Abstract
The therapeutic benefits offered by 5-fluorouracil (5-FU) are limited because of the acquisition of drug resistance, the main cause of treatment failure and metastasis. The ability of the cancer cells to undergo epithelial-mesenchymal transition (EMT) contributes significantly to cancer metastatic potential and chemo-resistance. However, the underlying molecular mechanisms of 5-FU-resistance have remained elusive. Here, we show that reactive oxygen species (ROS), produced by dual oxidase 2 (DUOX2), promote 5-FU-induced EMT. First, we showed that 5-FU–resistant SNUC5 colon cancer cells (SNUC5/FUR cells) undergo EMT by analyzing the expression of EMT markers such as N-cadherin, vimentin and E-cadherin. In addition, we found that the resistant cells expressed higher levels of Snail, Slug, Twist and Zeb1, which are all critical EMT regulators and had enhanced migratory and invasive capabilities. Furthermore, SNUC5/FUR cells had increased level of DUOX2, resulting in increased ROS level. This effect was due to the enhanced binding of the ten eleven translocation 1 (TET1) demethylase to the DUOX2 promoter in the SNUC5/FUR cells. Importantly, silencing of TET1 reversed the effects of 5-FU on the cells. Finally, the antioxidant N-acetylcysteine attenuated the effects of 5-FU on EMT and metastasis. Our study demonstrates the existence of a TET1/DUOX2/ROS/EMT axis that could play a role in colon cancer chemo-resistance and the aggressiveness of this cancer.
Abbreviations: [Ca2+]I, intracellular calcium concentration; 5-FU, 5-fluorouracil; BAPTA-AM, 1,2-bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid; ChIP, chromatin immune-precipitation; Ct, cycle threshold; DAPI, 4,6-diamidino-2-phenylindole; DCF-DA, dichlorodihydrofluorescein diacetate; DUOX, dual oxidase; EDTA, ethylenediaminetetraacetic acid; EGTA, ethylene-bis(oxyethylenenitrilo)tetraacetic acid; EMT, epithelial-mesenchymal transition; FITC, fluorescein isothiocyanate; GAPDH, glyceraldeyde 3-phosphate dehydrogenase; IC50, half maximal inhibitory concentration; Ig, immunoglobulin; MMP, matrix metalloproteinase; MTT, 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide; NAC, N-acetylcysteine; NOX, NADPH oxidase; OD, optical density; PBS, phosphate-buffered saline; PCR, polymerase chain reaction; qMSP, quantitative methylation specific PCR; qPCR, quantitative PCR; ROS, reactive oxygen species; SDS-PAGE, sodium dodecyl sulfate polyacrylamide gel electrophoresis; shRNA, short hairpin RNA; siRNA, small interfering RNA; TET, ten eleven translocation; TIMP, tissue inhibitor of metalloproteinase; Zeb1, zinc finger E-box-binding homeobox 1
Keywords: 5-FU resistance, DNA demethylase, DUOX2, Epithelial-mesenchymal transition, Metastasis
Highlights
-
•
5-Fluorouracil (FU)-resistance is associated with epithelial-mesenchymal transition.
-
•
ROS play a key role in EMT induction in 5-FU resistant colon cancer cells.
-
•
5-FU-resistant cells overexpress the DNA demethylase TET1, which binds to the promoter of the NADPH oxidase DUOX2 and induces its expression.
1. Introduction
Resistance to anticancer drugs, such as 5-fluorouracil (5-FU), is an important cause of treatment failure in colon cancer [1]. The mechanisms of drug resistance are complex and have yet to be fully elucidated. The epithelial to mesenchymal transition (EMT) is the change from an epithelial to a mesenchymal cell phenotype; notably, this process has been implicated in various biological processes, including gastrulation and early steps of cancer metastasis [2]. Cancer metastasis is a complex process involving infiltration of primary cancer cells into surrounding tissues, followed by extravasation into distant organs, and metastatic colonization [3], [4]. The loss of E-cadherin expression is the most predominant characteristic of EMT and many transcription factors, such as Twist, Snail, and Zinc finger E-box-binding homeobox (Zeb), have been shown to directly or indirectly repress the activity of E-cadherin promoter and subsequently induce EMT [5].
Many sources of reactive oxygen species (ROS) exist in the cell. This includes the NADPH oxidase (NOXs) family (NOX1–5), dual oxidase (DUOX) 1 and 2, and the mitochondria [6], [7]. The cells have evolved a homeostatic system to eliminate ROS and thus maintain the cellular redox balance. This equilibrium can be disrupted by chemotherapeutic treatments [8].
Many studies have shown a strong relationship between oxidative stress and increased cancer cell growth [9], [10], or chemo-resistance [11]. Additional studies have indicated that the altered production of ROS is associated with EMT and metastasis [12], [13].
The purpose of this study was to determine whether 5-FU-resistant colon cancer cells undergo EMT and induce metastasis and decipher the underlying mechanisms. Additionally, we investigated whether ROS are involved in the regulation of EMT induced in 5-FU-resistant cells.
2. Materials and methods
2.1. Cell culture
SNUC5 colon cancer cells were from the Korean Cell Line Bank (Seoul, Republic of Korea). The SNUC5 were obtained from colon of a female patient (77 years old) with colorectal cancer. The histopathology of this cell line is adenocarcinoma. They grow in a monolayer and are poorly differentiated. They were cultured in RPMI-1640 medium (Invitrogen, Grand Island, NY, USA) containing 10% heat-inactivated fetal bovine serum (Sigma-Aldrich Co., St. Louis, MO, USA) at 37 °C in a 5% CO2 atmosphere. SNUC5 resistant to 5-FU (SNUC5/FUR cells) were obtained from the Research Center for Resistant Cells of Chosun University (Gwangju, Republic of Korea). They were obtained by culturing SNU5 cells in 140 μM 5-FU for more than six months.
2.2. Cell viability assay
Cells were seeded at a density of 1.2 × 105 cells/mL in 96-well plates and treated with 5-FU. After 48 h, 3-(4,5-dimethyl-2-thiazolyl)− 2,5-diphenyl-2H-tetrazolium bromide (MTT) was added and after 4 h, the absorbance was measured at 540 nm.
2.3. Western blot analysis
Cell lysates were separated by 8–12% sodium dodecyl sulfate polyacrylamide gel electrophoresis, transferred to a nitrocellulose membrane, immunoblotted with the indicated antibodies and detected with enhanced chemiluminescent detection. The antibodies used in this study were against Snail, Slug, Twist, Zeb1, E-cadherin, N-cadherin and β-actin (Santa Cruz Biotechnology, Santa Cruz, CA, USA), and against matrix metalloproteinase (MMP)-2, MMP-9, tissue inhibitor of metalloproteinase (TIMP)-1 and TIMP-2 (Cell Signaling Technology Inc., Danvers, MA, USA).
2.4. Immunocytochemistry
Cells, seeded on chamber slides, were fixed, incubated for 2 h with the indicated primary antibody and the primary antibody was detected using a fluorescein isothiocyanate (FITC)- or Alexa 594-conjugated secondary antibody (Santa Cruz Biotechnology). Stained cells were mounted on microscope slides in 4′,6-diamidino-2-phenylindole (DAPI) containing mounting medium and imaged on a Zeiss confocal microscope using the LSM 510 software (Zeiss, Oberkochen, Germany).
2.5. In vitro wound healing assays
Cell migration was analyzed using wound-healing assays. The cells (1 × 105 cells/well) were placed for 24 h in 6-well plates and, at confluence, a wound was made using a sterile plastic pipette tip. Free-floating cells and debris were removed by washing the cells with culture medium. Representative scratch zones were photographed 48 h after the scratch wounding using a microscope (Olympus, Japan). For both cell lines (drug-sensitive and -resistant), the distance between the wound edges was measured. Cell lines to be compared were processed in parallel to prevent differences to environmental cues in wound healing responses.
2.6. Cell invasion assay
Experiments were carried out using the Cytoselect 24-well cell invasion assay (Cell Biolabs, Inc., San Diego, CA, USA). Cells (1 × 106 cells/mL) were cultured in the upper chamber of the inserts overnight. After removal of all non-invasive cells, the ones that had passed through the membrane were quantified using OD560 nm in a microplate reader (Thermo Scientific, MA, USA).
2.7. SDS-PAGE zymography
SDS-PAGE Gelatin zymography was used to measure MMP-2 and MMP-9 enzymatic activity in cell lysates. To this aim, we used the Novex™ In-gel zymography system (Invitrogen, Carlsbad, CA, USA), according to the manufacturer's instructions. The equal amounts of protein (30 μg/lane) were combined with sample buffer and loaded on a Novex™ 10% zymogram (Gelatin) gel (Invitrogen) for electrophoresis. Gels were then incubated with Novex™ zymogram renaturing and developing buffer (overnight incubation). The gels were stained with SimplyBlue™ Safestain (Invitrogen) and destained appropriately. The gelatinolytic activity was visualized as a transparent band against a blue background. Gelatinolytic bands were measured densitometrically with an image analyzer.
2.8. RNA interference
For short hairpin RNA (shRNA) transfection, cells were seeded at a density of 1.5 × 105 cells/well in 24-well plates. The cells were transfected when they reached ~50% confluency. shControl RNA and shTET1 RNA (shRNA directed against TET1) were purchased from Santa Cruz Biotechnology. Lentiviruses expressing control or TET1 shRNA were diluted in OptiMEM containing 6 μg/mL polybrene and added to the SNUC5/FUR cells. After 72 h, transduced cells were selected using puromycin (5 μg/mL). Clones were cultured up to three weeks, and silencing of TET1 was verified by measuring the level of TET1. The human shTET1 RNA sequence was 5′-UUGUGUCACGGCCAUCUGCdTdT-3′ and the shControl RNA sequence was 5′-TTCTCCGAACGTGTCACGT-3′. In addition, small interfering RNAs (siRNAs) specific for DUOX2 (siRNA Nos. 1044506 and 1044512, Bioneer, Daejeon, Republic of Korea) were used according to the manufacturer's protocol. For transfection, SNUC5/FUR cells were transfected using two different specific siRNAs or one non-targeting control siRNA using Lipofectamine® RNAiMAX (Invitrogen), according to the manufacturer's protocol.
2.9. Detection of ROS
ROS detection in the cells was performed by confocal microscopy or flow cytometry after staining with dichlorodihydrofluorescein diacetate (DCF-DA, Sigma-Aldrich, MO, USA). Cells were seeded in 6-well plates at a density of 3 × 105 cells/well. After 24 h at 37 °C, the cells were treated with 5-FU for various amounts of time. Cells were treated with 25 µM DCF-DA, trypsinized, and analyzed using a flow cytometer (Becton Dickinson, CA, USA) and the CellQuest™ software (Becton Dickinson) or confocal microscopy.
H2O2 detection in the cells was performed by confocal microscopy after staining with Amplex® red reagent (Invitrogen). Cells were seeded and, after 16 h, the dye (50 μM of Amplex® red reagent and 0.1 U/mL of horseradish peroxidase in phosphate buffer) was added to each well to a final volume of 100 μl and samples were incubated for 30 min in the dark. Fluorescence was monitored at excitation/emission values of 485 nm/580 nm in a microplate reader (Thermo Scientific).
2.10. Chromatin immune-precipitation (ChIP) sequencing
ChIP sequencing was conducted by Genomictree Inc. (Daejeon, Republic of Korea). For the ChIP-sequencing analysis, reads were mapped to the UCSC hg19 human referenced genome. Cells were cross-linked with 1% formaldehyde for 10 min at room temperature, and neutralized with 0.125 M glycine. DNA was sonicated to 300–500 bp fragments in SDS-lysis buffer (50 mM Tris-HCl, 1% SDS, 10 mM ethylenediaminetetraacetic acid (EDTA), pH 8.1), using 15 cycles (burst 30 s, with a repetition 30 s), at 320 W of power. Chromatin was then immune-precipitated using Dynabeads Protein G (Invitrogen) pre-treated with a ChIP grade TET1 antibody (Abcam, Cambridge, MA, USA). To construct the sequencing library, the enriched DNA fragments were blunted using the NEXTflex Chip-Seq library prep kit (BIOO Scientific, Texas, USA), ligated to the sequencing adapter and subjected to polymerase chain reaction (PCR) amplification and purified. Pair-end ChIP and input DNA libraries were sequenced using the Illumina HiSeq. 2500 system (Illumina, San Diego, CA, USA) according to the manufacturer's instructions. Initial quality-control analysis and adapter of raw data was performed using the Cutadapt v1.15 [14], Trim Galore v0.4.5 (http://bioinformatics.babraham.ac.uk) and FastQC v0.11.7 (http://bioinformatics.babraham.ac.uk). Finally, about 28 million mapped reads by BWA aligner v0.7.12 [15] for each ChIP group were analyzed using the HOMER v4.7 software [16] (http://homer.ucsd.edu/homer/) for peak calling, annotation, Gene Ontology, and signal pathway analyses, including 30 million mapped input reads as control. Both IPA (http://www.ingenuity.com) and DAVID softwares were used for the ontology analysis of the TET1 occupied targets.
2.11. ChIP-quantitative PCR (qPCR)
Cells were first cross-linked with 1% formaldehyde. Chromatin was prepared and digested with nuclease (12 min at 37 °C). Immunoprecipitation was performed with an antibody against TET1 and mouse immunoglobulin G (IgG) with constant rotation overnight at 4 °C. Immune complexes were then captured using ChIP-grade protein G magnetic beads. The beads were washed and eluted with ChIP elution buffer. The DNA/protein complexes were reversed by incubation at 65 °C for 30 min followed by 2 h incubation with Proteinase K at 65 °C. Spin columns were used to purify the the immune-precipitated DNA fragments. DNA was then subjected to 35 cycles of PCR, to amplify the DUOX2 promoter region across the TET1 binding sites, using the following primers: (forward, [F]) 5′-GAAGGGCGCCATCTGT-3′ and (reverse, [R]) 5′-GGCTGAGCTTCCGAAAA-3′. The PCR products were separated on 2% agarose gels, and DNA bands were visualized using the ImageJ (National Institute of Health, Bethesda, MD, USA) software. In addition, the ChIPed DNA was sequenced by Genomictree.
2.12. Analysis of gene expression
For detection of gene expression, we performed reverse transcription (RT)-PCR and quantitative real-time (qRT)-PCR. SNUC5 and SNUC5/FUR cells were seeded at a density of 500 cells per 60 mm dish and cultured for 2 days with either PBS as a control or with 10 μM of 5-FU in SNUC5 cell. For RT-PCR, total RNA was isolated from cells using easy-BLUE (Intron, Daegeon, Republic of Korea). cDNA was prepared from 0.3 to 4 μg RNA using MMLV reverse transcriptase enzyme and oligo dT as primers (Invitrogen, Carlsbad, CA, USA). Amplification reactions were performed in a total volume of 25 μl, which contained 200 ng of cDNA samples, primers, dNTPs, and 0.5U of Taq DNA polymerase. The PCR conditions for DUOX2 and the housekeeping gene glyceraldeyde 3-phosphate dehydrogenase (GAPDH) were as follows: 35 cycles of 94 °C for 45 s, 52 °C for 1 min, and 72 °C for 1 min. Primer pairs (Bionics, Seoul, Republic of Korea) were as follows: DUOX2, (F) 5′- CCGGCAATCATCTATGGAGGT-3′ and (R) 5′-TTGGATGATGTCAGCCAGCC-3′; (F) GAPDH, 5′-AAGGTCGGAGTCAACGGATTT-3′ and (R) 5′-GCAGTGAGGGTCTCTCTCCT-3′. The amplified products were resolved by 1% agarose gel electrophoresis, stained with ethidium bromide, and photographed under UV illumination.
For qRT-PCR, cDNA was mixed with 1 × of Power SYBR green PCR master mix (Applied Biosystems, Foster City, CA, USA) and a couple of forward and reverse primers (300 nM). The primers used to amplify the NOX family cDNA were: NOX1 (F) 5′- CAATCTCTCTCCTGGAATGGCATCCT-3′, (R) 5′-CCTGCTGCTCGGATATGAATGGAGAA-3′; NOX2 (F) 5′- AAGGCTTCAGGTCCACAGAGGAAA-3′, (R) 5′-AGACTTTGTATGGACGGCCCAACT- 3′; NOX3 (F) 5′- ACCGTGGAGGAGGCAATTAGACAA-3′, (R) 5′- CAGGTTGAAGAAATGCGCCACGAT-3′; NOX4 (F) 5′- AGCAGAGCCTCAGCATCTGTTCTT-3′, (R) 5′-TGGTTCTCCTGCTTGGAACCTTCT-3′; NOX5 (F) 5′-CCTCCTCATGTTCATCTGCTCCAGTT-3′, (R) 5′-AGGAGGTAGGACAGGTGAGTCCAATA-3′; DUOX1 (F) 5′-GCAGGACATCAACCCTGCACTCTC-3′, (R) 5′-CTGCCATCTACCACACGGATCTGC-3′; DUOX2 (F) 5′-CCGGCAATCATCTATGGAGGT-3′, (R) 5′-TTGGATGATGTCAGCCAGCC-3′; DUOXA2 (F) 5′-GTCTTGGGGACTCTGGTTT-3′, (R) 5′-ACCCCAGTTCCCTATTGTCC-3′; Actin (F) 5′-CACCAACTGGGACGACAT-3′, (R) 5′-ACAGCCTGGATAGCAACG-3′. PCR conditions were as follows: 95 °C for 10 min followed by 40 cycles of 95 °C for 15 s, and 59–61 °C for 1 min. PCR reactions were performed in 96-well plates in Amplification was performed with a Bio-Rad iQ5 real-time PCR detector system (Bio-Rad, Richmond, CA, USA). Data analysis was performed using Bio-Rad iQ5 Optical System Software V1.0.
2.13. Quantitative methylation specific PCR (MSP) using qPCR and bisulfite sequencing analysis
The MethPrimer software (http://www.urogene.org/methprimer/) [17] was employed to design primers for the human DUOX2 promoter, which contains abundant CpG sites (potential DNA methylation targets) around the transcription start site [18].
For methylation analysis, a standard phenol-chloroform method was used to extract DNA. The EZ DNA methylation kit (Zymo Research, Irvine, CA, USA) was used for Bisulfite modification of genomic DNA. The Maxima SYBR Green qPCR kit (Fermentas, Carlsbad, CA, USA) and the CFX96 qPCR detection system (Bio-Rad) were used for quantitative analyses. The amplification consisted of an initial 10 min denaturation (95 °C), followed by 40 cycles of denaturation at 95 °C (15 s) and annealing and extension for 30 s and 60 s, respectively. For quantification, the comparative cycle threshold (Ct) method was used. The Ct values for the methylated DUOX2 were normalized to the Ct values of unmethylated DUOX2. For MSP analysis, the primer set for unmethylated DUOX2 were: (F) 5′-AGTAGTGGAATGTTGAAGTTTGTG-3′ and (R) 5′-ACTAACTTACCTACCCACCTACATA-3′; for methylated DUOX2: (F) 5′-AGTAGTGGAACGTTGAAGTTTGC-3′ and (R) 5′-CTAACTTACCTACCCGCCTACGTA-3′ [19].
For bisulfite sequence analysis, PCR amplicons were separated by electrophoresis on 2% agarose gel followed by purification with a gel extraction kit (Qiagen GmbH, Hilden, Germany). The amplicons were then and cloned in the TOPO TA vector system (Invitrogen). NucleoSpin plasmid isolation kit (Macherey-Nagel, Düren, Germany) was used to isolate and purify DNA from individual clones. M13F primer was used to sequence randomly selected positive clones (10–15 from each sample) and the methylation status of each CpG dinucleotide was analyzed. For quantification of DUOX2 methylation level, quantitative MSP amplification was performed on bisulfite-treated samples and normalized based on the amplification of the Alu element. A CFX96TM system (Bio-Rad) was used to perform qPCR using the primer set (F) 5′-TTTGTTTTGGGTTTTTTAGGAGATA-3′ and (R) 5′-CCCCAACTTACTAACTTACCTACCC-3′ [20].
2.14. Cytosolic Ca2+ detection
Cells were seeded on a culture plate at a concentration of 2 × 105 cells/mL. After 24 h, cells were treated with 10 µM of fluo-4 A.M. (Thermo Scientific) for 30 min at 37 °C, and fluorescence of the cells was measured using the flow cytometer (Becton Dickinson) and analyzed using the CellQuest™ (Becton Dickinson) software or confocal microscopy.
2.15. Statistical analysis
All experiments were made at least in triplicate. Results are presented as the mean ± the standard error of the mean (SEM). The SigmaStat software v12 (SPSS, Chicago, IL, USA) was used for statistical analysis. Data were analyzed using the analysis of variance as one-way ANOVA by Tukey's post-hoc test. Differences were considered statistically significant at p < 0.05.
3. Results
3.1. 5-FU-resistant SNUC5 colon cancer cells undergo EMT
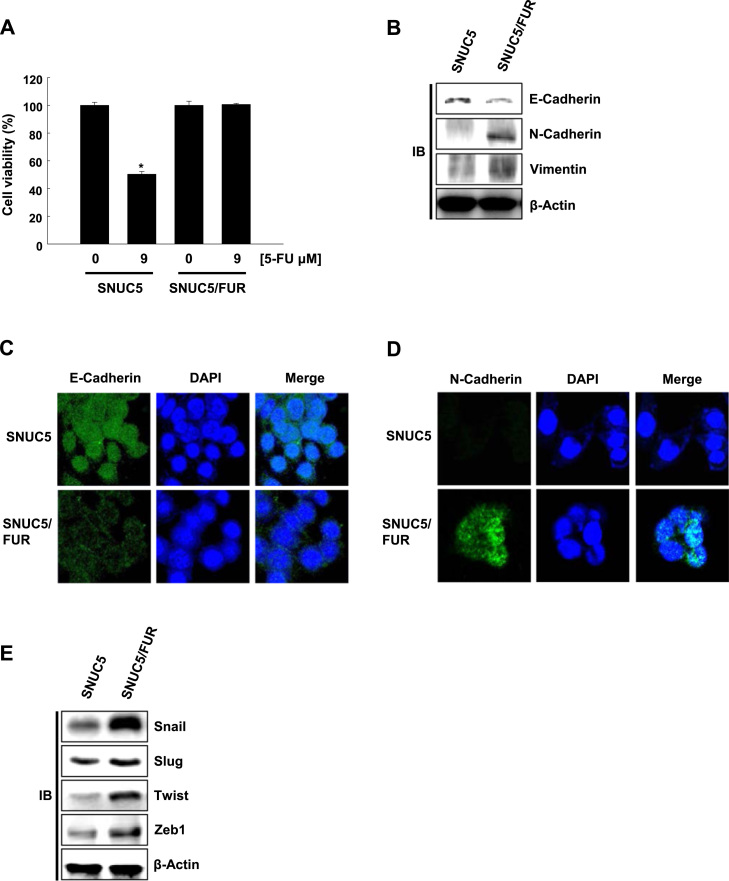
The 5-FU-resistant cell line SNUC5/FUR has previously been generated by culturing SNUC5 cells in the continuous presence of 5-FU [21], [22]. As expected, the resistant cells viability was not affected by 9 μM 5-FU, which caused 50% growth inhibition (IC50) in SNUC5 parental cells (Fig. 1A). Next, we compared the expression level of EMT markers in the two cell lines. SNUC5/FUR cells showed the large reduction of E-cadherin expression (epithelial marker) and the overexpression of N-cadherin and vimentin (mesenchymal markers, Fig. 1B). These changes were confirmed by immunofluorescence: as shown in Fig. 1C and D, E-cadherin staining showed weaker expression in SNUC5/FUR than in SNUC5 cells, whereas N-cadherin exhibited much higher expression in SNUC5/FUR than in SNUC5 cells. Finally, we analyzed the expression of critical transcription factors involved in EMT namely Snail, Slug, Twist, and Zeb1 in both cell lines. These factors have been shown to bind to E-box elements located in the promoter region of E-cadherin. This leads to the transcriptional repression of E-cadherin and the induction of N-cadherin [23], [24]. The expression of Snail, Slug, Twist, and Zeb1 was significantly higher in SNUC5/FUR than in SNUC5 (Fig. 1E). Taken together, our results suggested that resistance to 5-FU is associated with EMT in the colon cancer cell line SNUC5.
Fig. 1.
5-Fluorouracil (FU)-resistant SNUC5 colon cancer cells undergo epithelial-mesenchymal transition. (A) The viability of SNUC5 and 5-fluorouracil (FU)-resistant (SNUC5/FUR) cells was assessed after two days of 5-FU treatment. *p<0.05 vs. control cells. (B) Cell lysates were analyzed for the expression of E-cadherin, N-cadherin, and vimentin. (C, D) Immuno-fluorescence of cells stained with (C) anti-E-cadherin and (D) anti-N-cadherin antibodies. Nuclei were stained with DAPI. (E) Cell lysates were analyzed for the expression of Snail, Slug, Twist, and Zeb1. β-Actin was used as loading control.
3.2. 5-FU-resistance is associated with enhanced migratory and invasive ability
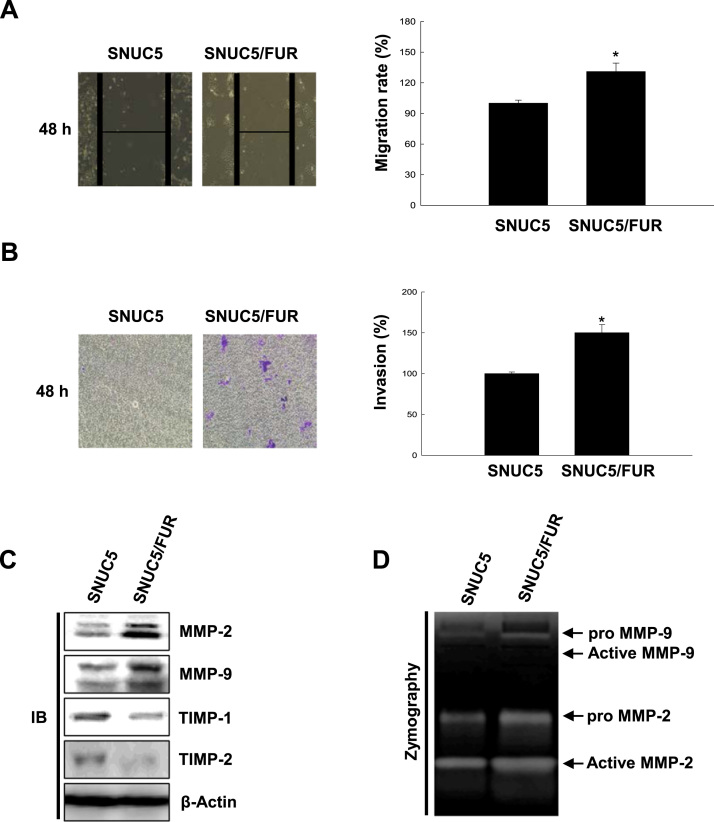
Epithelial cells that have undergone EMT have augmented migratory and invasive ability. This change is associated with increased expression and activity of metalloproteases, such as MMP-2 and MMP-9, which degrade the extracellular matrix and thus promote tumor invasion [25]. In wound healing (Fig. 2A) and trans-well migration assays (Fig. 2B), we found that SNUC5/FUR cells have enhanced migratory and invasive properties compared to the parental cell line. This correlated with increased expression of MMP-2 and MMP-9 in SNUC5/FUR compared to SNUC5 cells, as well as decreased expression of the metalloprotease inhibitors TIMP-1 and TIMP-2 (Fig. 2C). MMP-2 and MMP-9 are synthesized as proenzymes in cells, and are then cleaved to generate the secreted catalytically active enzymes. Using gelatin zymography, we showed that resistance to 5-FU is associated with increased MMP-2 and MMP-9 activity (Fig. 2D). Altogether, our results suggest that chemo-resistant colon cancer cells have enhanced invasive abilities.
Fig. 2.
5-FU–resistant SNUC5 colon cancer cells have enhanced migratory and invasive properties. (A) Wound healing assays were performed in SNUC5 and SNUC5/FUR cells. Images were taken 48 h after the scratch and quantified. Representative images are shown. *p < 0.05 vs. control cells. (B) Cell invasion assays were performed with the Cytoselect 24-well cell invasion assay kit. Cells were cultured in the upper chamber overnight and the ones that passed through the membrane were quantified at an optical density of 560 nm. *p < 0.05 vs. control cells. (C) Cell lysates were analyzed for the expression of MMP-2, MMP-9, TIMP-1, and TIMP-2. (D) MMP-2 and MMP-9 enzymatic activity in cell lysates was determined by gelatin zymography.
3.3. TET1 overexpression is involved in EMT and in 5-FU-resistant SNUC5 cells
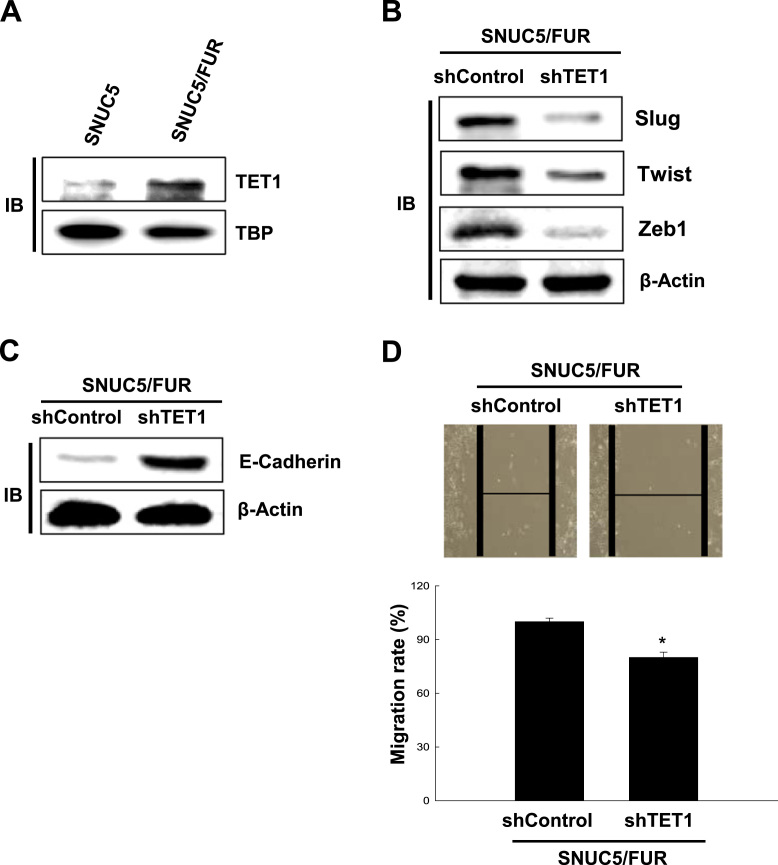
In line with our previous report [21], SNUC5/FUR cells highly express the DNA demethylase TET compared to SNUC5 cells (Fig. 3A). We investigated whether TET1 overexpression in SNU5/FUR cells plays a role in their EMT. We found that interference with TET1 expression was associated with lower expression levels of Slug, Twist, and Zeb1 and higher expression of E-cadherin (Fig. 3B and C). These changes in EMT-related protein expression were associated with reduced migration capacities upon TET1 knockdown (Fig. 3D).
Fig. 3.
The TET1 DNA demethylase is involved in the epithelial-mesenchymal transition of 5-FU-resistant SNUC5 colon cancer cells. (A) Cell lysates from SNUC5 and SNUC5/FUR cells were analyzed for the expression of TET1. (B, C) SNUC5/FUR cells overexpressing short hairpin RNA targeting TET1 (shTET1) or control shRNA (shControl) were analyzed for the expression of (B) Slug, Twist, and Zeb1 or (C) E-cadherin and β-actin was used as loading control. (D) SNUC5/FUR cells overexpressing shTET1 or shControl were subjected to wound healing assays. Images were taken 48 h after the scratch and quantified. Representative images are shown. *p < 0.05 vs. control cells.
3.4. Overproduction of ROS induces EMT in 5-FU-resistant SNUC5 colon cancer cells and is dependent on TET1 expression
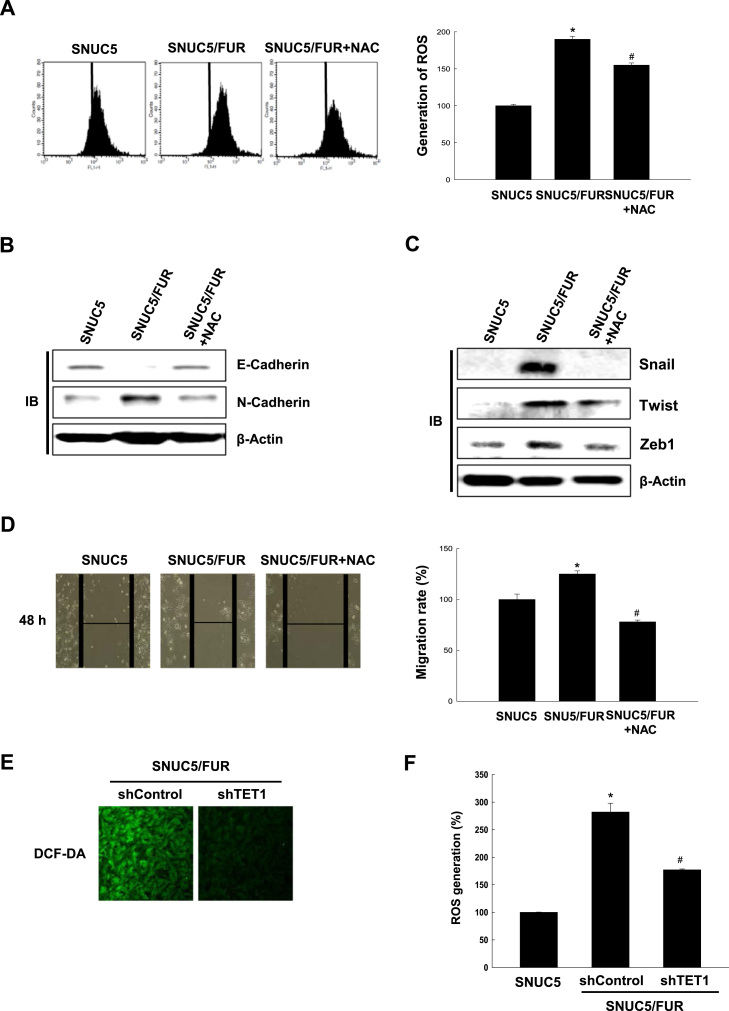
We have previously reported that SNUC5/FUR cells produce higher levels of ROS than their parental cells [21], [22]. Accordingly, SNUC5/FUR cells showed enhanced DCF-DA staining (specific for intracellular ROS detection) than SNUC5 cells, and the intensity of the staining decreased upon treatment with the antioxidant N-acetylcysteine (NAC, Fig. 4A). Interestingly, in SNUC5/FUR cells, NAC treatment restored the expression of EMT markers and EMT-associated transcription factors to the levels found in parental cells (Fig. 4B and C) and negatively affected cell migration (Fig. 4D). In addition, TET1 knockdown in SNUC5/FUR cells was associated with lower ROS generation compared with control cells (Fig. 4E and F). These results suggested that increased ROS production participates in EMT and enhanced migration of SNUC5/FUR cells and that TET1 is involved in ROS over-production.
Fig. 4.
Reactive oxygen species (ROS) are required for cell migration during 5-FU-induced epithelial-mesenchymal transition. (A) SNUC or SNUC5/FUR cells were cultured in the absence or presence of N-acetylcysteine (NAC) for 48 h. The ROS levels were measured by flow cytometry after staining with DCF-DA. *p < 0.05 vs. SNUC5 cells. #p < 0.05 vs. SNUC5/FUR cells. (B, C) Lysates from cells as in (A) were analyzed for the expression of (B) E-cadherin and N-cadherin or (C) Snail, Twist, Zeb1. (D) The cell migration of cells as in (A) was measured in a wound-healing assay. Images were taken 48 h after the scratch and quantified. Representative images are shown. *p < 0.05 vs. SNUC5 cells. #p < 0.05 vs. SNUC5/FUR cells. (E, F) Total ROS levels was measured by (E) confocal microscopy and (F) flow cytometry after staining the SNUC5/FUR cells overexpressing short interfering RNA targeting TET1 (shTET1) or control shRNA (shControl) with DCF-DA. *p < 0.05 vs. SNUC5 cells.#p < 0.05 vs. SNUC5/FUR shControl cells.
3.5. Overexpression of TET1 leads to the induction of the NADPH-oxidase DUOX-2 in 5-FU-resistant SNUC5 cells
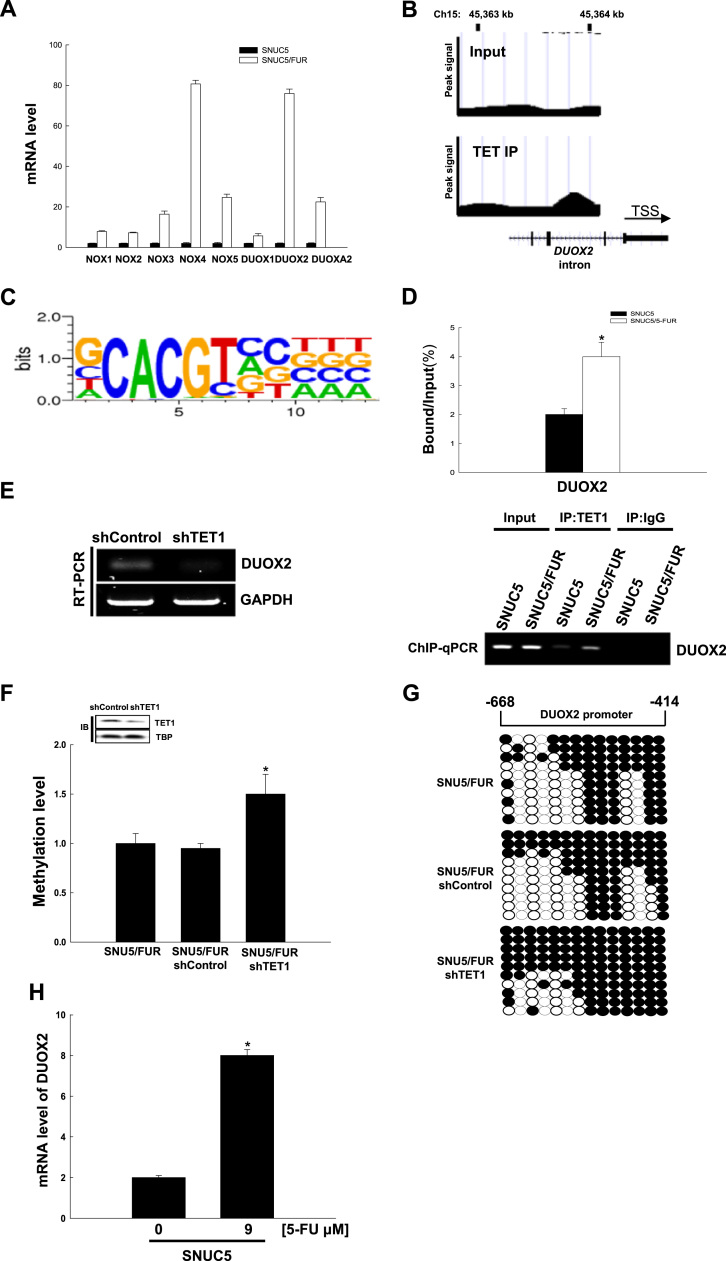
To identify the source of ROS in SNUC5/FUR cells, we first compared the expression of all NADPH oxidases, a major source of non-mitochondrial ROS. We found that, of all NOX isoforms, NOX4 and DUOX2 were the more significantly over-expressed in resistant compared to sensitive cell lines (Fig. 5A). Interestingly, we found that DUOXA2, a protein required for DUOX2 activation was also increased. Using ChIP-sequencing, we found that TET1 binds to the proximal promoter of the NADPH-oxidase DUOX2 (Fig. 5B) but not on the NOX4 gene, suggesting that DUOX2 might be responsible for TET1-induced ROS production. In addition, the binding sequence of TET1 on the DUOX2 promoter belongs to the TET1 binding consensus motif (CxxCxxC, Fig. 5C). Furthermore, using ChIP-qPCR assays, we confirmed that the binding of TET1 to the DUOX2 locus is higher in SNUC5/FUR cells than in parental cells (Fig. 5D). Importantly, TET1-silenced cells showed lower expression levels of DUOX2 (Fig. 5E).
Fig. 5.
The TET1 DNA demethylase activates the transcription of dual oxidase 2 (DUOX2) in 5-FU-resistant SNUC5 colon cancer cells. (A) qRT-PCR analysis was assessed for the expression of all NOX isoforms in SNUC5 and SNUC5/FUR. (B) ChIP-sequencing analysis was performed to assess TET1 distribution on the chromatin: the presence of TET1 on DUOX2 was confirmed. (C) The TET1-binding sequence on DUOX2 corresponds to the most enriched sequence found in the TET1 ChIP-sequencing assays. (D) ChIP-qPCR assay in SNUC5 or SNU5/FUR cells using the TET1 antibody. The enrichment of DUOX2 promoter sequences in the immune-precipitated DNA was assessed by qPCR and normalized to the input DNA. *p < 0.05 vs. SNUC5 cells. (E-G) SNUC5/FUR cells were transduced with lentiviruses overexpressing short hairpin RNA targeting TET1 (shTET1) or control shRNA (shControl). (E) The mRNA expression of DUOX2 was assessed by RT-PCR. (F) Quantitative methylation levels were assessed and normalized to the Alu element. *p < 0.05 vs. SNUC5/FUR shControl cells.(G) Bisulfite sequencing analysis of the DUOX2 promoter. Black circles represent methylated cytosine residues; white circles represent unmethylated cytosine residues. (H) The mRNA expression of DUOX2 was assessed by qRT-PCR after treatment of SNUC5 cells with 9 µM for 48 h. *p < 0.05 vs. control cells.
We then addressed whether TET1 might regulate DNA methylation of the DUOX2 promoter. For this purpose, we performed qMSP analysis and bisulfite-sequencing to measure DUOX2 promoter methylation status in SNUC5/FUR cells in which TET1 had been silenced or in control cells (transfected with control shRNA). As shown in Fig. 5F, the DNA methylation levels of the DUOX2 promoter region were significantly higher in TET1-silenced SNUC5/FUR cells compared with the control cells. This was confirmed by bisulfite sequencing analysis of the DUOX2 promoter region, which showed a higher methylation status (83% vs. 55%) in TET1-silenced cells compared with control cells (Fig. 5G). Finally, since we have previously shown that 5-FU induces the expression of TET1 in SNUC5 cells [21], we investigated the effect of 5-FU on DUOX2 mRNA expression in these cells. It showed that 9 μM 5-FU, the 5-FU IC50 in SNUC5, induced the expression of DUOX2 in SNUC5 cells (Fig. 5H). Therefore, our data suggested that TET1 overexpression, upon 5-FU treatment or in 5-FU resistant cells, leads to the transcriptional induction of the NADPH oxidase DUOX2 by regulating the methylation of its promoter.
3.6. Cytosolic [Ca2+]i accumulation, induced by 5-FU-resistance, activates DUOX2, which is responsible for ROS production
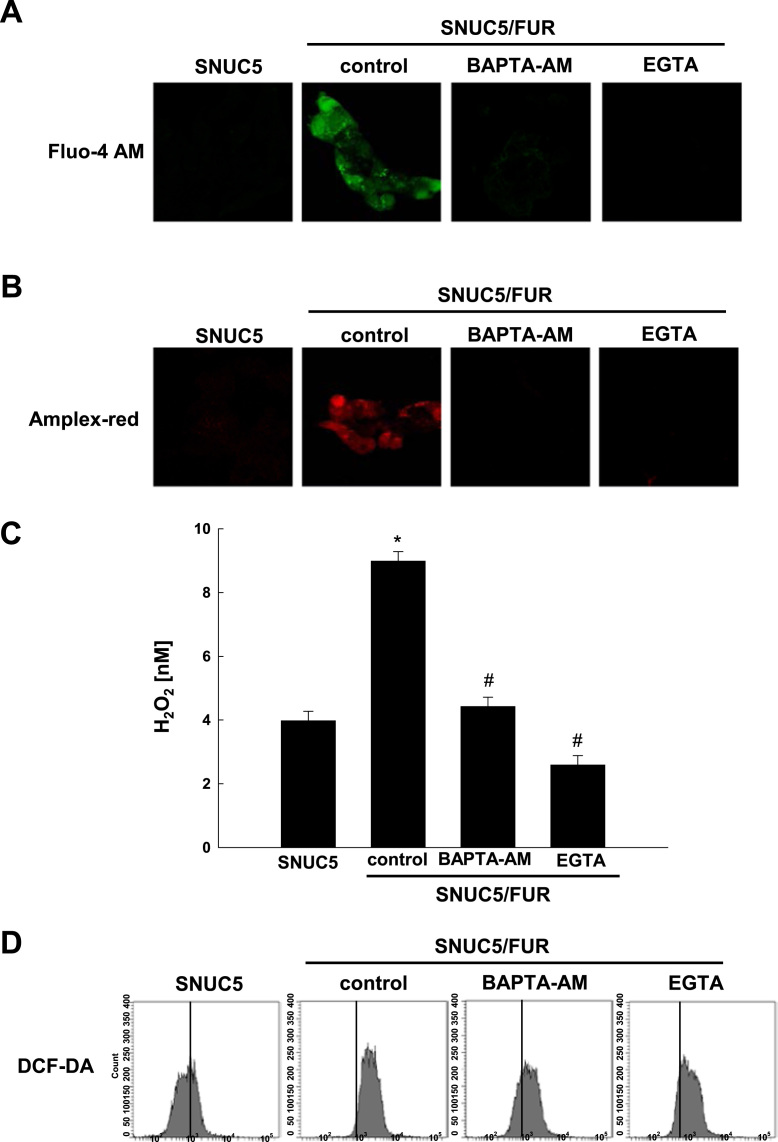
Human DUOX enzymes contain both an extracellular peroxidase-like domain and a canonical calcium-binding helix-loop-helix-hands [26]. Moreover, ROS generation by DUOX2 is dependent on Ca2+ mobilization [27]. To assess whether calcium-dependent DUOX2 activation might participate to the increased ROS generation in SNUC5/FUR cells, we first analyzed the intracellular calcium concentration ([Ca2+]i). The [Ca2+]i in SNUC5/FUR cells largely increased compared to the parental cells, while the Ca2+ scavenger 1,2-bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid (BAPTA-AM), and the chelating agent ethylene glycol tetraacetic acid or egtazic acid (EGTA) significantly reduced [Ca2+]i in SNUC5/FUR cells (Fig. 6A). Notably, the scavenging of Ca2+ led to a dramatic decrease in both H2O2, which is detected by confocal microscopy (Fig. 6B) and fluorescence spectrometer (Fig. 6C) after staining with Amplex red, as well as total ROS production measured by DCFDA staining (Fig. 6D) in the SNU5/FUR cells.
Fig. 6.
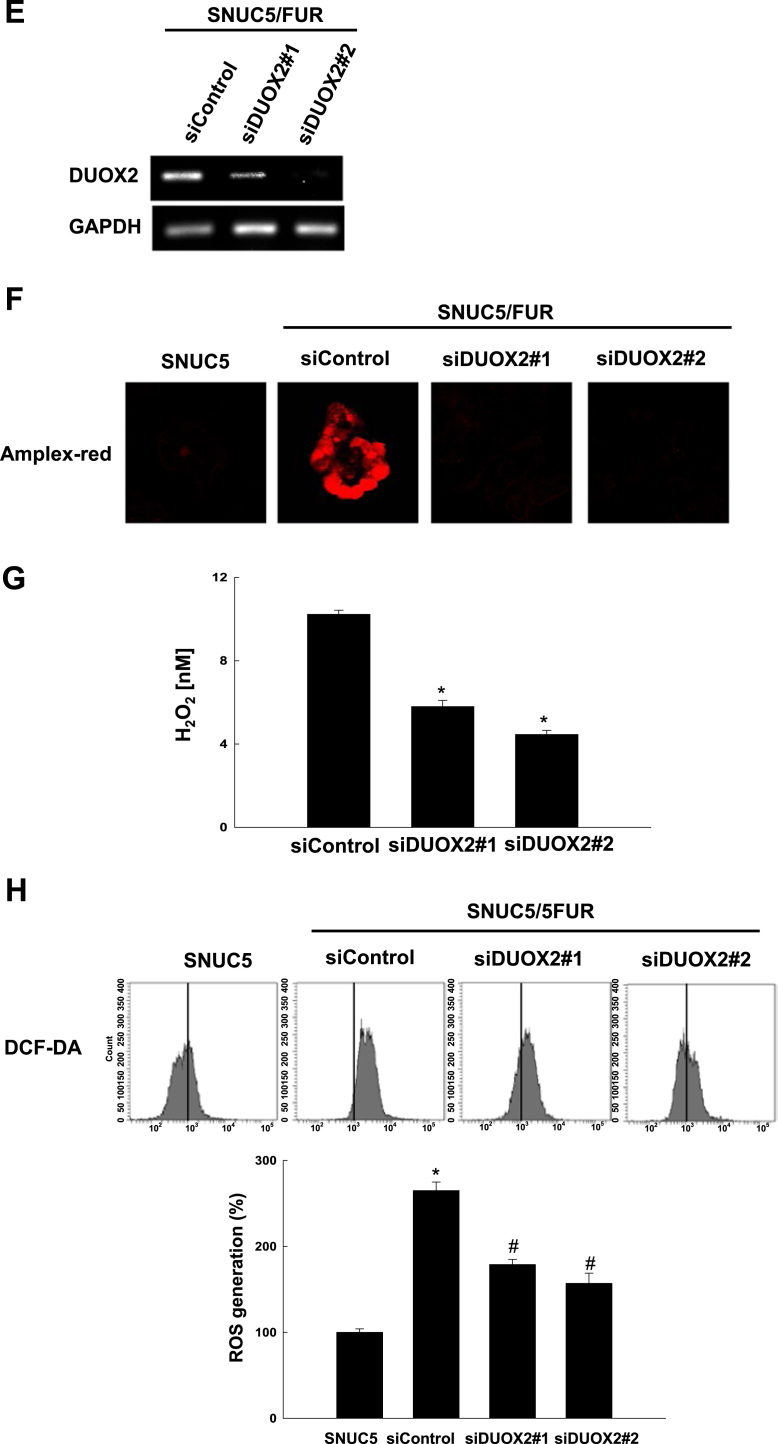
DUOX2 is activated via the induction of intracellular Ca2+ in 5-FU-resistant SNUC5 colon cancer cells. (A-D) SNUC5 or SNU5/FUR cells were treated for 24 h with BAPTA-AM (10 μM), or EGTA (1 mM). (A) The cytosolic [Ca2+] levels were assessed using fluorescence microscopy after staining with Fluo-4 A.M. (B, C) The levels of H2O2 were measured in a Amplex red hydrogen peroxide assay using (B) confocal microscopy and (C) fluorescence spectrometer at 485 nm excitation and 595 nm emission. (D) Total ROS levels were measured by flow cytometry after staining with DCF-DA. *p < 0.05 vs. SNUC5 cells.#p < 0.05 vs. SNUC5/FUR cells.(E) mRNA level of DUOX2 as assessed by qPCR in SNUC5/FUR cells after transfection with siRNA targeting DUOX2#1 and DUOX2#2 or control siRNA. (F, G) The H2O2 level using Amplex Red was measured by (F) confocal microscopy and (G) spectroscopy in SNUC5/FUR cells transfected with siDUOX2 or siControl. *p < 0.05 vs. siControl cells. (H) The total ROS levels were measured in SNU5/FUR cells transfected with siDUOX2 or siControl by flow cytometry after staining with DCF-DA. *p<0.05 vs. SNUC5 cells. #p <0.05 vs. SNUC5/FUR siControl cells.
This suggested that the increased [Ca2+]i in the resistant cells participates in the activation of DUOX2 and thus in the higher production of ROS. To further prove the involvement of DUOX2 in increased ROS production in SNUC5/FUR cells, we silenced DUOX2 expression in SNUC5/FUR cells using two siRNA against DUOX2#1 and DUOX2#2. The two siRNAs decreased DUOX2 mRNA levels (Fig. 6E) and strongly limited H2O2 generation (Fig. 6F and G) and intracellular ROS levels (Fig. 6H) compared to control cells (transfected with siRNA control). Altogether, these data suggested that DUOX2 plays a critical role in the elevated production of ROS in 5-FU-resistant colon cancer cells.
4. Discussion
Although 5-FU-based chemotherapies benefit colon cancer patients, a large fraction of them relapse [28], [29]. Chemo-resistance and metastasis remain a major problem in cancer treatment [30], [31]. EMT has recently been identified as a critical player in 5-FU resistance and metastasis [32], [33]. In agreement with previous reports, our study demonstrates that 5-FU resistance is associated with EMT (through the modulation of the expression of EMT-related proteins) and enhanced cell migration and invasion. Colon cancer cells have an initial oxidative stress response to 5-FU, which leads to cancer cell death, by activating the DNA damage checkpoints. Some cells, however, might escape this control, by adapting to oxidative stress and therefore becoming resistant to chemo- and radio-therapy [34]. In addition, oxidative stress has been proposed to participate in cancer metastasis, which is a multi-factorial process that includes EMT, migration, invasion of the cancer cells, but also angiogenesis around the tumor [35]. In this study, we explored the mechanisms underlying the increased ROS abundance in 5-FU-resistant SNUC5 colon cancer cells. We demonstrated that the mRNA levels of DUOX2, a ROS-producing NAPDH oxidase, are highly increased in 5-FU-resistant cells compared to SNUC5 cells. Several studies indicate that DUOX2 is involved in carcinogenesis, metastasis as well as resistance to chemotherapeutic drugs [36]. Our work has revealed a mechanism through which 5-FU resistance promotes ROS, which participate in the induction of EMT in colon cancer cells. It has been reported that oxaliplatin-induced increase in ROS induces Akt phosphorylation and enhances the expression of Snail, which results in promotion of EMT and metastasis [37]. In line with these observations, we found that increased EMT in 5-FU resistant colon cancer cells involves the production of ROS. This suggests that the increased levels of ROS in 5-FU-resistant cells could play a role in metastasis of colon cancer cells. Recently, we have reported that 5-FU-induced oxidative stress is involved in the transcriptional induction of the DNA demethylase TET1, which participates to 5-FU resistance [22]. In addition, TET1 increases migration and invasion of the cells by demethylating the vimentin promoter in the A2780 cisplatin-resistant ovarian cancer cells [38]. Our results show that TET1 binds to the promoter of DUOX2 and, by decreasing its methylation, enhances DUOX2 expression in SNUC5/FUR cells. It has been reported that DUOX2 is localized in plasma membrane in CaCo-2, a colon cancer cells [39]. DUOX2 protein possesses two canonical EF-hand motifs and its activation requires Ca2+ binding [40]. In addition to its increased expression, the increased intracellular Ca2+ mobilization in SNUC5/FUR cells, participates in the activation of DUOX2 and ROS generation in the resistant cells. Finally, we have shown that ROS lead to enhanced EMT-related protein expression. Specifically, we found that inhibition of TET1 expression by RNA interference was associated with the increase in E-cadherin, reduction in N-cadherin, and reduced expression of EMT-related transcription factors.
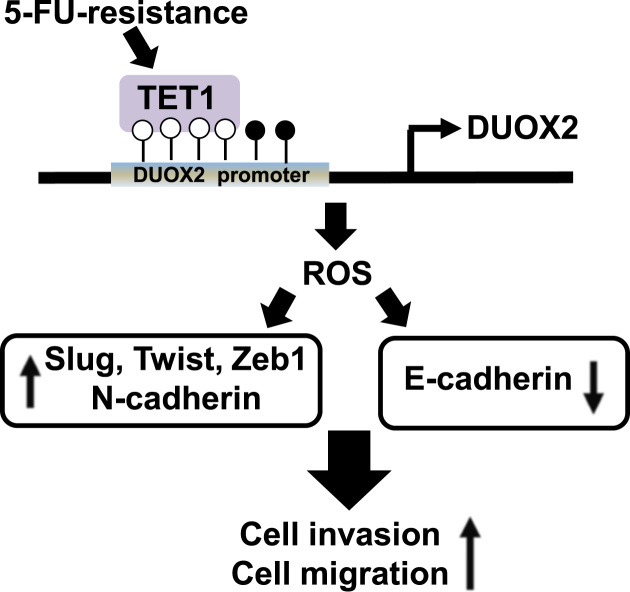
In summary, our findings suggest that the resistance to the chemotherapeutic agent 5-FU is associated with and increased production of cellular ROS and subsequent EMT in colon cancer cells (Fig. 7). Although we did not show that DUOX2 overexpression is itself responsible for the increased EMT in the resistant cells, our date suggests that it is the main enzyme involved in ROS production in these cells and might thus participate in this process. Thus, targeting the TET1/DUOX2/ROS/EMT axis may open a new avenue for therapeutic intervention in colon cancer. Such therapeutic strategies will however first require to be validated in primary colon cancer models, including in relevant mouse models.
Fig. 7.
Proposed model of TET1 and DUOX2 role in 5-FU-induced EMT. 5-FU-resistance increases TET1 expression, a DNA demethylase. TET1 binds to the promoter region of DUOX2 in SNUC5/FUR cells and induces the expression of DUOX2, which in turn increases the level of ROS and induces EMT. Specifically, ROS induce the expression of transcription factors such as Slug, Twist1, Zeb1, and N-cadherin in SNUC5/FUR cells, and repress E-cadherin expression. Therefore, ROS, increased by TET1-mediated DUOX2 activation in SNUC5/FUR cells, confer increased migratory and invasive properties to these cells.
Acknowledgements
Kyoung Ah Kang, Hee Kyoung Kang, Young Sang Koh, Guillaume Bossis, Sung Young Yoon, Seong Bong Kim and Jin Won Hyun conceived and designed the experiments. Kyoung Ah Kang, Yea Seong Ryu, Mei Jing Piao and Joo Mi Yi performed the experiments. Kyoung Ah Kang, Joo Mi Yi, Kristina Shilnikova, Mathias Boulanger, Rosa Paolillo and Jin Won Hyun analyzed the data and wrote the paper.
Acknowledgments
Funding
The work was supported by grant from the Basic Research Laboratory Program [NRF-2017R1A4A1014512], the Science and Technology Amicable Relationships [NRF-2017K1A3A1A21013557] and NRF-2016R1A2B4007934 by the National Research Foundation of Korea [NRF] grant funded by the Korea government. The study was also supported by the R&D Program of Plasma Advanced Technology for Agriculture and Food (Plasma Farming) via the National Fusion Research Institute of Korea (NFRI), funded by the Korea Government.
References
- 1.Kang K.A., Hyun J.W. Oxidative stress, Nrf2, and epigenetic modification contribute to anticancer drug resistance. Toxicol. Res. 2017;33:1–5. doi: 10.5487/TR.2017.33.1.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Serrano-Gomez S.J., Maziveyi M., Alahari S.K. Regulation of epithelial-mesenchymal transition through epigenetic and post-translational modifications. Mol. Cancer. 2016;15:18–32. doi: 10.1186/s12943-016-0502-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Steeg P.S. Targeting metastasis. Nat. Rev. Cancer. 2016;16:201–218. doi: 10.1038/nrc.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lambert A.W., Pattabiraman D.R., Weinberg R.A. Emerging biological principles of metastasis. Cell. 2017;168:670–691. doi: 10.1016/j.cell.2016.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lamouille S., Xu J., DerXu Jynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2014;15:178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dan Dunn J., Alvarez L.A., Zhang X., Soldati T. Reactive oxygen species and mitochondria: a nexus of cellular homeostasis. Redox Biol. 2015;6:472–485. doi: 10.1016/j.redox.2015.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kovac S., Angelova P.R., Holmstrom K.M., Zhang Y., Dinkova-Kostova A.T., Abramov A.Y. Nrf2 regulates ROS production by mitochondria and NADPH oxidase. Biochim. Biophys. Acta. 1850;2015:794–801. doi: 10.1016/j.bbagen.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Acharya A., Das I., Chandhok D., Saha T. Redox regulation in cancer: a double-edged sword with therapeutic potential. Oxid. Med. Cell Longev. 2010;3:23–34. doi: 10.4161/oxim.3.1.10095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prasad S., Gupta S.C., Tyagi A.K. Reactive oxygen species (ROS) and cancer: role of antioxidative nutraceuticals. Cancer Lett. 2017;387:95–105. doi: 10.1016/j.canlet.2016.03.042. [DOI] [PubMed] [Google Scholar]
- 10.Zhang L., Li J., Zong L., Chen X., Chen K., Jiang Z., Nan L., Li X., Li W., Shan T., Ma Q., Ma Z. Reactive oxygen species and targeted therapy for pancreatic cancer. Oxid. Med. Cell Longev. 2016;2016:1616781. doi: 10.1155/2016/1616781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Z., Duan Q., Zhao H., Liu T., Wu H., Shen Q., Wang C., Yin T. Gemcitabine treatment promotes pancreatic cancer stemness through the Nox/ROS/NF-κB/STAT3 signaling cascade. Cancer Lett. 2016;382:53–63. doi: 10.1016/j.canlet.2016.08.023. [DOI] [PubMed] [Google Scholar]
- 12.Nikitovic D., Corsini E., Kouretas D., Tsatsakis A., Tzanakakis G. ROS-major mediators of extracellular matrix remodeling during tumor progression. Food Chem. Toxicol. 2013;61:178–186. doi: 10.1016/j.fct.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 13.Jiang J., Wang K., Chen Y., Chen H., Nice E.C., Huang C. Redox regulation in tumor cell epithelial-mesenchymal transition: molecular basis and therapeutic strategy. Signal Transduct. Target Ther. 2017;2:17036. doi: 10.1038/sigtrans.2017.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011;17:10–12. [Google Scholar]
- 15.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler Transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li L.C., Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002;18:1427–1431. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- 18.Heyn H., Moran S., Hernando-Herraez I., Sayols S., Gomez A., Sandoval J., Monk D., Hata K., Marques-Bonet T., Wang L., Esteller M. DNA methylation contributes to natural human variation. Genome Res. 2013;23:1363–1372. doi: 10.1101/gr.154187.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herman J.G., Graff J.R., Myöhänen S., Nelkin B.D., Baylin S.B. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc. Natl. Acad. Sci. USA. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McGarvey K.M., Greene E., Fahrner J.A., Jenuwein T., Baylin S.B. DNA methylation and complete transcriptional silencing of cancer genes persist after depletion of EZH2. Cancer Res. 2007;67:5097–5102. doi: 10.1158/0008-5472.CAN-06-2029. [DOI] [PubMed] [Google Scholar]
- 21.Kang K.A., Piao M.J., Kim K.C., Kang H.K., Chang W.Y., Park I.C., Keum Y.S., Surh Y.J., Hyun J.W. Epigenetic modification of Nrf2 in 5-fluorouracil-resistant colon cancer cells: involvement of TET-dependent DNA demethylation. Cell Death Dis. 2014;5:e1183. doi: 10.1038/cddis.2014.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kang K.A., Piao M.J., Ryu Y.S., Kang H.K., Chang W.Y., Keum Y.S., Hyun J.W. Interaction of DNA demethylase and histone methyltransferase upregulates Nrf2 in 5-fluorouracil-resistant colon cancer cells. Oncotarget. 2016;7:40594–40620. doi: 10.18632/oncotarget.9745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gagliano N., Celesti G., Tacchini L., Pluchino S., Sforza C., Rasile M., Valerio V., Laghi L., Conte V., Procacci P. Epithelial-to-mesenchymal transition in pancreatic ductal adenocarcinoma: characterization in a 3D-cell culture model. World J. Gastroenterol. 2016;22:4466–4483. doi: 10.3748/wjg.v22.i18.4466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matos M.L., Lapyckyj L., Rosso M., Besso M.J., Mencucci M.V., Briggiler C.I., Giustina S., Furlong L.I., Vazquez-Levin M.H. Identification of a novel human E-cadherin splice variant and assessment of its effects upon EMT-related events. J. Cell Physiol. 2017;232:1368–1386. doi: 10.1002/jcp.25622. [DOI] [PubMed] [Google Scholar]
- 25.Webb A.H., Gao B.T., Gold smith Z.K., Irvine A.S., Saleh N., Lee R.P., Lendermon J.B., Bheemreddy R., Zhang Q., Brennan R.C., Johnson D., Steinle J.J., Wilson M.W., Morales-Tirado V.M. Inhibition of MMP-2 and MMP-9 decreases cellular migration, and angiogenesis in in vitro models of retinoblastoma. BMC Cancer. 2017;17:434. doi: 10.1186/s12885-017-3418-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burtenshaw D., Hakimjavadi R., Redmond E.M., Cahill P.A. Nox, reactive oxygen species and regulation of vascular cell fate. Antioxidants. 2017;6:E90. doi: 10.3390/antiox6040090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Joo J.H., Ryu J.H., Kim C.H., Kim H.J., Suh M.S., Kim J.O., Chung S.Y., Lee S.N., Kim H.M., Bae Y.S., Yoon J.H. Dual oxidase 2 is essential for the toll-like receptor 5-mediated inflammatory response in airway mucosa. Antioxid. Redox Signal. 2012;16:57–70. doi: 10.1089/ars.2011.3898. [DOI] [PubMed] [Google Scholar]
- 28.Che J., Pan L., Yang X., Liu Z., Huang L., Wen C., Lin A., Liu H. Thymidine phosphorylase expression and prognosis in colorectal cancer treated with 5-fluorouracil-based chemotherapy: a meta-analysis. Mol. Clin. Oncol. 2017;7:943–952. doi: 10.3892/mco.2017.1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kugimiya N., Nishimoto A., Hosoyama T., Ueno K., Takemoto Y., Harada E., Enoki T., Hamano K. JAB1-STAT3 activation loop is associated with recurrence following 5-fluorouracil-based adjuvant chemotherapy in human colorectal cancer. Oncol. Lett. 2017;14:6203–6209. doi: 10.3892/ol.2017.6994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo C., Ma J., Deng G., Qu Y., Yin L., Li Y., Han Y., Cai C., Shen H., Zeng S. ZEB1 promotes oxaliplatin resistance through the induction of epithelial - mesenchymal transition in colon cancer cells. J. Cancer. 2017;8:3555–3566. doi: 10.7150/jca.20952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garg M. Epithelial plasticity and cancer stem cells: major mechanisms of cancer pathogenesis and therapy resistance. World J. Stem Cells. 2017;9:118–126. doi: 10.4252/wjsc.v9.i8.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun L., Ke J., He Z., Chen Z., Huang Q., Ai W., Wang G., Wei Y., Zou X., Zhang S., Lan P., Hong C. HES1 promotes colorectal cancer cell resistance to 5-Fu by inducing of EMT and ABC transporter proteins. J. Cancer. 2017;8:2802–2808. doi: 10.7150/jca.19142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dinicola S., Pasqualato A., Proietti S., Masiello M.G., Palombo A., Coluccia P., Canipari R., Catizone A., Ricci G., Harrath A.H., Alwasel S.H., Cucina A., Bizzarri M. Paradoxical E-cadherin increase in 5FU-resistant colon cancer is unaffected during mesenchymal-epithelial reversion induced by γ-secretase inhibition. Life Sci. 2016;145:174–183. doi: 10.1016/j.lfs.2015.12.048. [DOI] [PubMed] [Google Scholar]
- 34.Yang Z., Chen J.S., Wen J.K., Gao H.T., Zheng B., Qu C.B., Liu K.L., Zhang M.L., Gu J.F., Li J.D., Zhang Y.P., Li W., Wang X.L., Zhang Y. Silencing of miR-193a-5p increases the chemosensitivity of prostate cancer cells to docetaxel. J. Exp. Clin. Cancer Res. 2017;36:178. doi: 10.1186/s13046-017-0649-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Z., Tu K., Wang Y., Yao B., Li Q., Wang L., Dou C., Liu Q., Zheng X. Hypoxia accelerates aggressiveness of hepatocellular carcinoma cells involving oxidative stress, epithelial-mesenchymal transition and non-canonical Hedgehog signaling. Cell Physiol. Biochem. 2017;44:1856–1868. doi: 10.1159/000485821. [DOI] [PubMed] [Google Scholar]
- 36.Shu Y., Liu Y., Li X., Cao L., Yuan X., Li W., Cao Q. Aspirin-triggered resolvin d1 inhibits TGF-β1-induced EndMT through increasing the expression of smad7 and is closely related to oxidative stress. Biomol. Ther. 2016;24:132–139. doi: 10.4062/biomolther.2015.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiao L., Li D.D., Yang C.L., Peng R.Q., Guo Y.Q., Zhang X.S., Zhu X.F. Reactive oxygen species mediate oxaliplatin-induced epithelial-mesenchymal transition and invasive potential in colon cancer. Tumour Biol. 2016;37:8413–8423. doi: 10.1007/s13277-015-4736-9. [DOI] [PubMed] [Google Scholar]
- 38.Han X., Zhou Y., You Y., Lu J., Wang L., Hou H., Li J., Chen W., Zhao L., Li X. TET1 promotes cisplatin-resistance via demethylating the vimentin promoter in ovarian cancer. Cell Biol. Int. 2017;41:405–414. doi: 10.1002/cbin.10734. [DOI] [PubMed] [Google Scholar]
- 39.Lipinski S., Till A., Sina C., Arlt A., Grasberger H., Schreiber S., Rosenstiel P. DUOX2-derived reactive oxygen species are effectors of NOD2-mediated antibacterial responses. J. Cell Sci. 2009;122:3522–3530. doi: 10.1242/jcs.050690. [DOI] [PubMed] [Google Scholar]
- 40.Razzell W., Evans I.R., Martin P., Wood W. Calcium flashes orchestrate the wound inflammatory response through DUOX activation and hydrogen peroxide release. Curr. Biol. 2013;23:424–429. doi: 10.1016/j.cub.2013.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]