ABSTRACT
Although Salmonella enterica can produce life-threatening colitis in horses, certain serotypes are more commonly associated with clinical disease. Our aim was to evaluate the proportional morbidity attributed to different serotypes, as well as the phenotypic and genotypic antimicrobial resistance (AMR) of Salmonella isolates from patients at an equine referral hospital in the southern United States. A total of 255 Salmonella isolates was obtained from clinical samples of patients admitted to the hospital between 2007 and 2015. Phenotypic resistance to 14 antibiotics surveilled by the U.S. National Antimicrobial Resistance Monitoring System was determined using a commercially available panel. Whole-genome sequencing was used to identify serotypes and genotypic AMR. The most common serotypes were Salmonella enterica serotype Newport (18%), Salmonella enterica serotype Anatum (15.2%), and Salmonella enterica serotype Braenderup (11.8%). Most (n = 219) of the isolates were pansusceptible, while 25 were multidrug resistant (≥3 antimicrobial classes). Genes encoding beta-lactam resistance, such as blaCMY-2, blaSHV-12, blaCTX-M-27, and blaTEM-1B, were detected. The qnrB2 and aac(6′)-Ib-cr genes were present in isolates with reduced susceptibility to ciprofloxacin. Genes encoding resistance to gentamicin (aph(3′)-Ia, aac(6′)-IIc), streptomycin (strA and strB), sulfonamides (sul1), trimethoprim (dfrA), phenicols (catA), tetracyclines [tet(A) and tet(E)], and macrolides [ere(A)] were also identified. The main predicted incompatibility plasmid type was I1 (10%). Core genome-based analyses revealed phylogenetic associations between isolates of common serotypes. The presence of AMR Salmonella in equine patients increases the risk of unsuccessful treatment and causes concern for potential zoonotic transmission to attending veterinary personnel, animal caretakers, and horse owners. Understanding the epidemiology of Salmonella in horses admitted to referral hospitals is important for the prevention, control, and treatment of salmonellosis.
IMPORTANCE In horses, salmonellosis is a leading cause of life-threatening colitis. At veterinary teaching hospitals, nosocomial outbreaks can increase the risk of zoonotic transmission, lead to restrictions on admissions, impact hospital reputation, and interrupt educational activities. The antimicrobials most often used in horses are included in the 5th revision of the World Health Organization's list of critically important antimicrobials for human medicine. Recent studies have demonstrated a trend of increasing bacterial resistance to drugs commonly used to treat Salmonella infections. In this study, we identify temporal trends in the distribution of Salmonella serotypes and their mechanisms of antimicrobial resistance; furthermore, we are able to determine the likely origin of several temporal clusters of infection by using whole-genome sequencing. These data can be used to focus strategies to better contain the dissemination and enhance the mitigation of Salmonella infections and to provide evidence-based policies and guidelines to steward antimicrobial use in veterinary medicine.
KEYWORDS: Salmonella, antibiotic resistance, antimicrobial agents, horses, nosocomial, whole-genome sequencing
INTRODUCTION
Salmonellosis is a leading cause of life-threatening colitis in horses, and it results in economic losses related to treatment costs, morbidity, and mortality (1). Although the trends in dominant serotypes may have varied over the years, the most common serotypes reported in clinical cases from horses in the United States are Salmonella enterica serotype Typhimurium, Salmonella enterica serotype Newport, Salmonella enterica serotype Javiana, and Salmonella enterica serotype Anatum (2). S. Newport is one of the most-reported serotypes associated with nosocomial Salmonella enterica outbreaks in horses in the United States (3). The most common antimicrobials used for salmonellosis treatment in horses are ceftiofur, enrofloxacin, and gentamicin; however, the use of such drugs may favor the persistence of the organism in the intestines following recovery because antimicrobial drugs do not kill all of the Salmonella bacteria in the intestine and may disrupt the normal intestinal microbiota that normally competes with Salmonella for nutrients and helps exclude the pathogen in healthy animals (4, 5). Recent studies have demonstrated increased resistance of Salmonella to antimicrobials; in turn, this resistance impedes the effective treatment of infections both in horses (6, 7) and in humans (8). Characterization of Salmonella isolates through serotyping and antimicrobial susceptibility testing can help trace the origin of the infections (9). Whole-genome sequencing (WGS) provides more accurate identification of strain variation in pathogens than other phenotypic and genotypic assays (10), thus making it ideal for outbreak investigations (11–13). Outbreaks of equine salmonellosis usually develop in locations with large groups of horses (1), as is often the situation among patients in veterinary hospitals (14). Although previous studies have focused on outbreaks caused by Salmonella in horses (15), there are few investigations that describe serotype and antimicrobial resistance (AMR) trends of Salmonella from equine teaching veterinary hospitals using WGS. The aim of this research was to describe the diversity of Salmonella serotypes isolated from patients at an equine referral hospital in the southern United States to develop a better understanding of the isolates associated with clinical salmonellosis, their transmission, and their association with AMR.
RESULTS
The 255 Salmonella isolates used in this study were obtained from fecal (98.4%; n = 251) and blood samples (1.6%; n = 4) between 8 January 2007 and 4 November 2015. The mean age of the horses, based on 233 available records, was 7 years (range, 1 month to 24 years). Horses were classified into 4 groups for further analysis, according to age, as follows: (i) foals (less than 1 year; n = 57), (ii) juveniles (1 to 2 years; n = 7), (iii) adults (greater than 2 and less than 20 years; n = 163), and (iv) seniors (greater than or equal to 20 years of age; n = 6). The most widely recorded complaints in horses were colic (47.4%; n = 121), followed by diarrhea (21.6%; n = 55), and then by other general symptoms (20.4%; n = 52), such as fever, dehydration, weight loss, or anorexia. Some of the samples (2.8%; n = 7) came directly from necropsy cases, which were either dead on arrival or else were euthanized.
Serotypes.
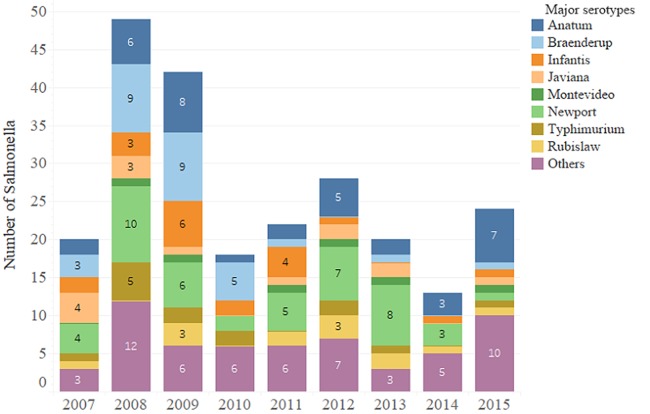
A total of 46 different serotypes were identified from case admission data; these identifications were performed at a national reference laboratory, using the White-Kauffmann-Le Minor scheme. Later, stored isolates were shared with our research laboratory, where we used WGS and SeqSero for the same purpose (16). In the end, when comparing both techniques, six isolates (2%) were not identified as the expected serotype, possibly due to a mixture of two serotypes present in the original patient sample or stored isolate, clerical errors in data entry in either the clinical or reference microbiology laboratory, or else errors in storing the strains at the time of original isolation or in reviving the isolates for this study. Subsequently, inferential results presented in this paper were analyzed and then reported based on the serotyping by the SeqSero scheme, since these data were directly aligned with the isolate preparations and yielded contemporaneous genotypic and phenotypic resistance and phylogenetic data. Overall, the most frequent serotype reported in the clinical data set was S. Newport (18.4%; n = 46), followed by S. Anatum (15.3%; n = 39), S. Braenderup (11.8%; n = 30), S. Infantis (8.2%; n = 21), S. Javiana (5.5%; n = 14), S. Typhimurium (6.27%; n = 16), S. Rubislaw (4.7%; n = 12), and S. Montevideo (2.4%; n = 6). The frequency of detection of the major serotypes varied somewhat across the years (Fig. 1). The remaining percentage of isolates (30.3%) was comprised of 38 additional serotypes (see Table S1 in the supplemental material, WGS data with footnote descriptions of isolates within assay disagreement).
FIG 1.
Frequency of detection of the main serotypes (as determined via the White-Kauffman-LeMinor method) throughout the years of the study (2007 to 2015).
Genome quality.
The mean of the assembly size in the isolates was 4,768,172 bp, with a range of 4,485,055 to 5,504,808 (see Fig. S1 in the supplemental material). The genome size of the isolates fit into the range of 4 to 6 Mb, allowing some latitude for accessory gene content on plasmids (17) and with upper and lower limits based on Salmonella genome sizes reported in NCBI databases (18). The mean of coverage depth was 74.6×, with a range of 16.1× to 196.3× (see Fig. S2 in the supplemental material). Although 15× is a good coverage depth, the recommendation for single-nucleotide polymorphism (SNP) analysis is to have 42× (19); our phylogenetic analysis was made with serotypes with very good coverage. The mean number of contigs was 68.3, with a range of 21 to 455, while <500 contigs indicates good quality (17). The mean size of the largest contig was 708,197.6 bp, with a range of 84,715 bp to 2,030,748 bp. The mean of the N50 was 297,234.9 bp, with a range of 19,493 bp to 732,857 bp; an N50 of >15,000 bp generally indicates a good-quality sequence, but a minimum size of 30,000 bp is often preferred (20).
Plasmid analysis.
Fifteen different incompatibility (Inc) plasmid replicon types were identified (IncA/C2, IncCOL, IncFIA, IncFIB, IncFII, IncFIIS, IncHI1, IncHI2, IncI1, IncI2, IncQ, IncR, IncU, IncX, and IncY); meanwhile, the predominant type was I1, at ∼10% (n = 25 isolates). A total of 96 (38%) of the Salmonella isolates harbored at least one plasmid. Replicons detected by each Salmonella serotype are shown in Fig. S3 in the supplemental material. Analysis to identify those plasmids carrying antimicrobial resistance genes (ARGs) was performed. From the 96 plasmids identified by ResFinder, 76 were detected by Bioinformatics Application for Navigating De novo Assembly Graphs Easily (Bandage), and 15 of these were carrying ARGs (see Table S3 in the supplemental material).
Evaluation of AMR patterns.
The MIC was for each Salmonella isolate determined via broth microdilution using the Sensititre NARMS plate CMV3AGNF, which includes 14 drugs for which resistance is monitored by the U.S. National Antimicrobial Monitoring System (NARMS). The proportion of tested isolates resistant to any individual antimicrobial ranged from a low of 0% (ciprofloxacin) to a high of 10.2% (sulfisoxazole). Out of 255 isolates, 219 (85.9%) were pansusceptible (susceptible to all the antimicrobials), 10 (3.9%) were resistant to fewer than 3 classes of antimicrobials, and 26 (10.2%) were multidrug resistant (MDR; resistant to 3 or more classes of antimicrobials). Among isolates exhibiting resistance to at least one antimicrobial, a total of 20 distinct patterns were found among 14 different serotypes. The specific phenotypic AMR patterns grouped by the number of antimicrobial classes to which resistance occurred are shown in Table 1.
TABLE 1.
Phenotypic antimicrobial resistance patterns for Salmonella serotypesa
| No. of antimicrobials | Antimicrobial resistance pattern | Serotype | % (no.) of resistant isolatesb | No. of isolates with this profile |
|---|---|---|---|---|
| 10 | AMC-AMP-FOX-XNL-CRO-CHL-GEN-STR-FIS-TET | Kiambu | 50 (1/2) | 2 |
| Oranienburg | 25 (1/4) | |||
| AMP-AZM-XNL-CRO-CHL-GEN-STR-FIS-TET-SXT | Agona | 100 (4/4) | 4 | |
| 9 | AMP-CRO-CHL-GEN-NAL-STR-FIS-TET-SXT | Anatum | 3 (1/37) | 1 |
| AMP-XNL-CRO-CHL-GEN-STR-FIS-TET-SXT | 4,12:i:− | 33 (1/3) | 4 | |
| Anatum | 3 (1/37) | |||
| Rubislaw | 15 (2/13) | |||
| AMC-AMP-FOX-XNL-CRO-CHL-STR-FIS-TET | Braenderup | 3 (1/29) | 3 | |
| Typhimurium | 7 (1/14) | |||
| Minnesota | 100 (1/1) | |||
| 8 | AMP-AZM-GEN-NAL-STR-FIS-TET-SXT | Anatum | 3 (1/37) | 1 |
| AMP-XNL-CRO-GEN-STR-FIS-TET-SXT | Rubislaw | 8 (1/13) | 1 | |
| AMC-AMP-FOX-XNL-CRO-GEN-FIS-SXT | Braenderup | 14 (4/29) | 5 | |
| Rubislaw | 8 (1/13) | |||
| 7 | AMP-CHL-GEN-STR-FIS-TET-SXT | Newport | 2 (1/46) | 1 |
| 6 | AMP-AZM-GEN-NAL-TET-SXT | Anatum | 3 (1/37) | 1 |
| 5 | AMP-CHL-GEN-FIS-TET | Anatum | 3 (1/37) | 1 |
| 3 | AMP-XNL-CRO | Braenderup | 3 (1/29) | 1 |
| FIS-TET-SXT | Infantis | 5 (1/20) | 1 | |
| STR-FIS-TET | Muenster | 33 (1/3) | 1 | |
| 2 | STR-FIS | Newport | 2 (1/46) | 1 |
| 1 | FIS | Newport | 2 (1/46) | 1 |
| GEN | Anatum | 3 (1/37) | 1 | |
| STR | Anatum | 3 (1/37) | 2 | |
| Kentucky | 33 (1/3) | |||
| TET | Anatum | 5 (2/37) | 3 | |
| Kentucky | 33 (1/3) | |||
| Montevideo | 17 (1/6) |
Serotypes were based on the White-Kauffman-LeMinor method and were isolated from equine samples submitted to the clinical microbiology laboratory between 8 January 2007 and 4 November 4 2015, which were tested for susceptibility to 14 antimicrobials. AMC, amoxicillin-clavulanic acid (2:1 ratio); AMP, ampicillin; AZM, azithromycin; CRO, ceftriaxone; CHL, chloramphenicol; FIS, sulfisoxazole; FOX, cefoxitin; GEN, gentamicin; NAL, nalidixic acid; SXT, trimethoprim-sulfamethoxazole; STR, streptomycin; TET, tetracycline; and XNL, ceftiofur.
Number of Salmonella isolates exhibiting a resistance pattern/total number of Salmonella per serotype.
ARGs.
Out of 255 isolates, 236 Salmonella (92.5%) carried at least a single resistance gene (Table 2; note that most of these single genes, i.e., aac(6′)-Iaa or aac(6′)-Iy, are cryptic, meaning that they do not produce phenotypic resistance because they have reached their evolutionary limit [21]). Meanwhile, only 19 (7.4%) did not harbor any ARG. A total of 38 different ARGs were identified. Functional genes based on the sequence were associated with phenotypic resistance (CLSI/NARMS breakpoints), while other ARGs were associated with decreased susceptibility, e.g., to ciprofloxacin. A heat map of plasmid replicon type by serotype among 36 antimicrobial-resistant Salmonella isolates is illustrated in Fig. S4 in the supplemental material.
TABLE 2.
List and frequencies of resistance genes identified by ResFinder
| Antimicrobial or antimicrobial family | Antimicrobial resistance gene | % (no.) of isolatesa |
|---|---|---|
| Aminoglycosidesb | aac(3′)-IId | 0.4 (1) |
| aac(3′)-VIa | 0.8 (2) | |
| aacA4 | 2.8 (7) | |
| aac(6′)-IIc | 4.4 (11) | |
| aada1 | 0.4 (1) | |
| aadA2 | 2.8 (7) | |
| aadA7 | 0.4 (1) | |
| aph(3′)-Ia | 4 (10) | |
| aph(3′)-Ic | 0.8 (2) | |
| rmtE | 1.6 (4) | |
| strA | 7.1 (18) | |
| strB | 7.5 (19) | |
| Beta-lactams/beta-lactam inhibitors | blaTEM-1B | 5 (13) |
| blaSHV-12 | 4 (10) | |
| blaCMY-2 | 4 (10) | |
| blaCTX-M-27 | 0.4 (1) | |
| Sulfisoxazole | sul1 | 8 (20) |
| sul2 | 5 (13) | |
| Trimethoprim-sulfamethoxazole | dfrA18 | 3.9 (10) |
| dfrA15 | 1.9 (5) | |
| dfrA14 | 0.4 (1) | |
| dfrA12 | 0.4 (1) | |
| Macrolides | ere(A) | 3.9 (10) |
| mphA | 2.8 (7) | |
| Phenicols | catA2 | 4.1 (10) |
| floR | 2 (5) | |
| cmlA1 | 2 (5) | |
| Quinolones | aac(6′)-Ib-cr | 3 (8) |
| qnrB2c | 2 (5) | |
| Ansamycin | arr-3 | 0.8 (2) |
| arr-6 | 1.2 (3) | |
| Tetracyclines | tet(A) | 1.9 (5) |
| tet(B) | 2.4 (6) | |
| tet(C) | 0.4 (1) | |
| tet(D) | 1.6 (4) | |
| tet(E) | 2.4 (6) |
No. of Salmonella isolates with the presence of the specified antimicrobial resistance gene (ARG).
The most common aminoglycosides genes were aac(6′)-Iaa and aac(6′)-Iy; however, these genes do not appear to encode for resistance.
Isolates carrying the qnrB2 gene typically show reduced susceptibility to ciprofloxacin; however, this is below the breakpoint MIC value established for resistance.
Using the Comprehensive Antimicrobial Resistance Database (CARD), we identified multiple different antimicrobial resistance mechanisms. The most common genes encoding MDR efflux pumps and reduced permeability to antimicrobials were soxS and marA. High prevalences of the glpT gene, encoding reduced susceptibility to fosfomycin, and the aac(6′)-Iy gene (a cryptic aminoglycoside resistance gene) were found. Antimicrobial target protection was conferred by qnrB20 in 2% of isolates. The dfrA12, dfrA14, dfrA15, dfrA19, sul1, and sul2 genes encoded resistance to diaminopyrimidine, sulfonamides, and sulfones. Detailed information about the antimicrobial resistance gene mechanisms and their prevalence is shown in Table S2 in the supplemental material. It should be noted that clinically defined phenotypic resistance more closely aligned with observed phenotypic resistance when utilizing ResFinder (22) database results versus those from CARD (23).
Agreement between detected ARGs and MIC values.
There was substantive agreement (kappa, 0.61 to 0.94) between phenotypic antimicrobial resistance and the presence of at least one ARG encoding resistance to aminoglycosides, beta-lactams, cephems, penicillins, folate pathway inhibitors, phenicols, and macrolides. There was good agreement (kappa, 0.76) between phenotypic and genotypic resistance to tetracycline. The intermediate MIC (i.e., 0. 5 to 1 μg/ml) of ciprofloxacin presented moderate agreement (kappa = 0.48) with the presence of plasmid-mediated quinolone resistance (PMQR) genes (encoding reduced susceptibility). Phenotypic resistance and genotypic resistance (analysis restricted to horizontally acquired resistance genes) to nalidixic acid had very poor agreement (kappa, −0.02); however, detection of chromosomal point mutations in gyrA and parC genes were identified using the PointFinder tool (24) and Resistance Gene Identifier (RGI) software (23), increasing the agreement considerably (kappa, 0.39). Detailed information on the agreement between ARGs (found by the ResFinder database) and phenotypic resistance is shown in Table 3.
TABLE 3.
Agreement between genotypes and phenotypes of 255 Salmonella strainsa
| Antimicrobial family or agent | No. of test results for: |
Agreement (%) | Kappa | |||
|---|---|---|---|---|---|---|
| Genotype for resistant phenotype: |
Genotype for susceptible phenotype: |
|||||
| Resistantb | Susceptible | Resistantb | Susceptible | |||
| Aminoglycosides | ||||||
| Gentamicin | 18 | 4 | 3 | 230 | 97.25 | 0.8222 |
| Streptomycin | 17 | 4 | 2 | 232 | 97.65 | 0.8373 |
| Beta-lactams/beta-lactam inhibitors | ||||||
| Amoxicillin-clavulanic acid | 9 | 1 | 0 | 245 | 99.61 | 0.9453 |
| Cephems | ||||||
| Cefoxitin | 9 | 1 | 0 | 245 | 99.61 | 0.9453 |
| Ceftiofur | 19 | 1 | 3 | 232 | 98.43 | 0.8962 |
| Ceftriaxone | 19 | 1 | 3 | 232 | 98.43 | 0.8962 |
| Penicillins | ||||||
| Ampicillin | 22 | 3 | 0 | 230 | 98.82 | 0.9297 |
| Folate pathway inhibitors | ||||||
| Sulfisoxazole | 22 | 5 | 1 | 227 | 97.65 | 0.8670 |
| Trimethoprim-sulfamethoxazole | 13 | 6 | 0 | 236 | 97.65 | 0.8004 |
| Macrolides | ||||||
| Azithromycin | 5 | 1 | 2 | 246 | 98.43 | 0.7064 |
| Phenicols | ||||||
| Chloramphenicol | 14 | 2 | 1 | 238 | 98.82 | 0.8970 |
| Quinolones | ||||||
| Ciprofloxacinc | 4 | 2 | 6 | 249 | 96.86 | 0.4848 |
| Nalidixic acidc | 0 | 3 | 10 | 242 | 94.9 | −0.0184 |
| Tetracyclines | ||||||
| Tetracycline | 18 | 7 | 3 | 227 | 96.08 | 0.7612 |
Isolated from equine samples submitted to the clinical laboratory between 8 January 2007 and 4 November 2015.
Antimicrobial resistance genes detected by ResFinder.
Analysis using MIC classified as reduced susceptibility instead of resistance.
Phylogenetic analysis.
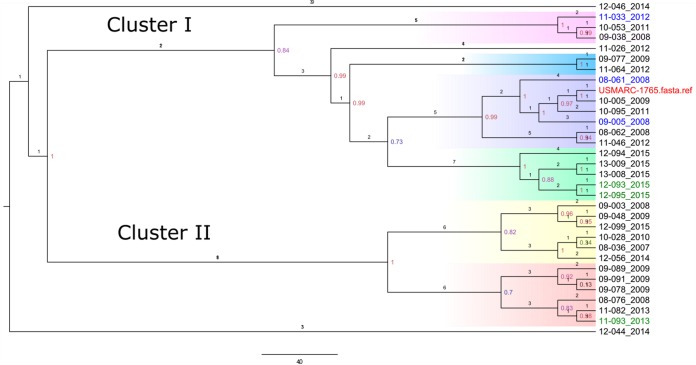
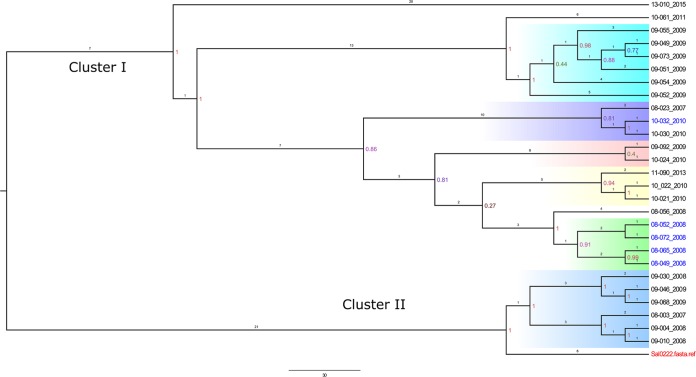
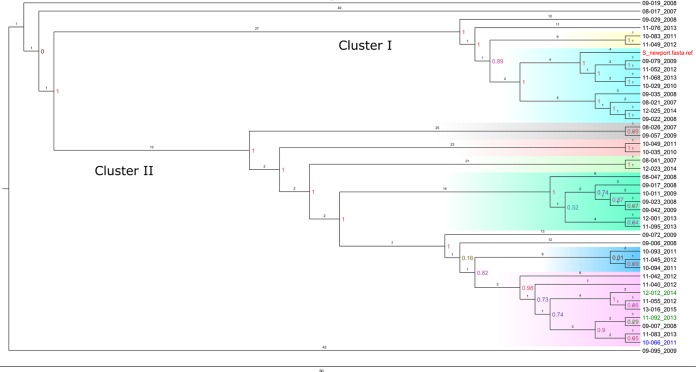
Salmonella Anatum (33 genomes) (Fig. 2), S. Braenderup (27 genomes) (Fig. 3), and S. Newport (42 genomes) (Fig. 4) were further explored to identify genetic associations that might be suggestive of nosocomial transmission or outbreak in the hospital. Genomes with a maximal unique match (MUM) index of >0.01 were excluded from the analysis (25).
FIG 2.
Whole-genome SNP-based phylogenetic tree of 31 Salmonella Anatum isolates and a single S. Anatum reference strain. The branch lengths are expressed in terms of changes per number of SNPs. The numbers in color show the bootstrap corresponding to the specific internal node. Strain names are marked with the colors red (reference), blue (MDR), green (resistant), and black (susceptible). Strain names are labeled with the year of admission of the patient to the hospital. Clusters are colored according to the phylogenetic group (clade). Cluster I includes subclusters I-a (pink), I-b (blue), I-c (purple), and I-d (green). Cluster II includes subclusters II-a (yellow) and II-b (red). The scale bar shows the estimated number of substitutions per SNP.
FIG 3.
Whole-genome SNP-based phylogenetic tree of 27 Salmonella Braenderup isolates and an S. Braenderup reference strain. The branch lengths are expressed in terms of changes per number of SNPs. The numbers in color show the bootstrap corresponding to the specific internal node. Strain names are marked with the colors red (reference), blue (resistant), and black (susceptible). Strain names are labeled with the year of admission of the patient to the hospital. Clusters are colored according to the phylogenetic group (clade). Cluster I includes subcluster I-a (cyan blue), I-b (purple), I-c (pink), I-d (yellow), and I-e is (green). Clade II, light violet. The scale bar shows the estimated number of substitutions per SNP.
FIG 4.
Whole-genome SNP-based phylogenetic tree of 42 Salmonella Newport isolates and an S. Newport reference strain. The branch lengths are expressed in terms of changes per number of SNPs. The tree was visualized using FigTree. Red (reference strain), blue stars (MDR), and black (susceptible). Strain names are labeled with the year of admission of the patient to the hospital. Clusters are colored according to the phylogenetic group (clade). Cluster I includes subclusters I-a (yellow) and I-b (cyan blue). Cluster II includes subclusters II-a (gray), II-b (rose), II-c (light green), II-d (emerald green), II-e (blue), and II-f (pink). The scale bar shows the estimated number of substitutions per SNP.
Salmonella Anatum.
Two main clusters were detected, cluster I and cluster II, with two cases from 2014 falling outside the main clusters. Cluster I was subdivided into four subclusters (I-a to I-d). In 2008, subcluster I-c had two MDR isolates; one was isolated in February and the other was isolated in May from a patient who was present in the hospital shortly after the first patient carrying the MDR isolate was admitted. Both patients originated from different farm locations, and their stays in the hospital did not directly overlap. In 2015, subcluster I-d contained five cases of adult horses seen between April and May. Out of isolates from these five cases, two isolates from patients at the same farm were tetracycline resistant, each carrying tet(B) genes, while three isolates were pansusceptible. These results suggest that these horses likely arrived at the hospital with the infection, rather than acquiring infection from nosocomial spread. Cluster II contained 2 subclusters (II-a and II-b). In 2009, subcluster II-b represented three cases occurring between April and September in two adults and one foal. These cases came from different farms. The foal's stay occurred in April and did not overlap those of the adult horses. One of the adult horses was hospitalized for 2 months for treatment of clostridial myositis and secondary salmonellosis; this patient was diagnosed before the admission of the second adult horse, which was euthanized upon presentation due to poor prognosis. As the two horses were in the hospital at the same time, this case was possibly due to nosocomial transmission; however, the immediate euthanasia of the second horse upon arrival makes this much less likely. Detailed information concerning the cases within each S. Anatum cluster is shown in Table S4 in the supplemental material.
Salmonella Braenderup.
There were two main clusters, cluster I and cluster II. Cluster I was further subdivided into five subclusters (I-a to I-e). In 2009, subcluster I-a consisted of 6 cases, with four foals and two mares (boarding) presented to the hospital as a result of an acknowledged outbreak at a local farm. In 2010, subcluster I-b contained 2 cases, one in a foal and the other in the associated mare (dam boarding in the hospital with the foal). The isolate from the foal was an MDR S. Braenderup isolate carrying a rarely identified extended-spectrum beta-lactamase (ESBL) gene (bla-CTX-M-27) associated to the chromosome, as detected by analysis with Bandage software. In 2010, subcluster I-d contained two adult cases from April to May. Between March and April of 2008, subcluster I-e contained 5 cases. Four identical S. Braenderup MDR (to 8 antimicrobials) isolates with the same three ARGs (rmtE, blaCMY-2, and sul1) were isolated from concurrently hospitalized patients, suggesting a possible outbreak at the hospital; in addition, one isolate was pansusceptible, with four more nucleotide substitutions per hundred nucleotide sites than the multidrug-resistant isolates. Cluster II contained 6 cases from 2007 (1 case), 2008 (3 cases), and 2009 (2 cases). In 2008, two foal cases between May and June were related. Between January and March of 2009, one case from a foal and another from a mare with dystocia were clustered. Detailed information concerning the cases per each S. Braenderup cluster is shown in Table S5 in the supplemental material.
Salmonella Newport.
There were two main clusters, cluster I, with two subclusters, and cluster II, with six subclusters. Isolates collected from two patients in 2008 between August and November comprised subcluster I-b, which was grouped with the clinically unrelated reference genome. Subcluster II-d included three cases from between August and December of 2008; however, the associated patients were not hospitalized at the same time and there were no data to link the cases directly to each other. Two isolates in this cluster were collected in September and October 2013; however, the patients were not hospitalized concurrently. Subcluster II-b contained two cases, one foal and one adult. In 2011, subcluster II-e contained 2 cases occurring between September and October in adult horses. The first horse was euthanized at presentation. The second horse was admitted 12 days later, and the Salmonella isolate came from a fecal sample collected within 24 h of admission. Subcluster II-f contained 3 cases from August to October of 2008. Two cases from 2013 were related, a foal and its dam. Detailed information concerning the cases per each S. Newport cluster is shown in Table S6 in the supplemental material.
DISCUSSION
Monitoring and surveillance of Salmonella infections in horses are essential for improving knowledge concerning the epidemiology, diagnosis, treatment, and identification of common-source outbreaks, and to distinguish whether they are likely to be of nosocomial origin (9). Our study is pioneering because it uses whole-genome sequencing (WGS) to analyze Salmonella isolates from clinical cases of horses arising in a referral hospital over an extended period of time. WGS is an accurate molecular epidemiological tool that, when combined with in silico prediction of phenotype (serotype identification and acquired antimicrobial resistance mechanisms), helps to track and surveil the evolution and spread of resistant pathogens. Although pulsed-field gel electrophoresis (PFGE) was, until recently, the gold standard for performing strain-typing epidemiological investigations, it remains insufficient to separate very closely related strains (26). Meanwhile, WGS and core genome SNP analysis help to provide differentiation in those PFGE-indistinguishable strains due to the increased efficiencies to identify SNPs, inversions, insertions, and deletions (25).
Identification of Salmonella serotypes helps in outbreak investigations to track isolates to their sources. Increased numbers of cases of a specific serotype can generate suspicion of an outbreak (https://www.cdc.gov/salmonella/reportspubs/salmonella-atlas/serotyping-importance.html). Serotyping is mainly accomplished using the White-Kauffmann-Le Minor (WKL) scheme; however, serotyping results can take several weeks and serotyping is often performed offsite at reference laboratories. Although performed retrospectively here on an existing set of characterized isolates, when conducted in real-time, WGS allows rapid characterization and comparisons of isolates as they are collected (27). The most common Salmonella serotype in this study was S. Newport, as reported in previous studies (6, 28, 29). Additional major serotypes were S. Anatum, followed by S. Braenderup, S. Infantis, S. Javiana, and S. Typhimurium; in turn, these results also agree with previous reports (9).
There remain six isolates for which incongruous results were obtained by traditional serotyping (historical data set) versus whole-genome sequencing performed in the present day (Fig. 1 and Table S1). These mismatches most likely arose during clinical laboratory reporting or clerical/data transcription sometime in the past (as far back as 2007). We opted to include the results of the inferential analyses performed on these six isolates via WGS and our own phenotypic resistance profiling. We have no additional information that would readily persuade us to exclude them from our reporting (i.e., they are Salmonella enterica isolates and they came from horses at this hospital). Other than the initial summary and description of the distribution of traditionally serotyped isolates (used to describe the clinical data set), all subsequent analyses relating serotype to (i) resistance (genotype and phenotype), (ii) plasmid types, and (iii) phylogenetics within serotype were conducted using the data generated through WGS.
In our study, the most frequent plasmid replicon type was IncI, which was also previously reported in Salmonella (30). IncI1 has been recognized as harboring ESBL genes (31, 32). IncF was the second most common replicon detected in this study; it is frequent in Salmonella, often harboring beta-lactamases and acc(6′)-Ib-cr genes. Furthermore, IncF has often been detected in the intestinal microflora (33) in earlier work. IncHI1 and IncHI2 have been associated with resistance genes, while the first has been related to bacterial proliferation and persistence (34). Genes on IncCOL plasmids can encode colicins that help to kill (or, suppress) other bacteria (35). In our study, IncI1, Incq, IncI1, IncI2, IncAC2 IncFII, IncHI1, IncCOL, and IncFI mainly harbored genes encoding aminoglycoside resistance. We used Bandage software to identify plasmids and chromosome in the bacterial genome, which makes it easier to identify problematic parts of the assembly; however, there remains much manual work with the aid of other software to comprehensively perform the analysis (36). While there has been improvement in techniques to monitor the dispersion of resistance genes by plasmids, existing algorithms often fail to reconstruct plasmids from short-read WGS files. Most of the Salmonella isolates in this study were pansusceptible; meanwhile, the highest prevalences of resistance were to sulfisoxazole, ampicillin, and tetracycline. These results are somewhat consistent with the antimicrobials used in horses in the United States (gentamicin, potentiated sulfonamides, and doxycycline) (37). In a recent study on Salmonella involving horses at the Cornell University Animal Hospital, isolates reflecting similar serotypes to our study were more often resistant to amoxicillin-clavulanic acid, ampicillin, cefazolin, cefoxitin, ceftiofur, chloramphenicol, and tetracycline than to drugs of other classes (6). Salmonella recovered from horse diagnostic samples at four state veterinary diagnostic laboratories (Arizona, Missouri, North Carolina, and Tennessee) presented increased prevalences of resistance to ampicillin, chloramphenicol, and sulfamethoxazole (38). Similar results were reported in the Netherlands among 232 Salmonella isolates from horses exhibiting resistance to tetracycline (53%) and ampicillin (34%) (39). In our study, 10% of the isolates were MDR (≥3 classes of antimicrobials). These MDR Salmonella isolates usually harbored plasmids that could transfer the resistance genes (40). Isolates of the serotype S. Anatum were the most resistant (by count of classes), followed by those of S. Rubislaw and then of S. Braenderup. Cummings et al. (6) reported that the most resistant serotypes were S. Newport, S. Oranienburg, and S. Typhimurium; however, in another study at a veterinary hospital in Florida (USA), S. Java, S. Typhimurium var. Copenhagen, S. Javiana, and S. Newport (29) dominated. The most resistant serotype in a similar study in the Netherlands was S. Typhimurium (39).
Most of the Salmonella isolates carried at least one aminoglycoside resistance gene (aac(6′)-Iaa or aac(6′)-Iy); however, these genes appear to have reached their evolutionary limits and no longer encode resistance (21). Instead, the aac(6′)-Iy gene has been associated with carbohydrate transport or endogenous metabolism in Salmonella (41). Streptomycin resistance genes (aadA1 and aadA2) also were detected, as reported in previous studies (38). Aminoglycosides are high-priority, critically important antimicrobials used to treat septicemia and digestive, respiratory, and urinary diseases in horses (http://www.oie.int/fileadmin/Home/eng/Our_scientific_expertise/docs/pdf/Eng_OIE_List_antimicrobials_May2015.pdf). In addition, they are used to treat infections caused by Pseudomonas aeruginosa, and Enterobacteriaceae can exchange resistance genes with this bacterial species (42).
In this study, acquired phenotypic resistance in Salmonella presented almost perfect agreement with known resistance genes [that is, when ignoring the aac(6′)-Iaa or aac(6′)-Iy genes]. Nonetheless, there remained discrepancies for quinolones, tetracycline, and macrolides. However, those discrepancies could be resolved by evaluating sequences for the presence of other antimicrobial resistance mechanisms, such as the presence of efflux pumps that when overexpressed can result in MDR (43, 44). Without another antimicrobial resistance mechanism present, the efflux pumps do not confer high-level resistance to antimicrobials; however, the bacteria have the potential to mutate the genes encoding target sites (45). We tested our hypothesis that phenotypic resistance without the presence of horizontally transferable genes encoding resistance to quinolones was likely conferred by quinolone resistance-determining regions (QRDRs) of the chromosome; to pursue this, mutations were detected using the PointFinder tool (24) and CARD (23). Resistant genotypes with susceptible phenotypes were associated with plasmid-mediated quinolone resistance (PMQR) (e.g., qnrB) but do not exhibit clinical levels of resistance to quinolones unless QRDR mutations or additional PMQR genes are present. While fluoroquinolones are recommended to treat salmonellosis in humans because fluoroquinolones are lipid soluble and Salmonella are facultative intracellular organisms, in horses, the oral use of ciprofloxacin can cause colitis and in foals can affect cartilage development (46). This class of antimicrobials is included on the World Health Organization list of highest priority, critically important antimicrobials for human medicine (47).
Tetracycline is considered a highly important antimicrobial for human medicine (http://www.who.int/mediacentre/news/releases/2017/bacteria-antibiotics-needed/en/). Tetracycline resistance genes [tet(A), tet(B), tet(C), tet(D), and tet(E)] were present in Salmonella in this study. The common tetracycline genes reported in Salmonella are tet(A), tet(B), tet(C), tet(D), and tet(G) (48). Salmonella isolates in this study occasionally carried mphA, and this gene has been reported in E. coli and Shigella species resistant to azithromycin (49); in addition, ere(A) is frequently associated with Citrobacter, Enterobacter, Escherichia, Klebsiella, Pantoea, Pseudomonas, Proteus, Serratia, Stenotrophomonas, Vibrio, and Staphylococcus (50). In horses, macrolides are not used to treat Salmonella infection because they induce acute colitis by disrupting the microflora (51, 52); generally speaking, macrolides are not currently used to treat nontyphoidal Salmonella infection. The presence of these genes can generate public health consequences because E. coli, Shigella species, and Salmonella enterica can exchange plasmids (49, 53).
In our study, beta-lactamase (blaTEM-1B), plasmid-mediated AmpC cephamycinase (blaCMY-2), and extended-spectrum beta-lactamase (ESBL) (blaSHV-12 and blaCTX-M-27) genes were found. When blaTEM-1B and ESBL genes are present in the same isolate, blaTEM-1B produces resistance to narrow-spectrum beta-lactams, while the ESBL gene is expected to produce resistance to extended-spectrum beta-lactams, such as third-generation cephalosporins (54). Interestingly, almost all Salmonella isolates with the blaSHV-12 gene (a purported ESBL) contained the blaTEM-1B gene as well; however, many of these failed to exhibit an ESBL phenotype (Table S3). Other researchers have reported the presence of AmpC in an S. Newport isolate related to a veterinary teaching hospital outbreak that involved a high case fatality rate (15). In 2003, an outbreak in equines with S. Newport MDR-AmpC carrying blaTEM-1B and blaSHV-12 genes led to closure of that veterinary teaching hospital for 3 months. Beta-lactamase resistance in Salmonella isolates is not restricted to the United States. In Argentina, there was a case reported of S. Typhimurium carrying the blaCMY-2 gene in a race horse (55). In foals, it is recommended to use extended-spectrum cephalosporins, or else ampicillin-sulbactam, alone or with an aminoglycoside (gentamicin or streptomycin) (46); however, the presence of resistance genes may result in treatment failure.
Phylogenetic analysis of the presumptively related Salmonella genomes was performed. Salmonella Anatum and S. Braenderup were associated with likely nosocomial transmission at the hospital and also with an outbreak at local horse farms. In general, the closely related S. Newport genomes did not come from the same farm, thus necessitating further research into the potential common sources of this frequent and dispersed (geographically and temporally) serotype. WGS was useful in the detection of unsuspected epidemiological connections that—along with complementary metadata—helped to explore outbreaks at both hospitals and farms.
MATERIALS AND METHODS
Sample set and isolates.
The sample set included 255 Salmonella isolates, each derived from the first clinical sample collected and submitted to the clinical microbiology laboratory from any horse admitted to the veterinary teaching hospital with a differential diagnosis of salmonellosis between 8 January 2007 and 4 November 2015. Samples were initially inoculated into tetrathionate broth and incubated overnight at 37°C before DNA extraction, or else cultured on XLT-4 and MacConkey agar. Before 2010, samples were subjected only to culture; after 2010, patient samples were first tested by PCR for the spaQ gene, which encodes the surface presentation of antigenic protein SpaQ (transmembrane) (56), and then cultured only if positive by PCR. Tetrathionate broths (Difco, Becton Dickinson, Franklin Lakes, NJ) that tested positive by direct PCR were subsequently struck onto XLT-4 (Difco, Becton Dickinson, Franklin Lakes, NJ) and MacConkey agar (Difco, Becton Dickinson, Franklin Lakes, NJ). Hydrogen sulfide-producing colonies from XLT-4 agar and lactose-negative colonies from MacConkey agar were subcultured on Trypticase soy agar (TSA) supplemented with 5% sheep blood (Difco, Becton Dickinson, Franklin Lakes, NJ) and grown overnight at 37°C. Isolates were then tested for oxidase production. Oxidase-negative isolates were then presumptively identified as Salmonella, based on production of characteristic biochemical reactions when grown on triple sugar iron agar (TSI) (Difco, Becton Dickinson, Franklin Lakes, NJ), lysine iron agar (LIA) (Difco, Becton Dickinson, Franklin Lakes, NJ), Christensen's urea agar (Difco, Becton Dickinson, Franklin Lakes, NJ), and tryptophan broth (Difco, Becton Dickinson, Franklin Lakes, NJ), and agglutination with polyvalent antisera (Difco, Becton Dickinson, Franklin Lakes, NJ). These isolates were sent to the National Veterinary Services Laboratory (NSVL-USDA) in Ames, IA, for serotyping according to the White-Kauffmann-Le Minor scheme. For further analysis, some isolates were preserved in lysogeny broth (Difco, Becton Dickinson, Franklin Lakes, NJ) supplemented with glycerol at 10%, and others were preserved on cryopreservation beads at −80°C. Clinical and microbiological data were entered into a central veterinary hospital management database as they were reported and then extracted later for the purposes of this study.
Antimicrobial susceptibility testing.
The determination of the antimicrobial susceptibility of Salmonella isolates was performed using the broth microdilution method via the Sensititre system (TREK, Thermo Scientific Microbiology, Oakwood Village, OH) to identify the MICs. This test was performed using the National Antimicrobial Resistance Monitoring System (NARMS) custom plate CMV3AGNF (TREK, Thermo Scientific Microbiology, Oakwood Village, OH), which has 14 antimicrobials covering 9 classes of antimicrobials. The guidelines of surveillance from NARMS were followed to generate comparable data procedures and interpretations with representative antimicrobial families. Beta-lactams, including ampicillin, amoxicillin-clavulanic acid, cefoxitin, ceftiofur, and ceftriaxone; quinolones, such as nalidixic acid and ciprofloxacin; folic acid inhibitors, like sulfisoxazole and trimethoprim-sulfamethoxazole; and aminoglycosides, like streptomycin and gentamicin, were part of the observation. Lastly, azithromycin (macrolides), chloramphenicol (phenicols), and tetracycline (tetracyclines) also were evaluated. The Sensititre system provided a simple, practical, and quantitative method that measured the MIC and provided clinical interpretative criteria (resistant, intermediate, or susceptible) based on the Clinical and Laboratory Standards Institute (CLSI) breakpoints (57) or on NARMS consensus breakpoints (58) where CLSI interpretative criteria were missing. A detailed description of the protocol followed is given by Ohta et al. (59).
DNA extraction for whole-genome sequencing (WGS).
In the present study, DNA from stored Salmonella isolates was extracted using the QIAamp 96 DNA QIAcube HT kit (Qiagen, Valencia, CA) in the QIAcube HT instrument (Qiagen, Valencia, CA). One Salmonella colony from a fresh culture per isolate was suspended into 5 ml of Trypticase soy broth (Difco, Becton Dickinson, Franklin Lakes, NJ) and incubated overnight at 37°C. A detailed description of the protocol is given in Ohta et al. (59).
WGS using the Illumina MiSeq platform.
WGS was performed, using the Illumina MiSeq platform (Illumina, San Diego, CA) to determine serotypes, plasmid replicons, and antimicrobial resistance genes and to conduct phylogenetic analysis of the Salmonella isolates. Libraries for 32 Salmonella DNA samples were multiplexed using the Illumina Nextera XT kit, following the manufacturer's instructions, using the Qiagility robot (Qiagen, Valencia, CA) to set up the reactions and using a thermocycler (Eppendorf) for amplification. The quality and quantity of the libraries were checked on the Fragment Analyzer instrument (Advanced Analytical, Ankeny, IA). The libraries were run on the MiSeq automated DNA sequencer, using the MiSeq reagent kit v3 (Illumina, San Diego, CA) with paired-end 2 × 300-bp reads.
Bioinformatics analysis of WGS data.
Raw sequence data (fastq files for reverse and forward ends) were obtained from the Illumina MiSeq runs and used for further analysis after the sequencing run was complete. De novo genome assembly was performed by the Velvet software package using SRST2 (a short-read sequence typing pipeline) (60) on the Illumina BaseSpace platform and also by the SPAdes software package using the Pathosystems Resource Integration Center (PATRIC) information system and by the Center for Genomic Epidemiology (CGE) website (61).
Quality of the sequence.
The quality of the genomes was assessed using FastQC and the Quality Assessment Tool for Genomic Assemblies (QUAST) software. FastQC makes a simple quality control analysis to guarantee that the raw data are good and there are no problems or biases that will affect the analysis (62). QUAST is an assembly quality assessment tool that evaluates different metrics, even without a reference genome (63). Summary results of the fastq files and the assembled fasta files are provided in Fig. S1 and S2 and Table S7 in the supplemental material.
Serotype analysis.
The SeqSero pipeline was used for serotype identification (16). On the Web-based tool, raw sequence reads derived from the Illumina MiSeq analysis were uploaded. We used raw data for the SeqSero analysis because it reduces the false-positive calls, since draft assemblies (fasta files) are not considered to be high-quality assemblies (16). The somatic (O) group was determined by analysis of wzx and wzy genes and by analysis of the rfb cluster; meanwhile, flagellar (H) phases were determined by analysis of the fliC and fljB genes combined in the same H antigen database. Mapping was run 3 times for the final serotype call.
Validation of WGS serotypes through comparison with serotyping using the White-Kauffmann-Le Minor scheme.
All Salmonella isolates were previously sent to the National Veterinary Services Laboratories (NSVL-USDA) and serotyped using the White-Kauffmann-Le Minor scheme; in turn, those results were compared with the results obtained by the SeqSero pipeline, and any notable differences and potential biases were explored.
Plasmid analysis.
Genomes were analyzed through SRST2 on the Illumina BaseSpace platform and by the Center for Genomic Epidemiology (CGE), using the PlasmidFinder database (64) for the determination of plasmid types. To localize the resistance genes into the plasmids, BLAST was used to find hits of sequences, and the Bandage tool was used to visualize the bacterial genome (36).
Resistance genes.
The genomes were analyzed for the presence of horizontally acquired resistance genes through the ResFinder tool using the SRST2 application in the Illumina BaseSpace platform and via the Center for Genomic Epidemiology Web page (http://www.genomicepidemiology.org/). The PointFinder tool was used to detect possible chromosomal point mutations in isolates that have phenotypic resistance without the presence of horizontally acquired resistance genes (24). The genes detected using the ResFinder database were subjected to a BLAST search against the assembled genome and were reported if they covered at least 2/5 the length of the resistance genes in the database (22). RGI software was used to complement the detection of antibiotic resistance genes. This software integrates ARO (antibiotic resistance ontology), bioinformatics models, and molecular reference (23). Bandage, a graphical user interface (GUI) application, was used to load and display the de novo assembly graph for a bacterial genome, helping to localize the resistance genes into the bacterial chromosome and plasmids (36).
Phylogenetic analysis.
The analysis was conducted in only three of the most common serotypes that presented temporal clusters of cases admitted to the hospital (Salmonella Anatum, Salmonella Braenderup, and Salmonella Newport) to explore the epidemiology of possible outbreaks or the presence of circulating nosocomial isolates at the hospital.
Assembled genomes (contigs) from the Salmonella genome of the three main serotypes were used to construct the massive core-genome alignments into Parsnp in the Harvest package (25). Parsnp is useful in intraspecific genome analysis (i.e., outbreak analysis). The phylogenetic tree was visualized with FigTree (65). Phylogenetic trees were evaluated by each serotype, along with the information available per case (year, antimicrobial susceptibility, presenting complaint, age, and farm location); however, the information about farm location is not shown here to ensure confidentiality. The complete genome sequence of Salmonella enterica subsp. enterica serotype Anatum strain USDA-ARS-USMARC-1765 (GenBank accession number NZ_CP014659.2) was used as a reference to assemble the phylogenetic tree; notably, this strain is from a human salmonellosis case and was published as part of a study that compared the genomes of Salmonella Anatum from human and bovine sources (66). The complete genome sequence of S. Braenderup Sal_JBP_2011K-0222 from the U.S. Centers for Disease Control and Prevention (CDC) was obtained through personal communication with Henk den Bakker from the University of Georgia; this genome sequence was used to develop the phylogenetic analysis. The complete genome sequence of Salmonella enterica subsp. enterica serotype Newport strain 0007-33 (GenBank accession number NZ_CP013685.1) was used as a reference to analyze the phylogenetic tree; this strain is from a bovine gastroenteritis case and was collected and curated by the University of Pennsylvania Salmonella Reference Center (67).
Statistical analysis.
The epidemiological unit for analysis was the isolate (individual serotype) recovered from the first sample per patient admission to the hospital. Information from each isolate was recovered from the clinical laboratory and hospital databases and saved as a spreadsheet in Excel format. The data were imported to Stata software version 14.0 (StataCorp, College Station, TX) and were used to generate variables to perform descriptive analyses.
The antimicrobial susceptibility MIC data were imported into Stata software version 14.0 (StataCorp, College Station, TX) in Trek SWIN (CSV) format. Intermediate resistance was collapsed into the “susceptible” category when resistance (encoded as a binary variable) was portrayed. The presence or absence of a known resistance gene was compared with the interpretation of resistant or susceptible to a given corresponding antimicrobial. Measures of agreement between the phenotypic and genotypic results were made using the kappa (κ) statistic (68).
Accession number(s).
A total of 255 assembled genome sequences have been submitted to the National Center for Biotechnology Information GenBank database under BioProject accession number PRJNA433689.
Supplementary Material
ACKNOWLEDGMENTS
Funding was provided as part of a startup package provided to H. M. Scott.
We acknowledge and thank the technical staff of the Clinical Microbiology Laboratory of the Veterinary Medical Teaching Hospital and Roberta Pugh and staff at the Molecular Epidemiology and Microbial Ecology Laboratory at the Veterinary Pathobiology department at Texas A&M University. Writing assistance was provided by Sarah Murray and Daniel Garcia. Portions of this research were conducted with the advanced computing resources and consultation provided by Texas A&M High Performance Research Computing.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02829-17.
REFERENCES
- 1.Chapman AM. 2006. Characterizing Salmonella fecal shedding among racehorses in Louisiana. MSc thesis Louisiana State University, Baton Rouge, LA. [Google Scholar]
- 2.Astorga R, Arenas A, Tarradas C, Mozos E, Zafra R, Perez J. 2004. Outbreak of peracute septicaemic salmonellosis in horses associated with concurrent Salmonella enteritidis and Mucor species infection. Vet Rec 155:240–242. doi: 10.1136/vr.155.8.240. [DOI] [PubMed] [Google Scholar]
- 3.Steneroden KK, Van Metre DC, Jackson C, Morley PS. 2010. Detection and control of a nosocomial outbreak caused by Salmonella Newport at a large animal hospital. J Vet Intern Med 24:606–616. doi: 10.1111/j.1939-1676.2010.0484.x. [DOI] [PubMed] [Google Scholar]
- 4.Feary DJ, Hassel DM. 2006. Enteritis and colitis in horses. Vet Clin North Am Equine Pract 22:437–479, ix. doi: 10.1016/j.cveq.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 5.Oliver OE, Stampfli H. 2006. Acute diarrhea in the adult horse: case example and review. Vet Clin North Am Equine Pract 22:73–84. doi: 10.1016/j.cveq.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 6.Cummings KJ, Perkins GA, Khatibzadeh SM, Warnick LD, Aprea VA, Altier C. 2016. Antimicrobial resistance trends among Salmonella isolates obtained from horses in the northeastern United States (2001–2013). Am J Vet Res 77:505–513. doi: 10.2460/ajvr.77.5.505. [DOI] [PubMed] [Google Scholar]
- 7.Dargatz DA, Traub-Dargatz JL. 2004. Multidrug-resistant Salmonella and nosocomial infections. Vet Clin North Am Equine Pract 20:587–600. doi: 10.1016/j.cveq.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 8.CDC. 2016. National Antimicrobial Resistance Monitoring System for Enteric Bacteria (NARMS): 2014 human isolates surveillance report. CDC, Atlanta, GA: https://www.cdc.gov/narms/pdf/2014-Annual-Report-narms-508c.pdf. [Google Scholar]
- 9.Hernandez JA, Long MT, Traub-Dargatz JL, Besser TE. 2014. Salmonellosis, p 321.e4–333.e4. In Sellon DC, Long M (ed), Equine infectious diseases, 2nd ed W.B. Saunders, St. Louis, MO. [Google Scholar]
- 10.Bergholz TM, Moreno Switt AI, Wiedmann M. 2014. Omics approaches in food safety: fulfilling the promise? Trends Microbiol 22:275–281. doi: 10.1016/j.tim.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hendriksen RS, Leekitcharoenphon P, Lukjancenko O, Lukwesa-Musyani C, Tambatamba B, Mwaba J, Kalonda A, Nakazwe R, Kwenda G, Jensen JD, Svendsen CA, Dittmann KK, Kaas RS, Cavaco LM, Aarestrup FM, Hasman H, Mwansa JC. 2015. Genomic signature of multidrug-resistant Salmonella enterica serovar Typhi isolates related to a massive outbreak in Zambia between 2010 and 2012. J Clin Microbiol 53:262–272. doi: 10.1128/JCM.02026-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leekitcharoenphon P, Nielsen EM, Kaas RS, Lund O, Aarestrup FM. 2014. Evaluation of whole genome sequencing for outbreak detection of Salmonella enterica. PLoS One 9:e87991. doi: 10.1371/journal.pone.0087991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pedersen K, Sorensen G, Lofstrom C, Leekitcharoenphon P, Nielsen B, Wingstrand A, Aarestrup FM, Hendriksen RS, Baggesen DL. 2015. Reappearance of Salmonella serovar Choleraesuis var. Kunzendorf in Danish pig herds. Vet Microbiol 176:282–291. doi: 10.1016/j.vetmic.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 14.Schott HC II, Ewart SL, Walker RD, Dwyer RM, Dietrich S, Eberhart SW, Kusey J, Stick JA, Derksen FJ. 2001. An outbreak of salmonellosis among horses at a veterinary teaching hospital. J Am Vet Med Assoc 218:1152–1159, 1100. doi: 10.2460/javma.2001.218.1152. [DOI] [PubMed] [Google Scholar]
- 15.Dallap Schaer BL, Aceto H, Rankin SC. 2010. Outbreak of salmonellosis caused by Salmonella enterica serovar Newport MDR-AmpC in a large animal veterinary teaching hospital. J Vet Intern Med 24:1138–1146. doi: 10.1111/j.1939-1676.2010.0546.x. [DOI] [PubMed] [Google Scholar]
- 16.Zhang S, Yin Y, Jones MB, Zhang Z, Deatherage Kaiser BL, Dinsmore BA, Fitzgerald C, Fields PI, Deng X. 2015. Salmonella serotype determination utilizing high-throughput genome sequencing data. J Clin Microbiol 53:1685–1692. doi: 10.1128/JCM.00323-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robertson J, Yoshida C, Kruczkiewicz P, Nadon C, Nichani A, Taboada EN, Nash JHE. 2018. Comprehensive assessment of the quality of Salmonella whole genome sequence data available in public sequence databases using the Salmonella in silico Typing Resource (SISTR). Microb Genom. doi: 10.1099/mgen.0.000151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.NCBI. 2018. Genome assembly and annotation report: Salmonella enterica. https://www.ncbi.nlm.nih.gov/genome/genomes/152?.
- 19.Fu S, Octavia S, Tanaka MM, Sintchenko V, Lan R. 2015. Defining the core genome of Salmonella enterica serovar Typhimurium for genomic surveillance and epidemiological typing. J Clin Microbiol 53:2530–2538. doi: 10.1128/JCM.03407-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ellington MJ, Ekelund O, Aarestrup FM, Canton R, Doumith M, Giske C, Grundman H, Hasman H, Holden MTG, Hopkins KL, Iredell J, Kahlmeter G, Köser CU, MacGowan A, Mevius D, Mulvey M, Naas T, Peto T, Rolain JM, Samuelsen Ø, Woodford N. 2017. The role of whole genome sequencing in antimicrobial susceptibility testing of bacteria: report from the EUCAST Subcommittee. Clin Microbiol Infect 23:2–22. doi: 10.1016/j.cmi.2016.11.012. [DOI] [PubMed] [Google Scholar]
- 21.Salipante SJ, Hall BG. 2003. Determining the limits of the evolutionary potential of an antibiotic resistance gene. Mol Biol Evol 20:653–659. doi: 10.1093/molbev/msg074. [DOI] [PubMed] [Google Scholar]
- 22.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jia B, Raphenya AR, Alcock B, Waglechner N, Guo P, Tsang KK, Lago BA, Dave BM, Pereira S, Sharma AN, Doshi S, Courtot M, Lo R, Williams LE, Frye JG, Elsayegh T, Sardar D, Westman EL, Pawlowski AC, Johnson TA, Brinkman FSL, Wright GD, McArthur AG. 2017. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res 45:D566–D573. doi: 10.1093/nar/gkw1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zankari E, Allesoe R, Joensen KG, Cavaco LM, Lund O, Aarestrup FM. 2017. PointFinder: a novel web tool for WGS-based detection of antimicrobial resistance associated with chromosomal point mutations in bacterial pathogens. J Antimicrob Chemother 72:2764–2768. doi: 10.1093/jac/dkx217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Treangen TJ, Ondov BD, Koren S, Phillippy AM. 2014. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol 15:524. doi: 10.1186/s13059-014-0524-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Foley SL, Zhao S, Walker RD. 2007. Comparison of molecular typing methods for the differentiation of Salmonella foodborne pathogens. Foodborne Pathog Dis 4:253–276. doi: 10.1089/fpd.2007.0085. [DOI] [PubMed] [Google Scholar]
- 27.Gilmour MW, Graham M, Reimer A, Van Domselaar G. 2013. Public health genomics and the new molecular epidemiology of bacterial pathogens. Public Health Genomics 16:25–30. doi: 10.1159/000342709. [DOI] [PubMed] [Google Scholar]
- 28.Ernst NS, Hernandez JA, MacKay RJ, Brown MP, Gaskin JM, Nguyen AD, Giguere S, Colahan PT, Troedsson MR, Haines GR, Addison IR, Miller BJ. 2004. Risk factors associated with fecal Salmonella shedding among hospitalized horses with signs of gastrointestinal tract disease. J Am Vet Med Assoc 225:275–281. doi: 10.2460/javma.2004.225.275. [DOI] [PubMed] [Google Scholar]
- 29.Vetro Widenhouse TS. 2004. Equine salmonellosis—molecular epidemiology of clinical isolates and the effect of antibiotics on the cecal microenvironment with particular reference to short-chain fatty acids and the Salmonella plasmid virulence (spv) genes. PhD dissertation. University of Florida, Gainesville, FL. [Google Scholar]
- 30.Rychlik I, Gregorova D, Hradecka H. 2006. Distribution and function of plasmids in Salmonella enterica. Vet Microbiol 112:1–10. doi: 10.1016/j.vetmic.2005.10.030. [DOI] [PubMed] [Google Scholar]
- 31.Castellanos LR, Donado-Godoy P, León M, Clavijo V, Arevalo A, Bernal JF, Timmerman AJ, Mevius DJ, Wagenaar JA, Hordijk J. 2017. High heterogeneity of Escherichia coli sequence types harbouring ESBL/AmpC genes on IncI1 plasmids in the Colombian poultry chain. PLoS One 12:e0170777. doi: 10.1371/journal.pone.0170777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bortolaia V, Guardabassi L, Trevisani M, Bisgaard M, Venturi L, Bojesen AM. 2010. High diversity of extended-spectrum beta-lactamases in Escherichia coli isolates from Italian broiler flocks. Antimicrob Agents Chemother 54:1623–1626. doi: 10.1128/AAC.01361-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carattoli A. 2013. Plasmids and the spread of resistance. Int J Med Microbiol 303:298–304. doi: 10.1016/j.ijmm.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 34.Phan MD, Wain J. 2008. IncHI plasmids, a dynamic link between resistance and pathogenicity. J Infect Dev Ctries 2:272–278. [DOI] [PubMed] [Google Scholar]
- 35.Fredericq P. 1958. Colicins and colicinogenic factors. Symp Soc Exp Biol 12:104–122. [PubMed] [Google Scholar]
- 36.Wick RR, Schultz MB, Zobel J, Holt KE. 2015. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics 31:3350–3352. doi: 10.1093/bioinformatics/btv383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barr BS, Waldridge BM, Morresey PR, Reed SM, Clark C, Belgrave R, Donecker JM, Weigel DJ. 2013. Antimicrobial-associated diarrhoea in three equine referral practices. Equine Vet J 45:154–158. doi: 10.1111/j.2042-3306.2012.00595.x. [DOI] [PubMed] [Google Scholar]
- 38.Zhao S, McDermott PF, White DG, Qaiyumi S, Friedman SL, Abbott JW, Glenn A, Ayers SL, Post KW, Fales WH, Wilson RB, Reggiardo C, Walker RD. 2007. Characterization of multidrug resistant Salmonella recovered from diseased animals. Vet Microbiol 123:122–132. doi: 10.1016/j.vetmic.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 39.van Duijkeren E, Wannet WJB, Heck MEOC, van Pelt W, Sloet van Oldruitenborgh-Oosterbaan MM, Smit JAH, Houwers DJ. 2002. Sero types, phage types and antibiotic susceptibilities of Salmonella strains isolated from horses in The Netherlands from 1993 to 2000. Vet Microbiol 86:203–212. doi: 10.1016/S0378-1135(02)00007-X. [DOI] [PubMed] [Google Scholar]
- 40.Nikaido H. 2009. Multidrug resistance in bacteria. Annu Rev Biochem 78:119–146. doi: 10.1146/annurev.biochem.78.082907.145923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Magnet S, Courvalin P, Lambert T. 1999. Activation of the cryptic aac(6′)-Iy aminoglycoside resistance gene of Salmonella by a chromosomal deletion generating a transcriptional fusion. J Bacteriol 181:6650–6655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chandler PM, Krishnapillai V. 1974. Phenotypic properties of R factors of Pseudomonas aeruginosa: R factors readily transferable between Pseudomonas and the Enterobacteriaceae. Genet Res 23:239–250. doi: 10.1017/S0016672300014890. [DOI] [PubMed] [Google Scholar]
- 43.Thanassi DG, Cheng LW, Nikaido H. 1997. Active efflux of bile salts by Escherichia coli. J Bacteriol 179:2512–2518. doi: 10.1128/jb.179.8.2512-2518.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Webber MA, Piddock LJV. 2003. The importance of efflux pumps in bacterial antibiotic resistance. J Antimicrob Chemother 51:9–11. doi: 10.1093/jac/dkg050. [DOI] [PubMed] [Google Scholar]
- 45.Oethinger M, Kern WV, Jellen-Ritter AS, McMurry LM, Levy SB. 2000. Ineffectiveness of topoisomerase mutations in mediating clinically significant fluoroquinolone resistance in Escherichia coli in the absence of the AcrAB efflux pump. Antimicrob Agents Chemother 44:10–13. doi: 10.1128/AAC.44.1.10-13.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Papich MG. 2003. Antimicrobial therapy for gastrointestinal diseases. Vet Clin North Am Equine Pract 19:645–663. doi: 10.1016/j.cveq.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 47.WHO. 2013. Integrated surveillance of antimicrobial resistance—guidance from a WHO advisory group. WHO, Geneva, Switzerland. [Google Scholar]
- 48.McDermott PF, Tyson GH, Kabera C, Chen Y, Li C, Folster JP, Ayers SL, Lam C, Tate HP, Zhao S. 2016. Whole-genome sequencing for detecting antimicrobial resistance in nontyphoidal Salmonella. Antimicrob Agents Chemother 60:5515–5520. doi: 10.1128/AAC.01030-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Phuc Nguyen MC, Woerther P-L, Bouvet M, Andremont A, Leclercq R, Canu A. 2009. Escherichia coli as reservoir for macrolide resistance genes. Emerg Infect Dis 15:1648–1650. doi: 10.3201/eid1510.090696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.White DGPD, Alekshun MN, McDermott PF, Levy SB. 2005. Frontiers in antimicrobial resistance: a tribute to Stuart B. Levy. American Society for Microbiology, Washington, DC. [Google Scholar]
- 51.Gustafsson A. 2004. Antibiotic associated diarrhea in horses. PhD thesis Swedish University of Agricultural Sciences, Uppsala, Sweden. [Google Scholar]
- 52.Weese JS, Staempfli HR, Prescott JF. 2000. Isolation of environmental Clostridium difficile from a veterinary teaching hospital. J Vet Diagn Invest 12:449–452. doi: 10.1177/104063870001200510. [DOI] [PubMed] [Google Scholar]
- 53.O'Brien TF. 2002. Emergence, spread, and environmental effect of antimicrobial resistance: how use of an antimicrobial anywhere can increase resistance to any antimicrobial anywhere else. Clin Infect Dis 34:S78–S84. doi: 10.1086/340244. [DOI] [PubMed] [Google Scholar]
- 54.Matagne A, Misselyn-Bauduin AM, Joris B, Erpicum T, Granier B, Frere JM. 1990. The diversity of the catalytic properties of class A beta-lactamases. Biochem J 265:131–146. doi: 10.1042/bj2650131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dominguez JE, Gutkind GO, Di Conza JA, Mercado EC. 2015. Occurrence of plasmidic AmpC β-lactamase in a Salmonella Typhimurium isolate of equine origin: first report of CMY-2 in animals in Argentina. J Glob Antimicrob Resist 3:315–316. doi: 10.1016/j.jgar.2015.09.006. [DOI] [PubMed] [Google Scholar]
- 56.Gentry-Weeks C, Hutcheson HJ, Kim LM, Bolte D, Traub-Dargatz J, Morley P, Powers B, Jessen M. 2002. Identification of two phylogenetically related organisms from feces by PCR for detection of Salmonella spp. J Clin Microbiol 40:1487–1492. doi: 10.1128/JCM.40.4.1487-1492.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Clinical and Laboratory Standards Institute. 2015. Performance standards for antimicrobial disk susceptibility tests; approved standard—twelfth edition. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 58.CDC. 2013. 2012–2013 integrated report data tables. CDC, Atlanta, GA: https://www.fda.gov/downloads/AnimalVeterinary/SafetyHealth/AntimicrobialResistance/NationalAntimicrobialResistanceMonitoringSystem/UCM453387.pdf. [Google Scholar]
- 59.Ohta N, Norman KN, Norby B, Lawhon SD, Vinasco J, den Bakker H, Loneragan GH, Scott HM. 2017. Population dynamics of enteric Salmonella in response to antimicrobial use in beef feedlot cattle. Sci Rep 7:14310. doi: 10.1038/s41598-017-14751-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Inouye M, Dashnow H, Raven LA, Schultz MB, Pope BJ, Tomita T, Zobel J, Holt KE. 2014. SRST2: rapid genomic surveillance for public health and hospital microbiology labs. Genome Med 6:90. doi: 10.1186/s13073-014-0090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nurk S, Bankevich A, Antipov D, Gurevich A, Korobeynikov A, Lapidus A, Prjibelsky A, Pyshkin A, Sirotkin A, Sirotkin Y, Stepanauskas R, McLean J, Lasken R, Clingenpeel SR, Woyke T, Tesler G, Alekseyev MA, Pevzner PA. 2013. Assembling genomes and mini-metagenomes from highly chimeric reads, p 158–170. In Deng M, Jiang R, Sun F, Zhang X (ed), Research in computational molecular biology: 17th Annual International Conference, RECOMB 2013, Beijing, China, April 7–10, 2013. Proceedings. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 62.Babraham Bioinformatics. 2018. FastQC/0.11.6-Java-1.8.0. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- 63.Gurevich A, Saveliev V, Vyahhi N, Tesler G. 2013. QUAST: quality assessment tool for genome assemblies. Bioinformatics 29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Carattoli A, Zankari E, Garcia-Fernandez A, Voldby Larsen M, Lund O, Villa L, Moller Aarestrup F, Hasman H. 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rambaut A. 2009. FigTree v1.4.2. Molecular evolution, phylogenetics and epidemiology. http://tree.bio.ed.ac.uk.
- 66.Nguyen SV, Harhay DM, Bono JL, Smith TP, Fields PI, Dinsmore BA, Santovenia M, Kelley CM, Wang R, Bosilevac JM, Harhay GP. 2016. Complete and closed genome sequences of 10 Salmonella enterica subsp. enterica serovar Anatum isolates from human and bovine sources. Genome Announc 4:e00447-16. doi: 10.1128/genomeA.00447-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rankin SC, Aceto H, Cassidy J, Holt J, Young S, Love B, Tewari D, Munro DS, Benson CE. 2002. Molecular characterization of cephalosporin-resistant Salmonella enterica serotype Newport isolates from animals in Pennsylvania. J Clin Microbiol 40:4679–4684. doi: 10.1128/JCM.40.12.4679-4684.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Viera AJ, Garrett JM. 2005. Understanding interobserver agreement: the kappa statistic. Fam Med 37:360–363. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.