Abstract
We show for the first time that, in contrast to other glutathione transferases and peroxidases, deletion of microsomal glutathione transferase 1 (MGST1) in mice is embryonic lethal. To elucidate why, we used zebrafish development as a model system and found that knockdown of MGST1 produced impaired hematopoiesis. We show that MGST1 is expressed early during zebrafish development and plays an important role in hematopoiesis. High expression of MGST1 was detected in regions of active hematopoiesis and co-expressed with markers for hematopoietic stem cells. Further, morpholino-mediated knock-down of MGST1 led to a significant reduction of differentiated hematopoietic cells both from the myeloid and the lymphoid lineages. In fact, hemoglobin was virtually absent in the knock-down fish as revealed by diaminofluorene staining. The impact of MGST1 on hematopoiesis was also shown in hematopoietic stem/progenitor cells (HSPC) isolated from mice, where it was expressed at high levels. Upon promoting HSPC differentiation, lentiviral shRNA MGST1 knockdown significantly reduced differentiated, dedicated cells of the hematopoietic system. Further, MGST1 knockdown resulted in a significant lowering of mitochondrial metabolism and an induction of glycolytic enzymes, energetic states closely coupled to HSPC dynamics. Thus, the non-selenium, glutathione dependent redox regulatory enzyme MGST1 is crucial for embryonic development and for hematopoiesis in vertebrates.
Abbreviations: GST, glutathione transferase; HSC, hematopoietic stem cell; HSPC, hematopoietic stem/progenitor cells; hpf, hours post fertilization; MGST1, microsomal glutathione transferase 1; zf, zebrafish
Keywords: Embryonic development, Hematopoiesis, Redox regulation, Microsomal glutathione transferase/peroxidase
Graphical abstract
Highlights
-
•
A redox enzyme, MGST1, is indispensable for mouse embryonic development.
-
•
Knock-down experiments in zebrafish embryos revealed a critical role for MGST1 in hematopoiesis.
-
•
A function in hematopoiesis was confirmed in a mammalian cell model.
1. Introduction
Higher organisms have multiple defenses against toxic chemical insult and oxidative stress that include thioredoxin and glutathione dependent enzymes of which some are selenium dependent [1], [2]. These enzymes reduce hydrogen peroxide and/or lipid hydroperoxides, catalytic activities that are also emerging as important processes in biological redox regulation [3]. The enzymes that perform these functions are plentiful and organized in several families (thioredoxin reductases [4], Se-dependent glutathione peroxidases [5], Se-independent glutathione peroxidases (i.e. glutathione transferases) and peroxiredoxins [6]). As the enzymes are organized in families and have overlapping functions most knockout animals created to date have been found to be viable, albeit more sensitive to toxic challenge [7], [8], [9]. There are however, notable exceptions, including embryonic lethality of GPX4 (Se-dependent glutathione peroxidase) [10] and thioredoxin system deletions in mice [11]. It is thought that vital developmental and cellular processes are regulated by these enzymes via redox control [9] accounting for their absolute necessity. GPX4 forms part of a system where lipid hydroperoxides are generated enzymatically by lipoxygenases and reduced by GPX4 to affect various cellular outcomes [12], [13], [14]. Microsomal glutathione transferase 1 (MGST1) shares an important unique characteristic with GPX4 [15], namely the ability to reduce lipid hydroperoxides directly in membranes [16]. MGST1 knockout mice have not so far been investigated. We therefore generated knockout mice of MGST1 to determine whether this enzyme is also necessary for embryonic development. Indeed, we show here that knockouts are embryonic lethal. In order to gain insight into the function of MGST1 in development, we have carried out zebrafish knockdown experiments and subsequently identified a role in hematopoiesis that was confirmed in a mouse hematopoiesis stem cell system. Redox regulation of hematopoiesis or other aspects of development by different selenium- and GSH dependent redox systems (e.g. TrxR2 [17] and GPX4 [18]) has previously been demonstrated. Thus, lipid peroxide signaling and attenuation by MGST1 in this case, is suggested as a mechanism in embryonic development redox biology affecting hematopoiesis.
2. Materials and methods
2.1. Mouse knockouts
We previously described the isolation and sequencing of MGST1 gene from a 129/SvJ genomic P1 plasmid [19]. The targeting vector pGKneob46 (4570 bp) was obtained from the UCSD Transgenic Core Services. A fragment of the murine MGST1 gene from nucleotides 412–9743 and containing exon 2 was inserted 5′ to the neo selection marker, and a fragment from nucleotides 9743–12,884 containing exon 3 was inserted downstream of the marker (all nucleotide numbering pertains to Genbank Accession number AY329626.1). A 1200 bp insert containing Thymidine-kinase promoter linked to Diptheria Toxin-A (YK-DTa) was then inserted at the NotI restriction site as a negative selection marker to allow the enrichment of homologous recombinants prior to screening [20]. The entire vector was sequenced to confirm nucleotide identity.
Production and breeding of mice missing the MGST1 gene was by conventional methods [21], [22], [23] by the UCSD Transgenic Animal Core Services. Mice were maintained under standard 12-h light/12-h dark cycle with access to standard rodent food and given water ad libitum. All protocols for mouse experiments were approved by the Institutional Animal Care and Use Committee at the University of California San Diego. In summary, following electroporation of the targeting construct into 129 J ES cells and selection of clones with G418, clones were analyzed by Southern blotting after Sac I digestion as described below. Selected ES clones were injected into C57BL/6 blastocysts. Male chimaeras were then backcrossed to C57BL/6 females to generate heterozygous founders (MGST1+/-). Confirmation of deletion of the MGST1 gene was performed by Southern Blot analysis using DNA recovered from tail tips (SacI, XbaI, HindIII digest, etc.) and the following probes: Probe A containing nucleotides 6042–6215 (upstream of exon 2); Probe B containing nucleotides 11,152–11,431 and containing exon 2; Probe C containing nucleotides 11,520–12,802 and containing exon 3 (see Fig. S1 for an example). Animals deemed partially heterozygous for MGST1 deletion were retained for breeding purposes, and a colony containing animals deemed heterozygous for the MGST1 gene was eventually obtained.
2.1.1. Analysis of embryos to identify MGST1 -/- mice
A total of 48 embryos were harvested from female heterozygous mice crossbred to male heterozygous mice in the following number and times: 9 embryos at day 7.5; 5 embryos at day 8.5; 5 embryos at day 11.5; 8 embryos at day 8.0; 8 embryos at day 8.5; 4 embryos at day 14.5; and 9 embryos at day 15. No embryos were recovered with a DNA profile consistent with a knockout or null MGST1 genotype (-/-). A representative Southern blot of embryo DNA analysis is provided (Fig. S2).
2.1.2. Expression of MGST1 protein in mice
Relative expression of MGST1 protein was studied by Western Blot analysis. Briefly, 2 μg of rough liver endoplasmic reticulum protein was loaded per lane. For detection a 1:2000 dilution of rabbit anti-MGST1 polyclonal antibody was used, followed by a 1:10,000 dilution of ECL-anti-rabbit horseradish peroxidase labeled antibody.
2.2. Zebrafish husbandry and injections
Zebrafish were housed in standard conditions and embryos staged according to established protocols [24]. All experiments were performed in compliance with national ethical guidelines. The morpholinos knocking down MGST1a and b were designed and obtained from Genetools (www.gene-tools.com) and targeted against the ATG codon. A scrambled morpholino was used as a control. The sequences can be found in Supplementary Table S1. Morpholinos were injected as described before [25]. Morphants were viable during the 96 h observation window (after this the morpholino gets diluted and less efficient).
2.3. In situ hybridization and hemoglobin staining
Diaminofluorene staining for hemoglobin was performed as described previously [26]. Synthesis of DIG-labeled RNA probes and in situ hybridization was carried out using standard protocols [27]. Plasmids encoding the transcription factors (gata1: NM_131234; fog1: AY515850; mpx: NM_212779; mpeg1.1: NM_212737; rag1: NM_131389) were generously provided by the zebrafish community. Statistical analysis is not provided on staining methods, but rather on the quantification through RTqPCR.
2.4. Antibody production, immunohistochemistry and in situ hybridization
Antibodies recognizing zfMGST1a and b respectively were generated in guinea pigs and obtained from Thermofisher. Anti zfcmyb antibodies were obtained from Anaspec and used in a 1/250 dilution. Secondary antibodies were purchased from Sigma and used in a 1/2000 dilution.
2.5. GST activity measurements
50–60 embryos were collected and transferred to 400 µL of ice cold 0.1 M potassium phosphate buffer pH 6.5 containing 1% Triton X-100 followed by gentle homogenization on ice using a glass-glass homogenizer. Homogenates were sonicated on ice for 3 × 10 s with 10 s cooling in between (Branson 2.200 sonifier) and analyzed directly. The fluorimetric GST activity assay was performed in a total volume of 100 µL at 22 °C in potassium phosphate buffer pH 6.5, 1% Triton X-100 with 5 mM GSH and 2.5 µM 2,4-dinitrobenzene sulfonamide-acetylrhodamine 110 (DNS-AcRh), using excitation at 490 nm and emission at 522 nm as previously described [28]. The enzymatic reaction was started by adding 25 µL of homogenate. The non-enzymatic background reaction rate was subtracted. Homogenate samples treated with N-ethyl maleimide (2 mM final concentration) were incubated on ice for 5 min before assayed.
2.6. Quantitative real-time PCR
Quantification of gene expression was performed using the Maxima SYBR Green qPCR Master Mix (Thermofisher) with detection on an AB 7500 Real-Time PCR System (Applied Biosystems) or a Rotorgene (Qiagen). The reaction cycles used were 95 °C for 2 min followed by 40 cycles at 95 °C for 15 s and 60 °C for 1 min followed by melt curve analysis. Relative gene expression quantification was based on the comparative threshold cycle method (2−ΔΔCt) using beta actin as an endogenous control gene. Verified primers from KICqStart (Sigma) were used. Experiments were performed in triplicate and at least three biological replicates were evaluated.
2.7. Lentivirus transfection
HEK-293T cells (1 × 106) were seeded in 6 cm cell culture plates in Dulbecco's modified Eagle medium (DMEM; ATCC, Manassas, VA), supplemented with 10%FBS, and incubated at 37 °C with 5% CO2 for 24 h. When the cells reached 50–80% confluence, pLKO.5 non-target (scrambled) shRNA or pLKO.5 MGST1 shRNA plasmids (both from Sigma-Aldrich, St. Louis, MO), together with psPAX2 and pMD2. G packaging plasmids were transfected into the cells using FuGENE 6. After 48 h and 72 h of transfection, the lentiviral particle solutions were collected.
Hematopoietic stem and progenitor cells (HSPC) were generated according to previous reported procedures [29] and maintained in StemSpan™ Serum-Free Expansion Medium (StemCell technologies, Vancouver, BC, Canada), supplemented with 10% FBS, 100 ng/ml recombinant mouse stem cell factor (Sigma, Saint Louis, MO), thrombopoietin, and Flt3L (both from BioAbChem, Ladson, SC) (HSPC medium). For lentivirus infections, HSPC were seeded in 24 well culture plates in HSPC medium and lentiviral particle solution (2:1) containing 8 µg/ml polybrene, and incubated at 37 °C with 5% CO2 for 24 h. Puromycin (2 µg/ml) was used to select the infected cells. After 96 h of infection, the HSPC, control or MGST1 knockdown (KD) cells, were collected and used for the following experiments.
2.8. Colony forming unit assay
Colony forming unit (CFU) assays were performed in complete M3434 methylcellulose medium (StemCell technologies, Vancouver, BC, Canada) following the manufacturer's instructions. Briefly, control or MGST1 KD HSPC (1000 cells/1.5 ml/well) were seeded in SmartDish™ 6-well culture plates in complete M3434 methylcellulose medium, and colonies of CFU-granulocyte macrophage (GM), burst-forming unit-erythroid (BFU-E), and CFU-granulocyte, erythrocyte, monocyte, and megakaryocyte (GEMM) were counted on day 6, 8 and 11, respectively.
2.9. Immunoblotting
Total soluble protein was quantitated by bicinchoninic acid protein assay (Pierce, Rockford, IL). Cell lysates were resolved in an SDS-loading buffer (80 mM Tris-HCl, pH 6.8, 2% SDS, 10% glycerol, 0.02% bromophenol blue, 5 mM tris (2-carboxyethyl) phosphine) and heated to 95 °C for 5 min. Equal amounts of protein were electrophoretically separated by SDS-PAGE (BioRad, Hercules, CA) and transferred onto low fluorescent polyvinylidene fluoride membranes (Millipore, Billerica, MA) by the Trans-Blot Turbo Transfer System (BioRad, Hercules, CA). PVDF membranes were incubated in the Odyssey blocking buffer (LI-COR) for 1 h and then probed with MGST1 or actin antibodies (both from Abcam, Cambridge, MA) at 4 °C overnight. Immunoblots were developed with infrared (IR) fluorescence IRDye secondary antibodies (LI-COR) at a dilution of 1:15,000, imaged with a two-channel (red and green) IR fluorescent Odyssey CLx imaging system (LI-COR) and quantified with Image Studio 4.0 software (LI-COR).
2.10. Microscopy, image processing, and statistics
Living specimens were mounted in low-melting agarose, fixed embryos in glycerol. Laser scanning microscopy images were taken with a Zeiss LSM700 confocal microscope and bright-field pictures with a Leica MZ16 microscope equipped with a Leica DFC300FX camera. Images were processed with ImageJ and Gimp (www.gimp.org) without obscuring original data. All data were expressed as mean ± SD. Statistical significance was calculated using two-tailed Student´s t-test. *: p < 0.05; **: p < 0.01; ***: p < 0.001.
3. Results
3.1. Knockout of MGST1 in mice is embryonic lethal
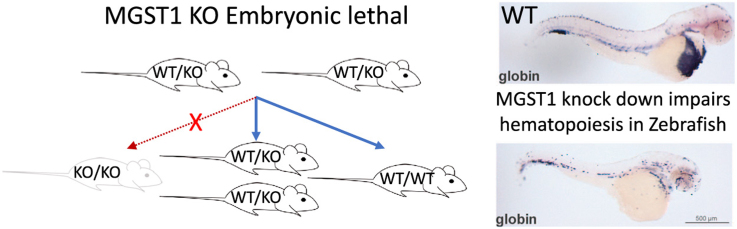
We generated mice that lacked the MGST1 gene by conventional methodology [21], [22], [23]. Upon breeding heterozygous mice, the offspring did not contain any homozygous null mice. Of 562 offspring 369 and 193 were heterozygous and wild type respectively. Heterozygous mice expressed about half the amount of MGST1 protein compared to wildtype (Fig. S3). Judging from the 2:1 proportion, heterozygous mice were not compromised in terms of embryonic development whereas homozygous embryos were resorbed prior to embryonic development stage E 7.5–10 (see Section 2 and Fig. S2). To investigate the functional role of MGST1 in development we decided to use zebrafish, a model system previously shown to be relevant for mammalian physiology and pathology.
3.2. Zebrafish contain two homologs of MGST1
To use zebrafish as our experimental model we first confirmed the presence of the gene examining the annotated zebrafish genome (GRCz10, www.ensembl.org). Two potential homologs of human MGST1 were identified. The isoforms (NM_001005957, termed MGST1a; and NM_001002215, termed MGST1b) are located on chromosome 4 and share ~ 56% identity at the amino acid level with the human homologue. Residues critical for activity [30] are conserved between fish and human (see Fig. S4) indicating similar functions. Our previous data showed that MGST1 from the freshwater pike (Esox lucius, 82% identical to the zebrafish MGST1b enzyme, comparing the 33 N-terminal amino acids) catalyzes reactions identical to the human enzyme, albeit with higher catalytic efficiency [31]. To date, fish MGST1 has been found to be one of the most efficient GSTs described.
3.3. MGST1 peaks during development and is strongly expressed in the intermediate cell mass, co-localized with markers of hematopoietic stem cells
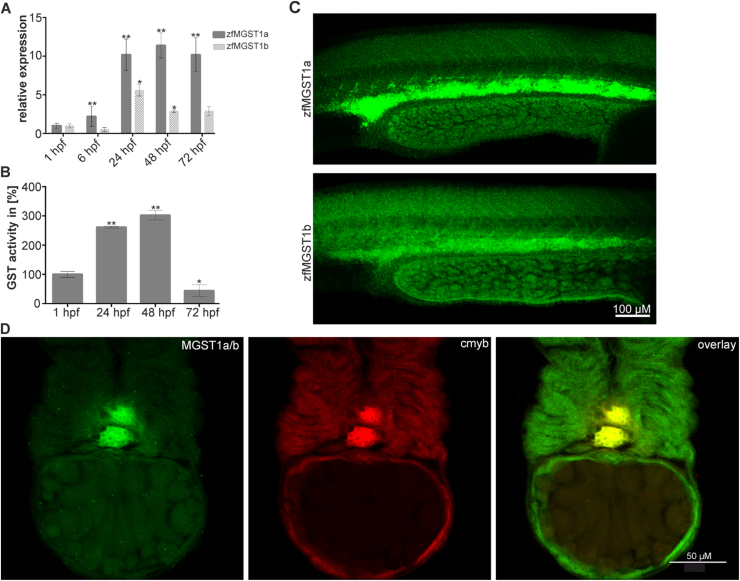
Since the zebrafish develops very rapidly with almost all major organs being formed during the first 3 days of development, we sought to study the expression profile of MGST1a and b during this developmental window. RTqPCR revealed that the expression of both isoforms increases during embryogenesis, reaching a maximum between 24 h post-fertilization (hpf) and 48 hpf (n = 6, 20 embryos/group) (see Fig. 1A). We also measured the total GST enzymatic activity in embryo lysates at these developmental stages, and found it to be highest at 48 hpf, confirming the mRNA expression data (n = 3, 50 embryos/group; see Fig. 1B). To obtain insight into what developmental process(es) could be regulated by MGST1 we next determined the location of the enzyme.
Fig. 1.
Expression of MGST1 during zebrafish embryo development. (A) Relative expression of zfMGST1a/b during zebrafish embryonic development. (B) Relative activity of GST enzymes during zebrafish embryonic development. (C) Expression of MGST1a and MGST1b in the intermediate cell mass of 24 hpf embryos. (D) MGST1a/b protein colocalizes with cmyb, a marker specific for HSCs, in 48 hpf embryos. Confocal images were taken on a Leica LSM700 with a 20× lens; Alexa 488 and 555 filters were used; images stacks were produced with ImageJ, GIMP was used to adjust the gamma channel.
We raised antibodies specific for zfMGST1a/b. Immunocytochemistry revealed that both isoforms are highly expressed in the intermediate cell mass at 24 hpf, the primary site of primitive hematopoiesis in the zebrafish embryo (see Fig. 1C). Concordantly, at 48 hpf we found MGST1 to co-localize with Cmyb and Runx, two known markers of hematopoietic stem cells, indicating an associative role for this GST in vertebrate hematopoiesis (see Figs. 1D and S5).
3.4. Knockdown of MGST1 confirmed hematopoietic stem cell localization and revealed that MGST1 is responsible for the majority of fish GST activity during development
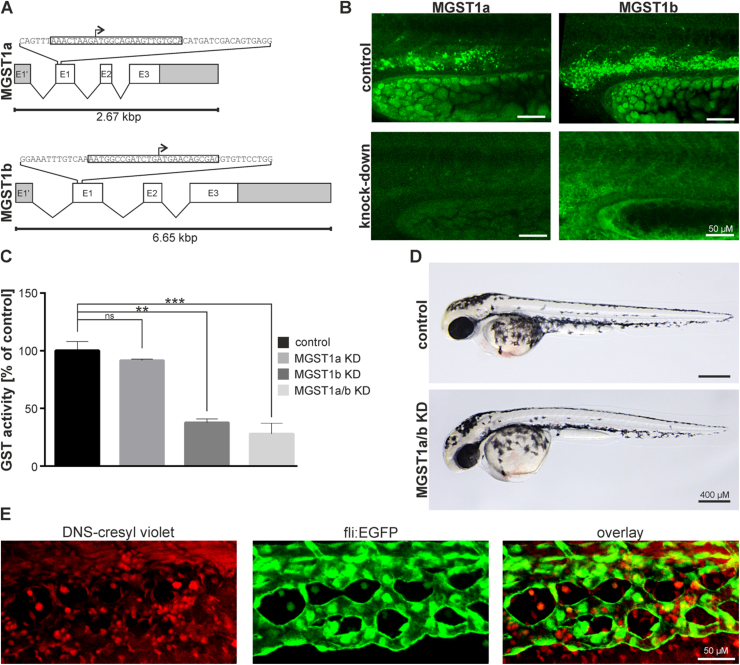
To understand the mechanistic importance of MGST1 in the hematopoietic process, we designed antisense morpholinos blocking the translational start site of the two isoforms (see Fig. 2A). Whereas knockdown of MGST1a alone did not significantly reduce GST activity in zebrafish embryos (92 ± 1% compared to control), knock-down of MGST1b or both isoforms together significantly decreased the enzymatic activity of GSTs to (38 ± 2%, p = 0.002 and 28 ± 5%, p = 0.0001, respectively; n = 4; 20 embryos/group) (see Fig. 2C). By using whole-mount immunohistochemistry we confirmed at the protein level that depletion of MGST1a and b occurred following morpholino injection (see Fig. 2B). Depletion of functional MGST1 did not alter the gross morphology (see Fig. 2D). Because simultaneous knockdown of both isoforms led to the highest depletion of GST activity in vivo we used the combination for all further experiments.
Fig. 2.
Morpholino induced knock-down of MGST1 in zebrafish. (A) Genomic organization of zfMGST1 and location of morpholino attachment sites targeting transcription. (B) Immunohistochemistry in embryos injected with morpholinos knocking down MGST1a/b. (C) Enzymatic activity of GST enzymes in extracts of embryos injected with morpholinos knocking-down MGST1a, MGST1b or both. (D) Gross morphology of embryos injected with morpholinos knocking-down MGST1a/b. (E) DNS-cresyl violet staining indicating MGST1 positive cells in the caudal hematopoietic tissue, see also Supplementary movie S1. Brightfield images were taken on a Leica MZ16 microscope equipped with a Leica DFC300FX camera; Confocal images were taken on a Leica LSM700 with a 20× lens; Alexa 488 and 555 filters were used; images stacks were produced with ImageJ, GIMP was used to adjust the gamma channel.
Fish MGST1 is extremely sensitive to sulfhydryl modification by N-ethylmaleimide [31] and as expected, we show here that only the N-ethylmaleimide sensitive enzymatic activity is decreased by the MGST1 morpholino. This confirmed the specificity of the morpholino and also revealed that about half of the total GST activity in the fish embryo can be accounted for by MGST1 (see Fig. S6). This is quite a remarkable result considering that the enzyme is present in only a limited subset of cells.
We confirmed expression of MGST1 in the caudal hematopoietic tissue of live specimens by using the fluorogenic substrate 2,4-dinitrobenzene sulfonamide cresyl violet (fig. S7). When cleaved by GSTs this substrate releases highly fluorescent cresyl violet that is retained only in those cells where it is formed [28]. We also observed that some of the stained cells were motile in the intermediate cell mass and entered the blood stream (see Fig. 2E and Video S1), a critical characteristic of HSC [32].
Movie S1.
48 h old zebrafish embryos were mounted in low melting agarose and incubated with 2.5 µM fluorogenic GST substrate 2,4-dinitrobenzene sulfonamide cresyl violet. Images were recorded with excitation at 540 nm and emission at 620 nm. ISV: intersegmental vessels; DA: dorsal aorta; CV: caudal vein. Arrow follows GST stained and mobile cell.
3.5. MGST1 influences both myeloid and lymphoid lineages in hematopoiesis
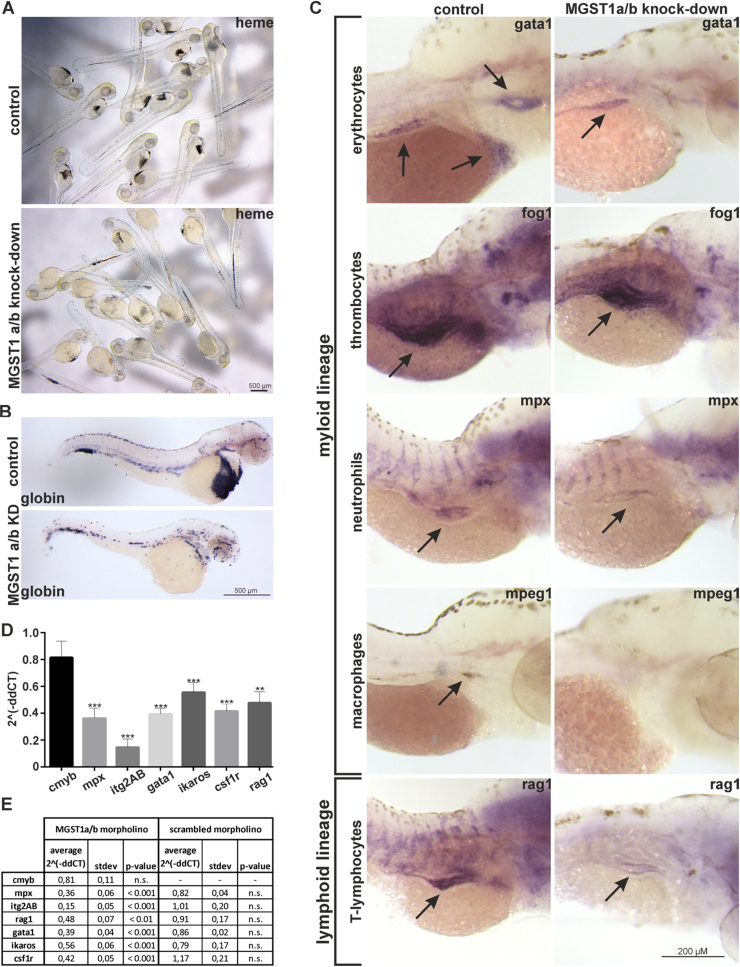
We analyzed the hematopoietic system of morphants by using diaminofluorene staining in living embryos and found that loss of MGST1 was followed by a striking decrease in hemoglobin content compared to control embryos at 2 days post injection (see Fig. 3A; n = 3; 20 embryos/group). In situ hybridization against globin, a marker for erythrocytes, revealed that indeed the number of red blood cells was dramatically decreased in MGST1 morphants (see Fig. 3B; n = 3; 20 embryos/group). Intrigued by these initial data, we continued with a thorough analysis of the hematopoietic system in embryos injected with MGST1a/b morpholinos. At 96 hpf, after onset of definitive hematopoiesis, MGST1 morphant embryos displayed a reduced staining for representative cells of both the myeloid and lymphoid lineages (see Fig. 3C; n = 3; 20 embryos/group). Confirming our previous histological approaches, transcripts for marker genes of both the myeloid as well as the lymphoid lineage were significantly lower in embryos lacking MGST1 (n = 30/group; 3 independent groups/experiment (see Fig. 3D, E)). A transcript (cmyb) specific for hematopoietic stem cells were not reduced in MGST1 morphants and critically, injection of a scrambled control morpholino did not have any effect on the hematopoietic system (see Fig. 3D). We did not observe increased cell death upon knock-down of MGST1, confirming that differentiation of HSC is indeed blocked (as opposed to being simply the result of increased HSC cell death, see Fig. S8).
Fig. 3.
Loss of functional MGST1 blocks differentiation of hematopoietic stem cells in zebrafish. (A) Histochemical staining of hemoglobin in living zebrafish embryos. (B) Whole-mount in situ hybridization in 48 hpf embryos staining globin transcripts specific for erythrocytes. (C) Whole-mount in situ hybridization against marker genes for different myloid and lymphoid lineages in 96 hpf embryos. (D) RTqPCR quantifying transcripts specific for different myloid and lymphoid lineages in 96 hpf embryos. (E) Quantification and statistics for RTqPCR. Brightfield images were taken on a Leica MZ16 microscope equipped with a Leica DFC300FX camera; images stacks were produced with ImageJ, GIMP was used to adjust the gamma channel.
3.6. MGST1 is highly expressed in mouse hematopoietic stem and progenitor cells (HSPC) and knockdown inhibits differentiation
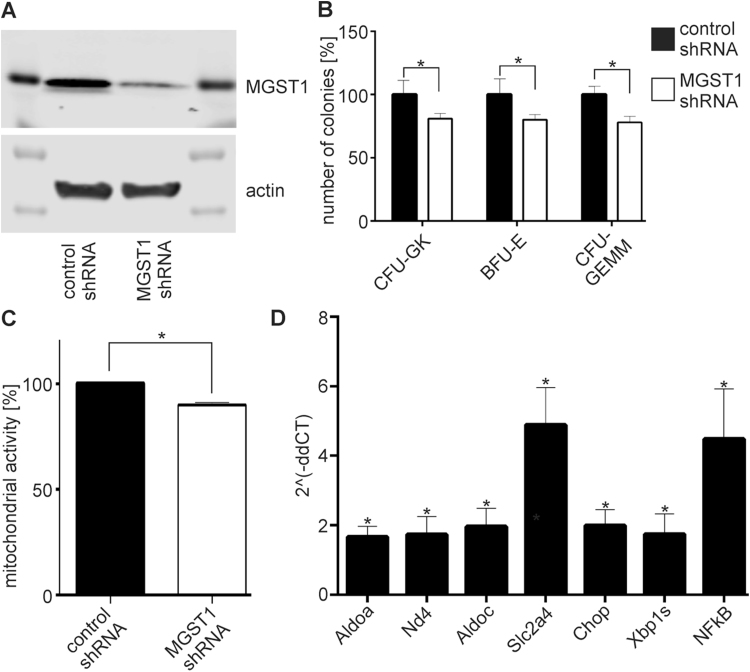
To confirm that MGST1 influences hematopoiesis in species other than zebrafish, and in particular mammals (hematopoiesis is highly conserved between zebrafish and mammals [33], [34]), we isolated hematopoietic stem and progenitor cells from mice and detected MGST1 at high levels in these cells (see Fig. 4A). We then developed lentiviral vectors, shRNA, against mouse MGST1 to determine if MGST1 also influences hematopoiesis in this species. Treatment of HSPC with MGST1 shRNA for 72 h resulted in a significant reduction of MGST1 (see Fig. 4A). MGST1 knockdown per se did not decrease cell viability. After inducing differentiation of the HSPC with M3434 methylcellulose medium we observed that the differentiation process after MGST1 knockdown was significantly impaired when compared with cells receiving control, scrambled shRNA (see Fig. 4B). These data support the hypothesis that MGST1 is critical to embryonic blood cell development.
Fig. 4.
Loss of functional MGST1 blocks differentiation of HSPC cells and induces metabolic changes. (A) HSPC cells were transfected with a lentiviral-scrambled (control) shRNA or a lentiviral-shRNA against MGST1. Immunoblot analysis indicated that shRNA against MGST1 significantly reduced the protein levels in HSPC. (B) MGST1 knock-down in HSPC resulted in decreased differentiation. Colony forming unit (CFU) assays were used and CFU-granulocyte macrophage (GM), burst forming unit erythroid (BFU-E) and CFU- granulocyte, erythrocyte, monocyte and megakaryocyte (GEMM) colonies were counted. (C) Mitochondrial activity was assessed with the MTT assay. (D) Quantitative RT-PCR analysis of the expression of key genes associated with glycolysis, HIF1 controlled genes and NFkB. Data are presented as MGST1 knock-down HSPC relative to controls. Data are means (+/− SD) for at least three experiments *p < 0.05.
3.7. MGST1 knockdown preserves the HSPC phenotype
Redox signaling mechanisms are known to be important in regulating “stemness” and differentiation [35], [36]. Moreover, HSPCs are usually located in hypoxic environments and rely on anaerobic glycolysis for energy production, while their subsequent differentiation is accompanied by a metabolic switch to oxidative phosphorylation [37]. We found that shRNA mediated knock-down of MGST1 in HSPC was accompanied by enhanced gene expression of aldolase A and C, key enzymes in glycolysis and Slc2a4 (Solute carrier family 2, member 4) a protein critical to the transport of glucose (see Figs. 4D, S9). Consistent with this, the colorimetric MTT assay results indicated that loss of functional MGST1 suppressed mitochondrial metabolism (see Figs. 4C, S9). The metabolic switch of HSCs is controlled by, amongst others, the transcription factor HIF1 (hypoxia inducible factor 1) [38]. When we knocked down MGST1, we observed an up-regulation of HIF1 regulated ER genes including Xbp1 and Chop, as well as an increase of NfkB transcripts, previously implicated in HSC self-renewal (see Fig. 4D) [39]. These results imply that MGST1 is a likely part of the HIF1 signaling axis that controls HSC differentiation, perhaps through the influence on energy metabolism.
4. Discussion
Our finding that MGST1 knockout mice are not viable is quite unexpected as knockout of cytosolic glutathione transferases [8] and membrane bound enzymes related to MGST1 (MGST2 [40], LTC4S [41], MPGES1 [42]) are not embryonic lethal. In fact, most enzymes with functions similar to MGST1 in redox biology (e.g. glutathione peroxidases, superoxide dismutase, catalase) can be deleted from mice producing viable offspring (reviewed in [9]). However there is one enzyme, GPX4, sharing functional characteristics with MGST1 (catalyzing reduction of lipid hydroperoxides directly in membranes) where knockout also results in embryonic lethality (at development stage E7.5–E8.5 [9]). GPX4 has been suggested to be important for redox control of development, amongst others for cardiovascular development [9].
Our data from morpholino knockdown of MGST1 in zebrafish suggest that the enzyme plays a vital role in the development of hematopoiesis that can explain mouse embryo lethality. Critically, MGST1 is responsible for a major part of total GST enzyme activity in the developing zebrafish embryo, concentrated in areas and cells implied in hematopoiesis, and peaks during development. These conclusions are supported by several methodological approaches including, hemoglobin staining, enzyme activity measurements, live embryo activity staining, immunohistochemistry and RTqPCR. The peak of MGST1 expression is supported by other studies in zebrafish [43]. In addition, an enhanced sensitivity to pro-oxidants has been described at 72 hpf [44], in zebrafish, an observation that fits well with the noted MGST1 decline. With few exceptions, other GSTs do not show a developmental peak in zebrafish development [45].
In zebrafish morphants we noted a decline in transcripts for both myeloid and lymphoid lineages, whereas markers of hematopoietic stem cells were not decreased, indicating a role for MGST1 in cell differentiation. To our knowledge, MGST1 is the only redox protein for which such a role has been directly observed in zebrafish. We could confirm this role in mouse HSPC and suggest that MGST1 driven negative control of HIF1 signaling, affecting energy balance that impacts on glycolysis, promotes differentiation. In this regard, we have previously found that the null phenotype of another member of the GST family, GSTP1, plays a role in regulating bone marrow proliferation [46], [47], migration of HSPC [29] and glycolytic functions in bone marrow dendritic cells [48]. In these dendritic cells, the GSTP1 null phenotype imbues differential S-glutathionylation of estrogen receptor alpha and this serves to emphasize the critical nature of the redox differences in regulating gene expression in bone marrow cells [48].
MGST1 is highly expressed in human embryonic stem cells [49], [50], [51], consistent with a role also in human early embryonic development. Our data show that hematopoiesis is one such role. Redox regulatory processes are highly important in the determination of cell fate in hematopoiesis [52], [53] and in a fashion similar to GPX4, by affecting lipid hydroperoxide levels, MGST1 can influence the redox status. Conditional deletion of GPX4 in hematopoietic cells predisposes mice to anemia [54] whereas GPX4 null mice die before embryonic stage E8.5 [9]. This timing is similar to MGST1 and a mouse model with APE1 deficiency, where lack of its redox regulatory function leads directly to lethality at E9 [53]. It appears that redox control compromised mouse embryos die at similar developmental stages. The data presented here show that redox regulation by MGST1 promotes HSC differentiation to more mature and dedicated hematopoietic cells. Knock down of MGST1 induces expression of HIF1 downstream target genes that impact glycolysis and “stemness” thereby preventing differentiation. In this regard, impaired bone marrow function may be a determinant factor in MGST1 null phenotype lethality.
5. Conclusions
-
•
A redox enzyme, MGST1, is indispensable for mouse embryonic development.
-
•
Knock-down experiments in zebrafish embryos revealed a critical role in hematopoiesis, a function confirmed in a mammalian cell model.
Acknowledgements
We thank the zebrafish facility of the Karolinska Institutet for excellent service. We acknowledge Elias Arnér for helpful discussions. We appreciate very much the help of the zebrafish community sending templates, especially the Paw, Thisse, Boehm and Lieschke laboratories. This work was supported by Karolinska Institutet (Ph.D. fellowship (KID) Dnr. 2379/07-225 to LB), the Swedish Society for Medical Research (postdoctoral fellowships to LB and KJ), The Swedish Research Council (2015-03955) and The Swedish Foundation for Strategic Research (MDB10-0025). Further support (KDT) by grants from the National Institutes of Health (CA08660, CA117259, NCRR P20RR024485 - COBRE in Oxidants, Redox Balance and Stress Signaling) and support from the South Carolina Centers of Excellence program and was conducted in a facility constructed with the support from the National Institutes of Health, Grant no. C06 RR015455 from the Extramural Research Facilities Program of the National Center for Research Resources. The Analytical Redox Core Facility of the COBRE grant also contributed (DMT).
Acknowledgments
Author contributions
LB, JZ, KD, LS and MJK performed and analyzed experiments. LB, RM, KD, KJ, JZ and MJK analyzed data. AH, KDT, DMT and HA contributed experimental systems, facilities and expertise. LB, KD, RM, KJ, JZ, KDT and MJK wrote the manuscript. All authors approved the final version.
Conflict of interest disclosures
The authors declare that they have no competing interests.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.redox.2018.04.013.
Appendix A. Supplementary material
Supplementary material
References
- 1.Hayes J.D., Flanagan J.U., Jowsey I.R. Glutathione transferases. Annu. Rev. Pharmacol. Toxicol. 2005;45:51–88. doi: 10.1146/annurev.pharmtox.45.120403.095857. [DOI] [PubMed] [Google Scholar]
- 2.Rinaldi R., Eliasson E., Swedmark S., Morgenstern R. Reactive intermediates and the dynamics of glutathione transferases. Drug Metab. Dispos. 2002;30(10):1053–1058. doi: 10.1124/dmd.30.10.1053. [DOI] [PubMed] [Google Scholar]
- 3.Sies H., Berndt C., Jones D.P. Oxidative stress. Annu. Rev. Biochem. 2017;86:715–748. doi: 10.1146/annurev-biochem-061516-045037. [DOI] [PubMed] [Google Scholar]
- 4.Arner E.S. Focus on mammalian thioredoxin reductases–important selenoproteins with versatile functions. Biochim. Biophys. Acta. 2009;1790(6):495–526. doi: 10.1016/j.bbagen.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 5.Brigelius-Flohe R., Maiorino M. Glutathione peroxidases. Biochim. Biophys. Acta. 2013;1830(5):3289–3303. doi: 10.1016/j.bbagen.2012.11.020. [DOI] [PubMed] [Google Scholar]
- 6.Perkins A., Nelson K.J., Parsonage D., Poole L.B., Karplus P.A. Peroxiredoxins: guardians against oxidative stress and modulators of peroxide signaling. Trends Biochem Sci. 2015;40(8):435–445. doi: 10.1016/j.tibs.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Board P.G. The use of glutathione transferase-knockout mice as pharmacological and toxicological models. Expert Opin. Drug Metab. Toxicol. 2007;3(3):421–433. doi: 10.1517/17425255.3.3.421. [DOI] [PubMed] [Google Scholar]
- 8.Henderson C.J., Wolf C.R. Knockout and transgenic mice in glutathione transferase research. Drug Metab. Rev. 2011;43(2):152–164. doi: 10.3109/03602532.2011.562900. [DOI] [PubMed] [Google Scholar]
- 9.Ufer C., Wang C.C., Borchert A., Heydeck D., Kuhn H. Redox control in mammalian embryo development. Antioxid. Redox Signal. 2010;13(6):833–875. doi: 10.1089/ars.2009.3044. [DOI] [PubMed] [Google Scholar]
- 10.Yant L.J., Ran Q., Rao L. The selenoprotein GPX4 is essential for mouse development and protects from radiation and oxidative damage insults. Free Radic. Biol. Med. 2003;34(4):496–502. doi: 10.1016/s0891-5849(02)01360-6. [DOI] [PubMed] [Google Scholar]
- 11.Conrad M. Transgenic mouse models for the vital selenoenzymes cytosolic thioredoxin reductase, mitochondrial thioredoxin reductase and glutathione peroxidase 4. Biochim. Biophys. Acta. 2009;1790(11):1575–1585. doi: 10.1016/j.bbagen.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 12.Friedmann Angeli J.P., Schneider M., Proneth B. Inactivation of the ferroptosis regulator Gpx4 triggers acute renal failure in mice. Nat. Cell Biol. 2014;16(12):1180–1191. doi: 10.1038/ncb3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kagan V.E., Mao G., Qu F. Oxidized arachidonic and adrenic PEs navigate cells to ferroptosis. Nat. Chem. Biol. 2017;13(1):81–90. doi: 10.1038/nchembio.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schneider M., Wortmann M., Mandal P.K. Absence of glutathione peroxidase 4 affects tumor angiogenesis through increased 12/15-lipoxygenase activity. Neoplasia. 2010;12(3):254–263. doi: 10.1593/neo.91782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ursini F., Maiorino M., Gregolin C. The selenoenzyme phospholipid hydroperoxide glutathione peroxidase. Biochim. Biophys. Acta. 1985;839:62–70. doi: 10.1016/0304-4165(85)90182-5. [DOI] [PubMed] [Google Scholar]
- 16.Mosialou E., Ekstrom G., Adang A.E., Morgenstern R. Evidence that rat liver microsomal glutathione transferase is responsible for glutathione-dependent protection against lipid peroxidation. Biochem. Pharmacol. 1993;45(8):1645–1651. doi: 10.1016/0006-2952(93)90305-g. [DOI] [PubMed] [Google Scholar]
- 17.Conrad M., Jakupoglu C., Moreno S.G. Essential role for mitochondrial thioredoxin reductase in hematopoiesis, heart development, and heart function. Mol. Cell. Biol. 2004;24(21):9414–9423. doi: 10.1128/MCB.24.21.9414-9423.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ufer C., Wang C.C. The roles of glutathione peroxidases during embryo development. Front. Mol. Neurosci. 2011;4:12. doi: 10.3389/fnmol.2011.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kelner M.J., Bagnell R.D., Morgenstern R. Structural organization of the murine microsomal glutathione S-transferase gene (MGST1) from the 129/SvJ strain: identification of the promoter region and a comprehensive examination of tissue expression. Biochim. Biophys. Acta. 2004;1678(2–3):163–169. doi: 10.1016/j.bbaexp.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 20.Yanagawa Y., Kobayashi T., Ohnishi M. Enrichment and efficient screening of ES cells containing a targeted mutation: the use of DT-A gene with the polyadenylation signal as a negative selection maker. Transgenic Res. 1999;8(3):215–221. doi: 10.1023/a:1008914020843. [DOI] [PubMed] [Google Scholar]
- 21.Conner D.A. Transgenic mouse production by zygote injection. Curr. Protocols Mol. Biol. 2004;23 doi: 10.1002/0471142727.mb2309s68. (23.29.21–23.29.29) [DOI] [PubMed] [Google Scholar]
- 22.Conner D.A. Transgenic mouse colony management. Curr. Protocols Mol. Biol. 2005;23:23.10.21–23.10.28. doi: 10.1002/0471142727.mb2310s71. [DOI] [PubMed] [Google Scholar]
- 23.Mortensen R. Production of a heterozygous mutant cell line by homologous recombination (single knockout) Curr. Protocols Mol. Biol. 2008;23 doi: 10.1002/0471142727.mb2305s82. (23.25. 21–23.25.11) [DOI] [PubMed] [Google Scholar]
- 24.Kimmel C.B., Ballard W.W., Kimmel S.R., Ullmann B., Schilling T.F. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995;203(3):253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 25.Brautigam L., Schutte L.D., Godoy J.R. Vertebrate-specific glutaredoxin is essential for brain development. Proc. Natl. Acad. Sci. USA. 2011;108(51):20532–20537. doi: 10.1073/pnas.1110085108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weinstein B.M., Schier A.F., Abdelilah S. Hematopoietic mutations in the zebrafish. Development. 1996;123:303–309. doi: 10.1242/dev.123.1.303. [DOI] [PubMed] [Google Scholar]
- 27.Hauptmann G., Gerster T. Multicolor whole-mount in situ hybridization. Methods Mol. Biol. 2000;137:139–148. doi: 10.1385/1-59259-066-7:139. [DOI] [PubMed] [Google Scholar]
- 28.Zhang J., Shibata A., Ito M. Synthesis and characterization of a series of highly fluorogenic substrates for glutathione transferases, a general strategy. J. Am. Chem. Soc. 2011;133(35):14109–14119. doi: 10.1021/ja205500y. [DOI] [PubMed] [Google Scholar]
- 29.Zhang J., Ye Z.W., Gao P. Glutathione S-transferase P influences redox and migration pathways in bone marrow. PLoS One. 2014;9(9):e107478. doi: 10.1371/journal.pone.0107478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuang Q., Purhonen P., Alander J. Dead-end complex, lipid interactions and catalytic mechanism of microsomal glutathione transferase 1, an electron crystallography and mutagenesis investigation. Sci. Rep. 2017;7(1):7897. doi: 10.1038/s41598-017-07912-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bresell A., Weinander R., Lundqvist G. Bioinformatic and enzymatic characterization of the MAPEG superfamily. Febs J. 2005;272(7):1688–1703. doi: 10.1111/j.1742-4658.2005.04596.x. [DOI] [PubMed] [Google Scholar]
- 32.Orkin S.H., Zon L.I. Hematopoiesis: an evolving paradigm for stem cell biology. Cell. 2008;132(4):631–644. doi: 10.1016/j.cell.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martin C.S., Moriyama A., Zon L.I. Hematopoietic stem cells, hematopoiesis and disease: lessons from the zebrafish model. Genome Med. 2011;3(12):83. doi: 10.1186/gm299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kwan W., North T.E. Netting novel regulators of hematopoiesis and hematologic malignancies in zebrafish. Curr. Top. Dev. Biol. 2017;124:125–160. doi: 10.1016/bs.ctdb.2016.11.005. [DOI] [PubMed] [Google Scholar]
- 35.Ito K., Suda T. Metabolic requirements for the maintenance of self-renewing stem cells. Nat. Rev. Mol. Cell Biol. 2014;15(4):243–256. doi: 10.1038/nrm3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yusuf R.Z., Scadden D.T. Fate through fat: lipid metabolism determines stem cell division outcome. Cell Metab. 2012;16(4):411–413. doi: 10.1016/j.cmet.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang C.C., Sadek H.A. Hypoxia and metabolic properties of hematopoietic stem cells. Antioxid. Redox Signal. 2014;20(12):1891–1901. doi: 10.1089/ars.2012.5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Semenza G.L. Hypoxia-inducible factors in physiology and medicine. Cell. 2012;148(3):399–408. doi: 10.1016/j.cell.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao C., Xiu Y., Ashton J. Noncanonical NF-kappab signaling regulates hematopoietic stem cell self-renewal and microenvironment interactions. Stem Cells. 2012;30(4):709–718. doi: 10.1002/stem.1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dvash E., Har-Tal M., Barak S., Meir O., Rubinstein M. Leukotriene C4 is the major trigger of stress-induced oxidative DNA damage. Nat. Commun. 2015;6:10112. doi: 10.1038/ncomms10112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kanaoka Y., Maekawa A., Penrose J.F., Austen K.F., Lam B.K. Attenuated zymosan-induced peritoneal vascular permeability and IgE-dependent passive cutaneous anaphylaxis in mice lacking leukotriene C4 synthase. J. Biol. Chem. 2001;276(25):22608–22613. doi: 10.1074/jbc.M103562200. [DOI] [PubMed] [Google Scholar]
- 42.Kamei D., Yamakawa K., Takegoshi Y. Reduced pain hypersensitivity and inflammation in mice lacking microsomal prostaglandin e synthase-1. J. Biol. Chem. 2004;279(32):33684–33695. doi: 10.1074/jbc.M400199200. [DOI] [PubMed] [Google Scholar]
- 43.Glisic B., Hrubik J., Fa S., Dopudj N., Kovacevic R., Andric N. Transcriptional profiles of glutathione-S-Transferase isoforms, Cyp, and AOE genes in atrazine-exposed zebrafish embryos. Environ. Toxicol. 2016;31(2):233–244. doi: 10.1002/tox.22038. [DOI] [PubMed] [Google Scholar]
- 44.Sant K.E., Hansen J.M., Williams L.M. The role of Nrf1 and Nrf2 in the regulation of glutathione and redox dynamics in the developing zebrafish embryo. Redox Biol. 2017;13:207–218. doi: 10.1016/j.redox.2017.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tierbach A., Groh K.J., Schonenberger R., Schirmer K., Suter M.J. Glutathione S-transferase protein expression in different life stages of zebrafish (Danio rerio) Toxicol. Sci. 2018 doi: 10.1093/toxsci/kfx293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gate L., Majumdar R.S., Lunk A., Tew K.D. Increased myeloproliferation in glutathione S-transferase pi-deficient mice is associated with a deregulation of JNK and Janus kinase/STAT pathways. J. Biol. Chem. 2004;279(10):8608–8616. doi: 10.1074/jbc.M308613200. [DOI] [PubMed] [Google Scholar]
- 47.Ruscoe J.E., Rosario L.A., Wang T. Pharmacologic or genetic manipulation of glutathione S-transferase P1-1 (GSTpi) influences cell proliferation pathways. J. Pharmacol. Exp. Ther. 2001;298(1):339–345. [PubMed] [Google Scholar]
- 48.Zhang J., Ye Z.W., Chen W., Manevich Y., Mehrotra S., Ball L., Janssen-Heininger Y., Tew K.D., Townsend D.M. S-Glutathionylation of estrogen receptor α affects dendritic cell function. J. Biol. Chem. 2018;293(12):4366–4380. doi: 10.1074/jbc.M117.814327. (Epub 2018 Jan 26) PMID: 29374060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Assou S., Le Carrour T., Tondeur S. A meta-analysis of human embryonic stem cells transcriptome integrated into a web-based expression atlas. Stem Cells. 2007;25(4):961–973. doi: 10.1634/stemcells.2006-0352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zeng X., Miura T., Luo Y. Properties of pluripotent human embryonic stem cells BG01 and BG02. Stem Cells. 2004;22(3):292–312. doi: 10.1634/stemcells.22-3-292. [DOI] [PubMed] [Google Scholar]
- 51.Dormeyer W., van Hoof D., Braam S.R., Heck A.J., Mummery C.L., Krijgsveld J. Plasma membrane proteomics of human embryonic stem cells and human embryonal carcinoma cells. J. Proteome Res. 2008;7(7):2936–2951. doi: 10.1021/pr800056j. [DOI] [PubMed] [Google Scholar]
- 52.Grek C.L., Townsend D.M., Tew K.D. The impact of redox and thiol status on the bone marrow: pharmacological intervention strategies. Pharmacol. Ther. 2011;129(2):172–184. doi: 10.1016/j.pharmthera.2010.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zou G.M., Luo M.H., Reed A., Kelley M.R., Yoder M.C. Ape1 regulates hematopoietic differentiation of embryonic stem cells through its redox functional domain. Blood. 2007;109(5):1917–1922. doi: 10.1182/blood-2006-08-044172. [DOI] [PubMed] [Google Scholar]
- 54.Canli O., Alankus Y.B., Grootjans S. Glutathione peroxidase 4 prevents necroptosis in mouse erythroid precursors. Blood. 2016;127(1):139–148. doi: 10.1182/blood-2015-06-654194. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material