (Amst.) 2008;7(9):1413–1425. doi: 10.1016/j.dnarep.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 58.Kuraoka I., Bender C., Romieu A., Cadet J., Wood R.D., Lindahl T. Removal of oxygen free-radical-induced 5',8-purine cyclodeoxynucleosides from DNA by the nucleotide excision-repair pathway in human cells. Proc. Natl. Acad. Sci. USA. 2000;97(8):3832–3837. doi: 10.1073/pnas.070471597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang J., Clauson C.L., Robbins P.D., Niedernhofer L.J., Wang Y. The oxidative DNA lesions 8,5'-cyclopurines accumulate with aging in a tissue-specific manner. Aging Cell. 2012;11(4):714–716. doi: 10.1111/j.1474-9726.2012.00828.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhu X.D., Niedernhofer L., Kuster B., Mann M., Hoeijmakers J.H., de Lange T. ERCC1/XPF removes the 3' overhang from uncapped telomeres and represses formation of telomeric DNA-containing double minute chromosomes. Mol. Cell. 2003;12(6):1489–1498. doi: 10.1016/s1097-2765(03)00478-7. [DOI] [PubMed] [Google Scholar]
- 61.Rai R., Chang S. Probing the Telomere Damage Response. Methods Mol. Biol. 2017;1587:133–138. doi: 10.1007/978-1-4939-6892-3_13. [DOI] [PubMed] [Google Scholar]
- 62.de Vries A., van Oostrom C.T., Hofhuis F.M., Dortant P.M., Berg R.J., de Gruijl F.R., Wester P.W., van Kreijl C.F., Capel P.J., van Steeg H., Verbeek S.J. Increased susceptibility to ultraviolet-B and carcinogens of mice lacking the DNA excision repair gene XPA. Nature. 1995;377(6545):169–173. doi: 10.1038/377169a0. [DOI] [PubMed] [Google Scholar]
- 63.Marnett L.J. Lipid peroxidation-DNA damage by malondialdehyde. Mutat. Res. 1999;424(1–2):83–95. doi: 10.1016/s0027-5107(99)00010-x. [DOI] [PubMed] [Google Scholar]
- 64.Frijhoff J., Winyard P.G., Zarkovic N., Davies S.S., Stocker R., Cheng D., Knight A.R., Taylor E.L., Oettrich J., Ruskovska T., Gasparovic A.C., Cuadrado A., Weber D., Poulsen H.E., Grune T., Schmidt H.H., Ghezzi P. Clinical Relevance of Biomarkers of Oxidative Stress. Antioxid. Redox Signal. 2015;23(14):1144–1170. doi: 10.1089/ars.2015.6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Harris C.M., Massey V. The reaction of reduced xanthine dehydrogenase with molecular oxygen. Reaction kinetics and measurement of superoxide radical. J. Biol. Chem. 1997;272(13):8370–8379. doi: 10.1074/jbc.272.13.8370. [DOI] [PubMed] [Google Scholar]
- 66.Vida C., Corpas I., De la Fuente M., Gonzalez E.M. Age-related changes in xanthine oxidase activity and lipid peroxidation, as well as in the correlation between both parameters, in plasma and several organs from female mice. J. Physiol. Biochem. 2011;67(4):551–558. doi: 10.1007/s13105-011-0100-8. [DOI] [PubMed] [Google Scholar]
- 67.Son J.M., Sarsour E.H., Kakkerla Balaraju A., Fussell J., Kalen A.L., Wagner B.A., Buettner G.R., Goswami P.C. Mitofusin 1 and optic atrophy 1 shift metabolism to mitochondrial respiration during aging. Aging Cell. 2017;16(5):1136–1145. doi: 10.1111/acel.12649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Muller F. The nature and mechanism of superoxide production by the electron transport chain: its relevance to aging. J. Am. Aging Assoc. 2000;23(4):227–253. doi: 10.1007/s11357-000-0022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Brace L.E., Vose S.C., Stanya K., Gathungu R.M., Marur V.R., Longchamp A., Trevino-Villarreal H., Mejia P., Vargas D., Inouye K., Bronson R.T., Lee C.H., Neilan E., Kristal B.S., Mitchell J.R. Increased oxidative phosphorylation in response to acute and chronic DNA damage. NPJ Aging Mech. Dis. 2016;2:16022. doi: 10.1038/npjamd.2016.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Harper M.E., Monemdjou S., Ramsey J.J., Weindruch R. Age-related increase in mitochondrial proton leak and decrease in ATP turnover reactions in mouse hepatocytes. Am. J. Physiol. 1998;275(2 Pt 1):E197–E206. doi: 10.1152/ajpendo.1998.275.2.E197. [DOI] [PubMed] [Google Scholar]
- 71.Hekimi S., Lapointe J., Wen Y. Taking a "good" look at free radicals in the aging process. Trends Cell Biol. 2011;21(10):569–576. doi: 10.1016/j.tcb.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Maher P. The effects of stress and aging on glutathione metabolism. Ageing Res. Rev. 2005;4(2):288–314. doi: 10.1016/j.arr.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 73.Fink M.P., Macias C.A., Xiao J., Tyurina Y.Y., Jiang J., Belikova N., Delude R.L., Greenberger J.S., Kagan V.E., Wipf P. Hemigramicidin-TEMPO conjugates: novel mitochondria-targeted anti-oxidants. Biochem. Pharmacol. 2007;74(6):801–809. doi: 10.1016/j.bcp.2007.05.019. [DOI] [PubMed] [Google Scholar]
- 74.Kagan V.E., Bayir A., Bayir H., Stoyanovsky D., Borisenko G.G., Tyurina Y.Y., Wipf P., Atkinson J., Greenberger J.S., Chapkin R.S., Belikova N.A. Mitochondria-targeted disruptors and inhibitors of cytochrome c/cardiolipin peroxidase complexes: a new strategy in anti-apoptotic drug discovery. Mol. Nutr. Food Res. 2009;53(1):104–114. doi: 10.1002/mnfr.200700402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Trnka J., Blaikie F.H., Smith R.A., Murphy M.P. A mitochondria-targeted nitroxide is reduced to its hydroxylamine by ubiquinol in mitochondria. Free Radic. Biol. Med. 2008;44(7):1406–1419. doi: 10.1016/j.freeradbiomed.2007.12.036. [DOI] [PubMed] [Google Scholar]
- 76.Xun Z., Rivera-Sanchez S., Ayala-Pena S., Lim J., Budworth H., Skoda E.M., Robbins P.D., Niedernhofer L.J., Wipf P., McMurray C.T. Targeting of XJB-5-131 to mitochondria suppresses oxidative DNA damage and motor decline in a mouse model of Huntington's disease. Cell Rep. 2012;2(5):1137–1142. doi: 10.1016/j.celrep.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fink M.P., Macias C.A., Xiao J., Tyurina Y.Y., Delude R.L., Greenberger J.S., Kagan V.E., Wipf P. Hemigramicidin-TEMPO conjugates: novel mitochondria-targeted antioxidants. Crit. Care Med. 2007;35(9 Suppl):S461–S467. doi: 10.1097/01.CCM.0000279192.96303.E7. [DOI] [PubMed] [Google Scholar]
- 78.Kagan V.E., Wipf P., Stoyanovsky D., Greenberger J.S., Borisenko G., Belikova N.A., Yanamala N., Samhan Arias A.K., Tungekar M.A., Jiang J., Tyurina Y.Y., Ji J., Klein-Seetharaman J., Pitt B.R., Shvedova A.A., Bayir H. Mitochondrial targeting of electron scavenging antioxidants: regulation of selective oxidation vs random chain reactions. Adv. Drug Deliv. Rev. 2009;61(14):1375–1385. doi: 10.1016/j.addr.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gregg S.Q., Gutierrez V., Robinson A.R., Woodell T., Nakao A., Ross M.A., Michalopoulos G.K., Rigatti L., Rothermel C.E., Kamileri I., Garinis G.A., Stolz D.B., Niedernhofer L.J. A mouse model of accelerated liver aging caused by a defect in DNA repair. Hepatology. 2012;55(2):609–621. doi: 10.1002/hep.24713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sawada M., Carlson J.C. Changes in superoxide radical and lipid peroxide formation in the brain, heart and liver during the lifetime of the rat. Mech. Ageing Dev. 1987;41(1–2):125–137. doi: 10.1016/0047-6374(87)90057-1. [DOI] [PubMed] [Google Scholar]
- 81.Sohal R.S., Sohal B.H. Hydrogen peroxide release by mitochondria increases during aging. Mech. Ageing Dev. 1991;57(2):187–202. doi: 10.1016/0047-6374(91)90034-w. [DOI] [PubMed] [Google Scholar]
- 82.Sullivan-Gunn M.J., Lewandowski P.A. Elevated hydrogen peroxide and decreased catalase and glutathione peroxidase protection are associated with aging sarcopenia. BMC Geriatr. 2013;13:104. doi: 10.1186/1471-2318-13-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhu Y., Carvey P.M., Ling Z. Age-related changes in glutathione and glutathione-related enzymes in rat brain. Brain Res. 2006;1090(1):35–44. doi: 10.1016/j.brainres.2006.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Baker D.J., Wijshake T., Tchkonia T., LeBrasseur N.K., Childs B.G., van de Sluis B., Kirkland J.L., van Deursen J.M. Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature. 2011;479(7372):232–236. doi: 10.1038/nature10600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chang J., Wang Y., Shao L., Laberge R.M., Demaria M., Campisi J., Janakiraman K., Sharpless N.E., Ding S., Feng W., Luo Y., Wang X., Aykin-Burns N., Krager K., Ponnappan U., Hauer-Jensen M., Meng A., Zhou D. Clearance of senescent cells by ABT263 rejuvenates aged hematopoietic stem cells in mice. Nat. Med. 2016;22(1):78–83. doi: 10.1038/nm.4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhu Y., Tchkonia T., Pirtskhalava T., Gower A.C., Ding H., Giorgadze N., Palmer A.K., Ikeno Y., Hubbard G.B., Lenburg M., O'Hara S.P., LaRusso N.F., Miller J.D., Roos C.M., Verzosa G.C., LeBrasseur N.K., Wren J.D., Farr J.N., Khosla S., Stout M.B., McGowan S.J., Fuhrmann-Stroissnigg H., Gurkar A.U., Zhao J., Colangelo D., Dorronsoro A., Ling Y.Y., Barghouthy A.S., Navarro D.C., Sano T., Robbins P.D., Niedernhofer L.J., Kirkland J.L. The Achilles' heel of senescent cells: from transcriptome to senolytic drugs. Aging Cell. 2015;14(4):644–658. doi: 10.1111/acel.12344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Maciejczyk M., Mikoluc B., Pietrucha B., Heropolitanska-Pliszka E., Pac M., Motkowski R., Car H. Oxidative stress, mitochondrial abnormalities and antioxidant defense in Ataxia-telangiectasia, Bloom syndrome and Nijmegen breakage syndrome. Redox Biol. 2017;11:375–383. doi: 10.1016/j.redox.2016.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Weyemi U., Redon C.E., Aziz T., Choudhuri R., Maeda D., Parekh P.R., Bonner M.Y., Arbiser J.L., Bonner W.M. NADPH oxidase 4 is a critical mediator in Ataxia telangiectasia disease. Proc. Natl. Acad. Sci. USA. 2015;112(7):2121–2126. doi: 10.1073/pnas.1418139112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ewald C.Y., Hourihan J.M., Bland M.S., Obieglo C., Katic I., Moronetti Mazzeo L.E., Alcedo J., Blackwell T.K., Hynes N.E. NADPH oxidase-mediated redox signaling promotes oxidative stress resistance and longevity through memo-1 in C. elegans. Elife. 2017;6 doi: 10.7554/eLife.19493. e19493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.D'Souza A.D., Parish I.A., Krause D.S., Kaech S.M., Shadel G.S. Reducing mitochondrial ROS improves disease-related pathology in a mouse model of ataxia-telangiectasia. Mol. Ther. 2013;21(1):42–48. doi: 10.1038/mt.2012.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Alexander A., Cai S.L., Kim J., Nanez A., Sahin M., MacLean K.H., Inoki K., Guan K.L., Shen J., Person M.D., Kusewitt D., Mills G.B., Kastan M.B., Walker C.L. ATM signals to TSC2 in the cytoplasm to regulate mTORC1 in response to ROS. Proc. Natl. Acad. Sci. USA. 2010;107(9):4153–4158. doi: 10.1073/pnas.0913860107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kuang X., Yan M., Ajmo J.M., Scofield V.L., Stoica G., Wong P.K. Activation of AMP-activated protein kinase in cerebella of Atm-/- mice is attributable to accumulation of reactive oxygen species. Biochem. Biophys. Res. Commun. 2012;418(2):267–272. doi: 10.1016/j.bbrc.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Okuno Y., Nakamura-Ishizu A., Otsu K., Suda T., Kubota Y. Pathological neoangiogenesis depends on oxidative stress regulation by ATM. Nat. Med. 2012;18(8):1208–1216. doi: 10.1038/nm.2846. [DOI] [PubMed] [Google Scholar]
- 94.Fang E.F., Kassahun H., Croteau D.L., Scheibye-Knudsen M., Marosi K., Lu H., Shamanna R.A., Kalyanasundaram S., Bollineni R.C., Wilson M.A., Iser W.B., Wollman B.N., Morevati M., Li J., Kerr J.S., Lu Q., Waltz T.B., Tian J., Sinclair D.A., Mattson M.P., Nilsen H., Bohr V.A. NAD+ Replenishment Improves Lifespan and Healthspan in Ataxia Telangiectasia Models via Mitophagy and DNA Repair. Cell Metab. 2016;24(4):566–581. doi: 10.1016/j.cmet.2016.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fang E.F., Scheibye-Knudsen M., Brace L.E., Kassahun H., SenGupta T., Nilsen H., Mitchell J.R., Croteau D.L., Bohr V.A. Defective mitophagy in XPA via PARP-1 hyperactivation and NAD(+)/SIRT1 reduction. Cell. 2014;157(4):882–896. doi: 10.1016/j.cell.2014.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Perez V.I., Bokov A., Van Remmen H., Mele J., Ran Q., Ikeno Y., Richardson A. Is the oxidative stress theory of aging dead? Biochim. Biophys. Acta. 2009;1790(10):1005–1014. doi: 10.1016/j.bbagen.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Perez V.I., Van Remmen H., Bokov A., Epstein C.J., Vijg J., Richardson A. The overexpression of major antioxidant enzymes does not extend the lifespan of mice. Aging Cell. 2009;8(1):73–75. doi: 10.1111/j.1474-9726.2008.00449.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jiang J., Kurnikov I., Belikova N.A., Xiao J., Zhao Q., Amoscato A.A., Braslau R., Studer A., Fink M.P., Greenberger J.S., Wipf P., Kagan V.E. Structural requirements for optimized delivery, inhibition of oxidative stress, and antiapoptotic activity of targeted nitroxides. J. Pharmacol. Exp. Ther. 2007;320(3):1050–1060. doi: 10.1124/jpet.106.114769. [DOI] [PubMed] [Google Scholar]
- 99.Buettner G.R., Ng C.F., Wang M., Rodgers V.G., Schafer F.Q. A new paradigm: manganese superoxide dismutase influences the production of H2O2 in cells and thereby their biological state. Free Radic. Biol. Med. 2006;41(8):1338–1350. doi: 10.1016/j.freeradbiomed.2006.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Qian S.Y., Buettner G.R. Iron and dioxygen chemistry is an important route to initiation of biological free radical oxidations: an electron paramagnetic resonance spin trapping study. Free Radic. Biol. Med. 1999;26(11–12):1447–1456. doi: 10.1016/s0891-5849(99)00002-7. [DOI] [PubMed] [Google Scholar]
- 101.Kujoth G.C., Hiona A., Pugh T.D., Someya S., Panzer K., Wohlgemuth S.E., Hofer T., Seo A.Y., Sullivan R., Jobling W.A., Morrow J.D., Van Remmen H., Sedivy J.M., Yamasoba T., Tanokura M., Weindruch R., Leeuwenburgh C., Prolla T.A. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science. 2005;309(5733):481–484. doi: 10.1126/science.1112125. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material