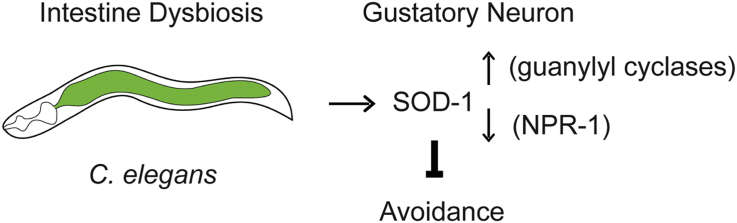
Abstract
Superoxide dismutase, an enzyme that converts superoxide into less-toxic hydrogen peroxide and oxygen, has been shown to mediate behavioral response to pathogens. However, it remains largely unknown how superoxide dismutase is regulated in the nervous system amid pathogen-induced gut dysbiosis. Although there are five superoxide dismutases in C. elegans, our genetic analyses suggest that SOD-1 is the primary superoxide dismutase to mediate the pathogen avoidance response. When C. elegans are fed a P. aeruginosa diet, the lack of SOD-1 contributes to enhanced lethality. We found that guanylyl cyclases GCY-5 and GCY-22 and neuropeptide receptor NPR-1 act antagonistically to regulate SOD-1 expression in the gustatory neuron ASER. After C. elegans ingests a diet that contributes to high levels of oxidative stress, the temporal regulation of SOD-1 and the SOD-1–dependent response in the gustatory system demonstrates a sophisticated mechanism to fine-tune behavioral plasticity. Our results may provide the initial glimpse of a strategy by which a multicellular organism copes with oxidative stress amid gut dysbiosis.
Graphical abstract
1. Introduction
Gut dysbiosis is caused by an imbalance of beneficial and harmful microbes in the intestine [1], [2]. In addition to digestive tract–related morbidity [2], [3], [4], [5], [6], recent studies have revealed that dysbiosis of gut microbiota plays a role in emotional and cognitive behaviors [7], [8], [9], [10] and is found in patients with neurodegenerative diseases [11], [12], [13].
In the laboratory, C. elegans is often reared on a petri dish that contains a lawn of non-pathogenic Escherichia coli OP50 [14]. Recent studies reveal that pathogenic bacteria Pseudomonas aeruginosa, which are frequently found in the habitats of nematodes [15], [16], can colonize the intestine of C. elegans [17], [18], [19], [20]. Switching C. elegans from a diet of E. coli to a diet of P. aeruginosa creates a condition that mimics gut dysbiosis [21], [22], [23]. The change in the microbial composition in the intestine activates host responses [22], [24], [25], [26]. For example, intestinal accumulation of P. aeruginosa activates the production of reactive oxygen species (ROS) [22], [27], [28], [29]. The ROS burst further elevates the expression of antioxidant enzymes, including superoxide dismutase SOD-1 [22]. SOD-1 is an enzyme that converts superoxide into less-toxic hydrogen peroxide and oxygen [30], [31], [32]. Indeed, animals carrying a sod-1 deletion elicit a strong aversive response to P. aeruginosa, presumably due to their reduced capacity to ameliorate elevated ROS [22].
ASE neurons are a pair of chemosensory neurons defined by their ability to detect water-soluble cues and transmit this information to evoke a specific behavioral response [33]. Therefore, they are often regarded as the gustatory (taste) neurons in C. elegans. Although ASE neuron cell bodies are symmetrically positioned in the lateral ganglia region of C. elegans brain, the molecular compositions of left (ASEL) and right (ASER) gustatory neurons are not identical [34], [35]. ASEL and ASER gustatory neurons express distinct members of a putative chemoreceptor gene family and respond in distinct manners to different cues [36], [37]. For instance, animals devoid of the ASER neuron elicited a heightened aversive response to P. aeruginosa [22]. In contrast, animals lacking the ASEL neuron did not show the heightened pathogen avoidance response. SOD-1 is present in the nervous system, and SOD-1 expression is elevated in the ASER neuron by P. aeruginosa. It is possible that ASER-specific chemosensory receptors activate SOD-1, but experimental data have yet to support this notion.
Another poorly understood phenomenon concerns the following observation: After extended feeding on P. aeruginosa, ASER-specific SOD-1 elevation becomes diminished. Reduction of SOD-1 is coupled with the initiation of aversive behavior to P. aeruginosa [22]. However, the mechanism of SOD-1 down-regulation after extended P. aeruginosa feeding remains unknown. It has been shown that deletion in neuropeptide receptor NPR-1 results in extended feeding on P. aeruginosa [21], [26]. Neuropeptide receptor NPR-1 is C. elegans neuropeptide Y receptor, which regulates food satiety and stress response [21], [26], [38], [39]. The NPR-1 receptor is present in the ASER neuron [40]. We hypothesize that NPR-1 may play a role in modulating the SOD-1–dependent behavioral response to P. aeruginosa by down-regulating SOD-1 expression in the ASER neuron.
There are three zinc–copper superoxide dismutase isoforms (SOD-1, SOD-4, and SOD-5) and two manganese superoxide dismutase isoforms (SOD-2 and SOD-3) in C. elegans [41]. Based on their subcellular localization, the zinc–copper isoforms are further classified as cytoplasmic (i.e., SOD-1 and SOD-5) and extracellular/secreted (i.e., SOD-4) superoxide dismutases. Previous studies suggest these isoforms are expressed rather ubiquitously, including in the nervous system. For example, SOD-1 appears to be expressed in most cells of the worm [42], [43]. SOD-5 expression is inducible and has been detected in a small subset of neurons [42]. SOD-1 has been previously suggested to act in the gustatory neuron ASER to regulate C. elegans behavioral response to P. aeruginosa [22]. Do additional superoxide dismutases in C. elegans mediate the behavioral response to P. aeruginosa? And what are the molecular mechanisms that regulate the superoxide dismutase–dependent response to pathogen-induced gut dysbiosis?
In this study, we found that SOD-1 and SOD-5 are both present in the gustatory neuron ASER. Our genetic analyses suggest that SOD-1 plays a primary role in regulating the behavioral response to P. aeruginosa. SOD-1 is induced in the nervous system and in the intestine by oxidative stress. Lack of SOD-1 contributes to enhanced lethality after C. elegans is fed the P. aeruginosa diet. ASER-specific guanylyl cyclases GCY-5 and GCY-22 activate SOD-1 expression, whereas neuropeptide receptor NPR-1 promotes the reduction of SOD-1 in the ASER neuron. Therefore, we have identified the regulatory mechanisms that contribute to the activation and reduction of superoxide dismutase. Our results also suggest that zinc–copper superoxide dismutase plays a major role in mediating the behavioral response amid pathogen-induced gut dysbiosis.
2. Results
2.1. Superoxide dismutase isoform SOD-1 plays a primary role in behavioral response to P. aeruginosa
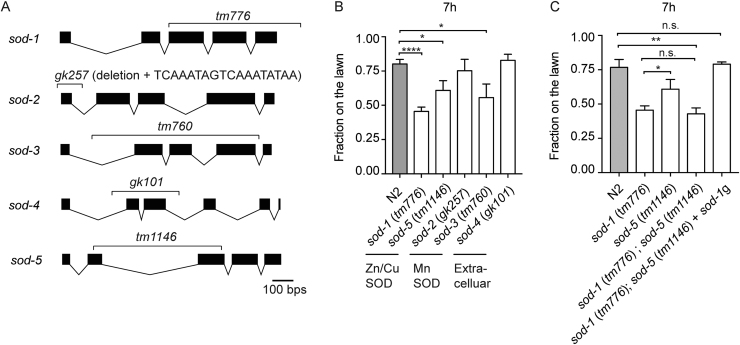
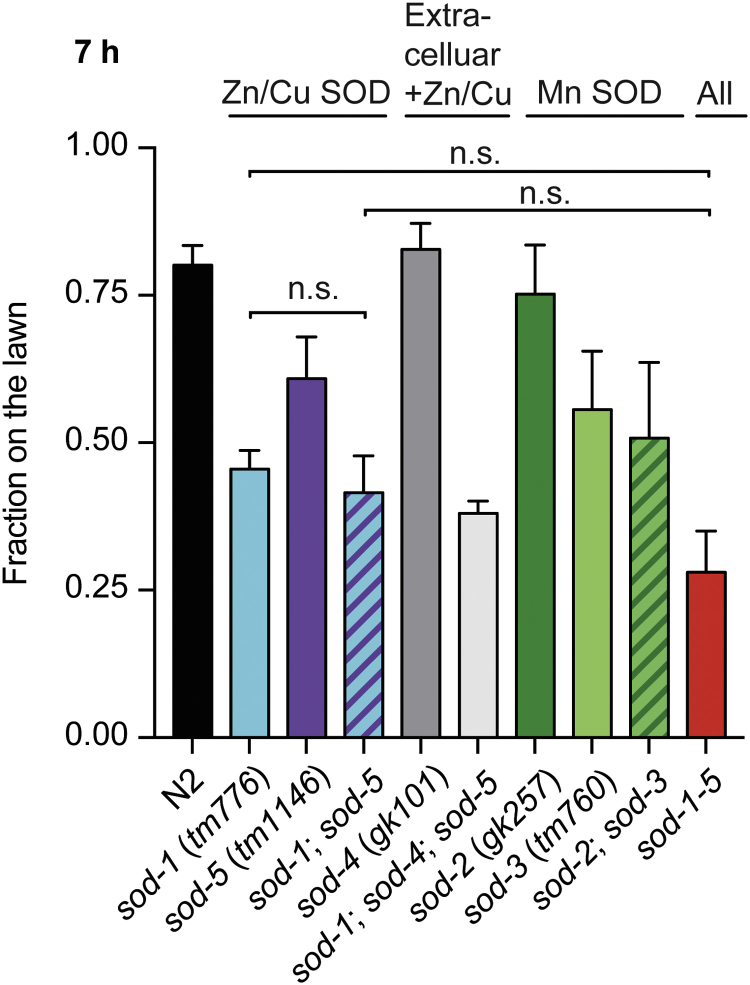
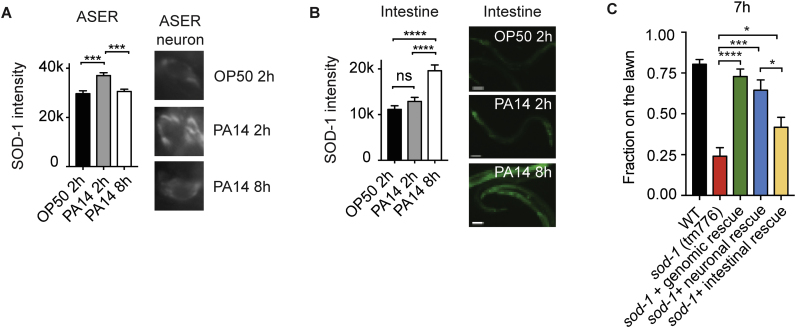
We began our study by investigating the role of additional superoxide dismutases in C. elegans. Deletion in sod-1 contributes to a heightened pathogen avoidance response [22]. Therefore, we first obtained deletion mutations of sod-2, sod-3, sod-4, and sod-5 (Fig. 1A). We then exposed the mutant animals to a lawn of P. aeruginosa on a petri dish and compared their P. aeruginosa lawn avoidance phenotypes (Fig. 1B). We found that sod-2 (gk257) and sod-4 (gk101) elicited phenotypes similar to wild type. In contrast, we found that sod-3 (tm760) and sod-5 (tm1146) showed enhanced pathogen avoidance responses at 7 h. However, the P. aeruginosa avoidance responses of sod-3 (tm760) and sod-5 (tm1146) were not as strong as that of sod-1 (tm776) (Fig. 1B). Since both zinc–copper superoxide dismutase SOD-1 and SOD-5 play roles in regulating C. elegans avoidance response to P. aeruginosa, we generated a sod-1 and sod-5 double mutant and fed the double mutant P. aeruginosa. We found that sod-1 (tm776); sod-5 (tm1146) elicited a pathogen avoidance response similar to that elicited by sod-1 (tm776) alone (Fig. 1C). We then introduced a 2.9 kb sod-1 genomic fragment that encompasses the coding and the 5′ and 3′ regulatory regions of sod-1 [22] into the sod-1 (tm776); sod-5 (tm1146) double mutant. We were able to rescue the pathogen avoidance response of the sod-1 (tm776); sod-5 (tm1146) double mutant to the wild type level (Fig. 1C). Because a genomic fragment of sod-1 rescues the pathogen avoidance response of sod-1; sod-5, SOD-5 may act as an auxiliary isoform to regulate C. elegans pathogen response.
Fig. 1.
Deletions in superoxide dismutase isoforms elicit heightened avoidance response to P. aeruginosa PA14. (A) The deleted regions of the sod-1, sod-2, sod-3, sod-4, and sod-5 mutations used in this study are indicated. Black box indicates an exon. Connected lines indicate the region of an intron. (B) After 7 h exposure to P. aeruginosa, sod-1 (tm776), sod-5 (tm1146), and sod-3 (tm760) single mutants elicit heightened avoidance responses to a lawn of P. aeruginosa. (C) sod-1(tm776); sod-5(tm1146) double mutant does not elicit enhanced pathogen avoidance response compared to sod-1 (tm776) single mutant alone. The behavioral response of sod-1 (tm776); sod-5 (tm1146) is rescued by a 2.9 kb sod-1 genomic DNA fragment (sod-1g). Error bar represents s.e.m. * represents p < 0.05. ** represents p < 0.01. **** p represents< 0.0001. n.s.: not significant. Determined by one-way ANOVA Tukey's multiple comparison.
Finally, we compared the pathogen avoidance phenotype of a sod-1; sod-2; sod-3; sod-4; sod-5 quintuple mutant to that of the sod-1; sod-5 double mutant and to the sod-1 single mutant. We found there are no statistically significant differences between the quintuple mutant, the sod-1; sod-5 double mutant, and the sod-1 single mutant with respect to avoidance responses to P. aeruginosa (Supplementary Fig. 1). Since the quintuple mutant does not further enhance sod-1 single mutant phenotype, this suggests that SOD-1 is the primary superoxide dismutase to regulate the behavioral avoidance response to P. aeruginosa.
2.2. SOD-1 and SOD-5 are present in overlapping amphid sensory neurons
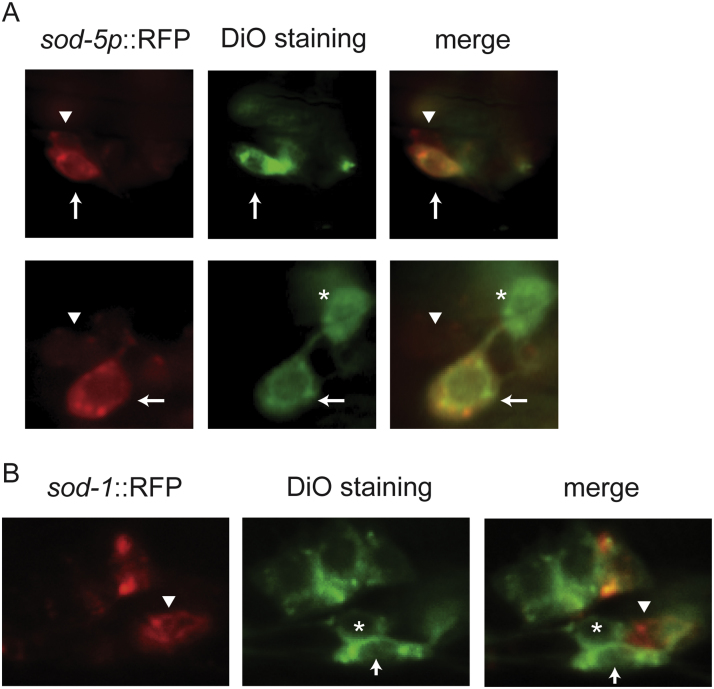
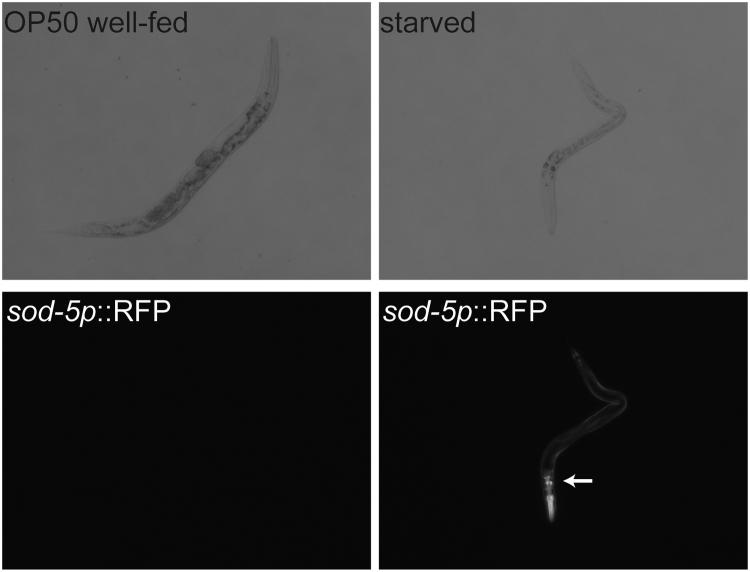
Our data indicate that SOD-5 may act as a redundant zinc–copper superoxide dismutase in the context of pathogen avoidance. If this is the case, SOD-5 is likely present in a similar set of neuronal cells to SOD-1. We therefore sought to investigate the expression pattern of SOD-5. We first generated an integrated reporter strain that contains an RFP reporter driven by the promoter of sod-5 (sod-5p::RFP). We examined the fluorescence signals of sod-5p::RFP and detected no obvious sod-5p::RFP fluorescence signals when the animals were reared under normal growth condition. However, when the animals were starved, we found sod-5p::RFP fluorescence signals in the pharynx and in the nervous system (Supplementary Fig. 2). The fluorescence signals of sod-5p::RFP are present in the neuron ganglion posterior to the anterior bulb of the pharynx. Several of the neurons have projections extending anteriorly to the tip of the nose, a morphology that resembles amphid sensory neurons. To determine if SOD-5 is present in the amphid sensory neurons, we performed dye-filing experiments using starved sod-5p::RFP animals. We found that sod-5p::RFP fluorescence is present in the ASJ neurons. We also observed a weak signal of sod-5p::RFP in the ASE neuron pair (Fig. 2A). To further compare the expression pattern of SOD-5 to that of SOD-1, we performed dye-filling experiments using the sod-1p::sod-1 cDNA::RFP reporter strain. Similar to prior results [22], we found that SOD-1 is present in the ASER neuron (Fig. 2B). These results suggest that SOD-1 and SOD-5 are both present in ASER gustatory neurons.
Fig. 2.
SOD-1 and SOD-5 are present in gustatory neuron ASE. Integrated sod-1p::sod-1 cDNA::RFP and sod-5p::RFP reporters were stained by DiO fluorescence dye. (A) Fluorescence micrographs of sod-5p::RFP transgenic animals. sod-5p::RFP signals are present in ASE (arrowhead, weak expression) and ASJ (arrow), but not in ASH (star) sensory neurons. Green: DiO staining of amphid sensory neurons. Anterior is to the right. Dorsal is at the top. (B) Fluorescence micrographs of sod-1p::sod-1 cDNA::RFP transgenic animals. SOD-1::RFP signals are present in ASER (arrowhead) and ASJ (arrow), but not in ASH (star) sensory neurons. Green: DiO staining of amphid sensory neurons. Anterior is to the left. Dorsal is at the top.
Previous investigation suggests that SOD-5 expression is inducible [42]. Consistent with the prior result, our data show that sod-5p::RFP is at undetectable levels under the normal growth condition. We found that the SOD-5 reporter is induced in the nervous system amid starvation and that SOD-5 is present at low levels in the ASE neuron pair. Previous work has also shown that SOD-1 contributes almost 80% of the total sod mRNA expression and 80% of the total SOD activities [42]. Together with our results, we reason that SOD-5 likely plays an auxiliary role in mediating the zinc–copper superoxide dismutase–dependent response to P. aeruginosa.
2.3. SOD-1 is induced by reactive oxygen species in the nervous system and intestine
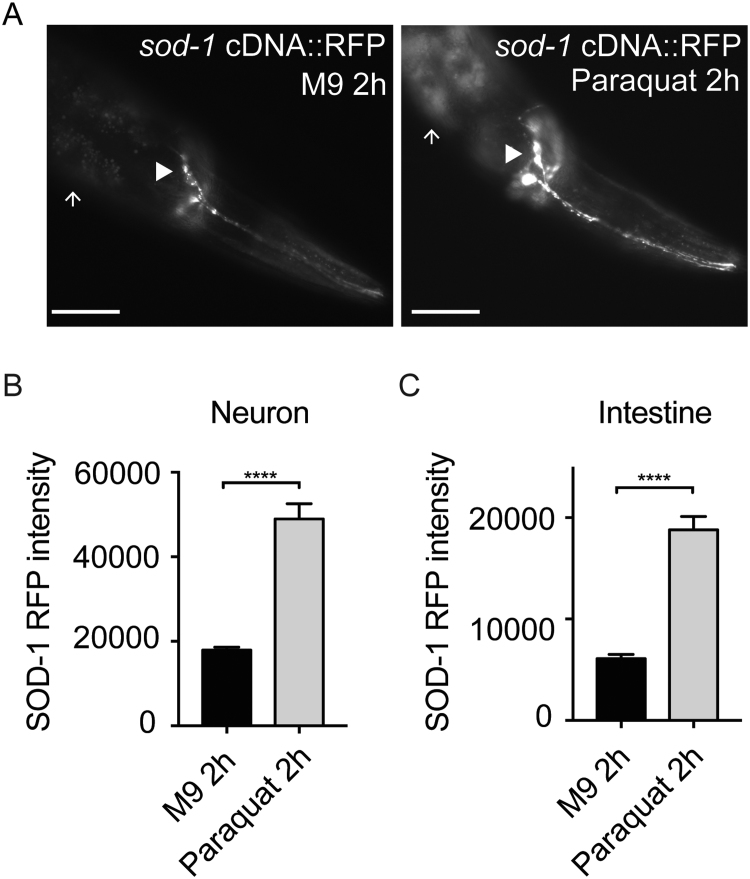
SOD-1 is an enzyme that converts superoxide into less-toxic hydrogen peroxide and oxygen. We hypothesized that SOD-1 expression is activated by an increase in ROS. To test this, we exposed the SOD-1 fluorescence reporter strain (sod-1p::sod-1 cDNA::RFP) to 100 μM paraquat and measured the fluorescence intensity of SOD-1::RFP. We found that SOD-1 is elevated by paraquat in the nervous system and in the intestine (Fig. 3). We also tested whether exposing C. elegans to P. aeruginosa induces SOD-1 expression. Similar to previous report [22], we found that the fluorescence signals of SOD-1::GFP are elevated in the ASER neuron after 2 h P. aeruginosa exposure. In contrast, intestinal SOD-1::GFP is elevated after 8 h (Supplementary Fig. 3A and B). These results suggest that SOD-1 may act in the nervous system and/or in the intestine in response to an increase in pathogen-induced oxidative stress.
Fig. 3.
Reactive oxygen species induce SOD-1 expression in the nervous system and intestine. (A) Fluorescence micrographs of integrated sod-1p::sod-1 cDNA::RFP transgenic animals. SOD-1::RFP level is elevated in the nervous system and intestine by a 2 h treatment with 100 μM paraquat. Arrow: intestine. Arrowhead: ASER neuron. Scale bar represents 50 µm. (B–C) Average fluorescence intensity of SOD-1::RFP in the nervous system (ASER neuron) and intestine. For each experiment in (B) and (C), N > 30. **** represents p < 0.0001. Determined by one-way ANOVA Tukey's multiple comparison. Error bar: s.e.m.
2.4. SOD-1 alleviates lethality triggered by pathogen-induced oxidative stress
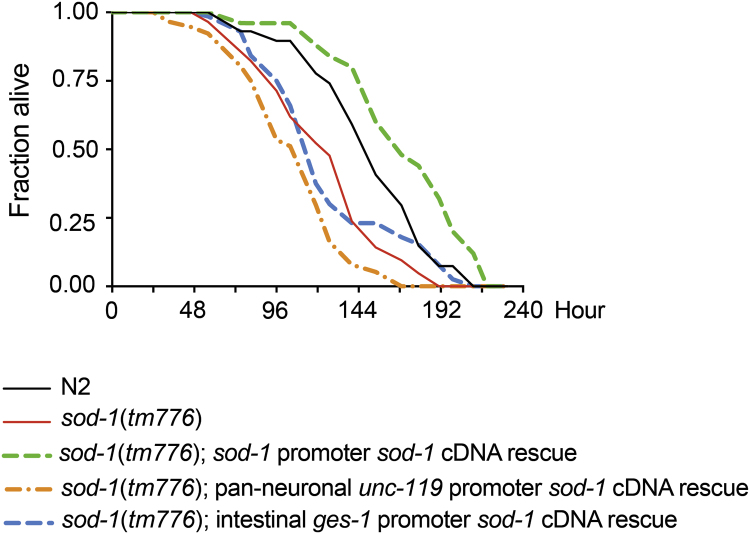
Exposure to P. aeruginosa elevates oxidative stress [22], [29] and contributes to early demise of C. elegans [21], [44], [45]. To determine the tissue-specific requirements of SOD-1 in alleviating pathogen-induced oxidative stress, we performed the P. aeruginosa survival assay [21]. We introduced wild type, sod-1 deletion, neuron-specific SOD-1 rescue, intestine-specific SOD-1 rescue, and sod-1 promoter SOD-1 rescue strains onto a petri dish that contained a lawn of P. aeruginosa. We monitored C. elegans survival over time (Fig. 4). We found that sod-1 (tm776) died faster than wild type. Expressing SOD-1 under its endogenous promoter (by which SOD-1 is present both in the gut and in the nervous system) in the sod-1(tm776) mutant restored and extended the survival phenotype. Intestinal SOD-1 rescue extended the survival of C. elegans only at later time points. Although pan-neuronal over expression of SOD-1 in the nervous system rescues the heightened avoidance response of sod-1 mutant (Supplementary Fig. 3C), constitutive pan-neuronal over expression of SOD-1 exacerbated lethality (Fig. 4). This suggests that constitutive SOD-1 elevation in the nervous system is detrimental to C. elegans. This also indicates that the levels of neuronal SOD-1 expression need to be carefully controlled.
Fig. 4.
Tissue-specific requirement of SOD-1 in survival after P. aeruginosa exposure. After being exposed to P. aeruginosa PA14, sod-1 (tm776) mutant animals (red) die faster than wild type (black). Endogenous expression of SOD-1 rescues the sod-1 (tm776) lethality phenotype and promotes survival (dotted green). Constitutive neuronal expression of SOD-1 (dotted orange) exacerbates the lethality of sod-1 (tm776). Intestinal SOD-1 expression (dotted blue) rescues the sod-1 (tm776) lethality phenotype at later time points. For each experiment, N > 75.
2.5. SOD-1 is activated by guanylyl cyclases GCY-5 and GCY-22 in the ASER neuron
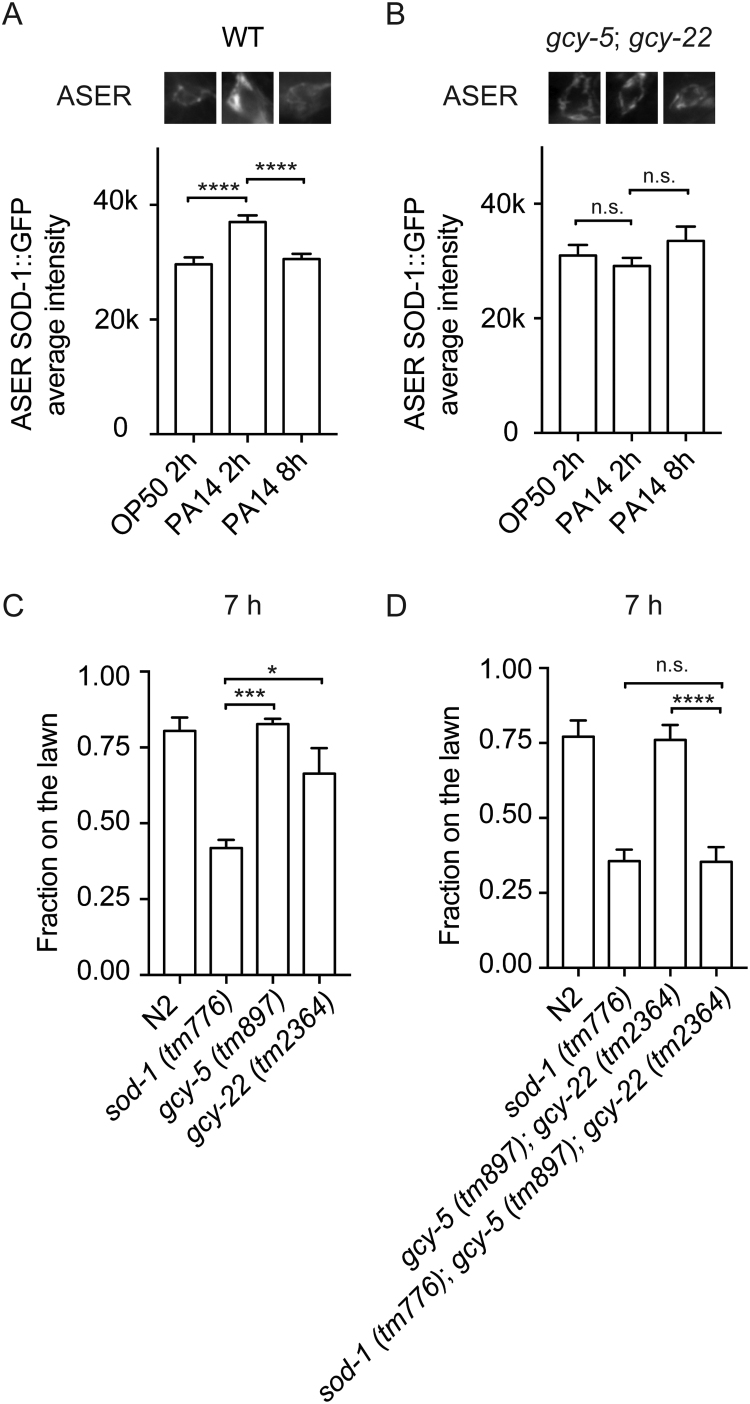
After transferring C. elegans to a P. aeruginosa diet, SOD-1 expression becomes elevated in the ASER neuron (Fig. 5A). However, the mechanism involved in ASER-specific SOD-1 activation remains unknown. Since the right and left gustatory neurons each express their own set of guanylyl cyclases, we sought to examine whether ASER-specific guanylyl cyclase plays a role in SOD-1 activation. GCY-5 and GCY-22 are guanylyl cyclases uniquely expressed in the ASER neuron [36], [37], [46]. GCY-5 and GCY-22 each contain a single transmembrane domain and a guanylyl cyclase domain. The guanylyl cyclase domain is located near the carboxyl terminus and catalyzes the formation of cyclic GMP. GCY-5 and GCY-22 are proposed to act as gustatory receptors and are responsible for activating the signaling cascade after ligand binding [36], [37]. We introduced a gcy-5(tm897); gcy-22(tm2364) double mutant and a sod-1p::sod-1 cDNA::GFP reporter into the same strain. We then switched the strain to a P. aeruginosa diet and measured the fluorescence signals of SOD-1::GFP at 2 h and 8 h. We found that the ASER-specific SOD-1::GFP activation is abolished in the gcy-5; gcy-22 double mutant background (Fig. 5B). We then examined whether GCY-5 and GCY-22 regulate C. elegans behavioral response to P. aeruginosa. We found that neither the single mutants of gcy-5 and gcy-22 nor the double mutant of gcy-5; gcy-22 elicited a heightened aversive response to P. aeruginosa (Fig. 5C and D). Since the gcy-5; gcy-22 double mutant abolished ASER-specific induction of SOD-1, we reasoned that SOD-1 likely acts downstream of GCY-5 and GCY-22. Therefore, we generated the sod-1(tm776); gcy-5(tm897); gcy-22(tm2364) triple mutant. We compared the pathogen avoidance phenotype of the sod-1; gcy-5; gcy-22 triple mutant to that of the sod-1 single mutant. If SOD-1 acts downstream of GCY-5 and GCY-22, introducing gcy-5 and gcy-22 mutations into the sod-1 background will not change the behavioral response. Indeed, the sod-1; gcy-5; gcy-22 triple mutant showed a similar P. aeruginosa avoidance response to the sod-1 single mutant. Our results indicate that GCY-5 and GCY-22 both play a role in activating SOD-1 expression in the ASER neuron when C. elegans are exposed to P. aeruginosa. However, GCY-5 and GCY-22 do not directly participate in the behavioral response to P. aeruginosa.
Fig. 5.
ASER-specific guanylyl cyclases GCY-5 and GCY-22 play a role in SOD-1 induction. (A) SOD-1 expression is elevated in the ASER neuron by P. aeruginosa at 2 h. SOD-1 expression returns to the baseline at 8 h. (B) gcy-5 (tm857) and gcy-22 (tm2364) double mutant blocks SOD-1 elevation at 2 h. (C) Pathogen avoidance phenotypes of gcy-5 (tm857) and gcy-22 (tm2364) single mutants. (D) The pathogen avoidance response of sod-1 (tm776); gcy-5 (tm857); gcy-22 (tm2364) triple mutant is similar to that of sod-1 (tm776) single mutant. Error bar represents s.e.m. * represents p < 0.05. *** represents p < 0.001. **** represents p < 0.0001. n.s.: not significant. Determined by one-way ANOVA Tukey's multiple comparison. N > 30 for each experiment.
2.6. NPR-1 promotes SOD-1 attenuation in the nervous system
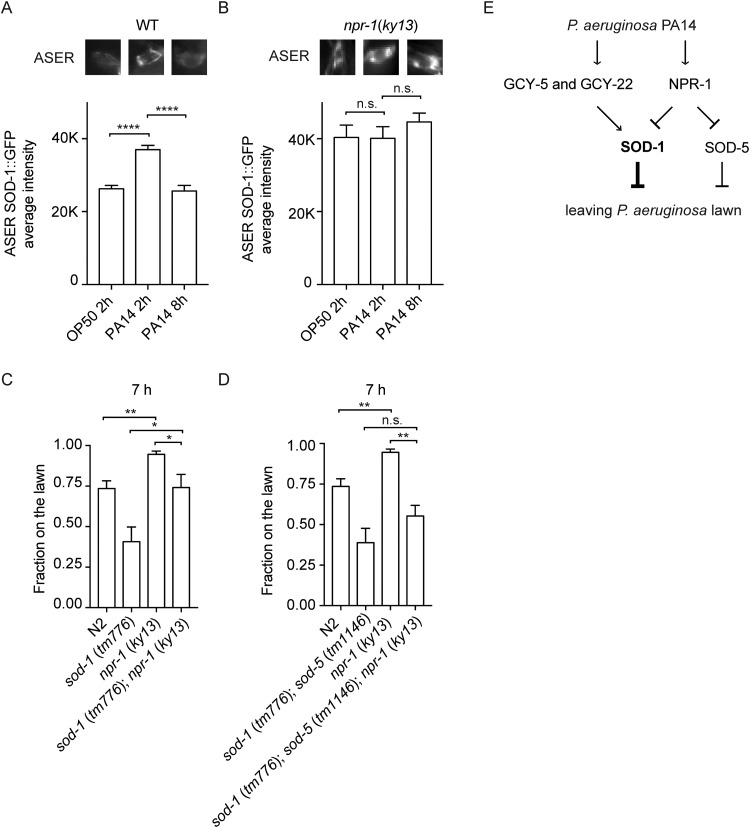
Neuropeptide receptor NPR-1 is present in the gustatory neuron ASER [40]. We first tested whether NPR-1 regulates SOD-1 expression in the ASER neuron. We introduced the npr-1 (ky13) mutation into the sod-1p::sod-1 cDNA::GFP reporter line, fed the animals P. aeruginosa, and measured the SOD-1::GFP signals. In wild type, SOD-1 expression becomes elevated in the ASER neuron. After 8 h, SOD-1 expression returns to the baseline in the ASER neuron (Fig. 6A). In contrast, the temporal regulation of SOD-1 is abolished by the lack of NPR-1. In the npr-1 (ky13) background, SOD-1 remained constantly elevated throughout the time course of our experiments (Fig. 6B). These results suggest that NPR-1 plays a role in promoting SOD-1 reduction.
Fig. 6.
Neuropeptide receptor NPR-1 modulates superoxide dismutase levels and superoxide dismutase–dependent pathogen avoidance response. (A) SOD-1 is elevated in the gustatory neuron ASER by P. aeruginosa at 2 h. SOD-1 expression returns to the baseline at 8 h. (B) The temporal regulation of SOD-1 is abolished by the lack of NPR-1. In npr-1 (ky13) null mutant, SOD-1 level remains constantly elevated in the ASER neuron. (C) sod-1 (tm776); npr-1 (ky13) double mutant elicits intermediate pathogen avoidance response compared to sod-1 (tm776) and npr-1 (ky13) single mutants. (D) sod-1 (tm776); sod-5 (tm1146); npr-1 (ky13) triple mutant elicits a similar pathogen avoidance response to sod-1 (tm776); sod-5 (tm1146) double mutant. (E) Guanylyl cyclases (GCY-5 and GCY-22) and neuropeptide receptor (NPR-1) act antagonistically to regulate Zn/Cu superoxide dismutase expression in the ASER neuron. Error bar represents s.e.m. * represents p < 0.05. ** represents p < 0.01. **** p represents < 0.0001. n.s.: not significant. Determined by one-way ANOVA Tukey's multiple comparison. N > 30 for each experiment.
2.7. NPR-1 inhibits SOD-1– and SOD-5–dependent behavioral response to pathogen
SOD-1 deletion and NPR-1 null mutations elicit opposite P. aeruginosa avoidance phenotypes [21], [22], [26]. To determine whether NPR-1 and SOD-1 have a genetic epistasis relationship in C. elegans behavioral response to P. aeruginosa, we generated npr-1 (ky13); sod-1 (tm776) double mutants, fed the double mutants P. aeruginosa, and observed the P. aeruginosa avoidance behavior. Compared to sod-1 and npr-1 single mutants, the npr-1; sod-1 double mutant elicited an intermediate behavioral response at 7 h (Fig. 6C). Our data suggest that SOD-5 acts as an auxiliary isoform of zinc–copper superoxide dismutase in C. elegans (Figs. 1C and 2). We reasoned that the intermediate pathogen avoidance response of the npr-1 and sod-1 double mutant might be contributed by the remaining superoxide dismutase activity of SOD-5. Therefore, we generated a sod-1(tm776); sod-5(tm1146); npr-1(ky13) triple mutant. We found that the sod-1; sod-5; npr-1 triple mutant elicited a behavioral response comparable to that elicited by the sod-1; sod-5 double mutant (Fig. 6D). This suggests that zinc–copper superoxide dismutase isoforms SOD-1 and SOD-5 act downstream of NPR-1 to regulate the behavioral response to pathogen (Fig. 6E).
Together, our results demonstrate that superoxide dismutase mediates C. elegans pathogen avoidance behavior. Among all five isoforms of superoxide dismutase in C. elegans, SOD-1 plays a primary role in mediating the behavioral response to P. aeruginosa (Fig. 1). SOD-1 is elevated in the intestine and in the nervous system by oxidative stress and P. aeruginosa (Fig. 3 and Supplementary Fig. 3). Constitutive neuronal expression of SOD-1 exacerbates the lethality after prolonged exposure to P. aeruginosa (Fig. 4). SOD-1 expression becomes elevated in the ASER neuron after 2 h exposure to P. aeruginosa. After 8 h, SOD-1 expression returns to the baseline (Supplementary Fig. 3 and Fig. 5A). Although ASER-specific guanylyl cyclases GCY-5 and GCY-22 do not directly mediate the behavioral response to pathogen, GCY-5 and GCY-22 activate superoxide dismutase SOD-1 expression in the ASER neuron (Fig. 5). In contrast, neuropeptide receptor NPR-1 reduces SOD-1 expression in the ASER neuron (Fig. 6). The zinc–copper superoxide dismutase isoform SOD-5 plays a role in mediating pathogen avoidance response (Fig. 1). Because SOD-5 is normally expressed at an undetectable level (Fig. 2 and Supplementary Fig. 2), SOD-5 is likely acting as an auxiliary zinc–copper superoxide dismutase.
3. Discussion
The data presented here establish a central role for zinc–copper superoxide dismutase in the pathogen-induced oxidative stress response amid gut dysbiosis. When C. elegans are switched to the P. aeruginosa diet, both SOD-1 and SOD-5 promote P. aeruginosa feeding (Fig. 1). SOD-1 and SOD-5 are present in overlapping sensory neurons, including ASER, a gustatory neuron that has been previously shown to play a role in extending P. aeruginosa feeding [22] (Fig. 2). We found that sod-1 mutant animals showed a stronger aversive response to P. aeruginosa than sod-5 mutant animals. The sod-1; sod-5 double mutant did not show an enhancement of the aversive behavior when compared to the sod-1 single mutant. Most importantly, we were able to rescue the sod-1; sod-5 double mutant phenotype using a sod-1 genomic fragment (Fig. 1). In addition, we found that SOD-5 is normally expressed at low levels (Supplementary Fig. 2). Previous research has found that SOD-1 contributes almost 80% of the total sod mRNA expression as well as 80% of the total SOD activity in C. elegans [42]. Together, these data suggest that SOD-1 is the primary zinc–copper superoxide dismutase isoform to mediate the behavioral response to pathogen-induced oxidative stress.
Oxidative stress activates SOD-1 expression in the nervous system and in the intestine (Fig. 3). However, our results show that constitutive SOD-1 overexpression in the nervous system has an adverse effect on animal survival (Fig. 4). This is likely because SOD-1 acts in the nervous system to delay the aversive response to P. aeruginosa [22]. This suggests that neuronal expression of SOD-1 has to be carefully controlled. Indeed, we found that SOD-1 is initially elevated by P. aeruginosa via ASER-specific GCY-5 and GCY-22 guanylyl cyclases (Fig. 5). After extended feeding on P. aeruginosa, SOD-1 expression returns to the baseline (Figs. 5A and 6A). The down-regulation of SOD-1 is mediated by neuropeptide receptor NPR-1 (Fig. 6). Based on these results, we propose that superoxide dismutase SOD-1 is activated by P. aeruginosa in the gustatory neuron via guanylyl cyclases GCY-5 and GCY-22. The increase in SOD-1 expression allows C. elegans to cope with elevated oxidative stress. However, prolonged SOD-1 elevation in the nervous system extends P. aeruginosa feeding and is detrimental to C. elegans. It is possible that intestine-derived signals, such as neuropeptides, may act through neuropeptide receptor NPR-1 to facilitate the reduction of SOD-1 and to further fine-tune the behavioral response to pathogens (Fig. 6E).
After extended feeding, P. aeruginosa is accumulated in the intestine [26]. We reason that pathogen-induced oxidative stress likely originates in the intestine [2], [27], [29]. Unexpectedly, intestinal SOD-1 overexpression only marginally extended the survival of C. elegans (Fig. 4). We found that only by expressing SOD-1 in the nervous system and intestine simultaneously (using the sod-1 promoter) could we restore the SOD-1 dependent phenotype. These results suggest that there is a possible interplay between the gut and the nervous system in the SOD-1–dependent pathogen response. The underlined molecular mechanism remains to be identified in future studies.
Together, our findings demonstrate that zinc–copper superoxide dismutase SOD-1 plays a primary role in mediating the behavioral response amid gut dysbiosis. SOD-1 expression is regulated by guanylyl cyclases GCY-5 and GCY-22 and neuropeptide receptor NPR-1 in the gustatory neuron ASER. The fine-tuning of behavioral plasticity by the gustatory system via the regulation of superoxide dismutase amid microbe-induced oxidative stress demonstrates a novel strategy to benefit survival. Recent investigations have suggested that consumption of diets that are rich in sugar and fat often contributes to gut dysbiosis. In turn, the sugar- and fat-rich diets activate metabolic endotoxemia and oxidative stress and further negatively impact the function of the brain [47], [48]. The superoxide dismutase–dependent molecular machinery offers the initial evidence for how the nervous system responds to oxidative stress amid gut dysbiosis. Our findings may provide the molecular mechanisms that are conserved across different species.
4. Materials and methods
4.1. Strains
C. elegans strains were maintained at 20 °C using standard methods [14]. Strains were maintained at 20 °C, then shifted to 22.5 °C for P. aeruginosa lawn avoidance assays. All of the superoxide dismutase deletion mutants, gcy-5(tm897), and gcy-22(tm2364) were obtained from the Caenorhabditis Genetics Center and backcrossed six times to N2 prior to analysis. Transgenic strains were isolated by microinjecting plasmids (typically at 50–100 μg/mL), together with one of the following co-injection markers—myo-2p::mstrawberry, unc-122p::GFP, and unc-122p::mcherry—in wild-type or mutant animals. UV integration of the extrachromosomal array was performed following the protocol originated by S. Mitani [49]. The integrated lines were then backcrossed six times to N2 prior to the analysis.
4.2. P. aeruginosa avoidance assay
A 100 mL solution of LB was inoculated with a single colony of P. aeruginosa PA14 and grown overnight without shaking at 37 °C until O.D. reached 0.2–0.3. 30 μL of this culture was used to seed the center of the 100 mm NGM plate. Seeded plates were incubated for 24 h at room temperature (22.5 °C) prior to the experiment. Approximately 30 animals (young adults) were transferred onto plates containing the P. aeruginosa PA14 lawn at 22.5 °C, and lawn occupancy was measured at the indicated times. Three plates of each genotype were performed in each experiment, and all experiments were performed at least three times. Upon being transferred to the P. aeruginosa–containing plates, animals explored the plate for about 10–15 min until they found the bacterial lawn and then remained in the lawn. Subsequently, lawn occupancy was measured over time as the lawn avoidance behavior was observed [21].
4.3. P. aeruginosa pathogenesis survival assay
Survival of C. elegans on P. aeruginosa PA14 was set up as described previously [21] with the following modifications: 10 μL of the P. aeruginosa PA14 culture was used to seed 35-mm Slow-Killing Assay plates. Seeded plates were incubated for 24 h at 22.5 °C. Young adult animals were then transferred to the P. aeruginosa PA14 plates, which were maintained at 22.5 °C throughout the survival assay. Animals were transferred to fresh plates every two days that were seeded at the beginning of the experiments. Because of potential adverse effects [50], we did not add 5-fluorodeoxyuridine to our culture media. Animals were scored as dead when they no longer responded to the repeated touch of a platinum wire.
4.4. SOD-1 reporter strain activation by paraquat
sod-1p::sod-1 cDNA::RFP was cloned as described previously [22]. The plasmid was injected, integrated, and backcrossed six times to N2 prior to the analysis. The sod-1p::sod-1 cDNA::RFP reporter strain was soaked with 100 μM paraquat in 500 μL M9 buffer for 2 h in a 1.5 mL tube shaking on a nutating mixer. The animals were immediately mounted on slides for imaging after three quick rinses using 500 μL M9 buffer.
4.5. Molecular cloning
The genomic region of sod-1 was amplified by PCR using primers 5′- GAACACCAAACCGGACTGACCAAGT − 3′ and 5′- GTTTATGACGCAAAGCGTACGGACAATCTC − 3′. The 2.9 kb genomic fragment was cloned into a Topo® (Invitrogen) vector. The sod-5 promoter region and part of the first exon of sod-5 were amplified by using a 5′ primer containing 5′- GGAAACATCTTTCACGCTGCTGCAACAC − 3′ and a 3′ primer containing 5′- CTGAT ATTGCCAATGCCGTTCTT CCA CA − 3′. The 873 bp fragment was subsequently cloned using SphI/KpnI sites into the modified pPD95.75 vector (Fire Lab Vector Kit, Addgene) that contains RFP to generate the sod-5 translational RFP reporter. sod-1 cDNA clones were gifts from Y. Kohara (yk524g1, yk593d7, yk1381e03, and yk1715f05). Detailed primer sets and methods used for cloning are available upon request.
4.6. Microscopy
Animals were mounted in M9 with levamisole (10 mM) onto slides with a 3% agarose pad. The slides were viewed using an AxioImager Z1 fluorescence microscope (Zeiss) with 10x/0.25, 40x/0.75, and 63x/1.4 (oil) objectives. The fluorescence signals were recorded by a CCD camera in a 16-bit format without saturation. The images were captured and analyzed by ProgRes imaging software. Fluorescence intensity was measured and calculated using Image-Pro software.
4.7. Statistical analysis
Statistical analysis was performed using GraphPad Prism software.
Acknowledgement
We thank the Caenorhabditis Genetics Center (CGC), which is supported by the NIH (P40 OD010440), to provide us many strains. We thank Abu Rahat for backcrossing mutant alleles and integrated transgenic lines.
Acknowledgments
Contributions
A.M.H. performed the experiments, analyzed the results and wrote the draft of manuscript. H.C.C. designed and performed the experiments, analyzed the results and wrote the paper.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.redox.2018.05.007.
Appendix A. Supporting information
Supp. Fig. 1.
P. aeruginosa avoidance response of all superoxide dismutase mutants at 7 h. N > 30 for each experiment. n.s.: not significant, as determined by one-way ANOVA Tukey's multiple comparison.
Supp. Fig. 2.
SOD-5 expression is elevated in the nervous system by starvation. Arrow indicates head neurons. Scale bar is 200 µm.
Supp. Fig. 3.
SOD-1 is elevated by P. aeruginosa in the nervous system and in the intestine. (A) SOD-1 is elevated by P. aeruginosa in the ASER neuron after 2 h. But the expression returns to the baseline at 8 h. (B) SOD-1 is elevated by P. aeruginosa in the intestine after 8 h. Scale bar is 50 µm. (C) Pan-neuronal expression of SOD-1 rescues sod-1 (tm776) heightened behavior response to P. aeruginosa. Error bar represents s.e.m. * represents p < 0.05. *** represents p < 0.001. **** represents p < 0.0001. n.s.: not significant. Determined by one-way ANOVA Tukey's multiple comparison. N > 30 for each experiment.
References
- 1.Honda K., Littman D.R. The microbiota in adaptive immune homeostasis and disease. Nature. 2016;535:75–84. doi: 10.1038/nature18848. [DOI] [PubMed] [Google Scholar]
- 2.Hooper L.V., Littman D.R., Macpherson A.J. Interactions between the microbiota and the immune system. Science. 2012;336:1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jostins L. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ley R.E., Turnbaugh P.J., Klein S., Gordon J.I. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 5.Nicholson J.K. Host-gut microbiota metabolic interactions. Science. 2012;336:1262–1267. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- 6.Qin J. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490:55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- 7.Biagi E. Gut microbiota and extreme longevity. Curr. Biol. 2016;26:1480–1485. doi: 10.1016/j.cub.2016.04.016. [DOI] [PubMed] [Google Scholar]
- 8.Bravo J.A. Ingestion of Lactobacillus strain regulates emotional behavior and central GABA receptor expression in a mouse via the vagus nerve. Proc. Natl. Acad. Sci. USA. 2011;108:16050–16055. doi: 10.1073/pnas.1102999108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cryan J.F., Dinan T.G. Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour. Nat. Rev. Neurosci. 2012;13:701–712. doi: 10.1038/nrn3346. [DOI] [PubMed] [Google Scholar]
- 10.Diaz Heijtz R. Normal gut microbiota modulates brain development and behavior. Proc. Natl. Acad. Sci. USA. 2011;108:3047–3052. doi: 10.1073/pnas.1010529108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buffington S.A. Microbial reconstitution reverses maternal diet-induced social and synaptic deficits in offspring. Cell. 2016;165:1762–1775. doi: 10.1016/j.cell.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hasegawa S. Intestinal dysbiosis and lowered serum lipopolysaccharide-binding protein in Parkinson's disease. PLoS One. 2015;10:e0142164. doi: 10.1371/journal.pone.0142164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sampson T.R. Gut microbiota regulate motor deficits and neuroinflammation in a model of parkinson's disease. Cell. 2016;167:1469–1480. doi: 10.1016/j.cell.2016.11.018. (e1412) (e1412) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dirksen P. The native microbiome of the nematode Caenorhabditis elegans: gateway to a new host-microbiome model. BMC Biol. 2016;14:38. doi: 10.1186/s12915-016-0258-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Samuel B.S., Rowedder H., Braendle C., Felix M.A., Ruvkun G. Caenorhabditis elegans responses to bacteria from its natural habitats. Proc. Natl. Acad. Sci. USA. 2016;113:E3941–E3949. doi: 10.1073/pnas.1607183113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Labrousse A., Chauvet S., Couillault C., Kurz C.L., Ewbank J.J. Caenorhabditis elegans is a model host for Salmonella typhimurium. Curr. Biol. 2000;10:1543–1545. doi: 10.1016/s0960-9822(00)00833-2. [DOI] [PubMed] [Google Scholar]
- 18.Mahajan-Miklos S., Tan M.W., Rahme L.G., Ausubel F.M. Molecular mechanisms of bacterial virulence elucidated using a Pseudomonas aeruginosa-Caenorhabditis elegans pathogenesis model. Cell. 1999;96:47–56. doi: 10.1016/s0092-8674(00)80958-7. [DOI] [PubMed] [Google Scholar]
- 19.Irazoqui J.E. Distinct pathogenesis and host responses during infection of C. elegans by P. aeruginosa and S. aureus. PLoS Pathog. 2010;6:e1000982. doi: 10.1371/journal.ppat.1000982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Couillault C., Ewbank J.J. Diverse bacteria are pathogens of Caenorhabditis elegans. Infect. Immun. 2002;70:4705–4707. doi: 10.1128/IAI.70.8.4705-4707.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang H.C., Paek J., Kim D.H. Natural polymorphisms in C. elegans HECW-1 E3 ligase affect pathogen avoidance behaviour. Nature. 2011;480:525–529. doi: 10.1038/nature10643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horspool A.M., Chang H.C. Superoxide dismutase SOD-1 modulates C. elegans pathogen avoidance behavior. Sci. Rep. 2017;7:45128. doi: 10.1038/srep45128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee K., Mylonakis E. An intestine-derived neuropeptide controls avoidance behavior in caenorhabditis elegans. Cell Rep. 2017;20:2501–2512. doi: 10.1016/j.celrep.2017.08.053. [DOI] [PubMed] [Google Scholar]
- 24.Cabreiro F. Metformin retards aging in C. elegans by altering microbial folate and methionine metabolism. Cell. 2013;153:228–239. doi: 10.1016/j.cell.2013.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.MacNeil L.T., Watson E., Arda H.E., Zhu L.J., Walhout A.J. Diet-induced developmental acceleration independent of TOR and insulin in C. elegans. Cell. 2013;153:240–252. doi: 10.1016/j.cell.2013.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reddy K.C., Andersen E.C., Kruglyak L., Kim D.H. A polymorphism in npr-1 is a behavioral determinant of pathogen susceptibility in C. elegans. Science. 2009;323:382–384. doi: 10.1126/science.1166527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chavez V., Mohri-Shiomi A., Garsin D.A. Ce-Duox1/BLI-3 generates reactive oxygen species as a protective innate immune mechanism in Caenorhabditis elegans. Infect. Immun. 2009;77:4983–4989. doi: 10.1128/IAI.00627-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ha E.M. Coordination of multiple dual oxidase-regulatory pathways in responses to commensal and infectious microbes in drosophila gut. Nat. Immunol. 2009;10:949–957. doi: 10.1038/ni.1765. [DOI] [PubMed] [Google Scholar]
- 29.Hoeven R., McCallum K.C., Cruz M.R., Garsin D.A. Ce-Duox1/BLI-3 generated reactive oxygen species trigger protective SKN-1 activity via p38 MAPK signaling during infection in C. elegans. PLoS Pathog. 2011;7:e1002453. doi: 10.1371/journal.ppat.1002453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muller F.L. Absence of CuZn superoxide dismutase leads to elevated oxidative stress and acceleration of age-dependent skeletal muscle atrophy. Free Radic. Biol. Med. 2006;40:1993–2004. doi: 10.1016/j.freeradbiomed.2006.01.036. [DOI] [PubMed] [Google Scholar]
- 31.Oeda T. Oxidative stress causes abnormal accumulation of familial amyotrophic lateral sclerosis-related mutant SOD1 in transgenic Caenorhabditis elegans. Hum. Mol. Genet. 2001;10:2013–2023. doi: 10.1093/hmg/10.19.2013. [DOI] [PubMed] [Google Scholar]
- 32.Valentine J.S., Doucette P.A., Zittin Potter S. Copper-zinc superoxide dismutase and amyotrophic lateral sclerosis. Annu Rev. Biochem. 2005;74:563–593. doi: 10.1146/annurev.biochem.72.121801.161647. [DOI] [PubMed] [Google Scholar]
- 33.Bargmann C.I., Horvitz H.R. Chemosensory neurons with overlapping functions direct chemotaxis to multiple chemicals in C. elegans. Neuron. 1991;7:729–742. doi: 10.1016/0896-6273(91)90276-6. [DOI] [PubMed] [Google Scholar]
- 34.Chang S., Johnston R.J., Jr, Frokjaer-Jensen C., Lockery S., Hobert O. MicroRNAs act sequentially and asymmetrically to control chemosensory laterality in the nematode. Nature. 2004;430:785–789. doi: 10.1038/nature02752. [DOI] [PubMed] [Google Scholar]
- 35.Hobert O. Development of left/right asymmetry in the Caenorhabditis elegans nervous system: from zygote to postmitotic neuron. Genesis. 2014;52:528–543. doi: 10.1002/dvg.22747. [DOI] [PubMed] [Google Scholar]
- 36.Ortiz C.O. Searching for neuronal left/right asymmetry: genomewide analysis of nematode receptor-type guanylyl cyclases. Genetics. 2006;173:131–149. doi: 10.1534/genetics.106.055749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ortiz C.O. Lateralized gustatory behavior of C. elegans is controlled by specific receptor-type guanylyl cyclases. Curr. Biol. 2009;19:996–1004. doi: 10.1016/j.cub.2009.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Bono M., Bargmann C.I. Natural variation in a neuropeptide Y receptor homolog modifies social behavior and food response in C. elegans. Cell. 1998;94:679–689. doi: 10.1016/s0092-8674(00)81609-8. [DOI] [PubMed] [Google Scholar]
- 39.Gruner M. Feeding state, insulin and NPR-1 modulate chemoreceptor gene expression via integration of sensory and circuit inputs. PLoS Genet. 2014;10:e1004707. doi: 10.1371/journal.pgen.1004707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Macosko E.Z. A hub-and-spoke circuit drives pheromone attraction and social behaviour in C. elegans. Nature. 2009;458:1171–1175. doi: 10.1038/nature07886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Y., Branicky R., Noe A., Hekimi S. Superoxide dismutases: dual roles in controlling ROS damage and regulating ROS signaling. J. Cell Biol. 2018 doi: 10.1083/jcb.201708007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Doonan R. Against the oxidative damage theory of aging: superoxide dismutases protect against oxidative stress but have little or no effect on life span in Caenorhabditis elegans. Genes Dev. 2008;22:3236–3241. doi: 10.1101/gad.504808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yanase S., Onodera A., Tedesco P., Johnson T.E., Ishii N. SOD-1 deletions in Caenorhabditis elegans alter the localization of intracellular reactive oxygen species and show molecular compensation. J. Gerontol. A Biol. Sci. Med Sci. 2009;64:530–539. doi: 10.1093/gerona/glp020. [DOI] [PubMed] [Google Scholar]
- 44.Tan M.W., Mahajan-Miklos S., Ausubel F.M. Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proc. Natl. Acad. Sci. USA. 1999;96:715–720. doi: 10.1073/pnas.96.2.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tan M.W., Rahme L.G., Sternberg J.A., Tompkins R.G., Ausubel F.M. Pseudomonas aeruginosa killing of Caenorhabditis elegans used to identify P. aeruginosa virulence factors. Proc. Natl. Acad. Sci. USA. 1999;96:2408–2413. doi: 10.1073/pnas.96.5.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith H.K. Defining specificity determinants of cGMP mediated gustatory sensory transduction in Caenorhabditis elegans. Genetics. 2013;194:885–901. doi: 10.1534/genetics.113.152660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cani P.D. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes. 2008;57:1470–1481. doi: 10.2337/db07-1403. [DOI] [PubMed] [Google Scholar]
- 48.Noble E.E., Hsu T.M., Kanoski S.E. Gut to brain dysbiosis: mechanisms linking western diet consumption, the microbiome, and cognitive impairment. Front Behav. Neurosci. 2017;11:9. doi: 10.3389/fnbeh.2017.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kage-Nakadai E., Imae R., Yoshina S., Mitani S. Methods for single/low-copy integration by ultraviolet and trimethylpsoralen treatment in Caenorhabditis elegans. Methods. 2014;68:397–402. doi: 10.1016/j.ymeth.2014.02.036. [DOI] [PubMed] [Google Scholar]
- 50.Van Raamsdonk J.M., Hekimi S. FUdR causes a twofold increase in the lifespan of the mitochondrial mutant gas-1. Mech. Ageing Dev. 2011;132:519–521. doi: 10.1016/j.mad.2011.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]