ABSTRACT
Recombination is a major driver of evolution in bacterial populations, because it can spread and combine independently evolved beneficial mutations. Recombinant lineages of bacterial pathogens of plants are typically associated with the colonization of novel hosts and the emergence of new diseases. Here we show that recombination between evolutionarily and phenotypically distinct plant-pathogenic lineages generated recombinant lineages with unique combinations of pathogenicity and virulence factors. Xanthomonas euvesicatoria and Xanthomonas perforans are two closely related lineages causing bacterial spot disease on tomato and pepper worldwide. We sequenced the genomes of atypical strains collected from tomato in Nigeria and observed recombination in the type III secretion system and effector genes, which showed alleles from both X. euvesicatoria and X. perforans. Wider horizontal gene transfer was indicated by the fact that the lipopolysaccharide cluster of one strain was most similar to that of a distantly related Xanthomonas pathogen of barley. This strain and others have experienced extensive genomewide homologous recombination, and both species exhibited dynamic open pangenomes. Variation in effector gene repertoires within and between species must be taken into consideration when one is breeding tomatoes for disease resistance. Resistance breeding strategies that target specific effectors must consider possibly dramatic variation in bacterial spot populations across global production regions, as illustrated by the recombinant strains observed here.
IMPORTANCE The pathogens that cause bacterial spot of tomato and pepper are extensively studied models of plant-microbe interactions and cause problematic disease worldwide. Atypical bacterial spot strains collected from tomato in Nigeria, and other strains from Italy, India, and Florida, showed evidence of genomewide recombination that generated genetically distinct pathogenic lineages. The strains from Nigeria and Italy were found to have a mix of type III secretion system genes from X. perforans and X. euvesicatoria, as well as effectors from Xanthomonas gardneri. These genes and effectors are important in the establishment of disease, and effectors are common targets of resistance breeding. Our findings point to global diversity in the genomes of bacterial spot pathogens, which is likely to affect the host-pathogen interaction and influence management decisions.
KEYWORDS: Xanthomonas, bacterial spot, effectors, evolution, horizontal gene transfer, recombination, tomato
INTRODUCTION
Homologous and nonhomologous recombination are major drivers of evolution in bacterial populations (1–5). Horizontal gene transfer (HGT; also called nonhomologous recombination or lateral gene transfer) occurs between and within species (6). Transformation, transduction, and plasmid-mediated gene transfers are all mechanisms through which HGT is known to occur in bacteria (7). The rate of HGT can be high, leading to substantial variation in gene content even among members of a taxonomic group (2, 8, 9). Both homologous and nonhomologous recombination can spread beneficial mutations, which are otherwise at risk of elimination due to clonal interference, and bring together independently evolved beneficial mutations (3). Recombination can result in bacterial chromosomes that are mosaics of several closely related bacterial sequences (10). Epidemic expansion of recombinant strains generates new clonal lineages (11).
Homologous recombination requires some level of DNA sequence similarity. According to the model prevailing previously, a high degree of sequence identity was important, and the efficiency of recombination decreased greatly as identity decreased (12). However, it has been shown that the level of sequence identity does not need to be constant across the gene length; rather, it is most important within minimum efficiently processed segments (MEPS), which are present in the regions flanking the inserted DNA (4). In fact, the regions between MEPS may not share sequence similarity. It has been shown, for example, that a minimum of 96 bp of flanking DNA similarity is required to initiate homologous recombination in Xylella fastidiosa (13). The need for some level of identity means that homologous recombination is highest among close relatives and across regions of the “core” genome where similarities in sequences are retained (14, 15).
Advancements in DNA sequencing technology have led to genome-based analyses of bacterial recombination. Studies of major bacterial pathogens of humans have shown the importance of recombination in their evolution (16–18). In plant pathogens, recombination has likely played a role in host shifts (19–21), the evolution of new lineages (11), and adaptation to agricultural crops (20). Exploring genomic recombination could therefore uncover how new lineages evolve and how such evolution may have impacted key pathogenicity genes. This knowledge could improve plant disease management decisions, because the mode and tempo of evolution of pathogen populations can determine the fate and durability of plant resistance genes (22, 23).
Bacterial spot disease of tomato and pepper is found around the world and is caused by four Xanthomonas species: Xanthomonas euvesicatoria, Xanthomonas perforans, Xanthomonas vesicatoria, and Xanthomonas gardneri (24, 25). Strains belonging to X. euvesicatoria and X. perforans are closely related (26) and may constitute a single species (27). Yet X. euvesicatoria and X. perforans possess different sets of secreted effectors that elicit hypersensitive reactions on differential tomato lines (26, 28). Type III secretion system effectors (T3SEs) are essential virulence factors that are secreted through the highly conserved type III secretion system (T3SS) and translocated into host plants, where they interfere with host immunity (29). Within X. perforans, phylogenetically distinct groups of strains have variable T3SE contents (30, 31). Group 1A is a monophyletic lineage of strains isolated in 2012. The reference strain for X. perforans, 91-118, represents one of multiple lineages in the paraphyletic group 1B, together with strains isolated in 2006. Group 2 is another monophyletic lineage that contains the majority of all strains sequenced from tomato in Florida, including strains isolated in 2006, 2010, 2012, and 2013 (31). Group 2 strains contain alleles of housekeeping and effector genes that are consistent with homologous recombination with X. euvesicatoria (30). This result suggests that recombination could be contributing to the genetic variation observed in X. perforans in Florida and raises the question of whether recombinant lineages of X. perforans could be found in other tomato-growing regions.
Previously, we reported the occurrence in Nigeria of atypical Xanthomonas strains causing bacterial spot of tomato (32). Routine procedures used to differentiate X. perforans strains from X. euvesicatoria strains include race typing on differential tomato lines, sequencing of the conserved T3SS gene hrcN, and tests of amylolytic and pectolytic activity (33). Strains collected in Nigeria in 2014 were identified as X. perforans because reactions on differential tomato lines indicated the presence of the X. perforans effector protein AvrXv3, sequencing of the highly conserved gene hrcN showed 100% sequence identity to hrcN in X. perforans strains, and, like X. perforans strains, these strains were amylolytic and pectolytic (32). Therefore, we were surprised when multilocus sequence analysis (MLSA) using partial sequences of six housekeeping genes clustered these Nigerian strains with X. euvesicatoria. Specifically, the gltA, lacF, and gapA sequences were identical or nearly identical to those of X. euvesicatoria, yet the fusA and gyrB genes were identical to alleles found in X. perforans group 1 strains, and the lepA allele was distinct from those in X. euvesicatoria and X. perforans (30, 32). These strains from 2014 are represented by strain NI1. NI1-like strains were isolated from the same tomato field the following year, but we also identified a second strain type in 2015. Differential reactions indicated that this second variant in our 2015 collection contained the X. euvesicatoria effector protein AvrRxv, but the hrcN gene sequence placed the strains in X. perforans (M. O. Jibrin, P. D. Roberts, J. B. Jones, and G. V. Minsavage, unpublished data). This was surprising, because no X. perforans field strain has been reported to contain the AvrRxv gene. The presence of the AvrRxv gene in representative strain NI38 was confirmed by Sanger sequencing (Jibrin et al., unpublished). In summary, genotypic and phenotypic tests typically used to assign bacterial spot strains to species could not assign the Nigerian strains to a Xanthomonas species due to conflicting race, hrcN, and/or MLSA data.
Other studies have found atypical bacterial spot strains in Europe, Asia, and Africa (34–36). The genomes of two of these strains have been sequenced previously: X. perforans strain 4P1S2, from Italy, and X. euvesicatoria strain LMG918, from India (36, 37). Preliminary analysis of the genome of strain 4P1S2 showed that it contained regions that were identical in sequence to regions of genomes of other Xanthomonas species, suggesting recombination (34). Strain LMG918 was identified as X. euvesicatoria but is amylolytic, which is not typical of X. euvesicatoria strains, and sequencing of its genome revealed unique effector gene content relative to other X. euvesicatoria strains (37). Together with our initial analysis of the strains from Nigeria, these findings suggest that there are genetic groups of X. euvesicatoria and X. perforans that are not represented in genome studies of U.S. strains.
The objectives of this study were to resolve the conflicting assignments of the Nigerian strains and to evaluate their genomic relatedness to other X. perforans and X. euvesicatoria strains. Two representative strains from the Nigerian collections, NI1 and NI38, were newly sequenced. We compared the genomes of NI1 and NI38 to 70 previously published genomes of X. euvesicatoria and X. perforans strains (26, 31, 34, 36–39). We hypothesized that homologous recombination has generated genetically diverse phylogenetic lineages of X. perforans and X. euvesicatoria, based on results from multilocus sequencing and previous whole-genome comparisons (30, 31, 37). We further hypothesized that this recombination affected T3SS and T3SE genes. We were especially interested in T3SS effector profiles, because effectors are targets of resistance breeding and are used to monitor pathogen population shifts (28). We also examined lipopolysaccharide (LPS) clusters, which can act as pathogen-associated molecular patterns (PAMPs) that elicit PAMP-triggered immunity (40–43). Our results show that recombination has driven the evolution of genetically distinct lineages of bacterial spot pathogens in Nigeria and elsewhere, suggesting that these widely distributed pathogens may show high levels of genetic diversity across global populations. Furthermore, the observed shuffling of effectors between phylogenetic lineages affects resistance breeding strategies that target specific effectors.
RESULTS
ANI and phylogenetic reconstruction.
In an initial attempt to assign the sequenced Nigerian strains to X. perforans and X. euvesicatoria, we calculated average nucleotide identity using BLAST (ANIb) for comparisons of NI1 and NI38 to X. perforans and X. euvesicatoria strains with published draft genomes (see Table S1 in the supplemental material). Average nucleotide identity (ANI) values were >99% among strains within species (within X. perforans and within X. euvesicatoria) and <99% in cross-species (X. perforans versus X. euvesicatoria) comparisons (see Table S2 in the supplemental material). The Nigerian strain NI1 showed ANI values of 98.9% and 98.5% in pairwise comparisons to the X. perforans and X. euvesicatoria reference genomes, respectively (Table 1), and <99% in comparisons to other X. perforans and X. euvesicatoria strains (see Table S2 in the supplemental material). NI38 showed ANI values typical of an X. euvesicatoria strain: >99% in comparisons to X. euvesicatoria genomes and <99% in comparisons to X. perforans genomes.
TABLE 1.
Average nucleotide identities of genomes of Nigerian, X. perforans, and X. euvesicatoria strains to the genomes of the X. perforans reference strain 91-118 and the X. euvesicatoria reference strain 85-10a
| Strain(s) | ANIbb (%) to: |
|
|---|---|---|
| Xp91-118 | Xe85-10 | |
| Nigerian strains | ||
| NI1 | 98.92 | 98.58 |
| NI38 | 98.48 | 99.79 |
| X. perforans | ||
| Xp91-118 | 100 | 98.48 |
| Group 1A strains | 99.87–99.90 | 98.47–98.50 |
| Group 1B strains | 99.70–99.86 | 98.22–98.55 |
| Group 2 strains | 99.67–99.78 | 98.44–98.62 |
| Xp4P1S2 | 99.59 | 98.44 |
| X. euvesicatoria | ||
| Xe85-10 | 98.48 | 100 |
| Other X. euvesicatoria strains | 98.32–98.44 | 99.66–99.95 |
For all pairwise comparisons, see Table S2 in the supplemental material.
ANIb, average nucleotide identity, calculated using BLAST.
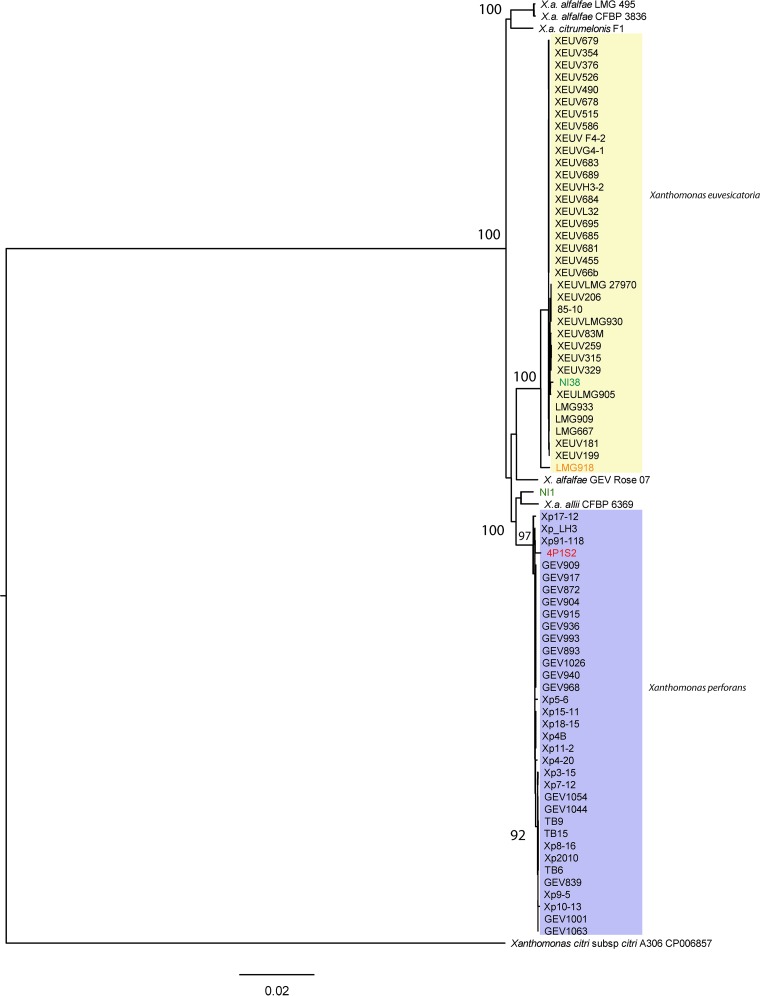
To examine the phylogenetic placement of strains NI1 and NI38, 2,205 core genes were used to construct a maximum likelihood phylogeny for X. euvesicatoria and X. perforans strains as well as other strains in the X. euvesicatoria species complex (35), rooted with Xanthomonas citri subsp. citri (Fig. 1). X. perforans and X. euvesicatoria each formed a monophyletic clade. However, the NI1 core genome was more similar to Xanthomonas axonopodis pv. allii strain CFBP6369 than to X. perforans strains (Fig. 1). NI1, X. axonopodis pv. allii CFBP6369, and X. perforans strains formed a strongly supported clade, but the phylogenetic relationships between NI1, X. axonopodis pv. allii, and X. perforans were not well supported by ultrafast bootstrap analysis. Xanthomonas alfalfae subsp. alfalfae strains LMG495 and CFBP3836 and Xanthomonas alfalfae subsp. citrumelonis strain F1 formed a third strongly supported clade, distinct from the X. euvesicatoria and the X. perforans–NI1–X. axonopodis pv. allii clade. A strain isolated from a rose plant grouped with X. euvesicatoria with poor (78%) ultrafast bootstrap support. NI38 appeared to be a typical X. euvesicatoria strain, while the core genome of the atypical Indian strain LMG918 was genetically distinct from those of all other X. euvesicatoria strains.
FIG 1.
Maximum likelihood phylogeny based on concatenated core genes from 65 genomes of X. euvesicatoria and X. perforans, rooted with Xanthomonas citri subsp. citri strain A306. X. euvesicatoria strains are highlighted in yellow and X. perforans strains in purple. Strains NI1 and NI38, from Nigeria, are shown in green letters, Italian strain 4P1S2 in red letters, and the atypical X. euvesicatoria strain LMG918 in orange letters. X. axonopodis pv. allii, and four X. alfalfae subsp. alfalfae strains have >96% ANI with X. euvesicatoria and X. perforans strains. Bar, substitutions per site. The tree was generated using IQ-TREE (v1.5.5) with the general time-reversible model and a proportion of invariant sites (GTR+I). Branch support was assessed by ultrafast bootstrap analysis using 1,000 replicates.
Pangenome variation.
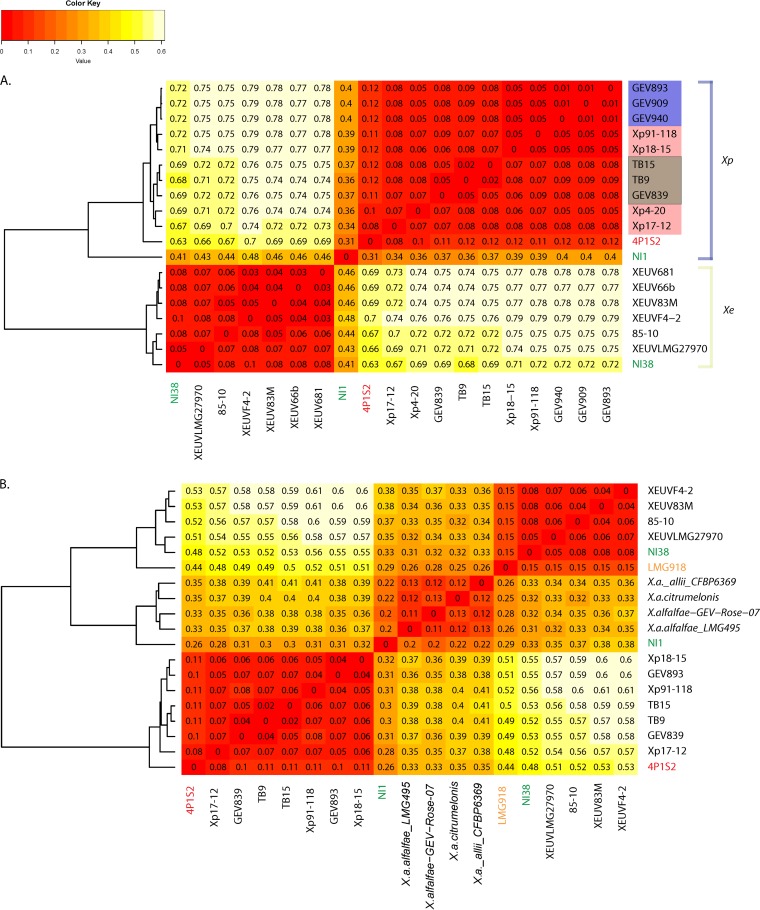
To further explore the genomic variation between Nigerian strains and other X. perforans and X. euvesicatoria strains, we examined variation in the pangenome based on our draft genome sequences. We produced a pangenome matrix and examined pairwise variation between strains using Gower's distance, which is zero when genomes have identical gene content and increases up to a maximum of 1 with increasing dissimilarity. Pangenome distances were less than 0.1 within X. perforans and within X. euvesicatoria, with a few exceptions (Fig. 2A and B). The X. perforans strain from Italy, 4P1S2, had slightly elevated distances of 0.11 to 0.12 from other known X. perforans strains (Fig. 2A), and the X. euvesicatoria strain LMG918 was also more distantly related to other X. euvesicatoria strains (distances of 0.15 [Fig. 2B]). NI38 again appeared to be a typical X. euvesicatoria strain, but the gene content of NI1 showed divergence from both X. euvesicatoria and X. perforans strains. The gene content of NI1 was only slightly more similar to that of X. perforans strains than to that of X. euvesicatoria strains (Fig. 2A) and more closely resembled that of the non-bacterial-spot taxa than that of X. perforans or X. euvesicatoria strains (Fig. 2B). While the non-bacterial-spot taxa grouped into three clades based on the core genome, they were relatively similar to each other in gene content, with Gower's distances of <0.13 (Fig. 2B).
FIG 2.
Comparison of gene content across strains. Gower's distance (dissimilarity), which is zero when genomes have identical gene content and is maximal at 1, is shown for each pairwise comparison of genomes. Pairwise distances were used to construct the dendrograms shown to the left of each heat map. (A) Comparison of genomes of Nigerian strains NI1 and NI38 (in green letters) to genomes from a subset of X. perforans and X. euvesicatoria strains. Previously described genetic groups of X. perforans are color coded: group 1A strains are highlighted in purple, group 1B strains in pink, and group 2 strains in brown. Other strain names are color coded as in Fig. 1. (B) Comparison of pangenomes between Nigerian strains and other closely related taxa, color coded as in Fig. 1.
To put these analyses into context, we examined overall heterogeneity in the core genomes and pangenomes of X. perforans and X. euvesicatoria. For both X. perforans and X. euvesicatoria, we observed a gradual decrease in core genome size with the addition of strains, and a wide distribution in core genome size depending on the genomes sampled (see Fig. S1a and c and Table S3 in the supplemental material). Likewise, pangenome size was highly dependent on the strains sampled (Fig. S1b and d; Table S3). The increasing sizes of the pangenomes as more strains were sampled indicate open pangenomes for both X. euvesicatoria and X. perforans.
Rates of homologous recombination in core genomes.
We used ClonalFrameML analyses to estimate rates of homologous recombination in the X. perforans and X. euvesicatoria lineages. Across all X. perforans and X. euvesicatoria strains, including NI1, we inferred that, overall, the rate of homologous recombination of imported DNA was less than half the rate of mutation (R/θ) (Table 2). The same result was obtained for X. euvesicatoria strains. In contrast, X. perforans strains from Florida exhibited recombination at 1.5 times the rate of mutation. When 4P1S2 and NI1 were included in the analysis of X. perforans strains, the estimated ratios of recombination to mutation rates were similar (R/θ = 0.85). In all cases, the impact of recombination on nucleotide variation was greater than the impact of mutation (R/m > 1).
TABLE 2.
Rates of recombination as calculated using ClonalFrameMLa
| Xanthomonas population | R/θ | δ (SD) | ν | R/m | Tr/Tv |
|---|---|---|---|---|---|
| All strains | 0.384 | 301 (5.0) | 0.0252 | 2.91 | 5.36 |
| All X. perforans strains (+ NI1) | 0.854 | 394 (7.7) | 0.0252 | 8.47 | 4.92 |
| Florida X. perforans strainsb | 1.535 | 696 (25.0) | 0.0245 | 26.23 | 3.47 |
| All X. euvesicatoria strains | 0.554 | 611 (13.0) | 0.0102 | 3.45 | 5.35 |
Values have been rounded for presentation. R/θ is the rate of recombination divided by the rate of mutation; δ is the size of the recombined fragment in base pairs; ν is the probability of recombination per site; R/m is the effect of recombination relative to that of mutation (or the ratio of per-base-pair substitution by recombination to that by mutation [R/m = R/θ × ν × δ]); Tr/Tv is the transition/transversion ratio used as input in the analysis.
NI1 (Nigeria) and 4P1S2 (Italy) were excluded.
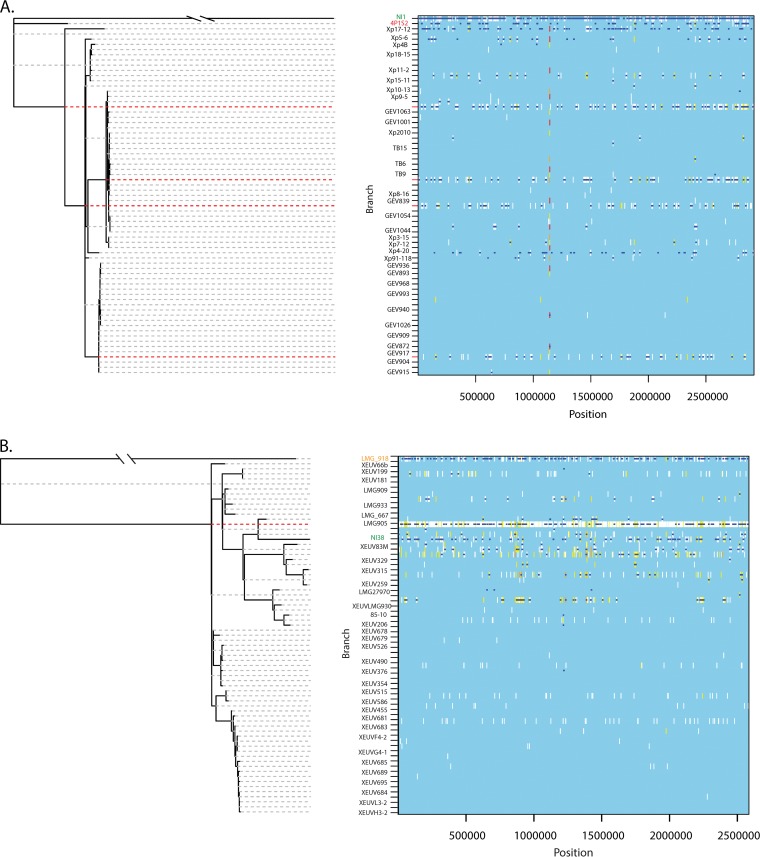
Within X. perforans, recombination events were generally inferred to have occurred in the ancestor of each clonal lineage (Fig. 3A, red dashed lines). Strain NI1 produced extensive regions of inferred recombination relative to X. perforans strains (shown by dark blue segments in Fig. 3). X. perforans strains 4P1S2 and Xp17-12 also formed distinct lineages in the clonal phylogeny and exhibited high numbers of inferred recombination events (Fig. 3A). X. euvesicatoria strain LMG918 was on a long branch in the recombination-corrected clonal phylogeny, as in the rooted core genome phylogeny, and extensive recombination was inferred (Fig. 3B). NI38 showed a longer branch in the recombination-corrected phylogeny (Fig. 3B) than in the rooted core genome phylogeny (Fig. 1), indicating both inferred recombination and mutation. However, regions of homoplasy (Fig. 3, yellow) were more common in X. euvesicatoria strains than in X. perforans strains, suggesting that there may be more undetected recombination events in X. euvesicatoria.
FIG 3.
Homologous recombination in the core genomes of X. perforans (A) and X. euvesicatoria (B), estimated by ClonalFrameML. Recombination-corrected maximum likelihood phylogenies are shown to the left. To the right of each tree, inferred recombination (dark blue horizontal lines) or mutation (vertical lines) is shown for each branch of the tree and each position across the alignment of core genes. Invariant sites are shown in light blue (i.e., the background). Polymorphic sites are shown in colors indicating their levels of homoplasy: white indicates no homoplasy, and the range from yellow to red represents increasing degrees of homoplasy (51). Strains are color coded as in Fig. 1. Rows that are not labeled represent branches (ancestral lineages) in the phylogeny. Ancestral lineages with many inferred recombination events are emphasized with red dashed lines. The corresponding analysis of all strains together is shown in Fig. S2 in the supplemental material.
Composition of LPS clusters.
Lipopolysaccharides (LPSs) are highly antigenic and often act as PAMPs, virulence factors, and defense response elicitors (39–41). LPS gene clusters in the genus Xanthomonas typically consist of 15 genes and are flanked by two conserved housekeeping genes, encoding cystathionine gamma lyase (metB) and electron transport flavoprotein (etfA) (42). A previous comparison of the LPS clusters of bacterial spot xanthomonads using genomes of reference strains showed that X. perforans has a unique LPS cluster, while X. euvesicatoria, X. vesicatoria, and X. gardneri have similar clusters (26). We observed a different gene content in the LPS cluster of NI1 than in those of X. perforans strains (Fig. 4a), which show high gene conservation and little variation among strains (Fig. 4b). For example, the NI1 LPS cluster contained two glycosyltransferase genes, which belong to a cluster of orthologous genes (COG1216, GT2 family) that is absent in typical X. perforans LPS clusters (Fig. 4a). BLAST searches of genes unique to the NI1 LPS cluster (Fig. 4a, green arrows) revealed similarity with the LPS cluster of Xanthomonas translucens pv. translucens, the causal agent of bacterial wilt and black chaff of barley (Hordeum vulgare). Of the two available X. translucens pv. translucens genome sequences, the NI1 LPS cluster was most similar to that of the X. translucens pv. translucens type strain, DSM18974, which was isolated from barley in the United States (44) (Fig. 4a). The NI38 LPS cluster was similar to that of other X. euvesicatoria strains.
FIG 4.
Variation in LPS clusters. Genes are represented by colored blocked arrows, with similar colors indicating genes belonging to the same COG (cluster of orthologous genes). (a) Comparison of NI1, NI38, X. euvesicatoria 85-10, X. perforans 91-118, and X. translucens pv. translucens DSM18974. (b) Comparison of representative LPS clusters for X. perforans groups 1A (GEV936 and GEV1026), 1B (Xp5-6, Xp91-118), and 2 (GEV1001) and the Italian strain 4P1S2. Orange arrows indicate the positions of etfA genes across the sets compared, while green arrows represent unique genes in NI1 and X. translucens pv. translucens.
Allelic variation in type III secretion system genes.
The T3SS is composed of the hypersensitivity response and pathogenicity (hrp) genes and the hrp conserved (hrc) genes (45, 46). Also present in the T3SS gene cluster are the hrp-associated (hpa) genes (45). Both Nigerian strains were identified as X. perforans based on a quantitative PCR (qPCR) assay using the hrcN gene (47); therefore, we compared gene sequences and constructed genealogies to examine allelic variation in this and other T3SS genes (Table 3; see also Table S4 and Fig. S3 in the supplemental material). NI1 generally contained X. perforans alleles for T3SS genes (Table 3). HrpB5 exhibited amino acid differences from other X. perforans strains, and hrpE, hpaB, and hrcQ showed differences at the nucleotide level. Given the similarity of the core genome and pangenome of NI1 to those of X. axonopodis pv. allii strain CFBP6369, we also compared the T3SS alleles of these strains. CFBP6369 alleles were substantially different from NI1 alleles and were more similar to those of X. alfalfae strains (Fig. S3b to e and g to j). Indeed, when NI1 and CFBP6369 shared an allele at a T3SS gene, other X. perforans isolates also exhibited the same allele (Fig. S3a and f). Strain NI38, which resembled a typical X. euvesicatoria strain in its core genome and pangenome, contained two X. perforans alleles in the hrp cluster and several unique alleles relative to both X. euvesicatoria and X. perforans strains, including the hrcN allele (Fig. S3a). X. perforans strain 4P1S2, which was divergent from other X. perforans strains in the recombination-corrected, clonal phylogeny (Fig. 3A), exhibited the most striking mixture of X. euvesicatoria and X. perforans T3SS alleles (Table 3; Fig. S3).
TABLE 3.
Allelic assignments of NI1, NI38, and 4P1S2 type III secretion system genes relative to those of the X. perforans reference strain 91-118 and the X. euvesicatoria reference strain 85-10a
| Secretion system geneb | Allele type |
|||
|---|---|---|---|---|
| NI1 | NI38 | 4P1S2 | X. axonopodis pv. allii | |
| Hrp genes | ||||
| hrpA | X. perforans | X. euvesicatoria | Unique | Unique |
| hrpB1 | Identical | Identical | Unique | Unique |
| hrpB2 | Identical | Identical | Identical | Unique |
| hrpB4 | X. perforans | X. perforans | X. perforans | Unique |
| hrpB5 | Unique | Identical | Identical | Unique |
| hrpB7 | Identical | Identical | Identical | Unique |
| hrpD6 | X. perforans | X. perforans | X. perforans | Unique |
| hrpE | Unique | Unique | X. euvesicatoria | Unique |
| hrpF | X. perforans | X. euvesicatoria | X. perforans | Unique |
| hrpG | X. perforans | X. euvesicatoria | Unique | X. perforans |
| hrpX | X. perforans | X. euvesicatoria | Unique | Unique |
| Hrc genes | ||||
| hrcA | X. perforans | X. euvesicatoria | X. perforans | Unique |
| hrcC | X. perforans | Unique | X. perforans | X. perforans |
| hrcT | Identical | Identical | Identical | Identical |
| hrcN | X. perforans | Unique | NI38 type | Unique |
| yscJ (hrcJ) | Identical | Identical | Unique | Unique |
| hrcU | Identical | Identical | Identical | Unique |
| hrcV | X. perforans | Unique | Unique | Unique |
| yscQ (hrcQ) | Unique | X. euvesicatoria | NI1 type | Unique |
| hrcR | X. perforans | X. euvesicatoria | X. euvesicatoria | Unique |
| hrcS | X. perforans | X. euvesicatoria | X. euvesicatoria | Unique |
| hrcD | X. perforans | X. euvesicatoria | X. euvesicatoria | Unique |
| Hpa genes | ||||
| hpa1 | X. perforans | X. perforans | X. perforans | Unique |
| hpa2 | X. perforans | X. perforans | X. perforans | Unique |
| hpaA | X. perforans | X. euvesicatoria | X. euvesicatoria | Unique |
| hpaB | X. perforans | X. euvesicatoria | X. euvesicatoria | X. perforans |
| hpaG | X. perforans | X. euvesicatoria | Unique | Unique |
| hpaP | X. perforans | Unique | X. perforans | Unique |
Allele types are based on nucleotide sequence identity. “Identical” indicates that the X. perforans and X. euvesicatoria alleles are identical to each other and thus cannot be distinguished. “Unique” indicates alleles not found in the reference strains. Allelic assignments based on amino acid sequence identity are shown in Table S3 in the supplemental material, because we observed both synonymous and amino acid replacement substitutions.
Hrp genes, hypersensitivity, resistance, and pathogenicity genes; Hrc genes, hypersensitivity, resistance, and conserved genes; Hpa genes, Hrp-associated genes.
Genealogies for hrpG and hrpX, which regulate the Hrp operon, were conserved in most X. perforans and X. euvesicatoria strains, but 4P1S2 and LMG918 had unique alleles for hrpG and hrpX, respectively (Fig. S3a and b in the supplemental material). We also uncovered evidence of recombination in hpa genes in other X. perforans and X. euvesicatoria strains. For hpaA, some X. perforans strains (Xp11-2, Xp4B, 4P1S2, Xp15-11, and Xp18-15) had a typical X. euvesicatoria allele, while LMG918 showed an X. perforans allele (Fig. S3e). For hpaB, we observed two major allele types. 4P1S2 shared an allele with nearly half of the X. euvesicatoria strains, including the reference strain, 85-10. The majority of X. perforans and X. euvesicatoria strains shared a different hpaB allele with X. axonopodis pv. allii CFBP3839 and the Xanthomonas alfalfae strain from a rose plant (Fig. S3f). In summary, recombination appears to have affected the T3SS genes and associated genes in NI38, 4P1S2, LMG918, and other strains.
Recombination and horizontal transfer of T3SS effector genes.
We examined both the presence or absence and allelic variation of T3SE genes, which are expected to be involved in virulence and host range. The NI1 genome contained all previously described X. perforans core effector genes (see Table S5 in the supplemental material) (26, 31). Among the effector genes that are shared by X. euvesicatoria and X. perforans but are absent in other bacterial spot xanthomonads, NI1 possessed all but the xopF2 effector (Table 4). Some effector genes have been described previously as species specific among the bacterial spot xanthomonads. NI1 had xopAJ, specific to X. euvesicatoria; xopC2, xopAE, and xopAF, specific to X. perforans; and the AvrHah1 gene and xopAQ, specific to X. gardneri. NI1 lacked xopJ4 (the AvrXv4 gene), which was found to be conserved among X. perforans strains in Florida (48). The absence of the AvrXv4 gene was confirmed in a tomato line carrying the Xv4 resistance gene (Fig. 5). Examination of allelic variation showed that NI1 had typical X. perforans alleles for the xopP, xopAK, xopF1, xopN, xopR, and xopX genes, while xopI was an X. euvesicatoria allele. For other genes, NI1 did not contain typical X. perforans or X. euvesicatoria alleles (see Fig. S4c, d, f, and i in the supplemental material). While xopAJ in NI1 shared 98.7% identity with the X. euvesicatoria allele, it was 100% identical to a copy of xopAJ found in Xanthomonas axonopodis pv. poinsettiicola. The NI1 allele of the AvrBs2 gene most closely resembled the X. axonopodis pv. allii CFBP6369 allele (Fig. S4a). X. perforans strain 4P1S2 was similar to NI1 in containing the AvrHah1 gene and xopAQ; unlike NI1, however, it contained xopJ4 (Table S5).
TABLE 4.
Effector profiles of NI1 and NI38 for species-specific effectors within the bacterial spot xanthomonads
| Effectora | Locus tagb in strain: |
|
|---|---|---|
| NI1 | NI38 | |
| Found uniquely in X. euvesicatoria | ||
| AvrBs1 | Absent | Absent |
| XopC1 | Absent | Ga0128168_10566 |
| XopJ1 | Absent | Ga0128168_10111 |
| XopJ3 (AvrRxv gene) | Absent | Ga0128168_12162 |
| XopO | Absent | Ga0128168_12403 |
| XopAA | Absent | Ga0128168_11918 |
| XopAJ | Ga0071335_10438 | Ga0128168_10823 |
| Found uniquely in X. perforans | ||
| XopC2 | Ga0071335_101102 | Ga0128168_101017, Ga0128168_101018 |
| XopJ4 | Absent | Absent |
| XopAE | Ga0071335_113188 | Ga0128168_11984, Ga0128168_11985 |
| XopAF (AvrXv3 gene) | Ga0071335_13475 | Absent |
| Found uniquely in X. gardneri | ||
| AvrBs1 | Absent | Absent |
| AvrHah1 | Ga0071335_1551/Ga0071335_1571c | Absent |
| XopAO | Absent | Absent |
| XopAQ | Ga0071335_1192U | Absent |
| XopAS | Absent | Absent |
| Found in X. euvesicatoria and X. perforans but absent in X. vesicatoria and X. gardneri | ||
| XopE1 | Ga0071335_10358U | Ga0128168_10456Xe, Ga0128168_109715U |
| XopF2 | Absent | Ga0128168_103921U |
| XopI | Ga0071335_12619Xe | Ga0128168_10765Xe |
| XopP | Ga0071335_101103Xp, Ga0071335_13041Xp, Ga0071335_1451U, Ga0071335_1461U | Ga0128168_101020U, Ga0128168_101021Xe |
| XopV | Ga0071335_13449U | Ga0128168_100834NI1 |
| XopAK | Ga0071335_111115U | Ga0128168_11919Xe |
| XopAP | Ga0071335_107226U | Ga0128168_100746Xe |
| Found in X. euvesicatoria, X. perforans, and X. vesicatoria: AvrBsT (YopJ) | Ga0071335_1442U | Absent |
| Found in X. gardneri and X. euvesicatoria but absent in X. perforans and X. vesicatoria: XopB | Absent | Ga0128168_114314 |
Effectors unique to other species combinations, such as X. gardneri and X. vesicatoria, which were not found in NI1 or NI38, are not shown.
Superscript Xe, Xp, U, and NI1 indicate the X. euvesicatoria, X. perforans, unique, and NI1 allele types, respectively. For effectors present only in one species, the allele type is that of the species unless otherwise indicated.
The coding region for the AvrHah1 gene is split into two locus tags located on different contigs.
FIG 5.

Differential reactions of tomato lines differential for AvrXv4 to Florida X. perforans strain Xp2010 (with the AvrXv4 gene) and a Nigerian X. perforans strain without the AvrXv4 gene. Xp2010 elicits a hypersensitive reaction on the differential line (left), while NI1 does not (right). For each strain, a bacterial suspension at a concentration of 108 CFU/ml was infiltrated on the abaxial side of the leaf, which was observed for hypersensitive reactions within 24 h.
NI38 had the bacterial spot core effector genes, together with xopE1 and xopP, which are effectors found exclusively in X. euvesicatoria and X. perforans among the bacterial spot species (see Table S6 in the supplemental material). Of the species-specific effector genes, NI38 had all the X. euvesicatoria-specific effector genes except the AvrBs1 gene, and two copies each of xopC2 and xopAE, specific to X. perforans (Table 4). Like other X. euvesicatoria strains, NI38 had xopB, the only effector shared by X. euvesicatoria and X. gardneri with 100% amino acid identity. The xopAJ allele in NI38 was identical to a copy of xopAJ in X. axonopodis pv. poinsettiicola but different from a copy of the xopAJ gene in NI1. The only X. euvesicatoria effector tentatively missing in the NI38 genome was xopG, encoding an effector common to X. euvesicatoria, X. vesicatoria, and X. gardneri. While NI38 had typical X. euvesicatoria alleles, it was strain LMG918 that showed divergence at multiple effector genes (Fig. S4 in the supplemental material).
DISCUSSION
Core genome phylogenies clarify taxonomic assignments of NI1 and NI38.
Whole-genome sequencing resolved previous conflicting multilocus sequence analyses and race differentiation tests of strains NI1 and NI38, which are representative of 2014 and 2015 collections from tomato fields in Nigeria. We identified NI38 as a typical X. euvesicatoria strain by use of core genome phylogenetic analysis and as a putatively novel lineage of X. euvesicatoria on the basis of a recombination-corrected genealogy and T3SS genes. Strain NI1 has a more complex evolutionary history. The rooted core genome phylogeny that included genomes from closely related strains showed NI1 to be more closely related to X. axonopodis pv. allii strain CFBP6369 than to X. perforans. The taxa X. axonopodis pv. allii, X. perforans, X. euvesicatoria, and X. alfalfae were previously classified into separate species, but a recent revision of the Xanthomonas axonopodis clade suggested that all be classified as pathovars of X. euvesicatoria based on ANI (26). NI1, X. perforans strains, and X. axonopodis pv. allii CFBP6369 descend from the same node, indicating recent common ancestry, but there was not strong support for the relationships among these lineages. Genealogies of T3SS and effector genes produced different evolutionary relationships between NI1 and X. axonopodis pv. allii alleles, suggesting that genes important in the host-pathogen interaction may have been horizontally transferred. We suggest that NI1 contains gene content from a Xanthomonas lineage closely related to, but distinct from, X. perforans and X. euvesicatoria. Recombination among genetically divergent lineages created a novel lineage of the bacterial spot pathogen. Indeed, NI1 has acquired effector genes from other bacterial spot pathogens: for example, the AvrHah1 gene and xopAQ from X. gardneri. NI1 has likely not shifted from onion, the host of X. axonopodis pv. allii, to tomato, because these strains are different in effector profiles and alleles of core effector genes.
Other published genomes of X. perforans and X. euvesicatoria strains showed genetic divergence in the core genome or T3SS genes from the X. perforans and X. euvesicatoria reference strains, which were collected in Florida in 1991 and 1985, respectively. Strain 4P1S2, isolated from a tomato plant with pith necrosis in Italy, showed slight divergence from other X. perforans strains in the pangenome and had a unique complement of effectors. LMG918 was isolated in India and has been described as an atypical X. euvesicatoria strain because it was positive for the pectolytic test and had only one X. euvesicatoria-specific effector, XopC1, and a X. perforans-specific effector, XopAE (37). We found that LMG918 is also a unique strain based on the core genome and effector alleles. Our recombination analyses showed that extensive recombination has likely shaped the genome of LMG918, including the T3SS and effector genes.
Recombination has shaped the evolution of the X. euvesicatoria and X. perforans lineages.
Our results show that recombination is a major driver of the evolution of both X. euvesicatoria and X. perforans. We found evidence of homologous recombination and horizontal gene transfer. Rates of homologous recombination differed within and between species; X. perforans showed more evidence of recombination than X. euvesicatoria, which is perhaps most obvious in the distinct, recombinant lineages of X. perforans. Our analysis also detected higher recombination rates within the Florida population of X. perforans than within X. euvesicatoria strains and the larger sample of X. perforans strains. Most of the X. euvesicatoria strains were collected in the southeastern United States, but over a longer span of time than the X. perforans strains (31). Homologous recombination is more likely to be detected in contemporaneous population samples, in which one is catching recombination in action, than in a time series, which also captures selection acting on the recombinant population (49). Recombination might be especially frequent in Florida populations, or our inclusion of single strains representing divergent lineages may have caused poor estimation of recombination in X. perforans as a whole. Additional strains from populations representing the divergent lineages will be required in order to determine if homologous recombination rates are as high in other regions.
Both X. perforans and X. euvesicatoria appear to have open and highly dynamic pangenomes, in contrast to the stability in gene composition implied when Xanthomonas bacterial spot pathogens are characterized by single reference genomes. Our data indicate that obtaining pangenomes for Xanthomonas populations will be important for understanding the ongoing evolution of these pathogens.
Horizontal gene transfer and recombination generate genome variation that likely contributes to adaptive divergence and the evolution of new bacterial spot lineages, as has been reported for other bacterial systems, such as Bartonella henselae and Clostridium difficile ST6 (50, 51). The observation of extensive chromosomal replacement in NI1 and other strains was unexpected and mirrors findings in Staphylococcus aureus, in which multiple chromosomal replacements were found in new strains relative to the reference strain, MRSA252 (51). The recombinant bacterial spot strains were collected from different continents than the reference strains of X. euvesicatoria and X. perforans, suggesting that these bacterial spot pathogens may have very dynamic populations across the globe.
Recombination shuffled secretion system and effector genes among lineages.
Recombination affected T3SS (hrp and hrc), associated (hpa), and effector genes in NI1 and NI38, as well as other X. euvesicatoria and X. perforans lineages. Generally, characterizations of secretion systems in bacterial species have focused on reference genomes (26, 52–54), and T3SS genes tend to be viewed as conserved elements of pathogenic bacteria. We found that recombination generates intraspecific variation in T3SS alleles, as indicated by genealogies that are incongruent among T3SS genes and with the core genome tree. The significance of the shuffling of secretion system genes between lineages remains to be experimentally tested for bacterial pathogens in general and bacterial spot xanthomonads in particular.
Recombination in T3SS genes explains why NI38 was initially identified as an X. perforans strain based on qPCR primers targeting the highly conserved hrcN gene (47). The hrcN allele in NI38 is distinct from those in X. euvesicatoria and X. perforans reference strains and is identical to that of 4P1S2; they both possess a single nucleotide substitution that is not present in X. perforans strains. The nucleotide sequences where the diagnostic oligonucleotides anneal in hrcN are identical for NI38 and X. perforans strains but differ by two nucleotides from those of other X. euvesicatoria strains. This study demonstrates that the primers have not correctly distinguished X. euvesicatoria and X. perforans strains, and it points to the limitations of pathogen identification using a single locus.
The recombination and presence/absence polymorphisms of T3SE genes in NI1, NI38, and 4P1S2 raise concerns regarding which effectors to use as targets for breeding durable resistance. One strategy being used to determine candidate targets for resistance breeding is to identify core, conserved effectors. Recently, comparison of sequenced genomes of X. perforans strains from Florida identified XopJ4 and AvrBST as putative targets for resistance breeding (48). The lack of XopJ4 in NI1 and of AvrBST in 4P1S2 makes this a less viable long-term strategy, due to the possibility of introduction of these strains into Florida. The observation of allelic variation in effector genes further complicates decisions regarding effector targets for resistance breeding, because the host resistance response may differ among alleles. For example, allelic variation in the Pseudomonas syringae HopZ family of T3SS effectors causes differential reactions in soybean and between soybean and Nicotiana benthamiana (55). New combinations of effectors and their alleles could be important for adaptation to local hosts and/or environments (11). Our findings suggest that bacterial spot xanthomonads may be evolving in response to conditions in local populations and that recombination potentially plays an important role in this evolution. Various effector gene profiles across tomato production regions suggest that global studies are needed in order to better understand the ecological and evolutionary processes that determine effector profiles and to identify stable targets for effector-based resistance.
Recombination as a diversifying and cohesive force in bacterial spot xanthomonad evolution.
The close genetic distance but distinct lineages of X. euvesicatoria and X. perforans are relevant to the ongoing debates on what defines a bacterial species. One view is that genetic exchange rarely hinders genetic divergence in bacterial populations (56). Rather, recombination fosters the acquisition of novel genes and operons that aid in adaptation, thereby promoting divergence (5, 57). A competing hypothesis suggests that recombination is a cohesive force that prevents bacterial diversification and maintains lineages by homogenizing populations (57, 58). Both processes may be occurring in these Xanthomonas populations. Thus far, most of the lineages of X. euvesicatoria and X. perforans can easily be assigned to species, while recombination has also led to the emergence of genetically distinct lineages within species. We also observed that genes involved in virulence are recombining among taxa, which may maintain ecological cohesion among diverging lineages of bacterial-spot pathogens. Exchange of ecologically and evolutionarily significant genes among phylogenetic lineages has been found to lend cohesion to the P. syringae species complex (59).
The sympatric distribution of NI1 and NI38 is surprising, because X. perforans strains are known to outcompete X. euvesicatoria strains under field conditions (60). Previously in Nigeria, typical X. euvesicatoria strains were found in adjoining pepper fields, not from tomato (61). The fact that both were isolated from tomato in the same field suggests that these strains may occupy slightly different ecological niches. Important phenotypic differences also remain between X. euvesicatoria and X. perforans strains. There are practical and academic needs to distinguish them. The ecotype model, which defines species by their ecology (56), may be an appropriate concept for X. euvesicatoria and X. perforans, if we define ecotype by multidimensional ecological niche, not just host range. The ecotype model essentially defines ecotypes by their ability to coexist on the same or similar ecological resources and to persist through periodic selection events. Additional studies of the genetic diversity of locally evolved lineages and larger population samples could provide insight into factors that drive and maintain multiple lineages within tomato fields.
MATERIALS AND METHODS
Sequencing of strains and calculation of ANI.
Strains NI1 and NI38 were selected as representative strains of the Nigerian population for whole-genome sequencing. After the extraction of genomic DNA (62), the Nextera library preparation kit (Illumina Inc., San Diego, CA) was used to prepare genomic libraries, which were subsequently sequenced using the Illumina MiSeq platform. NI1 was sequenced at Kansas State University, while NI38 was sequenced at the Interdisciplinary Center for Biotechnology Research, University of Florida. Draft genomes for NI1 and NI38 were assembled de novo using CLC Genomics Workbench, v5. Whole-genome information for the newly sequenced strains is given in Table S7 in the supplemental material. GenBank accession numbers for additional previously sequenced genomes used for this study are provided in Table S1 in the supplemental material. Pairwise average nucleotide identity (ANI) for the strains from Nigeria, based on BLAST, was calculated using JSpecies, v1.2.1 (63). ANI values of previously published genomes can be found in the work of Barak et al. (37) and Richard et al. (39).
Alignment and phylogenetic inference using core genomes.
For phylogenetic analysis of core genomes, core genes were extracted using GET_HOMOLOGUES (64) and were filtered using an in-house Python script to select for single-copy-number orthologous genes. Alignments were carried out for each gene individually using MUSCLE (65) before concatenation. Gaps were removed from the concatenated core genome alignment using trimAl utilizing the “-nogaps” option (66). We used the concatenated core genome alignment to infer a maximum likelihood phylogeny with IQ-TREE, using ModelFinder for the selection of the best DNA substitution model (67, 68). Based on the Bayesian information criterion and the corrected Akaike information criterion, the general time-reversible model with a proportion of invariant sites (GTR+I) had the best fit out of a total of 88 models compared. Branch support was assessed by ultrafast bootstrap analysis and a SH-aLRT test using 1,000 replicates (69). The maximum likelihood tree was visualized using FigTree, v1.4.3 (http://tree.bio.ed.ac.uk/software/figtree/). Similar analyses were carried out to generate genealogies for single orthologous genes that make up the T3SS and associated effectors.
Pangenome and core genome analyses.
Pan- and core genome inferences were carried out with the GET_HOMOLOGUE program, using default settings (70). All-against-all BLASTP, OrthoMCL, and COG (cluster of orthologous genes) clustering were carried out to generate pan- and core gene clusters before other functions within the program were used to analyze pangenomes and core genomes (71, 72). Pangenome matrices (gene presence/absence) were generated using the subset of strains shown in Fig. 1 (due to computational limitations) to compare the gene contents of NI1 and NI38 to those of other strains. From the pangenome matrix, Gower's distance was calculated and was visualized using a dendrogram and heat map generated using the hcluster_matrix.sh function in get_homologues.pl. Genomes were resampled to estimate the sizes of core genomes and pangenomes for X. euvesicatoria and X. perforans. For these analyses, strains were added in the order shown in Table S3 in the supplemental material. Fitted exponential-decay functions were applied to resampled genomes as described in references 8 and 72.
Analysis of homologous recombination.
We carried out analysis of homologous recombination using ClonalFrameML on X. perforans and X. euvesicatoria strains. ClonalFrameML models homologous recombination as imports from external populations and uses a hidden Markov model to estimate the influence of this recombination on nucleotide variation (51). The three major outputs of ClonalFrameML are a phylogeny whose branch lengths are corrected to account for recombination, a genomic map to show where recombination took place, and an estimation of key parameters of the recombination process (51). For ClonalFrameML analyses, we used maximum likelihood trees generated from IQ-TREE and core genome alignments as infiles. Transition/transversion ratios were determined by PhyML (73). We conducted analyses using different combinations of strains as shown in Table 2.
Genome comparisons: LPS clusters, effectors, and secretion systems.
Lipopolysaccharide (LPS) clusters, T3SS genes, and T3SEs were compared among strains at both the nucleotide and amino acid levels. Genes and proteins orthologous to known LPS clusters, T3SS genes, and effectors were identified by BLAST using the IMG platform (https://img.jgi.doe.gov/) (74), EDGAR (edgar.computational.bio.uni-giessen.de) (75), and local databases. Allele assignments followed those of previous studies of X. euvesicatoria and X. perforans (26, 31).
Accession number(s).
De novo-assembled draft genomes for NI1 and NI38 have been deposited in the NCBI database under BioProject accession numbers PRJNA389556 and PRJNA391473, respectively.
Supplementary Material
ACKNOWLEDGMENTS
We thank Frank White for support in generating genome sequence data, Xavier Didelot for additional help in interpreting the results of ClonalFrameML, Jose Huguet Tapia for Python scripts used in implementing get_homologues, and the reviewers for comments that improved the manuscript.
This work was funded in part by USDA NIFA award number 2015-51181-24312.
We declare no conflict of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00136-18.
REFERENCES
- 1.Shapiro BJ. 2016. How clonal are bacteria over time? Curr Opin Microbiol 31:116–123. doi: 10.1016/j.mib.2016.03.013. [DOI] [PubMed] [Google Scholar]
- 2.Didelot X, Maiden MCJ. 2010. Impact of recombination on bacterial evolution. Trends Microbiol 18:315–322. doi: 10.1016/j.tim.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chaguza C, Cornick JE, Everett DB. 2015. Mechanisms and impact of genetic recombination in the evolution of Streptococcus pneumoniae. Comput Struct Biotechnol J 13:241–247. doi: 10.1016/j.csbj.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hanage WP. 2016. Not so simple after all: bacteria, their population genetics, and recombination. Cold Spring Harb Perspect Biol 8:a018069. doi: 10.1101/cshperspect.a018069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Melendrez MC, Becraft ED, Wood JM, Olsen MT, Bryant DA, Heidelberg JF, Rusch DB, Cohan FM, Ward DM. 2015. Recombination does not hinder formation or detection of ecological species of Synechococcus inhabiting a hot spring cyanobacterial mat. Front Microbiol 6:1540. doi: 10.3389/fmicb.2015.01540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ambur OH, Engelstadter J, Johnsen PJ, Miller EL, Rozen DE. 2016. Steady at the wheel: conservative sex and the benefits of bacterial transformation. Philos Trans R Soc Lond B Biol Sci 371:0528. doi: 10.1098/rstb.2015.0528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thomas CM, Nielsen KM. 2005. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat Rev Microbiol 3:711–721. doi: 10.1038/nrmicro1234. [DOI] [PubMed] [Google Scholar]
- 8.Tettelin H, Masignani V, Cieslewicz MJ, Donati C, Medini D, Ward NL, Angiuoli SV, Crabtree J, Jones AL, Durkin AS, Deboy RT, Davidsen TM, Mora M, Scarselli M, Margarit y Ros I, Peterson JD, Hauser CR, Sundaram JP, Nelson WC, Madupu R, Brinkac LM, Dodson RJ, Rosovitz MJ, Sullivan SA, Daugherty SC, Haft DH, Selengut J, Gwinn ML, Zhou L, Zafar N, Khouri H, Radune D, Dimitrov G, Watkins K, O'Connor KJ, Smith S, Utterback TR, White O, Rubens CE, Grandi G, Madoff LC, Kasper DL, Telford JL, Wessels MR, Rappuoli R, Fraser CM. 2005. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome.” Proc Natl Acad Sci U S A 102:13950–13955. doi: 10.1073/pnas.0506758102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vos M, Hesselman MC, Beek TA, van Passel MWJ, Eyre-Walker A. 2015. Rates of lateral gene transfer in prokaryotes: high but why? Trends Microbiol 23:598–605. doi: 10.1016/j.tim.2015.07.006. [DOI] [PubMed] [Google Scholar]
- 10.Marttinen P, Hanage WP, Croucher NJ, Connor TR, Harris SR, Bentley SD, Corander J. 2012. Detection of recombination events in bacterial genomes from large population samples. Nucleic Acids Res 40:e6. doi: 10.1093/nar/gkr928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Monteil CL, Yahara K, Studholme DJ, Mageiros L, Méric G, Swingle B, Morris CE, Vinatzer BA, Sheppard SK. 2016. Population-genomic insights into emergence, crop adaptation and dissemination of Pseudomonas syringae pathogens. Microb Genom 2(10):e000089. doi: 10.1099/mgen.0.000089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Majewski J, Cohan FM. 1999. DNA sequence similarity requirements for interspecific recombination in Bacillus. Genetics 153:1525–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kung SH, Retchless AC, Kwan JY, Almeida RP. 2013. Effects of DNA size on transformation and recombination efficiencies in Xylella fastidiosa. Appl Environ Microbiol 79:1712–1717. doi: 10.1128/AEM.03525-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Everitt RG, Didelot X, Batty EM, Miller RR, Knox K, Young BC, Bowden R, Auton A, Votintseva A, Larner-Svensson H, Charlesworth J, Golubchik T, Ip CL, Godwin H, Fung R, Peto TE, Walker AS, Crook DW, Wilson DJ. 2014. Mobile elements drive recombination hotspots in the core genome of Staphylococcus aureus. Nat Commun 5:3956. doi: 10.1038/ncomms4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dixit PD, Pang TY, Studier FW, Maslov S. 2015. Recombinant transfer in the basic genome of Escherichia coli. Proc Natl Acad Sci U S A 112:9070–9075. doi: 10.1073/pnas.1510839112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Didelot X, Bowden R, Street T, Golubchik T, Spencer C, McVean G, Sangal V, Anjum MF, Achtman M, Falush D, Donelly P. 2011. Recombination and population structure in Salmonella enterica. PLoS Genet 7(7):e1002191. doi: 10.1371/journal.pgen.1002191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Driebe EM, Sahl JW, Roe C, Bowers JR, Schupp JM, Gillece JD, Kelley E, Price LB, Pearson TR, Hepp CM, Brzoska PM, Cummings CA, Furtado MR, Andersen PS, Stegger M, Engelthaler DM, Keim PS. 2015. Using whole genome analysis to examine recombination across diverse sequence types of Staphylococcus aureus. PLoS One 10(7):e0130955. doi: 10.1371/journal.pone.0130955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yahara K, Didelot X, Jolley KA, Kobayashi I, Maiden MCJ, Sheppard SK, Falush D. 2016. The landscape of realized homologous recombination in pathogenic bacteria. Mol Biol Evol 33:456–471. doi: 10.1093/molbev/msv237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gordon JL, Lefeuvre P, Escalon A, Barbe V, Cruveiller S, Gagnevin L, Pruvost O. 2015. Comparative genomics of 43 strains of Xanthomonas citri pv. citri reveals the evolutionary events giving rise to pathotypes with different host ranges. BMC Genomics 16:1098. doi: 10.1186/s12864-015-2310-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang CL, Pu PH, Huang HJ, Sung HM, Liaw HJ, Chen YM, Chen CM, Huang MB, Osada N, Gojobori T, Pai TW, Chen YT, Hwang CC, Chiang TY. 2015. Ecological genomics in Xanthomonas: the nature of genetic adaptation with homologous recombination and host shifts. BMC Genomics 16:188. doi: 10.1186/s12864-015-1369-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Midha S, Bansal K, Kumar S, Girija AM, Mishra D, Brahma K, Laha GS, Sundaram RM, Sonti RV, Patil PB. 2017. Population genomic insights into variation and evolution of Xanthomonas oryzae pv. oryzae. Sci Rep 7:40694. doi: 10.1038/srep40694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ryan RP, Vorhölter FJ, Potnis N, Jones JB, Van Sluys MA, Bogdanove AJ, Dow JM. 2011. Pathogenomics of Xanthomonas: understanding bacterium-plant interactions. Nat Rev Microbiol 9:344–355. doi: 10.1038/nrmicro2558. [DOI] [PubMed] [Google Scholar]
- 23.McDonald BA, Linde C. 2002. Pathogen population genetics, evolutionary potential, and durable resistance. Annu Rev Phytopathol 40:349–379. doi: 10.1146/annurev.phyto.40.120501.101443. [DOI] [PubMed] [Google Scholar]
- 24.Jones JB, Stall R, Bouzar H. 1998. Diversity among xanthomonads pathogenic on pepper and tomato. Annu Rev Phytopathol 36:41–58. doi: 10.1146/annurev.phyto.36.1.41. [DOI] [PubMed] [Google Scholar]
- 25.Jones JB, Lacy GH, Bouzar H, Stall RE, Schaad NW. 2004. Reclassification of the xanthomonads associated with bacterial spot disease of tomato and pepper. Syst Appl Microbiol 27:755–762. doi: 10.1078/0723202042369884. [DOI] [PubMed] [Google Scholar]
- 26.Potnis N, Krasileva K, Chow V, Almeida N, Patil P, Ryan R, Sharlach M, Behlau F, Dow JM, Momol M, White FF, Preston JF, Vinatzer BA, Koebnik R, Setubal JC, Norman DJ, Staskawicz BJ, Jones JB. 2011. Comparative genomics reveals diversity among xanthomonads infecting tomato and pepper. BMC Genomics 12:146. doi: 10.1186/1471-2164-12-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Constantin EC, Cleenwerck I, Maes M, Baeyen S, Van Malderghem C, De Vos P, Cottyn B. 2016. Genetic characterization of strains named as Xanthomonas axonopodis pv. dieffenbachiae leads to a taxonomic revision of the X. axonopodis species complex. Plant Pathol 65:792–806. doi: 10.1111/ppa.12461. [DOI] [Google Scholar]
- 28.Stall RE, Jones JB, Minsavage GV. 2009. Durability of resistance in tomato and pepper to xanthomonads causing bacterial spot. Annu Rev Phytopathol 47:265284. doi: 10.1146/annurev-phyto-080508-081752. [DOI] [PubMed] [Google Scholar]
- 29.White FF, Potnis N, Jones JB, Koebnik R. 2009. The type III effectors of Xanthomonas. Mol Plant Pathol 10:749–766. doi: 10.1111/j.1364-3703.2009.00590.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Timilsina S, Jibrin MO, Potnis N, Minsavage GV, Kebede M, Schwartz A, Bart R, Staskawicz BJ, Boyer C, Vallad GE, Pruvost O, Jones JB, Goss EM. 2015. Multilocus sequence analysis of xanthomonads causing bacterial spot of tomato and pepper plants reveals strains generated by recombination among species and recent global spread of Xanthomonas gardneri. Appl Environ Microbiol 81:1520–1529. doi: 10.1128/AEM.03000-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwartz AR, Potnis N, Timilsina S, Wilson M, Patané J, Martins J Jr, Minsavage GV, Dahlbeck D, Akhunova A, Almeida N, Vallad GE, Barak JD, White FF, Miller SA, Ritchie D, Goss EM, Bart RS, Setubal JC, Jones JB, Staskawicz BJ. 2015. Phylogenomics of Xanthomonas field strains infecting pepper and tomato reveals diversity in effector repertoires and identifies determinants of host specificity. Front Microbiol 6:535. doi: 10.3389/fmicb.2015.00535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jibrin MO, Timilsina S, Potnis N, Minsavage GV, Shenge KC, Akpa AD, Alegbejo MD, Beed F, Vallad GE, Jones JB. 2015. First report of atypical Xanthomonas euvesicatoria strains causing bacterial spot of tomato in Nigeria. Plant Dis 99:415. doi: 10.1094/PDIS-09-14-0952-PDN. [DOI] [PubMed] [Google Scholar]
- 33.Obradovic A, Mavridis A, Rudolph K, Janse JD, Arsenijevic M, Jones JB, Minsavage GV, Wang JF. 2004. Characterization and PCR-based typing of Xanthomonas campestris pv. vesicatoria from peppers and tomatoes in Serbia. Eur J Plant Pathol 110:285–292. doi: 10.1023/B:EJPP.0000019797.27952.1d. [DOI] [Google Scholar]
- 34.Vancheva T, Lefeuvre P, Bogatzevska N, Moncheva P, Koebnik R. 2015. Draft genome sequences of two Xanthomonas euvesicatoria strains from the Balkan Peninsula. Genome Announc 3(1):e01528-14. doi: 10.1128/genomeA.01528-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mbega ER, Mabagala RB, Adriko J, Lund OS, Wulff EG, Mortensen CN. 2012. Five species of xanthomonads associated with bacterial leaf spot symptoms in tomato from Tanzania. Plant Dis; 96:760. doi: 10.1094/PDIS-01-12-0105-PDN. [DOI] [PubMed] [Google Scholar]
- 36.Torelli E, Aiello D, Polizzi G, Firrao G, Cirvilleri G. 2015. Draft genome of a Xanthomonas perforans strain associated with pith necrosis. FEMS Microbiol Lett 362(4):1–3. doi: 10.1093/femsle/fnv001. [DOI] [PubMed] [Google Scholar]
- 37.Barak JD, Vancheva T, Lefeuvre P, Jones JB, Timilsina S, Minsavage GV, Vallad GE, Koebnik R. 2016. Whole-genome sequences of Xanthomonas euvesicatoria strains clarify taxonomy and reveal a stepwise erosion of type 3 effectors. Front Plant Sci 7:1805. doi: 10.3389/fpls.2016.01805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thieme F, Koebnik R, Bekel T, Berger C, Boch J, Büttner D, Caldana C, Gaigalat L, Goesmann A, Kay S, Kirchner O, Lanz C, Linke B, McHardy AC, Meyer F, Mittenhuber G, Nies DH, Niesbach K, Patschkowski T, Rückert C, Rupp O, Schneiker S, Schuster SC, Vorhölter FJ, Weber E, Pühler A, Bonas U, Bartels D, Kaiser O. 2005. Insights into genome plasticity and pathogenicity of the plant pathogenic bacterium Xanthomonas campestris pv. vesicatoria revealed by the complete genome sequence. J Bacteriol 187:7254–7266. doi: 10.1128/JB.187.21.7254-7266.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Richard D, Boyer C, Lefeuvre P, Canteros BI, Beni-Madhu S, Portier P, Pruvost O. 2017. Complete genome sequences of six copper-resistant Xanthomonas strains causing bacterial spot of solaneous plants, belonging to X. gardneri, X. euvesicatoria, and X. vesicatoria, using long-read technology. Genome Announc 5:e01693-16. doi: 10.1128/genomeA.01693-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dharmapuri S, Yashitola J, Vishnupriya MR, Sonti RV. 2001. Novel genomic locus with atypical G+C content that is required for extracellular polysaccharide production and virulence in Xanthomonas oryzae pv. oryzae Mol Plant Microbe Interact. 14:1335–1339. doi: 10.1094/MPMI.2001.14.11.1335. [DOI] [PubMed] [Google Scholar]
- 41.Keshavarzi M, Soylu S, Brown I, Bonas U, Nicole M, Rossiter J, Mansfield J. 2004. Basal defenses induced in pepper by lipopolysaccharides are suppressed by Xanthomonas campestris pv. vesicatoria. Mol Plant Microbe Interact 17:805–815. doi: 10.1094/MPMI.2004.17.7.805. [DOI] [PubMed] [Google Scholar]
- 42.Newman MA, Dow JM, Molinaro A, Parrilli M. 2007. Priming, induction and modulation of plant defense responses by bacterial lipopolysaccharides. J Endotoxin Res 13:68–79. doi: 10.1177/0968051907079399. [DOI] [PubMed] [Google Scholar]
- 43.Vorhölter FJ, Niehaus K, Puhler A. 2001. Lipopolysaccharide biosynthesis in Xanthomonas campestris pv. campestris: a cluster of 15 genes is involved in the biosynthesis of the LPS O-antigen and the LPS core. Mol Genet Genomics 266:79–95. doi: 10.1007/s004380100521. [DOI] [PubMed] [Google Scholar]
- 44.Jaenicke S, Bunk B, Wibberg D, Spröer C, Hersemann L, Blom J, Winkler A, Schatschneider S, Albaum SP, Kolliker R, Goesmann A, Puhler A, Overmann J, Vorhölter F-J. 2016. Complete genome sequence of the barley pathogen Xanthomonas translucens pv. translucens DSM 18974T (ATCC 19319T). Genome Announc 4(6):e01334-16. doi: 10.1128/genomeA.01334-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lohou D, Lonjon F, Genin S, Vailleau F. 2013. Type III chaperones & Co. in bacterial plant pathogens: a set of specialized bodyguards mediating effector delivery. Front Plant Sci 4:e435. doi: 10.3389/fpls.2013.00435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guo Y, Figueiredo F, Jones J, Wang N. 2011. HrpG and HrpX play global roles in coordinating different virulence traits of Xanthomonas axonopodis pv. citri. Mol Plant Microbe Interact 24:649–661. doi: 10.1094/MPMI-09-10-0209. [DOI] [PubMed] [Google Scholar]
- 47.Strayer AL, Jeyaprakash A, Minsavage GV, Timilsina S, Vallad GE, Jones JB, Paret ML. 2016. A multiplex real-time PCR assay differentiates four Xanthomonas species associated with bacterial spot of tomato. Plant Dis 100:1660–1668. doi: 10.1094/PDIS-09-15-1085-RE. [DOI] [PubMed] [Google Scholar]
- 48.Timilsina S, Abrahamian P, Potnis N, Minsavage GV, White FF, Staskawicz BJ, Jones JB, Vallad GE, Goss EM. 2016. Analysis of sequenced genomes of Xanthomonas perforans identifies candidate targets for resistance breeding in tomato. Phytopathology 106:1097–1104. doi: 10.1094/PHYTO-03-16-0119-FI. [DOI] [PubMed] [Google Scholar]
- 49.Martin DP, Lemey P, Posada D. 2011. Analysing recombination in nucleotide sequences. Mol Ecol Resour 11:943–955. doi: 10.1111/j.1755-0998.2011.03026.x. [DOI] [PubMed] [Google Scholar]
- 50.Guy L, Nystedt B, Sun Y, Näslund K, Berglund EC, Andersson SG. 2012. A genome-wide study of recombination rate variation in Bartonella henselae. BMC Evol Biol 12:65. doi: 10.1186/1471-2148-12-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Didelot X, Wilson DJ. 2015. ClonalFrameML: efficient inference of recombination in whole bacterial genomes. PLoS Comput Biol 11(2):e1004041. doi: 10.1371/journal.pcbi.1004041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vandroemme J, Cottyn B, Baeyen S, Vos PD, Maes M. 2013. Draft genome sequence of Xanthomonas fragariae reveals reductive evolution and distinct virulence-related gene content. BMC Genomics 14:829. doi: 10.1186/1471-2164-14-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Costa TR, Felisberto-Rodrigues C, Meir A, Prevost MS, Redzej A, Trokter M, Waksman G. 2015. Secretion systems in Gram-negative bacteria: structural and mechanistic insights. Nat Rev Microbiol 13:343–359. doi: 10.1038/nrmicro3456. [DOI] [PubMed] [Google Scholar]
- 54.Green ER, Mecsas J. 2016. Bacterial secretion systems: an overview. Microbiol Spectr doi: 10.1128/microbiolspec.VMBF-0012-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhou H, Morgan RL, Guttman DS, Ma W. 2009. Allelic variants of the Pseudomonas syringae type III effector HopZ1 are differentially recognized by plant resistance systems. Mol Plant Microbe Interact 22:176–189. doi: 10.1094/MPMI-22-2-0176. [DOI] [PubMed] [Google Scholar]
- 56.Cohan FM. 2002. What are bacterial species? Annu Rev Microbiol 56:457–487. doi: 10.1146/annurev.micro.56.012302.160634. [DOI] [PubMed] [Google Scholar]
- 57.Fraser C, Hanage WP, Spratt BG. 2007. Recombination and the nature of bacterial speciation. Science 315:476–480. doi: 10.1126/science.1127573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Doroghazi JR, Buckley DH. 2011. A model for the effect of homologous recombination on microbial diversification. Genome Biol Evol 3:1349–1356. doi: 10.1093/gbe/evr110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dillion MM, Thakur S, Almeida RND, Guttman DS. 2017. Recombination of ecologically and evolutionarily significant loci maintains genetic cohesion in the Pseudomonas syringae species complex. bioRxiv 10.1101/227413. [DOI] [PMC free article] [PubMed]
- 60.Tudor-Nelson S, Minsavage G, Stall R, Jones JB. 2003. Bacteriocin-like substances from tomato race 3 strains of Xanthomonas campestris pv. vesicatoria. Phytopathology 93:1415–1421. doi: 10.1094/PHYTO.2003.93.11.1415. [DOI] [PubMed] [Google Scholar]
- 61.Jibrin MO, Timilsina S, Potnis N, Minsavage GV, Shenge KC, Akpa AD, Alegbejo MD, Vallad GE, Jones JB. 2014. First report of Xanthomonas euvesicatoria causing bacterial spot disease in pepper in northwestern Nigeria. Plant Dis 98:10. doi: 10.1094/PDIS-04-13-0421-RE. [DOI] [PubMed] [Google Scholar]
- 62.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. 1994. Current protocols in molecular biology. John Wiley and Sons, New York, NY. [Google Scholar]
- 63.Richter M, Rosselló-Móra R. 2009. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci U S A 106:19126–19131. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Contreras-Moreira B, Vinuesa P. 2013. GET_HOMOLOGUES, a versatile software package for scalable and robust microbial pangenome analysis. Appl Environ Microbiol 79:7696–7701. doi: 10.1128/AEM.02411-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Capella-Gutiérrez S, Silla-Martinez JM, Gabaldon T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods 14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Minh BQ, Nguyen MAT, von Haeseler A. 2013. Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol 30:1188–1195. doi: 10.1093/molbev/mst024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Altschul SF, Madden TL, Schaer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li L, Stoeckert CJ Jr, Roos DS. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res 13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Willenbrock H, Hallin PF, Wassenaar TM, Ussery DW. 2007. Characterization of probiotic Escherichia coli isolates with a novel pan-genome microarray. Genome Biol 8(12):R267. doi: 10.1186/gb-2007-8-12-r267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Guindon S, Gascuel O. 2003. A simple, fast and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 74.Markowitz VM, Chen IM, Chu K, Szeto E, Palaniappan K, Pillay M, Ratner A, Huang J, Pagani I, Tringe S, Huntemann M, Billis K, Varghese N, Tennessen K, Mavromatis K, Pati A, Ivanova NN, Kyrpides NC. 2014. IMG/M 4 version of the integrated metagenome comparative analysis system. Nucleic Acids Res 42:D568–D573. doi: 10.1093/nar/gkt919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Blom J, Kreis J, Spänig S, Juhre T, Bertelli C, Ernst C, Goesmann A. 2016. EDGAR 2.0: an enhanced software platform for comparative gene content analyses. Nucleic Acids Res 44(W1):W22–W28. doi: 10.1093/nar/gkw255. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.