Abstract
Background
A dysbiosis in the intestinal microbiome plays a role in the pathogenesis of several immunological diseases. These diseases often show a sex bias, suggesting sex differences in immune responses and in the intestinal microbiome. We hypothesized that sex differences in immune responses are associated with sex differences in microbiota composition.
Methods
Fecal microbiota composition (MITchip), mRNA expression in intestinal tissue (microarray), and immune cell populations in mesenteric lymph nodes (MLNs) were studied in male and female mice of two mouse strains (C57B1/6OlaHsd and Balb/cOlaHsd). Transcriptomics and microbiota data were combined to identify bacterial species which may potentially be related to sex-specific differences in intestinal immune related genes.
Results
We found clear sex differences in intestinal microbiota species, diversity, and richness in healthy mice. However, the nature of the sex effects appeared to be determined by the mouse strain as different bacterial species were enriched in males and females of the two strains. For example, Lactobacillus plantarum and Bacteroides distasonis were enriched in B6 females as compared to B6 males, while Bifidobacterium was enriched BALB/c females as compared to BALB/c males. The strain-dependent sex effects were also observed in the expression of immunological genes in the colon. We found that the abundance of various bacteria (e.g., Clostridium leptum et rel.) which were enriched in B6 females positively correlated with the expression of several genes (e.g., Il-2rb, Ccr3, and Cd80) which could be related to immunological functions, such as inflammatory responses and migration of leukocytes. The abundance of several bacteria (e.g., Faecalibacterium prausnitzii et rel. and Coprobacillus et rel.- Clostridium ramosum et rel.) which were enriched in BALB/c males positively correlated to the expression of several genes (e.g., Apoe, Il-1b, and Stat4) related to several immunological functions, such as proliferation and quantity of lymphocytes. The net result was the same, since both mouse strains showed similar sex induced differences in immune cell populations in the MLNs.
Conclusions
Our data suggests a correlation between microbiota and intestinal immune populations in a sex and strain-specific way. These findings may contribute to the development of more sex and genetic specific treatments for intestinal-related disorders.
Electronic supplementary material
The online version of this article (10.1186/s13293-018-0186-6) contains supplementary material, which is available to authorized users.
Keywords: Microbiota, Sex differences, B6, BALB/c, Mesenteric lymph nodes, Colon, Intestinal immune populations, Mucosal immune populations
Background
The human gut harbors trillions of microbes [1]. The recent change of the traditional view that gut microbiota not only affect fermentation of food components, but also influence metabolism and immune status, has led to the realization that these microbes can impact health on different levels and that they are instrumental for maintaining health [2–4]. Microbes in the intestine can substantially be influenced by external factors such as diet and antibiotics, which may disturb the microbiota-host interactions in an undesirable way and can ultimately lead to disease [5]. However, these findings also demonstrate the potential to improve human health or to treat and prevent diseases by using nutrition or drugs [5, 6].
During recent years, a disbalance in intestinal microbiota communities (intestinal dysbiosis) has been found to play a significant role in the pathogenesis of a large number of immunological Western diseases, such as inflammatory bowel disease (IBD), other autoimmune diseases, and metabolic syndrome [7, 8]. This growing list of Western-world diseases correlates with changes in microbiota composition [9–11]. Microbiota-derived molecules, such as short-chain fatty acids (SCFAs), have been recognized to influence intestinal immune cells [12, 13]. For example, the SCFA butyrate has been shown to induce the differentiation of T regulatory (Tregs) in the colon [14]. Furthermore, some microbiota are involved in the generation of specific regulatory responses in T-cells [12, 15], while others stimulate specific T helper 17 (Th17) cell responses [16].
There is a sex bias in the prevalence of many of the aforementioned Western-world diseases [17–19]. It is currently unknown whether this sex bias is influenced by sex-dependent differences in immune modulating microbiota. Sex differences in peripheral immune responses are well known [20, 21], and in general, it is thought that females have a stronger innate and adaptive immune response as compared with males [22]. However, also here the influence of microbiota differences has gained minor attention. Although several studies showed the existence of sex differences in microbiota composition [23–28], minor knowledge is available on sex differences in intestinal immunology and on the influence of microbiota on sex-specific immune responses. Markle et al. and Yurkovetskiy et al. both showed that microbiota and sex hormones contribute to the effector mechanism of sex bias in type 1 diabetes in non-obese diabetic (NOD) mice [23, 27]. If sex-dependent microbiota differences underlie the differences in sex-specific immunity, it might open new venues for designing effective strategies to improve human health by manipulating microbiota and associated immune responses in a sex-specific way. Therefore, in this study, we investigated the relationship between sex differences in immune populations and sex differences in microbiota in healthy mice.
As various factors, such as the reproductive condition, genetic background, and diet, can interfere with the sex effects [25, 28, 29], we compared male and female mice from two different strains (C57B1/6OlaHsd (B6) and Balb/cOlaHsd (BALB/c)) with two difference genetic backgrounds under exactly the same reproductive and dietary conditions. The B6 and BALB/c strains were specifically chosen, because of their known difference in intestinal immune responses during Dextran sulfate sodium (DSS)-induced colitis [30, 31]. In mice from both sexes and mouse strains, we analyzed the microbiota composition and we performed a microarray on colonic tissue. We combined the transcriptomics data and the microbiota data and performed a bio-mathematical analysis, in order to find bacterial species, which may potentially be related to sex-specific differences in intestinal gene expression. Subsequently, key immunological changes found in the microarray were studied in the MLN using flow cytometry. The MLN are used as intestinal reference site, as this is the place where lymphocytes are primed and activated by intestinal DCs deriving from the gut [32].
Methods
Study design
This study was designed to assess the effect of sex on intestinal microbiota and intestinal immune cell composition in mice. Two different mice strains were used; C57B1/6OlaHsd (B6) and Balb/cOlaHsd (BALB/c). In both strains (n = 20 per strain) two groups were present; female and male mice (n = 10 per sex). Between an age of 11 and 23 weeks, all mice were sacrificed by cervical dislocation under anesthesia (isoflurane and oxygen). Table 1 provides an overview of the characteristics of all mice (n = 10 per group), and Table 2 provides an overview of the characteristics of the mice which were, per strain and sex, randomly selected from two cages for microbiota and microarray analysis (n = 5 per group). Subsequently, their mesenteric lymph nodes (MLN) were removed for immune cell analysis. During sacrifice, feces from the distal colon were collected for MITChip analysis. Approximately 1 cm of proximal colon was removed for microarray analysis. All female mice were sacrificed during the diestrus phase of their ovarian cycle to ensure low stable levels progesterone and estrogens.
Table 1.
Overview of mice characteristics. No significant differences in age at sacrifice were found between males and females within each mouse strain (Kruskal-Wallis test followed by Dunn’s multiple comparison test, p < 0.05)
| BALB/c male | BALB/c female | B6 male | B6 female | |
|---|---|---|---|---|
| Number of mice | 10 | 10 | 10 | 10 |
| Age at sacrifice (weeks) | 12.8 (2.4) | 15.3 (3.4) | 17.0 (3.4) | 18.8 (3.3) |
Table 2.
Overview of mice characteristics selected for microbiota and microarray analysis. No significant differences in age at sacrifice were found between males and females within each mouse strain (Kruskal-Wallis test followed by Dunn’s multiple comparison test, p < 0.05)
| BALB/c male | BALB/c female | B6 male | B6 female | |
|---|---|---|---|---|
| Number of mice | 5 | 5 | 5 | 5 |
| Age at sacrifice (weeks) | 13.9 (4.3) | 18.5 (3.5) | 19.7 (1.4) | 18.2 (0.4) |
Mice
Male and female wild-type B6 and BALB/c mice were purchased from Harlan (Harlan, Horst, the Netherlands) at an age of 8 weeks. Mice were co-housed (five mice per cage, according to sex and strain) in isolated ventilated cages to limit environmental influences. The animals had ad libitum access to food (D12450B diet from Research Diets Services, Wijk bij Duurstede, the Netherlands) and water.
Bacterial DNA extraction and microbiota profiling
Total DNA was extracted from the fecal samples (n = 5 mice per group, divided over at least two different cages) using the repeated bead-beating-plus column (RBB + C) method [33]. The microbiota composition was determined using the mouse intestinal tract chip (MITChip), a diagnostic 16S rRNA array, which consists of 3580 unique probes designed to profile murine intestinal microbiota [34]. Briefly, for MITChip, 16S rRNA gene amplification of the bacterial DNA, in vitro transcription, labeling, and hybridization were carried out as described previously [35]. Data were normalized and analyzed using a set of R-based scripts in combination with a custom-designed relational database, which operates under the MySQL database management system. For microbial profiling, the Robust Probabilistic Averaging (RPA) signal intensities of 2667 specific probes for the 94 genus-level bacterial groups detected on the MITChip, were used [36]. Diversity calculations were performed using a microbiome R-script package (https://github.com/microbiome). The redundancy analysis (RDA) was performed in Canoco 5.0, where variables were tested for their significance by the Monte Carlo permutation and visualized in triplots [37].
Intestinal microarray analysis
For microarray analysis, RNA was purified from the proximal colon of mice (n = 5 per group) using TRIzol (Life Technologies, Calsbad, CA, USA) followed by an additional round of purification with RNeasy Minikit columns (Qiagen, Venlo, the Netherlands). The quality of RNA was determined using RNA 6000 nanochips on the Agilent 2100 bioanalyzer (Agilent Technologies, Amsterdam, the Netherlands). Purified RNA (100 ng) was labeled with the Affymetrix WT PLUS reagent kit (Affymetrix, Santa Clara, CA, USA) and hybridized to an Affymetrix Mouse Gene 1.1 ST array plate (Affymetrix, Santa Clara, CA, USA). Hybridization, washing, and scanning were carried out on an Affymetrix GeneTitan platform according to the manufacturer’s instructions. Arrays were normalized using the robust multiarray average method [38, 39]. Probe sets were defined according to Dai et al. (2005) [40]. In this method, probes are assigned to Entrez IDs as a unique gene identifier. The p values were calculated using an intensity-based moderated t statistic (IBMT) [41]. Only probe sets with a fold-change of at least 1.2 (up/down) and a p value < 0.05 were considered to be significantly different. The microarray data was validated by real-time quantitative PCR (see Additional file 1 for the used method and results).
To gain insight into the biological role of the sexually dimorphically expressed genes, we investigated the functions in which these genes are involved using Ingenuity Pathway Analysis (IPA) (Ingenuity System). The IPA output includes biological functions and signaling pathways with statistical assessment of the significance of their representation based on Fisher’s exact test. Here, this test calculates the probability that genes participate in a given biological function relative to their occurrence in all other biological function annotations. Our IPA analyses included comparison of differentially regulated genes in the colon of males and females in both B6 and BALB/c mice.
Multivariate integration and correlation analysis
To gain insight in the relationship between the colonic gene expression and microbiota composition, the microarray and MITChip datasets were combined, using the linear multivariate method partial least squares (PLS) [42], as described previously [43]. This integration of datasets per individual mouse gives a direct correlation between gene expression and microbiota composition in these samples. For 15 mice, both gene expression and data on microbiota composition were available (n = 3–5 per group). Both datasets were log2 transformed before analysis, and the canonical correlation framework of PLS was used [44]. The correlation matrices were visualized in clustered image maps [45]. Analyses were performed in R using the library mixOmics [46]. A positive correlation between bacteria and genes indicates that a higher abundance of the bacteria is associated with a higher expression of the particular cluster of genes. A negative correlation between bacteria and genes indicates that a lower abundance of the bacteria is associated with a lower expression of the particular cluster of genes.
Mesenteric lymph node cell isolation
Single cell suspensions of the MLN were made by mechanical disruption of the tissues between two object glasses in 2 ml ice cold RPMI containing 10% heat inactivated fetal calf serum (FCS). Falcon tubes with cell strainer caps (Corning, Amsterdam, the Netherlands) (35 μm) were used to remove cell clumps before the cells were counted and used for staining.
Cell staining
MLN cells were stained for T lymphocytes (CD3+), T cytotoxic cells (CD8+) and T helper cells (CD4+). Expression of CD69, α4ß7, CD62L, and CD44 was measured within the CD8+ and CD4+ cell subsets. Specifications of the antibodies used are described in Table 3. All antibodies were diluted in a volume of 25 μl, supplemented to a volume of 25 μl with FACS buffer (PBS + 10% FCS (v/v)). Approximately 0.5 × 106 MLN cells were incubated for 20 min in FACS buffer (10% FCS (v/v)) containing 20% (v/v) normal rat serum (Jackson, Newmarket, UK) and 2% (v/v) Fc block (CD16/32) (Biolegend, Uithoorn, the Netherlands) to prevent non-specific antibody binding followed by incubation in the primary antibody mix for 30 min. Next, the cells were incubated with a biotinylated antibody (streptavidin-Pacific Orange) for 30 min and subsequently fixed in FACS lysing solution (BD Biosciences, Breda, the Netherlands) for 30 min. Washing was performed in between all incubation steps. The whole procedure was performed on ice and in the dark. Isotype control antibodies were used at the same concentration and purchased from the same company as the primary and secondary antibodies.
Table 3.
Antibody specifications
| Specificity | Clone name | Fluorchrome | Concentration | Dilutiona | Supplier |
|---|---|---|---|---|---|
| CD3 | 17A2 | Pacific blue | 0.5 mg/ml | 80× | Biolegend |
| CD8 | 53–6.7 | A700 | 0.5 mg/ml | 50× | Biolegend |
| CD4 | GK1.5 | PE-Cy7 | 0.2 mg/ml | 100× | Biolegend |
| CD69 | H1.2F3 | FitC | 0.5 mg/ml | 25× | Biolegend |
| α4β7 | DATK32 | APC | 0.2 mg/ml | 25× | Biolegend |
| CD62L | MEL-14 | Biotin | 0.5 mg/ml | 200× | Biolegend |
| Streptavidin-Pacific Orange | 1 mg/ml | 100× | ThermoFisher | ||
| CD44 | IM7 | APC-Cy7 | 0.2 mg/ml | 100× | Biolegend |
aDilution used in a total volume of 25 μl supplemented with PBS + 10% FCS
Flow cytometry
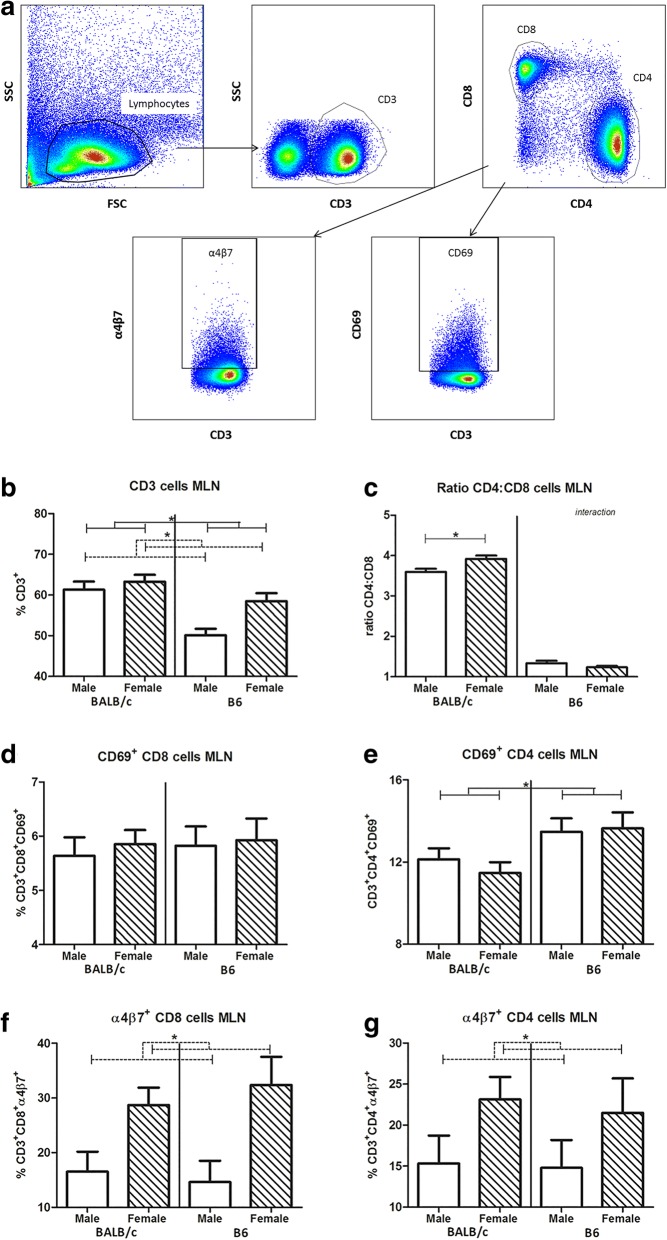
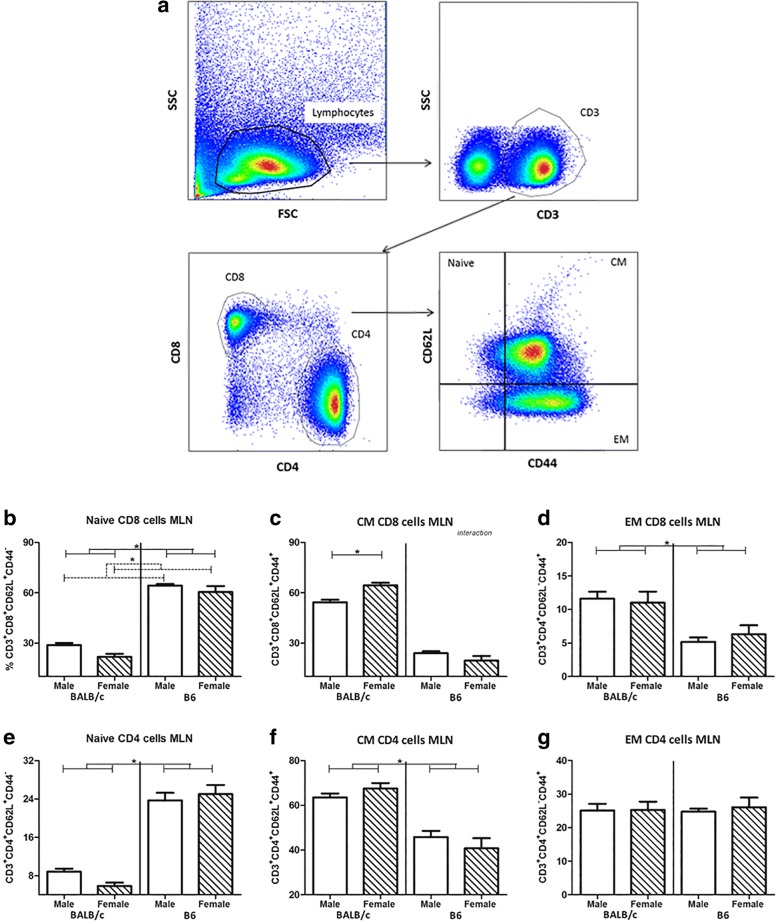
Cell samples were analyzed using the LSR-II Flow Cytometer system (BD Biosciences, Breda, the Netherlands) using FACS Diva software. Analysis was performed using FlowJo version 10 software (FlowJo, LLC, OR, USA). Lymphocytes were gated based on size in the forward side scatter plot, and T cells were determined by selecting CD3+ cells. Within the CD3+ cells, CD4+ and CD8+ cells were selected. Within both the CD4+ and CD8+ population, the percentage of cells expressing CD69, α4ß7, CD62L, and CD44 was measured. Therefore, all their isotype controls were set at 1% and these gates were copied to the samples with the antibody mix (see also Figs. 4 and 5).
Fig. 4.
Effect of sex and strain on T lymphocytes in the mesenteric lymph nodes. Gating strategy for determination of T cell subsets in the mesenteric lymph nodes (a). Lymphocytes were gated based on size and scatter in the forward side scatter plot. T cells were determined by selecting CD3+ cells. Within the CD3+ cells, CD8+ (Tc cells) and CD4+ (Th cells) cells were selected. Within both the CD8+ and CD4+ population, the percentage of CD69 and α4ß7 were measured. All isotype controls were set at 1%. Frequency of CD3+ T lymphocytes (b), the ratio of T helper cells (CD4)/T cytotoxic cells (CD8) (c), frequency of CD69+ CD8 (d), CD69+ CD4 (e), 4β7+ CD8 (f), and 4β7+ CD4 (g) in the mesenteric lymph nodes of male and female BALB/c and B6 mice (10 mice per group). T cytotoxic and T helper cells are expressed as the frequency of CD8+ and CD4+ cells within the CD3+ population, respectively. Results are shown as mean + SEM and were tested for overall strain and sex effects using a two-way ANOVA followed by a Bonferroni post hoc test to test for strain-specific sex effects when interaction was found. Significant strain effects are indicated with solid lines and significant sex effects are indicated with dashed lines (p < 0.05)
Fig. 5.
Effect of sex and strain on maturation of T lymphocytes in the mesenteric lymph nodes. Gating strategy for determination of T cell subsets in the mesenteric lymph nodes (a). Lymphocytes were gated based on size and scatter in the forward side scatter plot. T cells were determined by selecting CD3+ cells. Within the CD3+ cells, CD8+ (Tc cells) and CD4+ (Th cells) cells were selected. Within both the CD8+ and CD4+ population, the percentage of CD62L and CD44 were measured. All isotype controls were set at 1%. Frequency of CD62L+CD44− naive CD8 (b) and CD4 (e), CD62L+CD44+ central memory CD8 (c) and CD4 (f) and CD62L−CD44+ effector memory CD8 (d) and CD4 (g) in the mesenteric lymph nodes of male and female BALB/c and B6 mice (10 mice per group). T cytotoxic cells are expressed as the frequency of CD8+ cells within the CD3+ population, whereas T helper cells are expressed as the frequency of CD4+ cells within the CD3+ population. Results are shown as mean + SEM and were tested for overall strain and sex effects using a two-way ANOVA followed by a Bonferroni post hoc test to test for strain-specific sex effects when interaction was found. Significant strain effects are indicated with solid lines and significant sex effects are indicated with dashed lines (p < 0.05)
Statistical analysis
For flow cytometry data, Shannon diversity, microbiota richness and the Firmicutes/Bacteroidetes ratio, data are expressed as the mean with standard error of the mean (SEM). The Kolmogorov-Smirnov test was used to determine normal distribution of the data. When the data were not normally distributed a log transformation was performed before analysis. The data were analyzed with a two-way ANOVA followed by a Bonferroni post-test when interaction was found. For analyzing significant effects of sex and strain on the abundance of bacteria groups a Mann–Whitney U test was used. p values of 0.05 or smaller were considered statistically significant and p values between 0.05 and 0.1 were defined as a trend.
Results
Sex influenced intestinal microbiota composition in a mouse strain-dependent way
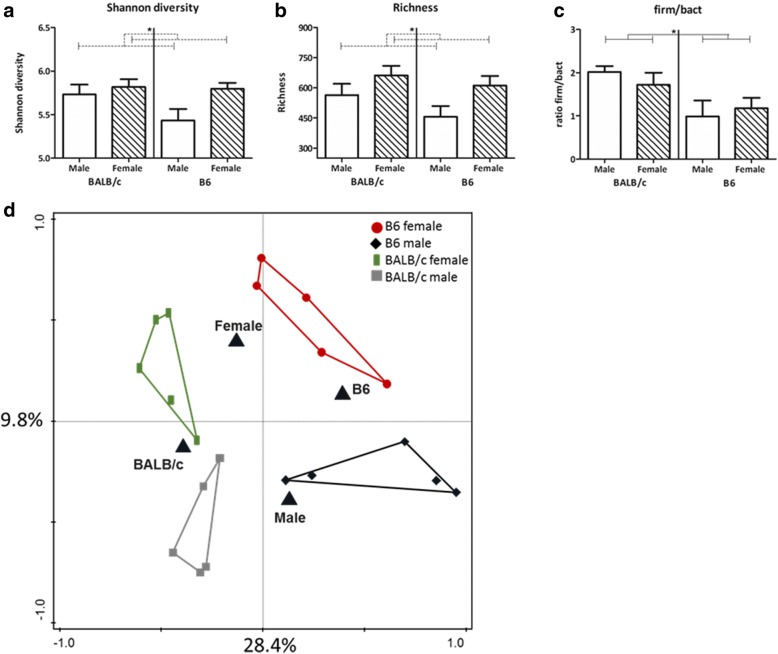
The microbial composition in the feces of males and females of both BALB/c and B6 mice was determined using the phylogenetic microarray, the mouse intestinal tract Chip (MITChip). Additionally, we determined the richness (number of unique species) and Shannon diversity (calculation between richness and evenness (abundances over species) of the microbiota composition. Overall, males had a lower diversity (two-way ANOVA, p = 0.046) and richness (two-way ANOVA, p = 0.027) than females (Fig. 1a, b), while there was no effect of strain. A higher ratio of Firmicutes/Bacteroidetes was found in BALB/c mice (two-way ANOVA, p = 0.009); however, this ratio was not influenced by sex (Fig. 1c).
Fig. 1.
Effect of sex and strain on fecal microbiota characteristics. Shannon diversity (a), richness (b), and the Firmicutes/Bacteroidetes (c) ratio in the fecal microbiota of male and female BALB/c and B6 mice (5 mice per group). Results are shown as mean + SEM and were tested for overall strain and sex effects using a two-way ANOVA followed by a Bonferroni post hoc test to test for strain-specific sex effects when interaction was found. Significant strain effects are indicated with solid lines and significant sex effects are indicated with dashed lines (p < 0.05). RDA plot showing the variation explained by the components genotype and sex (five mice per group) (d). The total variation that can be explained by the variables genotype (26.5%) and sex (11.6%) is 38.1%. Both variables are significant in explaining the variation (Monte Carlo permutation, p < 0.05)
Redundancy analysis showed that the total variation in microbiota composition explained by the variables genotype and sex is 38.1%. Sex explained 11.6% of the variance in microbiota composition (Fig. 1d). This was, however, mouse strain dependent, since the variable strain explained 26.5% of the variance in the microbial composition. Table 4 provides an overview of the relative abundance of the bacteria groups and the differences in relative abundance between males and females within each mouse strain.
Table 4.
Relative abundance of bacteria groups in BALB/c and B6 male and female mice (n = 5). Differences between males and females within each mouse strain were determined with a Mann–Whitney U test. Significant differences are highlighted in italics
| Bacteria group | BALB/c female | BALB/c male | p value | B6 female | B6 male | p value B6 |
|---|---|---|---|---|---|---|
| Acholeplasma et rel. | 0.0041% | 0.0061% | 0.095 | 0.0084% | 0.0096% | 0.690 |
| Aerococcus urinaeequi et rel. | 0.0074% | 0.0117% | 0.095 | 0.0098% | 0.0097% | 0.841 |
| Alistipes et rel. | 0.9156% | 0.3753% | 0.095 | 0.5414% | 0.3036% | 0.095 |
| Allobaculum et rel. | 0.2716% | 0.2299% | 0.548 | 4.0636% | 7.4252% | 0.095 |
| Anaerovorax et rel. | 0.0644% | 0.0841% | 0.222 | 0.0513% | 0.0578% | 0.548 |
| Atopobium | 0.0046% | 0.0078% | 0.032 | 0.0096% | 0.0150% | 0.310 |
| Bacteroides distasonis et rel. | 0.1100% | 0.0266% | 0.690 | 0.1126% | 0.0373% | 0.016 |
| Bacteroides vulgatus et rel. | 0.2397% | 0.0924% | 0.151 | 0.0631% | 0.0585% | 0.548 |
| Bifidobacterium | 0.0828% | 0.0250% | 0.008 | 1.8595% | 2.9444% | 1.000 |
| Bilophila et rel. | 0.0179% | 0.0196% | 0.421 | 0.0155% | 0.0138% | 1.000 |
| Catenibacterium | 0.0140% | 0.0202% | 0.056 | 0.0259% | 0.0330% | 0.548 |
| Clostridium difficile et rel. | 0.9357% | 0.7979% | 0.310 | 1.1169% | 0.2468% | 0.032 |
| Clostridium herbivorans et rel. | 0.0083% | 0.0134% | 0.095 | 0.0132% | 0.0143% | 0.690 |
| Clostridium lactifermentans et rel. | 0.3127% | 0.3251% | 0.841 | 0.1212% | 0.1025% | 0.421 |
| Clostridium leptum et rel. | 0.5591% | 0.4151% | 0.095 | 0.4190% | 0.1997% | 0.008 |
| Clostridium perfringens et rel. | 2.3301% | 2.7132% | 0.690 | 0.3328% | 0.0963% | 0.016 |
| Clostridium symbosium et rel. | 0.5136% | 0.5040% | 0.841 | 0.7587% | 0.3233% | 0.056 |
| Coprobacillus catenoformis et rel. | 0.0230% | 0.0483% | 0.008 | 0.0273% | 0.0410% | 0.222 |
| Coprobacillus et rel.- Clostridium ramosum et rel. | 0.1377% | 0.2504% | 0.032 | 0.2859% | 0.3510% | 0.690 |
| Corynebacterium et rel. | 0.0067% | 0.0104% | 0.056 | 0.0108% | 0.0128% | 0.548 |
| Desulfovibrio et rel. | 0.0789% | 0.0904% | 0.548 | 0.0454% | 0.0355% | 0.841 |
| Dialister et rel. | 0.0027% | 0.0044% | 0.095 | 0.0044% | 0.0048% | 0.548 |
| Dorea et rel. | 3.4135% | 8.6217% | 0.095 | 2.3603% | 2.4100% | 0.841 |
| Eggerthella et rel. | 0.2295% | 0.4959% | 0.016 | 0.2927% | 0.5021% | 0.032 |
| Enterococcus | 0.2846% | 0.4653% | 0.310 | 0.1058% | 0.0130% | 0.008 |
| Eubacterium cylindroides et rel. | 0.0141% | 0.0225% | 0.095 | 0.0232% | 0.0263% | 0.690 |
| Eubacterium hallii et rel. | 0.0026% | 0.0043% | 0.095 | 0.0043% | 0.0047% | 0.548 |
| Eubacterium siraeum et rel. | 0.0084% | 0.0139% | 0.095 | 0.0135% | 0.0145% | 0.548 |
| Faecalibacterium prausnitzii et rel. | 0.0029% | 0.0046% | 0.095 | 0.0051% | 0.0050% | 0.690 |
| Lachnobacillus bovis et rel. | 1.9742% | 2.1309% | 0.841 | 1.6788% | 1.0041% | 0.421 |
| Lachnospira pectinoschiza et rel. | 0.9649% | 0.4554% | 0.310 | 0.3867% | 0.3047% | 0.421 |
| Lactobacillus acidophilus et rel. | 0.0418% | 0.0592% | 0.222 | 0.4314% | 0.0518% | 0.016 |
| Lactobacillus delbrueckii et rel. | 0.0062% | 0.0097% | 0.222 | 0.0176% | 0.0100% | 0.548 |
| Lactobacillus paracasei et rel. | 0.0121% | 0.0182% | 0.095 | 0.0153% | 0.0129% | 0.690 |
| Lactobacillus plantarum et rel. | 0.0222% | 0.0300% | 0.222 | 0.3977% | 0.0452% | 0.032 |
| Lactobacillus salivarius et rel. | 0.5801% | 1.9226% | 0.095 | 1.1330% | 0.7385% | 1.000 |
| Lactococcus et rel. | 0.0029% | 0.0047% | 0.056 | 0.0048% | 0.0054% | 0.548 |
| Mucispirillum schaedleri et rel. | 0.1270% | 0.1483% | 0.690 | 0.0544% | 0.0260% | 0.222 |
| Papillibacter cinnamivorans et rel. | 0.1624% | 0.1898% | 0.548 | 0.0784% | 0.0734% | 0.841 |
| Propionibacterium | 0.0036% | 0.0055% | 0.056 | 0.0067% | 0.0082% | 0.548 |
| Roseburia intestinalis et rel. | 0.0210% | 0.0344% | 0.056 | 0.0324% | 0.0333% | 0.690 |
| Ruminobacter amylophilus et rel. | 0.0035% | 0.0056% | 0.056 | 0.0054% | 0.0056% | 0.548 |
| Ruminococcus callidus et rel | 0.0197% | 0.0268% | 0.095 | 0.0241% | 0.0242% | 0.548 |
| Ruminococcus obeum et rel. | 0.0063% | 0.0107% | 0.095 | 0.0091% | 0.0098% | 0.690 |
| Solobacterium moorei et rel. | 0.0116% | 0.0184% | 0.056 | 0.0231% | 0.0275% | 0.690 |
| Sporobacter termitidis et rel. | 17.1016% | 13.7342% | 0.421 | 8.8797% | 5.9207% | 0.222 |
| Staphylococcus aureus et rel. | 0.0171% | 0.0669% | 0.008 | 0.0216% | 0.0238% | 0.690 |
| Streptococcus intermedius et rel. | 0.0067% | 0.0112% | 0.032 | 0.0101% | 0.0085% | 0.690 |
| Subdoligranulum et rel. | 0.0046% | 0.0073% | 0.056 | 0.0061% | 0.0065% | 0.548 |
| Sutterella wadsworthia et rel. | 0.0127% | 0.0205% | 0.056 | 0.0300% | 0.1068% | 0.310 |
| Turicibacter et rel. | 0.6261% | 0.4110% | 0.421 | 0.3876% | 0.0261% | 0.008 |
| Unclassified Bacteroidetes | 0.0029% | 0.0044% | 0.095 | 0.0046% | 0.0049% | 0.548 |
| Unclassified Clostridiales I | 0.0617% | 0.1277% | 0.056 | 0.1199% | 0.1494% | 0.690 |
| Unclassified Clostridiales II | 0.1700% | 0.2517% | 0.032 | 0.1490% | 0.1350% | 1.000 |
| Unclassified Clostridiales XIVa–close to Anaerostipes caccae | 0.0210% | 0.0329% | 0.056 | 0.0305% | 0.0311% | 0.690 |
| Unclassified Clostridiales XVI | 0.0751% | 0.1497% | 0.056 | 0.1577% | 0.1977% | 0.548 |
| Unclassified Mollicutes | 0.1218% | 0.1663% | 0.016 | 0.2794% | 0.3999% | 0.548 |
| Unclassified Porphyromonadaceae | 34.4807% | 31.7121% | 0.841 | 44.8531% | 52.5080% | 0.421 |
| Unclassified Prevotella | 1.1225% | 0.3418% | 0.690 | 0.2821% | 0.2037% | 0.095 |
| Uncultured Clostridiales | 0.2248% | 0.1868% | 0.151 | 0.1670% | 0.1381% | 0.690 |
| Veilonella | 0.0026% | 0.0043% | 0.095 | 0.0044% | 0.0048% | 0.548 |
Sex influenced gene expression profiles in the colon in a mouse strain-dependent way
Next, we performed a microarray analysis on the colon of male and female mice. We performed this analysis in both BALB/c and B6 mice to identify potential mouse strain-dependent sex effects. In the colon, a total of 1110 genes in BALB/c mice and 3309 genes in B6 mice were differently expressed between males and females. The two mice strains shared 531 genes that were differently expressed between males and females in the colon (both up- and downregulated).
To gain insight into the biological role of the genes which were differently expressed between the sexes, we first studied the physiological activities and molecular and cellular functions per mouse strain in which these genes are involved using Ingenuity Pathway Analysis (IPA). We specifically focused on functions related to immunology. In the colon in both mouse strains, IPA showed enrichment for genes related to, among others, hematological system development and immune cell trafficking (Table 5).
Table 5.
The top physiological activities and molecular and cellular functions related to the sexually dimorphic expressed genes in the proximal colon of both BALB/c and B6 mice (fold-change of 1.2 (up/down) and a p value < 0.05)
| Group | Physiological system development and function | p value | # Mol.a |
|---|---|---|---|
| BALB/c | Hematological system development and function | 6.67E-04-1.74E-13 | 170 |
| Tissue morphology | 6.56E-04-4.43E-13 | 157 | |
| Cell-mediated immune response | 6.37E-04-3.59E-12 | 67 | |
| Immune cell trafficking | 6.67E-04-3.59E-12 | 110 | |
| B6 | Hematopoiesis | 6.44E-04-2.71E-09 | 80 |
| Tissue morphology | 2.67E-07-1.21E-29 | 614 | |
| Immune cell trafficking | 4.90E-07-3.62E-29 | 360 | |
| Hematological system development and function | 4.90E-07-3.07E-26 | 577 | |
| Cardiovascular system development and function | 2.56E-07-2.40E-24 | 424 | |
| Organismal development | 4.33E-07-2.40E-24 | 808 |
aNumber of molecules included in the indicated functions
Subsequently, we analyzed in more detail the functions of the genes that were differently expressed between the sexes and in both mouse strains. IPA listed 500 functions that were enriched, in both or one of the two strains, from the genes displaying sexually dimorphic expression. Again, we specifically focused on functions related to immune response (Table 6). We found that sex altered the expression of genes related to several immune functions in the colon in both mouse strains. Many functions were related to T lymphocytes, and more specifically to T cell activation, development, proliferation capacity, and homing and migration. However, for most functions, we found that sex did not have the same effect in both strains. For example, the quantity of T lymphocytes was increased in BALB/c males as compared with BALB/c females, while reduced in B6 males as compared with B6 females.
Table 6.
Selection of immunological functions that are related to the genes with a different expression in males and females in both BALB/c and B6 mice in the proximal colon. The z score gives an indication of the activation or inhibition of the functions in males versus females. The number of molecules includes the number of molecules involved in the indicated function (fold-change of 1.2 (up/down) and a p value < 0.05)
| Diseases or functions annotation | z score BALB/c | z score B6 | p value BALB/c | p value B6 | # Mol. BALB/ca | # Mol. B6a |
|---|---|---|---|---|---|---|
| Quantity of leukocytes | 2.743 | − 5.066 | 2.57E-11 | 4.64E-21 | 113 | 292 |
| Quantity of lymphocytes | 2.803 | − 5.853 | 2.71E-09 | 2.45E-14 | 87 | 215 |
| Quantity of T lymphocytes | 2.638 | −4.628 | 4.40E-07 | 1.46E-09 | 64 | 154 |
| Quantity of granulocytes | − 0.018 | 0.063 | 5.13E-07 | 4.60E-12 | 42 | 104 |
| Quantity of antigen presenting cells | 2.556 | − 1.933 | 6.91E-06 | 3.42E-08 | 35 | 82 |
| Quantity of macrophages | 1.925 | 1.03E-04 | 25 | |||
| Quantity of B lymphocytes | 1.750 | − 3.840 | 3.85E-05 | 3.11E-09 | 40 | 104 |
| Quantity of dendritic cells | 2.980 | 1.62E-04 | 17 | |||
| Proliferation of immune cells | 0.895 | − 2.700 | 5.17E-11 | 3.03E-12 | 100 | 228 |
| Proliferation of lymphocytes | 1.046 | − 1.941 | 5.22E-10 | 3.57E-11 | 91 | 208 |
| Proliferation of T lymphocytes | 0.311 | 9.85E-10 | 78 | |||
| Homing of leukocytes | 4.371 | − 4.686 | 1.81E-08 | 2.24E-12 | 52 | 122 |
| Homing of lymphocytes | 4.226 | 2.67E-08 | 28 | |||
| Homing of T lymphocytes | 3.798 | 4.00E-10 | 26 | |||
| Homing of helper T lymphocytes | 2.805 | 1.41E-06 | 9 | |||
| Homing of regulatory T lymphocytes | 1.980 | 1.69E-04 | 4 | |||
| Activation of leukocytes | 2.053 | − 3.085 | 1.15E-05 | 4.66E-18 | 74 | 225 |
| Activation of T lymphocytes | 1.303 | − 1.944 | 1.01E-04 | 1.88E-08 | 40 | 105 |
| Differentiation of neutrophils | 0.599 | 2.62E-04 | 8 | |||
| Differentiation of leukocytes | 2.423 | − 3.410 | 6.65E-04 | 2.40E-10 | 64 | 190 |
| Differentiation of T lymphocytes | 0.697 | 6.75E-04 | 39 | |||
| T cell development | 1.636 | 3.74E-04 | 51 | |||
| Inflammatory response | 3.496 | − 3.851 | 7.30E-10 | 1.19E-22 | 95 | 259 |
| Bacterial Infections | 0.179 | 2.86E-05 | 46 |
aNumber of molecules included in the indicated functions
Correlation between microbiota composition and gene expression profile in the colon
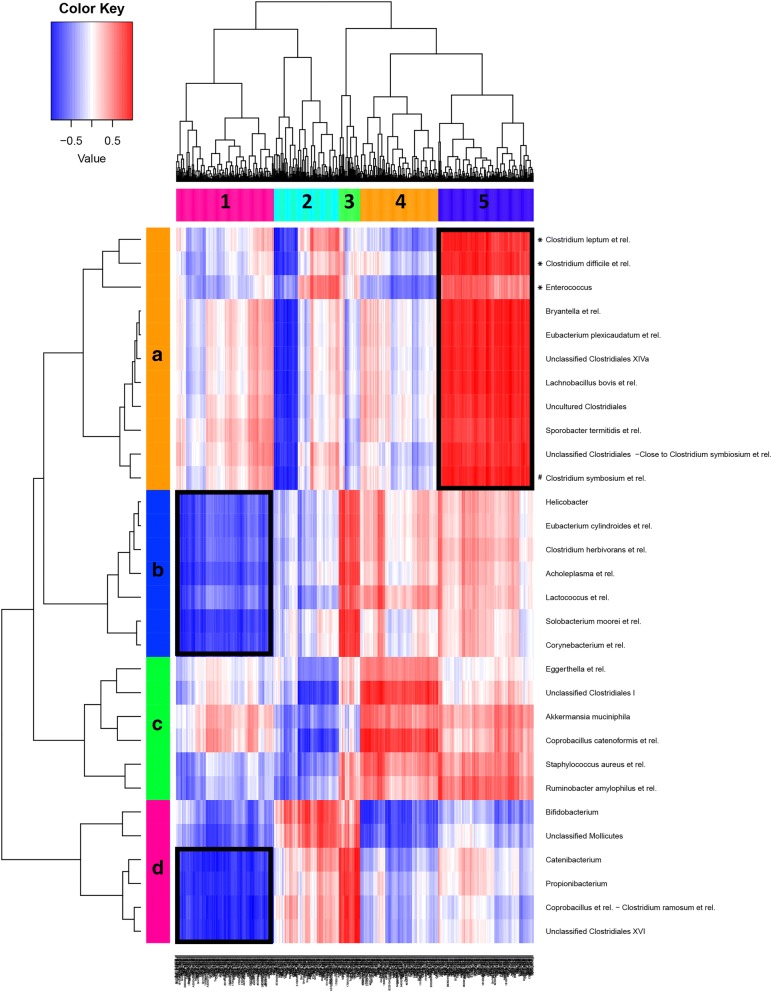
To investigate the relation between microbiota species and immunological gene expression, we combined microbiota and colonic gene expression data from each BALB/c and B6 male and female mice individually, to evaluate direct correlations between gene expression and microbiota composition in these samples. We performed two separate correlations within each mouse strain to determine the effect of sex independent of mouse strain (Figs. 2 and 3). We integrated these datasets using a PLS-based canonical correlation approach. In total, 600 genes and 30 bacterial groups were retained for the first three components, and clustering of the correlation coefficients revealed six main clusters of host genes that correlated positively (red) or negatively (blue) to specific bacteria.
Fig. 2.
Correlation between microbiota species and gene expression in the colon of B6 mice. Heatmap of correlation analysis of MITChip (vertical) and microarray (horizontal) datasets of male and female B6 mice. The integration of datasets was done per individual mouse (five mice per group) and gives the direct correlations between gene expression and microbiota composition from these samples. In deep red, the cluster of genes that most positively correlated with a respective group of bacteria. In deep blue, the cluster of genes that most negatively correlated with a respective group of bacteria. Five main gene clusters (1–5) and four main bacteria clusters (A-D) were identified. The cluster framed in black is discussed in more detail in the text. A positive correlation between bacteria and genes indicates that a higher abundance of the bacteria is associated with a higher expression of the particular cluster of genes. A negative correlation between bacteria and genes indicates that a lower abundance of the bacteria is associated with a lower expression of the particular cluster of genes. The functions to which these genes are related to are presented in Table 7. Note that the bacteria and intestinal genes that were selected for the correlation were the ones most explanatory for variation between sex and mouse strain, and therefore the genes in a specific cluster do not necessarily have a significantly different expression between the sexes. Moreover, a positive correlation between bacteria and genes not necessary indicates that the particular function related to these genes is upregulated, as the genes involved may also have a suppressive effect on the function. Asterisks (*) and hashtags (#) indicate that the specific bacteria has a significantly/or tend to have a higher abundance in B6 females as compared to B6 males, respectively
Fig. 3.
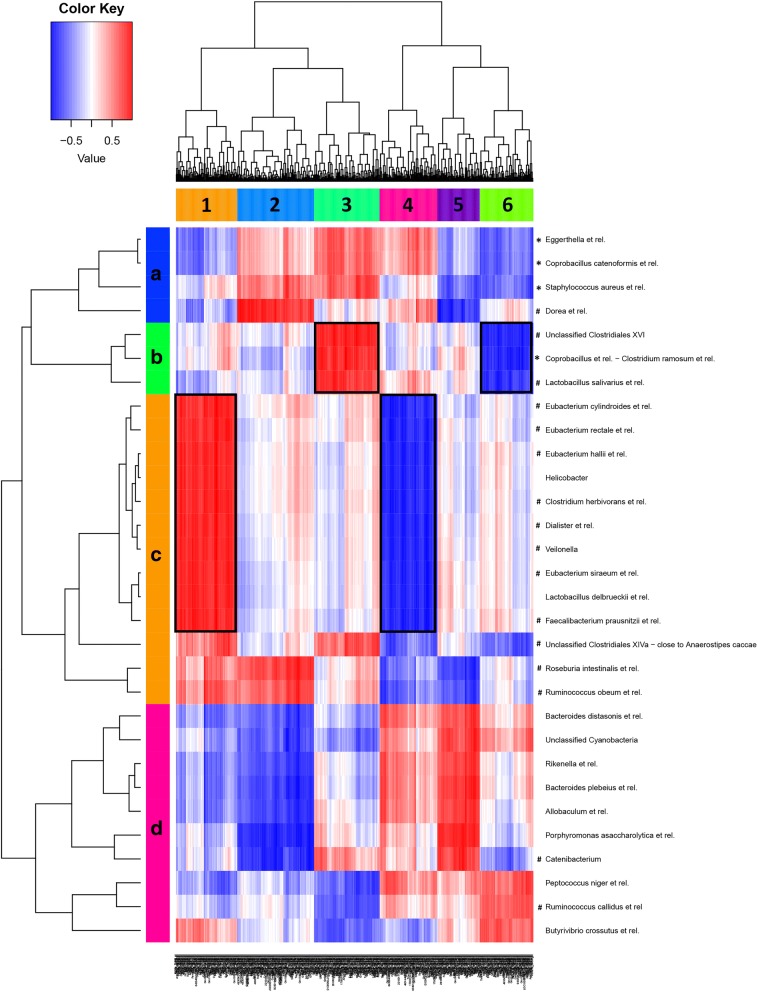
Correlation between microbiota species and gene expression in the colon of BALB/c mice. Heatmap of correlation analysis of MITChip (vertical) and microarray (horizontal) datasets of male and female BALB/c mice. The integration of datasets was done per individual mouse (five mice per group) and gives the direct correlations between gene expression and microbiota composition from these samples. In deep red, the cluster of genes that most positively correlated with a respective group of bacteria. In deep blue, the cluster of genes that most negatively correlated with a respective group of bacteria. Six main gene clusters (1–6) and four main bacteria clusters (A–D) were identified. The clusters framed in black are discussed in more detail in the text. A positive correlation between bacteria and genes indicates that a higher abundance of the bacteria is associated with a higher expression of the particular cluster of genes. A negative correlation between bacteria and genes indicates that a lower abundance of the bacteria is associated with a lower expression of the particular cluster of genes. The functions to which these genes are related to are presented in Table 7. Note that the bacteria and intestinal genes that were selected for the correlation were the ones most explanatory for variation between sex and mouse strain, and therefore the genes in a specific cluster do not necessarily have a significantly different expression between the sexes. Moreover, a positive correlation between bacteria and genes not necessary indicates that the particular function related to these genes is upregulated, as the genes involved may also have a suppressive effect on the function. Asterisks (*) and hashtags (#) indicate that the specific bacteria has a significantly/or tend to have a higher abundance in BALB/c males as compared to BALB/c females, respectively
Within the B6 strain (Fig. 2), a strong positive correlation was found between gene expression cluster 5 and several bacteria which were or tended to be enriched in the females (such as Clostridium leptum et rel., Clostridium difficile et rel., Enterococcus, and Clostridium symbosium et rel.). The genes in this cluster are related to, among others, inflammatory response and migration of leukocytes (Table 7). Within the BALB/c strain (Fig. 3), we found four gene expression clusters which strongly correlated to certain bacteria groups. Both gene expression clusters 1 and 3 showed a strong positive correlation with bacteria which tended to be enriched in males (Eubacterium cylindroides et rel., Eubacterium hallii et rel., Clostridium herbivorans et rel. Dialister et rel., Veilonella, Eubacterium siraeum et rel., and Faecalibacterium prausnitzii et rel. in cluster 1 and Unclassified Clostridiales XVI, Coprobacillus et rel.-Clostridium ramosum et rel., and Lactobacillus salivarius et rel. in cluster 3). The genes in cluster 1 are related to, among others, proliferation of lymphocytes and quantity of leukocytes, whereas the genes in cluster 3 are related to, among others, expansion of T (helper) lymphocytes (Table 7). The gene expression clusters 4 and 6 showed a strong negative correlation with bacteria which tended to be enriched in males. Gene cluster 4 is related to, among others, quantity of leukocytes and chemotaxis of phagocytes, whereas gene cluster 6 is not related to immune functions (Table 7). The bacteria and intestinal genes that were selected for the correlation were the ones most explanatory for variation between sex and mouse strain, and therefore the genes in a specific cluster do not necessarily have a significantly different expression between the sexes. Moreover, a positive correlation between bacteria and genes not necessarily indicates that the particular function related to these genes is upregulated, as the genes involved may also have a suppressive effect on the function.
Table 7.
Selection of immunological functions that are related to the genes in cluster 5 (B6 mice) and clusters 1, 3, and 6 (BALB/c mice) from the correlation analysis of MITChip and microarray
| Gene cluster | Diseases or Functions Annotation | p value | # Mol. |
|---|---|---|---|
| Cluster 5 (B6) | Inflammatory response | 6.46E-05 | 20 |
| Leukocyte migration | 3.55E-04 | 20 | |
| Infection of CD4+ T-lymphocytes | 4.81E-04 | 2 | |
| Quantity of leukocytes | 6.13E-04 | 20 | |
| Activation of leukocytes | 8.90E-04 | 16 | |
| Cluster 1 (BALB/c) | Proliferation of lymphocytes | 5.22E-05 | 34 |
| Quantity of leukocytes | 5.78E-05 | 40 | |
| Quantity of lymphoid cells | 7.54E-05 | 33 | |
| Quantity of myeloid cells | 1.24E-04 | 24 | |
| Quantity of mononuclear leukocytes | 1.53E-04 | 33 | |
| Cluster 3 (BALB/c) | Expansion of T lymphocytes | 1.60E-05 | 7 |
| Expansion of helper T lymphocytes | 2.12E-05 | 4 | |
| Immune response of leukocytes | 1.06E-04 | 9 | |
| Immune response of phagocytes | 1.64E-04 | 7 | |
| Leukocyte migration | 5.60E-04 | 15 | |
| Cluster 4 (BALB/c) | Cell death of chronic lymphocytic leukemia B cells | 1.65E-05 | 3 |
| Quantity of leukocytes | 2.83E-05 | 17 | |
| Chemotaxis of phagocytes | 3.32E-05 | 9 | |
| Quantity of mononuclear leukocytes | 1.17E-04 | 14 | |
| Quantity of phagocytes | 1.32E-04 | 10 |
aNumber of molecules included in the indicated functions
Both sex and strain influenced T cell activation, migration, and maturation in the mesenteric lymph nodes
The results of the microarray showed that the expression of several genes which can be related to immunological functions (e.g., T cell trafficking, activation, and maturation) were up- or downregulated by sex (Table 6). Therefore, we evaluated the effect of sex on T lymphocytes in the mesenteric lymph nodes (MLN) using flow cytometry. We chose the MLN, since this is an important site for the induction of intestinal immune responses [32]. We measured the percentages of T lymphocytes, T helper (CD4+) cells, and T cytotoxic (CD8+) cells. Furthermore, we measured their expression of the early activation marker (CD69), their expression of gut-homing receptor α4ß7 and their maturation status (CD62L and CD44). Overall, male mice had a lower percentage of T lymphocytes in their MLN than female mice (two-way ANOVA, p = 0.010) (Fig. 4a). Interaction between sex and strain was found in the CD4+/CD8+ ratio (two-way ANOVA, p = 0.004); BALB/c males had a lower CD4+/CD8+ ratio than BALB/c females (Bonferroni, p < 0.01), while no effect of sex was seen in the B6 strain (Fig. 4b).
The percentage of CD8+ or CD4+ cells expressing of CD69 was not affected by sex (Fig. 4c, d). The expression of integrin α4ß7 (homing marker) on CD8+ and CD4+ was lower in male mice as compared to female mice (two-way ANOVA, p = 0.001 and p = 0.046, respectively) (Fig. 4e, f). In addition, males showed an increased percentage of naïve CD8+ cells as compared to females (two-way ANOVA, p = 0.031) (Fig. 5a). Interaction was found between sex and strain in the percentage of central memory (CM) CD8+ cells (two-way ANOVA, p = 0.001); BALB/c males had a lower percentage of CM CD8+ cells than BALB/c females (Bonferroni, p < 0.01), while sex had no effect on the B6 strain (Fig. 5b). We observed no effect of sex on CD8+ effector memory (EM) cells (Fig. 5c). Sex did not influence the percentage of naïve, CM, or EM CD4+ cells (Fig. 5d–f).
Discussion
In this study, we demonstrated clear sex differences in intestinal microbiota, intestinal gene expression, and immune cell composition. We validated the sex effects by using two mouse strains with different genetic backgrounds and microbiota profiles [47]. Although sex significantly explained part of the variance in microbiota composition, this was mouse strain-dependent. Furthermore, we found that the expression of many colonic (mucosal) genes related to immunological functions (e.g., T cell trafficking, activation, and maturation) were up- or downregulated by sex, again in a mouse strain-dependent way. As sex effects in microbiota and sex effects in mucosal gene expression were both strain dependent, we correlated microbiota species with mucosal gene expression data per mouse strain. We found correlations between genes associated with immune populations and certain sex-specific bacteria. Despite these strain-dependent effects of sex on microbiota composition and mucosal immune responses, almost similar sex differences in immune cell populations in the MLN in the two mouse strains were found.
This study demonstrated sex differences in intestinal microbiota composition in healthy mice of two different genetic backgrounds. We showed that sex influenced the microbial diversity and richness and found that males had a lower microbial diversity and richness than females. These findings are in line with our previous study in which we, among others, focused on sex-specific effects of aging on the microbiota composition (B6 mice) [48]. Additionally, this corroborates the findings of Yurkovetskiy et al. and Xiao et al., who also found a higher microbiota diversity in female mice than in male mice [23, 26]. In general, it is assumed that a microbiome with a higher diversity and richness is beneficial for host health [49]. High microbial richness is linked to microbiota stability [49], whereas a lower microbial richness and diversity is linked to several disorders, including obesity [9, 11], and IBD [10]. Therefore, the reduced microbial diversity and richness in male mice in this study may support the results and conclusions of Bábíčková et al. (2015) who found that male mice (B6) have a higher sensitivity to develop DSS-induced colitis (used as IBD model) than female mice (B6) [50]. However, this needs to be confirmed in future studies.
When focusing on the microbiota composition at species-like level we also found sex-specific differences, these were also strain dependent. We found that B6 females had a relative higher abundance of, among others, Lactobacillus plantarum and Bacteroides distasonis et rel. as compared to B6 males. These species have been shown to influence immune responses, such as enhancing Tregs [51, 52]. BALB/c females had a relative higher abundance of Bifidobacterium as compared to BALB/c males. Bifidobacteria were shown to have beneficial effects against a variety of gastrointestinal disorders, including colitis [53], and have also been shown to be able to induce regulatory T cells [54]. Also other studies have shown sex-specific microbiota in mice [23, 26–28]. It is interesting to note that the study of Org et al. (2016) also showed that sex differences in the microbiota of mice depends on the genetic background [28]. Pertinent sex differences in microbiota composition in the various studies, however, are difficult to compare, since they are dependent on the strain [28], diet [25] and probably also vendor [55]. Additionally, stress and the gut microbiome also showed to interact which each other and the sensitivity for stress seems to be dependent on genetic background but also sex [56]. Moreover, maternal stress showed to modulate sex differences in the microbiota composition of the offspring [57]. Taken together, stress also may be an interfering factor on dimorphism in microbiota composition and may be either an underling factor or consequence. Our data show that the mouse strain-dependent sex differences in microbiota composition highlight the importance of considering the genetic background when selecting an animal model and the need for standardization on genetic background and other interfering factors in human studies.
The causes of the sex differences in the intestinal microbiota composition are probably multifold. Sex hormones may play a role, since differences in microbiota profiles between males and females in NOD mice disappeared after castration of the males [23], suggesting the involvement of testosterone. However, in view of the strain differences, it is likely that genetic differences also play an important role. Genetic differences might for instance be variations in mucus composition, which affect microbiota composition [58]. Also maternal differences in oligosaccharide composition of mother milk are genetically determined and affect the development of the microbiota composition of the offspring [59, 60]. Whether sex differences in the microbiome also appear in humans is difficult to conclude as human studies on sex differences are still scarce and influenced by many confounding factors. Some human studies found small sex differences in the microbiome [61–64], while others did not [65–67]. Confounding factors might be heterogeneity in genetics, but also the reproductive condition of females (e.g., menstrual cycle, the use of oral contraceptives, and menopause), which is often not taken into account. Such factors can not only interfere with immune responses [68, 69], and with microbiota composition, but may also modulate the sex effects.
A major goal of this study was to correlate sex-specific intestinal immune differences with specific microbiota. The bacteria and intestinal genes that were selected for this correlation were the most explanatory for variation between sex and mouse strain. For B6 mice, we showed that various female-specific bacteria positively correlated with one cluster of genes, which were, among others, associated with inflammatory responses and leukocyte migration. In BALB/c mice, we found four clusters of genes correlating with various male specific bacteria (bacteria which were or tended to be significantly increased in males), which were involved in, among others, differentiation of lymphocytes and expansion of helper T cells, while the clusters which were negatively correlating with the male bacteria were involved in, among others, chemotaxis and quantity of phagocytes. Our data do suggest that microbiota may influence immunological gene expression in the gut. Although genes in the pathway of expansion of T helper cells were positively correlated with various bacteria increased in BALB/c males, this does not necessarily mean that this is associated with increased numbers of T helper cells, since genes in this pathway may also inhibit expansion of T helper cells. Indeed, our flow cytometry data show decreased numbers of T helper cells in the MLN in BALB/c mice.
Our study was merely observational. However, another study from our lab showed that sex-specific microbiota may indeed affect immune responses. Fransen et al. (2017) performed a microbiota transfer study in germ-free mice by transferring male microbiota into female germ-free mice and female microbiota into germ-free male mice [70]. Fransen observed that germ-free male recipients of male microbiota had a higher percentages of RORγt+ Foxp3+ cells in the PPs and MLN as compared with germ-free male recipients of female microbiota, indicating that indeed sex differences in microbiota may induce sex differences in immune responses [70]. However, they also found that males in general had a higher percentages of conventional Tregs, independent of whether they received microbiota from male or female mice, suggesting not all sex differences in immune response are dependent on the microbiome [70].
We also found sex differences in immune cell populations in the MLN of both strains and despite the different sex effects in the microbiota and gene expression in the two strains, we observed similar sex differences in immune cell populations in the MLN in both strains, and also in the spleen (21). Analysis of the microarray data showed that the expression of several genes which can be related to immunological functions (e.g., T cell trafficking, activation, and maturation) were up- or downregulated by sex. Therefore, we focused on T cells and T cell functions in the MLN, and we found that females had a higher percentage of total T cells, with an increased percentage of these T cells expressing the homing receptor α4β7 (T cell trafficking) than males. Furthermore, female mice had a lower percentage of naive T cytotoxic cells (T cell maturation) than male mice. The higher percentage of T cells and lower percentage of naïve T cells fits with the general idea that females have a stronger adaptive immune arm than males [22]. It is unknown from this study how these changes are induced, but as indicated above, sex differences in microbiota species may be involved. Also in the MLN, we found that some of the sex effects in the immune cell composition were strain dependent, although less apparent: only the CD4+/CD8+ ratio and central memory CD8+ cells were affected by sex in a strain-dependent way. Similar results were recently found in the Peyer’s patches (PP) and the spleen: sex effects on immune cell populations were mainly strain independent [21].
Conclusions
This study demonstrated sex differences in intestinal microbiota species, diversity, and richness in healthy mice of two different mouse strains. The nature of the sex effects, however, appeared to be determined by the mouse strain, since different bacterial species were enriched in males and females in the two strains. The strain-dependent sex effects were also observed in the expression of immunological genes in the colon. The correlations we found between male and female specific bacteria with various immunological gene pathways, suggest that sex differences in the microbiome may be involved in sex differences in immune responses. To our opinion, this is an important observation and not only implies that preventive measures for disease development may require a sex- and genetic-specific approach, but it also shows that in microbiome studies, both sex and genetic background should be taken into consideration. Our study may also shed light on the conflicting results in studies with respect to sex differences in the microbiome, especially in human studies. Conflicting results found in human studies [25, 61–67] may be due to the lack of standardization with respect to sex and genetic background.
Additional file
Method and results RT qPCR for validation microarray. (DOCX 27 kb)
Acknowledgements
The authors would like to thank Jenny Jansen (Division of Human Nutrition, Wageningen University) for technical support in the microarray analysis.
Funding
This work was supported by a project from the Top Institute Food and Nutrition, Wageningen, The Netherlands (Grant number GH002). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Availability of data and materials
The datasets (microarray) supporting the conclusions of this article are available in the gene expression omnibus from NCBI under ID number GSE85913.
Abbreviations
- B6
C57B1/6OlaHsd mice
- BALB/c
Balb/cOlaHsd mice
- CM
Central memory cells
- DSS
Dextran sulfate sodium
- EM
Effector memory cells
- FCS
Fetal calf serum
- IBD
Inflammatory bowel disease
- IBMT
Intensity-based moderated t statistic
- IPA
Ingenuity pathway analysis
- MITChip
Mouse intestinal tract chip
- MLN
Mesenteric lymph nodes
- NOD
Non-obese diabetic
- PLS
Partial least squares
- PP
Peyer’s patches
- RBB + C
Bead-beating-plus column
- RDA
Redundancy analysis
- RPA
Robust probabilistic averaging
- SCFAs
Short-chain fatty acids
- Th
T helper cells
- Tregs
T regulatory cells
Authors’ contributions
ME, FH, CB, MB, AvB, HS, PdV, and MF helped conceive and design the experiments. ME, FH, AvB, and BdH performed the experiments. ME and F.H analyzed the data. ME, FH, PdV, and MF wrote the paper. All authors read and approved the final manuscript.
Ethics approval
This study was carried out in accordance with the recommendations of the Animal Care Committee of the Groningen University. The protocol was approved by the Animal Care Committee of the Groningen University (approval number 6349A).
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s13293-018-0186-6) contains supplementary material, which is available to authorized users.
Contributor Information
Marlies Elderman, Email: m.e.elderman@umcg.nl.
Floor Hugenholtz, Email: f.hugenholtz@amc.uva.nl.
Clara Belzer, Email: clara.belzer@wur.nl.
Mark Boekschoten, Email: mark.boekschoten@wur.nl.
Adriaan van Beek, Email: a.vanbeek@erasmusmc.nl.
Bart de Haan, Email: b.j.de.haan@umcg.nl.
Huub Savelkoul, Email: huub.savelkoul@wur.nl.
Paul de Vos, Email: p.de.vos@umcg.nl.
Marijke Faas, Email: m.m.faas@umcg.nl.
References
- 1.Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312(5778):1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cerf-Bensussan N, Gaboriau-Routhiau V. The immune system and the gut microbiota: friends or foes? Nat Rev Immunol. 2010;10:735–744. doi: 10.1038/nri2850. [DOI] [PubMed] [Google Scholar]
- 3.Marchesi JR, Adams DH, Fava F, Hermes GD, Hirschfield GM, Hold G, et al. The gut microbiota and host health: a new clinical frontier. Gut. 2016;65(2):330–339. doi: 10.1136/gutjnl-2015-309990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prakash S, Rodes L, Coussa-Charley M, Tomaro-Duchesneau C. Gut microbiota: next frontier in understanding human health and development of biotherapeutics. Biol Targets Ther. 2011;5:71–86. doi: 10.2147/BTT.S19099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Foxx-Orenstein AE, Chey WD. Manipulation of the gut microbiota as a novel treatment strategy for gastrointestinal disorders. Am J Gastroenterol Suppl. 2012;1:41–46. doi: 10.1038/ajgsup.2012.8. [DOI] [Google Scholar]
- 6.Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489(7415):220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parekh PJ, Balart LA, Johnson DA. The influence of the gut microbiome on obesity, metabolic syndrome and gastrointestinal disease. Clin Transl Gastroenterol. 2015;6 10.1038/ctg.2015.16. [DOI] [PMC free article] [PubMed]
- 8.Wu HJ, Wu E. The role of gut microbiota in immune homeostasis and autoimmunity. Gut Microbes. 2012;3(1):4–14. doi: 10.4161/gmic.19320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Norman J, Handley S, Baldridge M, Droit L, Liu C, Keller B, et al. Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell. 2015;160(3):447–460. doi: 10.1016/j.cell.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony G, et al. Richness of human gut microbiome correlates with metabolic markers. Nature. 2015;500:541–546. doi: 10.1038/nature12506. [DOI] [PubMed] [Google Scholar]
- 12.Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly- YM, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341(6145):569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vinolo MA, Rodrigues HG, Nachbar RT, Curi R. Regulation of inflammation by short chain fatty acids. Nutrients. 2011;3(10):858–876. doi: 10.3390/nu3100858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504(7480):446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 15.Lathrop SK, Bloom SM, Rao SM, Nutsch K, Lio CW, Santacruz N, et al. Peripheral education of the immune system by colonic commensal microbiota. Nature. 2011;478(7368):250–254. doi: 10.1038/nature10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ivanov II, Frutos Rde L, Manel N, Yoshinaga K, Rifkin DB, Sartor RB, et al. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe. 2008;4(4):337–349. doi: 10.1016/j.chom.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ngo ST, Steyn FJ, McCombe PA. Gender differences in autoimmune disease. Front Neuroendocrinol. 2014;35(3):347–369. doi: 10.1016/j.yfrne.2014.04.004. [DOI] [PubMed] [Google Scholar]
- 18.Pradhan AD. Sex differences in the metabolic syndrome: implications for cardiovascular health in women. Clin Chem. 2014;60(1):44–52. doi: 10.1373/clinchem.2013.202549. [DOI] [PubMed] [Google Scholar]
- 19.Zelinkova Z, der Woude CJ. Gender and inflammatory bowel disease. J Clin Cell Immunol. 2014;5(4):245–250. doi: 10.4172/2155-9899.1000245. [DOI] [Google Scholar]
- 20.Bouman A, Schipper M, Heineman M, Faas M. Gender difference in the non-specific and specific immune response in humans. Am J Reprod Immunol. 2004;52(1):19–26. doi: 10.1111/j.1600-0897.2004.00177.x. [DOI] [PubMed] [Google Scholar]
- 21.Elderman M, van Beek A, Brandsma E, de Haan B, Savelkoul H, de Vos P, et al. Sex impacts Th1, Treg, and DCs in both the intestinal and systemic immunity in a mouse strain and location dependent manner. Biol Sex Differ. 2016;7 10.1186/s13293-016-0075-9. [DOI] [PMC free article] [PubMed]
- 22.Klein SL, Flanagan KL. Sex differences in immune responses. Nat Rev Immunol. 2016;16(10):626–638. doi: 10.1038/nri.2016.90. [DOI] [PubMed] [Google Scholar]
- 23.Yurkovetskiy L, Burrows M, Khan A, Graham L, Volchkov P, Becker L, et al. Gender bias in autoimmunity is influenced by microbiota. Immunity. 2013;39(2):400–412. doi: 10.1016/j.immuni.2013.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bernbom N, Norrung B, Saadbye P, Molbak L, Vogensen FK, Licht TR. Comparison of methods and animal models commonly used for investigation of fecal microbiota: effects of time, host and gender. J Microbiol Methods. 2006;66(1):87–95. doi: 10.1016/j.mimet.2005.10.014. [DOI] [PubMed] [Google Scholar]
- 25.Bolnick DI, Snowberg LK, Hirsch PE, Lauber CL, Org E, Parks B, et al. Individual diet has sex-dependent effects on vertebrate gut microbiota. Nat Commun. 2014;5:4500. doi: 10.1038/ncomms5500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiao L, Feng Q, Liang S, Sonne SB, Xia Z, Qiu X, et al. A catalog of the mouse gut metagenome. Nat Biotechnol. 2015;33:1103–1108. doi: 10.1038/nbt.3353. [DOI] [PubMed] [Google Scholar]
- 27.Markle JGM, Frank DN, Mortin-Toth S, Robertson CE, Feazel LM, Rolle-Kamkczyk U, et al. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science. 2013;339:1084–1088. doi: 10.1126/science.1233521. [DOI] [PubMed] [Google Scholar]
- 28.Org E, Mehrabian M, Parks BW, Shipkova P, Liu X, Drake TA, et al. Sex differences and hormonal effects on gut microbiota composition in mice. Gut Microbes. 2016;7(4):313–322. doi: 10.1080/19490976.2016.1203502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Org E, Parks BW, Joo JW, Emert B, Schwartzman W, Kang EY, et al. Genetic and environmental control of host-gut microbiota interactions. Genome Res. 2015;25(10):1558–1569. doi: 10.1101/gr.194118.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Melgar S, Karlsson A, Michaëlsson E. Acute colitis induced by dextran sulfate sodium progresses to chronicity in C57BL/6 but not in BALB/c mice: correlation between symptoms and inflammation. Am J Physiol Gastrointest Liver Physiol. 2005;(6):1328–38. [DOI] [PubMed]
- 31.Tsuchiya T, Fukuda S, Hamada H, Nakamura A, Kohama Y, Ishikawa H, et al. Role of gamma delta T cells in the inflammatory response of experimental colitis mice. J Immunol. 2003;15:5507–5513. doi: 10.4049/jimmunol.171.10.5507. [DOI] [PubMed] [Google Scholar]
- 32.Macpherson AJ, Smith K. Mesenteric lymph nodes at the center of immune anatomy. J Exp Med. 2006;203(3):497–500. doi: 10.1084/jem.20060227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu Z, Morrison M. Improved extraction of PCR-quality community DNA from digesta and fecal samples. BioTechniques. 2004;36(5):808–812. doi: 10.2144/04365ST04. [DOI] [PubMed] [Google Scholar]
- 34.Geurts L, Lazarevic V, Derrien M, Everard A, Van Roye M, Knauf C, et al. Altered gut microbiota and endocannabinoid system tone in obese and diabetic leptin-resistant mice: impact on apelin regulation in adipose tissue. Front Microbiol. 2011;2:149. doi: 10.3389/fmicb.2011.00149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rajilic-Stojanovic M, Heilig HG, Molenaar D, Kajander K, Surakka A, Smidt H, et al. Development and application of the human intestinal tract chip, a phylogenetic microarray: analysis of universally conserved phylotypes in the abundant microbiota of young and elderly adults. Environ Microbiol. 2009;11(7):1736–1751. doi: 10.1111/j.1462-2920.2009.01900.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lahti L, Elo LL, Aittokallio T, Kaski S. Probabilistic analysis of probe reliability in differential gene expression studies with short oligonucleotide arrays. IEEE/ACM Trans Comput Biol Bioinform. 2011;8:217–225. doi: 10.1109/TCBB.2009.38. [DOI] [PubMed] [Google Scholar]
- 37.Braak CJF, Šmilauer P. Canoco Reference Manual and User’s Guide: Software for Ordination (Version 5.0) Ithaca: NH; 2012. [Google Scholar]
- 38.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31(4):e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 40.Dai M, Wang P, Boyd AD, Kostov G, Athey B, Jones EG, et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 2005;33(20):e175. doi: 10.1093/nar/gni179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sartor MA, Tomlinson CR, Wesselkamper SC, Sivaganesan S, Leikauf GD, Medvedovic M. Intensity-based hierarchical Bayes method improves testing for differentially expressed genes in microarray experiments. BMC Bioinformatics. 2006;19:538. doi: 10.1186/1471-2105-7-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boulesteix AL, Strimmer K. Partial least squares: a versatile tool for the analysis of high-dimensional genomic data. Brief Bioinform. 2007;8(1):32–44. doi: 10.1093/bib/bbl016. [DOI] [PubMed] [Google Scholar]
- 43.Lange K, Hugenholtz F, Jonathan MC, Schols HA, Kleerebezem M, Smidt H, et al. Comparison of the effects of five dietary fibers on mucosal transcriptional profiles, and luminal microbiota composition and SCFA concentrations in murine colon. Mol Nutr Food Res. 2015;59(8):1590–1602. doi: 10.1002/mnfr.201400597. [DOI] [PubMed] [Google Scholar]
- 44.Le Cao KA, Martin PG, Robert-Granie C, Besse P. Sparse canonical methods for biological data integration: application to a cross-platform study. BMC Bioinformatics. 2009:10–34. 10.1186/1471-2105-10-34. [DOI] [PMC free article] [PubMed]
- 45.Gonzalez I, Cao KA, Davis MJ, Dejean S. Visualising associations between paired ‘omics’ data sets. BioData Min. 2012;5(1):19. doi: 10.1186/1756-0381-5-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Le Cao KA, Gonzalez I, Dejean S. integrOmics: an R package to unravel relationships between two omics datasets. Bioinformatics. 2009;25(21):2855–2856. doi: 10.1093/bioinformatics/btp515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fransen F, Zagato E, Mazzini E, Fosso B, Manzari C, El Aidy S, et al. BALB/c and C57BL/6 mice differ in polyreactive IgA abundance, which impacts the generation of antigen-specific IgA and microbiota diversity. Immunity. 2015;43(3):527–540. doi: 10.1016/j.immuni.2015.08.011. [DOI] [PubMed] [Google Scholar]
- 48.Elderman M, Sovran B, Hugenholtz F, Graversen K, Huijskes M, Houtsma E, et al. The effect of age on the intestinal mucus thickness, microbiota composition and immunity in relation to sex in mice. PLoS One. 2017;12(9):e0184274. doi: 10.1371/journal.pone.0184274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tap J, Furet JP, Bensaada M, Philippe C, Roth H, Rabot S, et al. Gut microbiota richness promotes its stability upon increased dietary fibre intake in healthy adults. Environ Microbiol. 2015;17:4954–4964. doi: 10.1111/1462-2920.13006. [DOI] [PubMed] [Google Scholar]
- 50.Babickova J, Tothova L, Lengyelova E, Bartonova A, Hodosy J, Gardlik R, et al. Sex differences in experimentally induced colitis in mice: a role for estrogens. Inflammation. 2015;38(5):1996–2006. doi: 10.1007/s10753-015-0180-7. [DOI] [PubMed] [Google Scholar]
- 51.Smelt MJ, de Haan BJ, Bron PA, van Swam I, Meijerink M, Wells JM, et al. L. Plantarum, L. salivarius, and L. lactis attenuate Th2 responses and increase Treg frequencies in healthy mice in a strain dependent manner. PLoS One. 2012;7(10):e47244. doi: 10.1371/journal.pone.0047244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kverka M, Zakostelska Z, Klimesova K, Sokol D, Hudcovic T, Hrncir T, et al. Oral administration of Parabacteroides distasonis antigens attenuates experimental murine colitis through modulation of immunity and microbiota composition. Clin Exp Immunol. 2011;163(2):250–259. doi: 10.1111/j.1365-2249.2010.04286.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Picard C, Fioramonti J, Francois A, Robinson’ T, Neant F, Matuchansky C. Review article: bifidobacteria as probiotic agents -- physiological effects and clinical benefits. Aliment Pharmacol Ther. 2005;15:495–512. doi: 10.1111/j.1365-2036.2005.02615.x. [DOI] [PubMed] [Google Scholar]
- 54.Konieczna P, Groeger D, Ziegler M, Frei R, Ferstl R, Shanahan F, et al. Bifidobacterium infantis 35624 administration induces Foxp3 T regulatory cells in human peripheral blood: potential role for myeloid and plasmacytoid dendritic cells. Gut. 2012;61:354–366. doi: 10.1136/gutjnl-2011-300936. [DOI] [PubMed] [Google Scholar]
- 55.Ericsson AC, Davis JW, Spollen W, Bivens N, Givan S, Hagan CE, et al. Effects of vendor and genetic background on the composition of the fecal microbiota of inbred mice. PLoS One. 2015;10(2):e0116704. doi: 10.1371/journal.pone.0116704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rea K, Dinan TG, Cryan JF. The microbiome: a key regulator of stress and neuroinflammation. Neurobiol Stress. 2016;4:23–33. doi: 10.1016/j.ynstr.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jasarevic E, Howard CD, Misic AM, Beiting DP, Bale TL. Stress during pregnancy alters temporal and spatial dynamics of the maternal and offspring microbiome in a sex-specific manner. Sci Rep. 2017;7:44182. doi: 10.1038/srep44182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sovran B, Lu P, Loonen LM, Hugenholtz F, Belzer C, Stolte EH, et al. Identification of commensal species positively correlated with early stress responses to a compromised mucus barrier. Inflamm Bowel Dis. 2016;22(4):826–840. doi: 10.1097/MIB.0000000000000688. [DOI] [PubMed] [Google Scholar]
- 59.Fuhrer A, Sprenger N, Kurakevich E, Borsig L, Chassard C, Hennet T. Milk sialyllactose influences colitis in mice through selective intestinal bacterial colonization. J Exp Med. 2010;207(13):2843–2854. doi: 10.1084/jem.20101098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Monteagudo-Mera A, Arthur JC, Jobin C, Keku T, Bruno-Barcena JM, Azcarate-Peril MA. High purity galacto-oligosaccharides enhance specific Bifidobacterium species and their metabolic activity in the mouse gut microbiome. Benef Microbes. 2016;7(2):247–264. doi: 10.3920/BM2015.0114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dominianni C, Sinha R, Goedert JJ, Pei Z, Yang L, Hayes RB, et al. Sex, body mass index, and dietary fiber intake influence the human gut microbiome. PLOSone. 2015;10(4):1–14. doi: 10.1371/journal.pone.0124599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li M, Wang B, Zhang M, Rantalainen M, Wang S, Zhou H, et al. Symbiotic gut microbes modulate human metabolic phenotypes. Proc Natl Acad Sci U S A. 2008;105(6):2117–2122. doi: 10.1073/pnas.0712038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mueller S, Saunier K, Hanisch C, Norin E, Alm L, Midtvedt T, et al. Differences in fecal microbiota in different European study populations in relation to age, gender, and country: a cross-sectional study. Appl Environ Microbiol. 2006;72(2):1027–1033. doi: 10.1128/AEM.72.2.1027-1033.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Haro C, Rangel-Zuniga OA, Alcala-Diaz JF, Gomez-Delgado F, Perez-Martinez P, Delgado-Lista J, et al. Intestinal microbiota is influenced by gender and body mass index. PLoS One. 2016;11(5):e0154090. doi: 10.1371/journal.pone.0154090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dicksved J, Floistrup H, Bergstrom A, Rosenquist M, Pershagen G, Scheynius A, et al. Molecular fingerprinting of the fecal microbiota of children raised according to different lifestyles. Appl Environ Microbiol. 2007;73(7):2284–2289. doi: 10.1128/AEM.02223-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Human Microbiome Project Consortium Structure, function and diversity of the healthy human microbiome. Nature. 2012;486(7402):207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lay C, Rigottier-Gois L, Holmstrom K, Rajilic M, Vaughan EE, de Vos WM, et al. Colonic microbiota signatures across five northern European countries. Appl Environ Microbiol. 2005;71(7):4153–4155. doi: 10.1128/AEM.71.7.4153-4155.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Oertelt-Prigione S. The influence of sex and gender on the immune response. Autoimmun Rev. 2012;11(6–7):A479–A485. doi: 10.1016/j.autrev.2011.11.022. [DOI] [PubMed] [Google Scholar]
- 69.Auerbach L, Hafner T, Huber JC, Panzer S. Influence of low-dose oral contraception on peripheral blood lymphocyte subsets at particular phases of the hormonal cycle. Fertil Steril. 2002;78(1):83–89. doi: 10.1016/S0015-0282(02)03173-4. [DOI] [PubMed] [Google Scholar]
- 70.Fransen F, van Beek AA, Borghuis T, Meijer B, Hugenholtz F, van der Gaast-de Jongh C, et al. The impact of gut microbiota on gender-specific differences in immunity. Front Immunol. 2017;8:754. doi: 10.3389/fimmu.2017.00754. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Method and results RT qPCR for validation microarray. (DOCX 27 kb)
Data Availability Statement
The datasets (microarray) supporting the conclusions of this article are available in the gene expression omnibus from NCBI under ID number GSE85913.