Abstract
Mitochondrial quality is under surveillance by autophagy, the cell recycling process which degrades and removes damaged mitochondria. Inadequate autophagy results in deterioration in mitochondrial quality, bioenergetic dysfunction, and metabolic stress. Here we describe in an integrated work-flow to assess parameters of mitochondrial morphology, function, mtDNA and protein damage, metabolism and autophagy regulation to provide the framework for a practical assessment of mitochondrial quality. This protocol has been tested with cell cultures, is highly reproducible, and is adaptable to studies when cell numbers are limited, and thus will be of interest to researchers studying diverse physiological and pathological phenomena in which decreased mitochondrial quality is a contributory factor.
Keywords: Autophagy, Mitophagy, Neuron, Beta cells, Mitochondrial fragmentation, Bioenergetics, Metabolomics
Highlights
-
•
We outline methods to assess different facets of the control of mitochondrial health.
-
•
We describe biochemical and microscopic methods to assess mitophagy.
-
•
We describe methods to measure mtDNA and protein damage.
-
•
We describe methods for targeted metabolomics to measure TCA cycle metabolites.
-
•
We describe measurement of mitochondrial function in intact and permeabilized cells.
1. Introduction
Mitochondria are complex cellular organelles responsible for maintaining cellular energy status and homeostasis through production of ATP, cell signaling and generation of intermediary metabolites. Mitochondria are highly dynamic and in a constant state of structural flux through fission and fusion processes and have important roles in cellular signaling cascades. During the physiological process of metabolism, the components of the organelle including proteins, lipids and DNA become damaged and must be repaired to maintain normal function. This maintenance process requires the integrated participation of a number of key pathways and is generally termed mitochondrial quality control [1].
An important contributor to mitochondrial quality control is autophagy, which is an indispensable cellular process responsible for the clearance of cellular debris, organelles, and damaged proteins [2], [3]. This complex process is regulated by nutrient availability and response to stress. Specific recycling of mitochondria by autophagy, termed mitophagy, is important for maintaining optimal mitochondrial function [4], [5], [6].
Mitochondrial dysfunction is associated with aging and chronic pathologies including neurodegenerative diseases, cardiovascular disease and diabetes [7], [8]. Mitochondrial dysfunction is a hallmark of pathologies in tissues that are highly energetic, in particular the heart and brain but also in a broad range of cells including platelets and those controlling immunity. In the brain, mitochondrial dysfunction has been frequently implicated in a range of age-dependent neurodegenerative diseases, including Parkinson's disease (PD) and Alzheimer's disease (AD). In these diseases, protein aggregation and mitochondrial dysfunction interplay and ultimately both contribute to neurodegeneration [9], [10], [11], [12], [13], [14], [15]. Notable examples of the importance of mitochondria at the center of this disease are evidenced by toxin-induced models of PD through administration of rotenone, paraquat, and MPTP [16]. These compounds have been reported to act through either complex I inhibition or through redox cycling [17], [18]. Furthermore, the lipid peroxidation product, 4-hydroxynonenal (HNE) has been shown to accumulate in both PD and AD brains, and to significantly alter mitochondrial and autophagic function in diverse cell types [19], [20], [21], [22], [23], [24]. We have proposed that autophagy can serve as an essential antioxidant pathway to attenuate oxidative damage [25].
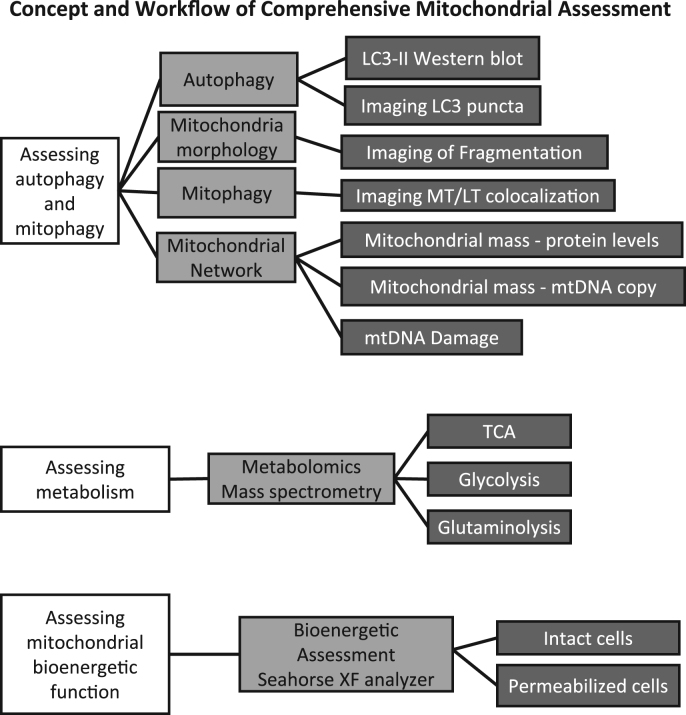
Given the importance of mitochondria in health and disease, here we have outlined the initial approaches that are accessible to most laboratories using existing technology to investigate different facets of mitochondrial health in a given disease paradigm (Fig. 1). We have grouped similar modalities together allowing investigators to select the appropriate methods and integrate them into a structured work flow. Noting the importance of mitochondria in chronic pathologies, we have used both primary cortical neurons and β-cells for the purposes of this manuscript.
Fig. 1.
Workflow of comprehensive mitochondrial assessment. Using primary neurons, assessment of mitochondrial function can be obtained by employing diverse techniques. Shown here is a list of experiments used to examine autophagy, mitophagy, mitochondrial damage, mitochondrial morphology, metabolism and mitochondrial bioenergetic function.
2. Protocol concept
The concept that autophagy, mitophagy, mitochondrial bioenergetics and mitochondrial quality control are linked has emerged over the last 10 years [26]. Although many studies have focused on individual elements of these processes, an integrated methodological approach to provide a comprehensive view of cellular and tissue-specific responses to physiological, pharmacological, and pathological perturbations has not been proposed. This is important since the assessment of mitochondrial quality requires standardized protocols that allow testing of key elements of this complex process using a broad range of biological read-outs. We provide here a streamlined comprehensive strategy to combine autophagy assessment with mitochondrial damage, mitochondrial dynamics and mitochondrial function, as well as cellular metabolism. Mitochondrial function can be assayed in real time to assess the consequence of exposures to nutrients, toxins and pharmacological agents. This approach will be of significant value to researchers over a broad area of biomedical research. The workflow is outlined in Fig. 1.
3. Applications and target audience
There are currently 4407 papers available in PubMed (surveyed on Mar 27th, 2018) discussing various aspects of autophagy and mitochondria. Of these, 1438 papers discussed mitochondrial bioenergetics, while 478 papers discussed mitochondrial quality control. Our protocols will be of use to researchers with established interest in a combined approach, as well as those who have been solely focused on one or two of these topics and wish to expand their repertoire of investigation, with the realization that these aspects of cellular function are fundamental to most biomedical problems. Researchers working on diverse biological challenges, including oxidative stress, mitochondrial biology, autophagy, mitophagy, metabolism and a broad range of pathologies are the target audience for the article.
4. Materials
Tables of needed chemicals, PCR primers, antibodies, consumables and equipment needed are listed in Supplemental Tables 1–5.
4.1. Protocol: steps and procedures
4.1.1. Neuron preparation
Primary neurons isolated from either rat or mice provide an excellent platform in which to test aspects of neuropathologies, especially before, during or after either neurotoxin or neuroprotective treatments administered through genetic and/or pharmacological means [27], [28]. While there are certainly some alterations of metabolism due to being cultured, primary neurons provide more relevant read-outs than immortalized cells. This is particularly the case when measuring parameters of mitochondrial metabolism and function.
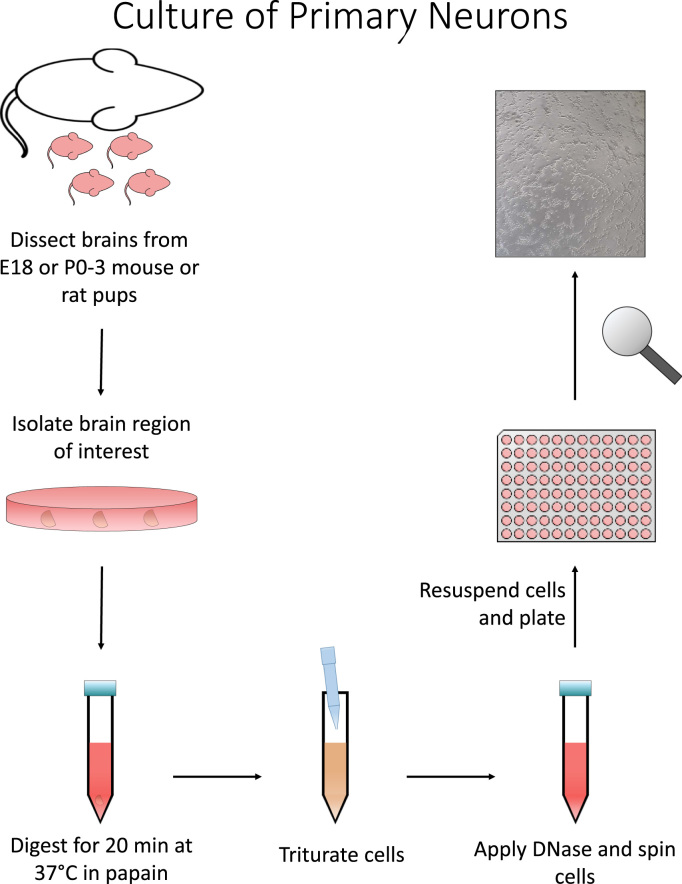
The isolation workflow is diagrammed in Fig. 2. The day before isolation, coat 24 well tissue plates for ≥1 h at room temperature in a poly-l-lysine coating solution (25 mL water, 12.5 mL 200 mM Boric Acid, 10 mL 50 mM Borax, 2.5 mL of 2 mg/mL filtered Poly-L-lysine). After coating, wash plates twice with ddH2O and allow to dry before storing at 4 °C until use. Brain tissue can then be isolated from E18 pups excised from Sprague-Dawley rats. Once cortices are removed from the brain, they are dissected in ice cold HBSS media and placed in a short-term storage solution (2 mL per brain) until further processing (500 mL HBSS supplemented with 250 µL 20% Glucose, 5 mL 1 M HEPES pH 7.3, 5 mL 100× pyruvate [29], [30]. Long exposures of the tissue to air should be avoided if possible during isolation.
Fig. 2.
Primary neuronal culture. Briefly, pups can be obtained from mice or rats ranging from E18 to p0–3 in age. Desired brain region can then be micro-dissected from the brain. Tissue is then digested using a papain solution for 20 min at 37 °C with gentle agitation every ~3 min. Cells are then triturated using 1 mL pipet tip with smooth edges. Remaining debris should be removed and DNase added. Cells are then centrifuged at ~250×g for 5 min. Cells are then to be resuspended, counted, and plated.
Next, decant storage media from cortex tissue and replace with 1 mL per brain of papain solution (activate papain for ~15 min at 37 °C prior to addition) and incubate for 20 min at 37 °C with gentle agitation every few min. After 20 min, add 2 mL of neurobasal media (NBM) supplemented with B27, pen/strep, and L-glutamine per brain to dilute digestion solution. Then wash brain tissue once with 1 mL per brain of NBM. Mechanically separate tissue by trituration in 4 mL per brain of NBM. After allowing a few minutes to settle, remove debris and add 10 µL per brain of 40 µg/mL DNase I. To remove cells from DNase solution, pellet the cells by centrifugation for 5 min at ~250×g. Decant the supernatant, and resuspend the cells in 2 mL of NBA per brain and count by hemocytometer. Neurons can then be plated at desired densities for analysis by western blot, seahorse, imaging or metabolomics approaches. This procedure takes about 6 h.
4.1.2. Exposing cells to biological or pharmacological reagents
Primary neurons can be maintained for several weeks in culture and as they age and mature, they extend their cellular processes and integrate into ever more reticulated networks. We have traditionally performed experiments between 7 and 14 days in vitro (DIV) and have performed all experiments at DIV7 for the purposes of this manuscript. For exposing neurons to pharmacological reagents, half of the media is removed and then replaced with NBM containing only pen/strep and L-glutamine. B27 is a cocktail of signaling molecules, antioxidants, and supplements, and has been omitted during our studies since these agents impact metabolism and energetics [30], [31].
4.1.3. Effects of lysosome inhibitors on autophagy
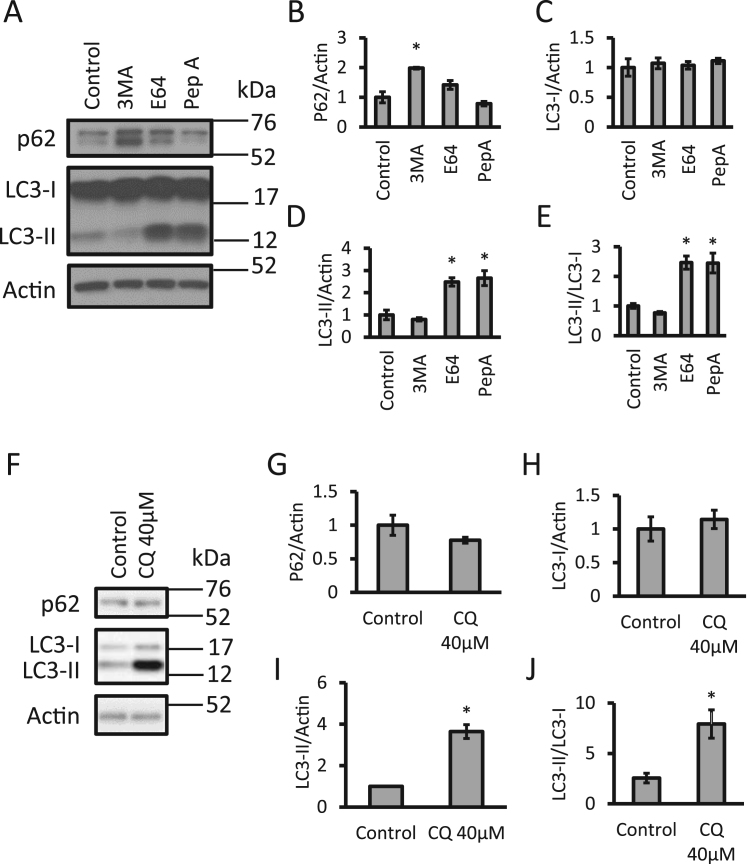
Given the importance of autophagy in maintaining healthy mitochondrial populations, an initial assessment of two key autophagy components can be utilized to determine if aberrations in autophagy alter mitochondrial quality. For western blot analyses, we routinely plate neurons in 24-well plates at 480,000 cells per well. In Fig. 3 we show the measurement of the scaffold, ubiquitin binding and autophagy substrate protein p62 as well as both cytosolic LC3-I and autophagosome incorporated LC3-II in response to various autophagy modulators by western blot analyses (antibodies see Table 2). In response to 3-methyladenine (3MA), an autophagy initiation inhibitor, LC3-II levels remained unchanged, but significant increases in p62 were observed, which is generally interpreted as decreased clearance through autophagy inhibition. The lysosomal inhibitors E64 and pepstatin A, are also shown as an additional example and showed no changes in p62 but significant increases in LC3-II.
Fig. 3.
Assessing autophagosomal LC3-II and autophagy adaptor/substrate p62 levels. (A-E) Primary cortical rat neurons were exposed to 3MA (10 mM), E64 (100 µM), or pepstatin A (100 µM) for 24 h. Levels of p62, LC3-I and LC3-II were measured by western blot analysis. (F-J) Primary cortical rat neurons exposed to 40 µM chloroquine (CQ) for 4 h and then analyzed by western blot for p62, LC3-I and LC3-II. *p < 0.05 compared to control, n = 3.
Furthermore, the LC3-II / LC3-I ratio is calculated, which measures autophagosomal LC3 versus cytosolic LC3 (Fig. 3A-E). Similar to other lysosome inhibitors, chloroquine (CQ) results in the accumulation of LC3-II and consequently increases the LC3-II/LC3-I ratio (Fig. 3F-J) [32], [33]. CQ is water soluble and inhibits lysosome enzyme function through increased pH, opposed to inhibition of specific lysosomal proteases as E64 and pepstatin A [3], [34].
4.1.4. Assessment of the mitochondrial network and mitophagy
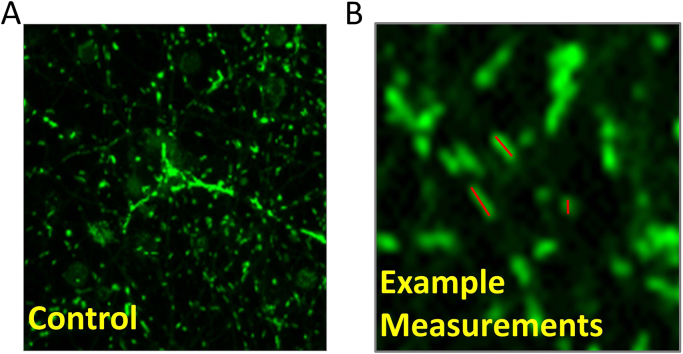
Confocal microscopy was used to measure both the morphology of the mitochondrial network, which undergoes alteration response to stress, and mitophagy. The mitochondrial network can change primarily through fission or fusion between individual organelles [35]. The level of fission/fragmentation in response to stress can be measured by quantification of the length of a cell's mitochondrial population. We have measured mitochondrial fragmentation. Neurons were plated on 8 well Nunc™ Lab-Tek™ Chambered coverglass plates at a density of 100,000 per well. MitoTracker Green FM (25 nM) was added to the cells for approximately 20 min before being washed three times with pre-warmed and equilibrated media. Images were taken with a Zeiss 700 laser-scanning microscope. Mitochondrial length was measured individually using the polygon-curve tool in proprietary ZEN Blue software (Fig. 4). By utilizing this tool to draw a line along the length of each mitochondrion, the length can be gathered for a population of cells per field. All measurements can then be exported to Microsoft Excel where additional analysis can be performed. Approximately 300–500 total mitochondria were counted from at least 3 images for each treatment group in each experimental replicate. Only mitochondria in-focus with defined borders were used for measurements. In this control sample the average mitochondrial length was 969.6 ± 78.2 nM.
Fig. 4.
Quantification of mitochondrial fragmentation. (A) Neurons were incubated with 25 nM MitoTracker Green for 15 min and imaged using laser scanning confocal microscopy. (B) Representative image of quantification of individual mitochondria using Zen Blue polygon curve tool.
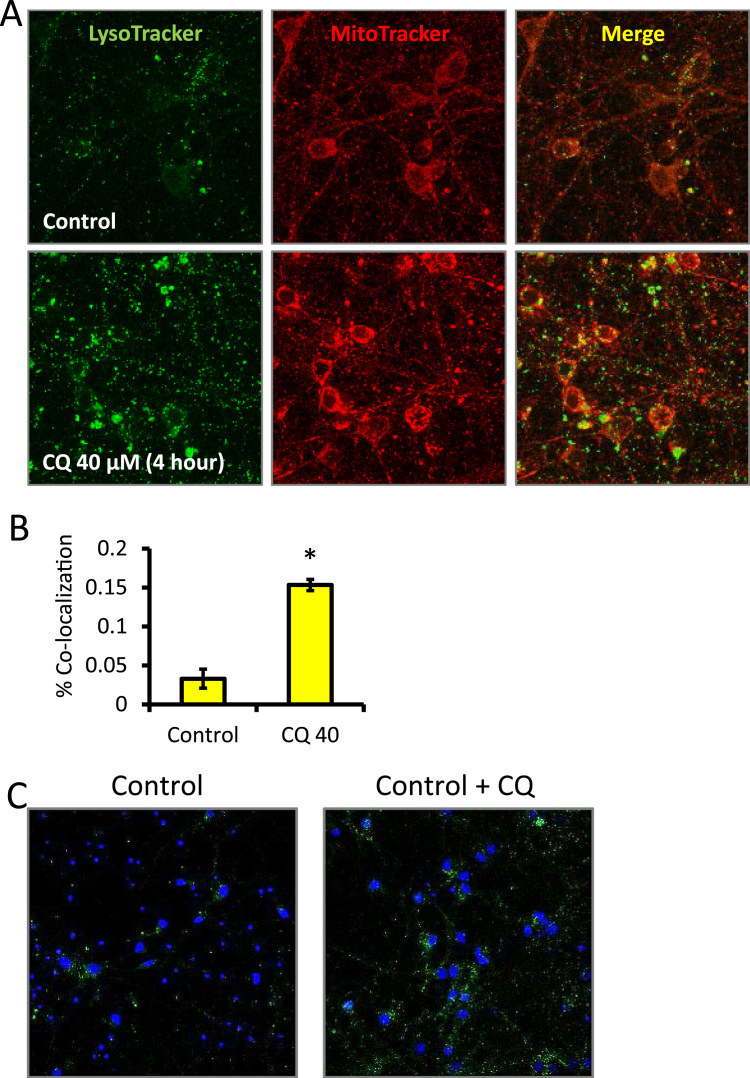
In addition to morphology, recycling of mitochondria by autophagy can be measured. Mitophagy events were determined from the co-localization between MitoTracker red and LysoTracker green. As shown, 100,000 cells per well were plated on 8-well Lab-Tek chambered cover-glass slides pre-coated coated with poly-L-lysine (Fig. 5). After 4 h CQ exposure, cells were washed with phenol-free media and incubated with 25 nM MitoTracker Red and 100 nM LysoTracker Green. After 30 min, cells were imaged using a Zeiss LSM 710 Confocal Microscope [28], [33]. The percentage of red pixels overlapping with green pixels normalized to total red pixels was quantified using ImageJ (NIH Bethesda, MD USA). One limitation of this technique is that an increase of co-localization between MitoTracker and LysoTracker does not distinguish between increases in mitophagy or a blockade in degradation of mitochondria by the lysosome (Fig. 5A-B). Therefore, an autophagic flux analyses, to quantify the percentage of red pixels overlapping with green pixels normalized to total red pixels in the presence and absence of chloroquine (CQ), can be performed for assessing increase or decrease of flux [3].
Fig. 5.
Quantification of MitoTracker and LysoTracker co-localization and LC3 puncta in the presence and absence of chloroquine (CQ). (A) Cells were exposed to vehicle versus 40 µM CQ for 4 h and probed with LysoTracker (green) and MitoTracker (red) then live cell imaged at 63×. (B) The co-localization was quantified as the percentage of red signal co-localized with the green signal per field. Data = mean ± SEM, n = 3. (C) Representative immunocytochemistry images of neurons plated at 250,000 per well on autoclaved glass coverslips and treated with either control or CQ, then probed for LC3 puncta. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.).
Fig. 5C shows primary neurons probed for LC3 after 24 h CQ exposure. This experiment was performed by plating neurons at 240,000 cells per well on autoclaved glass coverslips in 24 well plates. After treatment, cells were fixed using a mixture of 4% paraformaldehyde and 4% sucrose for 20 min followed by washes with PBS. After fixation, cells were blocked with 5% BSA and 10% horse serum and then probed for LC3. AlexaFluor 488 and Hoechst were added before slides were mounted and visualized with a Leica TCS SP5 V confocal laser scanning microscope. Quantification can be performed by counting puncta per cell or cells with puncta [35], [36]. Capturing images routinely takes 1–2 h, and analysis routinely takes 1 h per field.
4.1.5. Assessing changes in mitochondrial protein and DNA
Measurement of mitochondrial parameters can be performed under the same conditions used to assess autophagy. The protein complexes in the mitochondria can be turned over at different rates and a decrease in mitochondrial quality can be detected as decreased specific activity of the mitochondrial enzymes and damaged mitochondrial DNA.
The levels of all five mitochondrial complexes can be assessed by measuring representative subunits from each complex. This can be done individually by purchasing separate antibodies and probing each complex individually [37], [38]. Another approach is to use commercially available antibody cocktails that probe for all five complexes at once [35] (Table 2) (Fig. 6A, B). The advantage of this approach is that it is economical if sample size is limiting but can be challenging to optimize when the relative levels of individual complexes and antibody binding is very different as occurs in some cell types. This is evident in Fig. 6A-B in which the ATP synthase subunit gives a strong signal whereas complex I and III signals are relatively weak. It is important to note that the relative strength of the signals does not necessarily indicate the relative abundance of the respiratory chain complexes since the antibody affinity for each protein in specific respiratory complexes is unlikely to be the same. Furthermore, it is important that samples should not be boiled prior to western blotting of mitochondrial electron transport chain subunits as boiling cause irreversible aggregation of some the proteins and will hinder accurate assessment of their levels.
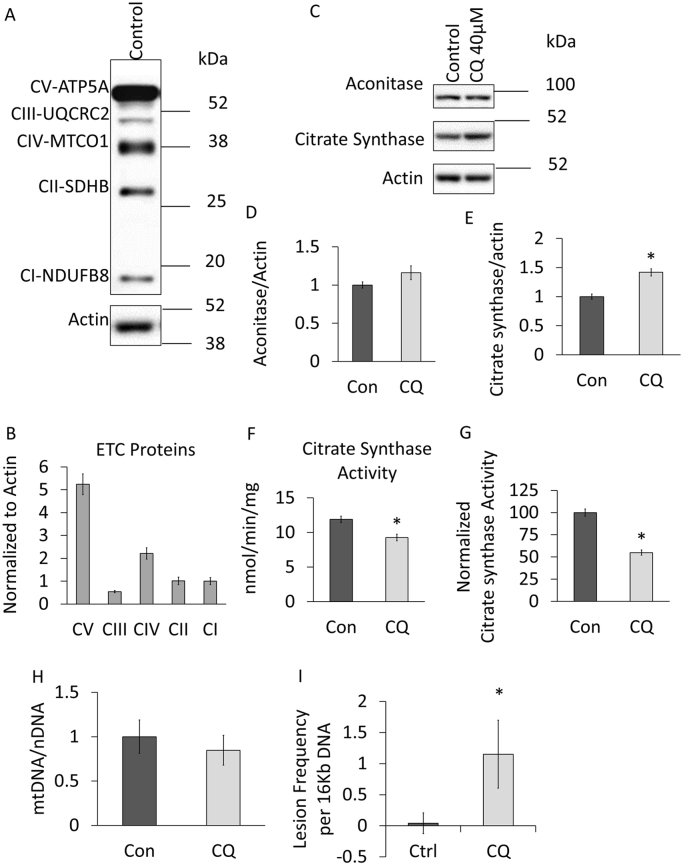
Fig. 6.
Assessment of mitochondrial proteins and DNA. (A-B) Levels of mitochondrial electron transport chain complex proteins were analyzed by western blot of extracts (samples mixed with sample buffers and at room temperature for 10 min without boiling) from primary cortical neurons by using Abcam's total OXPHOS rodent antibody cocktail, n = 3, normalized to CI, no statistical analysis performed. (C-I) Primary cortical neurons exposed for 24 h to 40 µM chloroquine (CQ). (C-E) Western blot analysis of two TCA cycle enzymes aconitase and citrate synthase. (F) Citrate synthase, a key enzyme in the TCA cycle, activity was measured by biochemical method and UV spectrometry. (G) The specific activity levels were then normalized to citrate synthase protein levels as determined in Fig. 6C, E. (H) Measurement of mtDNA copy number by normalizing mtDNA PCR to nDNA PCR. (I) Measurement of mtDNA damage. *p < 0.05 compared to control, n ≥ 3.
This approach can be extended to assess levels of mitochondrial proteins involved in other metabolic pathways such as the TCA cycle. In Fig. 6C-E we have performed western blot analyses (antibodies see Table 2) for aconitase and citrate synthase as representative TCA enzymes and observed modest but significant increase on citrate synthase while no changes were observed for aconitase in response to CQ. We further measured citrate synthase (CS) enzyme activity (Fig. 6F) and normalized to protein levels determined by western blotting (Fig. 6C, E) and found that although CS protein levels are increased by CQ, the specific activity is suppressed (Fig. 6G).
It is also important to measure mitochondrial DNA copy number as an additional indicator of mitochondrial mass. For DNA analyses, we routinely plate neurons in 24-well plates. This can be done by real-time PCR using mtDNA directed primers and then normalizing to nuclear DNA primers targeted to the 18S ribosome [28], [33] (Table 3). This is demonstrated in Fig. 6H, where no changes were observed after CQ exposure. We and others have used a mitochondrial DNA damage assay that calculates the amount of lesions per mitochondrial genome, providing an assessment of mitochondrial quality [31], [39], [40]. As shown in Fig. 6b, this assay reveals that CQ exposure results in an increase in mtDNA damage consistent with decreased mitochondrial quality.
Lesion frequency was calculated as follows [39]:
4.1.6. Measure changes in protein modification
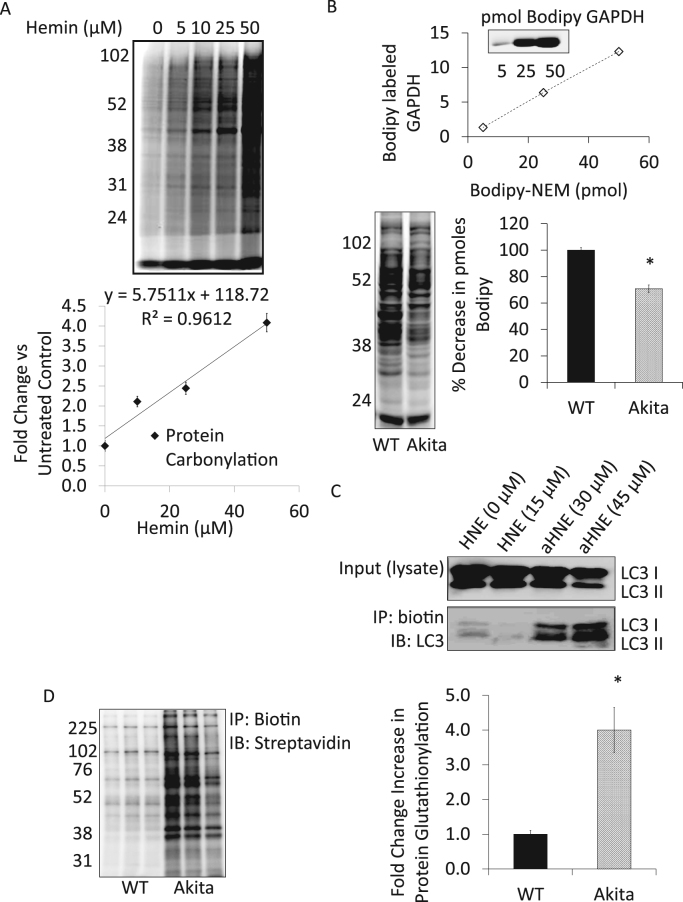
Protein modification can occur in response to cellular damage. These include carbonylation, thiol oxidation, HNE-protein adduct formation and protein glutathionylation. To determine protein carbonylation, samples can be derivatized with either Bodipy or biotin hydrazide and visualized by in-gel fluorescence or by western blot respectively. Fig. 7A shows an example of carbonyl detection in which cells were subjected to hemin, a pro-oxidant released from heme proteins during hemolysis for 4 h [41]. Post-treatment media was removed and the cells incubated with Bodipy hydrazide (1 mM) for 45 min at 37 °C in serum-free media after which protein lysates were prepared and protected from light [41]. To detect protein thiol modifications, intact cells can be incubated with 100 µM Bodipy-NEM for 15 min, a fluorophore-labeled alkylating agent that reacts specifically with thiol groups [42], [43]. Cell lysates can then be separated using 10% SDS-PAGE under non-reducing conditions, in-gel fluorescence imaging of the BODIPY signal is then used to visualize the thiol redox state [44]. An authentic standard curve can be made by incubating a known molar amount of a protein such as GAPDH with Bodipy hydrazide. The Bodipy-GAPDH standard can then be used for determining pmol thiol under basal conditions and under pharmacological stressors (Fig. 7B). Non-enzymatic lipid peroxidation protein adduct formation is a hallmark of oxidative stress and linked to several pathologies. Alkane HNE (aHNE) can be used as tool to identify key proteins that have been undergone an HNE modification [45]. A caveat to this approach is that the concentration of aHNE has to be determined experimentally to match results based on unmodified HNE. In the example shown, day 14 primary neurons (80 K) were incubated in the presence of HNE (15 µM) or aHNE (30 and 45 µM) for 4 h in a 96-well plate. Pooled cell lysates from 24 of these wells were harvested and post-labeled with biotin using the copper(I)-catalyzed azide/alkyne cycloaddition reaction (Click chemistry). Protein (200 µg) was incubated with NaBH4 (10 mM) for 1 h at RT followed by the addition of 2 mM ascorbate and 1 mM cupric sulfate and incubated at RT while mixing for at least 2 h in the dark (Samples can be stored at −80 °C at this point). Protein should be precipitated at this point to remove any remaining reactants and reconstituted using RIPA lysis buffer. Click chemistry labeling should be confirmed by running a streptavidin western blot. Once labeling has been confirmed, adducted proteins can be affinity enriched using neutravidin resin and the purified proteins validated by either western blot analysis and/or mass spectrometry. Fig. 7C shows an example of MAP1LC3A/B (microtubule-associated protein 1 light chain 3 alpha/beta) was modified at a concentration of 30–45 μM aHNE in primary neurons. Protein S-glutathionylation (PSSG) is a reversible post-translational protein modification resulting from increased levels of GSSG and has been shown to regulate enzyme activity and to protect critical thiol residues from irreversible oxidative damage [46]. As shown in Fig. 7D β-cells derived from Ins2Akita mice have increased levels of PSSG [47]. In this experiment shown, β-cells (3 × 106) were re-suspended in 100 µL of 1 mM GSH-biotin ethyl ester in serum-free medium and incubated O/N at 37 °C. Proteins were then extracted in lysis buffer containing 20 mM Tris, pH 7.4, 1% Triton x-100, 10 μM DTPA, 25 mM NEM, and protease inhibitors. To enrich for S-glutathiolated proteins, lysates were affinity purified using neutravidin beads and affinity purified lysates separated by SDS-PAGE and immunoblotted against streptavidin.
Fig. 7.
Analyses of protein oxidation. (A) Quantitative measurement of protein carbonylation. Bovine aortic endothelial cells were treated with increasing concentrations of hemin (0 – 50 µM) for 4 h. After media containing hemin was removed, the cells were treated with Bodipy Hydrazide (1 mM) for 45 min at 37 °C in serum-free media. (B) Detection of oxidative thiol protein modifications. WT and Akita+/Ins2 β-cells were treated with BODIPY-NEM (100 µM) for 15 min in the dark. Cell lysates were collected and separated using SDS-PAGE. Thiol modifications were visualized using in-gel fluorescence and scanned with a Typhoon fluorescence imager. BODIPY-GAPDH was used as a standard to determine pmol thiol content. Quantitation of free thiols was determined using ImageQuant software. n = 3. This study demonstrates that cysteine oxidation of proteins occurs in Akita β-cells and thus less BODIPY signal is detected. (C) Analyses of protein-HNE adduction. Primary neurons were treated with aHNE (30 and 45 µM) for 4 h prior to performing click chemistry. Western blot analysis with LC3 antibody of whole cell lysates (input) and biotin immunoprecipitation products. (D) Analyses of protein glutathionylation. WT and Akita+/Ins2-derived β-cells were treated with biotinylated glutathione ethyl ester (1 mM) in solution prior to affinity purification of S-glutathiolated proteins using streptavidin beads. Affinity purified proteins were separated using SDS-PAGE and probed for total glutathiolated proteins using streptavidin-HRP. Results are mean ± SEM, n = 3 per group, *p < 0.01 compared to WT β-cells.
4.1.7. Targeted metabolomics of TCA cycle metabolites
Metabolomics methods can be used to assess mitochondrial function [48]. There are various approaches to this which require varying levels of investment of resources to obtain the data and perform the analysis. As a first pass, we recommend some form of targeted metabolomics in which a limited number of analytes are assessed. The example shown here (Fig. 8) is a targeted metabolomics analysis of TCA cycle, glutaminolysis and glycolytic intermediates. For mouse cortical primary neurons, we plated 480,000 cells per well on 24-well plates. At the time for harvest, cells were washed with 1 mL of ice cold PBS, lysed and scraped in 500 µL of HPLC grade ice-cold methanol and incubated at −80 °C for 2 h. Then the plates were scraped again and cell lysates transferred to glass tubes and centrifuged at ~1000×g for 20 min at 4 °C. Deproteinated supernatants were then transferred to new glass tubes and stored at −80 °C [49].
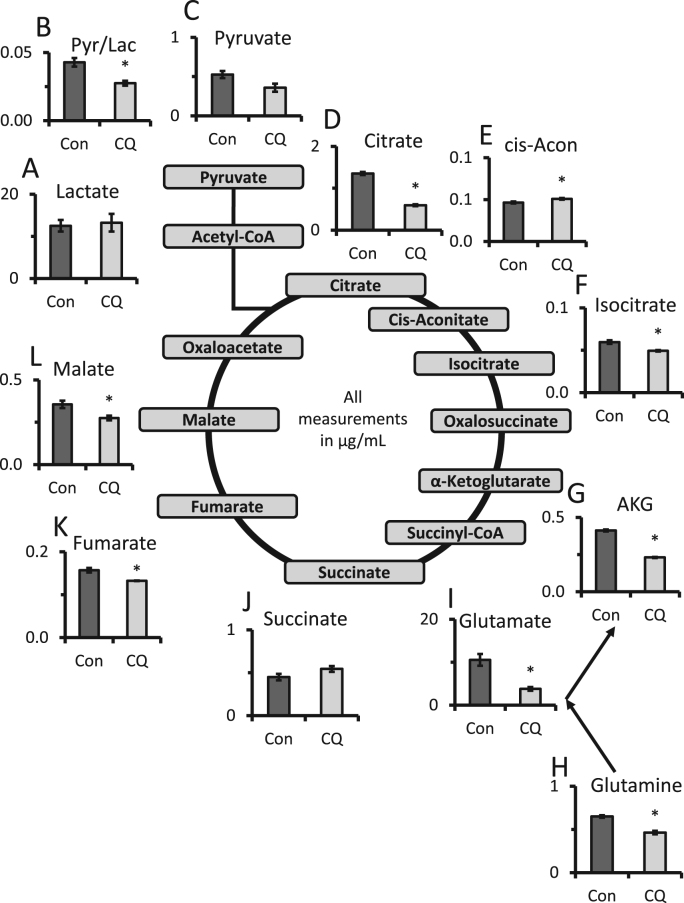
Fig. 8.
Targeted TCA metabolomics. (A-L) Targeted metabolomics of primary cortical rat neurons exposed to 40 µM chloroquine (CQ) for 24 h. Key TCA metabolites, including those important for glutaminolysis were measured by LC-multiple reaction ion monitoring-mass spectrometry. Metabolite levels are shown in μg/mL. *p < 0.05 compared to control.
For processing samples, a mix of 9 standards at 100 µg/mL in H2O was diluted to 10× of the final concentrations (0.05–10 µg/mL) before further diluted to 1× in methanol to a total volume of 1 mL and dried by a gentle stream of N2. For the experimental cell extracts, 1 mL of each were transferred to a glass tube and dried under a gentle stream of N2. Standards and samples were resuspended in 50 µL of 5% acetic acid, vortexed for 15 s. Amplifex™ Keto Reagent (SCIEX, Concord, Ontario, Canada) (50 µL) was added to each sample and incubated for 1 h at room temperature. Standards and samples were then dried again under a gentle stream of N2 and resuspended in 1 mL and 200 µL of 0.1% formic acid, respectively.
Samples were analyzed using LC-multiple reaction ion monitoring-mass spectrometry. Liquid chromatography was performed by LC20AC HPLC system (Shimadzu, Columbia, MD) with a Synergi Hydro-RP 4 µm 80 A 250 × 2 mm ID column (Phenomenex, Torrance, CA) with mobile phases of A) 0.1% formic acid and B) methanol/0.1% formic acid. Compounds were eluted using a 5–40% linear gradient of B from 1 to 7 min, followed by a column wash of 40–100% B from 7 to 10 min, and re-equilibrated at 5% B from 10.5 to 15 min. Eluants were passed into an electrospray ionization interface of an API 4000 triple-quadrupole mass spectrometer (SCIEX). The following mass transitions were monitored in the positive ion mode: m/z 261/118 for α-ketoglutarate, m/z 247/144 for oxaloacetate and m/z 204/144, 204/118 and 204/60 for pyruvate. In the negative mode, the following transitions are monitored: m/z 115/71 for fumarate, m/z 89/43 for lactate, m/z 117/73 for succinate, m/z 133/115 for malate, m/z 173/85 for cis-aconitate, m/z 191/87 for citrate, m/z 191/73 for isocitrate, m/z 147/129 for 2-hydroxyglutarate, m/z 146/102 for glutamate, m/z 145/42 for glutamine and m/z 132/88 for aspartate. The 16 transitions were each monitored for 35 ms, with a total cycle time of 560 ms. MS parameters were CAD 4, CUR 15, GS1 60, GS2 30, TEM 600, IS −3500 V for negative polarity mode and IS 4500 for positive polarity mode. Peak areas of metabolites in the sample extracts were compared in MultiQuant software (SCIEX) to those of the known standards to calculate metabolite concentrations.
Key steps of the TCA cycle and their products are outlined in Table 1. Under the condition of 24 h CQ exposure which decreased mitochondrial quality as assessed by increased mtDNA damage we observed marked decreases in citrate, cis-aconitate, and iso-citrate (Fig. 8). Key components of glutaminolysis including glutamine, glutamate, and α-ketoglutarate were also significantly decreased.
4.1.8. Assessment of mitochondrial function utilizing Seahorse extracellular flux
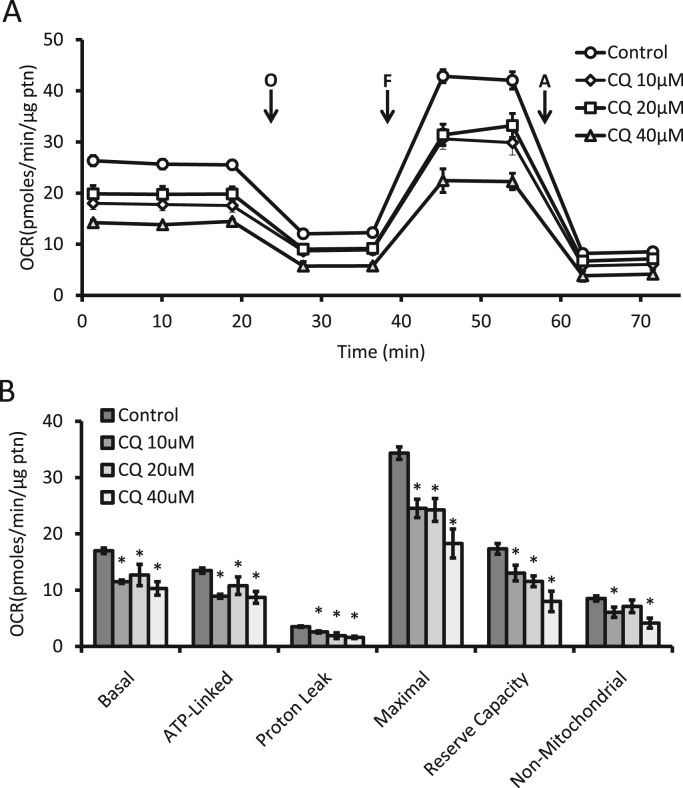
The development of Seahorse extracellular flux technology has allowed for the high throughput measurement of cellular bioenergetics and the activity of individual mitochondrial complexes within the cell and in isolated mitochondria [1], [24], [50], [51]. The principles and approaches of these methods have been outlined elsewhere, including how to optimize for plating densities and titrations for the optimal amount of oligomycin, FCCP, and antimycin A [1], [24]. Here we show how they can be employed as part of an integrated approach to assess mitochondrial quality control. For this analysis, we routinely plate neurons in XF96 plates with 5–10 replicated wells and then repeat experiments a minimum of 3 times using additional plates. In Fig. 9A, B, we have applied the mitochondria stress test, which consists of measurements of oxygen consumption rate (OCR) before and after the sequential injection of mitochondrial inhibitors. Oligomycin, FCCP, and antimycin A (Table 1) were used to measure basal (OCR before oligomycin minus OCR after antimycin), ATP-linked (OCR before oligomycin minus OCR after oligomycin), proton leak (OCR after oligomycin minus OCR after antimycin), maximal (OCR after FCCP minus OCR after antimycin), reserve capacity (OCR after FCCP minus OCR before oligomycin), and non-mitochondrial (OCR after antimycin) parameters of mitochondrial oxygen consumption. Here we show that upon exposure to increasing concentrations of CQ over a 4 h time span, mitochondrial OCR is diminished, with all parameters being significantly decreased.
Fig. 9.
Measurement of bioenergetic profile of intact cells. (A) Seahorse extracellular flux analysis of primary cortical rat neurons exposed for 4 h to increasing concentrations of chloroquine (CQ). Then the mitochondrial stress test was performed by measurements of OCR before and after sequential injections of oligomycin (O), FCCP (F), and antimycin A (A). (B) From panel A, basal, ATP-linked, proton leak, maximal, reserve capacity, and non-mitochondrial linked oxygen consumption rates were calculated. Shown is a representative experiment with data = mean ± SEM, n ≥ 3 independent samples, p < 0.05.
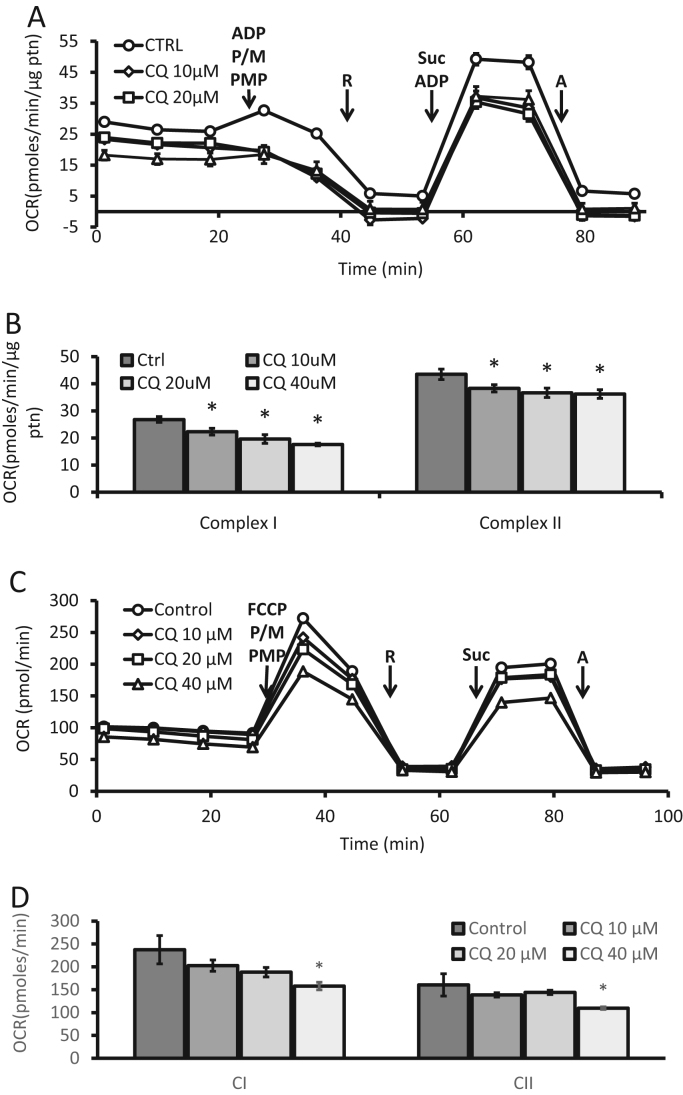
The intact-cell mitochondrial stress test used above is ideal for assessing the mitochondria's ability to provide energy in a cellular milieu. This can be complemented with the assay for the function of individual mitochondrial complexes. This is achieved by permeabilizing the cells with saponin, digitonin or other proprietary reagents [52] and providing specific substrates for each complex [52]. This has the advantage that since the mitochondria are assessed within the cell then selection bias for more robust mitochondria, which occurs in the more traditional methods which involve physical cell disruption, is avoided. In Fig. 10A, B, complex I and II substrate linked activities were measured by permeabilization with Plasma Membrane Permeabilizer (PMP; a proprietary reagent that permeabilizes intact cells in culture) and concurrent injection of pyruvate and malate that provides complex I substrates. ADP is also added as a substrate for complex V. The serial injection of rotenone followed by succinate and additional ADP, serves to inhibit complex I linked oxygen consumption and while providing substrates for complex II. Under these conditions in which ATP synthesis through complex V is rate limiting, both complex I and II linked oxygen consumption can be calculated. Using a 4 h CQ treatment we observed under cell permeabilized conditions, a significant decrease in both complex I and II linked activities.
Fig. 10.
Evaluation of complex I and II substrate linked activities in permeabilized cells. Seahorse extracellular flux analysis in PMP permeabilized primary cortical rat neurons exposed for 4 h to increasing concentrations of chloroquine (CQ). (A-B) ADP was added during permeabilization in the assay to determine OCR under ATP synthase limiting conditions. (C-D) FCCP was added during permeabilization in the assay to determine the maximum rate of respiration in the presence of substrates for each respective complex. Complex I substrate linked activities were measured by the addition of pyruvate (P) and malate (M) (calculated as OCR after these substrates minus OCR after rotenone (R). Complex II substrate linked activities were measured by the addition of succinate (suc) (calculated as OCR after substrates minus OCR after antimycin A (A)). Rotenone (R) was added to inhibit complex I before complex II measurements. Antimycin A (A) was added to inhibit respiration through electron transport chain. Shown is a representative experiment with data = mean ± SEM n ≥ 3 independent samples, p < 0.05.
In similar fashion to the mitochondrial stress test employed in Fig. 9 where FCCP was used to determine the maximal OCR, the same can be performed in permeabilized cells. In Fig. 10C, sequential injection of substrates and inhibitors were added as before, however, this time the injection of FCCP in the place of ADP allows for the maximal rate to be determined without limitation by complex V. Indeed, if the FCCP dependent activity is greater than ADP –dependent oxidation it suggests that complex V activity can be limiting for maximal mitochondrial function. In this example, we found that 40 µM CQ exposure for 4 h decreased both complex I and II substrate-linked activities in the presence of FCCP (Fig. 10D).
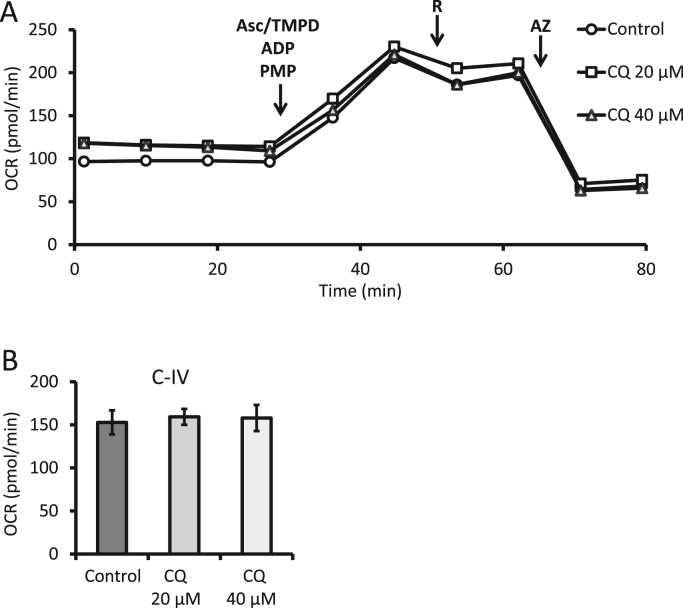
Complex IV substrate linked activities can be determined in similar fashion by using ascorbate and Tetramethyl-p-phenylenediamine (TMPD) allowing the flow of electrons through complex IV. Rotenone is used to mitigate the possible effect of reverse electron flow through complex I. Lastly, the addition of azide is used to terminate the experiment by inhibiting cytochrome c oxidase and is used as the baseline to correct of oxygen consumption by ascorbate/TMPD. As shown in Figs. 11, 4 h exposure to 40 µM CQ did not change complex IV substrate linked activities.
Fig. 11.
Evaluation of complex IV substrate linked activities. (A) Seahorse extracellular flux analysis of PMP permeabilized primary cortical rat neurons exposed for 4 h to increasing concentrations of chloroquine (CQ). (B) Complex IV substrate linked activities were calculated as OCR after substrates ascorbate (asc), TMPD, and rotenone (R), minus OCR after azide (AZ). Shown is a representative experiment with data = mean ± SEM n ≥ 3 independent samples, p < 0.05.
To determine whether any pharmacological reagent directly impacts mitochondrial bioenergetics, the compound can be directly injected into the assay wells in either intact or permeabilized cells. OCR can be observed as long as desired, then cells can be subjected to a mitochondrial stress test or complex activity assay at the end of the exposure. Preparation of each assay routinely takes 1–2 h.
5. Data interpretation
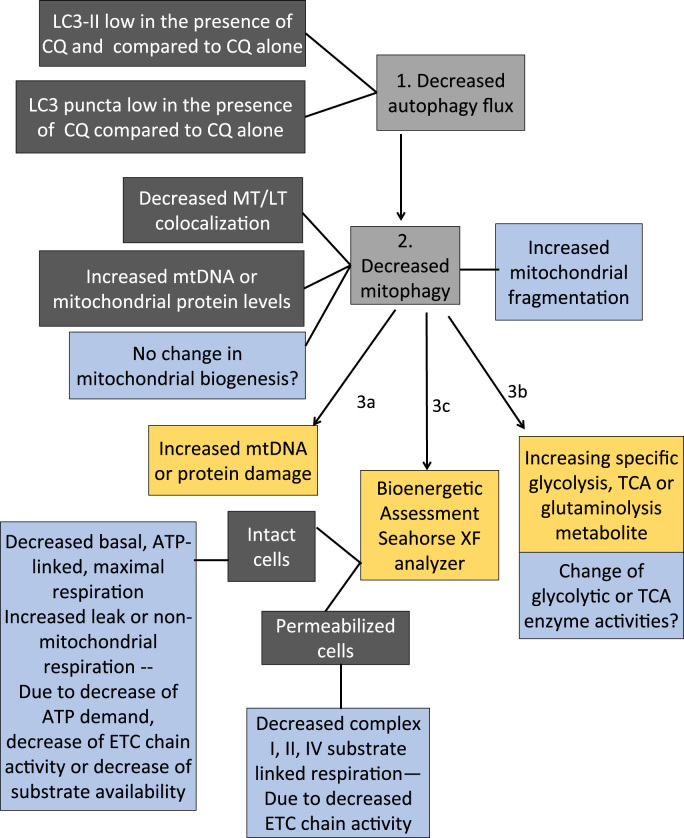
When experiments outlined here have been completed, it is then important to carefully analyze and compare the data obtained. Fig. 12 shows a decision tree for guidance in possible interpretation of possible observations. In assessing whether the mitochondrial quality control machinery is defective, autophagic flux measurement performed in the presence and absence of lysosomal disruptors (enzyme inhibitors or pharmacological neutralization of lysosomal pH), if LC3II protein and LC3 puncta are increased in the presence of reagents that block lysosomal mediated degradation (chloroquine, ammonium chloride, bafilomycin, or leupeptin), then autophagy is upregulated. If mitotracker and lysotracker colocalization or immuno-co-localization of mitochondrial and lysosomal proteins is increased, then mitophagy is increased. If mtDNA copy numbers normalized to nuclear DNA copy numbers, as well as mitochondrial proteins (VDAC, MnSOD, ETC subunits) normalized to cytosolic proteins (e.g. actin) are also decreased, then mitophagy is associated with decreased mitochondrial mass. To conclude this, additional measurements may be needed to determine whether mitochondrial biogenesis also contributes to this decrease. Insufficient mitophagy may decrease mitochondrial quality and can be assessed by mtDNA and protein damage and oxidation. Results from post-translational modifications (PTMs) such as protein carbonylation, protein thiol status, protein adduct formation and protein glutathionylation will give insight to the redox status of the cell. For example, increased protein carbonylation, glutathionylation and adduct formation are all indicative of oxidative stress indicating that the production of the PTMs has surpassed the cells ability to clear them. Decreased protein thiol content in the reduced form can be indicative of modification of critical cysteine residues which are important for enzyme function as well as signaling mechanisms.
Fig. 12.
Data interpretation decision tree for assessing mitochondrial quality control mechanisms and cellular consequences. 1) In autophagic flux measurements, if LC3II protein and LC3 puncta are decreased in the presence of reagents that block lysosomal mediated degradation (chloroquine, ammonium chloride, bafilomycin, or leupeptin), then these data indicate that autophagy is inhibited. 2) If mitotracker and lysotracker colocalization or immuno-co-localization of mitochondrial and lysosomal proteins is decreased, then the data indicate that mitophagy is inhibited. If mtDNA copy numbers normalized to nuclear DNA copy numbers, as well as mitochondrial proteins (VDAC, MnSOD, ETC subunits) normalized to cytosolic proteins (e.g. actin) are also increased, then the data is consistent with mitophagy being inhibited. However, to conclude this, additional measurements may be needed to determine whether mitochondrial biogenesis also contributes to this decrease. An increase of mitochondrial fragmentation may be a trigger for mitophagy initiation. However, a decrease of mitochondrial length would suggest that inhibition of mitophagy completion would lead to increased mitochondrial fragmentation and decreased mitochondrial networks. 3) a) Insufficient mitophagy may decrease mitochondrial quality and can be assessed by mtDNA and protein damage. b) Furthermore, if an increase of particular glycolysis or TCA cycle metabolites is determined by targeted metabolomics, this observation can suggest an increased synthesis of metabolites or decreased catabolism of metabolites. The activities of the enzymes catalyzing their synthesis and catabolism can then be determined to differentiate the two possibilities to determine the consequence of mitophagy inhibition on glycolysis and TCA cycle. c) Finally, in mitochondrial stress test in intact cells, inhibition of basal, ATP linked, or maximal respiration, or increase of leak or non-mitochondrial respiration suggest a decrease of ATP demand, substrate supply, or electron transport chain (ETC) activity, or increase of oxidase activities or uncoupling. Mitochondrial respiration assays in permeabilized cells in the presence of ADP versus FCCP will help distinguishing decreased substrate availability versus decreased complex I, II, IV or V activities. Combining these approaches can determine not only whether the mitochondrial quality control machinery is impaired, but also whether the inhibition of the quality control machinery results in perturbations of metabolism and bioenergetics, and increases of mitochondrial damage, which are related to disease pathologies.
A decrease of mitochondrial length suggests increased mitochondrial fragmentation and decreased mitochondrial networks which may be associated with decreased mitophagy. If an increase of particular glycolysis or TCA cycle metabolites is determined by targeted metabolomics, this can suggest an increased synthesis of metabolites or decreased catabolism of metabolites (altered flux). The activities of the enzymes catalyzing their synthesis and catabolism can then be measured to differentiate the two possibilities. Finally, changes of basal, ATP-linked, proton leak, maximal and non-mitochondrial respiration in the mitochondrial stress test can suggest whether ATP demand, turnover, substrate supply, uncoupling or ETC damage is involved in the altered mitochondrial quality control. Decreased complex I, II, and IV substrate linked oxygen consumption rates in permeabilized cells will directly assess the activities of the ETC complexes in excess of substrates. Combining these approaches can provide not only whether the mitochondrial quality control machinery is functioning, but also whether the increase or decrease of the quality control results in any changes in metabolism and bioenergetics, which are related to cellular and organismal health.
6. Advantages
Using the approach outlined in this article, we can assess mitochondrial quality and perturbation of the control mechanisms. Both biochemical and cellular biological methods are used that can detect the activities and the intracellular location of key processes. One can perform all the described experiments with consistent sample preparations and under well controlled laboratory conditions. The protocols will be detailed enough that one can easily teach a graduate student to perform these studies. The versatile integration of the combined approach will be useful for both preclinical animal and cell culture studies as well as of high potential for translational research including the applications related to precision medicine.
7. Limitations
As with any laboratory investigations, there are both conceptual and technical limitations. Technical limitations include the lack of spatial and temporal resolution when assessing enzymatic activities of each mitochondrial complexes and metabolites. Biochemical assessment of mitochondrial function and metabolism so far cannot be achieved effectively with a single mitochondrion, and thus can only assess the overall function in the entire population of mitochondria in a cell, and the cell populations in the assay unit. Targeted metabolomics analysis outline here can only give a snapshot of TCA, glycolytic and glutaminolysis intermediates. It cannot identify whether the increase or decrease of metabolites are the result of increased clearance or synthesis (flux) from enzymatic activities. Finally, when one decides to evaluate the reactive thiol proteome, considerations must be made as to the level of information needed to answer scientific questions. For example, biotin-NEM can be used instead of Bodipy-NEM with the use of reductants such ascorbate to delineate between cysteine oxidation states. Conceptually, this protocol does not integrate with proteasomal activity assays or reactive species detection, both of which can be important for mitochondrial quality control in health and diseases.
8. Concluding remarks
Taken together, we show here a workflow that allows researchers to investigate mitochondrial function and quality control that is adaptable to any cell-based system and should assist in helping researchers in addressing key questions of how post-translational modifications, mitochondrial function and autophagy are affected and interrelated in disease models.
Acknowledgements
This work was partially supported by UAB Health Service Foundation General Endowment Fund, UAB-UCSD O′Brien Acute Kidney Injury Center (P30 DK079337), UAB Blue Sky program, UAB AMC21 reload multi-investigator grant, Nathan Shock Center P30 G050886, and NIHR01-NS064090. The mass spectrometer was purchased with a grant from the UAB Health Services Foundation General Endownemt Fund.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.redox.2018.04.005.
Appendix A. Supplementary material
Supplementary material
References
- 1.Hill B.G., Benavides G.A., Lancaster J.R., Jr., Ballinger S., Dell'italia L., Zhang J., Darley-Usmar V.M. Integration of cellular bioenergetics with mitochondrial quality control and autophagy. Biol. Chem. 2012;393:1485–1512. doi: 10.1515/hsz-2012-0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang J. Teaching the basics of autophagy and mitophagy to redox biologists-Mechanisms and experimental approaches. Redox Biol. 2015;4C:242–259. doi: 10.1016/j.redox.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klionsky D.J., Abdelmohsen K., Abe A., Abedin M.J., Abeliovich H., Acevedo A.A., Adachi H., Adams C.M., Adams P.D., Adeli K., Adhihetty P.J., Adler S.G., Agam G., Agarwal R., Aghi M.K., Agnello M., Agostinis P., Aguilar P.V., guirre-Ghiso J., Airoldi E.M., it-Si-Ali S., Akematsu T., Akporiaye E.T., Al-Rubeai M., Albaiceta G.M., Albanese C., Albani D., Albert M.L., Aldudo J., Algul H., Alirezaei M., Alloza I., Almasan A., monte-Beceril M., Alnemri E.S., Alonso C., tan-Bonnet N., Altieri D.C., Alvarez S., varez-Erviti L., Alves S., Amadoro G., Amano A., Amantini C., Ambrosio S., Amelio I., Amer A.O., Amessou M., Amon A., An Z., Anania F.A., Andersen S.U., Andley U.P., Andreadi C.K., ndrieu-Abadie N., Anel A., Ann D.K., noopkumar-Dukie S., Antonioli M., Aoki H., Apostolova N., Aquila S., Aquilano K., Araki K., Arama E., Aranda A., Araya J., Arcaro A., Arias E., Arimoto H., Ariosa A.R., Armstrong J.L., Arnould T., Arsov I., Asanuma K., Askanas V., Asselin E., Atarashi R., Atherton S.S., Atkin J.D., Attardi L.D., Auberger P., Auburger G., Aurelian L., Autelli R., Avagliano L., Avantaggiati M.L., Avrahami L., Awale S., Azad N., Bachetti T., Backer J.M., Bae D.H., Bae J.S., Bae O.N., Bae S.H., Baehrecke E.H., Baek S.H., Baghdiguian S., Bagniewska-Zadworna A., Bai H., Bai J., Bai X.Y., Bailly Y., Balaji K.N., Balduini W., Ballabio A., Balzan R., Banerjee R., Banhegyi G., Bao H., Barbeau B., Barrachina M.D., Barreiro E., Bartel B., Bartolome A., Bassham D.C., Bassi M.T., Bast R.C., Jr., Basu A., Batista M.T., Batoko H., Battino M., Bauckman K., Baumgarner B.L., Bayer K.U., Beale R., Beaulieu J.F., Beck G.R., Jr., Becker C., Beckham J.D., Bedard P.A., Bednarski P.J., Begley T.J., Behl C., Behrends C., Behrens G.M., Behrns K.E., Bejarano E., Belaid A., Belleudi F., Benard G., Berchem G., Bergamaschi D., Bergami M., Berkhout B., Berliocchi L., Bernard A., Bernard M., Bernassola F., Bertolotti A., Bess A.S., Besteiro S., Bettuzzi S., Bhalla S., Bhattacharyya S., Bhutia S.K., Biagosch C., Bianchi M.W., Biard-Piechaczyk M., Billes V., Bincoletto C., Bingol B., Bird S.W., Bitoun M., Bjedov I., Blackstone C., Blanc L., Blanco G.A., Blomhoff H.K., Boada-Romero E., Bockler S., Boes M., Boesze-Battaglia K., Boise L.H., Bolino A., Boman A., Bonaldo P., Bordi M., Bosch J., Botana L.M., Botti J., Bou G., Bouche M., Bouchecareilh M., Boucher M.J., Boulton M.E., Bouret S.G., Boya P., Boyer-Guittaut M., Bozhkov P.V., Brady N., Braga V.M., Brancolini C., Braus G.H., Bravo-San Pedro J.M., Brennan L.A., Bresnick E.H., Brest P., Bridges D., Bringer M.A., Brini M., Brito G.C., Brodin B., Brookes P.S., Brown E.J., Brown K., Broxmeyer H.E., Bruhat A., Brum P.C., Brumell J.H., Brunetti-Pierri N., Bryson-Richardson R.J., Buch S., Buchan A.M., Budak H., Bulavin D.V., Bultman S.J., Bultynck G., Bumbasirevic V., Burelle Y., Burke R.E., Burmeister M., Butikofer P., Caberlotto L., Cadwell K., Cahova M., Cai D., Cai J., Cai Q., Calatayud S., Camougrand N., Campanella M., Campbell G.R., Campbell M., Campello S., Candau R., Caniggia I., Cantoni L., Cao L., Caplan A.B., Caraglia M., Cardinali C., Cardoso S.M., Carew J.S., Carleton L.A., Carlin C.R., Carloni S., Carlsson S.R., Carmona-Gutierrez D., Carneiro L.A., Carnevali O., Carra S., Carrier A., Carroll B. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2016;12:1–222. doi: 10.1080/15548627.2015.1100356. (3rd edition) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Redmann M., Darley-Usmar V., Zhang J. The role of autophagy, mitophagy and lysosomal functions in modulating bioenergetics and survival in the context of redox and proteotoxic damage: implications for neurodegenerative diseases. Aging Dis. 2016;7:150–162. doi: 10.14336/AD.2015.0820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Redmann M., Dodson M., Boyer-Guittaut M., Darley-Usmar V., Zhang J. Mitophagy mechanisms and role in human diseases. Int. J. Biochem. Cell Biol. 2014;53C:127–133. doi: 10.1016/j.biocel.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lemasters J.J. Selective mitochondrial autophagy, or mitophagy, as a targeted defense against oxidative stress, mitochondrial dysfunction, and aging. Rejuvenation. Res. 2005;8:3–5. doi: 10.1089/rej.2005.8.3. [DOI] [PubMed] [Google Scholar]
- 7.Wende A.R., Young M.E., Chatham J., Zhang J., Rajasekaran N.S., Darley-Usmar V.M. Redox biology and the interface between bioenergetics, autophagy and circadian control of metabolism. Free Radic. Biol. Med. 2016;100:94–107. doi: 10.1016/j.freeradbiomed.2016.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taegtmeyer H., Young M.E., Lopaschuk G.D., Abel E.D., Brunengraber H., Darley-Usmar V., Des Rosiers C., Gerszten R., Glatz J.F., Griffin J.L., Gropler R.J., Holzhuetter H.G., Kizer J.R., Lewandowski E.D., Malloy C.R., Neubauer S., Peterson L.R., Portman M.A., Recchia F.A., Van Eyk J.E., Wang T.J. American Heart Association Council on basic cardiovascular, S. Assessing cardiac metabolism: a scientific Statement From the American heart Association. Circ. Res. 2016;118:1659–1701. doi: 10.1161/RES.0000000000000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schapira A.H. Mitochondria in the aetiology and pathogenesis of Parkinson's disease. Lancet Neurol. 2008;7:97–109. doi: 10.1016/S1474-4422(07)70327-7. [DOI] [PubMed] [Google Scholar]
- 10.Mecocci P., MacGarvey U., Beal M.F. Oxidative damage to mitochondrial DNA is increased in Alzheimer's disease. Ann. Neurol. 1994;36:747–751. doi: 10.1002/ana.410360510. [DOI] [PubMed] [Google Scholar]
- 11.Swerdlow R.H. Pathogenesis of Alzheimer's disease. Clin. Interv. Aging. 2007;2:347–359. [PMC free article] [PubMed] [Google Scholar]
- 12.Swerdlow R.H., Burns J.M., Khan S.M. The Alzheimer's disease mitochondrial cascade hypothesis. J. Alzheimers Dis. 2010;20(Suppl. 2):S265–S279. doi: 10.3233/JAD-2010-100339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lustbader J.W., Cirilli M., Lin C., Xu H.W., Takuma K., Wang N., Caspersen C., Chen X., Pollak S., Chaney M., Trinchese F., Liu S., Gunn-Moore F., Lue L.F., Walker D.G., Kuppusamy P., Zewier Z.L., Arancio O., Stern D., Yan S.S., Wu H. ABAD directly links Abeta to mitochondrial toxicity in Alzheimer's disease. Science. 2004;304:448–452. doi: 10.1126/science.1091230. [DOI] [PubMed] [Google Scholar]
- 14.Hirai K., Aliev G., Nunomura A., Fujioka H., Russell R.L., Atwood C.S., Johnson A.B., Kress Y., Vinters H.V., Tabaton M., Shimohama S., Cash A.D., Siedlak S.L., Harris P.L., Jones P.K., Petersen R.B., Perry G., Smith M.A. Mitochondrial abnormalities in Alzheimer's disease. J. Neurosci. 2001;21:3017–3023. doi: 10.1523/JNEUROSCI.21-09-03017.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang X., Wang W., Li L., Perry G., Lee H.G., Zhu X. Oxidative stress and mitochondrial dysfunction in Alzheimer's disease. Biochim. Biophys. Acta. 2014;1842:1240–1247. doi: 10.1016/j.bbadis.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bove J., Perier C. Neurotoxin-based models of Parkinson's disease. Neuroscience. 2011 doi: 10.1016/j.neuroscience.2011.10.057. [DOI] [PubMed] [Google Scholar]
- 17.Dranka B.P., Zielonka J., Kanthasamy A.G., Kalyanaraman B. Alterations in bioenergetic function induced by Parkinson's disease mimetic compounds: lack of correlation with superoxide generation. J. Neurochem. 2012 doi: 10.1111/j.1471-4159.2012.07836.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giordano S., Lee J., Darley-Usmar V.M., Zhang J. Distinct effects of rotenone, 1-methyl-4-phenylpyridinium and 6-hydroxydopamine on cellular bioenergetics and cell death. PLoS. ONE. 2012;7:e44610. doi: 10.1371/journal.pone.0044610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dodson M., Liang Q., Johnson M.S., Redmann M., Fineberg N., Darley-Usmar V.M., Zhang J. Inhibition of glycolysis attenuates 4-hydroxynonenal-dependent autophagy and exacerbates apoptosis in differentiated SH-SY5Y neuroblastoma cells. Autophagy. 2013;9:1996–2008. doi: 10.4161/auto.26094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sayre L.M., Zelasko D.A., Harris P.L., Perry G., Salomon R.G., Smith M.A. 4-Hydroxynonenal-derived advanced lipid peroxidation end products are increased in Alzheimer's disease. J. Neurochem. 1997;68:2092–2097. doi: 10.1046/j.1471-4159.1997.68052092.x. [DOI] [PubMed] [Google Scholar]
- 21.Lovell M.A., Ehmann W.D., Mattson M.P., Markesbery W.R. Elevated 4-hydroxynonenal in ventricular fluid in Alzheimer's disease. Neurobiol. Aging. 1997;18:457–461. doi: 10.1016/s0197-4580(97)00108-5. [DOI] [PubMed] [Google Scholar]
- 22.Yoritaka A., Hattori N., Uchida K., Tanaka M., Stadtman E.R., Mizuno Y. Immunohistochemical detection of 4-hydroxynonenal protein adducts in Parkinson disease. Proc. Natl. Acad. Sci. USA. 1996;93:2696–2701. doi: 10.1073/pnas.93.7.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hill B.G., Dranka B.P., Zou L., Chatham J.C., Darley-Usmar V.M. Importance of the bioenergetic reserve capacity in response to cardiomyocyte stress induced by 4-hydroxynonenal. Biochem. J. 2009;424:99–107. doi: 10.1042/BJ20090934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dranka B.P., Benavides G.A., Diers A.R., Giordano S., Zelickson B.R., Reily C., Zou L., Chatham J.C., Hill B.G., Zhang J., Landar A., Darley-Usmar V.M. Assessing bioenergetic function in response to oxidative stress by metabolic profiling. Free Radic. Biol. Med. 2011;51:1621–1635. doi: 10.1016/j.freeradbiomed.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giordano S., Darley-Usmar V., Zhang J. Autophagy as an essential cellular antioxidant pathway in neurodegenerative disease. Redox Biol. 2014;2:82–90. doi: 10.1016/j.redox.2013.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hill B.G., Benavides G.A., Lancaster J.R., Jr., Ballinger S., Dell'Italia L., Jianhua Z., Darley-Usmar V.M. Integration of cellular bioenergetics with mitochondrial quality control and autophagy. Biol. Chem. 2012;393:1485–1512. doi: 10.1515/hsz-2012-0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Benavides G.A., Liang Q., Dodson M., Darley-Usmar V., Zhang J. Inhibition of autophagy and glycolysis by nitric oxide during hypoxia-reoxygenation impairs cellular bioenergetics and promotes cell death in primary neurons. Free Radic. Biol. Med. 2013;65:1215–1228. doi: 10.1016/j.freeradbiomed.2013.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Giordano S., Dodson M., Ravi S., Redmann M., Ouyang X., Darley-Usmar V.M., Zhang J. Bioenergetic adaptation in response to autophagy regulators during rotenone exposure. J. Neurochem. 2014;131:625–633. doi: 10.1111/jnc.12844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beaudoin G.M., III, Lee S.H., Singh D., Yuan Y., Ng Y.G., Reichardt L.F., Arikkath J. Culturing pyramidal neurons from the early postnatal mouse hippocampus and cortex. Nat. Protoc. 2012;7:1741–1754. doi: 10.1038/nprot.2012.099. [DOI] [PubMed] [Google Scholar]
- 30.Redmann M., Wani W.Y., Volpicelli-Daley L., Darley-Usmar V., Zhang J. Trehalose does not improve neuronal survival on exposure to alpha-synuclein pre-formed fibrils. Redox Biol. 2017;11:429–437. doi: 10.1016/j.redox.2016.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Redmann M., Benavides G.A., Berryhill T.F., Wani W.Y., Ouyang X., Johnson M.S., Ravi S., Barnes S., Darley-Usmar V.M., Zhang J. Inhibition of autophagy with bafilomycin and chloroquine decreases mitochondrial quality and bioenergetic function in primary neurons. Redox Biol. 2016;11:73–81. doi: 10.1016/j.redox.2016.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liang Q., Benavides G.A., Vassilopoulos A., Gius D., Darley-Usmar V., Zhang J. Bioenergetic and autophagic control by Sirt3 in response to nutrient deprivation in mouse embryonic fibroblasts. Biochem. J. 2013;454:249–257. doi: 10.1042/BJ20130414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boyer-Guittaut M., Poillet L., Liang Q., Bole-Richard E., Ouyang X., Benavides G.A., Chakrama F.Z., Fraichard A., Darley-Usmar V.M., Despouy G., Jouvenot M., Delage-Mourroux R., Zhang J. The role of GABARAPL1/GEC1 in autophagic flux and mitochondrial quality control in MDA-MB-436 breast cancer cells. Autophagy. 2014;10:986–1003. doi: 10.4161/auto.28390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Crabtree D., Dodson M., Ouyang X., Boyer-Guittaut M., Liang Q., Ballestas M.E., Fineberg N., Zhang J. Over-expression of an inactive mutant cathepsin D increases endogenous alpha-synuclein and cathepsin B activity in SH-SY5Y cells. J. Neurochem. 2014;128:950–961. doi: 10.1111/jnc.12497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dodson M., Wani W.Y., Redmann M., Benavides G.A., Johnson M., Ouyang X., Cofield S.S., Mitra K., Darley-Usmar V., Zhang J. Regulation of autophagy, mitochondrial dynamics and cellular bioenergetics by 4-hydroxynonenal in primary neurons. Autophagy. 2017 doi: 10.1080/15548627.2017.1356948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wani W.Y., Ouyang X., Benavides G.A., Redmann M., Cofield S., Shacka J.J., Darley-Usmar V., Chatham J.C., Zhang J. O-GlcNAc regulation of autophagy and α-synuclein homeostasis; implications for Parkinson's disease. Mol. Brain. 2017 doi: 10.1186/s13041-017-0311-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mitchell T., Johnson M.S., Ouyang X., Chacko B.K., Mitra K., Lei X., Gai Y., Moore D.R., Barnes S., Zhang J., Koizumi A., Ramanadham S., Darley-Usmar V.M. Dysfunctional mitochondrial bioenergetics and oxidative stress in Akita+/Ins2-derived beta-cells. Am. J. Physiol. Endocrinol. Metab. 2013;305:E585–E599. doi: 10.1152/ajpendo.00093.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schneider L., Giordano S., Zelickson B.R., Johnson S., Benavides A., Ouyang X., Fineberg N., Darley-Usmar V.M., Zhang J. Differentiation of SH-SY5Y cells to a neuronal phenotype changes cellular bioenergetics and the response to oxidative stress. Free Radic. Biol. Med. 2011;51:2007–2017. doi: 10.1016/j.freeradbiomed.2011.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Knight-Lozano C.A., Young C.G., Burow D.L., Hu Z.Y., Uyeminami D., Pinkerton K.E., Ischiropoulos H., Ballinger S.W. Cigarette smoke exposure and hypercholesterolemia increase mitochondrial damage in cardiovascular tissues. Circulation. 2002;105:849–854. doi: 10.1161/hc0702.103977. [DOI] [PubMed] [Google Scholar]
- 40.Fetterman J.L., Pompilius M., Westbrook D.G., Uyeminami D., Brown J., Pinkerton K.E., Ballinger S.W. Developmental exposure to second-hand smoke increases adult atherogenesis and alters mitochondrial DNA copy number and deletions in apoE(-/-) mice. PLoS ONE. 2013;8:e66835. doi: 10.1371/journal.pone.0066835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Higdon A.N., Benavides G.A., Chacko B.K., Ouyang X., Johnson M.S., Landar A., Zhang J., Darley-Usmar V.M. Hemin causes mitochondrial dysfunction in endothelial cells through promoting lipid peroxidation: the protective role of autophagy. Am. J. Physiol. Heart Circ. Physiol. 2012;302:H1394–H1409. doi: 10.1152/ajpheart.00584.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mitchell T., Johnson M.S., Ouyang X., Chacko B.K., Mitra K., Lei X., Gai Y., Moore D.R., Barnes S., Zhang J., Koizumi A., Ramanadham S., Darley-Usmar V.M. Dysfunctional mitochondrial bioenergetics and oxidative stress in Akita(+/Ins2)-derived beta-cells. Am. J. Physiol. Endocrinol. Metab. 2013;305:E585–E599. doi: 10.1152/ajpendo.00093.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oh J., Johnson M.S., Landar A. Methods for determining the modification of protein thiols by reactive lipids. Methods Cell Biol. 2007;80:417–434. doi: 10.1016/S0091-679X(06)80021-X. [DOI] [PubMed] [Google Scholar]
- 44.Hill B.G., Reily C., Oh J.Y., Johnson M.S., Landar A. Methods for the determination and quantification of the reactive thiol proteome. Free Radic. Biol. Med. 2009;47:675–683. doi: 10.1016/j.freeradbiomed.2009.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ravi S., Johnson M.S., Chacko B.K., Kramer P.A., Sawada H., Locy M.L., Wilson L.S., Barnes S., Marques M.B., Darley-Usmar V.M. Modification of platelet proteins by 4-hydroxynonenal: potential Mechanisms for inhibition of aggregation and metabolism. Free Radic. Biol. Med. 2016;91:143–153. doi: 10.1016/j.freeradbiomed.2015.10.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hill B.G., Higdon A.N., Dranka B.P., Darley-Usmar V.M. Regulation of vascular smooth muscle cell bioenergetic function by protein glutathiolation. Biochim. Biophys. Acta. 2010;1797:285–295. doi: 10.1016/j.bbabio.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cyr A.R., Brown K.E., McCormick M.L., Coleman M.C., Case A.J., Watts G.S., Futscher B.W., Spitz D.R., Domann F.E. Maintenance of mitochondrial genomic integrity in the absence of manganese superoxide dismutase in mouse liver hepatocytes. Redox Biol. 2013;1:172–177. doi: 10.1016/j.redox.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bernard K., Logsdon N.J., Benavides G.A., Sanders Y., Zhang J., Darley-Usmar V.M., Thannickal V.J. Glutaminolysis is required for transforming growth factor-beta1-induced myofibroblast differentiation and activation. J. Biol. Chem. 2018;293:1218–1228. doi: 10.1074/jbc.RA117.000444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith M.R., Vayalil P.K., Zhou F., Benavides G.A., Beggs R.R., Golzarian H., Nijampatnam B., Oliver P.G., Smith R.A., Murphy M.P., Velu S.E., Landar A. Mitochondrial thiol modification by a targeted electrophile inhibits metabolism in breast adenocarcinoma cells by inhibiting enzyme activity and protein levels. Redox Biol. 2016;8:136–148. doi: 10.1016/j.redox.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ravi S., Chacko B., Sawada H., Kramer P.A., Johnson M.S., Benavides G.A., O'Donnell V., Marques M.B., Darley-Usmar V.M. Metabolic plasticity in resting and thrombin activated platelets. PLoS One. 2015;10:e0123597. doi: 10.1371/journal.pone.0123597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Salabei J.K., Gibb A.A., Hill B.G. Comprehensive measurement of respiratory activity in permeabilized cells using extracellular flux analysis. Nat. Protoc. 2014;9:421–438. doi: 10.1038/nprot.2014.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Salabei J.K., Gibb A.A., Hill B.G. Comprehensive measurement of respiratory activity in permeabilized cells using extracellular flux analysis. Nat. Protoc. 2014;9:421–438. doi: 10.1038/nprot.2014.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material