ABSTRACT
Members of the bacterial genus Bifidobacterium generally dominate the fecal microbiota of infants. The species Bifidobacterium longum is prevalent, but the B. longum subsp. longum and B. longum subsp. infantis strains that are known to colonize the infant bowel are not usually differentiated in microbiota investigations. These subspecies differ in their capacities to metabolize human milk oligosaccharides (HMO) and may have different ecological and symbiotic roles in humans. Quantitative PCR provides a quick analytical method by which to accurately ascertain the abundances of target species in microbiotas and microcosms. However, amplification targets in DNA extracted from samples need to be dependably differential. We evaluated the tuf gene sequence as a molecular target for quantitative PCR measurements of the abundances of B. longum subsp. infantis and B. longum subsp. longum in fecal microbiotas. This approach resulted in the detection of a tuf gene variant (operational taxonomic unit 49 [OTU49]) in Chinese infants that has sequence similarities to both B. longum subsp. infantis and B. longum subsp. longum. We compared the genome sequence and growth and transcriptional characteristics of an OTU49 isolate cultured in HMO medium to those of other B. longum subsp. infantis cultures. We concluded from these studies that OTU49 belongs to B. longum subsp. infantis, that dependable quantitative PCR (qPCR) differentiation between the B. longum subspecies cannot be achieved by targeting tuf gene sequences, and that functional genes involved in carbohydrate metabolism might be better targets because they delineate ecological functions.
IMPORTANCE High-throughput DNA sequencing methods and advanced bioinformatics analysis have revealed the composition and biochemical capacities of microbial communities (microbiota and microbiome), including those that inhabit the gut of human infants. However, the microbiology and function of natural ecosystems have received little attention in recent decades, so an appreciation of the dynamics of gut microbiota interactions is lacking. With respect to infants, rapid methodologies, such as quantitative PCR, are needed to determine the prevalences and proportions of different bifidobacterial species in observational and microcosm studies in order to obtain a better understanding of the dynamics of bifidobacterial nutrition and syntrophy, knowledge that might be used to manipulate the microbiota and perhaps ensure the better health of infants.
KEYWORDS: Bifidobacterium longum subsp. infantis, microbiota, qPCR
INTRODUCTION
Bifidobacteria are commonly detected as predominant members of the bowel microbiota of infants, especially during the exclusively milk-fed period of life (1–11). The species Bifidobacterium longum is often detected in the infant fecal microbiota, but B. longum is composed of three subspecies, two of which are represented in the infant bowel (B. longum subsp. longum and B. longum subsp. infantis). Differentiation of B. longum into subspecies is difficult using 16S rRNA gene sequences (6, 12), resulting in relatively few reports of subspecies prevalences and abundances on a global scale (5, 11, 13). Knowledge of the occurrence and abundances of these subspecies is required so that a concept of the multispecies ecology, especially trophic interactions, of the bifidobacterial population of the infant bowel can be developed. Concept development requires an understanding of the roles of the different bifidobacterial species and subspecies in the infant bowel, how they form cooperative consortia, and how their activities impact infant health. Rapid quantitative methodologies would quicken the pace of this ecological research. Then, we may better understand the evolutionary importance of the infant-bifidobacterium relationship (14–17).
Quantitative PCR (qPCR) provides a quick analytical method by which to accurately ascertain the abundances of target species in the microbiota and in microcosms. However, amplification targets in DNA extracted from samples need to be dependably differential. Because of reported difficulties using 16S rRNA gene sequences as qPCR targets in relation to the differentiation of B. longum subsp. longum and B. longum subsp. infantis (6), the purpose of our study was to investigate tuf gene sequences (encoding elongation factor Tu) in this respect. These sequences have been reported to be useful in other species-specific assays (18). High-throughput sequencing (HTS) and qPCR experiments with fecal DNA from Chinese infants led to the recognition of a B. longum operational taxonomic unit (OTU), OTU49, in which the tuf gene had sequence characteristics of that in both B. longum subsp. longum and B. longum subsp. infantis. Additional goals of the work were therefore to assign the organism to the appropriate subspecies using human milk oligosaccharide (HMO) utilization/transcriptional profiles and genomic characterization of a cultured OTU49 isolate.
RESULTS
Recognition of OTU49 in the fecal microbiota of Chinese infants.
HTS analysis utilized scripts from QIIME version 1.4.0, with default settings. Thus, OTUs were generated de novo with the USEARCH (version 5.2.32) algorithm. This approach generated 9,891 OTU from 1,797,803 sequences. Of these, 78 OTU (clustered with a 97% identity threshold) were assigned to the species B. longum.
To derive a qPCR assay capable of discriminating between B. longum subsp. longum and B. longum subsp. infantis, the elongation factor Tu gene (tuf) was utilized as a target. Two TaqMan qPCR assays were developed that allowed absolute discrimination between the type strains of the two subspecies (see Tables S1 and S2 in the supplemental material) when genomic DNA from type cultures was used. The assays did not give a signal with DNA obtained from other bifidobacterial species also common in infant fecal microbiotas (Bifidobacterium bifidum and Bifidobacterium breve). The assays were then used to obtain information on the relative abundances and prevalences of both of the B. longum subspecies in a cohort of Chinese infants.
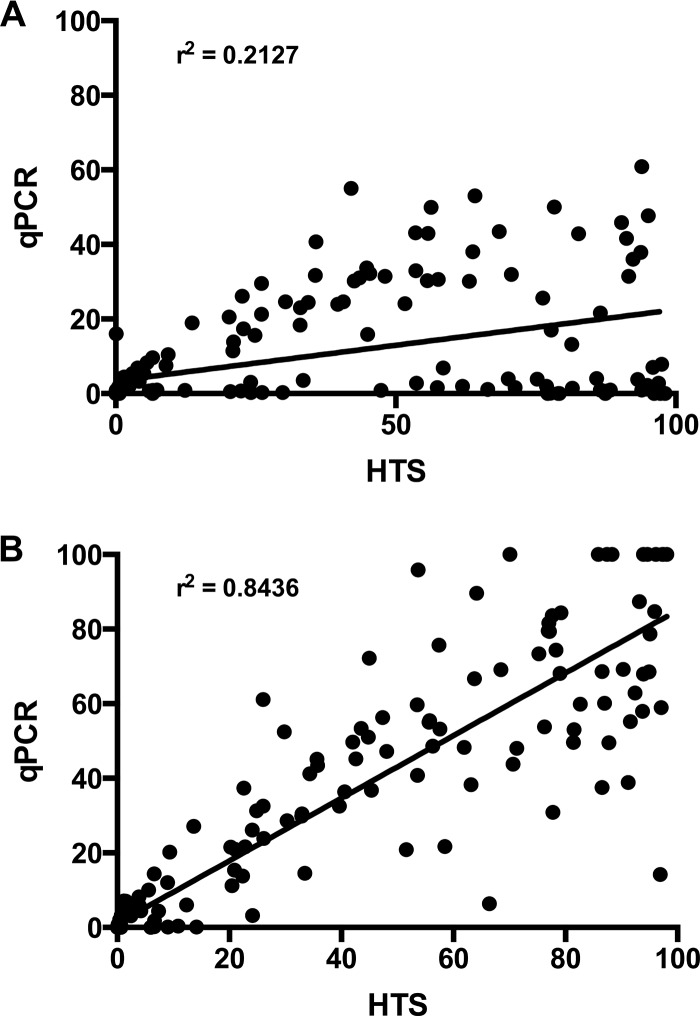
In a comparison of B. longum abundance estimates by tuf gene qPCR and HTS, it was clear that the combined B. longum subsp. longum and B. longum subsp. infantis abundances determined by tuf gene qPCR abundances did not correlate well (r2 = 0.2127) with HTS values (Fig. 1A). This result suggested that there was tuf gene sequence variation in the B. longum members of the microbiota. To confirm this opinion, a 16S rRNA gene qPCR method, developed in a previous study to differentiate B. longum subsp. longum and B. longum subsp. infantis (6), was used to obtain the relative abundances of the two subspecies. The 16S rRNA gene PCR primers were designed for PCR/gel electrophoretic differentiation of the subspecies (19, 20) and are reliable for this purpose. There is, however, a small amount of nonspecificity which is detected by qPCR, hence the reason for our search for better PCR targets. The combined relative abundances of B. longum subsp. longum and B. longum subsp. infantis obtained by the 16S rRNA gene qPCR correlated well with HTS data (r2 = 0.8436; Fig. 1B). This observation confirmed that the tuf gene qPCR was missing B. longum sequences in the fecal microbiota DNA.
FIG 1.
Correlation of B. longum group quantitation in Chinese infants by HTS and qPCR. The plot includes multiple time points for each individual. A linear regression line is shown along with the Pearson correlation r2 value. (A) Combined relative abundances for B. longum subsp. longum and B. longum subsp. infantis obtained by tuf gene-targeted qPCR assay versus HTS B. longum sequences. (B) Combined relative abundances for B. longum subsp. longum and B. longum subsp. infantis obtained by 16S rRNA gene-targeted qPCR assay versus HTS B. longum sequences.
An investigation of the DNA sequences from fecal samples showing a low abundance of B. longum in the tuf gene qPCR data but high abundance in the HTS data revealed an abundant OTU (26% of sequences) designated OTU49. The reference sequence for OTU49 was retrieved and used to search against the NCBI nt database using BLAST and revealed a closest match to a number of B. longum isolates (99% nucleotide identity over 485 bp). Several infants were predicted to have a high abundance of the bacterium associated with the OTU49 sequence, so bifidobacterial isolates were cultured from one of these fecal samples (see below). Sequencing of the 16S rRNA gene from one such isolate indicated that the culture was likely to belong to B. longum subsp. infantis (99.6% identity across the whole gene; 1,411 bp). Sequencing of the tuf gene from the culture indicated that its sequence was not identical to that of either of the type strains but had similarities to both (Fig. S1). The cultured OTU49 tuf gene sequence matched the B. longum subsp. infantis forward primer, B. longum subsp. longum probe, and B. longum subsp. longum reverse primer, thus explaining why this isolate had not been detected by the tuf gene qPCR assay targeting either subspecies (Fig. S1). It was then necessary to confirm by other methods that the cultured isolate of OTU49 belonged to B. longum subsp. infantis.
Characterization of B. longum subsp. longum and B. longum subsp. infantis strains by growth in media containing HMO.
There was a clear differentiation between the B. longum subspecies according to growth in medium containing HMO fractions or constituent carbohydrates of HMO (Fig. 2). B. longum subsp. longum strains grew well in medium containing lacto-N-tetraose (LNT), whereas other substrates gave growth similar to that in the control basal medium (no added carbohydrates). In contrast, OTU49 and B. longum subsp. infantis strains grew well on all substrates relative to the basal medium control, especially in media containing acidic human milk oligosaccharides (HMO-S), sialic acid, LNT, and lacto-N-neotetraose (LNnT).
FIG 2.
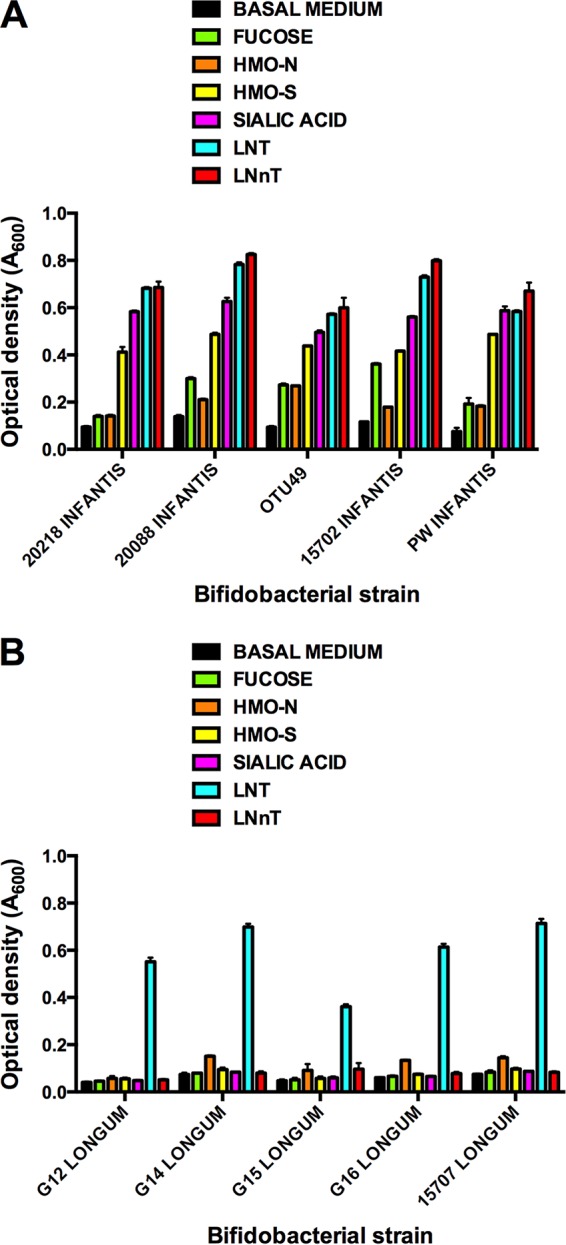
Growth of B. longum subsp. infantis (A) and B. longum subsp. longum (B) strains in basal medium or basal medium containing HMO fractions or component carbohydrates at 0.2% (wt/vol). Optical density (A600) of cultures was measured after 48 h of anaerobic incubation at 37°C.
Genomic comparison of OTU49 and reference strains.
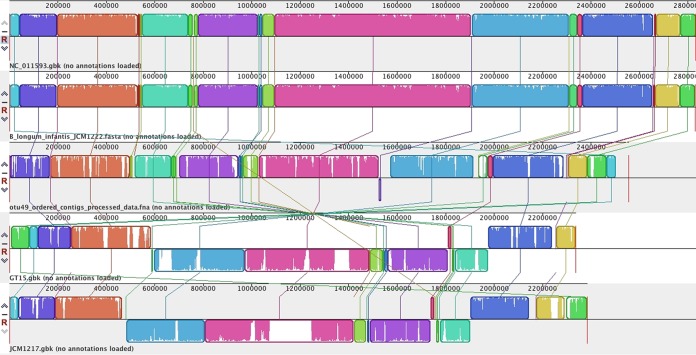
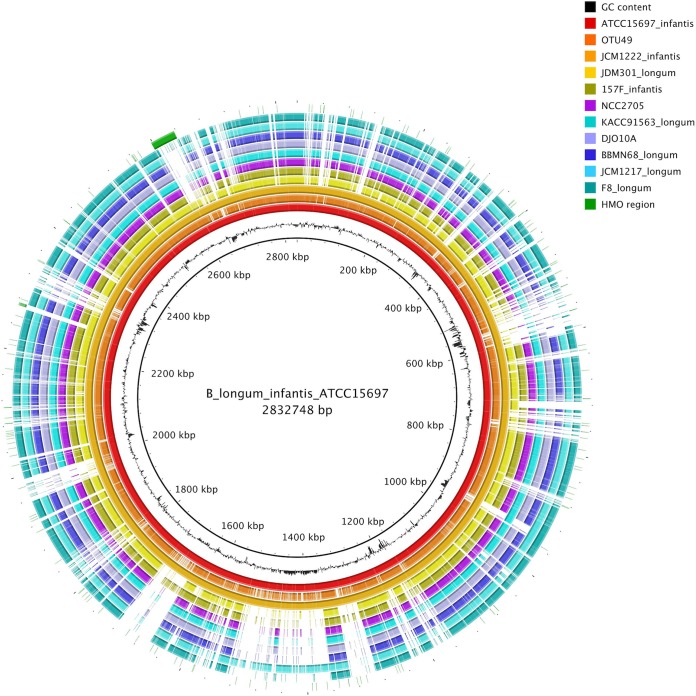
A draft genome sequence was generated for the OTU49 isolate and compared with available B. longum genome sequences. Alignment of multiple B. longum genomes showed that OTU49 had high homology and synteny with B. longum subsp. infantis isolates ATCC 15697 (DSM 20088T) and JCM1222. This alignment also revealed a large inverted region compared with the B. longum subsp. longum genomes (Fig. 3). The genomes were also compared using the BLAST-based BRIG software package. This analysis also indicated that the OTU49 genome was closely related to those of B. longum subsp. infantis isolates ATCC 15697 and JCM1222, including the HMO utilization region identified by Sela et al. (21) (Fig. 4). Regions where the OTU49 genome did not match well with the ATCC 15697 and JCM1222 genomes included insertion sequence and phage gene sequence regions (Tables S3 and S4). Finally, the HMO utilization region identified in B. longum subsp. infantis ATCC 15697 was compared with the same region in the OTU49 and JCM1222 genomes. This revealed a high degree of similarity between the three strains, although in comparison with ATCC 15697, JCM1222 was missing three open reading frames involved in solute binding and transport, and OTU49 was missing six open reading frames involved in solute binding, transport, and general function at the 3′ end of the region (Fig. 5).
FIG 3.
Multiple-genome alignment of B. longum subsp. longum, B. longum subsp. infantis, and OTU49 genomes. Locally colinear blocks (LCBs) are colored and include similarity shading (areas of white within colored blocks were not aligned and probably contain sequence elements specific to the particular genome). LCB connecting lines indicate regions where genomic rearrangement has occurred. The reference genome at the top shows B. longum subsp. infantis ATCC 15697 (NCBI RefSeq accession no. NC_011593), while the two lowest lines show B. longum subsp. longum genomes (GT15 and JCM1217).
FIG 4.
BLAST Ring Image Generator (BRIG) comparison of B. longum subsp. longum, B. longum subsp. infantis, and OTU49 genomes. B. longum subsp. infantis strain ATCC 15697 was used as a reference genome, and the HMO utilization cluster identified by Sela et al. (21) is included as an outer ring (green). Genome regions that do not align with the reference are indicated with a white space.
FIG 5.
Comparison of open reading frames (ORFs) within the HMO utilization cluster identified by Sela et al. (adapted from reference 21 [copyright 2008 National Academy of Sciences]). Greyed out ORF indicates absence in comparison to ATCC 15697 (NCBI RefSeq accession no. NC_011593.1; JCM1222 [NCBI RefSeq accession no. NC_017219.1]). Important carbohydrate utilization genes are named, and ORFs are colored according to function. SBP, solute binding protein; IS3, insertion sequence 3 family element.
Comparison of transcriptomic profiles of B. longum subsp. infantis strains DSM 20088T and OTU49.
RNA sequencing (RNA-seq) analysis was performed to compare the transcriptional levels of genes belonging to the HMO utilization region in B. longum subsp. infantis strains DSM 20088T and OTU49 when grown in medium containing HMO fractions. Paired-end reads from both strains were mapped to Bifidobacterium longum subsp. infantis ATCC 15697 (= DSM 20088T) reference genome (NCBI RefSeq accession no. GCF_000020425.1 [ASM2042v1]; BioProject accession number PRJNA17189). A total of 5.5 × 106 (DSM 20088T) and 5.2 × 106 (OTU49) paired-end reads mapped to the reference genome, providing 96% and 82% genome coverages for Bifidobacterium longum subsp. infantis DSM 20088T and OTU49, respectively (Table S5). A comparison of the normalized gene expression levels of open reading frames belonging to the HMO utilization cluster showed similar transcriptional levels for the two strains when grown in the presence of HMO (Fig. 6). Consistent with findings on the genomic structure of the HMO utilization cluster in B. longum subsp. infantis OTU49, the last six open reading frames showed no expression, confirming the absence of the last 6 genes of the HMO utilization region (BLON_RS12190, BLON_RS12195, BLON_RS12200, BLON_RS12205, BLON_RS12210, and BLON_RS12215) in the B. longum subsp. infantis OTU49 strain compared to the DSM 20088T strain.
FIG 6.
Comparison of transcriptomic profile of genes belonging to HMO gene utilization cluster in B. longum subsp. infantis DSM 20088T and OTU49 grown in basal medium containing the HMO-S fraction. Expression levels are indicated as log2 cpm.
DISCUSSION
The low evolutionary rate of the 16S rRNA gene allows closely related organisms to have highly similar sequences. Additionally, various analytical limitations are associated with HTS analysis of 16S rRNA genes (22), including the widely used clustering threshold of 97% identity (an approximation to species-level clusters), which can lead to poor discrimination between closely related organisms, as is the case with B. longum subspecies (9). Protein-encoding genes, however, tend to have a higher rate of evolutionary change and might provide better options for discrimination of subspecies or closely related species (23). Highly conserved protein-encoding genes, such as groEL (24), recA (25), and tuf (26), have been proposed as alternatives to 16S rRNA genes for phylogenetic discrimination of bacteria.
We investigated tuf gene sequences as a means of differentiating B. longum subspecies, but the detection of OTU49 showed that this is not a useful approach because OTU49 had tuf gene sequences resembling both subspecies. Examination of genetic, transcriptional, and growth characteristics of OTU49 showed that it is typical of B. longum subsp. infantis strains. The OTU49 genome was closely related to B. longum subsp. infantis isolates ATCC 15697 and JCM1222, including the presence of the HMO utilization region identified by Sela et al. (21), which was similarly transcribed to that of the type culture of B. longum subsp. infantis (DSM 20088T). The growth of OTU49 in medium containing human milk fractions or components of HMO was similar to that of other B. longum subsp. infantis cultures, whereas B. longum subsp. longum strains grew poorly in media containing HMO or most HMO components. These marked differences in HMO utilization by B. longum subspecies have been reported previously, but their implications in bifidobacterial ecology have been little discussed (27, 28).
OTU49 clearly belongs to B. longum subsp. infantis from a bacterial physiological point of view, but it has characteristics of both B. longum subsp. longum and B. longum subsp. infantis in terms of the housekeeping gene tuf, presumed previously to be highly conserved. These observations pose a dilemma for microbiologists wishing to study bifidobacterial ecology (i.e., the dynamic interactions of these bacteria) in vivo and in vitro, because recognition of the OTU49 tuf variant in Chinese infants may indicate a lack of dependable qPCR targets to discriminate B. longum subsp. longum and B. longum subsp. infantis when they are based on housekeeping genes, noncoding sequences, or other genes encoding proteins of unknown biological importance (10, 23–25, 29).
Reliable methodologies are required more than ever because new infant formulations (such as the addition of 2′-fucosyl-lactose [30]) and probiotic B. longum subsp. infantis products (29) are likely to soon be launched commercially. The impact of these new products on bifidobacterial ecology will need careful evaluation, with particular reference to the “gold standard” nutrition of feeding infants breast milk. Our results suggest that OTU49 would occupy the fundamental niche of HMO utilization in the infant gut because it has the genetic capacity and functional activity to do so. That seems to be the critical information required in ecological studies, knowing what does the species do and how does it do it. We have recently reported that qPCR targeting genes (based on type culture gene sequences) involved in the utilization of carbohydrates (sialidase and arabinose kinase) provides a qPCR method to differentiate B. longum subsp. longum and B. longum subsp. infantis (31). While further validation of the reliability of this methodology is required, it would have the benefit of providing information about the prevalence and abundance of ecologically important genes associated with bifidobacteria.
MATERIALS AND METHODS
Chinese infants.
The Chinese infants included in this study were part of Mead Johnson Nutrition clinical protocol no. 8602. Fecal samples were obtained from infants that were fed infant formula (n = 40) or were exclusively fed breast milk (n = 51). Fecal samples were obtained when the children were 6, 8, 10, and 12 weeks of age. Ethical approval for the work contained in this article was obtained and carried out in accordance with the ethics committee of Xin Hua Hospital, affiliated with the School of Medicine, Shanghai Jiao Tong University, China. Mothers/guardians of the infants gave informed consent.
Sequencing DNA extracted from infant feces.
The methodology for DNA extraction from feces was described by Tannock et al. (6) and included bead-beating to disrupt bacterial cells, phenol-chloroform treatment, and ethanol precipitation. Pyrosequencing of the V1 to V3 regions of 16S rRNA genes amplified from fecal genomic DNA of infants was carried out by Macrogen, Inc. (Seoul, South Korea) and utilized the Roche-454 FLX genome sequencer with Titanium chemistry, as described by Tannock et al. (6). Analysis of sequence data was carried out using the QIIME software package (version 1.4.0) (22) and incorporated de novo sequence clustering employing the UCLUST and USEARCH algorithms (32). In order to assign taxonomy to the species level, representative sequences from selected OTUs were searched against the NCBI nr database using BLAST (33).
Quantitative PCR differentiation of B. longum subsp. longum and B. longum subsp. infantis targeting the tuf or 16S rRNA gene.
Sequences of the tuf gene from B. longum subsp. longum and B. longum subsp. infantis, along with sequences from B. breve strains, were retrieved from the NCBI Nucleotide database and were aligned using Clustal W (34). Alignments were curated manually, and discriminatory polymorphisms were identified (Fig. S1). Several primer and probe combinations were designed in an attempt to discriminate between the two B. longum subspecies, without cross-reactivity to the closely related B. breve. Primers and TaqMan probes (Table 1) were purchased from Invitrogen and were tested for reaction efficiency and specificity using genomic DNA (gDNA) purified from B. breve (ATCC 15700T), B. bifidum (DSM 20456T), B. longum subsp. longum (ATCC 15707T), B. longum subsp. infantis (DSM 20088T), and Bifidobacterium animalis subsp. lactis (DSM 10140T) cultures on a Life Technologies ViiA7 real-time PCR system in MicroAmp Fast optical 96-well plates with optical adhesive film (Applied Biosystems, Carlsbad, CA). All reactions were carried out in a final volume of 20 μl containing 1× TaqMan Fast PCR mastermix (Applied Biosystems), 300 nM each primer, and 100 nM TaqMan probe. Template DNA was diluted to 1 ng/μl, and 2 ng was added to each reaction mixture. The thermocycling profile consisted of an initial activation of the polymerase at 95°C for 30 s, followed by 40 cycles of 95°C for 10 s and 60°C for 30 s. Fluorescence levels were measured after the 60°C annealing/extension step. Standard curves (to measure reaction efficiency) were generated using gDNA extracted from bifidobacterial strains Bifidobacterium breve (ATCC 15700T), Bifidobacterium longum subsp. longum (ATCC 15707T), and Bifidobacterium longum subsp. infantis (DSM 20088T) using the bead-beating phenol-chloroform-ethanol protocol described previously (6). The standard DNA was quantified spectrophotometrically using a NanoDrop 1000 spectrophotometer (Thermo Scientific) and diluted in 10-fold steps from 5 × 106 to 5 × 101 genomes/reaction, calculated using target gene copies per genome obtained from genome sequence information (NCBI). All reactions were carried out in duplicate and were run twice on separate plates. No-template controls were also included on each plate.
TABLE 1.
PCR primers and probes
| Target | Primer or probe | Sequence (5′ to 3′)a | Reference or source |
|---|---|---|---|
| B. longum subsp. longum tuf gene | tuf_Blon_F | GTATCCGTCCGACCCAGCAG | This study |
| tuf_Blon_P | FAM-CGTGGCCAGGTTGT-BHQ | This study | |
| tuf_Blon_R | GGTGACGGAGCCCGGCTTG | This study | |
| B. longum subsp. infantis tuf gene | tuf_Binf_2_Fb | CGTCACCGGTCGTGTCGAG | This study |
| tuf_Binf_6_Fc | GGTCGAGATCGTCGGCATCCGTCCGA | This study | |
| tuf_Binf_P | FAM-CTGCTGCGTGGCCT-BHQ | This study | |
| tuf_Binf_R | CTCGACATCCTCACGGCC | This study | |
| B. longum subsp. longum 16S rRNA gene | B_long_F | GTTCCCGACGGTCGTAGAG | 19 |
| B_long_R | GTGAGTTCCCGGCAYAATCC | ||
| B. longum subsp. infantis 16S rRNA gene | B_inf_F | CCATCTCTGGGATCGTCGG | 20 |
| B_inf_R | TATCGGGGAGCAAGCGTGA | ||
| Bifidobacterium 16S rRNA gene | g-Bif_F | CTCCTGGAAACGGGTGG | 46 |
| g-Bif_R1 | GGTGTTCTTCCCGATATCTACA | ||
| 16S rRNA gene (all bacteria) | Uni_F | ACTCCTACGGGAGGCAGCAGT | 47 |
| Uni_R | ATTACCGCGGCTGCTGGC |
FAM, 6-carboxyfluorescein; BHQ, black hole quencher.
Forward primer for original assay to discriminate between subspecies infantis and longum.
Forward primer for discrimination of OTU49 from subspecies infantis and longum.
For 16S rRNA gene-based detection, a previously described method for qualitative differentiation of B. longum subsp. longum and B. longum subsp. infantis was used with modification as to the concentrations of primers used (6). For quantitation of B. longum subsp. longum, primers B_long F and B_long R (Table 1) were used at final concentrations of 600 nM and 200 nM, respectively, while for the quantitation of B. longum subsp. infantis, primers B_inf F and B_inf R (Table 1) were used at final concentrations of 200 nM and 600 nM, respectively, in 20-μl reaction mixtures containing 1× Fast SYBR green PCR mastermix (Applied Biosystems). The thermocycling profile consisted of an initial activation of the polymerase at 95°C for 30 s, followed by 40 cycles of 95°C for 10 s, 65°C for 30 s, and 72°C for 30 s. Fluorescence levels were measured after the 65°C annealing step. A melt curve was generated to analyze product specificity. Genomic DNA from B. longum subsp. longum (ATCC 15707T) and B. longum subsp. infantis (DSM 20088T) were used as controls and for the generation of standard curves. While these reaction conditions improved the discrimination of the two subspecies, there was still some cross-reactivity. However, testing of serially diluted genomic DNA from both subspecies against both primer pairs indicated that a cycle threshold cutoff of 27 would be appropriate for use as a detection threshold for the presence of particular subspecies (see Table S1 in the supplemental material). Statistical analyses of data used GraphPad Prism 6, version 6.0f (GraphPad Software, Inc., La Jolla, CA).
Isolation of OTU49 from feces.
Bifidobacteria were cultured from a fecal sample collected from a Chinese baby that, according to HTS data, contained the OTU49 strain in abundance. A 10% (wt/vol) suspension of feces was prepared in prereduced Lactobacilli de Man-Rogosa-Sharpe (MRS) medium (Difco) using glass beads to aid homogenization. The suspension was diluted in 10-fold steps to 10−6 in the same medium, and aliquots of dilutions were spread on plates of prereduced Rogosa SL agar (Difco). The plates were incubated anaerobically for 3 days at 37°C. Bacterial colonies were picked and subcultured using Lactobacilli MRS broth and agar to obtain pure cultures. All culture manipulations were carried out in an anaerobic glove box. Identification of colonies as belonging to the OTU49 strain was achieved, as described in the next section.
Genomic sequencing of OTU49 and comparison with reference strains of B. longum subsp. infantis.
Genomic DNA was extracted from potential OTU49 cultures using the previously described bead-beating phenol-chloroform ethanol precipitation protocol (6). RNA was then removed by treatment with 2 μg of RNase A at room temperature for 30 min followed by cleanup of the gDNA through a NucleoSpin gDNA cleanup column, according to the manufacturer's protocol (Macherey-Nagel, Düren, Germany). Identification of extracted gDNA as belonging to OTU49 was achieved using TaqMan qPCR assays targeting the tuf gene (as described above with qPCR targeting the tuf gene), with the addition of a third reaction utilizing a combination of the tuf_Binf_6_F/tuf_Blon_R primers and the tuf_Blon_P probe (Table 1). This primer/probe combination allowed discrimination of OTU49 from the B. longum subsp. longum and B. longum subsp. infantis type strains (Table S2). Purified gDNA from isolates identified as OTU49 was sent to New Zealand Genomics Ltd. (Dunedin, New Zealand) for 250-base paired-end shotgun sequencing using a TruSeq DNA Nano library on an Illumina MiSeq platform. Sequencing data were returned as both unprocessed data and quality-screened data (checked with FastQC [www.bioinformatics.babraham.ac.uk/projects/fastqc/] and SolexaQA [35]) that had PhiX control library reads and adapter sequences removed using FASTQ-MCF (36). Analysis of processed reads followed procedures suggested by Edwards and Holt (37). Specifically, reads were assembled into contigs using Velvet (38), with a k-mer value of 151. Contigs were ordered against a reference genome sequence (B. longum subsp. infantis ATCC 15697) and visualized and aligned with available B. longum genomes using Mauve version 2.3.1 (39). The OTU49 draft genome was annotated using RAST version 4 (40). Reference-based comparisons of multiple genomes and identification of HMO utilization region positioning were achieved using BRIG version 0.95 (41).
Fractionation of human milk.
To obtain HMO for culture-based studies, expressed breast milk was obtained from the Newborn Intensive Care Unit of Dunedin Hospital with approval from the Lower South Ethics Committee. The milk was fractionated to obtain preparations containing predominantly acidic (HMO-S) or neutral (HMO-N) oligosaccharides. Methodology essentially as described by Ward (42) was used to prepare the fractions. Details are given in the supplemental material.
Growth of OTU49 and reference strains of B. longum subsp. infantis in HMO medium.
Growth of bifidobacterial strains was evaluated by measuring the optical density (A600) of cultures 24 h after a basal medium containing 0.2% (wt/vol) of either the HMO-N or HMO-S fraction or constituents of HMO (fucose [Sigma], sialic acid [Sigma], lacto-N-tetraose [LNT; Glycom], lacto-N-neotetraose [LNnT; Glycom]) were inoculated with bifidobacteria (1% [vol/vol]). Incubation of cultures, in triplicate, was performed under anaerobic conditions at 37°C. The basal medium consisted of Lactobacilli MRS medium that had been prepared from scratch with the omission of glucose (MRS-NO CHO). The growth of OTU49 was compared to that of B. longum subsp. longum strains previously isolated from infant feces (G12-16) and type culture ATCC 15707 and B. longum subsp. infantis strains DSM 20218, DSM 20088T, ATCC 15701, and PW (ingredient of the probiotic Infloran).
Comparison of transcriptomic profiles of B. longum subsp. infantis strains DSM 20088T and OTU49.
Bacterial cell pellets of 1-ml aliquots collected from cultures of B. longum subsp. infantis strain DSM 20088T and OTU49 grown in medium containing the HMO-S fraction were resuspended in RNAprotect bacterial reagent (Qiagen) and stored at −80°C. Total RNA was extracted using TRIzol reagent (Ambion; Life Technologies) with three bead-beating treatments at 5,000 rpm for 40 s to lyse the cells. Purification of total RNA was carried out using the RNeasy MinElute cleanup kit (Qiagen), according to the manufacturer's instructions. Total RNA quality was assessed using Agilent 2100 Bioanalyzer (Agilent) and NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific), and RNA concentration was measured with a Qubit fluorometer (Thermo Fisher Scientific).
For each bacterial strain, one replicate of purified total RNA samples with the best RNA integrity number (RIN) values was selected for sequencing and sent to New Zealand Genomics Ltd. (Dunedin, New Zealand). Total RNA samples were treated with the Ribo-Zero bacterial rRNA removal kit (Illumina) before library preparation with the TruSeq stranded total RNA dual-indexed library kit (Illumina). The Illumina HiSeq platform was used to perform 125-base paired-end sequencing. Adapter- and quality-trimmed data, checked with FastQC (www.bioinformatics.babraham.ac.uk/projects/fastqc/), were returned from the sequencing facility and used for analysis of gene expression levels. Reads were aligned to the genome of B. longum subsp. infantis ATCC 15697 (DSM 20088T; NCBI RefSeq accession no. GCF_000020425.1 [ASM2042v1]; BioProject accession number PRJNA17189) using Rockhopper version 2.0.3 (43). edgeR package version 3.16.5 (44, 45) was used to normalize raw counts and estimate gene expression levels, discarding transcripts with <0.5 counts per million (cpm) in at least one library. Log2 cpm values were used as normalized metrics to show the transcriptomic profiles of the two strains.
Accession number(s).
The OTU49 genome sequence and transcriptomic data have been deposited in the NCBI database under BioProject number PRJNA437004.
Supplementary Material
ACKNOWLEDGMENTS
Jun Watanabe was supported by a National Food Research Institute (Japan) fellowship. The OTU49 research was supported in part by funding from the Riddet Centre of Research Excellence.
We thank Karen Munro for extraction of DNA from feces and Charlotte Wilson for assistance with initial analysis of HTS data.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00336-18.
REFERENCES
- 1.Biavati B, Castagnoli P, Crociani F, Trovatelli LD. 1984. Species of the genus Bifidobacterium in the feces of infants. Microbiologica 7:341–345. [PubMed] [Google Scholar]
- 2.Favier CF, Vaughan EE, De Vos WM, Akkermans ADL. 2002. Molecular monitoring of succession of bacterial communities in human neonates. Appl Environ Microbiol 68:219–226. doi: 10.1128/AEM.68.1.219-226.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Young SL, Simon MA, Baird MA, Tannock GW, Bibiloni R, Spencely K, Lane JM, Fitzharris P, Crane J, Town I, Addo-Yobo E, Murray CS, Woodcock A. 2004. Bifidobacterial species differentially affect expression of cell surface markers and cytokines of dendritic cells harvested from cord blood. Clin Diagn Lab Immunol 11:686–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roger LC, Costabile A, Holland DT, Hoyles L, McCartney AL. 2010. Examination of faecal Bifidobacterium populations in breast- and formula-fed infants during the first 18 months of life. Microbiology 156:3329–3341. doi: 10.1099/mic.0.043224-0. [DOI] [PubMed] [Google Scholar]
- 5.Grönlund MM, Grzeœkowiak Ł, Isolauri E, Salminen S. 2011. Influence of mother's intestinal microbiota on gut colonization in the infant. Gut Microbes 2:227–233. doi: 10.4161/gmic.2.4.16799. [DOI] [PubMed] [Google Scholar]
- 6.Tannock GW, Lawley B, Munro K, Gowri Pathmanathan S, Zhou SJ, Makrides M, Gibson RA, Sullivan T, Prosser CG, Lowry D, Hodgkinson AJ. 2013. Comparison of the compositions of the stool microbiotas of infants fed goat milk formula, cow milk-based formula, or breast milk. Appl Environ Microbiol 79:3040–3048. doi: 10.1128/AEM.03910-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turroni F, Peano C, Pass DA, Foroni E, Severgnini M, Claesson MJ, Kerr C, Hourihane J, Murray D, Fuligni F, Gueimonde M, Margolles A, De Bellis G, O'Toole PW, van Sinderen D, Marchesi JR, Ventura M. 2012. Diversity of bifidobacteria within the infant gut microbiota. PLoS One 7:e36957. doi: 10.1371/journal.pone.0036957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Makino H, Kushiro A, Ishikawa E, Kubota H, Gawad A, Sakai T, Oishi K, Martin R, Ben-Amor K, Knol J, Tanaka R. 2013. Mother-to-infant transmission of intestinal bifidobacterial strains has an impact on the early development of vaginally delivered infant's microbiota. PLoS One 8:e78331. doi: 10.1371/journal.pone.0078331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bäckhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, Li Y, Xia Y, Xie H, Zhong H, Khan MT, Zhang J, Li J, Xiao L, Al-Aama J, Zhang D, Lee YS, Kotowska D, Colding C, Tremaroli V, Yin Y, Bergman S, Xu X, Madsen L, Kristiansen K, Dahlgren J, Wang J. 2015. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe 17:690–703. doi: 10.1016/j.chom.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 10.Milani C, Mancabelli L, Lugli GA, Duranti S, Turroni F, Ferrario C, Mangifesta M, Viappiani A, Ferretti P, Tett A, Segata N, van Sinderen D, Ventura M. 2015. Exploring vertical transmission of bifidobacteria from mother to child. Appl Environ Microbiol 81:7078–7087. doi: 10.1128/AEM.02037-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martin R, Makino H, Cetinyurek Yavuz A, Ben-Amor K, Roelofs M, Ishikawa E, Kubota H, Swinkels S, Sakai T, Oishi K, Kushiro A, Knol J. 2016. Early-life events, including mode of delivery and type of feeding, siblings and gender, shape the developing gut microbiota. PLoS One 11:e0158498. doi: 10.1371/journal.pone.0158498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Youn SY, Seo JM, Ji GE. 2008. Evaluation of the PCR method for identification of Bifidobacterium species. Lett Appl Microbiol 46:7–13. doi: 10.1111/j.1472-765X.2007.02263.x. [DOI] [PubMed] [Google Scholar]
- 13.Huda MN, Lewis Z, Kalanetra KM, Rashid M, Ahmad SM, Raqib R, Qadri F, Underwood MA, Mills DA, Stephensen CB. 2014. Stool microbiota and vaccine responses of infants. Pediatrics 134:e362–e372. doi: 10.1542/peds.2013-3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kunz C, Rudloff S, Baier W, Klein N, Strobel S. 2000. Oligosaccharides in human milk: structural, functional, and metabolic aspects. Annu Rev Nutr 20:699–722. doi: 10.1146/annurev.nutr.20.1.699. [DOI] [PubMed] [Google Scholar]
- 15.D'Aimmo MR, Mattarelli P, Biavati B, Carlsson NG, Andlid T. 2012. The potential of bifidobacteria as a source of natural folate. J Appl Microbiol 11:975–984. doi: 10.1111/j.1365-2672.2012.05261.x. [DOI] [PubMed] [Google Scholar]
- 16.Gordon JI, Dewey KG, Mills DA, Medzhitov RM. 2012. The human gut microbiota and undernutrition. Sci Transl Med 4:137ps12. doi: 10.1126/scitranslmed.3004347. [DOI] [PubMed] [Google Scholar]
- 17.Ruhaak LR, Stroble C, Underwood MA, Lebrilla CB. 2014. Detection of milk oligosaccharides in plasma of infants. Anal Bioanal Chem 406:5775–5784. doi: 10.1007/s00216-014-8025-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ventura M, Canchaya C, Meylan V, Klaenhammer TR, Zink R. 2003. Analysis, characterization, and loci of the tuf genes in Lactobacillus and Bifidobacterium species and their direct application for species identification. Appl Environ Microbiol 69:6908–6922. doi: 10.1128/AEM.69.11.6908-6922.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang RF, Cao WW, Cerniglia CE. 1996. PCR detection and quantitation of predominant anaerobic bacteria in human and animal fecal samples. Appl Environ Microbiol 62:1242–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roy D, Ward P, Champagne G. 1996. Differentiation of bifidobacteria by use of pulsed-field gel electrophoresis and polymerase chain reaction. Int J Food Microbiol 29:11–29. doi: 10.1016/0168-1605(95)00013-5. [DOI] [PubMed] [Google Scholar]
- 21.Sela DA, Chapman J, Adeuya A, Kim JH, Chen F, Whitehead TR, Lapidus A, Rokhsar DS, Lebrilla CB, German JB, Price NP, Richardson PM, Mills DA. 2008. The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome. Proc Natl Acad Sci U S A 2:18964–18969. doi: 10.1073/pnas.0809584105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Method 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Palys T, Berger E, Mitrica I, Nakamura LK, Cohan FM. 2000. Protein-coding genes as molecular markers for ecologically distinct populations: the case of two Bacillus species. Int J Syst Evol Microbiol 50:1021–1028. doi: 10.1099/00207713-50-3-1021. [DOI] [PubMed] [Google Scholar]
- 24.Sakamoto M, Ohkuma M. 2010. Usefulness of the hsp60 gene for the identification and classification of Gram-negative anaerobic rods. J Med Microbiol 59:1293–1302. doi: 10.1099/jmm.0.020420-0. [DOI] [PubMed] [Google Scholar]
- 25.Torriani S, Felis GE, Dellaglio F. 2001. Differentiation of Lactobacillus plantarum, L. pentosus, and L. paraplantarum by recA gene sequence analysis and multiplex PCR assay with recA gene-derived primers Appl Environ Microbiol 67:3450–3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mignard S, Flandrois J-P. 2007. Identification of Mycobacterium using the EF-Tu encoding (tuf) gene and the tmRNA encoding (ssrA) gene. J Med Microbiol 56:1033–1041. doi: 10.1099/jmm.0.47105-0. [DOI] [PubMed] [Google Scholar]
- 27.Ward RE, Ninonuevo M, Mills DA, Lebrilla CB, German JB. 2007. In vitro fermentability of human milk oligosaccharides by several strains of bifidobacteria. Mol Nutr Food Res 51:1398–1405. doi: 10.1002/mnfr.200700150. [DOI] [PubMed] [Google Scholar]
- 28.Locascio RG, Ninonuevo MR, Kronewitter SR, Freeman SL, German JB, Lebrilla CB, Mills DA. 2009. A versatile and scalable strategy for glycoprofiling bifidobacterial consumption of human milk oligosaccharides. Microb Biotechnol 2:333–342. doi: 10.1111/j.1751-7915.2008.00072.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frese SA, Hutton AA, Contreras LN, Shaw CA, Palumbo MC, Casaburi G, Ku G, Davis JCC, Lebrilla CB, Henrick BM, Freeman SL, Barile D, German JB, Mills DA, Smilowitz JT, Underwood MA. 2017. Persistence of supplemented Bifidobacterium longum subsp. infantis EVC–1 in breastfed infants. mSphere 2:e00501-. doi: 10.1128/mSphere.00501-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goehring KC, Marriage BJ, Oliver JS, Wilder JA, Barrett EG, Buck RH. 2016. Similar to those who are breastfed, infants fed a formula containing 2′-fucosyllactose have lower inflammatory cytokines in a randomized controlled trial. J Nutr 146:2559–2566. doi: 10.3945/jn.116.236919. [DOI] [PubMed] [Google Scholar]
- 31.Lawley B, Munro K, Hughes A, Hodgkinson AJ, Prosser CG, Lowry D, Zhou SJ, Makrides M, Gibson RA, Lay C, Chew C, Lee PS, Wong KH, Tannock GW. 2017. Differentiation of Bifidobacterium longum subspecies longum and infantis by quantitative PCR using functional gene targets. PeerJ 25: 5:e3375. doi: 10.7717/peerj.3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 33.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 34.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 35.Cox MP, Peterson DA, Biggs PJ. 2010. SolexaQA: at-a-glance quality assessment of Illumina second-generation sequencing data. BMC Bioinformatics 11:485. doi: 10.1186/1471-2105-11-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aronesty E. 2011. ea-utils: command-line tools for processing biological sequence data. https://github.com/ExpressionAnalysis/ea-utils.
- 37.Edwards DJ, Holt KE. 2013. Beginner's guide to comparative bacterial genome analysis using next-generation sequence data. Microb Inform Exp 3:2. doi: 10.1186/2042-5783-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zerbino DR, Birney E. 2008. Velvet algorithms for de novo short read assembly using de Bruijn graphs. Gen Res 18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rissman AI, Mau B, Biehl BS, Darling AE, Glasner JD, Perna NT. 2009. Reordering contigs of draft genomes using the Mauve aligner. Bioinformatics 25:2071–2073. doi: 10.1093/bioinformatics/btp356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA. 2011. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ward RE. 2009. Isolation of milk oligosaccharides using solid-phase extraction. Open Glycosci 2:9–15. doi: 10.2174/1875398100902010009. [DOI] [Google Scholar]
- 43.McClure R, Balasubramanian D, Sun Y, Bobrovskyy M, Sumby P, Genco CA, Vanderpool CK, Tjaden B. 2013. Computational analysis of bacterial RNA-seq data. Nucleic Acids Res 41:e140. doi: 10.1093/nar/gkt444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McCarthy DJ, Chen Y, Smyth GK. 2012. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res 40:4288–4297. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Matsuki T, Watanabe K, Fujimoto J, Kado Y, Takada T, Matsumoto K, Tanaka R. 2004. Quantitative PCR with 16S rRNA-gene-targeted species-specific primers for analysis of human intestinal bifidobacteria. Appl Environ Microbiol 70:167–173. doi: 10.1128/AEM.70.1.167-173.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hartman AL, Lough DM, Barupal DK, Fiehn O, Fishbein T, Zasloff M, Eisen JA. 2009. Human gut microbiome adopts an alternative state following small bowel transplantation. Proc Natl Acad Sci U S A 106:17187–17192. doi: 10.1073/pnas.0904847106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.