ABSTRACT
Protein haze formation in bottled wines is a significant concern for the global wine industry, and wine clarification before bottling is therefore a common but expensive practice. Previous studies have shown that wine yeast strains can reduce haze formation through the secretion of certain mannoproteins, but it has been suggested that other yeast-dependent haze protective mechanisms exist. On the other hand, the addition of chitin has been shown to reduce haze formation, likely because grape chitinases have been shown to be the major contributors to haze. In this study, Chardonnay grape must fermented by various yeast strains resulted in wines with different protein haze levels, indicating differences in haze-protective capacities of the strains. The cell wall chitin levels of these strains were determined, and a strong correlation between cell wall chitin levels and haze protection capability was observed. To further evaluate the mechanism of haze protection, Escherichia coli-produced green fluorescent protein (GFP)-tagged grape chitinase was shown to bind efficiently to yeast cell walls in a cell wall chitin concentration-dependent manner, while commercial chitinase was removed from synthetic wine in quantities that also correlated with the cell wall chitin levels of the strains. Our findings suggest a new mechanism of reducing wine haze, and we propose a strategy for optimizing wine yeast strains to improve wine clarification.
IMPORTANCE In this study, we establish a new mechanism by which wine yeast strains can impact the protein haze formation of wines, and we demonstrate that yeast cell wall chitin binds grape chitinase in a chitin concentration-dependent manner. We also show that yeast can remove this haze-forming protein from wine. Chitin has in the past been shown to efficiently reduce wine haze formation when added to the wine in high concentration as a clarifying agent. Our data suggest that the selection of yeast strains with high levels of cell wall chitin can reduce protein haze. We also investigate how yeast cell wall chitin levels are affected by environmental conditions.
KEYWORDS: chitin, haze protection, wine protein haze, wine yeast strains
INTRODUCTION
Bottled white wines can develop haze due to the precipitation of grape pathogenesis-related (PR) proteins that “survive” the fermentation process due to their stability under high ethanol conditions, low pH levels, and the proteolytic activities of grape and yeast proteases. While some contradictory findings have been made in the past regarding the exact causes of wine protein haze formation (1–3), several recent publications convincingly demonstrate that grape chitinases, and to a lesser degree thaumatin-like proteins, are the major contributors to the phenomenon (4–7). Other proteins, such as β-1,3-glucanases and the ripening-related protein Grip22 (5), have also been identified in wine haze particles but appear not to be causally responsible for its formation, or to be minor contributors at most (8).
The evidence for chitinases as a primary causative agent of protein wine haze is based on thermal unfolding studies of grape thaumatin-like protein and chitinase and on using differential scanning calorimetry. Falconer et al. (6) demonstrated that grape chitinases were the major players in heat-induced haze in unfined wines, as they have a low melting temperature. Studies by Marangon et al. (3) in simple model solutions further suggested that chitinases have a half-life in wine of 6 min at 55°C, thus extrapolating down to a half-life of 3 days at 35°C or 2 years at 25°C compared to the thaumatin-like protein, which had a melting temperature (Tm) of 62°C, with a calculated half-life of 300 years at 25°C. Moreover, the last stage of chitinase aggregation was demonstrated to be irreversible and strongly associated with visible haze compared to invertase and thaumatin-like protein. A linear correlation was also found to exist between chitinase content in wine and haze intensity (7, 9). Moreover, Palmisano et al. (10) observed that the glycoproteins identified in wine haze had hydrolase activity (38%) and chitinase activity (13%).
Several data sets have demonstrated that specific mannoproteins secreted or passively released by yeast during or after fermentation can reduce the formation of haze (10–13). Such proteins include the haze protection factors Hpf1 and Hpf2, as well as invertase and other extracellular enzymes. However, the quantities of mannoproteins released by current wine strains during wine making have been shown to be too low to have a significant impact on the technological and organoleptic properties of wine. Consequently, several strategies to increase mannoprotein production by wine yeast strains have been investigated (11–13).
Furthermore, several reports also suggest that factors other than mannoproteins may contribute to haze reduction by yeast. Indeed, the deletion of the haze protection factor genes, such as HPF1, HPF2, and HPF1′, results in strains with a barely reduced ability to impact wine haze formation (11). In addition, studies on wine aging have shown that a significant reduction in protein haze formation is observed in the presence of yeast lees, without noticeable increases in mannan levels (14, 15) or only a slight increase of up to 30 mg/liter in mannoproteins (15, 16). Dupin et al. (16) demonstrated that in order to reduce wine haze by 40%, 400 mg/liter of invertase would be required. This concentration is far beyond what is found in wine, even when aged on lees, thus demonstrating that the observed small increase in mannoproteins during aging may not be the sole cause for the observed haze reduction. Finally, Gonzalez-Ramos and Gonzalez (12) and Gonzalez-Ramos et al. (13) also observed no direct correlation between the amount of mannan polysaccharides released and the level of stabilization obtained, strongly suggesting the need to explore other possible protein haze-stabilizing factors. In line with this concept, Vincenzi et al. (17) observed that the addition of 20 g/liter chitin to unfined wine reduced wine haze by up to 80% of the total haze, while the addition of 1 g/liter chitin reduced wine haze by almost 50% with respect to the unfined wine. This haze reduction was directly connected to the removal of the class IV grape chitinase, which is a major contributor to wine haze.
In this study, we investigated the possibility that yeast cell wall chitin is involved in haze reduction through the binding of grape chitinases to the yeast cell wall. Yeast cell wall binding assays of grape chitinases and commercial chitinases were carried out. In addition, the cell walls of Saccharomyces cerevisiae BY4742 Δace2, BY4742 Δtus1, BY4742 Δygp1, and BY4742 Δirc8 mutants were observed to contain higher levels of chitin than those of the wild type, and the abilities of these strains to eliminate chitinases from solution were also evaluated concurrently with the commonly used S. cerevisiae wine yeast strains. Chitin evolution in yeast strains with responses to different environmental parameters was further explored in this study. The expression levels of genes involved in chitin biogenesis were evaluated under conditions that resulted in high chitin levels, such as exposure to elevated temperature and calcium addition into the growth media. Our findings indeed suggest a novel strategy not only for reducing wine haze employing yeast strains with higher cell wall chitin levels but also a strategy for producing wine yeast strains with high chitin levels for wine clarification purposes.
RESULTS
Protein stability.
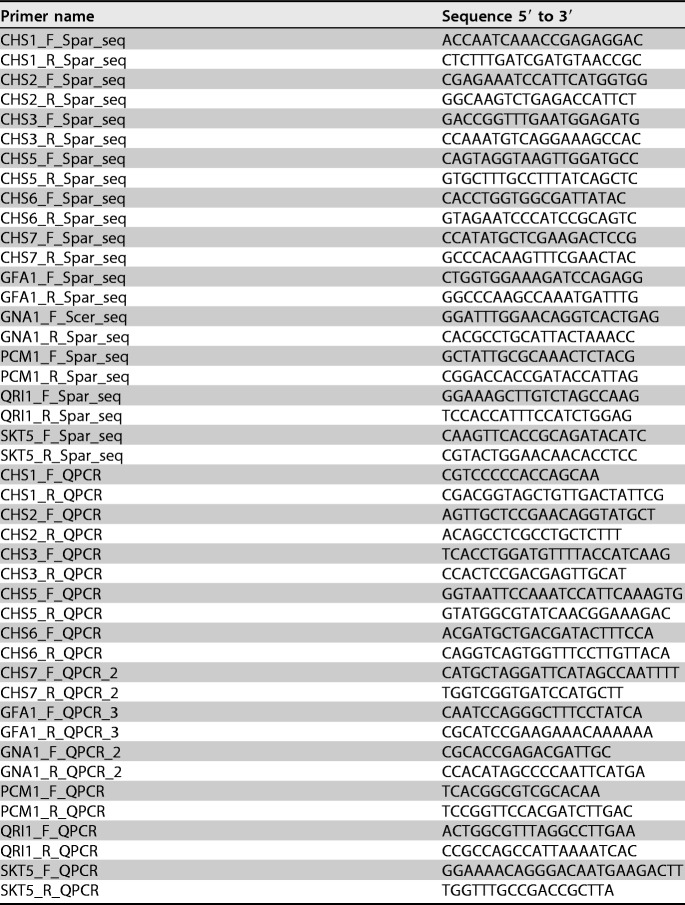
Heat tests were carried out in Chardonnay fermented grape must fermented to dryness using various wine yeast strains. Significant differences (P < 0.05) were observed in protein haze formed between the strains, with RO88, P01-167, and P01-146 showing strong haze-protective activities (Fig. 1). Similar differences were also observed between yeast strains when the experiment was repeated in Sauvignon Blanc grape must (data not shown).
FIG 1.
Wine haze levels in fermented Chardonnay must using S. cerevisiae and S. paradoxus wine yeast strains. Differences in haze levels (mean difference in absorbance before and after heating ± standard deviation of triplicate measurements) between S. cerevisiae, S. cerevisiae-S. paradoxus hybrid, and S. paradoxus yeast strains formed in fermented Chardonnay grape must juice at the end of fermentation are indicated.
Chitin levels of yeast strains.
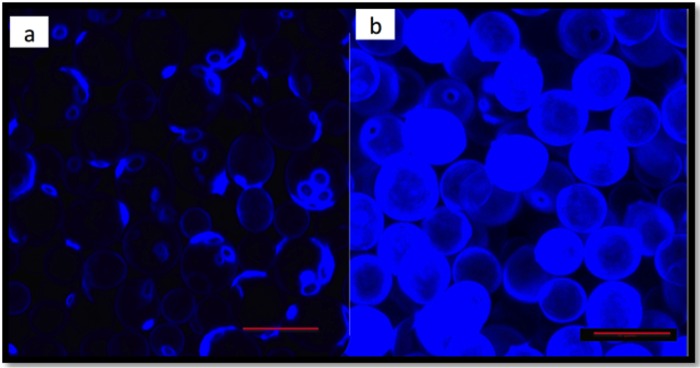
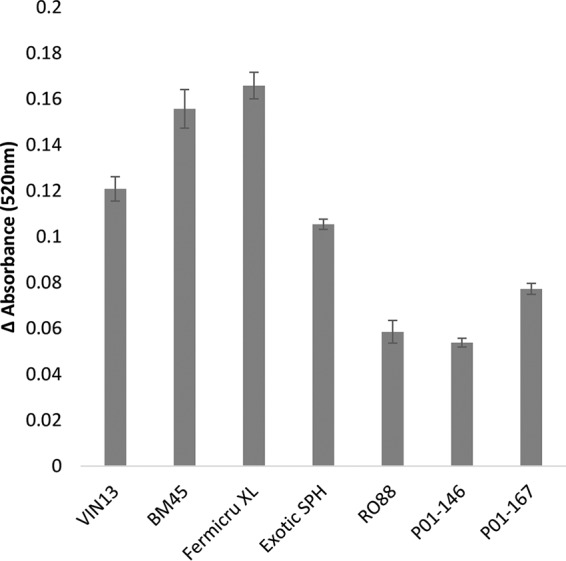
To assess the differences in cell wall chitin levels between the various yeast strains, cells grown under fermentative conditions were stained with calcofluor white. A visual inspection under the confocal fluorescence microscope suggested higher levels of fluorescence in yeast strains belonging to the Saccharomyces paradoxus species than in S. cerevisiae cells (Fig. 2). To confirm this observation, flow cytometry was used to quantify the chitin levels, and Fig. 3a shows the differences in chitin levels between various yeast strains measured using flow cytometry. RO88, P01-146, and P02-208 had significantly higher (P < 0.05) chitin levels than the S. cerevisiae wine strains used in the study. Figure 3b shows the correlation between the chitin levels and haze formation. A negative Pearson's r value of −0.832 (P < 0.05) was obtained, indicating that the higher the chitin levels are, the lower the protein haze level that was observed.
FIG 2.
S. cerevisiae (BM45) (a) and S. paradoxus (P02-208) (b) cells stained with calcofluor white stain. Cells were grown in YPD, as described by de Groot et al. (43), and washed in PBS buffer before staining and viewing under a Zeiss LSM 780 Elyra S1 confocal microscope.
FIG 3.
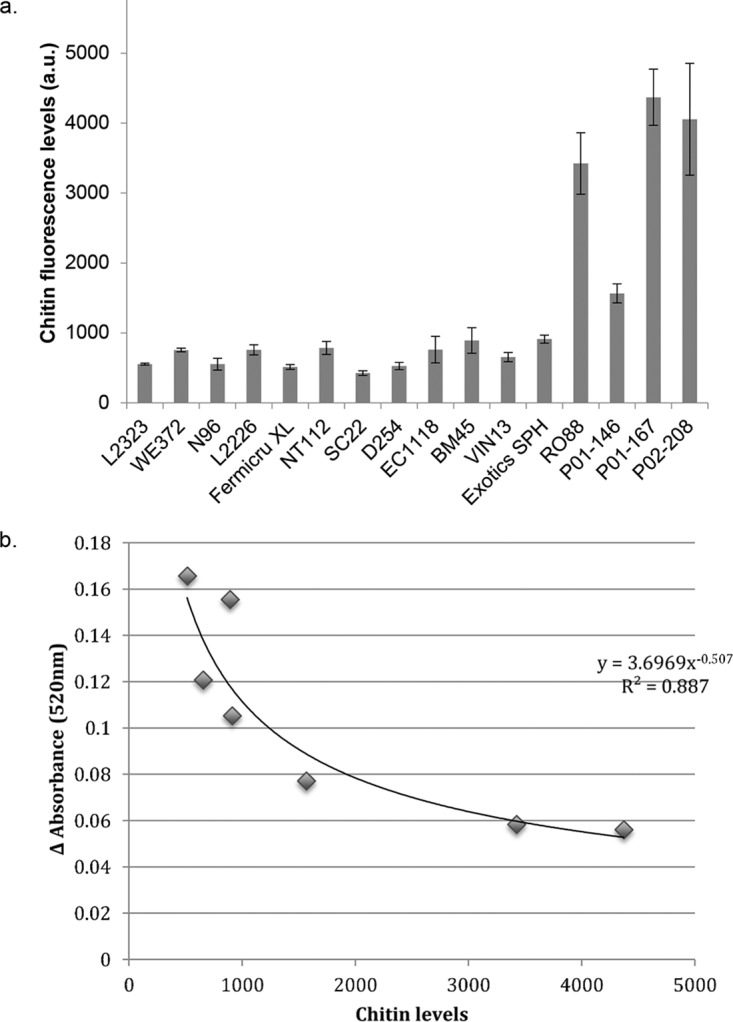
(a) Chitin levels quantified using flow cytometry after staining the cells with calcofluor white stain. Cells were grown overnight in YPD medium and a tenth of the overnight culture was preinoculated into fresh medium and grown for 5 h (43), reaching an OD of ∼7. Cells were stained with calcofluor white and further subjected to flow cytometry. Fluorescence intensity is expressed in arbitrary units (a.u.). (b) Scatter plot showing the correlation between wine haze levels and total cell wall chitin levels. Pearson's r value = −0.832 (P < 0.05). The data used for plotting were obtained from the haze formation of the 7 yeast strains appearing in Fig. 1 and the chitin level data from panel a.
GFP-tagged Vitis vinifera chitinase binds to yeast cell walls in a chitin-dependent manner.
In order to demonstrate the possibility that the high chitin levels found in cell walls of S. paradoxus strains could be responsible for the reduction of protein instability in wine, we developed a grape chitinase-yeast cell wall binding assay. V. vinifera chitinase class IVD (ChiivD) fused to the green fluorescent protein (GFP) was cloned in the pET14b vector and transformed in Escherichia coli Rosetta 2(DE3) pLysS. To characterize the expressed grape chitinase protein, the extracted crude protein extract was evaluated for chitinase enzyme activity. The data show that chitinase activity in the extract was the strongest when 4-nitrophenyl β-d-N,N′,N″-triacetylchitotriose was used as the substrate (Fig. 4), indicating that the overexpressed protein had the expected endochitinase activity (18). Three biological repeats were used for the assays. A chitinase-binding assay was carried out using this GFP-tagged grape chitinase. Figure 5 shows the differences in the fluorescence intensity as quantified by flow cytometry. These data clearly show that strains RO88, P01-167, P01-146, and P02-208 bound between 20 and 100% more GFP-tagged chitinase than the S. cerevisiae strains.
FIG 4.
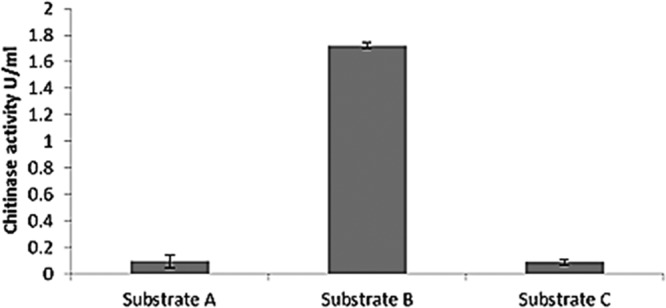
GFP-tagged chitinase activity from crude protein concentrate assayed in 3 different substrates suitable for exochitinase (substrate A), endochitinase (substrate B), and chitobiosidase (substrate C) activity detection supplied with the chitinase assay kit (catalog no. CS0980; Sigma-Aldrich), according to the manufacturer's instructions.
FIG 5.
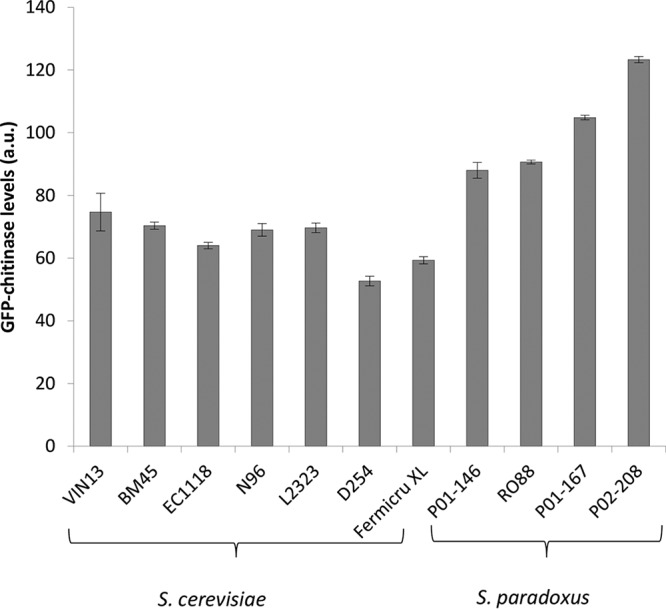
GFP-chitinase levels bound to different yeast strains, quantified using a BD FACSAria flow cytometer. Fluorescence levels shown are a result of subtracting the value corresponding to the control GFP protein bound to the cell walls from the total GFP-chitinase fluorescence levels. Cells were grown in YPD, as described by de Groot et al. (43), and equal amounts of cells based on the OD measurement at 600 nm were washed and resuspended in PBS buffer before adding GFP-tagged chitinase. Cells were further incubated for 2 h at room temperature with shaking before washing and resuspending in PBS buffer in preparation for quantification using a flow cytometer. Fluorescence intensity is expressed in arbitrary units (a.u.).
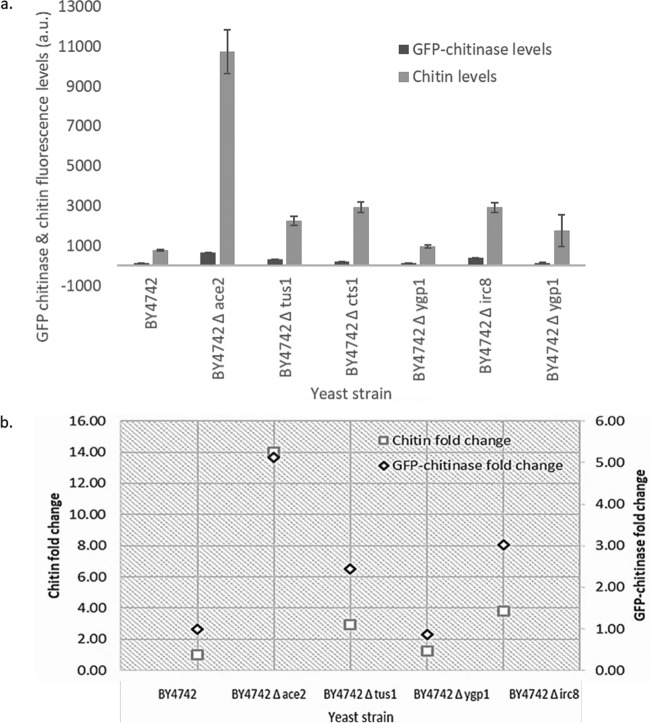
To evaluate the association between chitin levels and the amount of GFP-chitinase bound to cells, and to eliminate the possibility that the impact on chitinase binding was a species-specific feature of S. paradoxus, S. cerevisiae mutants exhibiting various chitin levels were used. A high-throughput selection screen for S. cerevisiae mutant strains with high cell wall chitin content was developed, and the yeast mutant deletion library (EUROSCARF [19]) was screened for mutants with high chitin levels. Mutant strains that showed higher chitin levels than the wild type encoded proteins linked to chitin synthesis or associated with cell wall biogenesis, morphogenesis, and signal transduction. Four such mutants (BY4742 Δace2, BY4742 Δtus1, BY4742 Δygp1, and BY4742 Δirc8) were selected for further analysis. Ace2p is a transcriptional activator of the CTS1 gene encoding chitinase, which is required for the degradation of the chitin ring that exists at the septum between mother and daughter cells (20, 21), and its deletion results in cells remaining attached at the chitin bud neck site (22). Tus1p, on the other hand, is a Rho1p exchange factor that contributes to the cell wall integrity-mediated modulation of Rho1p activity (23). Ygp1p is a highly glycosylated cell wall protein, and its synthesis is induced in response to cell wall disruptions and nutrient limitation (24). IRC8 has unknown function, and its product localizes in bud tips during cell division (25). An increase in chitin deposition has been observed as a compensation mechanism that mutants defective in cell wall synthesis or assembly activate to maintain cell integrity (26).
A positive correlation of 0.94 was observed between yeast cell wall chitin levels of S. cerevisiae mutant strains BY4742 Δace2, BY4742 Δtus1, BY4742 Δygp1, and BY4742 Δirc8 and wild-type strain BY4742 (Fig. 6). Figure 6b shows the fold changes observed for the mutant strains for both GFP-chitinase and chitin levels relative to the wild-type strain BY4742.
FIG 6.
(a) Bound GFP-chitinase levels and chitin levels of wild-type strain BY4742 and BY4742 Δygp1 and BY4742 Δtus1 mutant strains. GFP-chitinase levels bound to yeast strains and amount of cell wall chitin were quantified using a BD FACSAria flow cytometer. Cells were grown in YPD, as described by de Groot et al. (43), and equal amounts of cells based on OD measurement at 600 nm were washed and resuspended in PBS buffer before adding GFP-tagged chitinase or before staining with calcofluor white for chitin quantification. Fluorescence intensity is expressed in arbitrary units (a.u.). (b) Fold change of the deletion mutant strains relative to the wild-type strain BY4742. A correlation of 0.98 was obtained between the amount of yeast cell wall chitin and the amount of GFP-chitinase bound to the cell wall.
Yeast cells and cell wall extracts efficiently remove chitinases from model wine solutions.
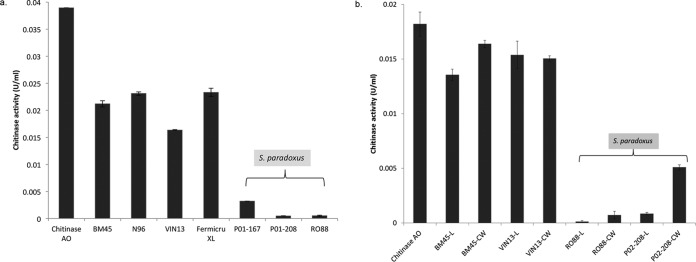
To further evaluate the abilities of different yeast strains to eliminate chitinases from solution, a commercial chitinase was used to assess the ability of yeast cells to reduce chitinase activity in a model wine solution. Commercial chitinase was dissolved in model wine solution containing 4 g/liter tartaric acid and 12% ethanol at pH 3.3 to a final concentration of 250 mg/liter. The chitinase-model wine solution was added to equal amounts of S. cerevisiae and S. paradoxus cells, mixed, and incubated with shaking. After centrifugation and elimination of the yeast cells, the activity in the supernatant was measured. The data showed that chitinase activity was reduced by 99.99% when P02-208 and RO88 live cells were used (Fig. 7a), while S. cerevisiae strains reduced chitinase levels by 46%, 41%, 58%, and 40%, for BM45, N96, VIN13, and Fermicru XL, respectively.
FIG 7.
(a) Chitinase activity levels of chitinase remaining in model wine solution (12% ethanol, 4 g/liter tartaric acid [pH 3.3]) not bound to the yeast cell wall after incubation with cells. (b) Chitinase activity levels of chitinase not bound to the yeast cell wall after incubation with live whole cells (L) and boiled cell wall extract (CW) from S. paradoxus and S. cerevisiae cells. Chitinase levels were quantified using the chitinase assay kit, according to the manufacturer's instructions. Commercial chitinase was used for the yeast cell wall binding assay in this experiment.
To evaluate the possible use of cell wall material of protein haze-protecting strains, the chitinase-binding ability of cell wall extract was evaluated. Similar to live cells, cell wall extracts from the S. paradoxus strains, especially RO88, revealed highly selective chitinase binding compared to S. cerevisiae strains (Fig. 7b). However, yeast cell wall extract generally bound less chitinase than live cells, suggesting that the dynamic cell wall of live cells appears to be better able to bind chitinases.
To rule out the possibility of cells releasing an enzyme inhibitor during incubation with chitinases, an investigation was carried out where commercial chitinase enzyme was added after removing the cells from the model wine solution following the incubation step. There were no significant differences in chitinase activity levels (P > 0.05) in all the strains used, including the control where no cells were added prior to incubation (see Fig. S2 in the supplemental material). These results strongly nullify the possibility of an enzyme inhibitor being released by the yeast cells during incubation.
Evaluation of the level of cell wall chitin in different wine yeast strains according to various environmental growth conditions.
The level of cell wall chitin in different wine yeast strains was investigated under various environmental growth conditions. As stated previously, RO88, P01-146, and P02-208 displayed significantly higher chitin levels than S. cerevisiae wine strains (Fig. 3). When investigating different environmental growth conditions, including growth of cells at 37°C, addition of calcium into the media, and changes in osmolarity, differences in chitin levels were observed and shown to be strain dependent. An increase in chitin levels was observed in all the wine yeast strains, with the S. paradoxus yeast strain P02-208 showing high chitin levels under all the different conditions investigated. Surprisingly, the domesticated strain BY4742 did not show a significant increase in chitin levels under all the growth conditions investigated in this study.
Gene expression.
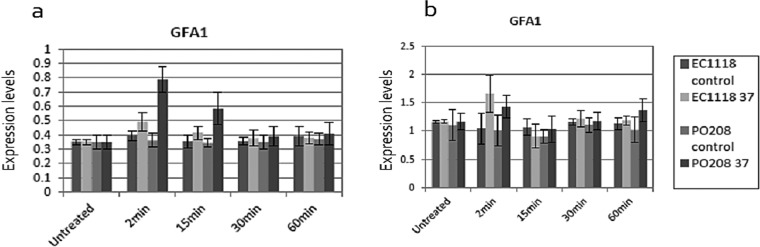
In order to assess whether the above-described observed phenotype was genetically determined, the expression levels of specific genes encoding proteins involved in chitin biosynthesis were investigated under the same conditions as those described above. The selected genes encode enzymes that are part of the chitin biosynthesis pathway from the precursor fructose-6-phosphate, including GFA1 and CHS1 to CHS3, as well as the CHS4 to CHS7 genes, which aid in the transportation of Chs1 to Chs3p. The GFA1 gene encodes glutamine:fructose-6-phosphate amidotransferase (glucosamine-6-phosphate synthase) catalyzing the first step of the hexosamine pathway required for the biosynthesis of cell wall precursors. No changes in the expression levels of the CHS1, CHS2, CHS3, CHS4, CHS5, CHS6, or CHS7 genes were observed despite the increase in chitin levels under different growth conditions, which were exposure of cells to 37°C and the addition of calcium to the growth medium. However, an increase in the expression levels of GFA1 was observed in both strains EC1118 and P02-208 (Fig. 8) under growth conditions shown to result in increased chitin levels, as described above.
FIG 8.
Quantitative real-time PCR (qRT-PCR) relative expression levels of chitin synthesis-related genes in S. cerevisiae EC1118 and S. paradoxus P02-208, after 0.1 M calcium was added to cells (a) and after cells were incubated at 37°C (b). Expression data were normalized to the expression levels of PDA1. Error bars indicate the standard deviation between three biological repeats.
DISCUSSION
Considering the negative impact of fining agents on wine quality, alternative haze protection methods, such as selection of yeast strains capable of reducing protein haze, are actively being sought after. Yeast strains had previously been shown to be able to impact the haze formation potential of wine. However, the data suggested that the mechanisms that had been proposed could not account for the full impact of these strains (11–13, 16).
Our data reveal a clear correlation between cell wall chitin levels and haze-protective capability. Interestingly, S. paradoxus strains contained significantly higher cell wall chitin levels than S. cerevisiae strains. To ensure that the correlation was not dependent on species-specific differences, the correlation was verified by using a set of S. cerevisiae mutants with known differences in chitin levels. Indeed, the industrial strains of S. cerevisiae evaluated in our laboratory tended to contain similar levels of cell wall chitin.
Since chitinases have been proposed as the major proteinaceous contributor to haze formation, two independent assays were designed to evaluate whether the cell wall chitin would impact levels of grape chitinases. Both assays suggest undeniably that yeast cells can bind chitinases in a chitin concentration-dependent manner, and that this ability can be used to reduce the levels of chitinases in liquid media. The data clearly support the hypothesis that this mechanism may indeed be a primary factor of yeast strain-dependent wine haze reduction.
The reduced wine haze levels and high chitinase levels bound by the RO88, P01-146, and P01-167 strains also concur with observations made by Manteau et al. (27). These authors observed the disappearance of chitinase from Champagne wine and attributed this observation to the likelihood that the chitinase was fixed on the cell wall of S. cerevisiae and that of the bacterium Oenococcus oeni during the aging period on lees. Furthermore, the grape chitinases have been demonstrated to maintain their activity in wine for at least a few months after the end of alcoholic fermentation (27, 28), thereby explaining the reduced haze levels observed during aging on lees with no observed increase in mannoprotein levels (data not shown).
In order to assess whether yeast cell wall extracts of strains displaying high chitin levels can be used as fining agents in cases where alcoholic fermentation is carried out using a strain with low chitin levels, yeast cell wall binding assays were carried out using yeast cell wall extracts. It was observed that similarly to live cells, RO88 and P02-208 cell wall extracts bound higher levels of chitinase than S. cerevisiae strains. These results further eliminate the possibility of S. paradoxus strains used in the study producing yeast acid proteases that may have hydrolyzed the grape chitinase (27). The differences in the amount of chitin bound to live and boiled yeast cells can be further explained by reduced cell surface or modified cell wall architectural structure of boiled yeast. Boiling cells results in a large number of wrinkles or folds on the cell walls. This is a cell structure similar to that of cells undergoing autolysis, as observed by Martínez-Rodríguez et al. (28). The reduced cell surface area in boiled cells may have consequently resulted in the slight reduction in the amount of chitinases bound to the cell wall.
It can also be further argued that in many wine haze studies where cell wall deletion mutants are used for haze assays (11–13), the observed wine haze reduction may not only be due to the greater release of mannoproteins but may also be due to increases in chitin levels in at least some of these mutants. Deletion studies of certain genes involved in cell wall structure and composition, such as GAS1, were indeed shown to result in high cell wall chitin levels (29). Differences in chitin levels have been reported for certain cell wall mutants, as is also seen in this study for BY4742 Δace2, BY4742 Δtus1, BY4742 Δygp1, and BY4742 Δirc8.
In this study, higher chitin levels were observed when cells were incubated at 15°C, which corroborates the results of studies by Gasch et al. (30) and Sahara et al. (31), who observed increased expression levels of CHS1, CHS3, and GNA1 at 10°C versus 25°C, thus indicating a potential increase in chitin levels at low temperatures. Surprisingly, no increases in the CHS1, CHS3, and GNA1 gene expression levels were observed in our study despite the increased chitin levels. Our results under hypo-osmotic conditions also correlate with those from a study conducted by Deshpande et al. (32), who observed a clear relationship between hypo-osmolarity and elevated extracellular chitin levels. The addition of calcium resulted in increased chitin levels, concurring with data obtained by Munro et al. (33), who also observed that elevated extracellular calcium levels increased cell wall chitin levels in Candida albicans and also stimulated chitin synthase (CHS) promoters.
Increases in chitin levels have been attributed to the spatial and temporal regulation of enzymes involved in chitin synthesis (34, 35). However, an increase in GFA1 expression levels observed in this study possibly indicates that the transcriptional regulation of this gene is directly correlated with an increase in chitin synthesis. An increase in GFA1 expression levels was also observed by Lagorce et al. (36) in cell wall mutants showing increased cell wall chitin levels. Furthermore, the same authors showed that overexpression of the GFA1 gene resulted in a 3-fold increase in cell wall chitin levels.
In conclusion, our findings strongly support the possibility of using yeast strains displaying high chitin levels to reduce wine haze formation. S. paradoxus has been evaluated under winemaking conditions and was found to possess good enological properties. For example, RO88 has been shown to have high polygalacturonase activity (37), influence the production of certain aroma compounds, and contribute positively to the final quality of wine (38, 39). S. paradoxus strains or specifically selected S. cerevisiae strains with high chitin levels may therefore be considered for use as starter cultures in wine fermentations and/or may be used for future yeast breeding projects with the aim of reducing wine haze formation. Alternatively, yeast cells showing high chitin levels should be investigated for use as clarifying agents and can be used in the form of yeast hulls.
MATERIALS AND METHODS
Fermentation.
Fermentations were carried out in triplicate at 21°C in either 120-ml or 2-liter bottles with a working volume of either 50 ml or 1.7 liters, respectively, without agitation, and fitted with air traps. Commercial wine strains (Table 1) were used to ferment chemically defined synthetic MS300 medium (40) and Sauvignon Blanc and Chardonnay grape juices to dryness. Residual glucose and fructose concentrations at the end of all fermentations were less than 5 g/liter, as measured using a d-glucose–d-fructose enzymatic kit (Amersham Biosciences, Freiburg, Germany).
TABLE 1.
S. cerevisiae, S. paradoxus, and hybrid yeast strains used in this study
| Strain | Description | Source or reference |
|---|---|---|
| S. cerevisiae | ||
| BM45 | Industrial wine yeast strain | Lallemand, Inc. (Montreal, Canada) |
| VIN13 | Commercial yeast strain | Anchor Yeast (Cape Town, South Africa) |
| L2323 | Industrial wine yeast strain | Lallemand, Inc. |
| WE372 | Industrial wine yeast strain | Anchor Yeast |
| N96 | Commercial yeast strain | Anchor Yeast |
| NT50 | Commercial yeast strain | Anchor Yeast |
| L2226 | Commercial yeast strain | Lallemand, Inc. |
| Fermicru XL | Commercial yeast strain | DSM Food Specialties B.V. (Flemingham, The Netherlands) |
| NT112 | Commercial yeast strain | Anchor Yeast |
| SC22 | Commercial yeast strain | University of California (Davis, CA, USA) |
| D254 | Commercial yeast strain | Lallemand, Inc. (Montreal, Canada) |
| EC1118 | Commercial yeast strain | Lallemand, Inc. (Montreal, Canada) |
| BY4742 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 | 29 |
| BY4742 Δirc8 | BY4742 irc8::kanMX4 | EUROSCARF deletion mutant library |
| BY4742 Δtus1 | BY4742 tus1::kanMX4 | EUROSCARF deletion mutant library |
| BY4742 Δygp1 | BY4742 ygp1::kanMX4 | EUROSCARF deletion mutant library |
| BY4742 Δace2 | BY4742 ace2::kanMX4 | EUROSCARF deletion mutant library |
| S. cerevisiae-S. paradoxus | ||
| Exotics SPH | Commercial yeast strain VIN13 × RO88 hybrid | 37 |
| S. paradoxus | ||
| P01-167 | Industrial wine yeast strain | Phaff Yeast Collection (University of California, Davis, CA) |
| P02-208 | Isolated from olive brine | Phaff Yeast Collection |
| P01-146 | Unspecified | Phaff Yeast Collection |
| RO88 | Industrial wine yeast strain | 48 |
Heat stability test.
The heat stability of wine samples was determined as described by Pocock and Waters (41), with all measurements made in triplicate with the appropriate controls. Briefly, the assay was carried out by centrifuging fermented Chardonnay grape must at 3,250 × g for 5 min to remove cells. After taking readings of the absorbance at 520 nm, the wine sample was heated at 80°C for 2 h and then cooled to 4°C for 16 h. The A520 was measured after acclimatization at room temperature for 30 min. Haze was measured by calculating the difference in absorbance before and after heating of the wine sample (42). Bentonite (0.5 g/liter) and S. cerevisiae and S. paradoxus yeast strains were added at the end of alcoholic fermentation in wines fermented using S. cerevisiae EC1118 after cell removal as a treatment.
Calcofluor white staining, fluorescence microscopy, and flow cytometry.
Cell staining with calcofluor white and fluorescence microscopy were adapted from de Groot et al. (43). Cells grown overnight at 30°C in yeast-peptone-dextrose (YPD) were either directly used for microscopy or inoculated into fresh YPD medium. To enhance the detection of cell wall-related phenotypes, the cells inoculated in the fresh medium were further incubated for 5 h at 37°C. About 200 μl of the cell culture was centrifuged, and the cells were washed with phosphate-buffered saline (PBS) (pH 7.4; Na2HPO4). Cells were stained with 10 μl calcofluor white after the addition of 10 μl of 10% KOH, according to the manufacturer's instructions (18909-100 ml; Fluka Analytical, Sigma-Aldrich). Z-sectioning image acquisition was performed on a Carl Zeiss confocal laser scanning microscope (LSM) 780 Elyra S1 with superresolution structured illumination microscopy (SR-SIM superresolution) platform. Z-series images were taken at 0.5-μm intervals through the specimens. The excitation laser used was the violet laser with 407-nm wavelength, and the emission filter used was the Pacific Blue channel with a 450/40 band-pass filter for calcofluor white-stained cells. Images were processed and background subtracted using the Zeiss Zen lite 2011 software and presented in a maximum-intensity projection.
For the quantification of chitin levels using flow cytometry, a BD FACSAria flow cytometer was used. The BD FACSDiva version 6.1.3 software was used for the data capture. The excitation and emission filter lasers used were the same as those described above. A total of 50,000 cells were used for the quantification of chitin levels.
High-throughput genome-wide screening.
For enrichment of deletion strains, a pooled EUROSCARF deletion mutant library (EUROSCARF, Frankfurt, Germany [19; http://web.uni-frankfurt.de/fb15/mikro/euroscarf/]) was labeled with calcofluor white, and chitin-rich mutants were selected using a BD FACSAria cell sorter. The cells with high cell wall chitin levels were enriched in 10 cycles through the selection of 5% of the cell population with highest chitin level and growing the selected population before a subsequent sorting round. The deletion mutant strains were identified and verified by sequencing the unique uptags and downtags coded in each deletion strain, as detailed by the Saccharomyces Genome Deletion project.
Impact of different environmental growth conditions on yeast cell wall chitin levels.
To assess chitin level at different temperatures, yeast cells were grown as described by de Groot et al. (43). Overnight precultures in YPD broth were cultured for 5 h at 15°C, 30°C, and 37°C.
In order to assess the impact of hypo-osmotic growth conditions on yeast cell wall chitin levels, changes in nutrient content after medium transfers were taken into account. An overnight YPD preculture of cells was transferred to 20% YPD diluted with 1 M sorbitol. In order to create hypo-osmotic stress, the cells were further transferred into 20% YPD after 5 h. For the controls, cells were grown continuously without transfers either in high-osmolarity medium containing 20% YPD with 1 M sorbitol or low-osmolarity medium in 20% YPD.
Quantitative PCR.
Quantitative PCRs (qPCRs) were performed in duplicate in a 7500 cycler (Applied Biosystems, Carlsbad, CA). The primers used are listed in Table 2 and were designed using Primer Express software version 3 (Applied Biosystems). Open reading frames of S. paradoxus P02-208 genes were sequenced prior to designing qPCR primers. Approximately 1 μg total RNA was used as the template for cDNA synthesis using the ImProm-II reverse transcriptase system (Promega, Madison, WI). RNA was treated with DNase I (Roche Diagnostics, Basel, Switzerland). The dye SYBR green was used for amplicon detection, and primers were diluted to a final concentration of 100 nM for the qPCR.
TABLE 2.
Sequences of primers used in quantitative real-time PCR analysis of both S. cerevisiae EC1118 and S. paradoxus P02-208
The conditions for the qPCR runs were 50°C for 2 min, 95°C for 10 min, 95°C for 15 s repeated for 40 cycles, and 60°C for 1 min; a dissociation curve analysis was added for confirmation of primer specificity. The Signal Detection Software (SDS) version 1.3.1 (Applied Biosystems) was used for initial analysis of data, where the relative expression value for each sample was defined as 2−CT(target), with CT(target) representing the cycle number at which a sample reaches a decided threshold value for the specific gene. The relative expression data were normalized to the value of the normalization gene PDA1 in each respective sample, thus giving normalized relative expression for a target gene as 2−CT(target)/2−CT(PDA1).
Overexpression of GFP-tagged Vitis vinifera chitinase in Escherichia coli.
General molecular biology techniques used in this study were as described by Sambrook et al. (44). E. coli DH5α (Gibco BRL/Life Technologies, Rockville, MD) was used as a host for all plasmid amplifications. Total grape berry RNA was isolated, as previously described by Schmitt et al. (45), and DNase I (Roche Diagnostics) treatment was used to eliminate DNA contamination. One microgram of the extracted RNA was used as the template for cDNA synthesis using the ImProm-II reverse transcription system, according to the manufacturer's instructions (Promega). Highly expressed grape chitinase gene was amplified from grape berry cDNA using chitinase forward primer 5′-CATATGGCAGCCAAGCTACTAACAGTC-3′ (NdeI) and chitinase reverse primer 5′-CTCGAGGCAAGTGAGGTTGTCACCA-3′ (XhoI) (where the underlining indicates restriction enzyme sites). The amplified chitinase class IVD fragment (accession number AF532966.1) was cloned into a shuttle vector pJet1.2/blunt (CloneJET PCR cloning kit; Thermo Fisher Scientific), according to the protocol described by the manufacturer, before subcloning into the pET14b vector (Novagen, Madison, WI, USA), also using the pET system manual. Green fluorescent protein was PCR amplified from pKEN mut 2 vector (Addgene, Cambridge, MA) using 5′-CTCGAGATGAGTAAAGGAGAAGAACTTTTCAC-3′ (XhoI) as the forward primer and 5′-GATCGGATCCTTATTTGTATAGTTCATCCATGCC-3′ (BamHI) as the reverse primer, cloned first in pJet1.2 vector (catalog no. K1232; Fermentas), according to the CloneJET PCR cloning kit instruction manual for further sequencing. The cassettes were then released from the pJet1.2/blunt cloning vector using Ndel and Xhol for the grape chitinase cassette and Xhol and BamHl for the GFP cassette into a shuttle vector before subcloning into the pET14b vector. The chitinase gene was cloned upstream of the GFP gene. Phusion high-fidelity DNA polymerase (catalog no. F530L; Finnzymes) was used for all the PCRs. The Finnzymes Phusion high-fidelity DNA polymerase manual was followed for the PCR program, and an annealing temperature of 60°C was used for all the primers. The final cassette in pET14b vector (pET-GFP-Chi) was sequenced to confirm the sequence.
A protocol described by Lee and Coleman (46) was used to overexpress the grape chitinase protein in E. coli Rosetta 2(DE3) pLysS. For the overexpression of GFP-tagged grape Vitis vinifera chitinase in E. coli, pET-GFP-Chi vector was transformed into E. coli Rosetta 2(DE3) pLysS. E. coli Rosetta 2(DE3) pLysS containing pET-GFP-Chi was grown in LB broth at 37°C until a cell density of ∼0.6 (optical density at 600 nm [OD600]) was reached. The cell culture was cooled to 25°C, and the grape chitinase expression was induced with 0.4 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at 25°C overnight. The cell pellet from the overnight culture was freeze-thawed once to rupture the bacterial cell membrane and allow the lysozyme, produced by E. coli Rosetta 2(DE3) pLysS, to degrade the bacterial cell wall. Lysozyme (10 mg/ml) was added to further lyse the cells. The cell pellet was resuspended in 50 mM potassium phosphate cell lysis buffer (pH 8.0) containing 300 mM KCl and 10% glycerol, based on 7 ml of lysis buffer per gram of wet cell pellet. DNase (0.1% [vol/vol]) and 0.1% (vol/vol) RNase were added to reduce the viscosity of the cell lysate. The soluble protein fraction (crude cell lysate) was separated from the cell debris by centrifugation at 9,500 × g. The crude protein extract was concentrated using Amicon Ultra-15 centrifugal filter columns (catalog no. UFC901096; Millipore, Merck, Ireland) with a cutoff of 10 kDa. The concentrated enzyme was dissolved in enzyme storage buffer (50 mM potassium phosphate buffer [pH 7.0] containing 150 mM KCl, 1 mM dithiothreitol [DTT], 1 mM EDTA, and 10% glycerol).
Chitinase assay.
A chitinase activity assay kit (catalog no. CS0980; Sigma-Aldrich, MO, USA) was used with 4-nitrophenyl N-acetyl-β-d-glucosaminide (a substrate suitable for exochitinase activity detection), 4-nitrophenyl β-d-N,N′,N″-triacetylchitotriose (a substrate suitable for endochitinase activity detection), and 4-nitrophenyl N,N′-diacetyl-β-d-chitobioside (a substrate suitable for exochitinase activity detection and chitobiosidase activity), as instructed by the manufacturer.
GFP-tagged grape chitinase-yeast cell wall binding assays.
Yeast cells were cultured in accordance with a method described by de Groot et al. (43). Equal amounts of cells based on a measurement of the OD at 600 nm were washed using 1× PBS buffer. One hundred microliters of crude GFP-tagged chitinase protein was added to the cells suspended in 200 μl PBS buffer. These were incubated for 2 h at either room temperature or 37°C with shacking. The cells were centrifuged at 3,250 × g, washed twice with PBS buffer, and visualized under a fluorescence microscope. In order to make sure that the yeast cell surfaces are not binding GFP, a control was run where GFP was used instead of GFP-chitinase. The excitation laser used was a solid-state sapphire laser at a wavelength of 488 nm, and the emission filters used consisted of a fluorescein isothiocyanate (FITC) channel, with 502 long-pass and 530/30 band-pass filters. To quantify the GFP-tagged grape chitinase bound to the yeast cell wall, flow cytometry (BD FACSAria flow) was performed using the above-described excitation and emission wavelengths. For the data analysis, the FITC-area geometric mean of fluorescence intensity was used for the quantification of fluorescence produced by 50,000 cells.
Yeast culturing and yeast cell wall extract preparation.
S. cerevisiae yeast strains VIN13, BM45, EC1118, N96, L2323, D254, and Fermicru XL and S. paradoxus strains P01-161, RO88, P01-167, and P02-208 (Table 1) were grown in synthetic medium MS300, and equal amounts of cells were centrifuged at 3,250 × g and washed with saline solution (0.9% NaCl). For the cell wall extract, yeast cells were grown in YPD broth overnight for 16 h, reaching an OD at 600 nm of ∼7. Equal amounts of cells of each strain were centrifuged at 3,250 × g, washed with saline water, and then boiled for 15 min at 100°C before washing again with saline solution.
Yeast cell wall binding assays.
Chitinase (Trichoderma viride; catalog no. C8241; Sigma-Aldrich) was dissolved in model wine solution of pH 3.3 containing 12% ethanol and 4 g/liter tartaric acid to a final concentration of 0.5 mg/ml. Yeast cells were suspended in 1 ml of this solution and incubated for 30 min with shaking at 37°C or for 2 h at room temperature. A negative control where no cells were added was also incubated. The suspension was centrifuged at 3,250 × g, and 30 μl of the supernatant was used for further chitinase activity analysis. The chitinase assay kit (catalog no. CS0980; Sigma-Aldrich) was used to assay for the remaining chitinase activity, as per the manufacturer's instructions.
A correlation was also determined between the levels of chitin and the amount of GFP-chitinase bound to the S. cerevisiae BY4742 Δtus1 and BY4742 Δygp1 mutant strains and the wild-type BY4742 strain. The BY4742 Δtus1 and BY4742 Δygp1 mutant strains were chosen, as they have various amounts of cell wall chitin levels. Lesage et al. (47) reported high levels of chitin deposition in the BY4742 Δtus1 mutant relative to the wild-type BY4742 strain. However, no increase/decrease in chitin levels has been reported for the BY4742 Δygp1 mutant strain relative to the wild-type strain BY4742.
In order to ensure that there were no enzyme inhibitors released by the cells, an assay was carried out where cells were incubated in a model wine solution for 2 h, after which they were removed through centrifugation. Chitinase was then added to the model wine without the cells, after which the level of chitinase activity was assayed.
Statistical analysis.
The data are expressed as the mean ± standard deviation from three biological repeats, while in the case of the heat test, the mean ± standard deviation were calculated from three biological and technical replicates. Statistical comparisons between values were performed with a multifactorial analysis of variance (ANOVA) using STATISTICA 10 (P < 0.05).
Supplementary Material
ACKNOWLEDGMENTS
This work was financially supported by the National Research Foundation (NRF) and the South African Wine Industry (Winetech).
We thank P. R. Young for supplying the Vitis vinifera grape cDNA. Flow cytometry and fluorescence microscopy experiments were carried out at the Central Analytical Facility (CAF, Stellenbosch University) with the help of B. Loos and L. Engelbrecht.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00668-18.
REFERENCES
- 1.Siebert KJ. 2009. Haze in beverages. Adv Food Nutr Res 57:53–86. doi: 10.1016/S1043-4526(09)57002-7. [DOI] [PubMed] [Google Scholar]
- 2.Batista L, Monteiro S, Loureiro VB, Teixeira AR, Ferreira RB. 2010. Protein haze formation in wines revisited. The stabilising effect of organic acids. Food Chem 122:1067–1075. doi: 10.1016/j.foodchem.2010.03.076. [DOI] [Google Scholar]
- 3.Marangon M, Van Sluyter S, Waters EJ, Herderich MJ, Pretorius IS. 2010. Recent advances help us understand wine haze more clearly. Aust N Z Wine Ind J 25:24–27. [Google Scholar]
- 4.Batista L, Monteiro S, Loureiro VB, Teixeira AR, Ferreira RB. 2009. The complexity of protein haze formation in wines. Food Chem 112:169–177. doi: 10.1016/j.foodchem.2008.05.070. [DOI] [Google Scholar]
- 5.Esteruelas M, Poinsaut P, Sieczkowski N, Manteau S, Fort MF, Canals JM, Zamora F. 2009. Characterization of natural haze protein in sauvignon white wine. Food Chem 113:28–35. doi: 10.1016/j.foodchem.2008.07.031. [DOI] [Google Scholar]
- 6.Falconer RJ, Marangon M, Van Sluyter SC, Neilson KA, Chan C, Waters EJ. 2010. Thermal stability of thaumatin-like protein, chitinase, and invertase isolated from Sauvignon Blanc and Semillon juice and their role in haze formation in wine. J Agric Food Chem 58:975–980. doi: 10.1021/jf902843b. [DOI] [PubMed] [Google Scholar]
- 7.Marangon M, Van Sluyter S, Neilson K, Chan C, Haynes P, Waters EJ, Falconer RJ. 2011. Roles of grape thaumatin-like protein and chitinase in white wine haze formation. J Agric Food Chem 59:733–7407. doi: 10.1021/jf1038234. [DOI] [PubMed] [Google Scholar]
- 8.Waters EJ, Shirley NJ, Williams PJ. 1996. Nuisance proteins of wine are grape pathogenesis-related proteins. J Agric Food Chem 44:3–5. doi: 10.1021/jf9505584. [DOI] [Google Scholar]
- 9.Marangon M, Van Sluyter SC, Chan C, Waters EJ, Falconer RJ. 2010. The different behaviours of thaumatin-like proteins and chitinases during white wine haze formation. 14th Australian Wine Industry Technical Conference, 3 to 8 July 2010, Adelaide, Australia. [Google Scholar]
- 10.Palmisano G, Antonacci D, Larse MR. 2010. Glycoproteomic profile in wine: a ‘Sweet’ molecular renaissance. J Proteome Res 9:6148–6159. doi: 10.1021/pr100298j. [DOI] [PubMed] [Google Scholar]
- 11.Brown SL, Stockdale VJ, Pettolino F, Pocock KF, Lopes MB, Williams PJ, Bacic A, Fincher GB, Høj PB, Waters EJ. 2007. Reducing haziness in white wine by overexpression of Saccharomyces cerevisiae genes YOL155c and YDR055w. Appl Microbiol Biotechnol 73:1363–1376. 12. doi: 10.1007/s00253-006-0606-0. [DOI] [PubMed] [Google Scholar]
- 12.Gonzalez-Ramos D, Gonzalez R. 2006. Genetic determinants of the release of mannoproteins of enological interest by Saccharomyces cerevisiae. J Agric Food Chem 54:9411–9416. doi: 10.1021/jf062196d. [DOI] [PubMed] [Google Scholar]
- 13.Gonzalez-Ramos D, Quiro M, Gonzalez R. 2009. Three different targets for the genetic modification of wine yeast strains resulting in improved effectiveness of bentonite fining. J Agric Food Chem 57:8373–8378. doi: 10.1021/jf901093v. [DOI] [PubMed] [Google Scholar]
- 14.Doco T, Quellec N, Moutounet M, Pellerin P. 1999. Polysaccharide patterns during the aging of Carignan noir red wines. Am J Enol Viticult 50:25–32. [Google Scholar]
- 15.Doco T, Vuchot P, Cheynier V, Moutounet M. 2003. Structural modification of wine arabinogalactans during aging on lees. Am J Enol Viticult 54:150–157. [Google Scholar]
- 16.Dupin IVS, McKinnon BM, Ryan C, Boulay M, Markides AJ, Jones GP, Williams PJ, Waters EJ. 2000. Saccharomyces cerevisiae mannoproteins that protect wine from protein haze: their release during fermentation and lees contact and a proposal for their mechanism of action. J Agric Food Chem 48:3098–3105. doi: 10.1021/jf0002443. [DOI] [PubMed] [Google Scholar]
- 17.Vincenzi S, Polesani M, Curioni A. 2005. Removal of specific protein components by chitin enhances protein stability in a white wine. Am J Enol Viticult 56:246–254. [Google Scholar]
- 18.Vincenzi S, Bierma J, Wickramasekara SI, Curioni A, Gazzola D, Bakalinsky AT. 2014. Characterization of a grape class IV chitinase. J Agric Food Chem 62:5660–5668. doi: 10.1021/jf501225g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giaever G, Nislow C. 2014. The yeast deletion collection: a decade of functional genomics. Genetics 197:451–465. doi: 10.1534/genetics.114.161620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dohrmann PR, Butler G, Tamai K, Dorland S, Greene JR, Thiele DJ, Stillman DJ. 1992. Parallel pathways of gene regulation: homologous regulators Swi5 and Ace2 differentially control transcription of HO and chitinase. Genes Dev 6:93–104. doi: 10.1101/gad.6.1.93. [DOI] [PubMed] [Google Scholar]
- 21.Voth WP, Olsen AE, Sbia M, Freedman KH, Stillman DJ. 2005. ACE2, CBK1, and BUD4 in budding and cell separation. Eukaryot Cell 4:1018–1028. doi: 10.1128/EC.4.6.1018-1028.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oud B, Guadalupe-Medina V, Nijkamp JF, de Ridder D, Pronk JT, van Maris AJA, Daran J-M. 2013. Genome duplication and mutations in ACE2 cause multicellular, fast-sedimenting phenotypes in evolved Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 110:E4223–E4231. doi: 10.1073/pnas.1305949110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmelzle T, Helliwell SB, Hall MN. 2002. Yeast protein kinases and the RHO1 exchange factor TUS1 are novel components of the cell integrity pathway in yeast. Mol Cell Biol 22:1329–1339. doi: 10.1128/MCB.22.5.1329-1339.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pardo M, Monteoliva L, Pla J, Sanchez M, Gil C, Nombela C. 1999. Two-dimensional analysis of proteins secreted by Saccharomyces cerevisiae regenerating protoplasts: a novel approach to study the cell wall. Yeast 15:459–472. doi:. [DOI] [PubMed] [Google Scholar]
- 25.Cherry JM, Hong EL, Amundsen C, Balakrishnan R, Binkley G, Chan ET, Christie KR, Costanzo MC, Dwight SS, Engel SR, Fisk DG, Hirschman JE, Hitz BC, Karra K, Krieger CJ, Miyasato SR, Nash RS, Park J, Skrzypek MS, Simison M, Weng S, Wong ED. 2012. Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic Acids Res 40:D700–D705. doi: 10.1093/nar/gkr1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Valdivieso MMH, Ferrario L, Vai M, Duran A, Popolo L. 2000. Chitin synthesis in a gas1 mutant of Saccharomyces cerevisiae. J Bacteriol 4752–4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manteau S, Lambert B, Jeandet P, Legendre L. 2003. Changes in chitinase and thaumatin-like pathogenesis-related proteins of grape berries during the champagne winemaking process. Am J Enol Viticult 54:267–272. [Google Scholar]
- 28.Martínez-Rodríguez AJ, Polo MC, Carrascosa AV. 2001. Structural and ultrastructural changes in yeast cells during autolysis in a model wine system and in sparkling wines. Int J Food Microbiol 71:45–51. doi: 10.1016/S0168-1605(01)00554-2. [DOI] [PubMed] [Google Scholar]
- 29.Popolo L, Gilardelli D, Bonfante P, Vai M. 1997. Increase in chitin as an essential response to defects in assembly of cell wall polymers in the ggp1Δ mutant of Saccharomyces cerevisiae. J Bacteriol 179:463–469. doi: 10.1128/jb.179.2.463-469.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gasch AP, Spellman TP, Kao CM, Carmel-Harel C, Eisen MB, Storz G, Botstein D, Brown PO. 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell 11:4241. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sahara T, Goda T, Ohgiya S. 2002. Comprehensive expression analysis of time-dependent genetic responses in yeast cells to low temperature. J Biol Chem 277:50015–50021. doi: 10.1074/jbc.M209258200. [DOI] [PubMed] [Google Scholar]
- 32.Deshpande MV, O'Donnell R, Gooday GW. 1997. Regulation of chitin synthase activity in the dimorphic fungus Benjaminiella poitrasii by external osmotic pressure. FEMS Microbiol Lett 152:327–332. doi: 10.1111/j.1574-6968.1997.tb10447.x. [DOI] [PubMed] [Google Scholar]
- 33.Munro CA, Selvaggini S, De Bruijn I, Walker L, Lenardon MD, Gerssen B, Milne S, Brown AJP, Gow NAR. 2007. The PKC, HOG and Ca2+ signaling pathways coordinately regulate chitin synthesis in Candida albicans. Mol Microbiol 63:1399–1413. doi: 10.1111/j.1365-2958.2007.05588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Valdivia RH, Baggott D, Chuang JS, Schekman RW. 2002. The yeast clathrin adaptor protein complex 1 is required for the efficient retention of a subset of late Golgi membrane proteins. Dev Cell 2:283–294. doi: 10.1016/S1534-5807(02)00127-2. [DOI] [PubMed] [Google Scholar]
- 35.Lesage G, Bussey H. 2006. Cell wall assembly in Saccharomyces cerevisiae. Microbiol Mol Biol Rev 70:317–343. doi: 10.1128/MMBR.00038-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lagorce A, Le Berre-Anton V, Aguilar-Uscanga B, Martin-Yken H, Dagkessamanskaia A, Francois J. 2002. Involvement of GFA1, which encodes glutamine-fructose-6-phosphate amidotransferase, in the activation of the chitin synthesis pathway in response to cell-wall defects in Saccharomyces cerevisiae. Eur J Biochem 269:1697–1707. doi: 10.1046/j.1432-1327.2002.02814.x. [DOI] [PubMed] [Google Scholar]
- 37.Mocke B. 2005. The breeding and characterization of starter culture yeast strains for red wine making. M.Sc. thesis, Stellenbosch University, Stellenbosch, South Africa. [Google Scholar]
- 38.Majdak A, Herjavec S, Orlić S, Redžepović S, Mirošević N. 2002. Comparison of wine aroma compounds produced by Saccharomyces paradoxus and Saccharomyces cerevisiae strains. Food Technol Biotechnol 40:103–109. [Google Scholar]
- 39.Orlic S, Redžepovic S, Jeromel A, Herjaveć S, Iacumin L. 2007. Influence of indigenous Saccharomyces paradoxus strains on Chardonnay wine fermentation aroma. Int J Food Sci Technol 42:95–101. doi: 10.1111/j.1365-2621.2006.01217.x. [DOI] [Google Scholar]
- 40.Bely M, Sablayrolles JM, Barre P. 1990. Automatic detection of assimilable nitrogen deficiencies during alcoholic fermentation in enological conditions. J Ferment Bioeng 70:246–252. doi: 10.1016/0922-338X(90)90057-4. [DOI] [Google Scholar]
- 41.Pocock KF, Waters EJ. 2006. Protein haze in bottled white wines: how well do stability tests and bentonite fining trials predict haze formation during storage and transport? Aust J Grape Wine Res 12:212–220. doi: 10.1111/j.1755-0238.2006.tb00061.x. [DOI] [Google Scholar]
- 42.Waters EJ, Wallace W, Williams PJ. 1992. Identification of heat-unstable wine proteins and their resistance to peptidases. J Agric Food Chem 40:1514–1519. doi: 10.1021/jf00021a008. [DOI] [Google Scholar]
- 43.de Groot PWJ, Ruiz C, de Aldana CRV, Duenas E, Cid VJ, Rey FD, Rodrıquez-Pena JM, Perez P, Andel A, Caubın J, Arroyo J, Garcıa JC, Gil C, Molina M, Garcıa LJ, Nombela C, Klis FM. 2001. A genomic approach for the identification and classification of genes involved in cell wall formation and its regulation in Saccharomyces cerevisiae. Comp Funct Genomics 2:124–142. doi: 10.1002/cfg.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sambrook KJ, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 45.Schmitt ME, Brown TA, Trumpower BL. 1990. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res 18:3091–3092. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee P, Colman RF. 2007. Expression, purification, and characterization of stable, recombinant human adenylosuccinate lyase. Prot Exp Purif 51:227–234. doi: 10.1016/j.pep.2006.07.023. [DOI] [PubMed] [Google Scholar]
- 47.Lesage G, Shapiro J, Specht CA, Sdicu A-M, Ménard P, Hussein S, Tong AHY, Boone C, Bussey H. 2005. An interactional network of genes involved in chitin synthesis in Saccharomyces cerevisiae. BMC Genet 6:8. doi: 10.1186/1471-2156-6-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Redžepović S, Orlić S, Majdak A, Kozina B, Volschenk H, Viljoen-Bloom M. 2003. Differential malic acid degradation by selected strains of Saccharomyces during alcoholic fermentation. Int J Food Microbiol 83:49–61. doi: 10.1016/S0168-1605(02)00320-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.