ABSTRACT
Integrative and conjugative elements (ICEs) of the SXT/R391 family are key drivers of the spread of antibiotic resistance in Vibrio cholerae, the infectious agent of cholera, and other pathogenic bacteria. The SXT/R391 family of ICEs was defined based on the conservation of a core set of 52 genes and site-specific integration into the 5′ end of the chromosomal gene prfC. Hence, the integrase gene int has been intensively used as a marker to detect SXT/R391 ICEs in clinical isolates. ICEs sharing most core genes but differing by their integration site and integrase gene have been recently reported and excluded from the SXT/R391 family. Here we explored the prevalence and diversity of atypical ICEs in GenBank databases and their relationship with typical SXT/R391 ICEs. We found atypical ICEs in V. cholerae isolates that predate the emergence and expansion of typical SXT/R391 ICEs in the mid-1980s in seventh-pandemic toxigenic V. cholerae strains O1 and O139. Our analyses revealed that while atypical ICEs are not associated with antibiotic resistance genes, they often carry cation efflux pumps, suggesting heavy metal resistance. Atypical ICEs constitute a polyphyletic group likely because of occasional recombination events with typical ICEs. Furthermore, we show that the alternative integration and excision genes of atypical ICEs remain under the control of SetCD, the main activator of the conjugative functions of SXT/R391 ICEs. Together, these observations indicate that substitution of the integration/excision module and change of specificity of integration do not preclude atypical ICEs from inclusion into the SXT/R391 family.
IMPORTANCE Vibrio cholerae is the causative agent of cholera, an acute intestinal infection that remains to this day a world public health threat. Integrative and conjugative elements (ICEs) of the SXT/R391 family have played a major role in spreading antimicrobial resistance in seventh-pandemic V. cholerae but also in several species of Enterobacteriaceae. Most epidemiological surveys use the integrase gene as a marker to screen for SXT/R391 ICEs in clinical or environmental strains. With the recent reports of closely related elements that carry an alternative integrase gene, it became urgent to investigate whether ICEs that have been left out of the family are a liability for the accuracy of such screenings. In this study, based on comparative genomics, we broaden the SXT/R391 family of ICEs to include atypical ICEs that are often associated with heavy metal resistance.
KEYWORDS: antibiotic resistance, conjugation, entry exclusion, integrative and conjugative element, Vibrio cholerae, genetic recombination, site-specific recombination
INTRODUCTION
Since the mid-1980s, multidrug resistance has considerably increased in toxicogenic Vibrio cholerae, the infectious agent of the acute diarrheal disease cholera (1, 2). The emergence and global spread of resistance in V. cholerae have been exacerbated by integrative and conjugative elements (ICEs) of the SXT/R391 family (3, 4). ICEs are mobile genetic elements that propagate by conjugative transfer, a process involving a direct contact between donor and recipient cells. ICEs are usually integrated into the chromosome of their host and are vertically inherited from one generation to another. Excision of SXT/R391 ICEs from the chromosome and their transfer to a new host are triggered by conditions that induce the host SOS response, including exposure to antibiotics (5).
The resistance profile and evolution of V. cholerae seventh-pandemic clones appear to have been shaped by two major independent events of ICE acquisition (4). First, in the mid-1980s, V. cholerae strains of the O1 El Tor serogroup gained ICEVchInd5, which is now globally distributed in seventh-pandemic clones. In addition to conferring multidrug resistance, ICEVchInd5 seems to have restricted horizontal gene transfer because it codes for the periplasmic DNA endonuclease IdeA, which has been shown to inhibit natural transformation (6, 7). The second event took place in 1992, when SXT was acquired by V. cholerae O139 or its El Tor progenitor, prior to the first O139 cholera outbreak in the Indian subcontinent in early 1993 (4, 8). SXT and ICEVchInd5 likely originate from environmental Vibrio spp. or other Gammaproteobacteria. Now, SXT/R391 ICEs are found in several other species of Vibrio, Proteus, Providencia, Alteromonas, Shewanella, and Marinomonas. Recently, such an ICE was even reported in a drug-resistant isolate of the Pasteurellaceae member Actinobacillus pleuropneumoniae recovered from a pneumonic pig in the United Kingdom (9).
SXT/R391 ICEs have been grouped together based on the conservation of a core set of 52 genes and their integration into the 5′ end of prfC, a gene coding for the peptide chain release factor 3 (10). Over the years, a function has been attributed to 30 of these genes. Hence, int and xis mediate integration into and excision from the chromosome at prfC (11, 12), srpR and srpM code for an active partitioning system (13), and mobI and traI initiate at the origin of transfer (oriT) a rolling-circle replication of the excised ICE (13, 14). Together with the putative type IV coupling protein TraD and the putative auxiliary relaxosome component TraJ, they also enable the conjugative translocation of the ICE into the recipient cell, via a type IV secretion system (T4SS) encoded by four operons of tra genes. bet and exo encode a λ Red-like homologous recombination system involved in ICE plasticity (15). Eex is the entry exclusion factor in the recipient cell that works jointly with TraG in the donor cell (16). setC and setD code for a class 2 transcriptional regulator that activates the expression of all the aforementioned operons except eex (17). Finally, setR and croS code for two transcriptional repressors that form a bistable switch controlling setCD expression in response to DNA damage (5, 18).
In recent years, two reports have described atypical ICEs that are closely related to SXT/R391 ICEs but differ in structure and/or specificity of integration. ICEVchBan8 of V. cholerae O37 carries alternative int and xis genes, is integrated at the 3′ end of a serine tRNA gene, and has undergone a large inversion between traG and srpM (19). Therefore, ICEVchBan8 was excluded from the SXT/R391 family (10). Whole-genome sequencing of Vibrio alginolyticus strains from China revealed three other atypical ICEs inserted in the same tRNA-Ser gene as ICEVchBan8 but lacking the large DNA inversion (20). These two reports show that int is not a conserved feature. Thus, the core set of conserved genes that was deduced from sequence comparison of a representative set of 13 ICEs integrated at prfC (10) and used to define the SXT/R391 family is likely smaller than previously described. Moreover, the lack of conservation of int suggests that its systematic use as a marker to detect SXT/R391 ICEs in epidemiological screenings needs to be reconsidered since atypical ICEs closely related to SXT/R391 are not detected when int is used as a marker, thereby jeopardizing the reliability of such screenings.
In this study, we conducted a comparative analysis of a large sample of representative elements that helped us redefine the core genome of SXT/R391 ICEs, which now excludes genes involved in integration/excision. We show that despite the change of integration/excision module, expression of the alternative integrase gene retains its dependency on the transcriptional activator SetCD. Based on these analyses and strict conservation of the regulatory functions, we propose a novel inclusive and comprehensive typing scheme for SXT/R391 ICEs.
RESULTS AND DISCUSSION
Conserved core of genes of the SXT/R391 family of ICEs.
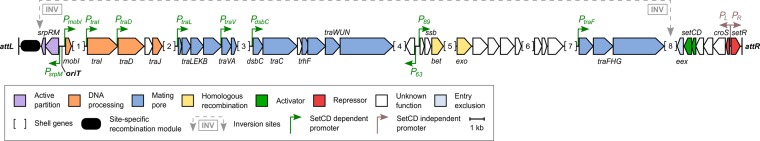
To assess the diversity of SXT/R391 ICEs, we searched the GenBank database using R997 from Proteus mirabilis as a reference. We also screened a collection of 275 Canadian Vibrio isolates using primers designed to detect setCD. One positive isolate, V. parahaemolyticus S107-1, was found, and its genome was sequenced and assembled. The sequence of ICEVpaCan1 was then extracted and included in this study. Overall, a set of 68 complete ICEs from different bacterial species and origins were recovered (Table 1). After manual curation of the annotated sequences to correct missing small open reading frames, such as mobI or xis, and inconsistent start codons, the soft core of SXT/R391-like ICEs was built based on genes present in 95% of all sampled ICEs (57 of 61 ICEs). Forty-three genes were found to be strictly conserved, including those involved in DNA recombination and repair (ssb-bet-exo, radC), initiation of conjugative rolling-circle replication (mobI and traI) and partition (srpRM), and conjugative DNA translocation (traDJ), as well as T4SS assembly (traLEKB, traVA, dsbC-traC-trhF-traWUN, and traFHG) and entry exclusion (eex) (Fig. 1). The regulation module that contains setCD, setR, and croS was also strictly conserved. Genes coding for a putative cobalamin synthase (cobS) and a putative lytic transglycosylase (s082) as well as 11 genes of unknown function (s091, s093, s063, s089, s088, s068, s069, s092, s072, s083, s084) are also part of the conserved soft core.
TABLE 1.
SXT/R391 ICEs used in the study
| Species | Strain | Origin | Yr | ICE name | Ta | Size (bp)b | Gc | Predicted resistance gene(s)d | Referencee | GenBank accession no. |
|---|---|---|---|---|---|---|---|---|---|---|
| Actinobacillus pleuropneumoniae | MIDG3553 | Pneumonic lung of a pig, UK | 2012 | ICEApl2 | 1R | 92,660 | + | sul2, floR, strAB, dfrA1 | 9 | MF187965 |
| Alteromonas macleodii | D7 | Adaman Sea, Thailand | 2000 | ICEAmaD7 | 1S | 117,055 | + | merTPCA, acrAB-tolC, zitB, zntA, copA | CP014323 | |
| Alteromonas mediterranea | MED64 | Aegean Sea, Lebanon | 2000 | ICEAmaMED64 | 1S | 80,715 | + | − | CP004848 | |
| Escherichia coli | HVH 177 | Human blood, Denmark | 2003 | ICEEcoHVH177 | 1S | 89,125 | + | − | AZJM01000017 | |
| Idiomarinaceae bacterium | HL-53 | Unknown | ND | ICEIbaHL53 | 1S | 71,962 | + | − | LN899469 | |
| Methylophaga frappieri | JAM7 | Seawater treatment plant, Montreal Biodome, Canada | ND | ICEMfrJAM7 | 1S | 109,780 | + | − | CP003380 | |
| Photobacterium damselae | PC554.2 | Senegalese sole, Galicia, Spain | 2003 | ICEPdaSpa1 | 1S | 102,985 | + | tetA | 10 | AJ870986 |
| Proteus mirabilis | TJ1809 | Stool, Tianjin, China | 2013 | ICEPmiCHN1809 | 1R | 76,218 | + | − | 36 | KX243413 |
| 09MAS2407 | Stool, Maansham, China | 2009 | ICEPmiCHN2407 | 1R | 97,078 | + | tetA, merTPCA, czcD | 36 | KX243405 | |
| 09MAS2410 | Stool, Maansham, China | 2009 | ICEPmiCHN2410 | 1R | 93,537 | + | merTPCA, czcD | 36 | KX243406 | |
| 09MAS2416 | Stool, Maansham, China | 2009 | ICEPmiCHN2416 | 1R | 92,556 | + | merTPCA, czcD | 36 | KX243407 | |
| TJ3237 | Stool, Tianjin, China | 2013 | ICEPmiCHN3237 | 1S | 87,215 | + | 36 | KX243414 | ||
| TJ3300 | Stool, Tianjin, China | 2013 | ICEPmiCHN3300 | 1R | 108,335 | + | sul2, floR, strAB, dfrG | 36 | KX243415 | |
| TJ3335 | Stool, Tianjin, China | 2013 | ICEPmiCHN3335 | 1S | 89,996 | + | sul2, floR, strAB, tetA | 36 | KX243416 | |
| MD20140901 | Stool, Beijing, China | 2014 | ICEPmiCHN901 | 1S | 89,493 | + | sul2, floR, strAB, dfrG | 36 | KX243408 | |
| MD20140902 | Stool, Beijing, China | 2014 | ICEPmiCHN902 | 1S | 89,096 | + | sul2, floR, strAB dfrG | 36 | KX243409 | |
| MD20140903 | Stool, Beijing, China | 2014 | ICEPmiCHN903 | 1S | 89,644 | + | sul2, floR, strAB, dfrG | 36 | KX243410 | |
| MD20140904 | Stool, Beijing, China | 2014 | ICEPmiCHN904 | 1S | 94,942 | + | sul2, floR, strAB, tetA, czcD | 36 | KX243411 | |
| MD20140905 | Stool, Beijing, China | 2014 | ICEPmiCHN905 | 1S | 94,956 | + | sul2, floR, strAB, tetA, czcD | 36 | KX243412 | |
| PM13C04 | Chicken fecal sample, Hubei, China | 2013 | ICEPmiChn1 | 1S | 92,752 | + | sul2, floR, strAB, tetA, czcD | 32 | KT962845 | |
| PM14C28 | Chicken fecal sample, Heibei, China | 2014 | ICEPmiJpn1 | 1S | 91,091 | + | blaCMY-2 | 30 | KT894734 | |
| HI4320 | Human urine, MA, USA | 1986 | ICEPmiUSA1 | 1S | 79,733 | + | − | 10 | AM942759 | |
| Unnamed | India | 1977 | R997 | 1S | 85,368 | + | blaHMS-1, czcD | 61 | KY433363.1 | |
| Proteus vulgaris | 08MAS2213 | Food, Maansham, China | 2008 | ICEPvuCHN2213 | 1S | 94,340 | + | sul2, floR, strAB | 36 | KX243403 |
| Providencia alcalifaciens | P-18 | Bangladesh | 1999 | ICEPalBan1 | 1R | 96,586 | + | sul2, floR, strAB, dfrA1 | 10 | GQ463139 |
| Providencia rettgeri | 107 | Pretoria, South Africa | 1967 | R391 | 1R | 88,532 | + | aph, merTPCA | 62 | AY090559 |
| Providencia stuartii | ATCC 33672 | ATCC, unknown | ND | ICEPst33672 | 1R | 75,588 | + | merTPCA | CP008920 | |
| Shewanella decolorationis | S12 | Wastewater treatment plant, Guangdong, China | 2012 | ICESdeCHNS12 | 1S | 70,735 | + | − | AXZL01000060 | |
| Shewanella sp. | W3-18-1 | Pacific Ocean | 2000 | ICESpuPO1 | 1S | 108,623 | + | acrAB-tolC, zitB, zntA, copA, czcA, czcD | 10 | CP000503 |
| Vibrio alginolyticus | A056 | Whiteleg shrimp, Guangdong, China | 2003 | ICEValA056-1 | 1S | 89,004 | + | sul2, strAB, acrAB-tolC | 20 | KR231688 |
| ICEValA056-2 | 2R | 103,826 | + | − | 20 | KR231689 | ||||
| E0601 | Seawater, Guangdong, China | 2006 | ICEValE0601 | 1R | 106,165 | + | − | 20 | KT072768 | |
| HN396 | Seawater, Guangxi, China | 2008 | ICEValHN396 | 2R | 86,687 | + | − | 20 | KT072770 | |
| HN437 | Seawater, Hainan, China | 2008 | ICEValHN437 | 2* | 94,920 | + | ugd | 20 | KT072771 | |
| HN492 | Seawater, Hainan, China | 2008 | ICEValHN492 | 1R | 106,164 | + | 20 | KT072769 | ||
| ZJ-T | Orange-spotted grouper, Guangdong, China | 2005 | ICEValZJT1 | 1S | >88,684 | sul2, strAB, acrAB-tolC, qnrVC5 | CP016224 | |||
| ICEValZJT2 | 2R | >100,515 | CP016224 | |||||||
| Vibrio cholerae O1 El Tor | 2055/98 | Bangladesh | 1998 | ICEVchBan5 | 1R | 102,131 | + | sul2, floR, strAB, dfrA1 | 10 | GQ463140 |
| MJ-1236 | Matlab, Bangladesh | 1994 | ICEVchBan9 | 1R | 106,124 | + | sul2, floR, strAB, tetA, dfrA1, zitB | 10 | CP001485 | |
| ICDC-2605 | Patient, Guizhou, China | 1998 | ICEVchCHN2605 | 1S | 98,458 | + | sul2, floR, strAB, dfrG | 37 | KT151661 | |
| ICDC-4210 | Patient, Jiangxi, China | 1999 | ICEVchCHN4210 | 1S | 110,349 | + | sul2, floR, strAB, tetA, dfrG | 37 | KT151662 | |
| ICDC-956 | Environment, Liaoning, China | 2001 | ICEVchCHN956 | 1S | 109,322 | + | sul2, floR, strAB, tetA, dfrG | 37 | KT151655 | |
| wujiang-2 | Patient, China | ND | ICEVchCHNwuji | 1R | 96,168 | + | sul2, strAB, tetA, dfrA1 | KT151664 | ||
| CO943 | Sevagram, India | 1994 | ICEVchInd5 | 1R | 97,847 | + | sul2, floR, strAB, dfrA1 | 10 | GQ463142 | |
| 2010EL-1786 | Patient, Artibonite, Haiti | 2010 | ICEVchHai1 | 1R | 97,847 | + | sul2, floR, strAB, dfrA1 | CP003069 | ||
| 2012HC-25 | Stool, Haiti | 2012 | ICEVch2012HC25 | 3R | >108,600 | + | − | JSTY01000047 | ||
| JSTY01000048 | ||||||||||
| V. cholerae O139 | AS207 | Clinical, Kolkata, India | 1997 | ICEVchInd4 | 1S | 95,491 | + | sul2, floR, strAB | 10 | GQ463141 |
| MO10 | Clinical, Chennai, India | 1992 | SXT | 1S | 99,452 | + | sul2, floR, strAB, dfr18 | 8 | DS990138 | |
| V. cholerae non-O1/O139 | HC-36A1 | Stool, Haiti | 2010 | ICEVchHai2 | 1S | 82,795 | + | − | 63 | AXDR01000007 |
| 2012Env-25 | Water, Haiti | 2012 | ICEVch2012Env25 | 3R | >108,600 | + | − | JSTE01000047 | ||
| JSTE01000048 | ||||||||||
| 1-010118-075 | Sewage, San Luis Potosi, Mexico | 2001 | ICEVchMex1 | 1R | 82,839 | + | − | 10 | GQ463143 | |
| V. cholerae O37 | MZO-3 | Clinical, Bangladesh | 2001 | ICEVchBan8 | 3R | 105,790 | + | acrAB-tolC | 10 | JQ345361 |
| V. cholerae O77 | 8-76 | Diarrhea, India | 1976 | ICEVch8-76-1 | 1S | >102,800 | ND | JIDN01000032 | ||
| JIDN01000021 | ||||||||||
| ICEVch8-76-2 | 3R | >122,200 | ND | JIDN01000012 | ||||||
| JIDN01000016 | ||||||||||
| V. cholerae | YB8E08 | Oyster Pond, MA, USA | 2009 | ICEVchYB8E08 | 3R | >104,300 | + | ND | LBGN01000012 | |
| LBGN01000009 | ||||||||||
| OYP6E07 | Oyster Pond, MA, USA | 2009 | ICEVchYP6E07 | 3* | 93,439 | + | acrAB-tolC | NMTB01000014 | ||
| 3272-78 | Water, MD, USA | 1977 | ICEVch3272-78 | 3R | 104,112 | + | acrAB-tolC | MIOZ01000052 | ||
| 3223-74 | Storm drain, Guam | 1974 | ICEVch3223-74 | 3R | 104,112 | + | acrAB-tolC | MIZG01000083 | ||
| Vibrio fluvialis | H-08942 | Infant diarrhea, Kolkata, India | 2002 | ICEVflInd1 | 1S | 102,862 | + | sul2, floR, strAB, dfr18 | 10 | KM213605 |
| Vibrio mytili | CAIM528 | Seawater, Spain | 1985 | ICEVmyCAIM528 | 2R | >63,900 | − | JXOK01000005 | ||
| JXOK01000034 | ||||||||||
| Vibrio parahaemolyticus | CHN25 | Shrimps, Shanghai, China | 2011 | ICEVpaCHN25 | 1R | 88,331 | + | sul2, strAB, tetA, dfrG | CP010883 | |
| S163 | Seafood, Malaysia | 2007 | ICEVpaS163 | 2R | >75,600 | + | − | AWHQ01000004 | ||
| AWHQ01000048 | ||||||||||
| S167 | Environment, China | 2007 | ICEVpaS167 | 2R | >82,000 | + | − | AWHM01000024 | ||
| AWHM01000222 | ||||||||||
| UCM-V493 | Sediment, Spain | 2002 | ICEVpaUCM493 | 1S | 111,578 | + | blaHMS-1 | CP007004 | ||
| S107-1 | Oyster, Ladysmith Harbor, BC, Canada | 2005 | ICEVpaCan1 | 1R | 81,255 | + | CP028481 | |||
| Vibrio vulnificus | SC9729 | Seawater, South Korea | 2011 | ICEVvuSC9729 | 4S | >85,200 | + | cbiO | JZEQ01000086 | |
| JZEQ01000106 | ||||||||||
| CladeA-yb158 | Tilapia fish, Israel | 2005 | ICEVvuCladeA158 | 4R | >87,800 | araJ | LBNN01000013 | |||
| LBNN01000014 | ||||||||||
| CG100 | Oyster, Taiwan | 1993 | ICEVvuCG100 | 4S | >105,300 | araJ | PDGD01000031 | |||
| PDGD01000050 |
Type of SXT/R391 ICEs corresponds to the integrase type (1 for intprfC and 2, 3, and 4 for inttRNA-Ser) and entry exclusion groups (S or R). An ambiguous entry exclusion group is indicated with an asterisk. Entry exclusion groups were inferred from phylogenetic analyses presented in Fig. 5 and 6.
For ICEs scattered over two contigs extracted from WGS data, an estimation of the minimum size is provided.
ICE included in the Get_Homologs analysis. Other ICEs were excluded due to poor sequencing quality or missing data.
ND, not determined; −, no resistance gene.
References are provided for ICEs that have already been described elsewhere.
FIG 1.
Genetic map of the soft core of conserved genes of 61 SXT/R391 ICEs. The soft core map is drawn to scale and based on conservation of genes in at least 57 of the 61 ICE representatives used in the analysis. Positions of the promoters are based on the work of Poulin-Laprade et al. (17, 18). Numbered loci in brackets contain shell genes. Locus 1 includes rumAB and its associated ISCR2 antibiotic resistance element, s024-s026, and the hot spot 5 (HS5) as described by Wozniak et al. (10). Loci 2, 3, and 4 correspond to variable DNA inserted at HS1, HS2, and HS4, respectively (10). Loci 5 and 6 correspond to orfZ and s070, respectively (15, 64). Locus 7 corresponds to s073 and variable DNA inserted at HS3, such as a trimethoprim resistance-conferring integron (10) or diguanylate cyclase genes (65). Locus 8 corresponds to the mercury resistance genes in R391.
Although int and xis are annotated in all sampled SXT/R391 ICEs, both genes are missing in the soft core (Fig. 1). This absence results from the low sequence identity between proteins encoded by the SXT-type int or xis genes and those encoded by ICEVchBan8-type int or xis genes. Int encoded by SXT is only 27% and 25% identical to Int encoded by ICEValA056-2 and ICEVchBan8, respectively, whereas these two alternative integrases share 90% identity.
Atypical ICEs integrated at the same insertion site as VPI-2.
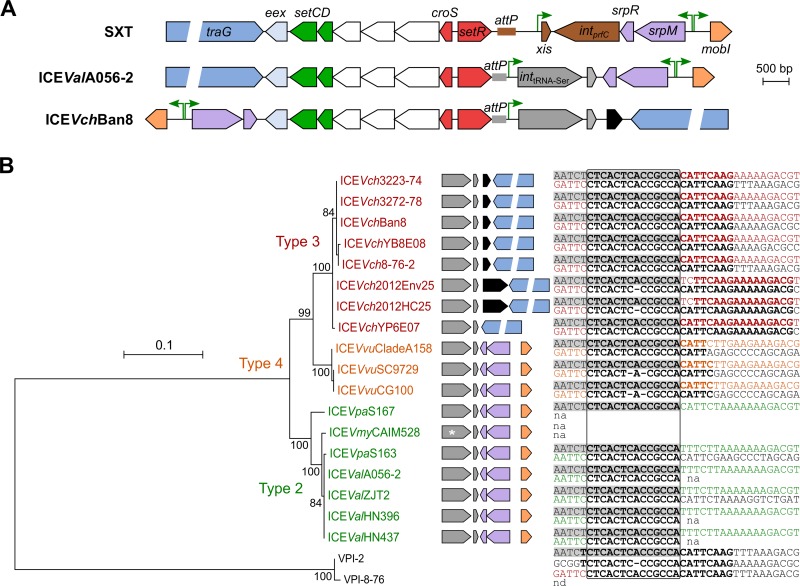
Eighteen of the 68 ICEs were found to carry an atypical site-specific recombination module and to be located within the 3′ end of a tRNA-Ser gene. Comparison of the attachment sites (attL and attR) that flanked these ICEs revealed that site-specific recombination for integration took place within a 14-bp repeated sequence (5′-CTCACTCACCGCCA-3′) corresponding to the 3′ end of the tRNA gene (Fig. 2B). In toxicogenic V. cholerae O1 and O139 isolates, this locus is the insertion site of Vibrio pathogenicity island 2 (VPI-2), which encodes sialic acid release, transport, and catabolism and a neuraminidase (21). VPI-2 is flanked by a 22-bp direct repeat (22) containing the same 14-bp conserved sequence (Fig. 2B). Although VPI-2 and atypical ICEs share the same integration site, their integrases share only 60% identity, thereby ruling out the emergence of atypical ICEs through direct substitution of the prfC recombination module of SXT/R391 ICEs by the tRNA-Ser recombination module of VPI-2. This idea is supported by Boyd et al. (23), who demonstrated that genomic island integrases are distinct from those encoded by phages, plasmids, integrons, and ICEs.
FIG 2.
Alternative integration/excision modules. (A) Structural comparison of the region surrounding the regulatory module of SXT from V. cholerae O139 MO10, ICEValA056-2 from V. alginolyticus A056, and ICEVchBan8 from V. cholerae O37 MZO-3. Maps of circularized ICEs are drawn to scale. Color keys are the same as for Fig. 1, except for gray and brown, indicating integration/excision, and black, indicating transposition. (B) Maximum likelihood phylogenetic analysis of 20 integrases targeting tRNA-Ser. The integrases of VPI-2 from V. cholerae N16961 (locus VC1758) and VPI-8-76 from V. cholerae O77 (locus DA89_2501) that target the same tRNA-Ser gene were used as the outgroup. Bootstrap supports, as percentages, are indicated at the branching points only when >80%. Branch length represents the number of substitutions per site over 406 amino acid positions. The genetic context of int genes in each taxon is represented on the right side of the tree (refer to panel A for color coding). An asterisk indicates the presence of a frameshift in int of ICEVmyCAIM528. Alignment of the attL and attR sites for each corresponding element is also indicated. Shaded sequences correspond to the 3′ end sequence of the serine tRNA gene. Nucleotides in bold show identity, while colors represent sequences internal to the elements, color coded by types. The region in which site-specific recombination likely takes place is indicated by a box.
Four types of SXT/R391 ICEs.
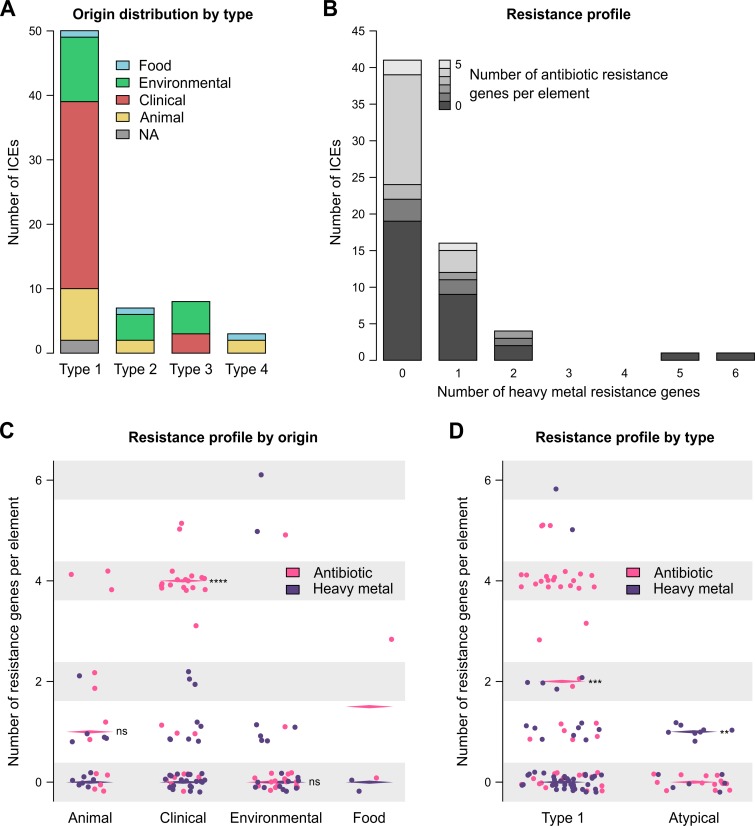
Phylogenetic analysis of the 18 atypical integrases suggests the existence of three types with two alternative ICE configurations (Fig. 2A and B). Therefore, SXT/R391 ICEs fall into four distinct types differing in insertion site, structural organization, prevalence, and distribution in bacterial species. Type 1 includes all ICEs inserted at the 5′ end of prfC, like SXT from V. cholerae O139 MO10. Type 1 ICEs are found in both Enterobacteriaceae and Vibrionaceae of clinical and environmental origin (Fig. 3A; Table 1). Type 1 ICEs of clinical origin are strongly associated with antibiotic resistance, significantly more than with heavy metal resistance (Fig. 3C and D). Interestingly, elements carrying multiple antibiotic resistance genes tend to carry fewer heavy metal resistance genes and vice versa (Fig. 3B). This observation likely reflects the selection of the most beneficial traits for a specific ecological niche. Thus, isolates from clinical settings have higher counts of antibiotic resistance genes (Fig. 3C), while isolates originating from wastewater or marine or freshwater sediments are more likely to be exposed to heavy metals such as zinc, cobalt, and cadmium.
FIG 3.
Distribution of ICEs and resistances. (A) Distribution of ICE types for each isolation niche. (B) Prevalence of heavy metal and antibiotic resistance genes. (C) Resistance profiles by niche of isolation. (D) Resistance profiles according to ICE types. Statistical analyses were performed using the Wilcoxon matched-pairs signed-rank test (two-tailed) to compare the medians of each resistance type for each isolation niche (animal, P = 0.1328; clinical, P < 0.0001; environmental, P = 0.2227) and according to the type (type 1, P = 0.0002; atypical, P = 0.0078).
Type 2, 3, and 4 ICEs are all inserted at the 3′ end of the tRNA-Ser gene and are found exclusively in isolates of Vibrio species that are mostly of environmental origin (Fig. 3A; Table 1). These ICEs do not carry antibiotic resistance genes but commonly encode a heavy metal efflux pump, which seems to be a distinctive trait of atypical ICEs (Fig. 3D). Type 2 ICEs, such as ICEValA056-2 from V. alginolyticus A056, retained the overall structure of SXT/R391 ICEs despite the alternative integration module. ICEVchBan8 from V. cholerae O37 MZO-3 provides an example of type 3 ICEs. In addition to the alternative integration module, type 3 ICEs hold another major rearrangement consisting in an inversion of the srpR-to-traG region (Fig. 2A). Currently, type 3 ICEs have been found only in V. cholerae, including environmental isolates recovered in the 1970s. Therefore, the emergence of type 3 ICEs in V. cholerae predates the spread of type 1 ICEs in toxigenic strains O1 and O139 of V. cholerae that began in the mid-1980s (4). Finally, three ICEs all found in Vibrio vulnificus belong to type 4. Although these ICEs retain the same structure as type 2, namely, lacking the large srpR-traG inversion, their integrases share the same common ancestor as type 3 ICEs. This suggests that types 3 and 4 would derive from a common ancestor sharing the same genetic structure as type 2 with a subsequent srpR-traG DNA inversion in the type 3 lineage. However, further analysis based on the soft-core genes suggests that V. vulnificus ICEs strongly diverge from other SXT/R391 ICEs (see below). Therefore, their type 4 integrase might be a more recent acquisition.
It is noteworthy that isolation biases exist in databases. For example, ICEs isolated in 2003 and 2005 were carried mostly by strains recovered from animals (see Fig. S1A in the supplemental material). Likewise, ICEs around the year 2000 belong mostly to type 1 (Fig. S1B). To better visualize how these elements cluster together based solely on the isolation perspective, a factorial analysis of mixed data (FAMD) revealed that ICEs found in clinical isolates belong mostly to type 1, whereas atypical ICEs are carried mostly by environmental strains (see Fig. S2 in the supplemental material).
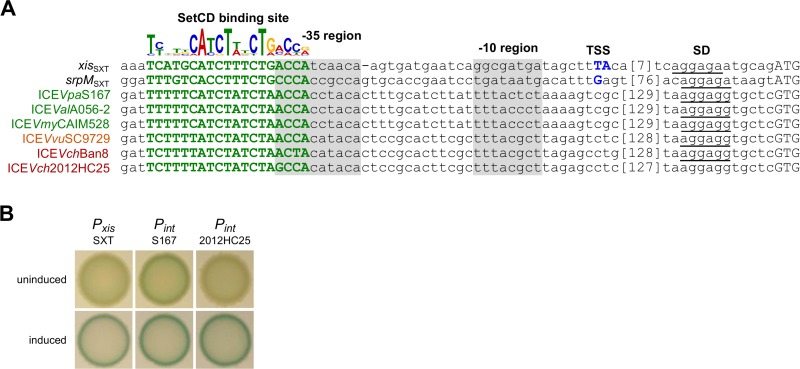
The int and xis genes of atypical ICEs remain under the control of the activator SetCD.
The site-specific recombination module of type 1 SXT/R391 ICEs consists of two convergent genes, int and xis, each preceded by the SetCD-dependent promoters Pxis and PsrpM, respectively (Fig. 2A). In atypical ICEs, int and xis likely form an operon under the control of a new promoter, Pint, that replaces Pxis. Bioinformatics analysis revealed the presence of a putative SetCD binding motif located 176 to 178 bp upstream of the GTG start codon of int (Fig. 4A). We introduced the promoter sequences Pint of type 2 ICEVpaS167 and type 3 ICEVch2012HC25 upstream of a promoterless lacZ reporter gene to test whether Pint is able to drive expression in a SetCD-dependent manner. β-Galactosidase assays confirmed that, like Pxis of type 1 ICEs, Pint of type 2 and 3 ICEs are constitutively off and activated by SetCD (Fig. 4B). We observed a 36-fold change in β-galactosidase activity (9.6 ± 6.3 arbitrary units [a.u.] versus 0.27 ± 0.25 a.u.) for type 2 Pint and a 51-fold change (15.4 ± 11.5 a.u. versus 0.30 ± 0.26 a.u.) for type 3 Pint upon induction of setCD expression with 0.2% arabinose.
FIG 4.
The integrase gene of atypical ICEs is under the control of SetCD. (A) Alignment of predicted SetCD-dependent promoters upstream of int in ICEs targeting the tRNA-Ser gene. The SetCD-binding logo and SetCD-dependent promoters of xis and srpM of SXT were characterized previously (17). SetCD boxes are shown in bold green capital letters. The position of known transcription start sites (TSS) is indicated in blue capital letters. Shine-Dalgarno sequences (SD) are underlined, while start codons are in capital letters. The approximate positions of the −35 and −10 regions are highlighted in gray. Numbers in brackets indicate the length in base pairs of spacers between the TSS region and the SD sequence. (B) The activity of two promoters representative of type 2 (ICEVch2012HC25) and 3 (ICEVpaS167) inttRNA-Ser genes was monitored from single-copy, chromosomally integrated lacZ transcriptional fusions in E. coli BW25113. Colorimetric assays of β-galactosidase activity were carried out on LB medium supplemented with or without arabinose to express setCD from PBAD on pGG2B (64).
Most atypical ICEs belong to the R entry exclusion group.
Entry exclusion is a mechanism by which DNA transport from the donor cell is blocked by the recipient cell to prevent redundant exchange of conjugative elements between cells containing elements belonging to the same exclusion group. For SXT/R391 ICEs, entry exclusion is mediated by two inner membrane proteins: Eex in the recipient and TraG in the donor. SXT/R391 ICEs have been shown to belong to either the S (SXT) or R (R391) entry exclusion group (24). ICEs of the S group exclude the entry of ICEs of the S group but not those of the R group and vice versa. ICEs that have been previously examined always code for matching pairs of Eex and TraG proteins belonging to the same exclusion group, i.e., EexS/TraGS or EexR/TraGR.
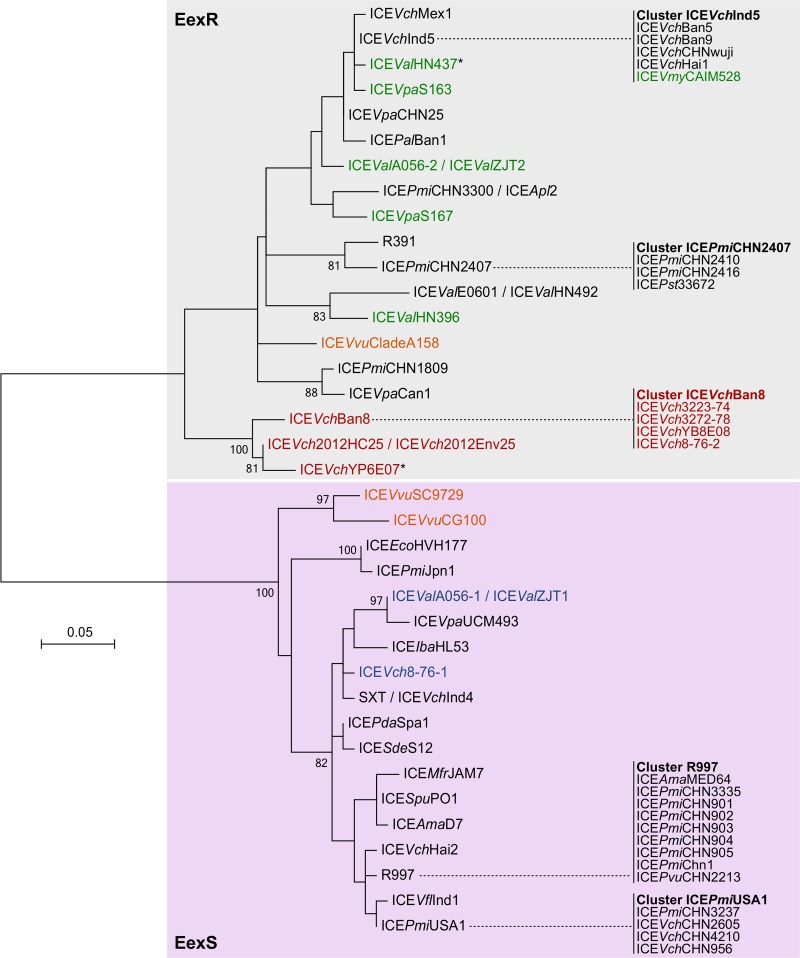
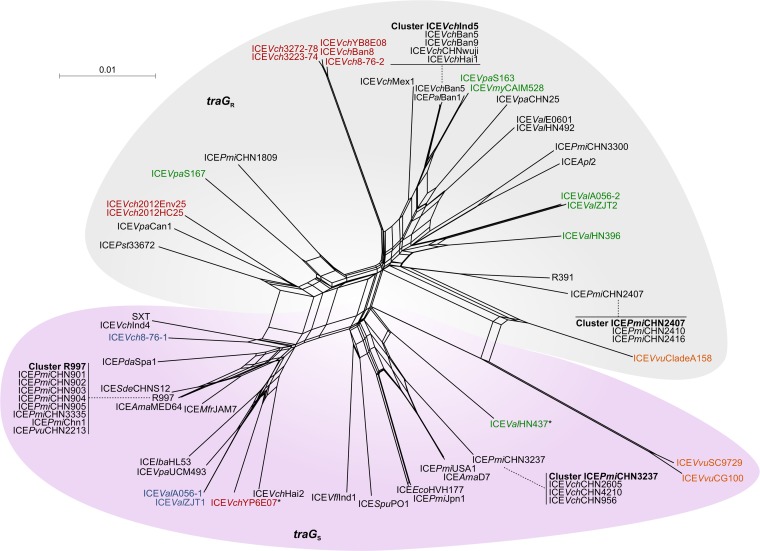
Phylogenetic analyses of the Eex and TraG proteins were carried out to determine the exclusion group of the 68 sampled ICEs. Eex proteins reliably clustered into the two main groups, S and R (Fig. 5). In contrast, TraG proteins did not provide a robust tree. While analysis of the conservation of an amino acid triplet at positions 606 to 608 allowed us to assign their exclusion group (PG[E/Q] for S, T[G/D]D for R) (24), TraG proteins of the same exclusion group did not cluster into consistent lineages (see Fig. S3 in the supplemental material). Considering that inter-ICE recombination events probably took place within traG and were blurring phylogenetic relationships, we built a phylogenetic network by using the DNA sequence of traG genes. This approach revealed a clear segregation between traG genes of the S and R groups (Fig. 6).
FIG 5.
Maximum likelihood phylogenetic analysis of 37 Eex orthologue proteins. Bootstrap supports, as percentages, are indicated at the branching points only when >80%. Branch length represents the number of substitutions per site over 143 amino acid positions. Taxa corresponding to type 1, 2, 3, and 4 ICEs are shown in black, green, dark red, and orange, respectively. Taxa highlighted in blue correspond to type 1 ICEs found in strains also bearing a coresident type 2 or 3 ICE. Asterisks indicate ICEs with an ambiguous entry exclusion group.
FIG 6.
NeigborNet phylogenetic network of 48 traG genes. Labels are color coded as described for Fig. 5. Asterisks indicate ICEs with an ambiguous entry exclusion group.
Except for type 4 ICEVvuSC9729 and ICEVvuCG100, atypical ICEs tended to cluster within the R exclusion group (Fig. 5 and 6). Interestingly, type 2 ICEVchYP6E07 and type 3 ICEValHN437 are the only ICEs of our sample set that could not be assigned to a specific entry exclusion group. Unlike any other SXT/R391 ICE, both encode an EexR entry exclusion protein and a TraGS subunit, thereby suggesting that they should exclude the entry of ICEs of the S group but not those of the R group or themselves. However, type 2 ICEVchYP6E07 and type 3 ICEValHN437 should be excluded by recipients containing an ICE of the R group.
Type 2 ICEs, which are found in multiple Vibrio species, are also uniformly distributed within the R exclusion group. In contrast, EexR proteins of type 3 ICEs from V. cholerae isolates recovered in the 1970s and 2000s exhibit little diversity and form a distinct lineage. The large traG-srpM inversion specific to the type 3 lineage might have isolated genetically these ICEs from other noninverted SXT/R391 ICEs by decreasing the opportunities of inter-ICE recombination events through loss of local synteny around eex (Fig. 2A). However, this isolation does not seem to occur for traG, as ICEVch2012Env25, ICEVch2012HC25 (traGR), and ICEVchYP6E07 (traGS) strongly diverge from other type 3 ICEs. These divergences also correlate with the presence or absence of a complete copy of an insertion sequence of the IS481 family (Fig. 2B, tnp) that is truncated or absent in the other type 3 ICEs. This observation suggests that the large traG-srpM inversion could have occurred more than once during the evolution of the type 3 lineage.
Overall, we found that exclusion groups are evenly distributed in the different ecological niches among which the ICE-containing isolates were found (Fig. S1C). Therefore, there is no association between a specific niche and a specific exclusion group.
Cohabitation of ICEs of distinct types in the same natural isolate.
Natural occurrences of isolates containing multiple tandem copies of SXT/R391 ICEs are strongly counterselected in the environment. Entry exclusion and RecA-dependent and -independent mechanisms prevent or resolve such occurrences (15, 16, 25). Homologous recombination plays a key role in destabilizing tandem arrays of SXT/R391 ICEs, reducing them to a singleton (25, 26). By doing so, it also considerably enhances their diversity by generating new recombinant ICEs (4, 10, 15). Nevertheless, a change of integration site through substitution of the int and xis genes seems to facilitate the cohabitation of a type 1 ICE with another type, likely by lowering the opportunities of homologous recombination. Indeed, two V. alginolyticus isolates and one V. cholerae isolate carry a combination of two ICEs: a type 1 ICE inserted at prfC, with either a type 2 or a type 3 ICE inserted at tRNA-Ser. In all instances, the type 1 ICE belongs to the S exclusion group whereas the second ICE belongs to the R exclusion group (Fig. 5 and 6).
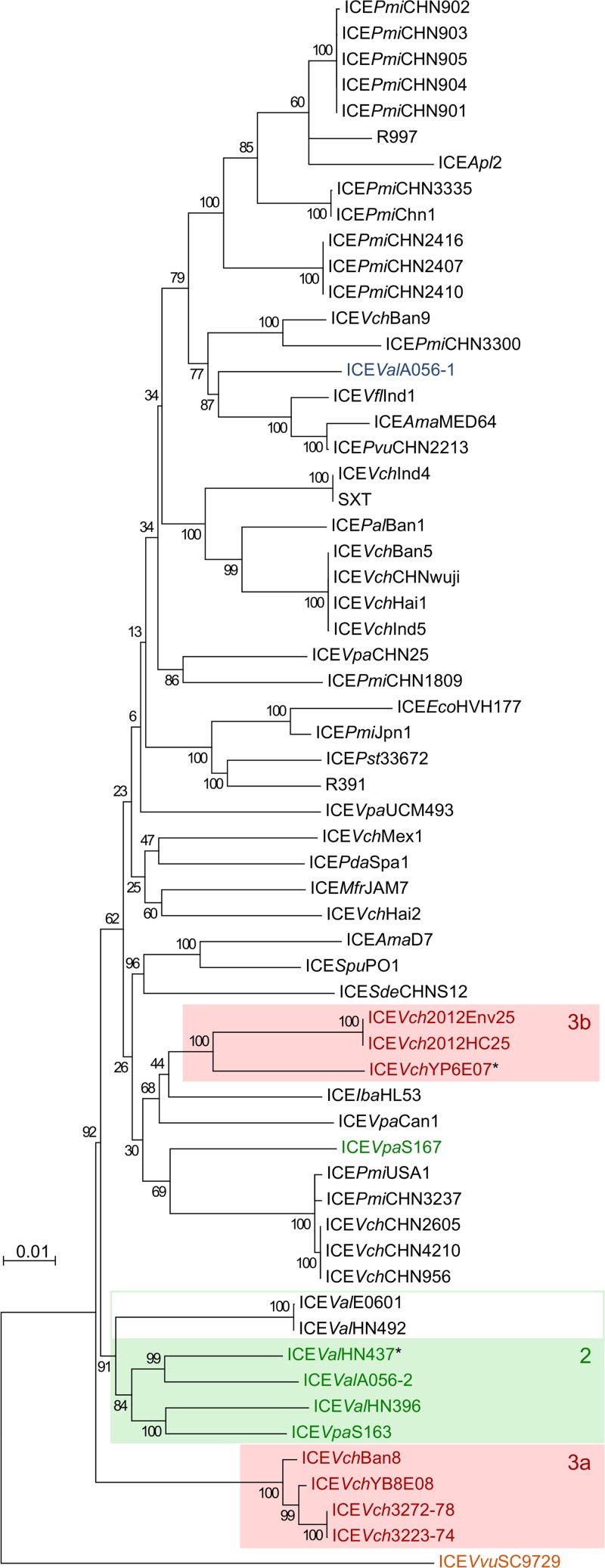
Atypical SXT/R391 ICEs form a polyphyletic group.
To better understand the relationship between typical and atypical SXT/R391 ICEs, we constructed a phylogenetic tree based on the alignment of the soft-core genes of 61 ICEs of our sample set (Fig. 7). We found that atypical ICEs do not cluster as a distinct lineage, thereby indicating that they form a polyphyletic group. Type 4 ICEs, represented by ICEVvuSC9729, seem to be the most divergent from all other types and constitute the outgroup of the tree. Furthermore, while a lineage of type 3 ICEs clearly diverges from all other ICEs (group 3a in Fig. 7), the other type 3 ICEs (group 3b in Fig. 7) are deeply rooted among type 1 ICEs. This observation is consistent with multiple occurrences of the large srpR-traG inversion. Except for ICEVpaS167, type 2 ICEs form a distinct cluster sharing a common ancestor with two type 1 ICEs found in V. alginolyticus. Altogether, these results indicate that atypical ICEs have undergone recombination events involving type 1 ICEs. Whether type 4 ICEVvuSC9729, the most distant lineage, is still capable of participating in such events remains to be determined.
FIG 7.
Maximum likelihood phylogenetic analysis of the soft core of 61 ICEs. Branch length represents the number of substitutions per site over 24,943 nucleotide positions. All gaps were removed prior to analysis. Labels are color coded as described for Fig. 5. Asterisks indicate ICEs with an ambiguous entry exclusion group.
Concluding remarks.
Based on the current definition of the SXT/R391 family, int has been routinely used as the initial and often sole marker for PCR screening of SXT/R391 ICEs in epidemiological surveys of multidrug-resistant environmental and clinical isolates of several pathogenic species (27–39). Our comparative genomics analyses revealed that the core set of genes conserved in SXT/R391 ICEs is smaller than previously described (17). Like entry exclusion, int, xis, and integration sites are variable features of this family. Nevertheless, we showed that the integration and excision functions remain under the control of the transcriptional activator SetCD that governs the expression of the conjugation genes. The lack of conservation of int makes it unfit for detection of SXT/R391 ICEs at large; however, int remains a suitable marker in epidemiological studies, as we showed that multidrug-resistant clinical isolates are associated mostly with type 1 ICEs. Nevertheless, we propose to researchers to renounce the systematic use of int as the first or sole marker for detection of SXT/R391 ICEs. Instead, setCD should be considered a more robust and specific marker given its key role and strict conservation. setCD will allow the comprehensive detection of all types of SXT/R391 ICEs in bacterial samples.
MATERIALS AND METHODS
Bacterial strains and media.
Bacterial strains were routinely grown in lysogeny broth (LB-Miller; EMD) at 37°C in an orbital shaker/incubator and were preserved at −80°C in LB broth containing 15% (vol/vol) glycerol. Antibiotics were used at the following concentrations: ampicillin (Ap), 50 μg/ml (Vibrio) and 100 μg/ml (Escherichia coli); nalidixic acid (Nx), 40 μg/ml (E. coli); and kanamycin (Kn), 10 μg/ml (E. coli).
Molecular biology.
lacZ reporter fusions were constructed by introducing the promoter sequences of int of ICEVpaS167 and ICEVch2012HC25 with primer pairs pOP-VpaS167F/pOP-VpaS167R and pOP-Vch12HCF/pOP-Vch12HCR, respectively (Table 2), into the PstI restriction site of pOPlacZ using the Q5 site-directed mutagenesis kit (New England BioLabs) according to the manufacturer's instructions. The resulting plasmids were confirmed by restriction profiling and DNA sequencing and then integrated in single copy into the chromosomal site attBλ of E. coli BW25113 using pINT-Ts (40, 41). Sequencing reactions (except PacBio sequencing) were performed by the Plateforme de Séquençage et de Génotypage du Centre de Recherche du CHUL (Québec, QC, Canada).
TABLE 2.
DNA sequences of the primers used in this study
| Primer name | Nucleotide sequence (5′ to 3′) |
|---|---|
| pOP-VpaS167F | TTTGCATCTTATTTTACTCTAAAAGTCGCACTGCAGATTCGACTCGAGCAGGA |
| pOP-VpaS167R | GTGTAGGTGGTTAGATAGATGAAAAAATCTAAGCTTGGCACTGGCCACGCA |
| pOP-Vch12HCF | TCCGCACTTCGCTTTACGCTTAGAGTCTCACTGCAGATTCGACTCGAGCAGGA |
| pOP-Vch12HCR | GTGTATGTGGCTAGATAGATAAAAGAATCTAAGCTTGGCACTGGCCACGCA |
| setDuF | ACAACCAATCARCAAGACTTCTATC |
| setCuR | ATTTAAGCTGAATGCGCTGTTG |
β-Galactosidase assays.
Qualitative assays on solid LB agar plate were done using 40 μg/ml 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) as the substrate with or without 0.02% arabinose. Plates were observed after overnight incubation at 37°C and storage at 4°C. Quantitative liquid assays using o-2-nitrophenyl-β-d-galactopyranoside (ONPG) as the substrate were done as previously described (42). Optical density at 420 nm (OD420) values were converted to log, and the slope of the linear regression was calculated for the early reaction. The slope was then normalized by the corresponding OD600 value and the reaction time.
ICE detection with setCD markers.
Primer pair setDuF and setCuR (Table 2) for setCD detection was designed using PrimerDesign-M (43) on a multiple-sequence alignment of setCD homologues. setCD loci were recovered from GenBank NT/NR and whole-genome shotgun contigs (WGS) databases restricted to Gammaproteobacteria (taxid:1236) using blastn with a 97% coverage threshold (44, 45). The resulting set of 521 sequences was aligned using the Muscle multiple-sequence alignment tool (46). A collection of 275 Canadian Vibrio strains (169 V. alginolyticus, 2 V. vulnificus, and 104 V. parahaemolyticus strains) were screened using the primers setDuF and setCuR to detect potential ICEs regulated by setCD. PCRs were carried out using the EasyTaq polymerase (Civic Bioscience) under the following reaction conditions: (i) 5 min at 94°C; (ii) 30 cycles of 30 s at 94°C, 30 s at the appropriate annealing temperature, and 30 s/kb at 72°C; and (iii) 1 min at 72°C.
PacBio sequencing.
Whole-genome sequencing of V. parahaemolyticus S107 containing ICEVpaCan1 was carried out from genomic DNA extracted from 2 ml of exponential-phase culture using the Gentra Puregene kit (Qiagen). PacBio RS II single-molecule real-time sequencing (Pacbio SMRTcell) and de novo genome assembly were performed at the McGill University and Génome Québec Innovation Centre.
ICE data set and comparative analysis.
ICE data sets were obtained by using the NCBI's blastn algorithm against several databases (NT/NR, WGS, Refseq genomic) on different integrase types and three other key sets of genes (croS-setR, setCD, and sprR-sprM). When complete ICE sequences were not available, the best blast hits found across contigs were extracted from NCBI's sequence set browser and were aligned against the adequate ICE type reference sequence using MUMmer3 (47). The following putative ICE draft sequences were then manually assembled and curated: ICEVch2012Env25, ICEVch2012HC25, ICEVch3223-74, ICEVch3272-78, ICEVchYB8E08, ICEVpaS163, ICEVpaS167, and ICEVvuSC9729. Whole ICE sequences were scanned to detect antibiotic and heavy metal resistances using CARD (48). The sequences of 61 assembled ICEs were submitted to the “Get_Homologues” software package (49) (80% minimum coverage in blastn pairwise alignments) to obtain homology information and isolate the soft-core genome (95% conservation). The 43 isolated soft-core genes of each ICE were extracted, reorganized to be syntenic, and concatenated using in-house scripts into 61 streamlined ICE sequences prior to their alignments using Clustal Omega (50).
Evolutionary analyses were conducted in MEGA7 (51) and PhyML (52) and inferred by using the maximum likelihood method based on the LG (InttRNA-Ser) or JTT (Eex or TraG proteins) matrix-based models (53, 54). Protein sequences were aligned with Muscle (46). Eex and TraG primary sequences were first clustered with CD-HIT (55) (sequence identity cutoff, 1) prior to alignment. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using a JTT model (Eex or TraG proteins) and then selecting the topology with the superior log likelihood value. A discrete gamma distribution was used to model evolutionary rate differences among sites (5 categories) for Eex or TraG proteins. A NeighborNet phylogenetic network was built for traG using SplitsTree4 (56) with default parameters (Uncorrected_P method for distances and EqualAngle drawing method) after clustering of the nucleotide sequences of traG genes recovered from the ICE sample set using CD-HIT-est (55) (sequence identity cutoff, 1). The 48 unique DNA sequences were aligned with Muscle prior to phylogenetic analysis. Phylogenetic analysis of the 61 streamlined ICEs was inferred by using the maximum likelihood method based on the general time-reversible (GTR) model (57). A discrete gamma distribution was used to model evolutionary rate differences among sites (5 categories). The rate variation model allowed for some sites to be evolutionarily invariable ([+I], 39.0911% sites). Versions Pfam 31.0, Uniprot 2017_11, and HMMER v3.1b2 were used.
Statistical analyses.
A factorial analysis of mixed data was performed on all 68 ICEs using 8 variables (origin, type, exclusion group, strain species, WHO region of isolation, year of isolation, number of antibiotic resistance genes, and number of heavy metal resistance genes). Data were imported in R version 3.4.3 (58). Missing values were handled with the missMDA package (59). The analysis itself was performed using FactoMineR package (60).
Accession number(s).
The whole-genome sequence of V. parahaemolyticus S107 was deposited in GenBank under accession numbers CP028481 and CP028482.
Supplementary Material
ACKNOWLEDGMENTS
We thank Kévin Huguet for technical assistance and Swapan Banerjee (Health Canada) for access to his collection of Canadian Vibrio isolates. We are grateful to Fanie Pelletier for her help with statistical analyses and Alain Lavigueur for critical reading of the manuscript.
Computations were made on the supercomputer Mp2 from Université de Sherbrooke, managed by Calcul Québec and Compute Canada. The operation of this supercomputer is funded by the Canada Foundation for Innovation (CFI), the ministère de l'Économie, de la science et de l'innovation du Québec (MESI) and the Fonds de recherche du Québec—Nature et technologies (FRQ-NT).
This work was supported by a Discovery Grant (2016-04365) from the Natural Sciences and Engineering Council of Canada (NSERC) and a Project Grant (PJT-153071) from the Canadian Institutes of Health Research (CIHR) to V.B.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00485-18.
REFERENCES
- 1.Ghosh A, Ramamurthy T. 2011. Antimicrobials & cholera: are we stranded. Indian J Med Res 133:225–231. [PMC free article] [PubMed] [Google Scholar]
- 2.Kitaoka M, Miyata ST, Unterweger D, Pukatzki S. 2011. Antibiotic resistance mechanisms of Vibrio cholerae. J Med Microbiol 60:397–407. doi: 10.1099/jmm.0.023051-0. [DOI] [PubMed] [Google Scholar]
- 3.Burrus V, Marrero J, Waldor MK. 2006. The current ICE age: biology and evolution of SXT-related integrating conjugative elements. Plasmid 55:173–183. doi: 10.1016/j.plasmid.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 4.Spagnoletti M, Ceccarelli D, Rieux A, Fondi M, Taviani E, Fani R, Colombo MM, Colwell RR, Balloux F. 2014. Acquisition and evolution of SXT-R391 integrative conjugative elements in the seventh-pandemic Vibrio cholerae lineage. mBio 5:e01356-14. doi: 10.1128/mBio.01356-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beaber JW, Hochhut B, Waldor MK. 2004. SOS response promotes horizontal dissemination of antibiotic resistance genes. Nature 427:72. doi: 10.1038/nature02241. [DOI] [PubMed] [Google Scholar]
- 6.Dalia AB, Seed KD, Calderwood SB, Camilli A. 2015. A globally distributed mobile genetic element inhibits natural transformation of Vibrio cholerae. Proc Natl Acad Sci U S A 112:10485–10490. doi: 10.1073/pnas.1509097112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ceccarelli D, Spagnoletti M, Bacciu D, Danin-Poleg Y, Mendiratta DK, Kashi Y, Cappuccinelli P, Burrus V, Colombo MM. 2011. ICEVchInd5 is prevalent in epidemic Vibrio cholerae O1 El Tor strains isolated in India. Int J Med Microbiol 301:318–324. doi: 10.1016/j.ijmm.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 8.Waldor MK, Tschäpe H, Mekalanos JJ. 1996. A new type of conjugative transposon encodes resistance to sulfamethoxazole, trimethoprim, and streptomycin in Vibrio cholerae O139. J Bacteriol 178:4157–4165. doi: 10.1128/jb.178.14.4157-4165.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Y, Li Y, Fernandez Crespo R, Leanse LG, Langford PR, Bossé JT. 2018. Characterization of the Actinobacillus pleuropneumoniae SXT-related integrative and conjugative element ICEApl2 and analysis of the encoded FloR protein: hydrophobic residues in transmembrane domains contribute dynamically to florfenicol and chloramphenicol efflux. J Antimicrob Chemother 73:57–65. doi: 10.1093/jac/dkx342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wozniak RAF, Fouts DE, Spagnoletti M, Colombo MM, Ceccarelli D, Garriss G, Déry C, Burrus V, Waldor MK. 2009. Comparative ICE genomics: insights into the evolution of the SXT/R391 family of ICEs. PLoS Genet 5:e1000786. doi: 10.1371/journal.pgen.1000786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burrus V, Waldor MK. 2003. Control of SXT integration and excision. J Bacteriol 185:5045–5054. doi: 10.1128/JB.185.17.5045-5054.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hochhut B, Waldor MK. 1999. Site-specific integration of the conjugal Vibrio cholerae SXT element into prfC. Mol Microbiol 32:99–110. doi: 10.1046/j.1365-2958.1999.01330.x. [DOI] [PubMed] [Google Scholar]
- 13.Carraro N, Poulin D, Burrus V. 2015. Replication and active partition of integrative and conjugative elements (ICEs) of the SXT/R391 family: the line between ICEs and conjugative plasmids is getting thinner. PLoS Genet 11:e1005298. doi: 10.1371/journal.pgen.1005298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ceccarelli D, Daccord A, René M, Burrus V. 2008. Identification of the origin of transfer (oriT) and a new gene required for mobilization of the SXT/R391 family of integrating conjugative elements. J Bacteriol 190:5328–5338. doi: 10.1128/JB.00150-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garriss G, Waldor MK, Burrus V. 2009. Mobile antibiotic resistance encoding elements promote their own diversity. PLoS Genet 5:e1000775. doi: 10.1371/journal.pgen.1000775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marrero J, Waldor MK. 2005. Interactions between inner membrane proteins in donor and recipient cells limit conjugal DNA transfer. Dev Cell 8:963–970. doi: 10.1016/j.devcel.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 17.Poulin-Laprade D, Matteau D, Jacques P-É, Rodrigue S, Burrus V. 2015. Transfer activation of SXT/R391 integrative and conjugative elements: unraveling the SetCD regulon. Nucleic Acids Res 43:2045–2056. doi: 10.1093/nar/gkv071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poulin-Laprade D, Burrus V. 2015. A λ Cro-like repressor is essential for the induction of conjugative transfer of SXT/R391 elements in response to DNA damage. J Bacteriol 197:3822–3833. doi: 10.1128/JB.00638-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taviani E, Spagnoletti M, Ceccarelli D, Haley BJ, Hasan NA, Chen A, Colombo MM, Huq A, Colwell RR. 2012. Genomic analysis of ICEVchBan8: an atypical genetic element in Vibrio cholerae. FEBS Lett 586:1617–1621. doi: 10.1016/j.febslet.2012.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luo P, He X, Wang Y, Liu Q, Hu C. 2016. Comparative genomic analysis of six new-found integrative conjugative elements (ICEs) in Vibrio alginolyticus. BMC Microbiol 16:79. doi: 10.1186/s12866-016-0692-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jermyn WS, Boyd EF. 2002. Characterization of a novel Vibrio pathogenicity island (VPI-2) encoding neuraminidase (nanH) among toxigenic Vibrio cholerae isolates. Microbiology 148:3681–3693. doi: 10.1099/00221287-148-11-3681. [DOI] [PubMed] [Google Scholar]
- 22.Carpenter MR, Rozovsky S, Boyd EF. 2015. Pathogenicity island cross talk mediated by recombination directionality factors facilitates excision from the chromosome. J Bacteriol 198:766–776. doi: 10.1128/JB.00704-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boyd EF, Almagro-Moreno S, Parent MA. 2009. Genomic islands are dynamic, ancient integrative elements in bacterial evolution. Trends Microbiol 17:47–53. doi: 10.1016/j.tim.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 24.Marrero J, Waldor MK. 2007. The SXT/R391 family of integrative conjugative elements is composed of two exclusion groups. J Bacteriol 189:3302–3305. doi: 10.1128/JB.01902-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burrus V, Waldor MK. 2004. Formation of SXT tandem arrays and SXT-R391 hybrids. J Bacteriol 186:2636–2645. doi: 10.1128/JB.186.9.2636-2645.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pembroke JT, Murphy DB. 2000. Isolation and analysis of a circular form of the IncJ conjugative transposon-like elements, R391 and R997: implications for IncJ incompatibility. FEMS Microbiol Lett 187:133–138. doi: 10.1111/j.1574-6968.2000.tb09149.x. [DOI] [PubMed] [Google Scholar]
- 27.Mohapatra H, Mohapatra SS, Mantri CK, Colwell RR, Singh DV. 2008. Vibrio cholerae non-O1, non-O139 strains isolated before 1992 from Varanasi, India are multiple drug resistant, contain intSXT, dfr18 and aadA5 genes. Environ Microbiol 10:866–873. doi: 10.1111/j.1462-2920.2007.01502.x. [DOI] [PubMed] [Google Scholar]
- 28.Taviani E, Ceccarelli D, Lazaro N, Bani S, Cappuccinelli P, Colwell RR, Colombo MM. 2008. Environmental Vibrio spp., isolated in Mozambique, contain a polymorphic group of integrative conjugative elements and class 1 integrons. FEMS Microbiol Ecol 64:45–54. doi: 10.1111/j.1574-6941.2008.00455.x. [DOI] [PubMed] [Google Scholar]
- 29.Ceccarelli D, Bani S, Cappuccinelli P, Colombo MM. 2006. Prevalence of aadA1 and dfrA15 class 1 integron cassettes and SXT circulation in Vibrio cholerae O1 isolates from Africa. J Antimicrob Chemother 58:1095–1097. doi: 10.1093/jac/dkl352. [DOI] [PubMed] [Google Scholar]
- 30.Harada S, Ishii Y, Saga T, Tateda K, Yamaguchi K. 2010. Chromosomally encoded blaCMY-2 located on a novel SXT/R391-related integrating conjugative element in a Proteus mirabilis clinical isolate. Antimicrob Agents Chemother 54:3545–3550. doi: 10.1128/AAC.00111-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodríguez-Blanco A, Lemos ML, Osorio CR. 2016. Unveiling the pan-genome of the SXT/R391 family of ICEs: molecular characterisation of new variable regions of SXT/R391-like ICEs detected in Pseudoalteromonas sp. and Vibrio scophthalmi. Antonie Van Leeuwenhoek 109:1141–1152. doi: 10.1007/s10482-016-0716-3. [DOI] [PubMed] [Google Scholar]
- 32.Lei C-W, Zhang A-Y, Wang H-N, Liu B-H, Yang L-Q, Yang Y-Q. 2016. Characterization of SXT/R391 integrative and conjugative elements in Proteus mirabilis isolates from food-producing animals in China. Antimicrob Agents Chemother 60:1935–1938. doi: 10.1128/AAC.02852-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Balado M, Lemos ML, Osorio CR. 2013. Integrating conjugative elements of the SXT/R391 family from fish-isolated Vibrios encode restriction-modification systems that confer resistance to bacteriophages. FEMS Microbiol Ecol 83:457–467. doi: 10.1111/1574-6941.12007. [DOI] [PubMed] [Google Scholar]
- 34.Badhai J, Kumari P, Krishnan P, Ramamurthy T, Das SK. 2013. Presence of SXT integrating conjugative element in marine bacteria isolated from the mucus of the coral Fungia echinata from Andaman Sea. FEMS Microbiol Lett 338:118–123. doi: 10.1111/1574-6968.12033. [DOI] [PubMed] [Google Scholar]
- 35.Rodríguez-Blanco A, Lemos ML, Osorio CR. 2012. Integrating conjugative elements as vectors of antibiotic, mercury, and quaternary ammonium compound resistance in marine aquaculture environments. Antimicrob Agents Chemother 56:2619–2626. doi: 10.1128/AAC.05997-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li X, Du Y, Du P, Dai H, Fang Y, Li Z, Lv N, Zhu B, Kan B, Wang D. 2016. SXT/R391 integrative and conjugative elements in Proteus species reveal abundant genetic diversity and multidrug resistance. Sci Rep 6:37372. doi: 10.1038/srep37372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang R, Yu D, Yue J, Kan B. 2016. Variations in SXT elements in epidemic Vibrio cholerae O1 El Tor strains in China. Sci Rep 6:22733. doi: 10.1038/srep22733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Song Y, Yu P, Li B, Pan Y, Zhang X, Cong J, Zhao Y, Wang H, Chen L. 2013. The mosaic accessory gene structures of the SXT/R391-like integrative and conjugative elements derived from Vibrio spp. isolated from aquatic products and environment in the Yangtze River estuary, China. BMC Microbiol 13:214. doi: 10.1186/1471-2180-13-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mantri CK, Mohapatra SS, Ramamurthy T, Ghosh R, Colwell RR, Singh DV. 2006. Septaplex PCR assay for rapid identification of Vibrio cholerae including detection of virulence and int SXT genes. FEMS Microbiol Lett 265:208–214. doi: 10.1111/j.1574-6968.2006.00491.x. [DOI] [PubMed] [Google Scholar]
- 40.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haldimann A, Wanner BL. 2001. Conditional-replication, integration, excision, and retrieval plasmid-host systems for gene structure-function studies of bacteria. J Bacteriol 183:6384–6393. doi: 10.1128/JB.183.21.6384-6393.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carraro N, Durand R, Rivard N, Anquetil C, Barrette C, Humbert M, Burrus V. 2017. Salmonella genomic island 1 (SGI1) reshapes the mating apparatus of IncC conjugative plasmids to promote self-propagation. PLoS Genet 13:e1006705. doi: 10.1371/journal.pgen.1006705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoon H, Leitner T. 2015. PrimerDesign-M: a multiple-alignment based multiple-primer design tool for walking across variable genomes. Bioinformatics 31:1472–1474. doi: 10.1093/bioinformatics/btu832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boratyn GM, Camacho C, Cooper PS, Coulouris G, Fong A, Ma N, Madden TL, Matten WT, McGinnis SD, Merezhuk Y, Raytselis Y, Sayers EW, Tao T, Ye J, Zaretskaya I. 2013. BLAST: a more efficient report with usability improvements. Nucleic Acids Res 41:W29–W33. doi: 10.1093/nar/gkt282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.NCBI Resource Collaborators. 2013. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 41:D8–D20. doi: 10.1093/nar/gks1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL. 2004. Versatile and open software for comparing large genomes. Genome Biol 5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jia B, Raphenya AR, Alcock B, Waglechner N, Guo P, Tsang KK, Lago BA, Dave BM, Pereira S, Sharma AN, Doshi S, Courtot M, Lo R, Williams LE, Frye JG, Elsayegh T, Sardar D, Westman EL, Pawlowski AC, Johnson TA, Brinkman FSL, Wright GD, McArthur AG. 2017. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res 45:D566–D573. doi: 10.1093/nar/gkw1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Contreras-Moreira B, Vinuesa P. 2013. GET_HOMOLOGUES, a versatile software package for scalable and robust microbial pangenome analysis. Appl Environ Microbiol 79:7696–7701. doi: 10.1128/AEM.02411-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sievers F, Higgins DG. 2017. Clustal Omega for making accurate alignments of many protein sequences. Protein Sci Publ Protein Soc 27:135–145. doi: 10.1002/pro.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 53.Jones DT, Taylor WR, Thornton JM. 1992. The rapid generation of mutation data matrices from protein sequences. Bioinformatics 8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 54.Le SQ, Gascuel O. 2008. An improved general amino acid replacement matrix. Mol Biol Evol 25:1307–1320. doi: 10.1093/molbev/msn067. [DOI] [PubMed] [Google Scholar]
- 55.Huang Y, Niu B, Gao Y, Fu L, Li W. 2010. CD-HIT Suite: a web server for clustering and comparing biological sequences. Bioinforma Oxf Engl 26:680–682. doi: 10.1093/bioinformatics/btq003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huson DH, Bryant D. 2006. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol 23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- 57.Nei M, Kumar S. 2000. Molecular evolution and phylogenetics. Oxford University Press, Oxford, England. [Google Scholar]
- 58.R Core Team. 2016. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 59.Josse J, Husson F. 2016. missMDA: a package for handling missing values in multivariate data analysis. J Stat Softw 70:1–31. doi: 10.18637/jss.v070.i01. [DOI] [Google Scholar]
- 60.Lê S, Josse J, Husson F. 2008. FactoMineR: an R package for multivariate analysis. J Stat Softw 25:1–18. doi: 10.18637/jss.v025.i01. [DOI] [Google Scholar]
- 61.Ryan MP, Armshaw P, O'Halloran JA, Pembroke JT. 2017. Analysis and comparative genomics of R997, the first SXT/R391 integrative and conjugative element (ICE) of the Indian sub-continent. Sci Rep 7:8562. doi: 10.1038/s41598-017-08735-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Böltner D, MacMahon C, Pembroke JT, Strike P, Osborn AM. 2002. R391: a conjugative integrating mosaic comprised of phage, plasmid, and transposon elements. J Bacteriol 184:5158–5169. doi: 10.1128/JB.184.18.5158-5169.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ceccarelli D, Spagnoletti M, Hasan NA, Lansing S, Huq A, Colwell RR. 2013. A new integrative conjugative element detected in Haitian isolates of Vibrio cholerae non-O1/non-O139. Res Microbiol 164:891–893. doi: 10.1016/j.resmic.2013.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Garriss G, Poulin-Laprade D, Burrus V. 2013. DNA-damaging agents induce the RecA-independent homologous recombination functions of integrating conjugative elements of the SXT/R391 family. J Bacteriol 195:1991–2003. doi: 10.1128/JB.02090-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bordeleau E, Brouillette E, Robichaud N, Burrus V. 2010. Beyond antibiotic resistance: integrating conjugative elements of the SXT/R391 family that encode novel diguanylate cyclases participate to c-di-GMP signalling in Vibrio cholerae. Environ Microbiol 12:510–523. doi: 10.1111/j.1462-2920.2009.02094.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.