Abstract
Cytochromes P450 play key roles in drug metabolism and disease by oxidizing a wide variety of natural and xenobiotic compounds. High-resolution crystal structures of P450cam bound to ruthenium sensitizer-linked substrates reveal an open conformation of the enzyme that allows substrates to access the active center via a 22-Å deep channel. Interactions of alkyl and fluorinated biphenyl linkers with the channel demonstrate the importance of exploiting protein dynamics for specific inhibitor design. Large changes in peripheral enzyme structure (F and G helices) couple to conformational changes in active center residues (I helix) implicated in proton pumping and dioxygen activation. Common conformational states among P450cam and homologous enzymes indicate that static and dynamic variability in the F/G helix region allows the 54 human P450s to oxidize thousands of substrates.
Cytochromes P450 comprise a family of monooxygenases that catalyze transformations of many diverse substrates (1). In particular, the ability of P450s to hydroxylate aliphatic carbon by generating a reactive heme–oxygen species has stimulated much research (R-H + O2 + 2H+ + 2e− → R-OH + H2O). Found in all phyla, P450s have the same protein fold and cysteine-ligated heme, despite low sequence similarity between some members (structurally similar P450cam and P450BM-3 have only 17% sequence identity) (2). Humans have at least 54 different P450 isozymes (http://drnelson.utmem.edu/CytochromeP450.html). The human P450 isozymes play key roles in steroid biosynthesis and arachidonic acid metabolism, as well as in the transformations of xenobiotics in detoxification and carcinogenesis (3). Particularly striking is the finding that P450 3A4 metabolizes up to half of all drugs in use (4). Despite broad substrate diversity, all P450s have significant structural constraints on their activity: P450s must control water access to the active center to avoid the conversion of activated dioxygen to superoxide or peroxide. Thus, the binding sites of P450 isozymes must be structurally diverse, yet conserve a mechanism of catalysis and solvent exclusion. An unanswered question is how thousands of substrates are metabolized by one enzyme family whose chemistry requires significant structural constraint.
We are investigating the archetypal P450, Pseudomonas putida P450cam, by using sensitizer-linked substrates that were designed to bind the enzyme selectively and communicate electronically with the heme center by way of tethered ruthenium tris(bipyridyl) [RuII(bpy)3]2+ complexes (5, 6). Herein, we describe the structures of two such Ru-substrate⋅P450cam conjugates. As predicted, the substrate moieties bind at the active center, and the Ru-sensitizers bind near the protein surface. Importantly, the enzyme changes conformation to accommodate the linkers. The open conformation we observe mimics structures of other P450 enzymes and reveals a likely path for substrates to access the active center. Notably, this rearrangement is coupled to conformational changes of catalytically important residues.
Materials and Methods
Crystallization and Data Collection.
Purification, crystallization and structure determination of P450cam⋅Ru-C9-Ad [C9 = (−CH2−)9, Ad = adamantane] have been described (5). P450cam⋅Ru-F8bp-Ad (F8bp = 4,4′-octafluorobiphenyl) seed crystals in the space group P1 [cell dimensions, 63.8 × 67.1 × 72.5 Å3; two molecules per asymmetric unit; Matthews coefficient (VM) = 2.56; solvent content = 51.9%] nucleated from C334A P450cam separated from camphor and complexed with stoichiometric Ru-F8bp-Ad. Hanging drops contained an equal volume mixture of reservoir and 396 μM P450⋅Ru-F8bp-Ad in 20 mM Hepes, 100 mM KCl, and 1 mM DTT, pH 7.5. The reservoir (pH 6.5) contained 0.1 M sodium cacodylate, 200 mM KCl, and 8–15% (wt/vol) polyethylene glycol (PEG; molecular weight = 8,000). Crystal nucleation was induced by setting the crystallization trays on ice for 30 min. The resulting temperature gradient causes partial dehydration of the hanging drops. The trays were then removed from the ice and stored at 4°C; seed crystal growth occurred overnight. Diffraction-quality crystals were grown over 24 h by moving seed crystals into sitting drops with reservoir PEG concentrations of 8–11%.
Two data sets were collected at the Stanford Synchrotron Research Laboratory (SSRL). Data set 1 (1.80-Å resolution) was collected at 100 K on beamline 9-2 (λ = 1.03 Å) at SSRL and processed with DENZO and SCALEPACK (7). Data set 2 (1.65-Å resolution) was collected at 100 K on beamline 9-1 (λ = 0.72 Å) and similarly processed (Table 1).
Table 1.
X-ray data collection and refinement
| Data set 1 | Data set 2 | |
|---|---|---|
| Unit cell | 64.0 67.3 72.5 Å | 63.8 67.1 72.5 Å |
| 71.3° 65.8° 62.4° | 71.2° 65.2° 62.3° | |
| Space group | P1 | P1 |
| Resolution, Å | 1.80 (1.86–1.80)* | 1.65 (1.71–1.65)* |
| Rsym† | 3.7 (25.6)* | 3.8 (29.2)* |
| Completeness, % | 96.8 (95.4)* | 97.8 (97.0)* |
| Wilson B, Å2 | 19.0 | |
| I/σ(I)‡ | 21.9 (3.80)* | 16.43 (1.99)* |
| No. molecules per unit cell | 2 | |
| Rfac§ | 21.0 (29.2)* | |
| Rfree¶ | 22.6 (28.7)* | |
| rms deviation bonds, angles‖ | 0.007 Å, 1.2° | |
| No. protein atoms, 〈B〉 | 6,569, 23.2 Å2 | |
| No. waters, 〈B〉 | 693, 34.3 Å2 | |
| No. Ru-F8bp-Ad atoms, 〈B〉 | 280, 25.7 Å2 | |
| Residues not modeled | A1–A9, B1–B9 | |
| Additional ligands | 5 cacodylate |
Highest resolution range for compiling statistics.
Rsym = ∑∑j |Ij − 〈I〉|/∑∑j|Ij|, Ij = intensity of observation j.
Intensity signal to noise.
R = ∑ ∥Fobs| − |Fcalc∥/∑ |Fobs| for all reflections (no σ cutoff).
Free R calculated against 7.4% of reflections removed at random.
rms deviations from ideal bond and angle restraints.
Structure Determination of P450⋅Ru-F8bp-Ad.
An initial molecular replacement solution (correlation coefficient = 46.1, and Rcryst = Σ ∥Fobs| - |Fcalc∥/Σ |Fobs| = 44.7%) for diffraction data set 1 (20.0- to 3.5-Å resolution) was found with AMORE (8) by using two probe molecules, each derived from the structure of camphor-bound P450cam (PDB code 2cpp). The initial model derived from molecular replacement on data set 1 was replaced with the protein coordinates from Ru-C9-Ad-bound P450cam (PDB code: 1qmq) by least-squares fitting and was further improved by simulated annealing. Ru-F8bp-Ad was positioned into the remaining difference density. Refinement was completed with data set 2 by iterative rounds of torsion-angle molecular dynamics and positional refinement with Crystallography & NMR System (9) and XFIT (10) amidst model rebuilding, water molecule placement, and resolution extension to 1.65 Å. Overall anisotropic thermal factor correction, bulk solvent correction, individual thermal factor refinement, and grouped occupancy refinement of Ru-F8bp-Ad produced the final model (7688 scatterers in the asymmetric unit, 2 P450⋅Ru-F8bp-Ad molecules, each containing a superposition of Ru-F8bp-Ad Δ and Λ stereoisomers; 18 residues in multiple conformations; 5 cacodylate molecules bound to cysteines 58A, 85A, 58B, 85B, and 136B; and 693 water molecules). Noncrystallographic symmetry restraints were not applied between the two molecules per asymmetric unit. The final model has excellent stereochemistry (Table 1) with 90.5% of all residues in the most favored regions of ϕ/ψ space as defined by PROCHECK (11). The residue Glu-94 falls outside the accepted regions of ϕ/ψ space because of steric interactions with the cacodylate bound to Cys-85. Figures were generated with BOBSCRIPT (12), MOLSCRIPT (13), RASTER3D (14), and INSIGHT II. Molecular surfaces were calculated with MSMS (15) and rendered with AVS (Advanced Visual Systems, Waltham, MA).
Results and Discussion
Ru-Substrate Binding Reveals a Substrate Access Channel in P450cam.
The P450cam complexes with Ru-C9-Ad and Ru-F8bp-Ad have strikingly similar protein conformations (Cα rms deviation = 0.7 Å) and sensitizer-linked substrate-binding modes, despite having crystallized in different space groups. Ru-C9-Ad and Ru-F8bp-Ad share the same [RuII(bpy)3]2+ and adamantyl functionalities, but are linked with a nine-carbon alkyl chain in Ru-C9-Ad and a 4,4′-substituted octafluorobiphenyl in Ru-F8bp-Ad.
The ruthenium complexes bind P450cam in a channel that likely gives natural substrates access to the buried active center (Fig. 1). Movement of the F (residues 173–185) and G (192) helices against the perpendicular I helix (234) retracts the F/G loop (185) from the enzyme's β-sheet domain and thereby opens an access channel to the heme that is 22 Å deep and 11 Å across (Fig. 2). In effect, the F and G helices translate relative to the protein core in a “shear” mechanism (17), whereas the core itself undergoes smaller motions to maintain hydrophobic interactions.
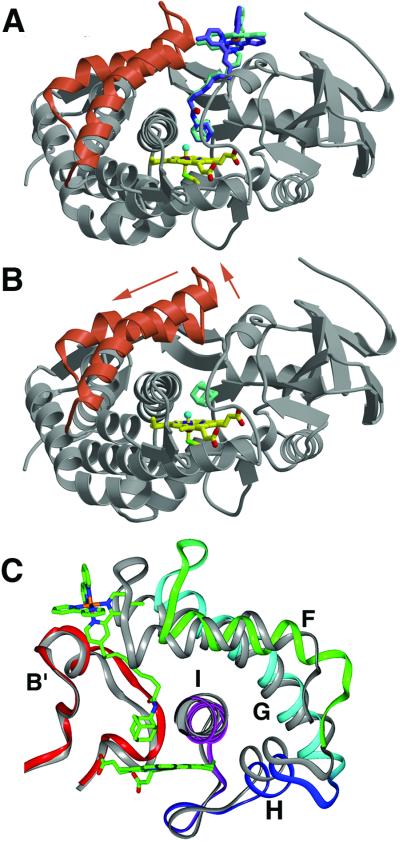
Figure 1.
Comparison of P450cam bound to Ru-C9-Ad (A) and adamantane (B) (16). On binding the Ru-substrate (Λ stereoisomer in blue, Δ stereoisomer in green) the F and G helices (red ribbons) retract from the P450cam β-sheet domain (gray ribbons). The adamantyl moiety binds in the same position above the heme (yellow) as free adamantane. (C) Movement of the F, G, H, and I helices (rotated ≈180° from A and B). For comparison, P450cam bound to camphor is shown in gray. Residues on the F/G loop move as much as 7.5 Å as the F and G helices slide approximately one helical turn (4.5 Å) across the I helix. The H helix (218) and the N terminus of the I helix (234) shift with the G helix to conserve interhelical contacts.
Figure 2.
Shape complementarity and hydrophobic interactions between Ru-F8bp-Ad and P450cam. The water molecules (red) hydrate newly exposed surface area in the P450cam⋅Ru-F8bp-Ad structure.
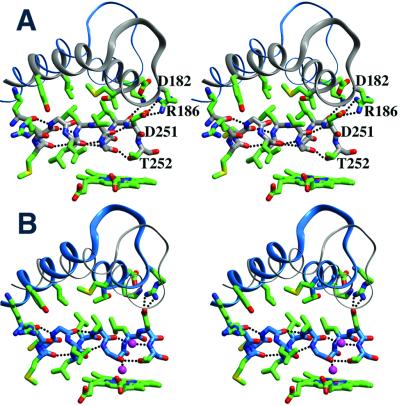
On opening of the access channel, the interactions of the F and G helices with the protein core manifest in two ways: (i) the making and breaking of salt bridges to stabilize helix juxtaposition; and (ii) slight distortion of the core backbone to conserve hydrophobic packing. Rearrangements of inter-residue salt bridges and hydrogen bonding interactions among the F helix, F/G loop, and the I helix facilitate sliding of the F helix relative to the I helix. These rearrangements (Fig. 3) either exploit the conformational flexibility of long side chains to maintain hydrogen bonding interactions (e.g., Glu-171 to Arg-161, Arg-186 to Asp-251) or involve the breaking and making of hydrogen bonds (e.g., Lys-178 to Asp-251 and Leu-250 exchanged for Lys-178 to Glu-156).
Figure 3.
Side-chain interactions in closed (A) and open (B) P450cam. The charged residues Lys-178 (F helix), Asp-182 (F helix), Thr-185 (F/G loop), and Arg-186 (F/G loop) alter their interactions with Asp-251, a key residue on the I helix implicated in delivering protons to activate heme-bound dioxygen. Alternate conformations of Arg-186 and Asp-251 are present in the Ru-C9-Ad complex, indicating conformational mobility. The N-terminal I helix segment translates and rotates to maintain a hydrophobic core of interdigitated branched hydrophobic residues (Leu-246, Leu-250, and Val-247) with the F (Leu-177, Thr-181, and Met-184) and G (Leu-200, Tyr-201, Leu-204, and Ile-208) helices.
The tendency to maintain hydrophobic packing interactions between the G helix and both the I helix and the B′ loop (residues 89–101) causes main-chain conformational distortions within the protein core in response to the new F/G helix positions. For example, the B′ loop moves to maintain packing of F87, Y96, and F98 with F193 and Y201 on the G helix. Similarly, the numerous contacts among the hydrophobic side chains of the F, G, and I helices cause the N-terminal half of the I helix to rotate in response to the translation of the F and G helices in the open structure. As discussed below, this change in I-helix main-chain conformation and hydrogen bonding in turn affects the conformation of the active site.
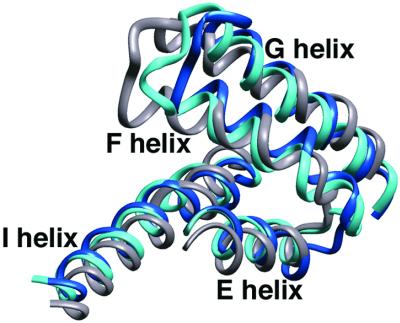
The position of the F and G helices in other P450s closely matches the conformations found in our open structures of P450cam. Substrate-free P450BM-3 crystallizes in an open form and P450NOR has a large, permanent access channel analogous to that observed in the Ru-substrate⋅P450cam structures (Fig. 4) (18–20). The structural similarity of the open P450cam structure with P450BM-3 and P450NOR suggests that the open conformation is important for substrate binding. The Ru-substrates stabilize a conformation that may exist only transiently for P450cam, but is clearly stable for other cytochromes P450. Thus, the P450 fold apparently allows an opening motion of the F and G helices with the relative stability of open and closed forms weighted differently among P450s.
Figure 4.
The F, G, and I helices of P450cam in its closed (gray) and open (blue) states compared with those of P450NOR (light blue).
The conformation of the F/G loop is similar in the open and closed structures of P450cam. However, mobility of the F/G loop is suggested by disorder in the crystal structures of P450terp and P450 2C5 (21, 22). Furthermore, the F/G loop of CYP 119 undergoes rearrangement on binding bulky substrates (23). Thus, F/G loop flexibility may also play an important role in P450 substrate binding.
Solution studies support a transient open state of P450cam. Photoacoustic calorimetry indicates that a short-lived (≈130 ns) intermediate of larger volume forms during the photolysis of heme-bound carbon monoxide and expulsion of camphor (24). Our structures confirm an earlier prediction based on photoacoustic spectroscopy that the residues Arg-186, Asp-251, Lys-178, and Asp-182 undergo rearrangement during substrate binding (25). Furthermore, tryptophan fluorescence quenching measurements show that substrate-free P450cam is conformationally more labile than the camphor-bound enzyme (26).
Indirect evidence also suggests an open/closed equilibrium in other P450s. Cooperative substrate hydroxylation, consistent with a flexible binding site, has been observed in P450 3A4, the most abundant hepatic P450 (27). Eukaryotic P450s, for instance P450scc, are known to exist in multiple conformational states (28). Drug resistance mutations in the fungal P450 CYP51 occur in the G and H helices, far from the active site (29). Finally, computer simulations support F/G helix fluctuations in both P450cam and P450BM-3 (30–32).
Interactions of Ru-Substrates with P450cam.
Two current problems in drug design are (i) how to avoid the deactivation of drugs by hepatic P450s and (ii) how to selectively inhibit a specific pathogenic P450 (33). Our Ru-substrate complexes bind with submicromolar dissociation constants, but are structurally very different from camphor. Thus, the interactions of the Ru-substrates with P450cam provide insight into why some P450s are promiscuous binders and suggest how to design specific P450 isozyme inhibitors.
Our structures provide examples of rarely characterized interactions among proteins, metal complexes, and fluorinated aromatics. Both Ru-substrates bind P450cam in a similar fashion. Notably, the ruthenium atom and adamantyl centroids are only 1.64 and 1.07 Å apart in the superimposed structures. In part, this is because of design: Ru-F8bp-Ad was synthesized after the crystal structure of Ru-C9-Ad was known. However, preferred interactions between the protein and Ru-substrates lead to similar structures.
Ru-F8bp–Ad Interactions.
Although direct contacts between [RuII(bpy)3]2+ and the protein are limited, both the Δ and Λ isomers of the complex could be discerned because of the rigidity of Ru-F8bp-Ad (Fig. 5). There are very few crystal structures of fluorinated aromatics bound to proteins. Phenyl and perfluorophenyl functionalized molecules are known to stack in the solid state because of favorable π–π interactions between the electron-rich phenyl groups and electron-poor perfluorophenyl groups (35). These attractive interactions, which have been estimated to be worth about 15 kJ/mol in vacuum, make the hydrophobic perfluorophenyl group a potentially useful functionality for drug design (36–40).
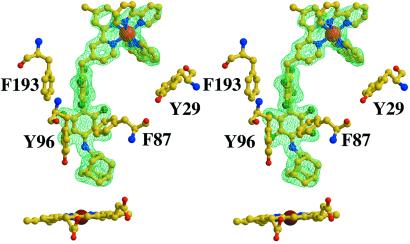
Figure 5.
Simulated-annealing omit map (Fobs − Fcalc) calculated with Ru-F8bp-Ad removed from Fcalc. Electron density is shown at 1.65-Å resolution and contoured at 2.5 σ. For clarity only one isomer is shown. The bipyridyl ring contacts Tyr-29. Phe-193 contacts one fluorinated ring with 3.4 Å between rings, consistent with the 3.4-Å face-to-face distance observed in the benzene–hexafluorobenzene crystal structure (34). Phe-87 contacts the perfluorobiphenyl unit in an edge-on fashion, with the ɛ carbon 3.5 Å from the face of the biphenyl unit. Tyr-96 packs against the biphenyl unit in an edge-on fashion, with fluorine–carbon contacts ranging from 3.2 to 3.9 Å.
The P450cam⋅Ru-F8bp-Ad complex shows both parallel and perpendicular stacking between the octafluorobiphenyl unit and aromatic residues (Figs. 2 and 5). The crystal structure of a matrix metalloproteinase inhibitor also shows a parallel stacking interaction (3.7 Å-separation) between a pentafluorophenyl group and a tyrosine, which contributes to the binding affinity of the inhibitor relative to the phenyl analog (39). In contrast, the crystal structure of a carbonic anhydrase inhibitor shows perpendicular stacking between a phenylalanine and a pentafluorophenyl group (41). Our results further demonstrate that the interaction between an aromatic electron donor and a fluorinated ring can be parallel or perpendicular and is influenced by both the intrinsic attraction and the structural constraints imposed by the tertiary structure.
Ru-C9-Ad Interactions.
Because of the flexibility of the alkyl chain, the bipyridyl ligands of Ru-C9-Ad were difficult to discern in the electron density. Anomalous scattering measurements revealed two distinct positions for the ruthenium atom separated by ≈1 Å in the access channel. The best fit to the electron density included both Δ and Λ isomers and interactions with Tyr-29 and Pro-187, as in the Ru-F8bp-Ad structure (Figs. 1 and 2). In addition, a bipyridine contacts Ala-92, and an acetate molecule (present in the crystallization solution) sandwiches between the [RuII(bpy)3]2+ unit and Phe-193. The hydrocarbon tether linking the ruthenium complex to the adamantyl unit winds across the side chains of Ile-395, Phe-193, Phe-87, and Tyr-96—the same residues that contact the fluorinated biphenyl unit in Ru-F8bp-Ad.
Tyr-96 is hydrogen bonded to the carbonyl of the Ru-C9-Ad amide bond as it is to the camphor ketone group in the substrate complex (42). The adamantyl unit binds in the same pocket as in the Ru-F8bp-Ad structure but enjoys more extensive hydrophobic interactions with Leu-244, Thr-101, Ile-395, Val-295, Thr-252, and the Gly-248 Cα. The strain induced by the short separation (3.00 Å) of the adamantyl unit and heme-bound water perhaps explains the partial low- to high-spin heme shift that occurs on binding (data not shown).
The [RuII(bpy)3]2+ moiety does not force the substrate access channel open as it is pulled in by the adamantyl group. If the interaction with the ruthenium complex was unfavorable the enzyme could push the complex into solution and close around the alkyl chain. Instead, Förster energy transfer experiments indicate that the ruthenium resides the same distance from the heme even when the linker is much longer than the access channel (5). Even in the Ru-C9-Ad structure the alkyl chain is not fully extended. Thus, favorable binding interactions between Ru-substrate and the enzyme likely stabilize an open conformation that already exists transiently under normal conditions. Our structures suggest that improved P450 inhibitors might be produced by taking advantage of the enzyme's intrinsic flexibility.
F/G Loop Movement Affects the P450cam Active Site.
The F/G loop movement in P450cam is coupled to changes in functionally important residues in the active center. I-helix residues 248–252 participate in dioxygen activation (1). In particular, Thr-252, Asp-251, and the Gly-248 peptide carbonyl play crucial roles in the conversion of heme-bound dioxygen to high-valent iron-oxo or peroxo species. The open structure reveals that I-helix residues also couple the coordination environment of the heme iron to enzyme tertiary structure peripheral to the active center.
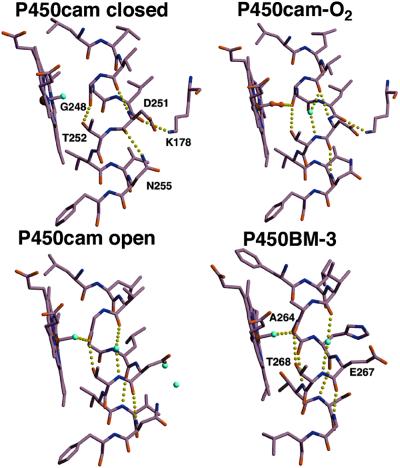
In closed P450cam, the I-helix segment adjacent to the heme iron bulges so that the peptide carbonyl groups of residues 248–251 do not form hydrogen bonds to C-terminal peptide nitrogens within the helix. A hydrogen bond between the Thr-252 hydroxyl and Gly-248 carbonyl stabilizes this bulge. In open P450cam, the bulge shifts toward the N terminus of the I helix. To effect this change, the peptide bonds between residues 250–251 and 251–252 rotate 90° relative to the closed structure and anneal back into the helix (Fig. 6), whereas the carbonyls of Leu-245 and Leu-246 are no longer hydrogen bonded within the helix but instead are bonded to a buried water molecule (Figs. 6 and 7). This shift in the I-helix bulge arises from a 1.5-Å translation of the N-terminal half of this helix that preserves hydrophobic contacts with the retracted conformations of the F and G helices (Figs. 3 and 7).
Figure 6.
The active sites of P450BM-3 and P450cam (closed, dioxygen-bound, and open conformations). Regularization of the I helix between Leu-250 and Asn-255 compensates for the loss of main-chain hydrogen bonds between Leu-245 and Leu-250 in the open P450cam structure. Interactions with the F and G helices break the hydrogen bond between the Asn-255 side-chain amide and the Asp-251 carbonyl, allowing the 251–252 peptide to flip down and hydrogen bond to the Asn-255 peptide amide. As in dioxygen-bound ferrous P450cam, this peptide flip is accompanied by the introduction of a helix-bridging water molecule (43). Movement of the F and G helices also breaks the hydrogen bond between Lys-178 and the peptide carbonyl of Leu-250, allowing the 250–251 peptide bond to flip 90° and anneal into the helix.
Figure 7.
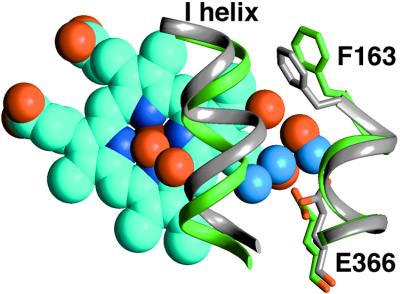
Buried water molecules facilitate the I helix rearrangement between open (green ribbon and red waters) and closed (gray ribbon and blue waters) states. Note the shift of the I helix bulge and concurrent rearrangement of the buried waters. Glu-366, a highly conserved residue among P450s, anchors the water molecules.
The altered interactions of the F and G helices with the I helix in open P450cam regularize the I helix to conformations similar to those found in other P450 structures. In P450BM-3 and P450NOR, the I-helix residues equivalent to P450cam 249–251 all have standard helical conformations. This is one more helical residue (249) than open P450cam, two more residues than O2–bound ferric P450cam (249 and 250), and three more residues than closed P450cam, where residues 249, 250, and 251 all form hydrogen bonds outside the I helix. Taken together, these structures show that the I-helix backbone adopts different conformations depending on the ligand bound. Importantly, the I-helix backbone conformation controls the water structure surrounding the heme iron (Fig. 6).
The I helix communicates changes in the F and G helices to the coordination environment above the heme. As a result of the I-helix conformational changes in the open structure, the Gly-248 carbonyl is even closer to the heme iron (4.8 Å) than in either the O2 complex (5.5 Å) or the low-spin closed conformation (6.4 Å). The resulting short hydrogen bond (2.6 Å) from the Gly-248 carbonyl to the iron-ligating water molecule stabilizes water-bound, low-spin, low-potential heme in the open form of the enzyme. Tilting the equilibrium toward water-ligated, ferric heme may help prevent heme reduction and the subsequent production of superoxide, peroxide, and other toxic forms of reduced dioxygen.
Solvation Changes Important for Substrate Binding and Catalysis.
Twenty-four additional ordered water molecules hydrate newly exposed surfaces in the Ru-bound structures of P450cam (Fig. 2). This number agrees well with earlier results that suggested the involvement of 28 water molecules in the catalytic cycle of the enzyme (44). Because of the motion of the F and G helices, 9 new ordered water molecules form hydrogen bonds to Asp-251, Arg-186, Asp-182, and Lys-178 between the F and I helices. In addition, the F helix residues Thr-185 and Thr-181 rotate in the open structure so that their hydroxyls can form hydrogen bonds to water. Although Asp-251 has been implicated in proton delivery to the active center, this residue is sequestered in the closed structure. Hydration of Asp-251 in the open structure suggests that the altered hydrogen bond patterns of this conformation are not only important for substrate binding but also in facilitating proton and/or water molecule exchange during catalysis.
Buried water molecules mediate conformational flexibility in proteins through their mobility and ability to switch hydrogen-bonding partners (45). Three conserved water molecules that have analogs in P450terp, P450eryF, and P450NOR stabilize the disrupted I helix i to i + 4 hydrogen bonds in both the open and closed conformations (Fig. 7) (21, 46, 47). The role of water molecules in facilitating the open/closed transition of P450cam is similar to that found in the facilitation of large-scale conformational fluctuations of acetylcholinesterase (48).
Structural Flexibility Makes Cytochrome P450 a Versatile Catalyst.
The motions of the F and G helices that we observed in the comparison of the open and closed P450cam structures, along with similar differences in structure between substrate-bound and free P450BM-3 (49), suggest an explanation for the extraordinary substrate diversity associated with human P450s. If P450cam, an enzyme specialized for a single small substrate, undergoes such large motions on substrate binding, many of the human liver isozymes may as well. In effect, the F and G helices act as a clamp, both to fix the substrate over the heme and to exclude excess water from the active site. Remarkably, P450cam hydroxylates Ru-substrates when suitable electron donors are provided (50). This observation further underlines the extraordinary ability of P450s to handle widely varying substrates.
The P450 cytochromes provide yet another demonstration of the importance of energetically low-lying conformational states in protein function. As in P450cam, these alternative conformations may be difficult to detect if they form and decay on a submicrosecond time scale. Our structures show that regions distant from the active center are critical for substrate binding and catalysis in cytochromes P450. Thus, although local structure tunes the reactivity of a metallo-cofactor, the entire polypeptide generates the dynamic properties necessary for enzymatic activity.
Acknowledgments
We thank Stanford Synchrotron Research Laboratory for access to data collection facilities and the staff of beamlines 9-1 and 9-2 for their assistance with data collection. This work was supported by the Fannie and John Hertz Foundation (to A.R.D.), the Helen Hay Whitney Foundation (to B.R.C.), the National Institutes of Health predoctoral program (to I.J.D.), the National Science Foundation, and the National Institutes of Health.
Abbreviations
- bpy
4,4′-bipyridine
- [RuII(bpy)3]2+
ruthenium tris(bipyridine)
- Ad
adamantane
- F8bp
4,4′-octafluorobiphenyl
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1K2O).
References
- 1.Ortiz de Montellano P R, editor. Cytochrome P450: Structure, Mechanism and Biochemistry. New York: Plenum; 1995. [Google Scholar]
- 2.Nelson D R. Arch Biochem Biophys. 1999;369:1–10. doi: 10.1006/abbi.1999.1352. [DOI] [PubMed] [Google Scholar]
- 3.Guengerich F P. In: Cytochrome P450: Structure, Mechanism and Biochemistry. Ortiz de Montellano P R, editor. New York: Plenum; 1995. pp. 473–536. [Google Scholar]
- 4.Guengerich F P. Annu Rev Pharmacol Toxicol. 1999;39:1–17. doi: 10.1146/annurev.pharmtox.39.1.1. [DOI] [PubMed] [Google Scholar]
- 5.Dmochowski I J, Crane B R, Wilker J J, Winkler J R, Gray H B. Proc Natl Acad Sci USA. 1999;96:12987–12990. doi: 10.1073/pnas.96.23.12987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilker J J, Dmochowski I J, Dawson J H, Winkler J R, Gray H B. Angew Chem Int Ed Engl. 1999;38:90–92. [Google Scholar]
- 7.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 8.Navaza J. Acta Crystallogr A. 1994;50:157–163. [Google Scholar]
- 9.Brunger A T, Adams P D, Clore G M, DeLano W L, Gros P, Grosse-Kunstleve R W, Jiang J S, Kuszewski J, Nilges M, Pannu N S, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 10.McRee D E. J Mol Graphics. 1992;10:44–46. [Google Scholar]
- 11.Laskowski R A, MacArthur M W, Moss D S, Thornton J M. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 12.Esnouf R M. J Mol Graphics. 1997;15:133–138. doi: 10.1016/S1093-3263(97)00021-1. [DOI] [PubMed] [Google Scholar]
- 13.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 14.Merritt E A, Bacon D J. Methods Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- 15.Sonner M F, Olson A J, Sphener J C. Biopolymers. 1996;38:305–320. doi: 10.1002/(SICI)1097-0282(199603)38:3%3C305::AID-BIP4%3E3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 16.Raag R, Poulos T L. Biochemistry. 1991;30:2674–2684. doi: 10.1021/bi00224a016. [DOI] [PubMed] [Google Scholar]
- 17.Gerstein M, Lesk A M, Chothia C. Biochemistry. 1994;33:6740–6749. doi: 10.1021/bi00188a001. [DOI] [PubMed] [Google Scholar]
- 18.Ravichandran K G, Boddupalli S S, Hasemann C A, Peterson J P, Deisenhofer J. Science. 1993;261:731–736. doi: 10.1126/science.8342039. [DOI] [PubMed] [Google Scholar]
- 19.Li H, Poulos T L. Nat Struct Biol. 1997;4:140–146. doi: 10.1038/nsb0297-140. [DOI] [PubMed] [Google Scholar]
- 20.Park S Y, Shimizu H, Adachi S, Nakagawa A, Tanaka I, Nakahara K, Shoun H, Obayashi E, Nakamura H, Iizuka T, et al. Nat Struct Biol. 1997;4:827–832. doi: 10.1038/nsb1097-827. [DOI] [PubMed] [Google Scholar]
- 21.Hasemann C A, Ravichandran K G, Peterson J A, Deisenhofer J. J Mol Biol. 1994;236:1169–1185. doi: 10.1016/0022-2836(94)90019-1. [DOI] [PubMed] [Google Scholar]
- 22.Williams P A, Cosme J, Sridhar V, Johnson E F, McRee D E. Mol Cell. 2000;5:121–131. doi: 10.1016/s1097-2765(00)80408-6. [DOI] [PubMed] [Google Scholar]
- 23.Yano J K, Koo L S, Schuller D J, Li H, Ortiz de Montellano P R, Poulos T L. J Biol Chem. 2000;275:31086–31092. doi: 10.1074/jbc.M004281200. [DOI] [PubMed] [Google Scholar]
- 24.DiPrimo C, Hoa G H B, Deprez E, Douzou P, Sligar S G. Biochemistry. 1993;32:3671–3676. doi: 10.1021/bi00065a021. [DOI] [PubMed] [Google Scholar]
- 25.Diprimo C, Deprez E, Sligar S G, Hoa G H B. Biochemistry. 1997;36:112–118. doi: 10.1021/bi961508a. [DOI] [PubMed] [Google Scholar]
- 26.Prasad S, Mazumdar S, Mitra S. FEBS Lett. 2000;477:157–160. doi: 10.1016/s0014-5793(00)01745-2. [DOI] [PubMed] [Google Scholar]
- 27.Ueng Y-F, Kuwabara T, Chun Y-J, Guengerich F P. Biochemistry. 1997;36:370–381. doi: 10.1021/bi962359z. [DOI] [PubMed] [Google Scholar]
- 28.Lepesheva G I, Strushkevich N V, Usanov S A. Biochim Biophys Acta. 1999;1434:31–43. doi: 10.1016/s0167-4838(99)00156-9. [DOI] [PubMed] [Google Scholar]
- 29.Podust L M, Poulos T L, Waterman M R. Proc Natl Acad Sci USA. 2001;98:3068–3073. doi: 10.1073/pnas.061562898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paulsen M D, Ornstein R L. Proteins. 1995;21:237–243. doi: 10.1002/prot.340210306. [DOI] [PubMed] [Google Scholar]
- 31.Li H, Poulos T L. Acta Crystallogr D. 1995;51:21–32. doi: 10.1107/S0907444994009194. [DOI] [PubMed] [Google Scholar]
- 32.Ludemann S K, Lounnas V, Wade R C. J Mol Biol. 2000;303:797–811. doi: 10.1006/jmbi.2000.4154. [DOI] [PubMed] [Google Scholar]
- 33.Rendic S, Dicarlo F J. Drug Metab Rev. 1997;29:413–580. doi: 10.3109/03602539709037591. [DOI] [PubMed] [Google Scholar]
- 34.Williams J H, Cockcroft J K, Fitch A N. Angew Chem Int Ed Eng. 1992;31:1655–1657. [Google Scholar]
- 35.Patrick C R, Prosser G S. Nature (London) 1960;187:1021. [Google Scholar]
- 36.Sakai T, Miki Y, Tsuboi M, Takeuchi H, Ema T, Uneyama K, Utaka M. J Org Chem. 2000;65:2740–2747. doi: 10.1021/jo9918551. [DOI] [PubMed] [Google Scholar]
- 37.Luhmer M, Bartik K, Dejaegere A, Bovy P, Reisse J. Bull Soc Chim Fr. 1994;131:603–606. [Google Scholar]
- 38.Bovy P R, Getman D P, Matsoukas J M, Moore G J. Biochim Biophys Acta. 1991;1079:23–28. doi: 10.1016/0167-4838(91)90019-v. [DOI] [PubMed] [Google Scholar]
- 39.Jacobsen E J, Mitchell M A, Hemdges S K, Belonga K L, Skaletzky L L, Stelzer L S, Lindberg T J, Fritzen E L, Schoztarez H J, O'Sullivan T J, et al. J Med Chem. 1999;42:1525–1536. doi: 10.1021/jm9803222. [DOI] [PubMed] [Google Scholar]
- 40.West A P, Mecozzi S, Dougherty D A. J Phys Org Chem. 1997;10:347–350. [Google Scholar]
- 41.Kim C-Y, Chang J S, Doyon J B, Baird T T, Jr, Fierke C A, Jain A, Christianson D W. J Am Chem Soc. 2000;122:12125–12134. [Google Scholar]
- 42.Poulos T L, Finzel B C, Gunsalus I C, Wagner G C, Kraut J. J Biol Chem. 1985;260:16122–16130. [PubMed] [Google Scholar]
- 43.Schlichting I, Berendzen J, Chu K, Stock A M, Maves S A, Benson D E, Sweet R M, Ringe D, Petsko G A, Sligar S G. Science. 2000;287:1615–1622. doi: 10.1126/science.287.5458.1615. [DOI] [PubMed] [Google Scholar]
- 44.Diprimo C, Sligar S G, Hoa G H B, Douzou P. FEBS Lett. 1992;312:252–254. doi: 10.1016/0014-5793(92)80946-e. [DOI] [PubMed] [Google Scholar]
- 45.Fischer S, Verma C S. Proc Natl Acad Sci USA. 1999;96:9613–9615. doi: 10.1073/pnas.96.17.9613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cupp-Vickery J R, Poulos T L. Nat Struct Biol. 1995;2:144–153. doi: 10.1038/nsb0295-144. [DOI] [PubMed] [Google Scholar]
- 47.Shimizu H, Obayashi E, Gomi Y, Arakawa H, Park S-Y, Nakamura H, Adachi S-i, Shoun H, Shiro Y. J Biol Chem. 2000;275:4816–4826. doi: 10.1074/jbc.275.7.4816. [DOI] [PubMed] [Google Scholar]
- 48.Koellner G, Kryger G, Millard C B, Siman I, Sussman J L, Steiner T. J Mol Biol. 2000;296:713–726. doi: 10.1006/jmbi.1999.3468. [DOI] [PubMed] [Google Scholar]
- 49.Li H, Poulos T L. Biochim Biophys Acta. 1999;1441:141–149. doi: 10.1016/s1388-1981(99)00161-4. [DOI] [PubMed] [Google Scholar]
- 50.Dmochowski I J. Ph.D. dissertation. Pasadena: California Institute of Technology; 2000. [Google Scholar]