Abstract
The general transcription factor, TFIID, consists of the TATA-binding protein (TBP) associated with a series of TBP-associated factors (TAFs) that together participate in the assembly of the transcription preinitiation complex. One of the TAFs, TAFII250, has acetyltransferase (AT) activity that is necessary for transcription of MHC class I genes: inhibition of the AT activity represses transcription. To identify potential cellular factors that might regulate the AT activity of TAFII250, a yeast two-hybrid library was screened with a TAFII250 segment (amino acids 848-1279) that spanned part of its AT domain and it's the domain that binds to the protein, RAP74. The TFIID component, TAFII55, was isolated and found to interact predominantly with the RAP74-binding domain. TAFII55 binding to TAFII250 inhibits its AT activity. Importantly, the addition of recombinant TAFII55 to in vitro transcription assays inhibits TAFII250-dependent MHC class I transcription. Thus, TAFII55 is capable of regulating TAFII250 function by modulating its AT activity.
Transcription mediated by RNA polymerase II (RNAP) requires the assembly of a preinitiation complex at the promoter. Assembly is initiated by the association of the general transcription factor (GTF), TFIID, with the promoter. Subsequently, the GTFs TFIIB, TFIIE, TFIIF, and TFIIH enter the complex; interactions between the GTFs and RNAP result in transcription initiation and elongation (1). Recruitment of RNAP to the promoter results in the phosphorylation of its carboxyl terminal domain (CTD) by TFIIH, which is required for initiation. Reinitiation depends on the prior dephosphorylation of the CTD by a phosphatase that is activated by TFIIF (2). More recently, it has been demonstrated that the function of the TFIID complex itself is regulated. TFIID consists of the TATA-binding protein (TBP) associated with a series of TBP-associated factors (TAFs) that, together, participate in the assembly of the transcription preinitiation complex. Binding of TBP to its site on DNA is inhibited by the interaction of TBP with N-terminal polypeptide of the TFIID component, TAFII250 (3). Thus, binding of TFIID to DNA seems to be regulated by allosteric changes in TAFII250 conformation that expose the TBP-binding site.
In addition to preinitiation complex assembly, transcription depends on a cascade of enzymatic activities. Included among these is the acetyltransferase (AT) activity of TAFII250 (4, 5). As we have shown, TAFII250 AT is essential for MHC class I promoter activity (4). In a temperature-sensitive TAFII250 mutant-cell line, the class I promoter is active at the permissive temperature, but inactive at the restrictive temperature where TAFII250 loses its AT activity. Furthermore, the HIV transactivator (Tat) represses transcription from the MHC class I promoter by binding to the AT domain of TAFII250 and inhibiting its AT activity (4). These findings suggested that the AT activity of TAFII250 might be a normal cellular target for regulation of transcription.
In the present study, we report that TAFII250 AT activity is regulated by TAFII55, which inhibits the AT activity upon binding to TAFII250 and represses class I transcription. Interestingly, TAFII55 binds more efficiently to the TAFII250 domain that binds the TFIIF component, RAP74 (RA4iD) than to the AT domain. We speculate that TAFII55 is a normal, cellular regulator of transcription through this interaction with TAFII250.
Materials and Methods
Constructs.
The (Gal4) DNA-binding domain TAFII250 vector was constructed by inserting the mouse TAFII250 fragment (amino acids 2541–3839) (4) into the NcoI/BamHI sites of the pAS1-CYH2 vector. The Gal4 activation domain-HeLa cell cDNA library was obtained from CLONTECH, and the mouse-thymus library kindly provided by A. Singer (National Institutes of Health, Bethesda, MD).
The Gal4 BD-AT and GAL4 BD-RAP74 interacting domain (RAPiD) vectors were generated by cloning into the NcoI and BamHI sites of pAS1-CYH2, the NcoI and BamHI linker-containing fragments of TAFII250 extending from nucleotides 2551 to 3361 and from 3363 to 3843, respectively. The subclones of the TAFII250 fragment, pCR3.1-TAFII250-AT(amino acids 848-1120) and pCR3.1-TAFII250-RAP74 (amino acids 1120–1279) were generated as described (6). The control SNAP23-expression plasmid was as described (7).
The pF:55–11d expressing the Flag-tagged TAFII55 was kindly provided by R. Roeder (Rockefeller Univ., New York). The ADTAFII55 clone was generated by PCR amplification of TAFII55 between the NdeI and BamHI sites, to generate a DNA fragment spanning the first 325 nucleotides of TAFII55. The MHC class I promoter construct, −313 chloramphenicol acetyltransferase (CAT), consisting of 313 bp of 5′ flanking sequences derived from the swine class I gene PD1 ligated to the CAT reporter gene, have been described (8). Cytomegalovirus (CMV)-promoter construct was provided with HeLa nuclear extract in vitro transcription system kit (Promega).
Yeast Two-Hybrid Screening.
Saccharomyces cerevisiae strain Y190 was sequentially transformed with the pAS1-TAFII250 bait vector and either the HeLa cDNA library or the mouse-thymus cDNA library, according to the Matchmaker yeast two-hybrid protocol (CLONTECH). Approximately 5 × 105 and 1.6 × 106 of each library were transformed into Y190 cells carrying the GAL BD-TAFII250 construct and were plated on selection medium lacking Trp, Leu, and His and supplemented with 50 mM 3-aminotriazole. Clones expressing His3 and β-galactosidase activity were isolated, and plasmid DNA was recovered and sequenced. The specificity of each clone was tested further by cotransformation into Y190 cells of the different clones with either Gal4 BD-TAFII250 or Gal4 BD-SNAP23 (as negative control), with reselection on medium lacking Trp and Leu. Clones were retested for His3 expression and β-galactosidase activity.
Isolation of Recombinant TAFII55 and ADTAFII55 Proteins.
Purification of both Flag-tagged proteins was as described (9). Briefly, bacterial cultures were induced with isopropyl β-d-thiogalactoside (IPTG) but without rifampicin. Bacterial pellets from 1-liter cultures were resuspended in 30 ml of lysis buffer (20 mM Tris, pH 7.9/20% (vol/vol) glycerol/500 mM NaCl/0.2 mM EDTA/10 mM 2-mercapto-ethanol/0.2 mM PMSF and Roche protease inhibitors tablets) and lysed by sonication. After centrifugation, the supernatant was incubated for 1 h at 4°C with 600 μl of M2 agarose beads (Sigma). The M2 agarose with the bound protein was washed four times in BC300 buffer (20 mM Tris, pH 7.9/20% (vol/vol) glycerol/300 mM KCl/0.2 mM EDTA/1 mM DTT/0.1% Nonidet/0.2 mM PMSF and Roche protease inhibitors tablets) and then incubated with 600 μl of Flag peptide (0.2 mg/ml in BC300 buffer) for 20 min. The elution was repeated four times. The eluate then was loaded at 4°C on a G25 Sephadex column to eliminate the Flag peptide. Fractions containing the purified protein were combined and dialyzed at 4°C with 20 mM Tris, pH 7.9/10% (vol/vol) glycerol/150 mM KCl/0.1 mM EDTA/1 mM DTT/0.2 mM PMSF.
Purification of dTAFII250.
dTAFII250 baculovirus stock was kindly provided by R. Tjian (Univ. of California, Berkeley). Insect SF9 cells infected by recombinant Flag-tagged dTAFII250 baculovirus were resuspended in 0.4 M KCl-HEMG buffer (including 0.1% Nonidet P-40, 1 mM DTT, 0.2 mM PMSF, and Roche inhibitors) and lysed with a tight Dounce. After centrifugation, the supernatant was incubated for 2 h at 4°C with 500 μl of M2 agarose beads, washed four times with 0.3 M KCl-HEMG, and the bound protein was eluted four times with 500 μl of Flag peptide (0.2 mg/ml in 0.3 M KCl-HEMG). The Flag peptide was eliminated either by gel filtration or dialysis with 20 mM Tris, pH 7.9/10% (vol/vol) glycerol/0.15 M KCl/0.1 mM EDTA/1 mM DTT/0.2 mM PMSF.
In Vitro Translation and Immunoprecipitations.
pCR3.1-TAFII250-AT and pCR3.1-TAFII250-RAP74 and SNAP23 were translated in vitro in the TnT-Coupled Reticulocyte Lysate System (Promega) from the T7 promoter with [35S]methionine (ICN). M2 agarose beads (Sigma) were prewashed in cold buffer B (20 mM Hepes, pH 7.9/100 mM KCl/12.5 mM MgCl2/0.1 mM DTT/0.2% Nonidet P-40/17% (vol/vol) glycerol) with 0.5 mg/ml of BSA, incubated for 2 h at 4°C with either buffer (control beads) or 2 μg of TAFII55 in buffer B. Either control or TAFII55/M2 beads were further incubated for 30 min in buffer B with 0.5 mg/ml BSA and washed. Aliquots were incubated for 1 h at 4°C with in vitro translated TAFII250 AT, TAFII250 RAPiD, or SNAP23 fragments. The complexes were washed four times in wash buffer (50 mM Tris, pH 7.9/150 mM NaCl/0.2% Nonidet P-40), and samples were resolved on a reducing SDS/15% PAGE gel and quantified by phosphorimaging (Amersham Pharmacia).
Histone Acetyltransferase (HAT) Assay.
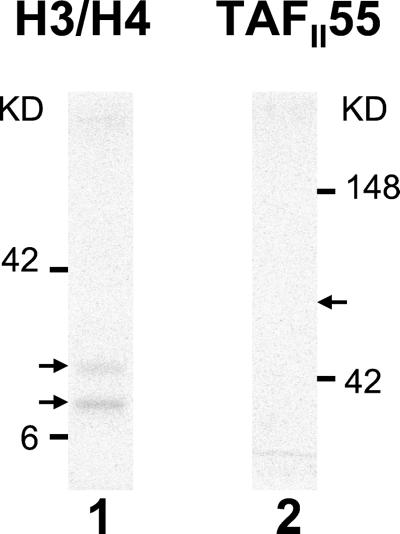
Histones H3 and H4 were prepared as described (10). The acetylation assays were a modification of the procedure described (11). dTAFII250 (250 ng) was incubated with 2 μg of histones H3/H4, 70 nCi (1 Ci = 37 GBq) of [14C]acetyl-CoA (60 mCi/mmol; Amersham Pharmacia) and 15 nCi of [3H]acetyl-Co (4.5 Ci/mmol; NEN). Reactions were performed at 30°C for 30 min in HAT buffer (25 mM Tris, pH 8/0.1 mM EDTA/1 mM DTT/10 mM butyric acid/10% (vol/vol) glycerol/0.2% PMSF). Where indicated, dTAFII250 was first preincubated with either 250 ng of control protein or 250 ng of TAFII55. Control protein in these assays was a fragment of PB1, a TAFII250 interacting protein that does not affect AT activity (A.G., et al., unpublished work). Yeast HAT1 protein was a generous gift of P. A. Wade (National Institutes of Health, Bethesda). The reactions were resolved on a reducing SDS/15% PAGE gel, processed, and quantified by phosphorimaging. In the work presented in Fig. 2, TAFII55 acetylation is performed as above, but in the absence of histones.
Figure 2.
TAFII55 is not acetylated by TAFII250. An AT assay was performed with 250 ng TAFII250 incubated in the presence of either 2 μg of histones H3/H4 (lane 1) or 0.5 μg of TAFII55 (lane 2). Arrowheads mark the positions of H3/H4 (lane 1) and TAFII55 (lane 2).
In Vitro Transcription Reactions.
In vitro transcription reactions contained 90 μg of HeLa nuclear extract and 0.8 mM rNTPs and were performed for 1 h at 20°C. TAFII55 or AD TAFII55 (250 ng, unless otherwise specified) were preincubated with the HeLa nuclear extract for 15 min at 20°C. The reaction was initiated by the addition of the promoter DNA. The conditions of the transcription reactions were optimized for each promoter. For MHC class I transcripts, 2 μg of −313 DNA was used in the presence of 0.8 mM MgCl2. For CMV transcripts, 1 μg of CMV DNA (Promega) was used in the presence of 0.3 mM MgCl2. The analysis of −313 MHC transcripts was by primer extension (4, 6). The analysis of CMV transcripts was by direct labeling of the transcripts by using 1 μl of [α-32P]UTP (NEN) in the in vitro transcription reaction according to the manufacturer's instructions.
Results
TAFII55 Binds to the TAFII250 Domain Spanning the RAP74-Binding Site.
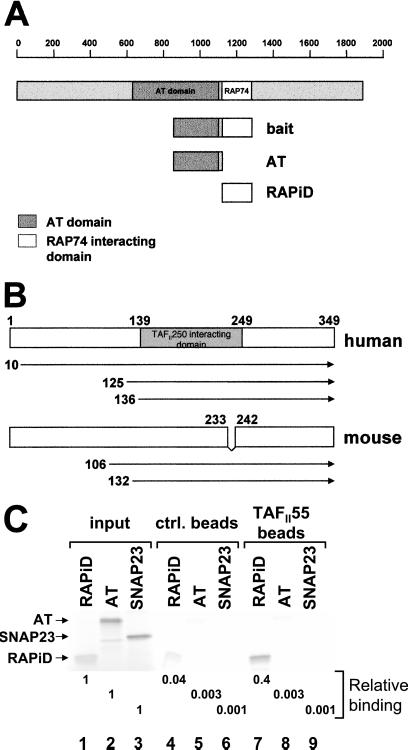
We have shown that the HIV Tat protein binds to the AT domain of TAFII250 and inhibits its activity, resulting in the repression of TAFII250-dependent MHC class I promoter activity (4). This finding raised the possibility that TAFII250 AT activity normally might be regulated by interactions with cellular proteins. To identify any such cellular proteins, we performed a yeast two-hybrid screen by using as bait the Tat-interacting domain of TAFII250 (amino acids 848-1279) fused to the Gal4 DNA-BD (Fig. 1A). Two-yeast cDNA libraries were screened: a human-HeLa library and a mouse-thymus library. Three clones were isolated from the human-HeLa library and two from the mouse-thymus library that reproducibly and specifically interacted with the TAFII250 bait and whose sequencing revealed that they encoded the TAFII55 gene. Previous studies reported that TAFII55 binds TAFII250 in vitro, but did not map the site of TAFII250 interaction (12, 13). By this screening, we show that this interaction occurs in vivo and, further, that TAFII55 binds TAFII250 in the 848-1279 region of TAFII250.
Figure 1.
TAFII55 binds TAFII250. (A) The TAFII250 fragment (amino acids 848-1279), spanning the AT and RAPiDs, was used as bait in a yeast two-hybrid screen to isolate TAFII55 clones. The location of the bait fragment, relative to the full-length molecule, is shown. TAFII250 AT domain (shaded box) is located approximately between amino acids 640 and 1093, which corresponds to the Drosophila AT domain (amino acids 612-1140) (18). (B) TAFII55 clones isolated in yeast two-hybrid screens. The locations of the five isolated TAFII55 clones are shown relative to the full-length human and mouse proteins. Although the clones differed in their 5′ termini, they shared a common 3′ end that extended to encode the carboxyl terminus of the TAFII55 protein. The mouse TAFII55 differs from the human in a deletion spanning amino acids 233–242. (C) TAFII55 binds to the RAPiD of TAFII250. Flag-tagged TAFII55 was used to assess its ability to bind to either [35S]methionine labeled, in vitro translated AT, or RAPiD domains, as shown in A (AT, lanes 2, 5, 8; RAPiD, lanes 1, 4, 7). In vitro translated SNAP23 protein was used as a control for nonspecific binding (lanes 3, 6, 9). The relative binding of the TAFII250 fragments to TAFII55 was quantitated and calculated relative to input, as shown beneath the lanes. Arrowheads mark the positions of input proteins.
The human TAFII55 is a protein of 349 amino acids (12, 13); mouse TAFII55 consists of 341 amino acids (14). TAFII55 homologues are found also in Drosophila and yeast (15, 16, 17). Of the three human TAFII55 clones isolated in the two-hybrid screen with the TAFII250 bait, one started at amino acid 10 (Fig. 1B), and the other two started at amino acids 125 and 136. Of the two mouse TAFII55 clones isolated, one began at amino acid 106 and the other at amino acid 132. All of the clones extended to the 3′ end of the coding sequence. This analysis maps a TAFII55 region of interaction with TAFII250 between amino acids 136 and 341, which is consistent with the earlier mapping of the binding of the central core of TAFII55 to TAFII250 (12). Because both the human and mouse clones bind to TAFII250, the TAFII55 segment between amino acids 233 and 242 in the human (which is missing in the mouse) is not necessary for the interaction.
The TAFII250 segment used as bait in the two-hybrid screen spans both part of the AT domain (amino acids 848-1120) and the domain RAPiD (amino acids 1120–1279). (In Fig. 1, the TAFII250 AT domain is located between amino acids 640 and 1093, corresponding to the Drosophila AT domain of amino acids 612-1140; ref. 18). To determine which of these TAFII250 domains interacted with TAFII55, we analyzed the ability of isolated TAFII250 AT and RAPiD domains to bind full-length human TAFII55 (12). In an in vitro pull-down assay, a bacterially expressed Flag-tagged TAFII55 protein bound stably to the RAPiD domain of TAFII250; binding to the AT domain was not detected above background under these conditions (Fig. 1C). In a direct in vivo yeast two-hybrid assay, TAFII55 (fused to the Gal4-activation domain) also interacted with the TAFII250 RAPiD domain fused to the Gal4 DNA-BD (data not shown). These results demonstrate that TAFII55 binds to the TAFII250 RAPiD.
In the functional studies reported below, the same full-length human TAFII55 protein is used (12).
TAFII55 Is Not Acetylated by TAFII250.
Although most ATs have been characterized by their ability to acetylate histones, there is an increasing recognition that non-histone proteins are also substrates for AT activity. For example, P300, a well characterized AT, has been shown to acetylate the transcription factor p53, thereby increasing its DNA-binding activity (19, 20). Similarly, acetylation of the transcription factor HNF-4 by CBP results in its enhanced retention in the nucleus and increased DNA binding (21). The finding that TAFII55 binds to TAFII250 raised the possibility that TAFII55 is a substrate for acetylation by TAFII250. However, under conditions in which purified TAFII250 acetylated histones H3 and H4, no acetylation of purified full-length TAFII55 protein could be detected (Fig. 2). (Increasing the amount of TAFII55 in the reaction did not reveal any acetylation; data not shown.) Therefore, TAFII55 is not a detectable substrate for the TAFII250 AT activity in vitro.
TAFII55 Inhibits TAFII250 HAT Activity.
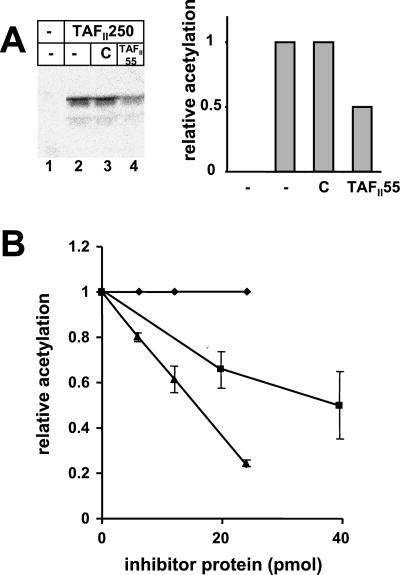
We next considered the possibility that TAFII55 binding could alter TAFII250 AT activity. To test this possibility, we measured the AT activity of TAFII250 in the presence of either TAFII55 or control protein by using histones H3/H4 as substrate. As shown in Fig. 3A, the presence of TAFII55 in the HAT assay reduced TAFII250 AT activity, whereas the control protein had no effect (Fig. 3A, compare lanes 3 and 4 and histogram). To extend this observation, we analyzed the dose–response of TAFII250 AT activity to TAFII55 (Fig. 3B). Inhibition of TAFII250 AT activity increased in the presence of increasing amounts of TAFII55; the control protein had no effect. (It is important to note that the histone substrate was present in a 15- to 20-fold molar excess relative to TAFII55. Therefore, it is unlikely that TAFII55 inhibited acetylation through an interaction with histones.)
Figure 3.
TAFII55 inhibits TAFII250 AT activity. (A) The AT activity of TAFII250 (250 ng) was assayed in the presence of 2 μg of histones H3/H4 with buffer (lane 2), 250 ng control protein (lane 3), or 250 ng TAFII55 (lane 4). Lane 1 contains histones incubated without TAFII250. Acetylation of histones was quantitated by PhosphorImager (Molecular Dynamics) and is plotted in the histogram relative to the level of acetylation observed in lane 2. (B) Increasing amounts of TAFII55 or the amino terminal fragment of TAFII55 (amino acids 1–109) result in increasing inhibition of TAFII250AT activity. TAFII250 AT activity was assayed on 2 μg of histones H3/H4 by using 250 ng TAFII250 in the presence of increasing amounts of TAFII55 (▴), the amino terminal fragment of TAFII55 (ADTAFII55, ■), or control protein (♦). The diagram represents the relative acetylation of histones in the presence of the respective proteins and summarizes seven different experiments performed in the presence of TAFII55 and four experiments performed in presence of AD TAFII55. Error bars show the standard deviation.
In contrast to TAFII250, the AT activity of yeast HAT1 is not affected by TAFII55, indicating that TAFII55 is not a histone deacetylase, nor is it a nonspecific inhibitor (A.G., data not shown). These findings are consistent with the conclusion that TAFII55 is a specific inhibitor of TAFII250 AT activity.
To map the domain of TAFII55 responsible for inhibition of AT activity, the ability of a TAFII55 fragment extending from amino acids 1 to 109 (ADTAFII55) to inhibit AT activity was examined. As shown in Fig. 3B, the amino terminal fragment of TAFII55, like the full-length molecule, was able to inhibit TAFII250 AT activity. However, approximately a 3-fold molar equivalent of ADTAFII55 was required to achieve the same level of inhibition as the full-length molecule. These findings demonstrate that TAFII55 modulates TAFII250 AT activity and maps the TAFII55 inhibitory domain to its amino terminus. Interestingly, although the amino terminus of TAFII55 contains the inhibitory activity of the molecule, and thus must interact with TAFII250, we have not been able to detect stable binding of the isolated domain to TAFII250 in pull-down assays. Rather, the major interaction of TAFII55 with TAFII250 is to the RAPiD and is mediated by the segment of TAFII55 between amino acids 139 and 249 (Fig. 1; ref. 12).
TAFII55 Represses TAFII250-Dependent MHC Class I Expression.
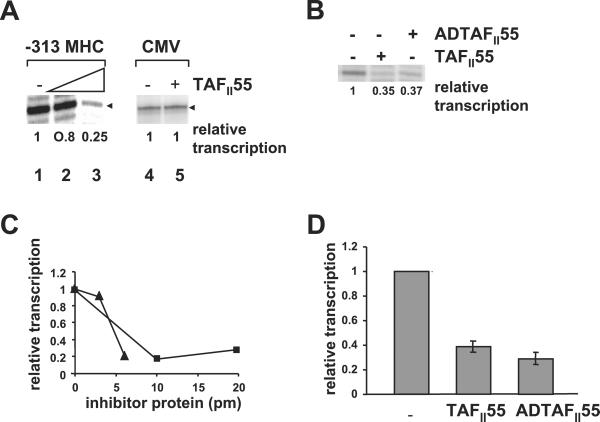
Because TAFII250 AT activity is necessary for transcription from the MHC class I promoter (4), we next examined the effect of TAFII55 on in vitro transcription of an MHC class I promoter (Fig. 4). Whereas HeLa nuclear extract alone supported in vitro transcription from the MHC class I promoter, the addition of increasing amounts of exogenous TAFII55 to the reaction decreased levels of transcription (Fig. 4A, compare lanes 1, 2, and 3; Fig. 4 C and D). To demonstrate that this observation reflects a specific effect of TAFII55 on TAFII250-dependent gene transcription and not a general repression of all transcription, we assessed the effect of TAFII55 on a promoter known to be TAFII250-independent, namely, the CMV promoter (22). As predicted, transcription from the CMV promoter was not affected by TAFII55 (Fig. 4A, lanes 4 and 5; ref. 4). Consistent with its ability to inhibit TAFII250 AT activity, the ADTAFII55 construct, containing only the amino terminal of amino acid 109, also represses transcription of the MHC class I promoter (Fig. 4 B–D). Transcription, like TAFII250 AT activity, is less efficiently inhibited by ADTAFII55 than by the full-length TAFII55 on a molar basis (Fig. 4C).
Figure 4.
TAFII55 represses in vitro transcription from the MHC class I promoter, but not the viral CMV promoter. (A) In vitro transcription reactions were performed by using either an MHC class I promoter (−313 MHC, lanes 1–3) or the viral CMV early promoter (lanes 4–5) in the absence (lanes 1, 4) or in the presence of TAFII55 (150 ng, lane 2; 250 ng, lane 3; 250 ng, lane 5). The MHC class I transcripts are detected by primer extension. The CMV transcripts are directly synthesized in the presence of [α-32P]UTP. They have been aligned for the purposes of presentation. The relative transcription is quantitated and reported below each lane. (B) Repression of transcription is mediated by the N-terminal domain of TAFII55. In vitro transcription reactions were performed by using the MHC class I promoter (−313 MHC) in the presence of buffer, TAFII55 (250 ng), or AD TAFII55 (250 ng). The relative transcription is quantitated and reported below each lane. (C) Titration of TAFII55 and ADTAFII55 inhibition of in vitro transcription of the MHC class I promoter. Increasing amounts of either TAFII55 (0, 3, or 6 pmol, ▴) or ADTAFII55 (0, 10, or 20 pmol, ■) were added to a standard in vitro transcription reaction with the −313 MHC class I promoter; the relative levels of transcription were quantitated by PhosphorImager analysis. (D) Summary of the TAFII55 and ADTAFII55 inhibition of in vitro transcription. The relative levels of transcription of −313 MHC class I promoter, in the presence or absence of 6 pmol of TAFII55 from seven different experiments or 20 pmol of ADTAFII55 from two experiments are compiled. Error bars represent standard deviation.
Taken together, these results indicate that repression of MHC class I transcription in the presence of TAFII55 is caused by the inhibition of TAFII250 AT activity by TAFII55.
Discussion
TAFII55 is a component of the TFIID complex that nucleates the preinitiation complexes associated with a large number of core promoters. In TFIID, TAFII55 interacts with other components: TAFII250, TAFII100, TAFII28, TAFII20, and TAFII18 (12, 13). In vitro, TAFII55 also interacts with a number of cellular transcription factors including USF, Sp1, and YY1, as well as viral transcription factors such as HIV Tat and E1A (12). Consistent with its association with TFIID, which is found in all cell types, TAFII55 is ubiquitously expressed (13). Despite the previous characterization of TAFII55 as a member of the TFIID complex, nothing was known about its function in transcription initiation.
In the present studies, we have identified functional activities of TAFII55. We have demonstrated that the binding of TAFII55 to TAFII250 results in the inhibition of the intrinsic AT activity of TAFII250. Most significantly, the binding of TAFII55 to TAFII250 results in the repression of the activity of an MHC class I promoter that is TAFII250-dependent. This repression is not seen with a TAFII250-independent promoter. Finally, we have mapped the regions of interaction of TAFII55 and TAFII250. Both inhibition of TAFII250 AT activity and transcription are mediated by the N-terminal of amino acids 1–109 of TAFII55. This segment of the protein contains two cysteines that could potentially form disulfide bonds and two regions capable of forming α-helices.
The mechanism by which TAFII55 regulates TAFII250 AT activity remains to be determined. Although there are multiple lysine residues in TAFII55, it is not acetylated as a result of its interaction with TAFII250. Because TAFII55 is not acetylated by TAFII250 AT activity, it is not simply competing with histone substrates; it may be acting as a competitive inhibitor. The precise mechanism by which TAFII55 represses transcription and the role of the N-terminal structure also remain to be established. We propose that repression of transcription by TAFII55 is a direct consequence of its inhibition of TAFII250 AT activity.
In previous studies, we have reported that Tat binds to TAFII250. As in the case of TAFII55, the binding of Tat results in the inhibition of the TAFII250 AT activity and the consequent repression of transcription of TAFII250-dependent promoters (4). Tat mediates repression through its C-terminal domain, which binds only weakly to the AT domain of TAFII250. It is the core of Tat that binds strongly to the RAPiD of TAFII250 that anchors Tat to TAFII250 (6). In the present study, we have found that the core of TAFII55, like the core of Tat, binds strongly to the RAPiD of TAFII250. Like Tat, the functionally active N-terminal segment of TAFII55, which inhibits the AT activity of TAFII250, must bind only weakly because binding is not detected in standard pull-down assays (data not shown). Given the striking parallels between HIV Tat and TAFII55, as shown here, we speculate that TAFII55 is a cellular analog of Tat that modulates TAFII250 function in vivo. Despite the apparent similarities in their interactions and inhibition of TAFII250 AT activity, TAFII55 and Tat have no primary amino acid sequence similarity. However, like Tat, the ability of TAFII55 to inhibit TAFII250 AT activity suggests that it plays a pivotal role in regulating normal cellular transcription from TAFII250-dependent promoters through its modulation of the AT activity.
What purpose would such regulation of TAFII250 function serve? Although basal transcription of MHC class I genes requires TAFII250, this dependence is context-dependent. Thus, it can be overcome by a variety of activators associated with upstream regulatory elements. For example, ligation of the simian virus (SV)40 viral 72-bp enhancer upstream of the class I promoter relieves it of its requirement for TAFII250 (22). As an aside, it is interesting to note that, whereas the viral SV40 promoter does not require TAFII250 in the presence of its strong viral 72-bp enhancer, removal of the enhancer renders the promoter TAFII250-dependent. Even more striking, activation of the native class I promoter by the coactivator CIITA occurs independently of the AT activity of TAFII250 (22). Thus, normal cellular mechanisms modulate the requirement for TAFII250, and TAFII55 may help to regulate this requirement.
Two distinct mechanisms by which TAFII55 could regulate transcription can be considered. In the first model, TAFII55 is constitutively associated with the TFIID complex during basal transcription but displaced by upstream activators that function through TAFII250; the displacement of TAFII55 would increase AT activity and transcription initiation. The in vitro association of TAFII55 with known activators, such as USF, is consistent with this model (12). Alternatively, in a second model, activators that bypass the requirement for TAFII250, such as CIITA, might recruit TAFII55 to the TFIID complex, thereby inhibiting TAFII250 AT activity and refocusing activity to alternative initiation complexes. In another system, Roeder and coworkers (23) have demonstrated that TFIID is not necessary for transcription in the presence of the liganded thyroid hormone receptor/TRAP coactivator complex. These findings raise the intriguing possibility of initiation complex selectivity by coactivator complexes, in which TAFII55 would block the function of TAFII250-containing complexes. Currently, we cannot distinguish between these two models. In either case, we propose that the function of TAFII55 is to regulate the activity of the TFIID complex through its inhibition of TAFII250 AT activity.
Acknowledgments
We gratefully acknowledge Drs. Kevin Howcroft, Aparna Raval, and Julie Lovchik for helpful discussions, and Heather Lazusky and Anita Hohenstein for technical assistance. We thank Drs. John Brady and Alfred Singer for critical review of the manuscript.
Abbreviations
- TBP
TATA binding protein
- TAF
TBP-associated factors
- AT
acetyltransferase
- Tat
an HIV transactivator
- RAP74
74-kd component of TFIIF
- BD
binding domain
- AD
activation domain
- CMV
cytomegalovirus
- GTF
general transcription factor
- CTD
carboxyl terminal domain
- RAPiD
RAP74 interacting domain
- HAT
histone acetyltransferase
References
- 1.Orphanides G, Lagrange T, Reinberg D. Genes Dev. 1996;10:2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- 2.Dahmus M E. J Biol Chem. 1996;271:19009–19012. doi: 10.1074/jbc.271.32.19009. [DOI] [PubMed] [Google Scholar]
- 3.Liu D, Ishima R, Tong K I, Bagby S, Kokubo T, Muhandiram D R, Kay L E, Nakatani Y, Ikura M. Cell. 1998;94:573–583. doi: 10.1016/s0092-8674(00)81599-8. [DOI] [PubMed] [Google Scholar]
- 4.Weissman J, Brown J, Howcroft T K, Hwang J, Chawla A, Roche P, Schiltz L, Nakatani Y, Singer D S. Proc Natl Acad Sci USA. 1998;95:11601–11606. doi: 10.1073/pnas.95.20.11601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dunphy E L, Johnson T, Auerbach S S, Wang E H. Mol Cell Biol. 2000;20:1134–1139. doi: 10.1128/mcb.20.4.1134-1139.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weissman J D, Hwang J R, Singer D S. Biochim Biophys Acta. 2001;1546:156–163. doi: 10.1016/s0167-4838(01)00135-2. [DOI] [PubMed] [Google Scholar]
- 7.Ravichandran V, Chawla A, Roche P A. J Biol Chem. 1996;271:13300–13303. doi: 10.1074/jbc.271.23.13300. [DOI] [PubMed] [Google Scholar]
- 8.Howcroft T K, Strebel K, Martin M, Singer D S. Science. 1993;260:91–93. doi: 10.1126/science.8493575. [DOI] [PubMed] [Google Scholar]
- 9.Chiang C M, Roeder R G. Pept Res. 1993;6:62–64. [PubMed] [Google Scholar]
- 10.Robyr D, Gegonne A, Wolffe A P, Wahli W. J Biol Chem. 2000;275:28291–28300. doi: 10.1074/jbc.M002726200. [DOI] [PubMed] [Google Scholar]
- 11.Brownell J E, Allis C D. Proc Natl Acad Sci USA. 1995;92:6364–6368. doi: 10.1073/pnas.92.14.6364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiang C, Roeder R. Science. 1995;267:531–536. doi: 10.1126/science.7824954. [DOI] [PubMed] [Google Scholar]
- 13.Lavigne A C, Mengus G, May M, Dubrovskaya V, Tora L, Chambon P, Davidson I. J Biol Chem. 1996;271:19774–19780. doi: 10.1074/jbc.271.33.19774. [DOI] [PubMed] [Google Scholar]
- 14.Wu S Y, Thomas M C, Hou S Y, Likhite V, Chiang C M. J Biol Chem. 1999;274:23480–23490. doi: 10.1074/jbc.274.33.23480. [DOI] [PubMed] [Google Scholar]
- 15.Adams M D, Celniker S E, Holt R A, Evans C A, Gocayne J D, Amanatides P G, Scherer S E, Li P W, Hoskins R A, Galle R F, et al. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 16.Moqtaderi Z, Yale J D, Struhl K, Buratowski S. Proc Natl Acad Sci USA. 1996;93:14654–14658. doi: 10.1073/pnas.93.25.14654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shibuya T, Tsuneyoshi S, Azad A K, Urushiyama S, Ohshima Y, Tani T. Genetics. 1999;152:869–880. doi: 10.1093/genetics/152.3.869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wassarman D A, Aoyagi N, Pile L A, Schlag E M. Proc Natl Acad Sci USA. 2000;97:1154–1159. doi: 10.1073/pnas.97.3.1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu L, Scolnick D M, Trievel R C, Zhang H B, Marmorstein R, Halazonetis T D, Berger S L. Mol Cell Biol. 1999;19:1202–1209. doi: 10.1128/mcb.19.2.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yuan Z M, Huang Y, Ishiko T, Nakada S, Utsugisawa T, Shioya H, Utsugisawa Y, Yokoyama K, Weichselbaum R, Shi Y, Kufe D. J Biol Chem. 1999;274:1883–1886. doi: 10.1074/jbc.274.4.1883. [DOI] [PubMed] [Google Scholar]
- 21.Soutoglou E, Katrakili N, Talianidis I. Mol Cell. 2000;5:745–751. doi: 10.1016/s1097-2765(00)80253-1. [DOI] [PubMed] [Google Scholar]
- 22.Weissman J D, Howcroft T K, Singer D S. J Biol Chem. 2000;275:10160–10167. doi: 10.1074/jbc.275.14.10160. [DOI] [PubMed] [Google Scholar]
- 23.Fondell J D, Guermah M, Malik S, Roeder R G. Proc Natl Acad Sci USA. 1999;96:1959–1964. doi: 10.1073/pnas.96.5.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]