SUMMARY
The HIV-1 envelope (Env) spike is a trimer of gp120/gp41 heterodimers that mediates viral entry. Binding to CD4 on the host cell membrane is the first essential step for infection but disrupts the native antigenic state of Env, posing a key obstacle to vaccine development. We locked the HIV-1 Env trimer in a pre-fusion configuration, resulting in impaired CD4 binding and enhanced binding to broadly neutralizing antibodies. This design was achieved via structure-guided introduction of neo-disulfide bonds bridging the gp120 inner and outer domains and was successfully applied to soluble trimers and native gp160 from different HIV-1 clades. Crystallization illustrated the structural basis for CD4-binding impairment. Immunization of rabbits with locked trimers from two different clades elicited neutralizing antibodies against tier-2 viruses with a repaired glycan shield regardless of treatment with a functional CD4 mimic. Thus, interdomain stabilization provides a widely applicable template for the design of Env-based HIV-1 vaccines.
In Brief/eTOC paragraph
Binding to CD4 compromises the native antigenic state of the HIV-1 envelope trimer. Zhang et al. rationally designed interdomain-locked trimers that elicited the production of tier-2 neutralizing antibodies against glycan-repaired virus and were resistant to CD4-induced disruption of immunogenicity. This strategy provides a widely applicable template for HIV-1 Env-based vaccines.
INTRODUCTION
The quest to develop a protective HIV-1 vaccine is facing unprecedented challenges. HIV-1 has evolved an extraordinary array of immune-evasion tactics, including the ability to integrate its proviral genome into host cell chromosomes, a marked antigenic variability of its exposed envelope (Env) domains, and an extensive glycan cloak covering almost the entire Env surface (Burton et al., 2012; Mascola and Haynes, 2013). These mechanisms help to explain why only a proportion of infected individuals develop broadly neutralizing antibodies (bNAbs), which are critical for protective immunity, and none of the vaccines tested so far has been successful in eliciting such antibodies (Excler and Michael, 2016; Haynes et al., 2012). Indeed, bNAbs emerge only after months or years of antigenic stimulation resulting from sustained viral replication, and are usually characterized by extensive variable-region hypermutation and peculiarly long CDR3 loops, which enable them to penetrate the protective glycan shield (Burton et al., 2012; Mascola and Haynes, 2013). Another mechanism of immune evasion is the remarkable flexibility of the HIV-1 Env spike, which is maintained in a metastable, high-energy state, yet it continuously explores different conformations, from the most antigenically protected to more open forms analogous to the low-energy, CD4-bound state (Munro et al., 2014). As a consequence, conserved neutralization epitopes are protected by conformational masking, which imposes a strong entropic penalty for antibody binding (Kwong et al., 2002).
The molecular anatomy of the HIV-1 Env spike has progressively been refined in recent years, providing a platform for the design of effective vaccine immunogens. Important advancements have been made with the design and crystallization of stabilized soluble gp140 trimers, either cleaved or cleavage-independent, which display antigenic features of the pre-fusion Env spike (Julien et al., 2013; Pancera et al., 2014; Sanders et al., 2013; Sharma et al., 2015), as well as with the progressive increase in imaging resolution of membrane-bound and soluble trimers by cryo-electron microscopy (Bartesaghi et al., 2013; Lee et al., 2016; Liu et al., 2008; Liu et al., 2017; Lyumkis et al., 2013; Ozorowski et al., 2017). In spite of their near-native antigenic profile, however, stabilized soluble trimers have not yielded the anticipated breakthrough in HIV-1 vaccine development, eliciting primarily autologous neutralizing antibodies in pre-clinical models (Klasse et al., 2016; Pauthner et al., 2017; Sanders et al., 2015). Moreover, these neutralizing antibodies were mainly directed against glycan holes present on the surface of the immunizing trimers, being largely ineffective against glycan-repaired versions of the same trimers (Klasse et al., 2016; McCoy et al., 2016). Glycan-hole targeting by neutralizing antibodies was confirmed using trimers with site-specific glycan deletions (Crooks et al., 2015; Zhou et al., 2017). In view of these limitations, major efforts are currently focused on improving the trimer design, with the primary aim of restricting trimer flexibility (Chuang et al., 2017; Guenaga et al., 2017; Kulp et al., 2017; Kwon et al., 2015; Sullivan et al., 2017; Torrents de la Pena et al., 2017). Most of these modified trimers have shown gains in expression efficiency, thermal stability and antigenicity, but the effects of these alterations on immunogenicity have been limited.
Another key obstacle to vaccine development is the ability of HIV-1 Env-based immunogens to bind the CD4 receptor, which triggers rapid and dramatic conformational changes in the trimer, compromising its native antigenic state. Binding to CD4 may also result in the sequestration of Env-based immunogens by CD4+ T cells, which lack antigen-presenting function, and occlusion of the CD4-binding site, which is a major immunogenic region targeted by bNAbs. The detrimental effects of CD4 binding have been difficult to verify in pre-clinical models, including rabbits and nonhuman primates, because their CD4 is unable to bind with high affinity to the HIV-1 Env (Finzi et al., 2012; Humes et al., 2012; Li et al., 2016). Previous studies have reported the design of trimers with reduced CD4-binding capacity through the introduction of a neo-disulfide bond (Kwon et al., 2015) or stabilizing point mutations (de Taeye et al., 2015). However, CD4 binding was not fully abrogated, and at least some CD4-induced conformational changes were still detectable with these variants. Only one trimer modification so far reported, obtained by introducing multiple point mutations in various Env domains, nearly eliminated CD4 binding, but it caused some alterations in the antigenic profile, and the potential benefits of CD4 resistance were not investigated (Kulp et al., 2017).
In this study, we used a structure-guided approach to stabilize the HIV-1 Env trimer in a native-like pre-fusion configuration by covalently locking the two cardinal structural elements, the inner and outer domains, of the gp120 glycoprotein. We designed and characterized interdomain-locked trimers that are selectively and efficiently targeted by potent bNAbs and are unable to physiologically interact with CD4. Stabilized trimers derived from two HIV-1 clades elicited in rabbits the production of neutralizing antibodies against tier-2 autologous viruses with a repaired glycan shield, averting the disruptive effects of CD4-induced antigenic remodeling on immunogenicity. This interdomain-locking strategy provides a common template for the design of potentially effective vaccine immunogens based on diverse HIV-1 Env forms.
RESULTS
Structure-guided design of interdomain locks to stabilize the HIV-1 Env trimer
Since a major source of Env flexibility is the reciprocal mobility of the inner vs. outer domains of the gp120 glycoprotein, we reasoned that introduction of a covalent bond between these two domains might stabilize the trimer in a native-like pre-fusion configuration. We focused on a region of molecular mimicry between gp120 and human CD4, a stretch of 5 identical residues present in both the gp120 inner-domain α1-helix (aa. 110-114) and the DE loop of CD4 domain 1 (aa. 60-64) (Zagury et al., 1992), whose structural and functional features will be described elsewhere. In high-resolution structures of the BG505 SOSIP.664 trimer (Julien et al., 2013; Pancera et al., 2014), the α1-helix residue D113 is involved in interdomain interaction with R429 in the outer-domain β20-β21 loop (Figure 1A). Based on this observation, we aimed at introducing a neo-disulfide bond between the inner and outer gp120 domains of the BG505 SOSIP.664 trimer by mutating both D113 and R429 to cysteines. Single-cysteine mutants displayed reduced binding to the trimer-preferring monoclonal antibodies (mAbs) PG16, PGT145 and 35O22 in ELISA, confirming that disruption of the D113-R429 salt bridge weakened the interdomain association; however, no rescue of trimer-preferring mAb reactivity was observed with the double mutant (BG505 113C-429C), suggesting that the disulfide bond had not formed (Table S1, upper panel). We hypothesized that this failure was due to excessive distance between the side chains of C113 and C429 and therefore introduced spacer residues (G or S) on both sides of C429, reasoning that their insertion into the β20-β21 loop would be less disruptive than into the more rigid α1-helix. The introduction of spacer residues indeed restored reactivity with trimer-preferring mAbs in all the designed mutants but one (Table S1, upper panel). The BG505 113C-429GCG design, which displayed the most favorable antigenic profile, was selected for further studies and extensively purified by size-exclusion chromatography and negative selection with the anti-V3 loop mAb 447-52D (Figure S1). Formation of the expected neo-disulfide bridge was confirmed by liquid chromatography-mass spectrometry (LC-MS) (Figure S2A).
Figure 1. Structure-based design of an interdomain-locked HIV-1 Env trimer.
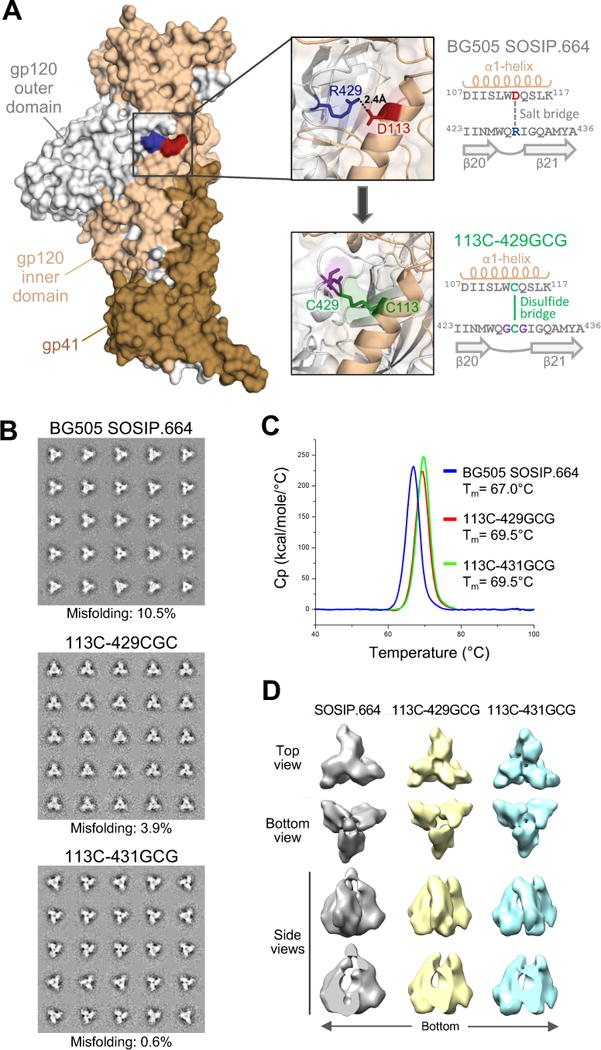
(A) Intramolecular interaction between the inner and outer domains of the gp120 Env glycoprotein. A single protomer from a crystal structure of the BG505 SOSIP.664 trimer (PDB ID: 4TVP) is shown in surface representation, highlighting the interdomain contact between R429 in the β20-21 loop (blue) and D113 in the α1-helix (red) of gp120. The upper inset shows a semi-transparent magnification of the interactive region with the side chains of R429 and D113 highlighted in stick representation, with the neighboring amino acid sequences reported in the scheme on the right. The design of an interdomain-locked mutant (D113C-R429GCG) bearing a neo-disulfide bridge (green) is illustrated in the lower inset, with the sequences reported in the scheme on the right.
(B) Negative-stain EM analysis of purified unmutated and interdomain-stabilized BG505 SOSIP.664 trimers. The 2D class averages are shown. The rate of misfolding was evaluated by analysis of loop movement, compactness and angles between individual protomers.
(C) Thermal stability of unmutated and interdomain-stabilized BG505 SOSIP.664 trimers as assessed by differential scanning calorimetry (DSC).
(D) Negative-staining electron microscopy (NS-EM) three-dimensional reconstructions of unmutated and interdomain-stabilized BG505 SOSIP.664 trimers. See also Figures S1, S2, S3 and S4, and Table S1.
Next, we examined the applicability of the same interdomain locking strategy to HIV-1 strains of other genetic subtypes, but we noticed that unlike D113, which is nearly universally conserved among group-M isolates (Figure S3A), R429 is conserved only in clades A and G, with clades B to F showing non-conservative substitutions (Figure S3B). However, the vast majority of group-M isolates that lack R429 contain a basic residue (K or R) at the neighboring position 432, which is also favorably positioned to establish a salt bridge with D113, as documented in high-resolution structures of a clade-B trimer, JR-FL SOSIP.664 (Figure S4). To determine the feasibility of this alternative disulfide-bridge partnering, a second set of BG505 mutants was generated by inserting a cysteine at position 432, with spacers of different length on either side. Most mutants were reactive with trimer-preferring bNAbs, validating the interchangeable function of positions 429 and 432 for interdomain interaction with D113 (Table S1, lower panel). The BG505 113C-431GCG mutant showed the best antigenic profile and was extensively purified using size-exclusion and 447-52D negative selection, confirming formation of the disulfide bond by LC-MS (Figures S1, S2B).
Increased stability of interdomain-locked trimers
To evaluate the morphology and homogeneity of interdomain-locked trimers, we performed negative-stain electron microscopy (NSEM) on purified BG505 113C-429GCG and BG505 113C-431GCG SOSIP.664 trimers. Both mutants displayed a more compact configuration, compared to the unmutated trimer, as well as a reduced propensity to generate misfolded trimers (Figure 1B). Differential scanning calorimetry demonstrated increased thermal stability for both locked trimers (Figure 1C). Furthermore, by three-dimensional reconstruction from NSEM data, stabilized trimers were more tightly packed than the original trimer, with an average volume of the internal trimer cavity of ~17 nm3 for each mutant vs. an average of 26 nm3 for the unmutated trimer (Figure 1D). These results confirmed that interdomain locks effectively stabilize the HIV-1 Env trimer in a tightly closed structure, limiting the spontaneous transition to more open conformational states.
Improved antigenic profile of interdomain-locked trimers
The antigenic profiles of BG505 113C-429GCG and BG505 113C-431GCG were extensively characterized by ELISA with a panel of anti-Env mAbs. All the bNAbs tested efficiently bound to both locked trimers (Figure 2A, upper panel). Strikingly, stabilized trimers exhibited greater reactivity with trimer-preferring bNAbs against V1V2 glycan-dependent epitopes at the tip of the trimer (i.e., PGT145, PG9, VRC26.03), compared to the unmutated trimer, in spite of the absence of stabilizing locks applied to this region (Figure 2B). In contrast, reactivity with weak or non-neutralizing mAbs, including those directed against CD4-induced (CD4i) epitopes tested in the presence or absence of soluble CD4 (sCD4), was abolished (Figure 2A, lower panel).
Figure 2. Antigenic profile and CD4 binding of interdomain-locked Env trimers.
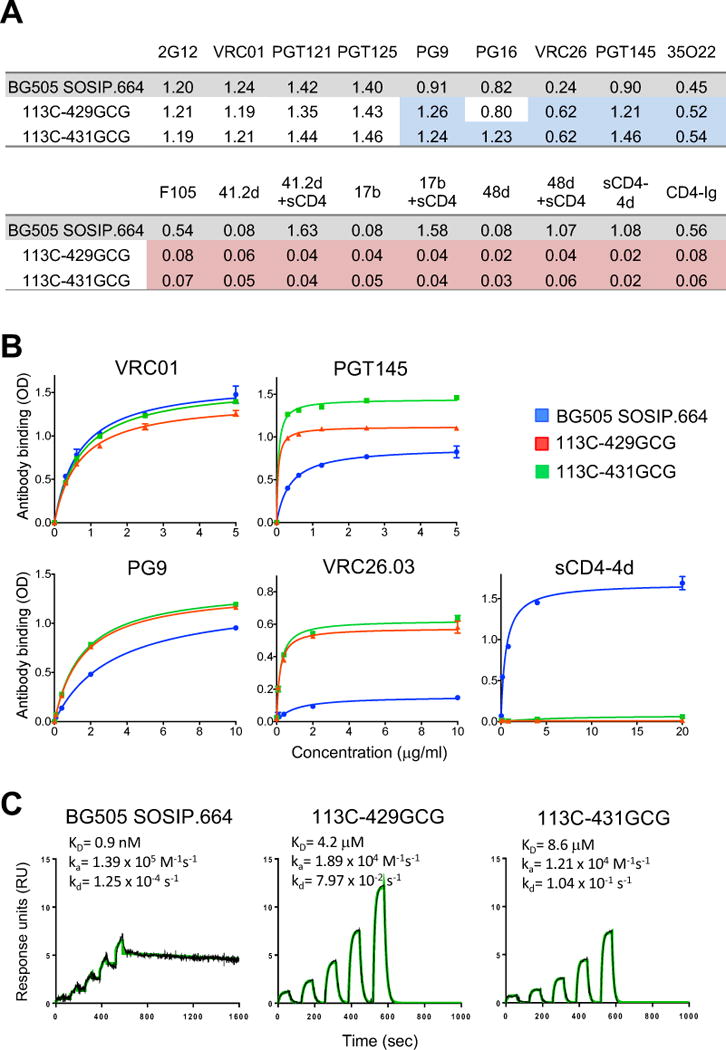
(A) Reactivity of unmutated and interdomain-stabilized BG505 SOSIP.664 trimers with a panel of anti-Env mAbs and sCD4 in ELISA. Mean optical density (OD) values at 450 nm for each mAb (complexed or not with sCD4) and for two different forms of sCD4 (sCD4-4d and CD4-Ig) are shown from a representative experiment performed in duplicate wells. The colors denote increased (blue) or decreased (red) binding to each stabilized trimer compared to the unmutated BG505 SOSIP.664 trimer.
(B) Dose-response reactivity of purified BG505 SOSIP.664 trimer and two interdomain-stabilized variants with a panel of anti-Env antibodies and 4-domain soluble CD4 (sCD4-4d) in ELISA. The data shown are mean ± SEM.
(C) Surface plasmon resonance (SPR) analysis of sCD4 binding to unmutated and interdomain-stabilized BG505 SOSIP.664 trimers. See also Figure S5 and Table S3.
Interdomain locking impairs CD4 binding
We hypothesized that structural fixation of the trimer via interdomain locks would reduce the conformational flexibility required for CD4 to gain access to its binding site. Thus, we measured the ability of the locked trimers to bind sCD4. Testing by ELISA showed no binding of either trimer to two forms of human sCD4, Ig-chimeric sCD4 (CD4-Ig) or 4-domain sCD4 (sCD4-4d) (Figures 2A and 2B). Ultra-sensitive analysis by single-cycle surface plasmon resonance (SPR) confirmed these findings by showing only a minimal residual sCD4-binding activity for both locked trimers, with a ~3-log reduction in binding affinity (Figure 2C). Due to the very rapid off rate, it is not surprising that this weak and transient CD4 interaction was completely lost using a steady-state assay like ELISA. However, potent bNAbs to the CD4 supersite such as VRC01 maintained unaltered their ability to bind to the locked trimers (Figure 2A and 2B), confirming that binding to CD4 and anti-CD4-supersite antibodies can be uncoupled (Liu et al., 2017).
To determine whether the low and transient CD4-binding activity measured by ultra-sensitive SPR was biologically relevant, we tested the ability of one stabilized trimer, BG505 113C-429GCG, to bind to freshly isolated human peripheral blood CD4+ T cells. While the original BG505 SOSIP.664 trimer efficiently bound to primary CD4+ T cells, no binding was detected with its locked counterpart (Figure S5A). Additionally, we tested the ability of BG505 113C-429GCG to block the infectivity of BG505 pseudovirus in TZM-bl cells via occupancy of the available CD4 receptor. As expected, the BG505 SOSIP.664 trimer potently reduced BG505 infectivity (>80%) by competing for CD4 occupancy, whereas the two trimers with reduced CD4-binding activity, DS-SOSIP and DS-SOSIP.6mut, inhibited to a lesser, albeit still significant (>50%), extent; in contrast, the locked BG505 113C-429GCG had no inhibitory effects on HIV-1 infectivity (Figure S5B). These results confirmed that the weak and transient CD4-binding activity detected by ultra-sensitive SPR does not reflect a biologically-relevant interaction of interdomain-locked trimers with CD4.
Crystal structure of an interdomain-locked HIV-1 trimer
To investigate the structural basis for the lack of CD4 interaction of interdomain-locked trimers, we solved the crystal structure of the BG505 113C-429GCG trimer in complex with bNAbs PGT122 and 35O22 at 4.3Å resolution (Figure 3 and Table S2). The neo-disulfide bridge was clearly visible in the electron density map. Overlap of the structure with a CD4-bound monomeric gp120 structure (Figure 3B) showed that multiple residues in the hydrophobic binding pocket for CD4 F43, including M426, W112 and W427, are differently oriented in the two configurations; in particular, in the CD4-bound structure M426 is pointing away from the F43-binding pocket (Figure 3B and 3C), while in the locked trimer M426 is positioned inside the pocket, creating a clash with the incoming CD4 F43 (Figure 3D). The disulfide bond precludes M426 from reorienting and constrains the distance of the β20-β21 loop from the α1-helix (Figures 3B-3D). Comparison with the only available CD4-bound trimeric structure (B41 SOSIP.664; PDB ID: 5VN3) (Ozorowski et al., 2017) revealed the same side-chain orientations as in CD4-bound monomeric gp120 (Figure S6B). Previous structures of CD4-unbound soluble trimers also showed M426 positioned inside the F43-binding pocket, while in CD4-bound gp120 M426 is invariably pointing outwards, without intereference with F43 insertion into the pocket (Figure S6). Binding to CD4 or CD4 mimics also induces the side chain of R429 (or K432) to flip outwards, interrupting the bond with D113, while the side chain of Q428 is reoriented toward the α1-helix (Figure S6B).
Figure 3. Crystal structure of an interdomain-locked HIV-1 Env trimer.
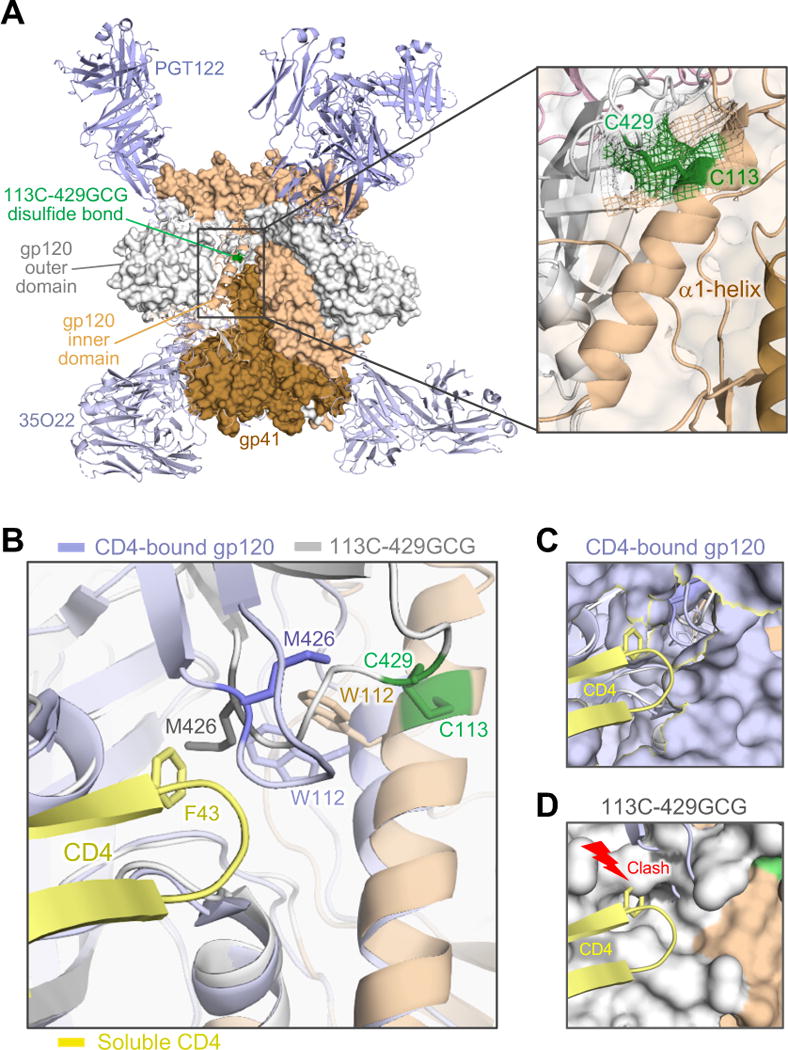
(A) The BG505 SOSIP.664 113C-429GCG trimer, bearing an engineered neo-disulfide bond between residues 113 and 429, was crystallized in complex with bNAbs PGT122 and 35O22. The overall structure, solved at 4.3 Å, is shown as protein surface with the α1-helix (beige) and the β20-21 loop (grey) of one protomer in cartoon representation. The neo-disulfide bond linking the two domains is highlighted in green. The inset shows a close-up of the stabilizing disulfide bond. Electron density (2Fo-Fc contoured at 1σ) clearly confirms the disulfide linkage.
(B) Overlap of the stabilized trimer structure (beige/grey) with a CD4-bound gp120 structure (light blue; 1RZJ). The F43 loop of CD4 (yellow) is shown aligned with the disulfide-stabilized trimer. Several residues in the hydrophobic pocket of CD4-bound gp120 are reoriented, including M426, W427 and W112. As in other CD4-bound structures (Huang et al., 2005; Kwong et al., 2000; Kwong et al., 1998; Pancera et al., 2010), including the recently reported structure of a CD4-bound SOSIP.664 trimer (PDB ID: 5VN3) (Ozorowski et al., 2017), Met426 is oriented away from the F43-binding pocket. In contrast, the disulfide bond precludes M426 from reorienting and constrains the distance of the β20-β21 loop from the α1-helix. (C) Surface representation of gp120 in CD4-bound conformation, with F43 finding its way into its hydrophobic binding pocket without interference from M426.
(D) In the locked trimer, M426 is positioned inside the pocket, as in other unbound trimer structures (4TVP, 5CEZ, 5FYL), creating a clash with the incoming F43. See also Figure S6.
Interdomain locks can be applied to both soluble trimers and membrane-expressed, native Env spikes from different HIV-1 clades
To verify the broad applicability of the interdomain-locking design to different strains and genetic subtypes of HIV-1, neo-disulfide bonds were introduced into both clade-B (JR-FL and B41) and clade-C (DU422) SOSIP.664 trimers. A schematic representation of JR-FL 113C-432GCG is presented in Figure S4. Extensive antigenic profiling demonstrated that the neo-disulfide bridges were effective in stabilizing these trimers in a manner similar to the clade-A BG505, as shown by preserved or increased binding to bNAbs, drastically reduced binding to weakly-neutralizing antibodies, and loss of CD4 binding (Table S3).
Since SOSIP trimers are truncated and artificially stabilized, it is still uncertain how precisely they represent the structure of the native pre-fusion HIV-1 Env spike. Thus, we investigated whether interdomain neo-disulfide bridges could stabilize the Env structure in the context of membrane-expressed gp160 derived from different HIV-1 strains and clades. Despite higher background signals than in ELISA, as commonly observed by flow cytometry, consistent antigenic profiles were observed for stabilized gp160s from clade A (BG505), clade B (BaL, JR-FL) and clade C (1086c), as shown by preserved or increased reactivity with bNAbs, compared to the wild-type (WT), and reduced binding to non-neutralizing mAbs as well as to two different forms of sCD4 (Figure 4A). As predicted by the lack of CD4 interaction, pseudoviruses expressing interdomain-locked Env were devoid of infectious activity, whereas individual cysteine mutants (113C or 429C) exhibited reduced infectious capacity, consistent with partial disruption of trimer stability (Figure 4B).
Figure 4. The interdomain-locking design can be successfully applied to membrane-expressed, native full-length gp160.
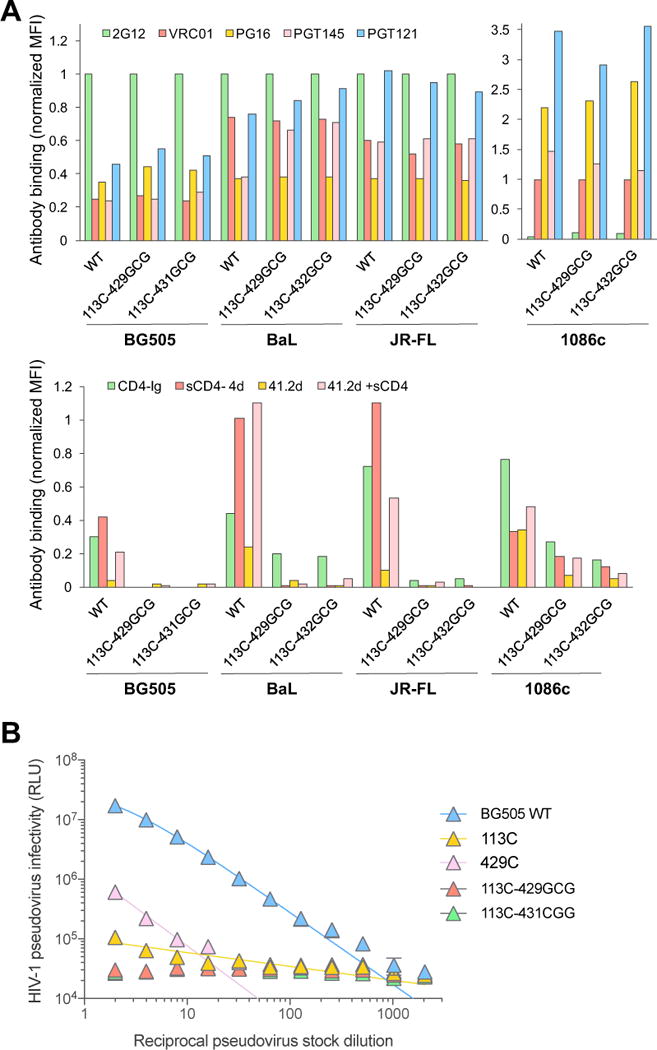
(A) Flow cytometry analysis of binding of a panel of anti-Env mAbs and sCD4 to cell-surface-expressed WT and interdomain-stabilized full-length HIV-1 Env (gp160) of different clades: BG505 (clade A), BaL and JR-FL (clade B) and 1086c (clade E). The HIV-1 gp160 were expressed on the surface of 293T cells and tested at 48-72 hours post-transfection. The results show normalized mean fluorescence intensity (MFI) values from a representative experiment, relative to the MFI of mAb 2G12 (VRC01 for Env 1086c, which is not reactive with 2G12).
(B) Infectivity of HIV-1 pseudoviruses bearing WT or mutated BG505 Env in TZM-bl cells. Hill slope nonlinear regression curves are shown for samples with detectable infectivity.
Interdomain-stabilized trimers are resistant to the effects of CD4 and a CD4 mimic
A potential drawback with the use of Env-based immunogens for human vaccination is binding to CD4 on the host cell membrane, which causes major conformational changes in the trimer, compromising its native antigenic state. Since current pre-clinical models, including macaques, lack a functional CD4 for high-affinity interaction with the HIV-1 Env (Finzi et al., 2012; Humes et al., 2012; Li et al., 2016), we devised a unique immunization protocol in order to simulate the conditions that will occur in human vaccination. Thus, soluble Env trimers were pre-exposed to a functional CD4 mimic, M48U1, which induces the same conformational changes as CD4 (Martin et al., 2003). We chose M48U1 because it lacks sequence homology with human CD4, and therefore its injection into heterologous hosts should not elicit anti-CD4 antibodies that may confound the interpretation of virus-specific neutralizing responses. As a preliminary step, the effects of M48U1 were evaluated in vitro on unmutated and interdomain-locked SOSIP.664 trimers derived from two HIV-1 strains of different clades: BG505 (clade A) and JR-FL (clade B). The results confirmed the ability of M48U1 to trigger the same conformational changes as sCD4 in both strains, while the respective interdomain-locked trimers were resistant to such changes (Figure 5). However, we found that JR-FL SOSIP.664 was fully responsive to M48U1 and sCD4, whilst the effects on BG505 SOSIP.664 were markedly less pronounced, indicating a partial resistance of the BG505 Env to CD4, in agreement with a recent report (Kumar et al., 2017). Thus, the discrepant behavior of BG505 and JR-FL upon M48U1 triggering gave us an opportunity to comparatively evaluate the immunogenicity of trimers with different degrees of CD4 responsiveness, along with their respective interdomain-locked counterparts.
Figure 5. CD4-induced conformational changes disrupt the native-like antigenic state of unmutated, but not of interdomain-locked HIV-1 Env trimers.
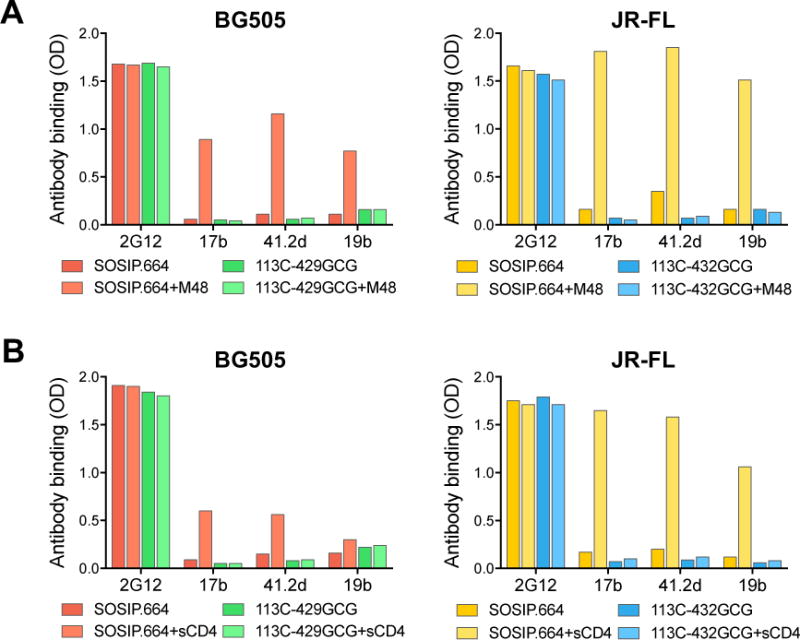
(A) Effect of the CD4-mimetic miniprotein M48U1 on the antigenicity of unmutated and interdomain-locked SOSIP.664 trimers from two HIV-1 strains (BG505, clade A, and JR-FL, clade B), as assessed by ELISA.
(B) Effect of soluble CD4 (sCD4) on the antigenicity of unmutated and interdomain-locked SOSIP.664 trimers from BG505 and JR-FL, as assessed by ELISA.
The results show mean optical density (OD) values at 450 nm for each mAb in the presence or absence of M48U1 or sCD4 from a representative experiment performed in duplicate wells.
Treatment with a functional CD4 mimic alters the immunogenicity of unmutated, but not of interdomain-locked HIV-1 Env trimers
Six groups of New Zealand white female rabbits (n=5 per group) were immunized with unmutated or interdomain-locked BG505 or JR-FL SOSIP.664 trimers: 4 groups received BG505-based trimers either alone (groups 1 and 3) or pre-complexed with M48U1 (groups 2 and 4); two groups received JR-FL-based trimers only in the presence of M48U1 (groups 5 and 6); and an additional group (group 7) received M48U1 alone. The rabbits received intramuscular injections of each Env trimer admixed with adjuvant (Adjuplex™) at weekly intervals during the first 3 weeks, and then 3 additional injections at 4-week intervals for a total of 6 immunizations (Figure 6A). The BG505 SOSIP.664 trimer, which lacks two conserved protective glycans at gp120 positions 241 and 289, was potently immunogenic in rabbits, eliciting high titers of trimer-binding antibodies, as measured by endpoint-dilution ELISA (Figure 6B). However, M48U1-complexed trimers elicited lower antibody titers, with a statistically significant difference against a glycan-repaired form of BG505 (241N 289N), which excludes antibodies directed against the 241/289 glycan hole, as well as against a heterologous trimer, JR-FL (also lacking the 241 glycan hole); conversely, M48U1-complexed trimers induced higher titers of antibodies against an autologous V3-loop peptide, compared to untriggered BG505 SOSIP.664, consistent with increased V3-loop exposure induced by the CD4 mimic (Figure 6B). In contrast, the interdomain-locked trimer (BG505 113C-429GCG SOSIP.664) elicited high antibody titers against both unmodified and glycan-repaired autologous and heterologous trimers, as well as low anti-V3 titers regardless of pre-treatment with the CD4 mimic (Figure 6B). Similar trends, although not statistically significant, were seen in rabbits immunized with M48U1-treated JR- FL-based trimers (Figure S7). These results provided in vivo evidence that CD4-induced conformational changes disrupt the immunogenicity of the HIV-1 Env trimers, while interdomain locking prevents such changes, thereby preserving the native-like pre-fusion antigenic state of the trimer.
Figure 6. Pre-exposure to a functional CD4 mimic alters the immunogenicity of unmutated, but not of interdomain-locked HIV-1 Env trimers.
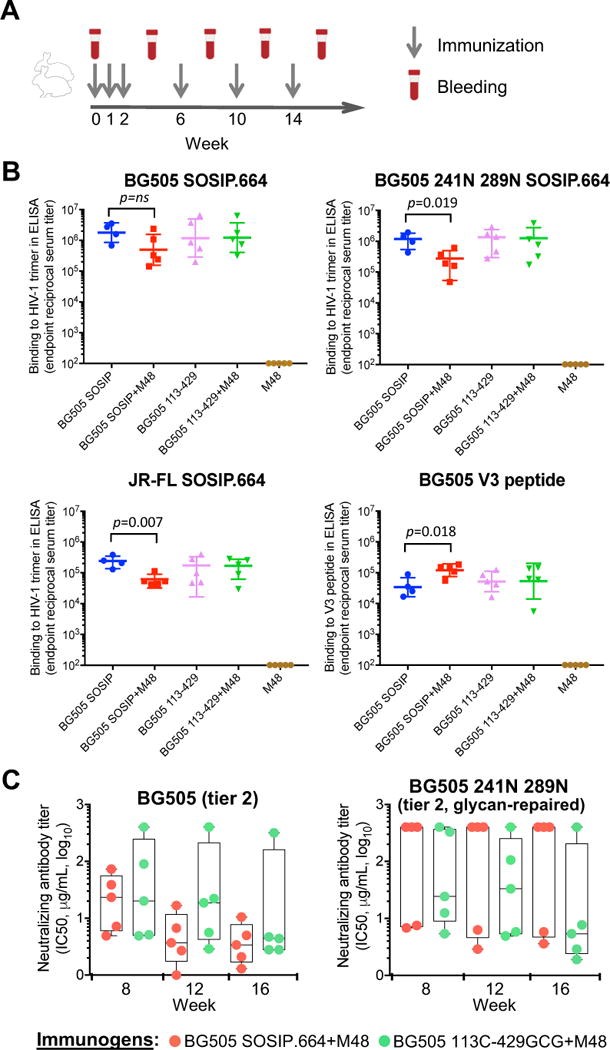
(A) Graphic scheme of the rabbit immunization protocol.
(B) Reactivity of immune rabbit sera with immobilized HIV-1 Env trimers or a V3 peptide, as tested by ELISA. Endpoint trimer- or peptide-binding titers are shown for week-16 sera obtained from rabbits immunized with unmutated or interdomain-locked HIV-1 BG505 SOSIP.664 trimers in the presence or absence of the CD4 mimic M48U1. Serially-diluted rabbit sera were tested on BG505 SOSIP.664, glycan-repaired BG505 (241N 289N) and JR-FL SOSIP.664 trimers, as well as on a BG505-derived linear V3 peptide. The data show endpoint titers for individual rabbits with geometric mean and standard deviation. P values were calculated using a two-tailed Mann-Whitney test using GraphPad Prism 7. Group 1 includes only 4 animals because one of them died from undetermined causes following the first immunization.
(C) Neutralization of HIV-1 pseudoviruses by purified serum IgG from rabbits immunized with unmutated or interdomain-stabilized HIV-1 BG505 SOSIP.664 trimers in the presence or absence of the CD4 mimic, M48U1. The data show half-maximal neutralizing IgG concentrations (IC50) for individual rabbits with means and minimum-to-maximum box and whiskers. Rabbit IgG were tested for neutralization of HIV-1 pseudoviruses bearing the wild-type BG505 Env (clade A, tier 2) or glycan-repaired BG505 Env (241N 289N, tier-2). Neutralization was assessed in TZM-bl cells. No statistical differences were found between the two groups of animals at any time point, as assessed using a two-tailed Mann-Whitney test. See also Tables S4 and S6.
Interdomain-locked trimers elicit neutralizing antibodies against autologous tier-2 viruses with a repaired glycan shield irrespective of treatment with a CD4 mimic
Sera from immunized rabbits were tested for the presence of neutralizing antibodies using the TZM-bl assay. To exclude the confounding effects of non-antibody factors present in rabbit serum, neutralization tests were performed using purified immunoglobulin G (IgG) fractions. Consistent with previous results (reviewed in Sanders and Moore, 2017), unmutated BG505 SOSIP.664 induced high titers of autologous neutralizing antibodies in all immunized rabbits regardless of M48U1 pre-treatment (9/9, 100%; IC50 range at week 16: 1.3-288 μg/mL of IgG; mean, 36.0 μg/mL, equivalent to ~1:350 serum dilution), but such antibodies predominantly targeted the 241/289 glycan hole, as shown by a marked drop in neutralization against glycan-repaired autologous virus (BG505 N241 N289), with only 4 out of 9 animals (44.4%) showing detectable neutralization; in contrast, the locked BG505 113C-429GCG trimer induced high titers of neutralizing antibodies against glycan-repaired virus in 8 out of 10 animals (80%; IC50 range at week 16: 1.1-54.5 μg/mL of IgG; mean, 10.4 μg/mL, equivalent to ~1:1200 serum dilution) (Table S4). Among animals receiving M48U1-complexed BG505-based trimers, neutralization of the glycan-repaired virus was detected in only in 2/5 (40.0%) immunized with the unmutated trimer, but in 4/5 (80.0%) immunized with the locked trimer, although the difference did not reach statistical significance, most likely reflecting the relative resistance of the BG505 Env to CD4-induced conformational changes (Figure 6C and Table S4). Further attesting to the only partial opening of the BG505 Env upon exposure to the CD4 mimic, relatively low levels of neutralization were observed against a heterologous tier-1b clade-B virus, BaL (Table S6).
Next, we analyzed the neutralizing antibody response in rabbits immunized with trimers based on JR-FL, which is fully responsive to CD4 and M48U1 (Figure 5). Previous studies had shown that, despite the lack of a protective glycan at position 197, the JR-FL SOSIP.664 trimer is a weak immunogen that elicits low and inconsistent titers of tier-2 autologous neutralizing antibodies (Feng et al., 2016; Ingale et al., 2016). In line with these observations, rabbits immunized with M48U1-treated unmutated JR-FL trimer developed only low and delayed titers of autologous neutralizing antibodies (IC50 range at week 16: 68.2->400 μg/mL of IgG; mean, >200 μg/mL, equivalent to <1:65 serum dilution); in contrast, rabbits immunized with the M48U1-treated locked trimer (JR-FL 113C-432GCG) developed significantly higher titers of neutralizing antibodies at all time points after week 8, with IC50 values at week 16 ranging between 7.1 and 65.3 μg/mL of IgG (mean, 41.1 μg/mL, equivalent to ~1:320 serum dilution) (Figure 7 and Table S5). To verify if these neutralizing antibodies were predominantly targeting the 197 glycan hole, we tested a glycan-repaired JR-FL pseudovirus (197N), but we obtained similar data, which were statistically significant at both week 12 and 16 (Figure 7 and Table S5), with week-16 IC50 values below 15 μg/mL (equivalent to >1:850 serum dilution) in 3 out of 5 animals, demonstrating that the elicited antibodies efficiently neutralized autologous tier-2 virus with a repaired glycan shield. At the same time, animals immunized with locked JR-FL trimers developed only low and inconsistent neutralizing antibody titers against the heterologous tier-1b virus, BaL, along with low titers of antibodies against the autologous V3-loop peptide, while all rabbits immunized with unlocked JR-FL developed high BaL-neutralizing titers with a significant difference at both week 12 and 16 (Figure 7). These results confirmed that M48U1 effectively triggered conformational changes in the unlocked JR-FL trimer, compromising its native antigenic state and leading to exposure of CD4-induced epitopes such as the V3 loop. In contrast, interdomain locking averted the CD4-induced conformational changes preserving the immunogenicity of the native-like trimer. No cross-reactive tier-2 neutralizing antibodies were detected in any of the study groups (Table S6). Altogether, these results demonstrated that interdomain-locked trimers have an improved immunogenicity over conventional trimers and prevent the disruption of immunogenicity induced by CD4 binding.
Figure 7. Interdomain-locked trimers elicit high titers of neutralizing antibodies against tier-2 virus with repaired glycan shield irrespective of pre-exposure to a CD4 mimic.
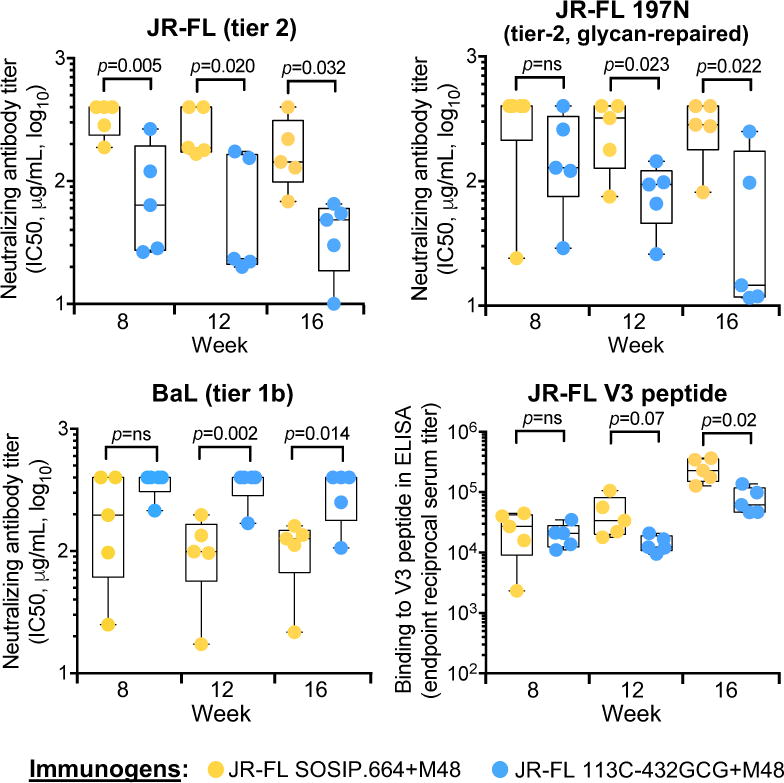
Neutralization of HIV-1 pseudoviruses by purified serum IgG from rabbits immunized with unmutated or interdomain-stabilized HIV-1 JR-FL SOSIP.664 trimers in the presence of the CD4 mimic, M48U1. The data show half-maximal neutralizing IgG concentrations (IC50) for individual rabbits with means and minimum-to-maximum box and whiskers. Rabbit IgG were tested for neutralization of HIV-1 pseudoviruses bearing the following Envs: wild-type JR-FL (clade B, tier 2), glycan-repaired JR-FL (tier-2) and BaL (clade B, tier 1b). Neutralization was assessed in TZM-bl cells. The ELISA reactivity of immune sera with an autologous V3-loop peptide was also tested. P values were calculated using a two-tailed Mann-Whitney test. See also Figure S7 and Table S5.
DISCUSSION
The present work provides a widely applicable template for the design of HIV-1 Env-based immunogens that overcomes two major obstacles in the development of a protective vaccine: i) the inherent flexibility of the HIV-1 Env trimer, which represents one of the most effective mechanisms of immune evasion enacted by this virus; and ii) the functional interaction with CD4, which causes a rapid and irreversible transition of the trimer from the pre-fusion state to a low-energy conformation that exposes vaccine-irrelevant epitopes at the expense of relevant epitopes targeted by bNAbs. Moreover, CD4 engagement may also reduce the efficacy of Env-based vaccines by other mechanisms, including immunogen sequestration by CD4+ T cells, which are not competent for antigen presentation, and occlusion of the CD4-binding site, which is a key immunogenic region targeted by protective antibodies. By introducing a covalent bond to reciprocally tie the two cardinal structural elements of gp120, the outer and inner domains, our design effectively stabilizes the HIV-1 Env in a native-like pre-fusion conformation associated with reduced structural flexibility and loss of biologically-relevant CD4 binding. Various strategies have been used by other groups with similar aims (de Taeye et al., 2015; Guenaga et al., 2017; Kulp et al., 2017; Kwon et al., 2015; Torrents de la Pena et al., 2017), but these trimer variants maintained the ability to bind CD4, resulting in at least some CD4-induced conformational changes. A substantial reduction of CD4-binding capacity has been reported in two recent studies, but neither the immunogenicity of these modified trimers nor the potential benefits deriving from the loss of CD4 interaction were investigated (Chuang et al., 2017; Kulp et al., 2017).
We solved the crystal structure of a stabilized trimer, which confirmed the formation of the expected interdomain bridge and identified a structural basis for CD4-binding impairment, revealing a potential molecular clash between the key F43 residue of CD4 and the side chain of M426 in gp120, which in the locked trimer is reoriented to partially occupy the F43-binding hydrophobic pocket, as opposed to its outward, non-interfering position in all reported CD4-bound structures. However, other factors may also contribute to hindering CD4 interaction. Binding of the HIV-1 Env to CD4 is an energetically unfavorable process that is hindered by conformational masking and imposes a substantial entropy penalty (Kwong et al., 2002). This implies that a tightly stabilized trimer should bind minimally, if at all, to CD4, while maintaining unaltered its binding activity to bNAbs such as VRC01, which have evolved to bypass conformational camouflage and engage the CD4 supersite with minimal entropic penalty. Thus, the inability of our mutants to bind CD4 is likely related to the structural rigidity achieved by mutual anchoring of the inner and outer domains of gp120. This concept is corroborated by recent studies on the HIV-1 entry inhibitor BMS-626529, which seems to interfere with CD4 binding through fixation of the α1-helix to the β20-β21 loop (Herschhorn et al., 2017; Pancera et al., 2017).
Since our locked mutants are stabilized in an antigenically native-like pre-fusion conformation that selectively presents the epitopes recognized by protective bNAbs, we studied their effectiveness as potential components of an HIV-1 vaccine in a pre-clinical rabbit model. To experimentally validate the advantage of using CD4-resistant Env immunogens, we devised a unique protocol whereby the trimers were pre-complexed with a CD4 miniprotein, M48U1, in order to mimic the encounter with CD4 that would occur in human vaccination. Indeed, animals immunized with conventional SOSIP.664 trimers pre-treated with M48U1 developed lower titers of neutralizing antibodies against tier-2 autologous virus, compared to those immunized with M48U1-untreated trimers, and higher titers of antibodies against tier-1 HIV-1 or the V3 loop, providing in vivo evidence for the detrimental effects of CD4 binding on Env-based immunization. In contrast, locked trimers induced high titers of neutralizing antibodies against tier-2 autologous viruses from two HIV-1 clades irrespective of M48U1 treatment. Moreover, the elicited antibodies did not target the glycan holes present on the immunizing trimers, as they neutralized viruses with a fully repaired glycan shield, a result that was only sporadically achieved in previous studies (Kulp et al., 2017; Sanders et al., 2015; Zhou et al., 2017). This result was more pronounced and statistically significant with the JR-FL trimer, which undergoes full conformational changes upon M48U1 engagement, unlike BG505 that was partially resistant to the CD4 mimic. The epitopes recognized by the elicited antibodies were not characterized in this study, although the lack of neutralization breadth suggests that they are not conserved across multiple tier-2 isolates. Of note, however, our locked JR-FL trimer induced markedly higher titers of autologous neutralizing antibodies than those previously reported for the conventional JR-FL trimer, which is considered a weak immunogen (Feng et al., 2016; Ingale et al., 2016), further supporting the improved immunogenicity of interdomain-locked trimers.
In summary, the present study provides a strategy to stabilize the HIV-1 Env trimer in order to steadily and selectively present to the immune system vaccine-relevant epitopes recognized by bNAbs. The lack of CD4 binding ensures that our locked immunogens would not undergo antigenic modifications or be lost by CD4+ T-cell sequestration after injection into human vaccine recipients. Moreover, the interdomain locks do not interfere with binding of potent bNAbs that recognize the CD4 supersite, maintaining this critical immunogenic site fully accessible for stimulation of specific antibody responses. Finally, our interdomain-locking design can be applied to both soluble trimers and full-length native Env spikes from diverse HIV-1 strains and genetic subtypes, thus providing a common template for the development of increasingly effective HIV-1 Env-based vaccines.
STAR★METHODS
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Monoclonal anti-HIV-1 Gp120 2G12 | NIH AIDS Reagent Program | Cat# 1476 |
| Monoclonal anti-HIV-1 Gp120 b12 | NIH AIDS Reagent Program | Cat# 2640 |
| Monoclonal anti-HIV-1 Gp120 VRC01 | NIH AIDS Reagent Program | Cat# 12033; RRID: AB_2491019 |
| Monoclonal anti-HIV-1 Gp120 F105 | NIH AIDS Reagent Program; | Cat# 857 |
| Monoclonal anti-HIV-1 Gp120 PGT121 | John R. Mascola, VRC, NIH | N/A |
| Monoclonal anti-HIV-1 Gp120 PG9 | John R. Mascola, VRC, NIH | N/A |
| Monoclonal anti-HIV-1 Gp120 PG16 | John R. Mascola, VRC, NIH | N/A |
| Monoclonal anti-HIV-1 Gp120 PGT145 | John R. Mascola, VRC, NIH | N/A |
| Monoclonal anti-HIV-1 Gp120 CH01 | NIH AIDS Reagent Program | Cat# 12561; RRID: AB_2491055 |
| Monoclonal anti-HIV-1 Gp120 CAP256.09 (VRC26.03) | John R. Mascola, VRC, NIH | N/A |
| Monoclonal anti-HIV-1 Gp120 PGT151 | John R. Mascola, VRC, NIH | N/A |
| Monoclonal anti-HIV-1 Gp120 35O22 | NIH AIDS Reagent Program | Cat# 12586 |
| Monoclonal anti-HIV-1 Gp120 8ANC195 | Michel C. Nussenzweig, Rockefeller University | RRID: AB_2491037 |
| Monoclonal anti-HIV-1 Gp120 447-52D | NIH AIDS Reagent Program | Cat# 4030 |
| Monoclonal anti-HIV-1 Gp120 19b | NIH AIDS Reagent Program | Cat# 11436 |
| Monoclonal anti-HIV-1 Gp120 48d | NIH AIDS Reagent Program | Cat# 1756 |
| Monoclonal anti-HIV-1 Gp120 412d | John R. Mascola, VRC, NIH | N/A |
| Goat anti-human IgG-PE | SouthernBiotech | Cat# 2040-09 |
| HRP-conjugated anti-human IgG, Fcr fragment Ab | Jackson ImmunoResearch | Cat# 109-035-008 |
| Peroxidase AffiniPure Donkey Anti-Rabbit IgG | Jackson ImmunoResearch | Cat# 711-035-152 |
| Bacterial and Virus Strains | ||
| NEB 5 – alpha Competent E. coli | New England BioLabs | Cat# C2987H |
| Biological Samples | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| Lectin from Galanthus nivalis | SIGMA-ALDRICH | Cat# L8275-5MG |
| BG505.T332N.SOSIP.664 gp140 trimer | This study | N/A |
| BG505.T332N.SOSIP.664.113C.429GCG gp140 trimer | This study | N/A |
| BG505.T332N.SOSIP.664.113C.431GCG gp140 trimer | This study | N/A |
| JR-FL.SOSIP.664 gp140 trimer | This study | N/A |
| JR-FL.SOSIP.664.113C.432GCG gp140 trimer | This study | N/A |
| DU422.SOSIP.664 gp140 trimer | This study | N/A |
| DU422.SOSIP.664.113C.432GCG gp140 trimer | This study | N/A |
| M48U1 | Loic Martin, CEA (Martin et., 2003) | N/A |
| Recombinant Human sCD4 Protein | R&D Systems | Cat# 514-CD |
| Recombinant CD4-Ig2 | NIH AIDS Reagent Program | Cat# 11780 |
| Critical Commercial Assays | ||
| Luciferase 1000 Assay System | Promega | Cat# E4550 |
| QuickChange II Site-Directed Mutagenesis Kit | Agilent | Cat# 200524 |
| Deposited Data | ||
| BG505.113C-429GCG bound to PGT122 Fab and 35O22 Fab | This study | PDB: 6DE7; Deposition ID: D_1000233685 |
| Experimental Models: Cell Lines | ||
| Human: HEK 293T, female | ATCC | Cat# CRL-3216 |
| Human: HeLa-derived TZM-bl, female | NIH AIDS Reagent Program | Cat# 8129-442 |
| Human: HEK 293FS (FreeStyle), female | Thermo Fisher | Cat# R79007 |
| Experimental Models: Organisms/Strains | ||
| New Zealand white rabbits, female | Purchased from USDA-licensed breeders by Pocono Farm & Laborator y | Animals were identified by implanted ear clips |
| Oligonucleotides | ||
| Recombinant DNA | ||
| Plasmids pPPI4-BG505.T332N.SOSIP.664 | John P. Moore, Weill-Cornell Medical College (Sanders et al., 2013) | N/A |
| Plasmids pPPI4-DU422.K295N.D386N.SOSIP.664 | John P. Moore, Weill-Cornell Medical College (Julien et al., 2015) | N/A |
| Plasmids pVRC8400-JR-FL.SOSIP.664 | Peter Kwong, NIH (Stewart-Jones et al., 2016) | N/A |
| Plasmids HIV-1 BG505 env expression vector | NIH AIDS Reagent Program | Cat# 11518 |
| Plasmids pCAGGS HIV-1 JR-FL gp160 expression plasmids | James Binley, San Diego Biomedical Research Institute (Binley et al., 2003) | N/A |
| Plasmids pCDNA3.1 HIV-1 clone BaL.26 | NIH AIDS Reagent Program | Cat# 11518 |
| Plasmids pSG3 ∆env | NIH AIDS Reagent Program | Cat# 11051 |
| Plasmids Panel of Global HIV 1 Env Clones | NIH AIDS Reagent Program | Cat# 12670 |
| Software and Algorithms | ||
| FlowJo v.9.9.4 | FlowJo, LLC | https://www.www.flowjo.com; RRID: SCR_008520 |
| IMGT/V-QUEST | International ImMunoGeneTics Information System; Marie-Paule Lefranc (marie-paule.lefranc@igh.cnrs.fr), University of Montpellier, France |
www.imgt.org; RRID: SCR_012780 |
| Pymol | The PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC. | https://pymol.org/2/ |
| Cloanalyst | Thomas B. Kepler (tbkepler@bu.edu), Boston University | http://www.bu.edu/computationalimmunology/research/software/ |
| HKL2000 Suite 6 | HKL Research | www.hkl-xray.com |
| Phenix | Paul D. Adams (pdadams@lbl.gov), Lawrence Berkeley Laboratory | www.phenix-online.org; RRID: SCR_014224 |
| Crystallographic Object-Oriented Toolkit (COOT) Software | Paul Emsley (paul.emsley@mrc-lmb.cam.ac.uk), Oxford University | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/; RRID: SCR_014222 |
| WebLogo | Steven Brenner, Berkeley (Crooks et al., 2004) | http://weblogo.berkeley.edu |
| PRISM 7 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| Other | ||
| TM Adjuvant: Adjuplex | Sigma-Aldrich | A0362 |
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents may be directed to and will be fulfilled by the lead contact, Paolo Lusso (plusso@niaid.nih.gov).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Ethics Statement
All animal studies were conducted following the guidelines of the US National Research Council Committee for the Care and Use of Laboratory Animals. The protocol was reviewed and approved by the Animal Care and Use Committee of Pocono Rabbit Farm & Laboratory, Canadensis, PA.
Rabbits
Two-month-old New Zealand White female rabbits were used for immunizations. The animals were housed in an AAALAC-accredited state-of-the-art indoor research animal facility at Pocono Rabbit Farm & Laboratory that meets or exceeds regulatory guidelines. They were maintained on a 12 hour light/dark cycle with unlimited access to water and food. Animal care was provided by specialized personnel following local, state and federal policies in accordance with the standards in the Guide for the Care and Use of Laboratory Animals, USDA, and OLAW.
Cells and Viruses
Human embryonic kidney (HEK) 293T cells, of presumable female origin as determined by the abundance of X chromosomes and absence of Y chromosomes, were obtained from the American Type Culture Collection (ATCC) and maintained in DMEM medium (Thermo Fisher) containing 10% fetal bovine serum (FBS, HyClone) at 37°C. HEK 293 free-style (FS) cells, a variant of HEK 293T, were originally purchased by the VRC, NIH, from Thermo Fisher Scientific and maintained in 293 Free-Style Expression Medium (Thermo Fisher) at 37°C. TZM-bl cells, obtained by stable transfection of the HeLa cell line originally derived from a female patient with cervical cancer, were obtained through the NIH AIDS Reagent Program and grown in DMEM with 10% FBS at 37°C. HIV-1 pseudoviruses expressing wild-type or mutated gp160 from HIV-1 BG505-T332N and derivative mutants were produced in HEK 293T cells by co-transfecting Env-expressing plasmids with a backbone plasmid, pSG3 env, expressing a full-length HIV-1 clone with a defective Env gene (obtained through the NIH AIDS Reagent Program). All cell lines were authenticated at the source and regularly monitored for consistency of appearance, growth efficiency and biological behavior.
METHOD DETAILS
Peptides and Proteins
A linear peptide representing the entire V3 loop sequence from BG505 gp120 (TRPNNNTRKSIRIGPGQAFYATGDIIGDIRQAH) was produced using solid-phase synthesis technology by Bio-Synthesis, Inc., Lewisville, TX. The peptide was purified to >95% and extensively quality-controlled by HPLC and Mass Spectrometry Electro-Spray. The peptide was dissolved in 10 nM NH4OH at 10 mg/mL and stored frozen until use. Human Fc-chimeric CD4-Ig (plasmid provided by Michael Farzan) was produced in 293FS cells and purified by protein-A affinity chromatography; two-domain sCD4 was obtained from the NIH AIDS Reagent Program; 4-domain sCD4 was purchased from R&D Systems; the CD4-M48U1 mini-protein was synthesized at Pepscan Presto Inc. (Lelystad, The Netherlands) by solid phase peptide synthesis and purified after refolding by RP-HPLC.
Mutagenesis, Protein Expression and Purification
Site-directed mutagenesis was carried out using QuikChange II Site-Directed Mutagenesis Kit (Agilent Technologies). The BG505 and JR-FL SOSIP.664 trimers were expressed by co-transfecting the relevant plasmids (BG505 from John P. Moore; JR-FL from the VRC) with a plasmid expressing the cellular protease furin into human embryonic kidney (HEK) 293 free style (FS) cells. Cell-free supernatants were harvested after 7 days, passed through a 0.22 μm filter, and loaded onto a Galanthus nivalis (GNA) lectin column (Vector laboratories). After repeated washing with PBS, bound proteins were eluted with 1 M methyl α-D-mannopyranoside in PBS, followed by dialysis toward PBS. Dialyzed samples were concentrated to 2 ml using Amicon Ultra-15 centrifugal filter units (MWCO 50,000, Millipore) and applied to a PBS pre-equilibrated Superdex 200 column. The peak corresponding to the trimeric form was collected and concentrated, followed by a second round of Superdex 200 purification and passed through a mAb 447-52D affinity column for negative selection. Finally, purified trimers were concentrated to 0.5-1 mg/ml and stored at −80°C.
Sequence Conservation Analysis
Sequence alignment of segments of the α1-helix region and of the β20-β21 loop of gp120 from the all the available group-M HIV-1 and SIVcpz isolates (including both subtypes A-K and recombinant forms) was obtained from the Los Alamos HIV database (http://www.hiv.lanl.gov). The sequence conservation logo was created using WebLogo (http://weblogo.berkeley.edu).
Mass-Spectrometry Analysis
High-resolution LC-MS and disulfide bond analysis of BG505 113C-429GCG and 113C-431GCG were performed as described (Go et al., 2014). Briefly, ~75 μg of each recombinant Env trimer protein were alkylated in the dark at room temperature with 4-vinylpyridine (10-fold molar excess) for 1 hour, then subjected to enzymatic deglycosylation with PNGase F (1 uL, 500,000 units/mL) at 37°C for one week. During both steps, the protein concentration was maintained at a minimum of 1 mg/mL in pH 6.5 citrate buffer. Subsequently, the protein was digested with trypsin overnight at 37°C at an enzyme-to-protein ratio of 1:30. To ensure that all results were reproducible, the sample preparation and analysis procedure was replicated on a different day, using fresh protein from the same batch. High-resolution LC/MS experiments were performed using an Acquity UPLC system (Waters, Milford, MA), coupled to an LTQ Velos Pro Orbitrap Mass Spectrometer, equipped with ETD, (ThermoScientific, San Jose, CA). The experiments employed mobile phases of 99.9% deionized water (with 0.1% formic acid) and 99.9 % CH3CN (with 0.1% formic acid), henceforth referred to as Mobile Phase A and Mobile Phase B, respectively. The digested samples (5 μL, 7 μM) were injected onto a C18 PepMap™ 300 column (300 μm i.d. × 15 cm, 300 Å Thermo Scientific, Sunnyvale, CA) at a flow rate of 5 μL/min, and the following liquid gradient was used: 5% B for 5 min, followed a linear increase to 40% B in 50 min, then a linear increase to 90% B in 10 min. The column was held at 95% B for 10 minutes before re-equilibration; a wash-and-blank run always followed every analytical run, in order to prevent carry-over. Both high-resolution MS data, along with low-resolution MS/MS data, were acquired in the positive ion mode throughout the chromatographic run; CID and ETD data were acquired by the data-dependent analysis routine embedded in the instrument software. Normalized collision energy of 35% was used for CID; for ETD, the ion-ion reaction between the precursor ion and fluoranthane was set at AGC target value of 2×105 and 100 msec ion-ion reaction time. To improve ETD efficiency, supplemental activation was turned on. Data analysis of the raw file was performed manually. All plausible disulfide-bonded peptides, including those that could arise from aberrant disulfide bonding or scrambling, were searched for in the data file. Each disulfide-bonded species verified to be present was confirmed by both high-resolution MS data along with MS/MS data.
Negative-Staining Electron Microscopy
Proteins were diluted to a concentration of about 0.02 mg/ml, adsorbed to freshly glow-discharged carbon-film grids for 15 seconds, washed with a buffer containing 10 mM HEPES, pH 7 and 150 mM NaCl, and stained with 0.7% uranyl formate. Images were collected semi-automatically with SerialEM (Mastronarde, 2005) on an FEI Tecnai T20 electron microscope operating at 200 kV and equipped with a 2k × 2k Eagle CCD camera at a nominal magnification of 100,000 and a pixel size of 0.22 nm. To estimate the fractions of aberrant trimers, particles were picked automatically and manually from micrographs of BG505 SOSIP.664, 113C-429GCG, and 113C-431GCG using e2boxer from the EMAN2 software package (Tang et al., 2007) and subjected to reference-free two-dimensional classification in EMAN2, SPIDER (Frank et al., 1996) (operation: AP C), and Xmipp (Scheres et al., 2008). The obtained class averages were assigned manually to either the native or aberrant trimer conformation based on expected sharp, symmetrical, propeller-like views typical of correctly folded HIV glycoproteins. The fractions of incorrectly formed trimers were then calculated based on numbers of particles forming the “native” and “aberrant” classes. The three software packages produced similar ratios, which were averaged to produce the final values.
For three-dimensional reconstructions, extended datasets were collected, each containing equal numbers of micrographs obtained at tilt angles of 0°, 15°, 30°, and 45°. Tilted micrographs were divided into two vertical slices each for more accurate contrast transfer function (CTF) correction. The initial datasets for BG505 SOSIP.664, 113C-429GCG and 113C-431GCG contained 25856, 22430, and 21234 particles, respectively. EMAN2 was used to estimate CTF parameters and perform CTF correction by phase flipping. Reference-free 2D class averages were obtained using SPIDER (Shaikh et al., 2008) by rotationally and translationally aligning all particles in a dataset and performing correspondence analysis and classification. Selected SPIDER class averages were then used to generate initial 3D maps in EMAN2 with C3 symmetry imposed. These maps were filtered to a resolution of 40 Å and used as initial models for three-dimensional reconstruction and refinement using reference projections in SPIDER. The C3 symmetry was used during the reconstruction and refinement. A limit was placed on the number of particles assigned to a single orientation to overcome the strong preferential orientation typical of HIV glycoproteins adsorbed to carbon. A total of 18142, 10370, and 10884 particles contributed to the final maps of BG505 SOSIP.664, 113C-429GCG and 113C-431GCG, respectively. The resolutions of the final maps were 17.6 Å, 18.9 Å, and 18.8 Å, respectively, when a Fourier shell correlation (FSC) threshold of 0.5 was used. The maps were visualized using UCSF Chimera (Pettersen et al., 2004).
To measure the volume of the internal cavity of the trimer glycoproteins, the maps were first low-pass filtered to a resolution of 19 Å, and the map thresholds were chosen such that the three maps had identical volumes as calculated using UCSF Chimera. The contrast of the original maps was then inverted (SPIDER’s operation “NEG”). Each inverted map was loaded in UCSF Chimera along with the original map, and the threshold of the inverted map was adjusted such that the positive density corresponding to the internal cavity slightly exceeded the size of the internal cavity of the original map. The inverted map was then segmented using the watershed algorithm implemented in Segger (Pintilie et al., 2010), a component of UCSF Chimera, and the density of the segment(s) corresponding to the internal cavity was extracted. The volume of this density was then measured in UCSF Chimera after adjusting the threshold such that this volume fit exactly in the internal cavity of the original map.
Crystal Structure
For molecular complex preparation, 3-fold molar excess of Fab fragments from antibodies PGT122 and 35O22 were added to a solution of size-exclusion chromatography (SEC)-purified BG505 SOSIP.113C.429GCG at room temperature. The ternary complex was passed over an SEC column, and the fractions of the complex were pooled, concentrated to 10 mg/ml, and screened against 576 crystallization conditions or flash frozen in liquid nitrogen and kept at −80°C until further use. Diffraction data were processed with the HKL2000 suite. The overall resolution was determined as the highest resolution for which the completeness was greater than 50% and the I/sigI was greater than 2.0. Thus, for SOSIP.113C.429GCG ternary complexes, the overall nominal resolution was 4.3 Å, however meaningful diffraction data was observed to 4.1 Å. The crystal diffraction data were then assessed for anisotropy through use of the Diffraction Anisotropy Server (http://services.mbi.ucla.edu/anisoscale/), which indicates the resolution at which F/sigma drops below 3.0 along a, b, and c axes; these were 5.1 Å, 5.1 Å, and 4.1 Å. Other investigators have sometimes determined overall resolutions by the following equation: Reseff (high resolution) (completeness)(−1/3), where Reseff is effective resolution. For the BG505 113C-429GCG complex, the untruncated data had a completeness of 75.6%, and the overall resolution is calculated as Reseff (4.1)(0.756)(−1/3) =4.5 Å; We also report the CC1/2 value (Pearson correlation coefficient between intensities averaged in two subsets of data) of the highest resolution shell at 0.733; CC1/2 values of 0.15 are reported to be significant.
The solution structure was obtained by molecular replacement with Phaser using the BG505 SOSIP.664 ternary complex structure (PDB accession number 4TVP) as a search model. Refinement was carried out with Phenix. Model building was carried out with Coot. The Ramachandran plot was determined by MolProbity. Data collection and refinement statistics are shown in Table S2. Structural figures were generated using Pymol.
Differential Scanning Calorimetry
A high-precision differential scanning VP-DSC microcalorimeter (GE Healthcare/MicroCal) was employed to measure the heat capacity of the trimers. In brief, samples were diluted to 0.3 mg/ml with PBS. Thermal denaturation scans were performed from 30°C to 110°C at a rate of 1°C/min.
Expression of Full-Length HIV-1 gp160
The gp160 genes from different HIV-1 strains were mutagenized as described above for the SOSIP trimer. HEK 293T cells were used to express cell-surface gp160 for flow cytometry and pseudovirus production or human CD4 for trimer-binding experiments. The cells were seeded into 6-well plates at 500,000 cells/well in 2 ml of DMEM containing 10% fetal bovine serum (DMEM 10%). After overnight incubation at 37 °C in humidified atmosphere with 5% CO2, the culture medium was replaced with 1.5 ml of fresh 10% DMEM. Transfection complexes were prepared by mixing 2 μg DNA with 6 μl Mirus293 Transfection Reagent (Mirus) in Opti-MEM medium (Gibco) to a final volume of 100 μl and incubated for 15 min at room temperature (RT). The cells were transfected by adding the DNA-Mirus293 complex dropwise to each well and harvested after 48-60 hours.
Flow Cytometry
HEK 293T cells expressing gp160 or CD4 were harvested by mechanical shaking and pipetting, washed with PBS and incubated with anti-CD4 or anti-gp120 antibodies (5 μg/ml) or serially diluted human Fc-chimeric CD4-Ig (produced in 293FS cells and purified by protein-A affinity chromatography; plasmid provided by Michael Farzan) at RT for 30 min. After washing with PBS twice, the cells were incubated with PE-conjugated sheep anti-human IgG (eBioscence) or goat anti-human IgG (Southern Biotech) at RT for 15 min. The cells were then washed once with PBS, fixed in 2% paraformaldehyde (PFA) and analyzed on a BD FACSCanto™ (BD Biosciences). Data analysis was performed using the FlowJo software.
Enzyme Immunoassays
All the antibodies and HIV-1 Env proteins used in ELISA experiments were diluted in 1X casein solution (R&D Systems) in PBS. All samples were washed 3 times after each step with 1X wash buffer (R&D Systems). For assessing endpoint binding titers in rabbit sera, 96-well ELISA plates (Corning) were coated with 4 μg/ml of Lectin Galanthus nivalis (Sigma) at 4 °C overnight. After blocking with 1X casein in PBS, purified trimers (5 μg/ml) were added and incubated for 2 hours at RT. After washing, serially diluted rabbit sera were added and incubated for 1 hour at RT. After washing, horseradish peroxidase (HRP)-conjugated mouse anti Rabbit IgG (LifeScience) was added for 1 hour at RT. The reaction was revealed by incubation with 50 μl of substrate (R&D Systems) for 5 min before addition of the stop solution. Light absorption was measured at a wavelength of 450 nm. All samples were tested in duplicate wells. To evaluate antibody binding, the trimers were captured on lectin-coated ELISA plates, and anti-gp120 antibodies (2 μg/ml or serial dilutions starting from 5-20 g/ml) were added to the wells and incubated for 1 hour, followed by 1-hour incubation with HRP-conjugated goat anti-human IgG antibody (Jackson ImmunoResearch); all incubations were done at RT. The subsequent steps were the same as described above. For all the mutants, relative binding compared to the unmutated trimer was calculated.
Surface Plasmon Resonance Analysis
Single-cycle kinetics surface plasmon resonance (SPR) analysis of 2-domain sCD4 binding to the BG505 SOSIP.664 and locked SOSIP.664 trimers was performed at 25 °C on a Biacore T200 (GE Healthcare) as described previously (Kwon et al., 2015). Briefly, MAb 2G12 was immobilized onto four flow cells of a CM5 sensor to ~2,000 response units (RU) using the amine-coupling method. Purified BG505 SOSIP.664 trimers were injected to two sample flow cells at a flow rate of 5 μl/min for two minutes, and captured to 300-400 RU. The other two flow cells were left blank as reference. Series dilutions of sCD4 (180 nM, 90 nM, 45 nM, 22.5 nM, 11.25 nM for BG505 SOSIP.664; 2000 nM, 1000 nM, 500 nM, 250 nM and 125 nM for locked BG505 SOSIP trimers) were injected to both the reference and sample flow cells at 50 μl/min in a single cycle, starting from the lowest concentration. Thirty minutes of dissociation phase was allowed after the last sCD4 injection. The same injections were carried out using HBS-EP buffer in order to obtain a reference curve. Data were fitted to 1:1 Langmuir model with Biacore T200 evaluation software.
Pseudovirus Preparation, Infectivity and Neutralization Assays
HIV-1 pseudoparticles expressing WT or mutated gp160 from HIV-1 BG505-T332N and derivative mutants were produced in HEK 293T cells by co-transfecting Env-expressing plasmids with a backbone plasmid, pSG3 env, expressing a full-length HIV-1 clone with a defective Env gene. To produce the pseudoviruses, 2 μg of each Env-expressing plasmid and 4 μg of the backbone plasmid were mixed in Opti-MEM medium (Gibco), and 24 μl of Mirus293 Transfection Reagent was added, followed by 15-min incubation at RT. DNA-Mirus293 complex was then added to the cells and incubated overnight at 37°C. After replacing the culture medium with 2 ml fresh 10% DMEM, the cells were incubated for 48 hours, and the supernatants containing pseudoviruses were harvested by centrifugation. For infection, TZM-bl cells (AIDS Reagent Program) were seeded into 96-well flat bottom plates at 10,000 cells/well in 100 μl 10% DMEM. After 30 min at 37°C, serial 3-fold dilutions of the pseudovirus stocks were added to the cells in a total volume of 200 μl/well. Reporter gene activation was detected 2 days later after removing 150 μl media and adding the 40 μl per well of Luciferase Assay Reagent (Promega). The cell lysates were transferred to lumino-plates and measured with a luminometer (PerkinElmer). Relative Light Units (RLU) were recorded, and the final values were normalized against the values obtained with the WT Env set at 100%. All the samples were tested in duplicate wells.
For neutralization assays using purified IgG from immunized rabbits, infectious HIV-1 pseudoviruses were prepared, titrated and used to infect TZM-bl target cells as described previously (Montefiori, 2009). Calculation of half-maximal inhibitory concentrations (IC50) and two-tailed t-tests were performed using Graphpad Prism 7 (Prism). All graphs were also plotted using Prism 7.
Rabbit Immunization
All rabbit immunizations were performed at Pocono Rabbit Farm & Laboratory. Two-month-old New Zealand White female rabbits were immunized intramuscularly at the hind leg muscles. Injections consisted of 40 μg of unmutated or interdomain-stabilized SOSIP.664 trimers pre-complexed or not with 10 μg of M48U1 miniprotein formulated in 250 μl of PBS with 150 μl of the adjuvant Adjuplex (Sigma). The immunizations were administered as two separate injections of 200 μl into each quadriceps muscle. Immunizations were performed at weeks 0, 1, 2, 6, 10 and 14. Blood draws for immunologic assessment included a pre-bleed week 0 and blood draws performed 2 weeks after each immunization starting at week 2. Animal experiments were reviewed and approved by the Animal Care and Use Committee of Pocono, and all animals were housed and cared for in accordance with local, state and federal policies. Collected sera were heat inactivated for 1 hour at 56°C before analysis.
Rabbit IgG Isolation
Rabbit sera were filtered through 0.22μm PVDF syringe drive filter (Millipore) and diluted with same volume of PBS (total volume is about 10 ml) and mixed with 5 ml of PBS pre-equilibrated Protein A-sepharose resin (GE) at RT for 3 hours. After washing the columns with PBS, bound IgG were eluted by IgG-elution buffer (ThermoFisher), followed by dialysis against PBS and concentration to ~ 50 mg/ml using Amicon Ultra-15 centrifugal filter units (MWCO 50,000, Millipore).
QUANTIFICATION AND STATISTICAL ANALYSIS
Calculation of half-maximal inhibitory concentrations (IC50) and statistical tests were performed using the Graphpad Prism 7 (Prism) software. All graphs were also plotted using Prism 7. Most data from in vitro experiments are presented as averages ± SEM from duplicate or triplicate samples, as indicated in the respective figure or table legends. For in vivo studies, 7 groups of 5 rabbits each (n = 5) were immunized; each data point in the figures denotes one rabbit tested at the indicated time point. Statistically significant differences were determined by Mann-Whitney U test or paired t test. Statistical details can be found directly in the figures or in the corresponding figure legends.
DATA AND SOFTWARE AVAILABILITY
Crystal structure coordinates have been deposited and validated by the PDB Protein Data Bank (PDB: 6DE7).
Supplementary Material
Highlights.
-
-
Interdomain locks stabilize soluble and membrane-bound HIV-1 Env trimers
-
-
Locked trimers maintain a native-like antigenicity but do not bind CD4
-
-
Locked trimers are resistant to CD4-induced disruption of immunogenicity
-
-
Immunized rabbits neutralized glycan-repaired autologous tier-2 viruses
Acknowledgments
We thank John P. Moore and Albert Cupo for providing plasmids for expression of the BG505, B41 and DU422 SOSIP.664 trimers and furin; Dennis R. Burton, Mark Connors and James E. Robinson for the gift of anti-Env monoclonal antibodies; Michael Farzan for the plasmid expressing human Fc-chimeric CD4-Ig; James Binley for the gift of glycan-repaired JR-FL N197 plasmid; Anthony S. Fauci for critical reading of the manuscript; and the AIDS Reagent Program for providing the reagents indicated in the Methods section. This research was supported by the Intramural Programs of the Division of Intramural Research, NIAID, NIH, and of the Vaccine Research Center, NIAID, NIH, by a strategic grant (to PL) from the Office of AIDS Research (OAR), NIAID, NIH, and by grant R01AI094797 (to HD).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental Information includes eight figures and six tables and can be found with this article online at http://doi-
AUTHORS CONTRIBUTIONS
P.Z. and P.L. designed the research; P.Z., H.G., Y.L., Q.L., T.A., A.K., M.P., D.G., F.U., C.G., R.C., H.M., R.C., C.G., K.M., G.Y.C. and P.L. performed biological and biochemical assays and analyzed the data; J.G. and P.D.K. performed crystallization and solved the structure; Y.T. performed NS-EM analysis and 3D-structure reconstruction; E.P.G. and H.D. performed Mass-Spectrometry analysis, L.M. provided the M48U1 miniprotein and participated in the study design; B.D., F.S., M.S.M. and E.A.B. participated in the early conception of the project and contributed to the study design; M.D. performed bioinformatics computational and structural analyses; J.R.M. contributed to the study design; P.Z. and P.L. wrote the manuscript; all of the authors contributed to manuscript revisions.
DECLARATION OF INTERESTS
PL and PZ have applied for a patent describing HIV-1 envelope trimers stabilized in a native-like, non-CD4-binding conformation (U.S. Provisional Patent Application No. 62/306,006, filed on March 9, 2016, entitled “RECOMBINANT HIV-1 ENVELOPE PROTEINS AND THEIR USE”). The authors declare no other competing interests.
References
- Bartesaghi A, Merk A, Borgnia MJ, Milne JL, Subramaniam S. Prefusion structure of trimeric HIV-1 envelope glycoprotein determined by cryo-electron microscopy. Nat Struct Mol Biol. 2013;20:1352–1357. doi: 10.1038/nsmb.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton DR, Ahmed R, Barouch DH, Butera ST, Crotty S, Godzik A, Kaufmann DE, McElrath MJ, Nussenzweig MC, Pulendran B, et al. A blueprint for HIV vaccine discovery. Cell Host Microbe. 2012;12:396–407. doi: 10.1016/j.chom.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang GY, Geng H, Pancera M, Xu K, Cheng C, Acharya P, Chambers M, Druz A, Tsybovsky Y, Wanninger TG, et al. Structure-based design of a soluble prefusion-closed HIV-1 Env trimer with reduced CD4 affinity and improved immunogenicity. J Virol. 2017;91:e02268. doi: 10.1128/JVI.02268-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks ET, Tong T, Chakrabarti B, Narayan K, Georgiev IS, Menis S, Huang X, Kulp D, Osawa K, Muranaka J, et al. Vaccine-elicited tier 2 HIV-1 neutralizing antibodies bind to quaternary epitopes involving glycan-deficient patches proximal to the CD4 binding site. PLoS Pathog. 2015;11:e1004932. doi: 10.1371/journal.ppat.1004932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Taeye SW, Ozorowski G, Torrents de la Pena A, Guttman M, Julien JP, van den Kerkhof TL, Burger JA, Pritchard LK, Pugach P, Yasmeen A, et al. Immunogenicity of stabilized HIV-1 envelope trimers with reduced exposure of non-neutralizing epitopes. Cell. 2015;163:1702–1715. doi: 10.1016/j.cell.2015.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excler JL, Michael NL. Lessons from HIV-1 vaccine efficacy trials. Curr Opin HIV AIDS. 2016;11:607–613. doi: 10.1097/COH.0000000000000312. [DOI] [PubMed] [Google Scholar]
- Feng Y, Tran K, Bale S, Kumar S, Guenaga J, Wilson R, de Val N, Arendt H, DeStefano J, Ward AB, et al. Thermostability of well-ordered HIV spikes correlates with the elicitation of autologous tier 2 neutralizing antibodies. PLoS Pathog. 2016;12:e1005767. doi: 10.1371/journal.ppat.1005767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finzi A, Pacheco B, Xiang SH, Pancera M, Herschhorn A, Wang L, Zeng X, Desormeaux A, Kwong PD, Sodroski J. Lineage-specific differences between human and simian immunodeficiency virus regulation of gp120 trimer association and CD4 binding. J Virol. 2012;86:8974–8986. doi: 10.1128/JVI.01076-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. Spider and web: Processing and visualization of images in 3d electron microscopy and related fields. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- Go EP, Hua D, Desaire H. Glycosylation and disulfide bond analysis of transiently and stably expressed clade C HIV-1 gp140 trimers in 293T cells identifies disulfide heterogeneity present in both proteins and differences in O-linked glycosylation. J Proteome Res. 2014;13:4012–4027. doi: 10.1021/pr5003643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenaga J, Garces F, de Val N, Stanfield RL, Dubrovskaya V, Higgins B, Carrette B, Ward AB, Wilson IA, Wyatt RT. Glycine substitution at helix-to-coil transitions facilitates the structural determination of a stabilized subtype C HIV envelope glycoprotein. Immunity. 2017;46:792–803. doi: 10.1016/j.immuni.2017.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes BF, Gilbert PB, McElrath MJ, Zolla-Pazner S, Tomaras GD, Alam SM, Evans DT, Montefiori DC, Karnasuta C, Sutthent R, et al. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. New Engl J Med. 2012;366:1275–1286. doi: 10.1056/NEJMoa1113425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herschhorn A, Gu C, Moraca F, Ma X, Farrell M, Smith AB, 3rd, Pancera M, Kwong PD, Schon A, Freire E, et al. The beta20-beta21 of gp120 is a regulatory switch for HIV-1 Env conformational transitions. Nat Commun. 2017;8:1049. doi: 10.1038/s41467-017-01119-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CC, Tang M, Zhang MY, Majeed S, Montabana E, Stanfield RL, Dimitrov DS, Korber B, Sodroski J, Wilson IA, et al. Structure of a V3-containing HIV-1 gp120 core. Science. 2005;310:1025–1028. doi: 10.1126/science.1118398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humes D, Emery S, Laws E, Overbaugh J. A species-specific amino acid difference in the macaque CD4 receptor restricts replication by global circulating HIV-1 variants representing viruses from recent infection. J Virol. 2012;86:12472–12483. doi: 10.1128/JVI.02176-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingale J, Stano A, Guenaga J, Sharma SK, Nemazee D, Zwick MB, Wyatt RT. High-density array of well-ordered HIV-1 spikes on synthetic liposomal nanoparticles efficiently activate B cells. Cell Rep. 2016;15:1986–1999. doi: 10.1016/j.celrep.2016.04.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien JP, Cupo A, Sok D, Stanfield RL, Lyumkis D, Deller MC, Klasse PJ, Burton DR, Sanders RW, Moore JP, et al. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science. 2013;342:1477–1483. doi: 10.1126/science.1245625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klasse PJ, LaBranche CC, Ketas TJ, Ozorowski G, Cupo A, Pugach P, Ringe RP, Golabek M, van Gils MJ, Guttman M, et al. Sequential and simultaneous immunization of rabbits with HIV-1 envelope glycoprotein SOSIP.664 trimers from clades A, B and C. PLoS Pathog. 2016;12:e1005864. doi: 10.1371/journal.ppat.1005864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulp DW, Steichen JM, Pauthner M, Hu X, Schiffner T, Liguori A, Cottrell CA, Havenar-Daughton C, Ozorowski G, Georgeson E, et al. Structure-based design of native-like HIV-1 envelope trimers to silence non-neutralizing epitopes and eliminate CD4 binding. Nat Commun. 2017;8:1655. doi: 10.1038/s41467-017-01549-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R, Ozorowski G, Kumar V, Holden LG, Shrivastava T, Patil S, Deshpande S, Ward AB, Bhattacharya J. Characterization of a stable HIV-1 B/C recombinant, soluble, and trimeric envelope glycoprotein (Env) highly resistant to CD4-induced conformational changes. J Biol Chem. 2017;292:15849–15858. doi: 10.1074/jbc.M117.803056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon YD, Pancera M, Acharya P, Georgiev IS, Crooks ET, Gorman J, Joyce MG, Guttman M, Ma X, Narpala S, et al. Crystal structure, conformational fixation and entry-related interactions of mature ligand-free HIV-1 Env. Nat Struct Mol Biol. 2015;22:522–531. doi: 10.1038/nsmb.3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong PD, Doyle ML, Casper DJ, Cicala C, Leavitt SA, Majeed S, Steenbeke TD, Venturi M, Chaiken I, Fung M, et al. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature. 2002;420:678–682. doi: 10.1038/nature01188. [DOI] [PubMed] [Google Scholar]
- Kwong PD, Wyatt R, Majeed S, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structures of HIV-1 gp120 envelope glycoproteins from laboratory-adapted and primary isolates. Structure. 2000;8:1329–1339. doi: 10.1016/s0969-2126(00)00547-5. [DOI] [PubMed] [Google Scholar]
- Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Ozorowski G, Ward AB. Cryo-EM structure of a native, fully glycosylated, cleaved HIV-1 envelope trimer. Science. 2016;351:1043–1048. doi: 10.1126/science.aad2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Wang S, Kong R, Ding W, Lee FH, Parker Z, Kim E, Learn GH, Hahn P, Policicchio B, et al. Envelope residue 375 substitutions in simian-human immunodeficiency viruses enhance CD4 binding and replication in rhesus macaques. Proc Natl Acad Sci U S A. 2016;113:E3413–3422. doi: 10.1073/pnas.1606636113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. Molecular architecture of native HIV-1 gp120 trimers. Nature. 2008;455:109–113. doi: 10.1038/nature07159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Acharya P, Dolan MA, Zhang P, Guzzo C, Lu J, Kwon A, Gururani D, Miao H, Bylund T, et al. Quaternary contact in the initial interaction of CD4 with the HIV-1 envelope trimer. Nat Struct Mol Biol. 2017;24:370–378. doi: 10.1038/nsmb.3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyumkis D, Julien JP, de Val N, Cupo A, Potter CS, Klasse PJ, Burton DR, Sanders RW, Moore JP, Carragher B, et al. Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science. 2013;342:1484–1490. doi: 10.1126/science.1245627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin L, Stricher F, Misse D, Sironi F, Pugniere M, Barthe P, Prado-Gotor R, Freulon I, Magne X, Roumestand C, et al. Rational design of a CD4 mimic that inhibits HIV-1 entry and exposes cryptic neutralization epitopes. Nat Biotechnol. 2003;21:71–76. doi: 10.1038/nbt768. [DOI] [PubMed] [Google Scholar]
- Mascola JR, Haynes BF. HIV-1 neutralizing antibodies: understanding nature’s pathways. Immunol Rev. 2013;254:225–244. doi: 10.1111/imr.12075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- McCoy LE, van Gils MJ, Ozorowski G, Messmer T, Briney B, Voss JE, Kulp DW, Macauley MS, Sok D, Pauthner M, et al. Holes in the glycan shield of the native HIV envelope are a target of trimer-elicited neutralizing antibodies. Cell Rep. 2016;16:2327–2338. doi: 10.1016/j.celrep.2016.07.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montefiori DC. Measuring HIV neutralization in a luciferase reporter gene assay. Methods Mol Biol. 2009;485:395–405. doi: 10.1007/978-1-59745-170-3_26. [DOI] [PubMed] [Google Scholar]
- Munro JB, Gorman J, Ma X, Zhou Z, Arthos J, Burton DR, Koff WC, Courter JR, Smith AB, 3rd, Kwong PD, et al. Conformational dynamics of single HIV-1 envelope trimers on the surface of native virions. Science. 2014;346:759–763. doi: 10.1126/science.1254426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozorowski G, Pallesen J, de Val N, Lyumkis D, Cottrell CA, Torres JL, Copps J, Stanfield RL, Cupo A, Pugach P, et al. Open and closed structures reveal allostery and pliability in the HIV-1 envelope spike. Nature. 2017;547:360–363. doi: 10.1038/nature23010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancera M, Lai YT, Bylund T, Druz A, Narpala S, O’Dell S, Schon A, Bailer RT, Chuang GY, Geng H, et al. Crystal structures of trimeric HIV envelope with entry inhibitors BMS-378806 and BMS-626529. Nat Chem Biol. 2017;13:1115–1122. doi: 10.1038/nchembio.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancera M, Majeed S, Ban YE, Chen L, Huang CC, Kong L, Kwon YD, Stuckey J, Zhou T, Robinson JE, et al. Structure of HIV-1 gp120 with gp41-interactive region reveals layered envelope architecture and basis of conformational mobility. Proc Natl Acad Sci U S A. 2010;107:1166–1171. doi: 10.1073/pnas.0911004107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancera M, Zhou T, Druz A, Georgiev IS, Soto C, Gorman J, Huang J, Acharya P, Chuang GY, Ofek G, et al. Structure and immune recognition of trimeric pre-fusion HIV-1 Env. Nature. 2014;514:455–461. doi: 10.1038/nature13808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauthner M, Havenar-Daughton C, Sok D, Nkolola JP, Bastidas R, Boopathy AV, Carnathan DG, Chandrashekar A, Cirelli KM, Cottrell CA, et al. Elicitation of robust tier 2 neutralizing antibody responses in nonhuman primates by HIV journaltrimer immunization using optimized approaches. Immunity. 2017;46:1073–1088e1076. doi: 10.1016/j.immuni.2017.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF chimera - A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Pintilie G, Zhang J, Goddard T, Chiu W, Gossard D. Quantitative analysis of cryo-EM density map segmentation by watershed and scale-space filtering, and fitting of structures by alignment to regions. J Struct Biol. 2010;170:427–438. doi: 10.1016/j.jsb.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders RW, Derking R, Cupo A, Julien JP, Yasmeen A, de Val N, Kim HJ, Blattner C, de la Pena AT, Korzun J, et al. A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog. 2013;9:e1003618. doi: 10.1371/journal.ppat.1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders RW, Moore JP. Native-like Env trimers as a platform for HIV-1 vaccine design. Immunol Rev. 2017;275:161–182. doi: 10.1111/imr.12481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders RW, van Gils MJ, Derking R, Sok D, Ketas TJ, Burger JA, Ozorowski G, Cupo A, Simonich C, Goo L, et al. HIV-1 neutralizing antibodies induced by native-like envelope trimers. Science. 2015;349:aac4223. doi: 10.1126/science.aac4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SHW, Nuñez-Ramirez R, Sorzano COS, Carazo JM, Marabini R. Image processing for electron microscopy single-particle analysis using XMIPP. Nat Protoc. 2008;3:977–990. doi: 10.1038/nprot.2008.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaikh TR, Gao H, Baxter WT, Asturias FJ, Boisset N, Leith A, Frank J. Spider image processing for single-particle reconstruction of biological macromolecules from electron micrographs. Nat Protoc. 2008;3:1941–1974. doi: 10.1038/nprot.2008.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma SK, de Val N, Bale S, Guenaga J, Tran K, Feng Y, Dubrovskaya V, Ward AB, Wyatt RT. Cleavage-independent HIV-1 Env trimers engineered as soluble native spike mimetics for vaccine design. Cell Reports. 2015;11:539–550. doi: 10.1016/j.celrep.2015.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart-Jones GB, Soto C, Lemmin T, Chuang GY, Druz A, Kong R, Thomas PV, Wagh K, Zhou T, Behrens AJ, et al. Trimeric HIV-1-Env structures define glycan shields from clades A, B, and G. Cell. 2016;165:813–826. doi: 10.1016/j.cell.2016.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan JT, Sulli C, Nilo A, Yasmeen A, Ozorowski G, Sanders RW, Ward AB, Klasse PJ, Moore JP, Doranz BJ. High-throughput protein engineering improves the antigenicity and stability of soluble HIV-1 envelope glycoprotein SOSIP trimers. J Virol. 2017;91 doi: 10.1128/JVI.00862-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. Eman2: An extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Torrents de la Pena A, Julien JP, de Taeye SW, Garces F, Guttman M, Ozorowski G, Pritchard LK, Behrens AJ, Go EP, Burger JA, et al. Improving the Immunogenicity of native-like HIV-1 envelope trimers by hyperstabilization. Cell Rep. 2017;20:1805–1817. doi: 10.1016/j.celrep.2017.07.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagury JF, Cantalloube H, Bernard J, Mornon JP, Bizzini B, Zagury D. Striking identity between HIV-1 envelope glycoprotein gp120 and its CD4 receptor. Lancet. 1992;340:483–484. doi: 10.1016/0140-6736(92)91803-g. [DOI] [PubMed] [Google Scholar]
- Zhou T, Doria-Rose NA, Cheng C, Stewart-Jones GBE, Chuang GY, Chambers M, Druz A, Geng H, McKee K, Kwon YD, et al. Quantification of the impact of the HIV-1-glycan shield on antibody elicitation. Cell Rep. 2017;19:719–732. doi: 10.1016/j.celrep.2017.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


