Abstract
Background
Tea plants [Camellia sinensis (L.) O. Kuntze] can produce one of the three most widely popular non-alcoholic beverages throughout the world. Polyphenols and volatiles are the main functional ingredients determining tea’s quality and flavor; however, the biotic or abiotic factors affecting tea polyphenol biosynthesis are unclear. This paper focuses on the molecular mechanisms of sucrose on polyphenol biosynthesis and volatile composition variation in tea plants.
Results
Metabolic analysis showed that the total content of anthocyanins, catechins, and proanthocyanidins(PAs) increased with sucrose, and they accumulated most significantly after 14 days of treatment. Transcriptomic analysis revealed 8384 and 5571 differentially expressed genes in 2-day and 14-day sucrose-treated tea plants compared with control-treated plants. Most of the structural genes and transcription factors (TFs) involved in polyphenol biosynthesis were significantly up-regulated after 2d. Among these transcripts, the predicted genes encoding glutathione S-transferase (GST), ATP-binding cassette transporters (ABC transporters), and multidrug and toxic compound extrusion transporters (MATE transporters) appeared up regulated. Correspondingly, ultra-performance liquid chromatography-triple quadrupole mass spectrometry (UPLC-QQQ-MS/MS) analysis revealed that the content of non-galloylated catechins and oligomeric PAs decreased in the upper-stem and increased in the lower-stem significantly, especially catechin (C), epicatechin (EC), and their oligomeric PAs. This result suggests that the related flavonoids were transported downward in the stem by transporters. GC/MS data implied that four types of volatile compounds, namely terpene derivatives, aromatic derivatives, lipid derivatives, and others, were accumulated differently after in vitro sucrose treatment.
Conclusions
Our data demonstrated that sucrose regulates polyphenol biosynthesis in Camellia sinensis by altering the expression of transcription factor genes and pathway genes. Additionally, sucrose promotes the transport of polyphenols and changes the aroma composition in tea plant.
Electronic supplementary material
The online version of this article (10.1186/s12870-018-1335-0) contains supplementary material, which is available to authorized users.
Keywords: Camellia sinensis, Polyphenol biosynthesis, Volatile; Sucrose induction; Transcriptomic and metabolic analysis
Background
The tea plant [Camellia sinensis (L.) O. Kuntze] is one of the most important economic crops cultivated in China, Japan, India, and other countries. Its leaves are used for making the tea beverage, one of three most widely consumed non-alcoholic beverages around the world because it contains abundant polyphenols, theanine, caffeine, and other secondary metabolites [1]. Among them, the polyphenol, also called tea polyphenol, is a collective term for phenolic acids and flavonoids including flavanols (catechins), anthocyanins, PAs (also named condensed tannins), and other special derivatives. Polyphenols account for 18–36% of the dry weight of tender leaves and are responsible for tea’s flavor [2–4]. Some studies have suggested that polyphenols play crucial roles in plant stress resistance. For example, they are crucial for protecting the tea plant against pathogens and insects [5, 6]. Additionally, polyphenols are the main functional ingredient in tea for preventing cancer, cardiovascular diseases, and obesity [7].
Studies have indicated that polyphenol biosynthesis in plants is influenced by chemical and physical factors, such as nutrients, hormones, and environmental conditions [8–13]. Among them, sucrose acts not only as carbon source for energy storage and sugar transportation, but also as a signal involved in metabolic processes such as anthocyanin synthesis in plants [14, 15]. Since the late twentieth century, the effects of sucrose on flavonoid and anthocyanin biosynthesis in grapes and radishes have been studied [16–18]. In Arabidopsis thaliana, sucrose induces anthocyanin biosynthesis through the upregulation of structural genes and positive transcription factors involved in the flavonoid biosynthesis pathway and potentially also through the concurrent down-regulation of the negative transcription factor, MYB-LIKE 2 (MYBL2) [19–21]. Previous studies also reported that sucrose could act as a signaling molecule, by first activating PRODUCTION OF ANTHOCYANIN PIGMENT 1 (PAP1) expression by a sucrose-specific signaling pathway and then triggering the expression of structural genes involved in anthocyanin and flavonoid biosynthesis [14, 19, 22, 23]. The sucrose-specific signaling pathway may be activated by different disaccharides, such as sucrose, maltose, and their breakdown products (glucose and fructose); however, sucrose is the most effective inducer of anthocyanin biosynthesis in Arabidopsis [23]. Liu et al. reported sucrose induction increases the content of non-galloylated catechins and up-regulates the expression of putative genes involved in their biosynthetic pathway in both tea callus and seedling [24]. Additionally, Wang et al. also reported sucrose up-regulates the expression of Camellia SINENSIS FLAVONOID 3′5′-HYDROXYLASE (CsF3′5′H), an important branch point gene involved in catechins biosynthesis [25]. In this study, test-tube tea plantlets were used to test for testing the effects of sucrose on polyphenol biosynthesis after 2, 7, 14, and 28d treatments. The results indicated that sucrose can increase the expression of structural genes involved in the biosynthesis of anthocyanins, catechins, and procyanidins. The sucrose specific induction machenism in tea plant is still unclear, one important reason is that we lack the information supported by accurate genome annotations.
Next-Generation Sequencing (NGS) based on the Illumina Hiseq 2000 platform provides a fast, cost-effective, and reliable approach to acquire abundant transcripts, especially for non-model organisms without reference genomic sequences [26]. In tea plants, the NGS technology has been used for analysis of putative genes associated with tea quality and stress response [27–29]. Here, it was performed to investigate the molecular mechanism of sucrose on polyphenol biosynthesis in tea plants and to provide a comprehensive analysis of the network of biochemical and cellular processes responding to sucrose.
In addition, we determined whether in vitro sucrose treatment affects the production of volatiles—the second group of compounds that affect tea taste and flavor in addition to polyphenols.
Results
Effects of sucrose on polyphenol accumulation
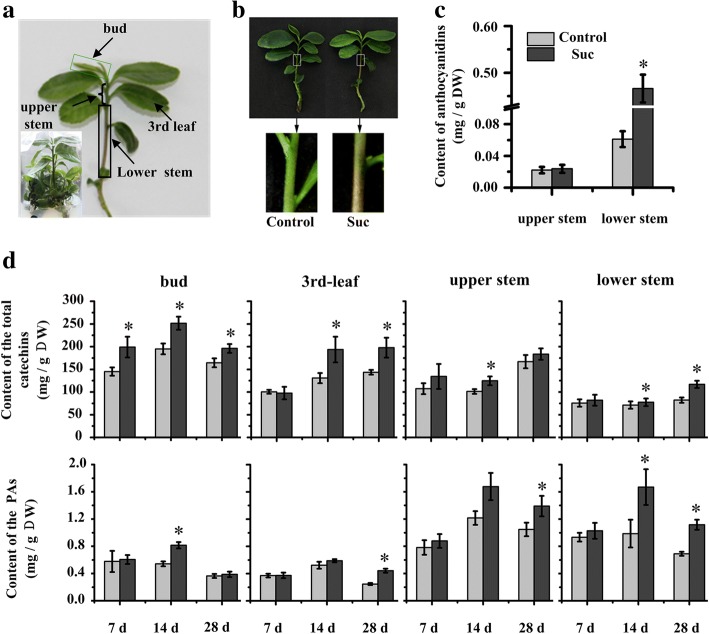
Similar sized test-tube tea plantlets were cultured on Murashige and Skoog standard medium (MS, Control) and MS supplemented with 90 mM sucrose (MS + 90 mM sucrose, Suc) for 28d (Fig. 1a). The stem of the plantlets grown on Suc for 9-14d began to turn red (Fig. 1b), while no red pigmentation was observed in the stem of the plantlets grown on MS or MS supplemented with 90 mM mannitol (data not shown). The anthocyanin levels were significantly different only in the lower part of the stem and were 7-fold higher than that in the control (Fig. 1c). Furthermore, the accumulation of total catechins and PAs in various organs of tea plants is affected by sucrose (Fig. 1d). The effects of sucrose treatment on polyphenol accumulation were observed after 7 and 14 days of treatment (Fig. 1d). However, the effects of sucrose on total catechins and PAs accumulation were not observed at 2d treatment (data not shown).
Fig. 1.
Effects of sucrose on polyphenol accumulation in test-tube tea plantlets. a. Test-tube tea plantlets; b. Red pigments accumulated in stems of plantlet after feeding sucrose; c. Anthocyanin levels are significantly different in the lower part of the stem; d. Accumulation of total catechins and PAs in various organs after 7, 14 and 28 d sucrose treatment. Note: * indicates significance at P < 0.05. The data represents the mean value of three biological replicates
Polyphenol, including phenolic acids, catechin monomers, oligomeric PAs, and flavonols, in different tissues of tea plantlets after 14d treatment was quantitatively measured using UPLC-QQQ-MS/MS (Table 1). Three types of phenolic acids were measured, including quinic acid, gallic acid derivatives (β-glucogallin, galloyl acid and galloylquinic acid), and hydroxycinnamic acid derivatives (caffeoylquinic acid and p-coumaroylquinic acid). The effect of sucrose on compound accumulation was different. For example, sucrose increased the content of galloylquinic acid, a special phenolic acid in the tea plant, increased in most parts of the plants, except for in the bud. However, the content of β-glucogallin, the precursor of galloylated catechins, significantly decreased by 84% in buds and by 71% in upper stems [30]. Monomers of flavanols (catechins) can be classified into non-galloylated and galloylated catechins and mainly exist in buds and upper stems. More non-galloylated catechins accumulated in buds and lower stems after sucrose treatment; however, their content in upper stems decreased significantly. Catechin (C) and epicatechin (EC) decreased by 69% in upper stems. The galloylated catechin content in buds and lower stems was not affected by sucrose, and its content in the 3rd leaf and upper stem decreased by 19%. Seven types of oligomeric PAs accumulated in the bud and 3rd leaf. Their content in lower stems increased 3-fold. However, their content in upper stems significantly decreased after sucrose treatment. For example, B2 (an oligomeric C or EC), decreased by 81%. The content of flavonols in the tea plant was also affected by sucrose. Among them, the flavonol with di-hydroxyl groups on the B-ring was significantly affected by sucrose, and its amount decreased by almost 40% in the third leaf and upper stems and by 14% in buds. However, its content increased by 1-fold in the lower stem.
Table 1.
Effects of sucrose on polyphenol accumulation in different tissues of tea plantlets after 14d treatment using UPLC-QQQ-MS/MS
| Compound | Control | Suc | ratio | Control | Suc | ratio | Control | Suc | ratio | Control | Suc | ratio |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| bud | bud | 3rd leaf | 3rd leaf | up-stem | up-stem | down-stem | down-stem | |||||
| Phenolic acid (mg/g) | ||||||||||||
| Quinic acid | 44.21 ± 2.01 | 86.06 ± 4.05 | 1.95 | 6.55 ± 0.23 | 7.45 ± 0.35 | 1.14 | 39.43 ± 1.89 | 40.19 ± 1.70 | 1.02 | 3.72 ± 0.15 | 6.19 ± 0.29 | 1.67 |
| Gallic acid derivatives | ||||||||||||
| β-glucogallin | 9.42 ± 0.41 | 1.47 ± 0.11 | 0.16 | 0.90 ± 0.05 | 0.97 ± 0.05 | 1.08 | 2.83 ± 0.12 | 0.81 ± 0.03 | 0.29 | 0.02 ± 0.00 | 0.01 ± 0.00 | 0.73 |
| galloyl acid | 0.38 ± 0.01 | 0.36 ± 0.02 | 0.95 | 0.08 ± 0.00 | 0.10 ± 0.01 | 1.19 | 0.25 ± 0.01 | 0.16 ± 0.01 | 0.65 | 0.20 ± 0.01 | 0.03 ± 0.00 | 0.13 |
| galloylquinic acid | 14.09 ± 0.9 | 13.29 ± 0.7 | 0.94 | 0.13 ± 0.01 | 0.47 ± 0.05 | 3.76 | 3.55 ± 0.16 | 6.71 ± 0. 32 | 1.89 | 0.09 ± 0.00 | 0.11 ± 0.01 | 1.15 |
| Summation | 23.88 ± 1.32 | 15.12 ± 0.83 | 0.63 | 1.11 ± 0.06 | 1.54 ± 0.11 | 1.40 | 6.64 ± 0.29 | 7.68 ± 0.36 | 1.16 | 0.31 ± 0.01 | 0.14 ± 0.01 | 0.47 |
| Hydroxycinnamic acids derivatives | ||||||||||||
| caffeoylquinic acid | 0.16 ± 0.01 | 0.14 ± 0.01 | 0.90 | 0.14 ± 0.01 | 0.02 ± 0.00 | 0.17 | 0.12 ± 0.01 | 0.06 ± 0.00 | 0.52 | ND | ND | |
| p-coumaroylquinic acid | 2.29 ± 0.12 | 3.44 ± 0.15 | 1.51 | ND | 0.51 ± 0.02 | 0.45 ± 0.04 | 2.11 ± 0.13 | 4.65 | ND | ND | ||
| Summation | 2.45 ± 0.13 | 3.59 ± 0.16 | 1.47 | 0.14 ± 0.01 | 0.53 ± 0.02 | 3.93 | 0.57 ± 0.05 | 2.17 ± 0.13 | 3.78 | ND | ND | |
| Flavanols (mg/g) | ||||||||||||
| NongalloylatedCatechins | ||||||||||||
| catechin | 2.79 ± 0.12 | 3.74 ± 0.16 | 1.34 | 0.86 ± 0.04 | 2.59 ± 0.13 | 3.02 | 5.51 ± 0.26 | 1.71 ± 0.08 | 0.31 | 0.99 ± 0.04 | 3.03 ± 0.13 | 3.06 |
| epicatechin | 3.64 ± 0.21 | 6.26 ± 0.29 | 1.72 | 3.37 ± 0.15 | 3.81 ± 0.19 | 1.13 | 8.73 ± 0.31 | 2.75 ± 0.11 | 0.31 | 3.02 ± 0.13 | 4.47 ± 0.15 | 1.48 |
| gallocatechin | 1.00 ± 0.06 | 2.66 ± 0.12 | 2.66 | 1.54 ± 0.08 | 2.30 ± 0.11 | 1.49 | 1.43 ± 0.06 | 1.34 ± 0.07 | 0.93 | 0.24 ± 0.01 | 1.05 ± 0.06 | 4.36 |
| epigallocatechin | 13.91 ± 0.8 | 26.89 ± 1.20 | 1.93 | 10.47 ± 0.62 | 7.93 ± 0.38 | 0.76 | 13.99 ± 0.80 | 12.11 ± 0.71 | 0.87 | 4.24 ± 0.15 | 3.19 ± 0.14 | 0.75 |
| Summation | 21.34 ± 1.19 | 39.55 ± 1.77 | 1.85 | 16.23 ± 0.89 | 16.63 ± 0.71 | 1.02 | 29.67 ± 1.36 | 17.90 ± 0.97 | 0.60 | 8.49 ± 0.33 | 11.74 ± 0.48 | 1.38 |
| Galloylatedcatechins | ||||||||||||
| epicatechingallate | 22.38 ± 1.09 | 20.75 ± 1.01 | 0.93 | 3.82 ± 0.15 | 3.58 ± 0.15 | 0.94 | 11.08 ± 0.84 | 9.29 ± 0.83 | 0.84 | 2.18 ± 0.13 | 2.05 ± 0.98 | 0.94 |
| epigallocatechingallate | 89.03 ± 4.21 | 95.88 ± 4.67 | 1.08 | 18.29 ± 0.95 | 14.26 ± 0.68 | 0.78 | 52.19 ± 2.65 | 42.19 ± 2.05 | 0.81 | 6.30 ± 0.31 | 6.46 ± 0.31 | 1.03 |
| Summation | 111.40 ± 5.30 | 116.63 ± 5.68 | 1.05 | 22.11 ± 1.10 | 17.84 ± 0.83 | 0.81 | 63.27 ± 3.49 | 51.48 ± 2.88 | 0.81 | 8.47 ± 0.44 | 8.51 ± 1.29 | 1.00 |
| total Catechins | 132.74 ± 6.49 | 156.18 ± 7.45 | 1.18 | 38.34 ± 1.99 | 34.47 ± 1.85 | 0.90 | 92.94 ± 4.85 | 69.38 ± 3.85 | 0.75 | 16.97 ± 0.77 | 20.25 ± 1.77 | 1.19 |
| Proanthocyanidins (area) | ||||||||||||
| m/z 865 | ND | ND | ND | ND | ND | ND | 490 ± 36 | 6057 ± 312 | 12.36 | |||
| m/z 577 PAs B2 | 33,626 ± 1670 | 52,158 ± 2600 | 1.55 | 17,040 ± 850 | 36,819 ± 1830 | 2.16 | 122,564 ± 6115 | 23,153 ± 1160 | 0.19 | 37,345 ± 1876 | 155,893 ± 7805 | 4.17 |
| m/z 729EC-ECG | 17,582 ± 880 | 18,560 ± 930 | 1.06 | 2125 ± 105 | 3947 ± 185 | 1.86 | 15,214 ± 755 | 7597 ± 380 | 0.50 | 2089 ± 117 | 10,067 ± 515 | 4.82 |
| m/z 593EC-EGC or ECDG | 2300 ± 110 | 6507 ± 320 | 2.83 | 5361 ± 260 | 17,280 ± 855 | 3.22 | 16,556 ± 815 | 5454 ± 267 | 0.33 | 3475 ± 184 | 20,418 ± 1015 | 5.88 |
| m/z 761EGC-EGCG | 11,308 ± 565 | 21,097 ± 1050 | 1.87 | 3841 ± 180 | 4698 ± 225 | 1.22 | 6627 ± 325 | 9111 ± 438 | 1.37 | 909 ± 55 | 4932 ± 264 | 5.43 |
| m/z 745 | 3570 ± 178 | 5468 ± 270 | 1.53 | 2062 ± 105 | 3916 ± 185 | 1.90 | 3806 ± 185 | 3133 ± 148 | 0.82 | ND | 3992 ± 196 | |
| m/z 609(EGC-EGC) | 3809 ± 190 | 11,528 ± 570 | 3.03 | 11,219 ± 550 | 32,040 ± 1505 | 2.86 | 11,924 ± 585 | 5195 ± 246 | 0.44 | 2501 ± 129 | 17,566 ± 868 | 7.02 |
| flavonols derivatives (area) | ||||||||||||
| tri-hydroxyl in B-ring | ||||||||||||
| myricetin 3-O-galactoside | 3929 ± 203 | 5100 ± 268 | 1.30 | 705 ± 42 | ND | ND | 2367 ± 123 | ND | 269 ± 12 | |||
| myricetin 3-O- glucoside | 6797 ± 346 | 6940 ± 359 | 1.02 | 1220 ± 58 | 1301 ± 72 | 1.07 | 3577 ± 185 | 3404 ± 164 | 0.95 | ND | 260 ± 10 | |
| Summation | 10,726 ± 549 | 12,040 ± 627 | 1.12 | 1925 ± 100 | 1301 ± 72 | 0.68 | 3577 ± 185 | 5771 ± 287 | 1.61 | ND | 529 ± 30 | |
| di-hydroxyl in B-ring | ||||||||||||
| quercetin 3-O-galactosylrutinoside | 2539 ± 136 | 2235 ± 126 | 0.88 | 780 ± 48 | 489 ± 34 | 0.63 | 2465 ± 131 | 1025 ± 55 | 0.42 | 806 ± 45 | 684 ± 28 | 0.85 |
| quercetin3-O-glucosylrutinoside | 9680 ± 496 | 8675 ± 456 | 0.90 | 3933 ± 208 | 2379 ± 126 | 0.60 | 5641 ± 291 | 3847 ± 184 | 0.68 | 793 ± 45 | 2704 ± 136 | 3.41 |
| quercetin 3-galactoside | 1404 ± 87 | 1367 ± 78 | 0.97 | 428 ± 30 | ND | 1376 ± 62 | 674 ± 31 | 0.49 | 290 ± 18 | 208 ± 12 | 0.72 | |
| quercetin 3-O-glucoside | 2465 ± 138 | 1630 ± 89 | 0.66 | 911 ± 42 | 850 ± 45 | 0.93 | 1284 ± 58 | 783 ± 45 | 0.61 | 168 ± 7 | 526 ± 32 | 3.14 |
| Summation | 16,088 ± 857 | 13,907 ± 749 | 0.86 | 6052 ± 328 | 3717 ± 201 | 0.61 | 10,766 ± 542 | 6330 ± 315 | 0.59 | 2056 ± 115 | 4122 ± 208 | 2.00 |
| mono -hydroxyl in B-ring | ||||||||||||
| kaempferol-3-O-galactosylrutinoside | 338,752 ± 16,950 | 290,468 ± 14,530 | 0.86 | 61,932 ± 3085 | 39,007 ± 1968 | 0.63 | 137,928 ± 6870 | 130,099 ± 6485 | 0.94 | 23,498 ± 1164 | 18,979 ± 1001 | 0.81 |
| kaempferol3-O-glucosylrutinoside | 853,325 ± 42,664 | 753,945 ± 37,665 | 0.88 | 206,694 ± 10,345 | 120,862 ± 6055 | 0.58 | 316,408 ± 15,808 | 334,177 ± 16,675 | 1.06 | 23,691 ± 1135 | 37,778 ± 1982 | 1.59 |
| kaempferol-3-O-galactoside | ND | 933 ± 59 | 154 ± 10 | 287 ± 28 | 1.86 | 447 ± 31 | 484 ± 30 | 1.08 | ND | 255 ± 10 | ||
| kaempferol-3-O-glucoside | ND | 20,072 ± 1008 | 1491 ± 85 | ND | 6994 ± 350 | 9054 ± 446 | 1.29 | 496 ± 71 | 199 ± 9 | 0.40 | ||
| Kaempferol-3-O-rhamnosylgalactoside | 25,559 ± 1289 | 26,315 ± 1315 | 1.03 | 11,173 ± 560 | 5296 ± 276 | 0.47 | 9333 ± 456 | 11,450 ± 564 | 1.23 | 1567 ± 76 | 2223 ± 124 | 1.42 |
| Summation | 1,217,636 ± 60,903 | 1,091,733 ± 54,577 | 0.90 | 281,445 ± 14,085 | 165,452 ± 8327 | 0.59 | 471,109 ± 23,515 | 485,263 ± 24,200 | 1.03 | 49,252 ± 2396 | 59,434 ± 3126 | 1.21 |
| total flavonols | 1,244,449 ± 62,309 | 1,117,680 ± 55,953 | 0.90 | 289,422 ± 14,513 | 170,470 ± 8600 | 0.59 | 485,453 ± 24,242 | 497,364 ± 24,802 | 1.02 | 51,309 ± 2511 | 63,556 ± 3334 | 1.24 |
Note: ND indicates that the polyphenol was not detected; the data represents the mean value of three biological replicates
Digit indicates the ratio of Suc / Control
Effects of sucrose on volatile compounds
Four types of volatile compounds were measured using GC/ MS, including terpene derivatives, aromatic derivatives, lipid derivative and other compounds, the effect of sucrose on their accumulation was different (see Additional file 1: Table S1). For example, the content of α-farnesene belonging to sesquiterpenoid diterpenoid increased 5.77-fold; the expression of one transcript (Unigene 46,443), which was predicted as the key biosynthetic gene encoding farnesene synthase, was significantly upregulated 3-fold after 2 and 14 days of sucrose treatment (see Additional file 2: Table S2). Here, 33 terpene derivatives were detected and classified into monoterpenoid sesquiterpenoid diterpenoid; these compounds were biosynthesized via methylerythritol phosphate (MEP) and mevalonate (MVA) pathways (see Additional file 3: Figure S1). The expression of HMGR (CL12062.Contig1) and DXS (Unigene57617) and DXR (Unigene46601) as the key genes involving in terpenoid backbone pathway were up-regulated by sucrose. The expression of one transcript (CL1850.Contig3 encoding linalool synthase) was not significantly affected by sucrose; and the content of linalool and geraniol in tea leaf only decreased by 4%. Additionally, the expression of 1 transcript (Unigene9305 encoding (E)-nerolidol synthase) was up-regulated by sucrose after 2d; however, its expression was down- regulated by sucrose after 14d; and the content of the (E)-nerolidol only decreased by 5%.
Effects of sucrose on the expression of key structural genes related to polyphenol biosynthesis using qRT-PCR
For further analysis of the effects of sucrose on polyphenol biosynthesis at the transcriptional level, Quantitative real-time-PCR (qRT-PCR) was used to test the expression of 11 key structural genes involved in the polyphenol biosynthetic pathway (Fig. 2). Their expression significantly increased 3-fold after 2d treatment. After 7d, the expression of Chalcone synthase (CHS), Flavanone 3-hydroxylase (F3H), Flavonoid 3′-hydroxylase (F3′H), Leucoanthocyanidin reductase (LAR), and Anthocyanidin reductase (ANR) increased 1-fold. After 14d, the effect of sucrose on the above genes was less noticeable.
Fig. 2.
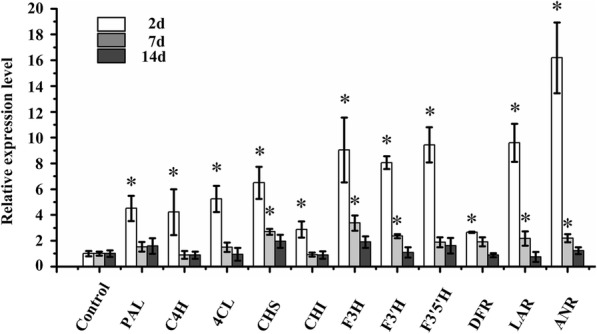
Effects of sucrose on expression of key structural genes involved in polyphenol biosynthesis using qRT-PCR. Note: * indicates significance with |log2 Ratio| ≥ 1. The data represents the mean value of three biological and three technical replicates
Sequencing, de novo assembly, and functional annotation
To obtain the overall transcriptional levels of genes in the tea plant treated by sucrose after 2 and 14d, four normalized cDNA libraries (2d: 2nd D Control and Suc; 14d: 14th D Control and Suc) were constructed for transcriptome sequencing. Based on the Illumina Hiseq 2000 platform, 21,381,193,620 nucleotide (nt) bases were generated from all libraries in total and about 237.6 million clean reads (94.94% of the raw reads) were achieved for de novo assembly (see Additional file 4: Table S3). Finally, a total of 118,843 transcripts were obtained with an average length of 1212 nt and a N50 of 1999 nt (see Additional file 5: Table S4).
To predict the functions of the assembly transcripts, a total of 82,459 transcripts (69.38% of all assembled Unigenes) were annotated using the NR (Non-redundant protein database), NT (Non-redundant nucleotide database), Swiss-Prot (Annotated protein sequence database), KEGG (Kyoto encyclopedia of genes and genomes), COG (Clusters of orthologous groups of protein), and GO (Gene ontology) databases based on two levels of sequence similarity, sequence-based and domain-based alignments, with an e-value<1e-5 (see Additional file 6: Table S5).
Analysis of DEGs responding to sucrose
Using the fragments per kb per million reads (FPKM) method, the DEGs between two samples were identified with a significant threshold of |log2 Ratio (FPKM Control-vs-Suc) | ≥ 1 and the false discovery rate (FDR) of ≤0.001 based on the P-value threshold set as ≤1e-5. A total of 8384 DEGs were detected in 2nd D Control-vs-Suc. Among them, 6187 DEGs (73.80% of the total DEGs) were up-regulated. A total of 5571 DEGs were detected in 14th D Control-vs-Suc, and only 2146 DEGs (38.52% of the total DEGs) were up-regulated (see Fig. 3).
Fig. 3.
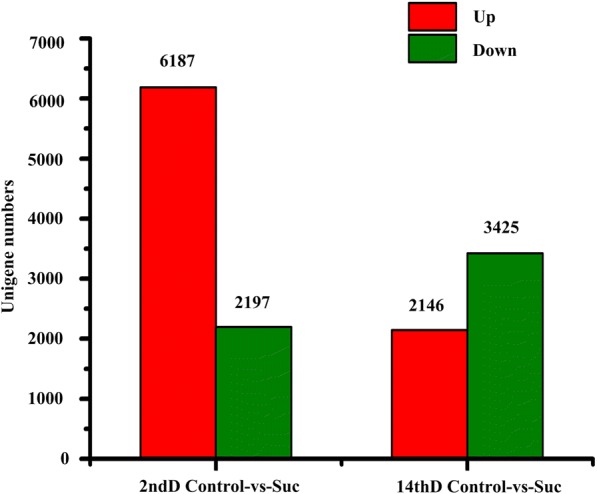
Statistics of DEGs from tea plants responding to sucrose. Note: DEGs were classified into two classes; the red bar indicates up-regulated and the green bar indicates down-regulated, the digit indicates the number of DEGs
GO function and KEGG pathways analysis of DEGs responding to sucrose
To better understand the biological functions of DEGs responding to sucrose, GO and KEGG analyses were performed for comparisons of 2nd D Control-vs-Suc and 14th D Control-vs-Suc. GO functional enrichment analysis indicated that 49 and 48 GO terms were classified into three ontologies which changed significantly between 2nd D and 14th D Control-vs-Suc (see Additional file 7: Figure S2).
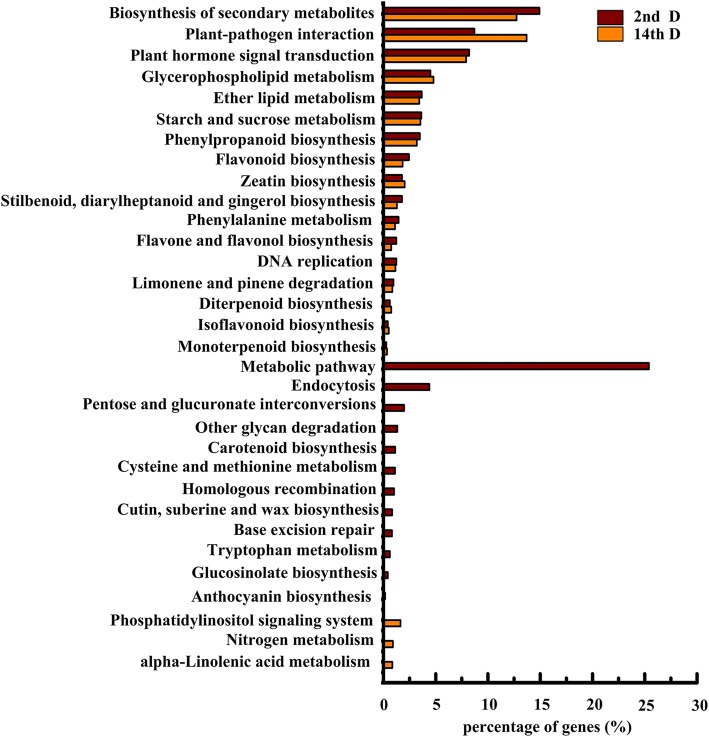
A total of 3553 DEGs (7.46% of all the transcripts aligned to the KEGG database) were annotated and 29 KEGG pathways were enriched significantly in the 2nd D Control-vs-Suc comparison based on a Q-value of ≤0.05. Among them, the most enriched pathway was “flavonoid biosynthesis” (Table 2). In 14th D Control-vs-Suc comparison, 2009 DEGs (4.22% of all the transcripts aligned to KEGG databases) were annotated and 20 KEGG pathways were significantly enriched with the same threshold. The most enriched pathway was that for “plant-pathogen interaction” (Table 3). A total of 17 KEGG-enriched pathways were common between second and fourteenth D Control-vs-Suc. Of the 12 KEGG pathways specific to the second D Control-vs-Suc comparison, one was the KEGG-enriched pathway for anthocyanin biosynthesis (Fig. 4).
Table 2.
Gene ontology analysis of DEGs obtained from tea plants treated by sucrose after 2d
| Pathway | DEGs genes | All genes | Q-value | |
|---|---|---|---|---|
| (3553) | (47655) | |||
| 1 | Flavonoid biosynthesis | 87 (2.45%) | 314 (0.66%) | 2.35E-25 |
| 2 | Biosynthesis of secondary metabolites | 530 (14.92%) | 4746 (9.96%) | 1.33E-20 |
| 3 | Phenylpropanoid biosynthesis | 124 (3.49%) | 653 (1.37%) | 1.76E-20 |
| 4 | Stilbenoid, diarylheptanoid and gingerol biosynthesis | 63 (1.77%) | 233 (0.49%) | 3.38E-18 |
| 5 | Flavone and flavonol biosynthesis | 44 (1.24%) | 165 (0.35%) | 1.41E-12 |
| 6 | Phenylalanine metabolism | 52 (1.46%) | 234 (0.49%) | 1.76E-11 |
| 7 | Plant hormone signal transduction | 291 (8.19%) | 2615 (5.49%) | 4.76E-11 |
| 8 | Zeatin biosynthesis | 63 (1.77%) | 365 (0.77%) | 5.88E-09 |
| 9 | Cutin, suberine and wax biosynthesis | 30 (0.84%) | 116 (0.24%) | 1.65E-08 |
| 10 | Pentose and glucuronateinterconversions | 70 (1.97%) | 452 (0.95%) | 6.26E-08 |
| 11 | DNA replication | 44 (1.24%) | 244 (0.51%) | 4.79E-07 |
| 12 | Carotenoid biosynthesis | 40 (1.13%) | 212 (0.44%) | 4.95E-07 |
| 13 | Limonene and pinene degradation | 34 (0.96%) | 170 (0.36%) | 1.05E-06 |
| 14 | Metabolic pathways | 902 (25.39%) | 10,454 (21.94%) | 1.79E-06 |
| 15 | Ether lipid metabolism | 130 (3.66%) | 1142 (2.4%) | 8.47E-06 |
| 16 | Starch and sucrose metabolism | 129 (3.63%) | 1141 (2.39%) | 1.24E-05 |
| 17 | Diterpenoid biosynthesis | 22 (0.62%) | 105 (0.22%) | 6.04E-05 |
| 18 | Tryptophan metabolism | 22 (0.62%) | 107 (0.22%) | 7.84E-05 |
| 19 | Other glycan degradation | 47 (1.32%) | 328 (0.69%) | 8.46E-05 |
| 20 | Endocytosis | 156 (4.39%) | 1526 (3.2%) | 2.40E-04 |
| 21 | Glycerophospholipid metabolism | 160 (4.5%) | 1577 (3.31%) | 2.69E-04 |
| 22 | Glucosinolate biosynthesis | 15 (0.42%) | 64 (0.13%) | 3.18E-04 |
| 23 | Isoflavonoid biosynthesis | 15 (0.42%) | 72 (0.15%) | 1.25E-03 |
| 24 | Plant-pathogen interaction | 309 (8.7%) | 3440 (7.22%) | 1.60E-03 |
| 25 | Monoterpenoid biosynthesis | 10 (0.28%) | 41 (0.09%) | 3.38E-03 |
| 26 | Anthocyanin biosynthesis | 6 (0.17%) | 20 (0.04%) | 1.26E-02 |
| 27 | Cysteine and methionine metabolism | 40 (1.13%) | 339 (0.71%) | 1.27E-02 |
| 28 | Base excision repair | 29 (0.82%) | 228 (0.48%) | 1.51E-02 |
| 29 | Homologous recombination | 36 (1.01%) | 323 (0.68%) | 4.46E-02 |
Table 3.
Gene Ontology analysis of DEGs obtained from tea plants treated by sucrose after 14d
| Pathway | DEGs genes | All genes | Q-value | |
|---|---|---|---|---|
| (2009) | (47655) | |||
| 1 | Plant-pathogen interaction | 275 (13.69%) | 3440 (7.22%) | 3.78E-23 |
| 2 | Phenylpropanoid biosynthesis | 64 (3.19%) | 653 (1.37%) | 3.04E-08 |
| 3 | Zeatin biosynthesis | 41 (2.04%) | 365 (0.77%) | 6.03E-07 |
| 4 | Flavonoid biosynthesis | 37 (1.84%) | 314 (0.66%) | 6.41E-07 |
| 5 | Plant hormone signal transduction | 159 (7.91%) | 2615 (5.49%) | 5.74E-05 |
| 6 | Stilbenoid, diarylheptanoid and gingerol biosynthesis | 26 (1.29%) | 233 (0.49%) | 1.37E-04 |
| 7 | Biosynthesis of secondary metabolites | 256 (12.74%) | 4746 (9.96%) | 3.87E-04 |
| 8 | Diterpenoid biosynthesis | 15 (0.75%) | 105 (0.22%) | 5.25E-04 |
| 9 | Glycerophospholipid metabolism | 96 (4.78%) | 1577 (3.31%) | 3.06E-03 |
| 10 | DNA replication | 23 (1.14%) | 244 (0.51%) | 3.55E-03 |
| 11 | Phenylalanine metabolism | 22 (1.1%) | 234 (0.49%) | 4.47E-03 |
| 12 | alpha-Linolenic acid metabolism | 17 (0.85%) | 164 (0.34%) | 5.98E-03 |
| 13 | Starch and sucrose metabolism | 71 (3.53%) | 1141 (2.39%) | 7.18E-03 |
| 14 | Isoflavonoid biosynthesis | 10 (0.5%) | 72 (0.15%) | 7.18E-03 |
| 15 | Limonene and pinene degradation | 17 (0.85%) | 170 (0.36%) | 7.18E-03 |
| 16 | Monoterpenoid biosynthesis | 7 (0.35%) | 41 (0.09%) | 1.12E-02 |
| 17 | Ether lipid metabolism | 69 (3.43%) | 1142 (2.4%) | 1.39E-02 |
| 18 | Nitrogen metabolism | 18 (0.9%) | 203 (0.43%) | 1.68E-02 |
| 19 | Phosphatidylinositol signaling system | 33 (1.64%) | 465 (0.98%) | 1.74E-02 |
| 20 | Flavone and flavonol biosynthesis | 15 (0.75%) | 165 (0.35%) | 2.62E-02 |
Fig. 4.
The pathways significantly enriched by DEGs after 2d and 14d sucrose treatment. Note: the horizontal coordinates indicate percent of DEGs, the vertical coordinates indicate significantly enriched pathways of differentially expressed genes
Effects of sucrose on polyphenol biosynthesis based on transcriptome sequencing
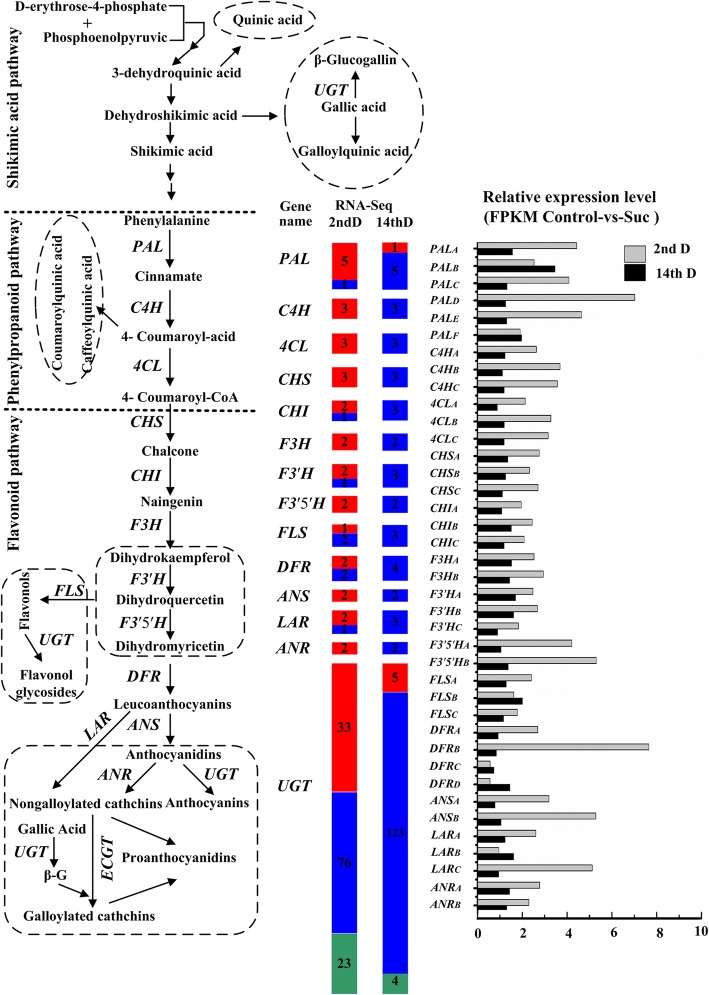
Based on the ratio of FPKM Control-vs-Suc, most of the transcripts involved in the phenylpropanoid and flavonoid pathways were up-regulated 2-fold or more after 2d of treatment. Additionally, the expression of transcripts annotated as Phenylalanine ammonialyase (PAL), Dihydroflavonol 4-reductase (DFR), LAR, and Anthocyanidin synthase (ANS) was notably up-regulated. After 14 days of treatment, the expression of only PALB increased 1-fold, whereas others were not affected by sucrose (Fig. 5). These results indicate that tea polyphenol biosynthesis is comprehensively affected by sucrose.
Fig. 5.
Effects of sucrose on the expression of structural genes related to polyphenol biosynthesis in tea plants after 2d and 14d. Note: Red indicates significant up-regulation, blue indicates no difference, green indicates significant down-regulation. Digit indicates the number of Unigenes
Effects of sucrose on the expression of transcription factors involved in polyphenol biosynthesis based on transcriptome sequencing
Polyphenol biosynthesis in plants is regulated by transcription factors (TFs) including R2R3-MYB, bHLH, and WD40 [31, 32]. In this study, 37 DEGs were predicted to be MYB members and were classified into three types: R1 (4 DEGs), R2R3 (29 DEGs), and R1R2R3 (4 DEGs). Most DEGs (23/37) were up-regulated after sucrose treatment for 2 days, and only five DEGs were up-regulated after sucrose treatment for 14 days (Table 4). Additionally, the phylogenetic tree, including 29 R2R3-MYBs and 126 Arabidopsis R2R3-MYBs, were classified into 13 subgroups (see Additional file 8: Figure S3). Phylogenetic analysis indicated that 33 bHLHs were dispersed into 15 subfamilies (see Additional file 9: Figure S4), and 21 of them were up-regulated after sucrose treatment for 2d (Table 5).
Table 4.
Analysis of DEGS-predicted as R2R3-MYB obtained from tea plants treated by sucrose
| Gene ID | Gene | 2ndD | 14thD | Type | Subgroups | Putative function clade and gene function |
|---|---|---|---|---|---|---|
| length | fold | fold | No. | |||
| CL5525.Contig4 | 955 | 476.9a | – | R2R3 | other | Trichome development-regulated: AtMYB82 [69] |
| Unigene18972 | 1084 | 17.02a | 0.41b | R1R2R3 | Unknown | |
| Unigene35962 | 3506 | 13.97a | 0.49b | R1R2R3 | Unknown | |
| Unigene12085 | 975 | 13.54a | 0.32b | R2R3 | 6 | Anthocyanin biosynthes-related: AtMYB75and AtMYB90 [54, 70, 71] |
| Unigene41846 | 938 | 4.98a | – | R2R3 | 6 | Secondary cell wall formation-related: AtMYB75 [72] |
| Unigene35958 | 3304 | 6.28a | – | R1R2R3 | Unknown | |
| CL8695.Contig1 | 1179 | 5.47a | – | R2R3 | 5 | Seed pigmentation biosynthesis -controlled: AtMYB123 [48, 73] |
| Unigene11002 | 1229 | 2.93a | – | R2R3 | 5 | |
| Unigene7972 | 1143 | 5.41a | – | R2R3 | 9 | Seed germination and reproductive development-related AtMYB17 [74, 75] |
| CL1441.Contig4 | 2364 | 2.85a | – | R2R3 | 9 | Petal development: AtMYB16 [76] |
| Repressor of cell outgrowth: AtMYB106 [77] | ||||||
| Unigene24177 | 714 | 4.91a | – | R2R3 | other | |
| Unigene20350 | 1829 | 2.20a | – | R2R3 | other | |
| CL12359.Contig1 | 3219 | 2.56a | – | R2R3 | other | |
| CL5017.Contig2 | 1322 | 4.04a | 0.34b | R2R3 | 1 | Hypersensitive response: AtMYB30Cooperates with BES1 to regulate |
| CL8708.Contig1 | 1933 | 2.91a | – | R2R3 | 1 | brassinosteroid-induced gene Expression; abiotic stress response, SA–mediated pathway AtMYB30 [77] |
| Unigene13855 | 767 | 3.84a | – | R2R3 | 15 | Epidermal cell fate specification: AtMYB23 [78] Trichome development: AtMYB0 and AtMYB23, |
| CL7877.Contig1 | 887 | 3.25a | – | R2R3 | 15 | Root hair patterning-controlled AtMYB66 [79] |
| Unigene1868 | 527 | 2.68a | – | R1 | Unknown | |
| Unigene16731 | 1118 | 2.41a | – | R2R3 | 14 | Axillary meristem initiation in roots-related: AtMYB36 [80] |
| CL3134.Contig13 | 4926 | 2.40a | – | R1R2R3 | Unknown | |
| CL13057.Contig1 | 995 | 2.31a | – | R2R3 | 4 | The battle against UV by repressing C4H: AtMYB4 [81] |
| CL13057.Contig2 | 827 | – | 2.64a | R2R3 | 4 | |
| CL2339.Contig1 | 1129 | 2.24a | – | R2R3 | 21 | Lignin, xylan and cellulose biosynthesis-regulated: AtMYB52, AtMYB54 and AtMYB69 [82] |
| Ovule and fruit development: AtMYB117 [83] | ||||||
| ABA hypersensitivity and drought tolerance: AtMYB52 [84] | ||||||
| CL8255.Contig3 | 1314 | – | 2.02a | R2R3 | 7 | Flavonol glycosides-related: AtMYB11, AtMYB12 and AtMYB111 [34] |
| CL6408.Contig3 | 1494 | 2.01a | – | R2R3 | 2 | Shoot apex morphogenesis: AtMYB13 [85] |
| CL9344.Contig1 | 1068 | – | 0.25b | R2R3 | 2 | Cold stress tolerance: AtMYB14 and AtMYB15 [86, 87] |
| CL6408.Contig1 | 1557 | – | 0.45b | R2R3 | 2 | |
| CL5350.Contig2 | 1322 | – | 0.16b | R2R3 | 2 | |
| Unigene48919 | 574 | 0.41b | – | R2R3 | 2 | |
| CL1581.Contig2 | 1552 | – | 0.18b | R1 | Unknown | |
| CL7764.Contig2 | 980 | – | 0.15b | R1 | Unknown | |
| Unigene6794 | 537 | – | 2.47a | R2R3 | other | |
| Unigene36358 | 1700 | – | 2.01a | R2R3 | other | AS1 leaf morphogenesis (polarity specificity) and plant immune response: AtMYB91 [88]; |
| Rough-sheath development: AtMYB91 [89] | ||||||
| Unigene11308 | 1618 | – | 2.10a | R2R3 | 13 | Stomatal closure: AtMYB61 [90]; |
| Multiple aspects of plant resource allocation-controled: AtMYB61 [91] | ||||||
| Unigene38120 | 1427 | – | 0.47b | R2R3 | 22 | Stomatal closure-regulated: AtMYB44,AtMYB70, AtMYB73 and AtMYB77 [92, 93] |
| Auxin signaling pathway- modulated: AtMYB77 [94]; | ||||||
| Unigene39226 | 735 | 0.49b | – | R2R3 | 20 | GA metabolism and signaling involved in regulation starvation responses:AtMYB62 [95]; |
| Cell separation processes-related: AtMYB116 [96] | ||||||
| Unigene2945 | 935 | 0.44b | – | R1 | Unknown | |
Note: “a”indicates significant up-regulation; “–”indicates no difference; “b”indicates significant down-regulation. Unknown and other indicate Unigene is not grouped
Table 5.
Analysis of DEGS-predicted as bHLH obtained from tea plants treated by sucrose
| GeneID | Gene | 2ndD | 14thD | Subfamily | Gene name | Putative function clade and gene function |
|---|---|---|---|---|---|---|
| length | fold | fold | No. | in Arabidopsis | ||
| Unigene60798 | 496 | 1967.8a | – | 3 | AtbHLH18 | |
| Unigene26720 | 1512 | 15.20a | – | AtbHLH25 | ||
| CL2783.Contig8 | 2320 | 280.50a | – | 25 | AtbHLH74 | Regulation root growth: AtbHLH74 [97] |
| CL4342.Contig3 | 2304 | 2.02a | – | |||
| CL9935.Contig2 | 1894 | 7.50a | 0.42b | 25 | AtbHLH137 | |
| Unigene21382 | 845 | 4.85a | – | 25 | AtbHLH63 | |
| Unigene29122 | 545 | 8.35a | 2.14a | 1 | AtbHLH33 | Cold tolerance: AtbHLH33,AtbHLH116(ICE1),AtbHLH61and AtbHLH93 [98] |
| AtbHLH116 | Stomatal differentiation: AtbHLH33(ICE2)and AtbHLH116 [99]; | |||||
| AtbHLH61 | Drought stress:AtbHLH116(ICE1) [100] | |||||
| ,AtbHLH93 | ||||||
| CL1034.Contig1 | 3358 | – | 0.30b | 1 | AtbHLH35 | |
| CL1034.Contig2 | 889 | – | 0.27b | AtbHLH27 | Drought stress:bHLH27 [100] | |
| CL1034.Contig5 | 942 | – | 0.27b | AtbHLH29 | Iron Uptake-regulated:AtBHLH29 [101] | |
| CL1768.Contig1 | 648 | 4.33a | – | 10 | AtbHLH57, | |
| AtbHLH67, | ||||||
| AtbHLH70 | ||||||
| CL12543.Contig1 | 1074 | 3.58a | – | 10 | AtbHLH71 | |
| CL9545.Contig2 | 1190 | 2.38a | – | 10 | AtbHLH94 | |
| CL9545.Contig1 | 813 | 2.31a | – | AtbHLH96 | ||
| Unigene17438 | 326 | 2.29a | – | |||
| CL13089.Contig1 | 2067 | 0.37b | – | 10 | AtbHLH57 | |
| Unigene32633 | 1085 | 3.54b | – | 9 | AtbHLH91 | |
| AtbHLH10 | ||||||
| AtbHLH89 | ||||||
| Unigene10835 | 1585 | 0.34b | – | 26 | AtbHLH69 | Female gametophyte development; |
| AtbHLH66 | Response to phosphate deficiency stress:AbHLH69, AbHLH66 [53] | |||||
| Unigene2520 | 732 | 2.89a | – | 16 | AtbHLH135 | |
| Unigene5385 | 844 | 2.74a | – | 5 | AtbHLH42 | Anthocyanin biosynthesis (GL3, EGL3, TT8) [53] |
| Unigene21617 | 2490 | 2.35a | – | Regulate proanthocyanidin biosynthesis [49, 51] | ||
| Unigene23312 | 1076 | 2.49a | – | 13 | AtbHLH106 | Abiotic stress-involved in cold, salt, ABA and drought stress: |
| AtbHLH107 | AtbHLH106 [102] | |||||
| Unigene47124 | 874 | 2.47↑ | 0.43b | 27 | AtbHLH128, | |
| Unigene39259 | 789 | – | 0.00b | AtbHLH129 | Regulation root elongation and ABA response:AtbHLH129 [103] | |
| AtbHLH80 | ||||||
| AtbHLH81 | ||||||
| AtbHLH122 | Drought and osmotic stress tolerance, ABA catabolism repression: AtbHLH122 [100] | |||||
| AtbHLH130 | ||||||
| Unigene28617 | 886 | 2.23a | – | 15 | AtbHLH133 | |
| AtbHLH68 | ||||||
| CL8951.Contig3 | 2042 | – | 0.30b | 15 | AtbHLH123 | |
| Unigene38437 | 809 | 2.20a | – | 19 | AtbHLH149 | |
| CL496.Contig1 | 889 | 2.19a | – | 31 | AtbHLH140 | |
| Unigene20853 | 1750 | – | 2.87a | 31 | AtbHLH87 | Flower and fruit development, initiation/maintenanceofaxillary meristems [53] |
| CL2917.Contig5 | 3168 | – | 0.28b | 2 | AtbHLH3 | Male fertility-affected:AtbHLH3(JAM3) [104] |
| Unigene63328 | 1505 | – | 4.65a | 2 | AtbHLH14 | |
| CL10048.Contig2 | 1395 | – | 0.05b | 7 | AtbHLH92 | Tolerance to NaCl and osmotic stresses: bHLH92 [105] |
| CL1061.Contig1 | 2440 | – | 0.10b | 7 | AtbHLH41 | |
Note:“a”indicates significant up-regulation; “–”no difference; “b” indicates significant down-regulation
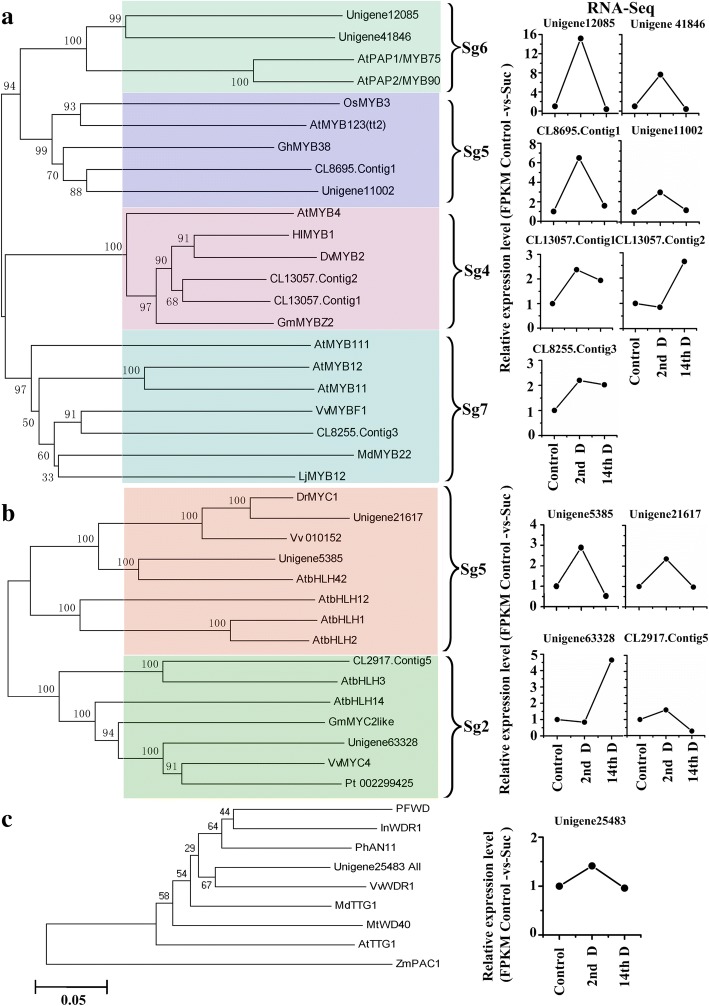
The R2R3-MYBs, bHLH, and WD40 TFs, could act as regulators of polyphenol biosynthesis individually or jointly. The R2R3-MYBs in Subgroup (Sg) 4 and Sg7 were predicted to be negative and positive regulators, respectively, for controlling the production of flavonols via regulating the up-stream genes involved in polyphenol biosynthetic pathway [33, 34]. However, the R2R3-MYBs in Sg5 and Sg6 require both bHLH (subfamily 2, 5, and 24) and WD40 for construction into a ternary complex MYB-bHLH-WD40 (MBW) for positively regulating down-stream genes involved in polyphenol biosynthetic pathway [31, 35, 36]. Here, 7 DEGs were classified into the above mentioned 4 subgroups of R2R3-MYBs. After 2d sucrose treatment, the expression of 3 DEGs (Unigene12085, Unigene 41,846 and CL8695 Contig1) in Sg6 and Sg5 were significantly up-regulated 6-fold; and the expression of CL13057.Contig2 in Sg4 was down-regulated significantly (Fig. 6a). Additionally, 2 DEGs (Unigene 21,617, Unigene 5385) in Subfamily 5 of bHLHs were up-regulated by sucrose (Fig. 6b). Based on the same method, only one transcript (Unigene25483) was predicted to be involved in the MBW complex, and its expression was not affected by sucrose (Fig. 6c).
Fig. 6.
Effects of sucrose on the expression of R2R3-MYB (a), bHLH (b) and WD40 (c) involved in polyphenol biosynthesis. Note: The phylogenetic tree was constructed based on amino acid sequences using MEGA5 according to the neighbor-joining method. GenBank accession numbers: MYB-Sg4: AtMYB4 (AEE86955), HlMYB1 (CAI46244), DvMYB2 (BAJ33514), GmMYBZ2 (ABI73970); MYB-Sg5: OsMYB3 (BAA23339), AtMYB123 (Q9FJA2), GhMYB38 (AAK19618); MYB-Sg6: AtMYB75 (AEE33419), AtMYB90 (AEE34503); MYB-Sg7: AtMYB11 (XP_002876680), AtMYB12 (O22264), AtMYB111 (XP_002865729), VvMYBF1 (ACV81697), MdMYB22 (AAZ20438), LjMYB12 (BAF74782). bHLH-Sg5: AtbHLH12 (Q8W2F1), AtbHLH42 (Q9FT81), AtbHLH1 (Q9FN69), AtbHLH2 (Q9CAD0), DrMYC1 (AEC03343), Vv_010152 (CAN62848.1); bHLH-Sg2: AtbHLH3 (O23487), AtbHLH14 (O23090), GmMYC2like (XP_003528771), VvMYC4 (XP_002279973), Pt_002299425 (XP_002299425). WD40: PFWD (BAB58883), InWDR (BAE94407), PhAN11 (AAC18914), VvWDR1 (NP_001268101), MdTTG1 (ADI58760), AtTTG1 (CAB45372), ZmPAC1 (AAM76742)
Effects of sucrose on the expression of genes involved in polyphenol transport
In plants, transporters (ABCs and MATEs), and GSTs are involved in polyphenol transporting. These transporters are found in many species including Arabidopsis TT19 and TT12 genes (AtTT19; AtTT12), the grape GST and ABCC1 genes (VvGST19; VvABCC1), the maize MRP3 gene (ZmMRP3), and the Medicago truncatula MATE (MtMATE) [37–42]. In the present study, 22, 15, and 21 DEGs were predicted to encode GST, ABC, and MATE-transporters, respectively. Phylogenetic analysis showed three transcripts closely corresponding to the above 3 transporters (Fig. 7). Among them, the expression of the ABC (CL11884.Contig7) and MATE (Unigene47970) decreases significantly by sucrose after 2d, and their expression increases after 14d (Additional file 10: Table S6). However, the expression of the GST (Unigene24131) responds to sucrose opposite of the above mentioned two transcripts (Additional file 10: Table S6). The above results indicate there could be different transporters and GSTS for transporting the polyphenol in tea plants.
Fig. 7.
Effects of sucrose on the expression of Unigenes encoding transporters related to flavonoid. a. Glutathione S-transferase;b. ABC transporters;c. mate transporters. Note: The phylogenetic tree was constructed based on amino acid sequences using MEGA5 according to the neighbor-joining method. All protein sequences used in this figure were provided in Additional file 13: Txt S1
Using qRT-PCR for transcriptome sequencing validation
To validate the results of transcriptome sequencing, 30 DEGS were randomly selected to be analyzed by qRT-PCR. We found that 83.33% of the total transcripts expression was consistent with the results from transcriptome sequencing, including 11 genes involved in polyphenol biosynthesis. Detailed information regarding the selected DEGs and 11 genes is presented in Additional file 11: Figure S5.
Discussion
The mechanisms of sucrose effects on tea polyphenol biosynthesis
In the past decades, exploration of tea polyphenol biosynthesis and their influencing factors have become a hotspot for research in plant secondary metabolism [30, 43]. Due to self-incompatibility, rich genetic diversity, and the large genome in tea plants, little genomic information is available and the molecular mechanisms of tea polyphenol biosynthesis are still unclear [44, 45]. Our previous research demonstrated tea polyphenol shared a similar biosynthetic pathway to other plants, such as shikimic acid, phenylpropanoid, and flavonoids synthetic pathways [2]. Its biosynthesis is also affected by sucrose, light, and other factors [24, 46].
Studies have demonstrated sucrose-specific transcriptional regulation of polyphenol biosynthesis in plants. For example, Boss et al. reported that the expression of DFR involved in anthocyanin and PAs biosynthesis in grape was induced by sucrose treatment, and they speculated that the accumulation of the two metabolites in grape berry skin could be attributed to sugar accumulation during grape berry development [47]. According to microarray data, it was revealed that anthocyanin biosynthesis in Arabidopsisis is stimulated by sucrose which acts as a signal to activate PAP1, a TF for activating the expression of structural genes involved in anthocyanin biosynthetic pathway, such as PAL, Cinnamate 4-hydroxylase (C4H), 4-coumaroyl-CoA ligase (4CL), and others [19, 23]. However, the structural gene F3′5′H and transcriptional factor PAP2 are not affected by sucrose [19]. In tea plants, Wang et al. found the expression of Cs F3′5′H increased 15-fold by feeding sucrose [25]. Liu et al. reported that sucrose induced the accumulation of catechins and upregulated the expression of putative genes involved in their biosynthetic pathway [24]. In this study, the total content of catechins and PAs significantly increases by sucrose induction for 7d and the accumulation of anthocyanin increases 7-fold in the stems of tea plantlets after 14d sucrose treatment. Only after 2d treatment, the expression of structural genes involved in their biosynthesis is up-regulated based on qRT-PCR and transcriptome sequencing. After 14d, the effects of sucrose were not detected.
In Arabidopsis, the correct expression of BANYULS (BAN) as a key gene of PAs biosynthesis is necessary for activation of TT2 (AtMYB123, an R2R3-MYB TF encoded by the TRANSPARENT TESTA2 gene) and TT8 (AtbHLH42, a bHLH TF encoded by the TRANSPARENT TESTA8 gene) together with TTG1 (AtTTG1, a WD-repeat protein encoded by the TRANSPARENTTESTA GLABRA1gene) [48–50]. TT2 cannot be replaced by any other AtMYB [51]. Additionally, the genes of Sg4, 5, 6, and 7 R2R3-MYB and the Subfamily2, 5, and 24 bHLH are all involved in flavonoid biosynthesis [35, 52]. Based on their amino acid sequence alignment, it was found that 7 R2R3-MYB and 4 bHLH are predicted to participate in flavonoid biosynthesis in tea plants [53]. In the present study, seven DEGs were classified into the aforementioned four subgroups of the R2R3-MYBs and four DEGs into bHLH subfamilies 5 and 2. Among them, the expression of 3 transcripts (Unigene12085, Unigene41846, and CL8695.Contig1) in R2R3-MYB Sg6 and Sg5 were up-regulated 6-fold; this finding is consistent with those of studies indicating that sucrose can induce the expression of PAP1/MYB75, which is essential for sucrose-induced anthocyanin biosynthesis [19, 23, 48, 54]. In addition, Unigene5385 corresponded to TT8 and its expression was significantly increased by sucrose treatment for 2d, indicating that it might be involved with others in regulating the accumulation of anthocyanins and PAs [55, 56]. Notably, only one transcript (Unigene25483) corresponds closely to AtTTG1, consistent with the results reported in C. sinensis [53]. However, it was not affected by sucrose, possibly because WD40 proteins have no catalytic activity and act as docking platforms for MYB and bHLH proteins in regulating flavonoid biosynthesis [48, 51, 53, 57].
As described above, it is inferred the accumulation of tea polyphenol might be directly due to high expression of their structural genes which could be synergistically regulated by TFs.
The mechanisms of sucrose effects on tea polyphenol transport
Based on analysis of UPLC-QQQ-MS/MS, the non-galloylated catechins and oligomeric PAs were significantly induced by sucrose in bud, 3rd leaf, and lower stems after 14d treatment; however, their content in upper stems decreased significantly, especially C, EC, and their oligomeric PAs. This suggests there was flavonoid transport in tea plants. Extensive research shows GST, ABC, and MATE transporters could be involved in flavonoid transport and there are at least three mechanisms, GST-linked, Vesicle trafficking (VT), and MATE transporters [38, 39, 42, 58–61]. In the present study, only three transcripts annotated as GST, ABC, and MATE were involved in flavonoid transport, and their expression was differently affected by sucrose. As described above, it is inferred that there are varieties of proteins for synergistically transporting tea polyphenol in tea plants. However, the molecular mechanisms remain unclear.
Impact of sucrose on the volatile
It is known that the flavor of tea is basically determined by taste (non-volatile compounds) and aroma (volatile compounds) [62]. The tea polyphenol is crucial for tea taste, and the terpene derivatives including monoterpenoid and sesquiterpenoid are important aroma ingredient due to their delectable fruit fragrance and low detection threshold [63]; for example, linalool and geraniol have fruity and sweet floral scents [62]. Previous research indicated that linalool, geraniol, nerolidol, ionone, and jasmone were identified as odour-active in many types of green teas [64, 65]. In the present study, (Z)-jasmone and β-ionone content increased by 2.63 and 0.57-fold, respectively; however, linalool, geraniol and nerolidol were not significantly affected by sucrose. As the biosynthetic pathway volatile compounds is complicated, and the molecular mechanisms involving in volatile compounds affected by sucrose need to be further studied.
Conclusions
In this paper, the test-tube tea plantlets were used for investigating the effects of sucrose on polyphenol biosynthesis. Metabolomics and transcriptomics analyses indicated that sucrose up-regulation of anthocyanins, catechins, and PAs biosynthesis. Sucrose controls the expression of structural and regulating genes. Additionally, sucrose promotes the transport of polyphenol in Camellia sinensis by the predicted transporters GST, ABC, and MATE involved in polyphenol transport. In summary, these results and analyses present valuable resources for better understanding the biosynthesis molecular mechanisms underlying the main characteristics of secondary metabolites in the tea plant and help improve the nutritional quality of tea.
Methods
Plant materials and cultivation conditions
The test-tube tea plantlets [Camellia sinensis (L.) O. Kuntzevar. cultivar Nongkangzao] were initially grown in vitro on classical solid MS medium and then transferred to solid MS supplemented with 90 mM sucrose for sucrose feeding studies with 10 h of light (42 μmol/m2 s) at 24 ± 1 °C. Correspondingly, similar sized test-tube tea plantlets were transferred to classical solid MS medium for the control under the same conditions. In the above experiments, the tea plantlets were incubated on MS supplemented with 90 mM mannitol for the osmotic control.
For metabolic analysis of polyphenol, the samples of different organs (the buds, third leaves, and the upper and lower stems) were collected from the tea plantlets cultivated after 2, 7, 14, and 28d. Meanwhile, samples of leaves were also collected from the tea plantlets cultivated after 2, 7, 14 and 28d for analysis of polyphenol biosynthesis at the transcriptional level. All the collected samples were immediately frozen in liquid nitrogen and stored at − 80°Cuntil use. In this study, approximately 10 independent tea plants were collected for one biological replicate; and three biological replicates were used for analysis.
Chemicals and reagents
The compounds viz., quinic acid, β-glucogallin, galloyl acid, galloylquinic acid, caffeoylquinic acid, p-coumaroylquinic acid, catechin, epicatechin, gallocatechin, epigallocatechin, epicatechingallate, epigallocatechingallate, procyanidin B2, myricetrin, quercitrin, and kaempferitrinwere obtained from Sigma (St Louis, MO, USA) and Axxora Co. and Ltd. (Lausanne,Switzerland). Cyanidin chloride was procured from Axxora Co. and Ltd. (Lausanne, Switzerland). HPLC grade acetic acid, methanol, and acetonitrile were bought from Tedia Co., Ltd. (Fairfield, OH, USA). Concentrated hydrochloric acid, vanillin, and other solvents used for extraction were acquired from Sinopharm Chemical Reagent Co., Ltd. (Shang-hai, China).
Extraction and quantitative analysis of the polyphenol
Extraction and quantitative analysis of the polyphenol was performed with UPLC-QQQ-MS/MS as suggested by Jiang et al. [2]. The total catechins were extracted and quantitatively analyzed using 1% vanillin–HCl (w/v) according to the methods described by Wang et al. [66].
Spectrophotometry analysis of anthocyanins was carried out as described by Pang et al. and the molar absorbance of cyanidin-3-O-glucoside was used for calculating the total anthocyanin concentration [67].
The total PAs were extracted and quantitatively analyzed using spectrophotometry by the methods reported by Jang et al. and their concentration was converted by using a standard curve of procyanidin B2 [2].
Extraction and analysis of the volatile compounds
Extraction and analysis of the volatile compounds collected from the samples of the leaves of tea plantlets cultivated after14 d were performed with a headspace-solid phase microextraction (HS-SPME) fiber, coupled with gas chromatography (Agilent 7697A) and mass spectrometry (Agilent 7890A) (GC/MS). In brief, 0.3 g of leaves samples were cut up and put in the 20 ml headspace bottle 4 mL by adding boiling double distilled water dissolved 0.8 g KCl. After incubation for 1.5 min, the volatile compounds were collected using a 50/30 μm DVB/CAR/PDMS SPME fiber (Supelco, PA, USA) for 50 min at 70 °C and then desorbed into the GC injection port at 250 °C for 5 min. Subsequently, the volatile compounds were resolved by BD-5 capillary column (30 m × 0.25 mm × 0.25 μm, Agilent) for GC/MS analysis according to Han et al. [64].
RNA extraction and qRT-PCR analysis
Total RNA was extracted as described by Zhao et al. [53]. The RNA concentration, quality, and integrity were measured by using spectrophotometry (Agilent2100) and gel electrophoresis. The single-stranded complementary deoxyribonucleic acid (cDNA) was synthesized using Prime-Script™ (Takara, Dalian, Code: DRR037A) for qRT-PCR analysis. All the primer sequences were designed using Primer Premier 6.0 and the selected Unigene IDs are detailed in the additional file (see Additional file 12: Table S7). The qRT-PCR assays were performed by using a CFX96™ optical reaction module (Bio-RAD, USA) and the detailed detection system was the same as previously described by Zhao et al. [53]. The resultant relative expression values were normalized against the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and evaluated from the mean value of three biological and three technical replicates by the 2-ΔΔCT method [68].
Library construction, RNA-seq and de novo assembly
Library Construction and de novo assembly were performed by Beijing Genome Institute (BGI; Shenzhen, China). Briefly, the specific operations are summarized as follows: the mRNA isolated from the total RNA was fragmented into smaller pieces to create templates for synthesizing the first-strand cDNA. Using the first-strand cDNA as templates, the double-stranded cDNA was produced with random primers (Japan, Takara). Subsequently, these cDNA fragments were processed by end repair using DNA polymerase and polynucleotide kinase and ligation of adapters to produce approximately 200 bp fragments. Finally, these fragments were purified by using Qiaquick Gel Extraction Kit (Qiagen) and enriched with PCR to construct cDNA libraries.
In this study, four cDNA libraries (2d: 2nd D Control and Suc; 14d: 14th D Control and Suc) were examined by using the Agilent 2100 Bioanalyzer and were sequenced using Illumina HiSeq™ 2000. The clean reads were obtained from the raw reads by removing the low-quality reads and the reads with adaptors or unknown nucleotides larger than 5%. Based on assembly of clean reads separately, Unigenes were the resulting sequences after removing redundancy and short contigs separately using the short reads assembling program–Trinity.
Bioinformatics analysis of the assembled Unigenes
By using BLASTx (E-value 10− 5) against the database of NR, NT, GO, Swiss-Prot, COG, and KEGG, the assembled Unigenes were annotated for functional analysis and their expression levels were calculated by the fragments per kb per million reads (FPKM). Differentially expressed genes (DEGs) were identified with a significant threshold of|log2 Ratio of FPKM (Control-vs-Suc)| ≥ 1 and FDR ≤ 0.001 based on the P-value threshold set as ≤1e− 5. Based on FDR ≤ 0.05, KEGG Pathway analysis was performed to ascertain the main biochemical and signal transduction pathways of DEGs.
Phylogenetic analysis of transcription factors and transport proteins involved in polyphenols
The phylogenetic trees for transcription factors and transport proteins were constructed according to the method as described by Zhao et al. [53]. Briefly, the MEGA 5.0 software was used for the phylogenetic analysis and the neighbor-joining statistical method was carried out based on amino acid sequences. The Bootstrap method with 1000 replicates was performed for evaluating the tree nodes. By using the p-distance method, evolutionary distances were computed. All the sequences used for the alignment were retrieved from The Arabidopsis Information Resource (TAIR, Carnegie Institution for Science Department of Plant Biology, USA), the UniProt Database (UniProt, Switzerland), and the National Center for Biotechnology Information (NCBI, USA).
Availability of supporting data
The transcriptome sequencing data based on the Illumina Hiseq 2000 platform obtained from leaves of Camellia sinensisare available in NCBI SRA (https://www.ncbi.nlm.nih.gov/sra/ with accessions SRR5427581,SRR5427580,SRR5427578 and SRR5427577.
Additional files
Table S1. Effects of sucrose on volatile compounds in leaves of tea plants using GC/ MS. Note: The data represents the mean value of three biological replications. The red indicates significant up-regulation; green indicates significant down-regulation; blue indicates no difference;. Digit indicates the ratio of Suc / Control. (DOCX 40 kb)
Table S2. Effects of sucrose on the expression of genes related to aroma. (DOCX 26 kb)
Figure S1. The pathway of terpenoids biosynthesis. (TIF 412 kb)
Table S3. Statistics of sequencing output. Note: Q20 percentage is the proportion of nucleotides with quality value larger than 20, N percentage is proportion of unknown nucleotides in clean reads, GC percentage is proportion of guanidine and cytosine nucleotides among total nucleotides. (DOCX 20 kb)
Table S4. Statistics of assembly quality. Note: Total Consensus Sequences represents the all assembled Unigenes, Distinct Clusters represents the cluster Unigenes; the same cluster contains some highly similar (more than 70%) Unigenes and these may come from same gene or homologous gene, Distinct Singletons represents Unigenes from a single gene. (DOCX 21 kb)
Table S5. Summary of Unigenes annotated to six databases. (DOCX 20 kb)
Figure S2. GO functional classification of DEGs obtained from tea plants treated by sucrose after 2d (A) and 14d (B). Note: GO functions are showed on X-axis, the right Y-axis shows the number of DEGs which have the GO function, the left Y-axis shows the percentage of DEGs. (TIF 27083 kb)
Figure S3. Evolutionary relationships of DEGs belong to R2R3-MYB obtained from tea plants treated by sucrose. Note: The phylogenetic tree was constructed based on amino acid sequences using MEGA5 per the neighbor-joining method, digit indicates subgroup, other indicates DEGs are not grouped. (TIF 18963 kb)
Figure S4. Evolutionary relationships of DEGs belong to bHLH obtained from tea plants treated by sucrose. Note: The phylogenetic tree was constructed based on amino acid sequences using MEGA5 according to the neighbor-joining method, digit indicates subfamily. (TIF 16115 kb)
Table S6. All expression data of contigs in Fig. 7. (XLSX 18 kb)
Figure S5. Validation of DEGs obtained from tea plants treated by sucrose using qRT-PCR. A. DEGs obtained from tea plants treated by sucrose after 2d; B. DEGs obtained from tea plants treated by sucrose after 14d. Note: The data of qRT-PCR represents the mean value of three biological and three technical replicates. (TIF 16000 kb)
Table S7. Primers used for qRT-PCR and detailed information regarding the selected DEGs. Note: “↑” indicates significant up-regulation; “–”no difference; “↓”indicates significant down-regulation. (DOCX 39 kb)
Txt S1. Protein sequences used in figure 7. (TXT 117 kb)
Acknowledgements
We would like to thank professor Frank Obrock and Katie Fonseca for professional writing services.
Funding
This work was funded in the framework of the Natural Science Foundation of China (31570694; 31470689; 31300577). LG, the funder of NSF (31570694) and TX, the funder of NSF (31470689) and Specialized Research Fund for the Doctoral Program of Higher Education (20133418130001) conceived and supervised this study. YQ, the funder of the Natural Science Foundation for Higher Education of Anhui Province (KJ2017A441) and the Natural Science Foundation of Suzhou University (2016jb02) performed, designed the experiments and wrote the manuscript. YJL, the funder of NSF(31270730) and Natural Science Foundation of Anhui Province, China (1408085QC51) revised the manuscript. The funders of the Special Foundation for Independent Innovation of Anhui Province, China (13Z03012), the Biology Key Subject Construction of Anhui and ‘Hundred Talents Program’ of the Chinese Academy of Sciences(39391503–7), Anhui Major Demonstration Project for the Leading Talent Team on Tea Chemistry and Health and the Innovative Research Team in University (IRT1101) had no role in the experiment design, data analysis, decision to publish or preparation of the manuscript but supported this study.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- 4CL
4-coumaroyl-CoA ligase
- ABCtransporter
ATP-binding cassette transporter
- ANR
Anthocyanidin reductase
- ANS
Anthocyanidinsynthase
- At
Arabidopsis thaliana
- C
Catechin
- C4H
Cinnamate 4-hydroxylase
- cDNA
Single-stranded complementary deoxyribonucleic acid
- CHI
Chalconeisomerase
- CHS
Chalcone synthase
- Cs
Camellia sinensis
- DEGs
Differentially expressed genes
- DFR
Dihydroflavonol 4-reductase
- EC
Epicatechin
- ECGT
Epicatechin:1-O-galloyl-β-D-glucose-O-galloyltransferase
- F3′5′H
Flavonoid3′,5′-hydroxylase
- F3′H
Flavonoid 3′-hydroxylase
- F3H
Flavanone 3-hydroxylase
- FDR
False discovery rate
- FLS
Flavonol synthase
- FPKM
Fragments per kb per million reads
- GAPDH
Glyceraldehyde-3-phosphate dehydrogenase
- GST
Glutathione S-transferase
- LAR
Leucoanthocyanidinreductase
- MATE transporter
Multidrug and toxic compound extrusion transporter
- MBW
MYB-bHLH-WD40
- MS
Murashige and Skoog standard medium
- NGS
The next-generation sequencing
- PAL
Phenylalanine ammonialyase
- PAP1
Production of anthocyanin pigment 1
- PAP2
Production of anthocyanin pigment 2
- PAs
Proanthocyanidins
- qRT-PCR
Quantitative real-time-PCR
- Sg
Subgroup
- TF
Transcription factor
- TT12
Transparent testa 12
- TT19
Transparent testa19
- TT2
Transparent testa 2
- TTG1
Transparent testa glabra1
- UGT
UDPG-glucosyltransferase
- UPLC-QQQ-MS/MS
Ultra-performance liquid chromatography-triple quadrupole mass spectrometry
Authors’ contributions
TX and LG conceived and supervised this study. YQ and SZ performed the experiments and designed the experiments. SY designed the GC/MS method and analysed the date. JX and YZL analyzed the data. YQ and YJL wrote and edited this manuscript. XD participated in sample collection. XJ designed the UPLC-QQQ-MS/MS method. WW and ML performed RNA preparation. All authors read and approved the final manuscript.
Competing interest
The authors declare that they have no competing interests.
Ethics approval and consent to participate
Not applicable.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Yumei Qian, Shuxiang Zhang and Shengbo Yao contributed equally to this work.
Electronic supplementary material
The online version of this article (10.1186/s12870-018-1335-0) contains supplementary material, which is available to authorized users.
Contributor Information
Yumei Qian, Email: qianym306@126.com.
Shuxiang Zhang, Email: zhangshuxiang90@126.com.
Shengbo Yao, Email: yaoandtea@163.com.
Jinxin Xia, Email: 15155137110@163.com.
Yanzhi Li, Email: zhizhikaoyan@163.com.
Xinlong Dai, Email: xinlongdai@163.com.
Wenzhao Wang, Email: wangwenzhao@ahau.edu.cn.
Xiaolan Jiang, Email: jiangxiaolan128@163.com.
Yajun Liu, Email: liuyajun1228@163.com.
Mingzhuo Li, Email: mingzhuo.1234@163.com.
Liping Gao, Email: gaolp62@126.com.
Tao Xia, Email: xiatao62@126.com.
References
- 1.Liang Y, Ma W, Lu J, Wu Y. Comparison of chemical compositions of Ilex latifolia thumb and Camellia sinensis L. Food Chem. 2001;75(3):339–343. doi: 10.1016/S0308-8146(01)00209-6. [DOI] [Google Scholar]
- 2.Jiang X, Liu Y, Li W, Zhao L, Meng F, Wang Y, Tan H, Yang H, Wei C, Wan X, et al. Tissue-specific, development-dependent phenolic compounds accumulation profile and gene expression pattern in tea plant [Camellia sinensis] PLoS One. 2013;8(4):e62315. doi: 10.1371/journal.pone.0062315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Winkel-Shirley B. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 2001;126(2):485–493. doi: 10.1104/pp.126.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xia T, Gao L. Advances in biosynthesis pathways and regulation of flavonoids and Catechins. Sci Agric Sin. 2009;8:2899–2908. [Google Scholar]
- 5.Treutter D. Significance of flavonoids in plant resistance and enhancement of their biosynthesis. Plant Biol. 2005;7(6):581–591. doi: 10.1055/s-2005-873009. [DOI] [PubMed] [Google Scholar]
- 6.Yang L, Zheng G, Liang L, Yang Y, Gao X. The research progress on pest and disease resistance of tea plant. Tea in Fujian. 2008;30(2):8–11. [Google Scholar]
- 7.Carmen C, Reyes A, Rafael G. Beneficial effects of green tea- a review. J Am Coll Nutr. 2006;25(2):79–99. doi: 10.1080/07315724.2006.10719518. [DOI] [PubMed] [Google Scholar]
- 8.Malusà E, Russo MA. Modification of secondary metabolism and flavonoid biosynthesis under phosphate deficiency in bean roots. J Plant Nutr. 2006;29(2):245–258. doi: 10.1080/01904160500474090. [DOI] [Google Scholar]
- 9.Santos RMFGAC, Ferri PH, Santos SC. Influence of foliar nutrients on phenol levels in leaves of Eugenia uniflora. Rev Bras. 2011;21(4):581–586. [Google Scholar]
- 10.Gumerova EA, Akulov AN, Rumyantseva NI. Effect of methyl jasmonate on growth characteristics and accumulation of phenolic compounds in suspension culture of tartary buckwheat. Russ J Plant Physiol. 2015;62(2):195–203. doi: 10.1134/S1021443715020077. [DOI] [Google Scholar]
- 11.Galieni A, Mattia CD, Gregorio MD, Speca S, Mastrocola D, Pisante M, Stagnari F. Effects of nutrient deficiency and abiotic environmental stresses on yield, phenolic compounds and antiradical activity in lettuce (Lactuca sativa L.) Sci Hortic. 2015;187:93–101. doi: 10.1016/j.scienta.2015.02.036. [DOI] [Google Scholar]
- 12.Wang YS, Gao LP, Shan Y, Liu YJ, Tian YW, Xia T. Influence of shade on flavonoid biosynthesis in tea (Camellia sinensis (L.) O. Kuntze) Sci Hortic. 2012;141(3):7–16. doi: 10.1016/j.scienta.2012.04.013. [DOI] [Google Scholar]
- 13.Wang YS, Gao LP, Wang ZR, Liu YJ, Sun ML, Yang DQ, Wei CL, Shan Y, Xia T. Light-induced expression of genes involved in phenylpropanoid biosynthetic pathways in callus of tea ( Camellia sinensis (L.) O. Kuntze) Sci Hortic. 2012;133(1):72–83. doi: 10.1016/j.scienta.2011.10.017. [DOI] [Google Scholar]
- 14.Horacio P, Martinez-Noel G. Sucrose signaling in plants: a world yet to be explored. Plant Signal Behav. 2013;8(3):e23316. doi: 10.4161/psb.23316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jia H, Wang Y, Sun M, Li B, Han Y, Zhao Y, Li X, Ding N, Li C, Ji W. Sucrose functions as a signal involved in the regulation of strawberry fruit development and ripening. New Phytol. 2013;198(2):453–465. doi: 10.1111/nph.12176. [DOI] [PubMed] [Google Scholar]
- 16.Boss PK, Davies C, Robinson SP. Expression of anthocyanin biosynthesis pathway genes in red and white grapes. Plant Mol Biol. 1996;32(3):565–569. doi: 10.1007/BF00019111. [DOI] [PubMed] [Google Scholar]
- 17.Larronde F, Krisa S, Decendit A, Chèze C, Deffieux G, Mérillon JM. Regulation of polyphenol production in Vitis vinifera cell suspension cultures by sugars. Plant Cell Rep. 1998;17(12):946–950. doi: 10.1007/s002990050515. [DOI] [PubMed] [Google Scholar]
- 18.Hara M, Oki K, Hoshino K, Kuboi T. Enhancement of anthocyanin biosynthesis by sugar in radish (Raphanus sativus) hypocotyl. Plant Sci. 2003;164(2):259–265. doi: 10.1016/S0168-9452(02)00408-9. [DOI] [Google Scholar]
- 19.Cinzia S, Alessandra P, Elena L, Amedeo A, Pierdomenico P. Sucrose-specific induction of the anthocyanin biosynthetic pathway in Arabidopsis. Plant Physiol. 2006;140(2):637–646. doi: 10.1104/pp.105.072579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dubos CGJ, Baudry A, Huep G, Lanet E, Debeaujon I, Routaboul JM, Alboresi A, Weisshaar LL. MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana. Plant J Cell Mol Biol. 2008;55(6):940–953. doi: 10.1111/j.1365-313X.2008.03564.x. [DOI] [PubMed] [Google Scholar]
- 21.Kyoko M, Yoshimi U, Masaru OT. AtMYBL2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in Arabidopsis. Plant J. 2008;55(6):954–967. doi: 10.1111/j.1365-313X.2008.03565.x. [DOI] [PubMed] [Google Scholar]
- 22.Smeekens S. Sugar-induced signal transduction in plants [review] Annu Rev Plant Physiol Plant Mol Biol. 2000;51(4):49–81. doi: 10.1146/annurev.arplant.51.1.49. [DOI] [PubMed] [Google Scholar]
- 23.Teng S, Keurentjes J, Bentsink L, Koornneef M, Smeekens S. Sucrose-specific induction of anthocyanin biosynthesis in Arabidopsis requires the MYB75/PAP1 gene. Plant Physiol. 2005;139(4):1840–1852. doi: 10.1104/pp.105.066688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Y, Wang Z, Sun M, Wang Y, Gao L, Xia T. Effect of sucrose on catechins biosynthesis in Camellia sinensis (L.) O. Kuntze. J Anhui Agricultural University. 2014;41(5):743–750. [Google Scholar]
- 25.Wang YS, Xu YJ, Gao LP, Yu O, Wang XZ, He XJ, Jiang XL, Liu YJ, Xia T. Functional analysis of flavonoid 3′,5′-hydroxylase from tea plant (Camellia sinensis): critical role in the accumulation of catechins. BMC Plant Biol. 2014;14:347. doi: 10.1186/s12870-014-0347-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang C, Wang Y, Fu J, Dong L, Gao S, Du D. Transcriptomic analysis of cut tree peony with glucose supply using the RNA-Seq technique. Plant Cell Rep. 2014;33(1):111–129. doi: 10.1007/s00299-013-1516-0. [DOI] [PubMed] [Google Scholar]
- 27.Zhang HB, Xia EH, Huang H, Jiang JJ, Liu BY, Gao LZ. De novo transcriptome assembly of the wild relative of tea tree (Camellia taliensis) and comparative analysis with tea transcriptome identified putative genes associated with tea quality and stress response. BMC Genomics. 2015;16(1):1–14. doi: 10.1186/1471-2164-16-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang XC, Zhao QY, Ma CL, Zhang ZH, Cao HL, Kong YM, Yue C, Hao XY, Chen L, Ma JQ. Global transcriptome profiles of Camellia sinensis during cold acclimation. BMC Genomics. 2013;14(1):1–15. doi: 10.1186/1471-2164-14-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shi CY, Yang H, Wei CL, Yu O, Zhang ZZ, Jiang CJ, Sun J, Li YY, Chen Q, Xia T. Deep sequencing of the Camellia sinensis transcriptome revealed candidate genes for major metabolic pathways of tea-specific compounds. BMC Genomics. 2011;12(1):1–19. doi: 10.1186/1471-2164-12-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Y, Gao L, Liu L, Yang Q, Lu Z, Nie Z, Wang Y, Xia T. Purification and characterization of a novel Galloyltransferase involved in Catechin Galloylation in the tea plant [Camellia sinensis] J Biol Chem. 2012;287(53):44406–44417. doi: 10.1074/jbc.M112.403071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mano H, Ogasawara F, Sato K, Higo H, Minobe Y. Isolation of a regulatory gene of anthocyanin biosynthesis in tuberous roots of purple-fleshed sweet potato. Plant Physiol. 2007;143(3):1252–1268. doi: 10.1104/pp.106.094425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nancy T, Laurent T, Agnès A, Sandrine V, Clotilde V, Véronique C, Charles R. Ectopic expression of VvMybPA2 promotes proanthocyanidin biosynthesis in grapevine and suggests additional targets in the pathway. Plant Physiol. 2009;149(2):1028–1041. doi: 10.1104/pp.108.131862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li M, Li Y, Guo L, Gong N, Pang Y, Jiang W, Liu Y, Jiang X, Zhao L, Wang Y. Functional Characterization of Tea (Camellia sinensis) MYB4a Transcription Factor Using an Integrative Approach. Frontiers in Plant Science. 2017;8:943. [DOI] [PMC free article] [PubMed]
- 34.Stracke R, Jahns O, Keck M, Tohge T, Niehaus K, Fernie AR, Weisshaar B. Analysis of PRODUCTION OF FLAVONOL GLYCOSIDES-dependent flavonol glycoside accumulation in Arabidopsis thaliana plants reveals MYB11-, MYB12-and MYB111-independent flavonol glycoside accumulation. New Phytol. 2010;188(4):985–1000. doi: 10.1111/j.1469-8137.2010.03421.x. [DOI] [PubMed] [Google Scholar]
- 35.Lorenzo CP, Anahit G, Irma RV, Martínez-García JF, Bilbao-Castro JR, Robertson DL. Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, poplar, rice, moss, and algae. Plant Physiol. 2010;153(3):1398–1412. doi: 10.1104/pp.110.153593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gonzalez A, Zhao M, Leavitt JM, Lloyd AM. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 2008;53(5):814–827. doi: 10.1111/j.1365-313X.2007.03373.x. [DOI] [PubMed] [Google Scholar]
- 37.Debeaujon I, Léon-Kloosterziel KM, Koornneef M. The TRANSPARENT TESTA12 gene of Arabidopsis encodes a multidrug secondary transporter-like protein required for flavonoid sequestration in vacuoles of the seed coat endothelium. Plant Cell. 2001;13(4):853–871. doi: 10.1105/tpc.13.4.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kitamura S, Shikazono N, Tanaka A. TRANSPARENT TESTA 19 is involved in the accumulation of both anthocyanins and proanthocyanidins in Arabidopsis. Tohoku J Agricultural Res. 2004;37(1):104–114. doi: 10.1046/j.1365-313x.2003.01943.x. [DOI] [PubMed] [Google Scholar]
- 39.Gomez C, Conejero G, Torregrosa L, Cheynier V, Terrier N, Ageorges A. In vivo grapevine anthocyanin transport involves vesicle-mediated trafficking and the contribution of anthoMATE transporters and GST. Plant J. 2011;67(6):960–970. doi: 10.1111/j.1365-313X.2011.04648.x. [DOI] [PubMed] [Google Scholar]
- 40.Francisco RM, Regalado A, Ageorges A, Burla BJ, Bassin B, Eisenach C, Zarrouk O, Vialet S, Marlin T, Chaves MM. ABCC1, an ATP binding cassette protein from grape berry, transports anthocyanidin 3-O-glucosides. Plant Cell. 2013;25(5):1840–1854. doi: 10.1105/tpc.112.102152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marrs KA, Alfenito MR, Lloyd AM, Walbot V. A glutathione S-transferase involved in vacuolar transfer encoded by the maize gene Bronze-2. Nature. 1995;375(6530):397–400. doi: 10.1038/375397a0. [DOI] [PubMed] [Google Scholar]
- 42.Zhao J, Dixon RA. MATE transporters facilitate vacuolar uptake of epicatechin 3'-O-glucoside for proanthocyanidin biosynthesis in Medicago truncatula and Arabidopsis. Plant Cell. 2009;21(8):2323–2340. doi: 10.1105/tpc.109.067819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jiang X, Liu Y, Wu Y, Tan H, Meng F, Ys W, Li M, Zhao L, Liu L, Qian Y, et al. Analysis of accumulation patterns and preliminary study on the condensation mechanism of proanthocyanidins in the tea plant [Camellia sinensis] Sci Rep. 2015;5:srep08742. doi: 10.1038/srep08742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tanaka J, Taniguchi F, Hirai N, Yamaguchi S. Estimation of the genome size of tea (Camellia sinensis), Camellia (C. Japonica), and their interspecific hybrids by flow cytometry. Chagyo Kenkyu Hokoku. 2006;101(101):1–7. doi: 10.5979/cha.2006.1. [DOI] [Google Scholar]
- 45.Zhao DW, Yang JB, Yang SX, Kato K, Luo JP. Genetic diversity and domestication origin of tea plant Camellia taliensis (Theaceae) as revealed by microsatellite markers. BMC Plant Biol. 2014;14(1):1–12. doi: 10.1186/1471-2229-14-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lu Z, Liu Y, Zhao L, Jiang X, Li M, Wang Y, Xu Y, Gao L, Xia T. Effect of low-intensity white light mediated de-etiolation on the biosynthesis of polyphenols in tea seedlings. Plant Physiol Biochem. 2014;80:328–336. doi: 10.1016/j.plaphy.2014.04.016. [DOI] [PubMed] [Google Scholar]
- 47.Boss PK, Davies C, Robinson SP. Analysis of the expression of anthocyanin pathway genes in developing Vitis vinifera L. cv shiraz grape berries and the implications for pathway regulation. Plant Physiol. 1996;111(4):1059–1066. doi: 10.1104/pp.111.4.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nesi N, Jond C, Debeaujon I, Caboche M, Lepiniec L. The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. Plant Cell. 2001;13(9):2099–2114. doi: 10.1105/TPC.010098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nesi N, Debeaujon I, Jond C, Pelletier G, Caboche M, Lepiniec L. The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. Plant Cell. 2000;12(10):1863–1878. doi: 10.1105/tpc.12.10.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Walker AR, Davison PA, Bolognesi-Winfield AC, James CM, Srinivasan N, Blundell TL, Esch JJ, Marks MD, Gray JC. The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell. 1999;11(7):1337–1349. doi: 10.1105/tpc.11.7.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baudry A, HM A, Bertr D, Michel C, Bernd W, Lo L. Iuml: TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J Cell Mol Biol. 2004;39(3):366–380. doi: 10.1111/j.1365-313X.2004.02138.x. [DOI] [PubMed] [Google Scholar]
- 52.Hichri I, Barrieu F, Bogs J, Kappel C, Delrot S, Lauvergeat V. Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. J Exp Bot. 2011;62(8):2465–2483. doi: 10.1093/jxb/erq442. [DOI] [PubMed] [Google Scholar]
- 53.Zhao L, Gao L, Wang H, Chen X, Wang Y, Yang H, Wei C, Wan X, Xia T. The R2R3-MYB, bHLH, WD40, and related transcription factors in flavonoid biosynthesis. Funct Integr Genomics. 2013;13(1):75–98. doi: 10.1007/s10142-012-0301-4. [DOI] [PubMed] [Google Scholar]
- 54.Borevitz JO, Xia Y, Blount J, Dixon RA, Lamb C. Activation tagging identifies a conserved MYB regulator of Phenylpropanoid biosynthesis. Plant Cell. 2000;12(12):2383–2394. doi: 10.1105/tpc.12.12.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Heim MA, Jakoby M, Werber M, Martin C, Weisshaar B, Bailey PC. The basic helix–loop–helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity. Mol Biol Evol. 2003;20(5):735–747. doi: 10.1093/molbev/msg088. [DOI] [PubMed] [Google Scholar]
- 56.Baudry A, Caboche M, Lepiniec L. TT8 controls its own expression in a feedback regulation involving TTG1 and homologous MYB and bHLH factors, allowing a strong and cell-specific accumulation of flavonoids in Arabidopsis thaliana. Plant J. 2006;46(5):768–779. doi: 10.1111/j.1365-313X.2006.02733.x. [DOI] [PubMed] [Google Scholar]
- 57.Pang Y, Wenger JP, Katie S, Peel GJ, Wen J, Huhman D, Allen SN, Tang Y, Cheng X, Tadege M. A WD40 repeat protein from Medicago truncatula is necessary for tissue-specific anthocyanin and proanthocyanidin biosynthesis but not for trichome development. Plant Physiol. 2009;151(3):1114–1129. doi: 10.1104/pp.109.144022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mueller LA, Goodman CD, Silady RA, Walbot V. AN9, a Petunia Glutathione S-Transferase Required for Anthocyanin Sequestration, Is a Flavonoid-Binding Protein1. Plant Physiol. 2000;123(4):1561–1570. doi: 10.1104/pp.123.4.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Goodman CD, Casati P, Walbot V. A multidrug resistance-associated protein involved in anthocyanin transport in Zea mays. Plant Cell. 2004;16(7):1812–1826. doi: 10.1105/tpc.022574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Narumi I, Kitamura S, Akita Y, Ishizaka H, Tanaka A. Molecular characterization of an anthocyanin-related glutathione S-transferase gene in cyclamen. J Plant Physiol. 2012;169(6):636–642. doi: 10.1016/j.jplph.2011.12.011. [DOI] [PubMed] [Google Scholar]
- 61.Zhao J, Dixon RA. The 'ins' and ‘outs’ of flavonoid transport. Trends Plant Sci. 2010;15(2):72–80. doi: 10.1016/j.tplants.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 62.Ho CT, Zheng X, Li S. Tea aroma formation. Food Sci Human Wellness. 2015;4(1):9–27. doi: 10.1016/j.fshw.2015.04.001. [DOI] [Google Scholar]
- 63.And CS, Schieberle P. Characterization of the key aroma compounds in the beverage prepared from Darjeeling black tea: quantitative differences between tea leaves and infusion. J Agricultural Food Chemistry. 2006;54(3):916–924. doi: 10.1021/jf052495n. [DOI] [PubMed] [Google Scholar]
- 64.Han ZX, Rana MM, Liu GF, Gao MJ, Li DX, Wu FG, Li XB, Wan XC, Wei S. Green tea flavour determinants and their changes over manufacturing processes. Food Chem. 2016;212:739–748. doi: 10.1016/j.foodchem.2016.06.049. [DOI] [PubMed] [Google Scholar]
- 65.Jumtee K, Komura H, Bamba T, Fukusaki E. Predication of Japanese green tea (Sen-cha) ranking by volatile profiling using gas chromatography mass spectrometry and multivariate analysis. J Biosci Bioeng. 2011;112(3):252–255. doi: 10.1016/j.jbiosc.2011.05.008. [DOI] [PubMed] [Google Scholar]
- 66.Wang Z, Liu Y, Gao L, Xia T, Sun M. Study on the detection of Catechins in tea plant with vanillin-acid reagent. J Anhui Agricultural University. 2010;37(4):675–681. [Google Scholar]
- 67.Pang Y, Peel GJ, Sharma SB, Tang Y, Dixon RA. A transcript profiling approach reveals an epicatechin-specific glucosyltransferase expressed in the seed coat of Medicago truncatula. Proc Natl Acad Sci. 2008;105(37):14210–14215. doi: 10.1073/pnas.0805954105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2 −ΔΔ C T method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 69.Liang G, Li Y, Ai Q, Yu D. MYB82 functions in regulation of trichome development in Arabidopsis. J Exp Bot. 2014;65(12):3215–3223. doi: 10.1093/jxb/eru179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zuluaga DL, Gonzali S, Loreti E, Pucciariello C, Degl'Innocenti E, Guidi L, Alpi A, Perata P. Arabidopsis thaliana MYB75/PAP1 transcription factor induces anthocyanin production in transgenic tomato plants. Funct Plant Biol. 2008;35(7):606–618. doi: 10.1071/FP08021. [DOI] [PubMed] [Google Scholar]
- 71.Sun B, Zhu Z, Cao P, Hao C, Chen C, Xin Z, Mao Y, Lei J, Jiang Y, Wei M. Purple foliage coloration in tea (Camellia sinensis L.) arises from activation of the R2R3-MYB transcription factor CsAN1. Sci Rep. 2016;6:srep32534. doi: 10.1038/srep32534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bery A. MYB75 functions in regulation of secondary cell wall formation in the Arabidopsis inflorescence stem. Plant Physiol. 2010;154(3):1428–1438. doi: 10.1104/pp.110.162735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lepiniec L, Debeaujon I, Routaboul J-M, Baudry A, Pourcel L, Nesi N, Caboche M. Genetics and biochemistry of seed flavonoids. Annu Rev Plant Biol. 2006;57:405–430. doi: 10.1146/annurev.arplant.57.032905.105252. [DOI] [PubMed] [Google Scholar]
- 74.Zhang Y, Cao G, Qu L-J, Gu H. Characterization of Arabidopsis MYB transcription factor gene AtMYB17 and its possible regulation by LEAFY and AGL15. J Genetics Genomics. 2009;36(2):99–107. doi: 10.1016/S1673-8527(08)60096-X. [DOI] [PubMed] [Google Scholar]
- 75.Pastore JJ, Limpuangthip A, Yamaguchi N, Wu M-F, Sang Y, Han S-K, Malaspina L, Chavdaroff N, Yamaguchi A, Wagner D. LATE MERISTEM IDENTITY2 acts together with LEAFY to activate APETALA1. Development. 2011;138(15):3189–3198. doi: 10.1242/dev.063073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Baumann K, Perez-Rodriguez M, Bradley D, Venail J, Bailey P, Jin H, Koes R, Roberts K, Martin C. Control of cell and petal morphogenesis by R2R3 MYB transcription factors. Development. 2007;134(9):1691–1701. doi: 10.1242/dev.02836. [DOI] [PubMed] [Google Scholar]
- 77.Jakoby MJ, Falkenhan D, Mader MT, Brininstool G, Wischnitzki E, Platz N, Hudson A, Hülskamp M, Larkin J, Schnittger A. Transcriptional profiling of mature Arabidopsis trichomes reveals that NOECK encodes the MIXTA-like transcriptional regulator MYB106. Plant Physiol. 2008;148(3):1583–1602. doi: 10.1104/pp.108.126979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kang YH, Kirik V, Hulskamp M, Nam KH, Hagely K, Lee MM, Schiefelbein J. The MYB23 gene provides a positive feedback loop for cell fate specification in the Arabidopsis root epidermis. Plant Cell. 2009;21(4):1080–1094. doi: 10.1105/tpc.108.063180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ambawat S, Sharma P, Yadav NR, Yadav RC. MYB transcription factor genes as regulators for plant responses: an overview. Physiol Mol Biol Plants. 2013;19(3):307–321. doi: 10.1007/s12298-013-0179-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Morant M, Ekstrøm C, Ulvskov P, Kristensen C, Rudemo M, Olsen CE, Hansen J, Jørgensen K, Jørgensen B, Møller BL. Metabolomic, transcriptional, hormonal, and signaling cross-talk in superroot2. Mol Plant. 2010;3(1):192–211. doi: 10.1093/mp/ssp098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jin H, Cominelli E, Bailey P, Parr A, Mehrtens F, Jones J, Tonelli C, Weisshaar B, Martin C. Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J. 2000;19(22):6150–6161. doi: 10.1093/emboj/19.22.6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhong R, Lee C, Zhou J, McCarthy RL, Ye Z-H. A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell. 2008;20(10):2763–2782. doi: 10.1105/tpc.108.061325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gomez MD, Urbez C, Perez-Amador MA, Carbonell J. Characterization of constricted fruit (ctf) mutant uncovers a role for AtMYB117/LOF1 in ovule and fruit development in Arabidopsis thaliana. PLoS One. 2011;6(4):e18760. doi: 10.1371/journal.pone.0018760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Park MY, J-y K, Kim SY. Over expression of AtMYB52 confers ABA hypersensitivity and drought tolerance. Mol Cells. 2011;31(5):447–454. doi: 10.1007/s10059-011-0300-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kirik V, Kölle K, Wohlfarth T, Miséra S, Bäumlein H. Ectopic expression of a novel MYB gene modifies the architecture of the Arabidopsis inflorescence. Plant J. 1998;13(6):729–742. doi: 10.1046/j.1365-313X.1998.00072.x. [DOI] [PubMed] [Google Scholar]
- 86.Chen Y, Chen Z, Kang J, Kang D, Gu H, Qin G. AtMYB14 regulates cold tolerance in Arabidopsis. Plant Mol Biol Report. 2013;31(1):87–97. doi: 10.1007/s11105-012-0481-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Agarwal M, Hao Y, Kapoor A, Dong C-H, Fujii H, Zheng X, Zhu J-K. A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J Biol Chem. 2006;281(49):37636–37645. doi: 10.1074/jbc.M605895200. [DOI] [PubMed] [Google Scholar]
- 88.Guo M, Thomas J, Collins G, Timmermans MC. Direct repression of KNOX loci by the ASYMMETRIC LEAVES1 complex of Arabidopsis. Plant Cell. 2008;20(1):48–58. doi: 10.1105/tpc.107.056127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Du H, Yang S-S, Liang Z, Feng B-R, Liu L, Huang Y-B, Tang Y-X. Genome-wide analysis of the MYB transcription factor superfamily in soybean. BMC Plant Biol. 2012;12:106. doi: 10.1186/1471-2229-12-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liang Y-K, Dubos C, Dodd IC, Holroyd GH, Hetherington AM, Campbell MM. AtMYB61, an R2R3-MYB transcription factor controlling stomatal aperture in Arabidopsis thaliana. Curr Biol. 2005;15(13):1201–1206. doi: 10.1016/j.cub.2005.06.041. [DOI] [PubMed] [Google Scholar]
- 91.Romano JM, Dubos C, Prouse MB, Wilkins O, Hong H, Poole M, Kang KY, Li E, Douglas CJ, Western TL. AtMYB61, an R2R3-MYB transcription factor, functions as a pleiotropic regulator via a small gene network. New Phytol. 2012;195(4):774–786. doi: 10.1111/j.1469-8137.2012.04201.x. [DOI] [PubMed] [Google Scholar]
- 92.Albert NW, Lewis DH, Zhang H, Schwinn KE, Jameson PE, Davies KM. Members of an R2R3-MYB transcription factor family in Petunia are developmentally and environmentally regulated to control complex floral and vegetative pigmentation patterning. Plant J. 2011;65:771–784. doi: 10.1111/j.1365-313X.2010.04465.x. [DOI] [PubMed] [Google Scholar]
- 93.Jung C, Seo JS, Han SW, Koo YJ, Kim CH, Song SI, Nahm BH, Do Choi Y, Cheong J-J. Overexpression of AtMYB44 enhances stomatal closure to confer abiotic stress tolerance in transgenic Arabidopsis. Plant Physiol. 2008;146(2):623–635. doi: 10.1104/pp.107.110981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shin R, Burch AY, Huppert KA, Tiwari SB, Murphy AS, Guilfoyle TJ, Schachtman DP. The Arabidopsis transcription factor MYB77 modulates auxin signal transduction. Plant Cell. 2007;19(8):2440–2453. doi: 10.1105/tpc.107.050963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Devaiah BN, Madhuvanthi R, Karthikeyan AS, Raghothama KG. Phosphate starvation responses and gibberellic acid biosynthesis are regulated by the MYB62 transcription factor in Arabidopsis. Mol Plant. 2009;2(1):43–58. doi: 10.1093/mp/ssn081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Agustí J, Gimeno J, Merelo P, Serrano R, Cercós M, Conesa A, Talón M, Tadeo FR. Early gene expression events in the laminar abscission zone of abscission-promoted citrus leaves after a cycle of water stress/rehydration: involvement of CitbHLH1. J Exp Bot. 2012;63(17):6079–6091. doi: 10.1093/jxb/ers270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bao M, Bian H, Zha Y, Li F, Sun Y, Bai B, Chen Z, Wang J, Zhu M, Ning H. miR396a-mediated basic helix–loop–helix transcription factor bHLH74 repression acts as a regulator for root growth in Arabidopsis seedlings. Plant Cell Physiol. 2014;55(7):1343–1353. doi: 10.1093/pcp/pcu058. [DOI] [PubMed] [Google Scholar]
- 98.Huang X-S, Wang W, Zhang Q, Liu J-H. A basic helix-loop-helix transcription factor, PtrbHLH, of Poncirus trifoliata confers cold tolerance and modulates peroxidase-mediated scavenging of hydrogen peroxide. Plant Physiol. 2013;162(2):1178–1194. doi: 10.1104/pp.112.210740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kanaoka MM, Lynn Jo P, Hiroaki F, Yuki Y, Bogenschutz NL, Junji T, Jian-Kang Z, Torii KU. SCREAM/ICE1 and SCREAM2 specify three cell-state transitional steps leading to arabidopsis stomatal differentiation. Plant Cell. 2008;20(7):1775–1785. doi: 10.1105/tpc.108.060848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rasheed S, Bashir K, Matsui A, Tanaka M, Seki M. Transcriptomic analysis of soil-grown Arabidopsis thaliana roots and shoots in response to a drought stress. Front Plant Sci. 2016;7:180. doi: 10.3389/fpls.2016.00180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhang J, Zhu HF, Liang H, Liu KF, Zhang AM, Ling HQ, Wang DW. Further analysis of the function of AtBHLH29 in regulating the iron uptake process in Arabidopsis thaliana. J Integr Plant Biol. 2006;48(1):75–84. doi: 10.1111/j.1744-7909.2006.00210.x. [DOI] [Google Scholar]
- 102.Ahmad A, Niwa Y, Goto S, Ogawa T, Shimizu M, Suzuki A, Kobayashi K, Kobayashi H. bHLH106 integrates functions of multiple genes through their G-box to confer salt tolerance on Arabidopsis. PLoS One. 2015;10(5):e0126872. doi: 10.1371/journal.pone.0126872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tian H, Guo H, Dai X, Cheng Y, Zheng K, Wang X, Wang S. An ABA down-regulated bHLH transcription repressor gene, bHLH129 regulates root elongation and ABA response when overexpressed in Arabidopsis. Sci Rep. 2015;5:srep17587. doi: 10.1038/srep17587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nakata M, Ohme-Takagi M. Two bHLH-type transcription factors, JA-ASSOCIATED MYC2-LIKE2 and JAM3, are transcriptional repressors and affect male fertility. Plant Signal Behav. 2013;8(12):e26473. doi: 10.4161/psb.26473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Jiang Y, Yang B, Deyholos MK. Functional characterization of the Arabidopsis bHLH92 transcription factor in abiotic stress. Mol Gen Genomics. 2009;282(5):503–516. doi: 10.1007/s00438-009-0481-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Effects of sucrose on volatile compounds in leaves of tea plants using GC/ MS. Note: The data represents the mean value of three biological replications. The red indicates significant up-regulation; green indicates significant down-regulation; blue indicates no difference;. Digit indicates the ratio of Suc / Control. (DOCX 40 kb)
Table S2. Effects of sucrose on the expression of genes related to aroma. (DOCX 26 kb)
Figure S1. The pathway of terpenoids biosynthesis. (TIF 412 kb)
Table S3. Statistics of sequencing output. Note: Q20 percentage is the proportion of nucleotides with quality value larger than 20, N percentage is proportion of unknown nucleotides in clean reads, GC percentage is proportion of guanidine and cytosine nucleotides among total nucleotides. (DOCX 20 kb)
Table S4. Statistics of assembly quality. Note: Total Consensus Sequences represents the all assembled Unigenes, Distinct Clusters represents the cluster Unigenes; the same cluster contains some highly similar (more than 70%) Unigenes and these may come from same gene or homologous gene, Distinct Singletons represents Unigenes from a single gene. (DOCX 21 kb)
Table S5. Summary of Unigenes annotated to six databases. (DOCX 20 kb)
Figure S2. GO functional classification of DEGs obtained from tea plants treated by sucrose after 2d (A) and 14d (B). Note: GO functions are showed on X-axis, the right Y-axis shows the number of DEGs which have the GO function, the left Y-axis shows the percentage of DEGs. (TIF 27083 kb)
Figure S3. Evolutionary relationships of DEGs belong to R2R3-MYB obtained from tea plants treated by sucrose. Note: The phylogenetic tree was constructed based on amino acid sequences using MEGA5 per the neighbor-joining method, digit indicates subgroup, other indicates DEGs are not grouped. (TIF 18963 kb)
Figure S4. Evolutionary relationships of DEGs belong to bHLH obtained from tea plants treated by sucrose. Note: The phylogenetic tree was constructed based on amino acid sequences using MEGA5 according to the neighbor-joining method, digit indicates subfamily. (TIF 16115 kb)
Table S6. All expression data of contigs in Fig. 7. (XLSX 18 kb)
Figure S5. Validation of DEGs obtained from tea plants treated by sucrose using qRT-PCR. A. DEGs obtained from tea plants treated by sucrose after 2d; B. DEGs obtained from tea plants treated by sucrose after 14d. Note: The data of qRT-PCR represents the mean value of three biological and three technical replicates. (TIF 16000 kb)
Table S7. Primers used for qRT-PCR and detailed information regarding the selected DEGs. Note: “↑” indicates significant up-regulation; “–”no difference; “↓”indicates significant down-regulation. (DOCX 39 kb)
Txt S1. Protein sequences used in figure 7. (TXT 117 kb)
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.