ABSTRACT
Of marine eubacteria, the genus Vibrio is intriguing because member species are relevant to both marine ecology and human health. Many studies have touted the relationships of Vibrio to environmental factors, especially temperature and salinity, to predict total Vibrio abundance but lacked the taxonomic resolution to identify the relationships among species and the key drivers of Vibrio dynamics. To improve next-generation sequencing (NGS) surveys of Vibrio, we have conducted both 16S small subunit rRNA and heat shock protein 60 (hsp60) amplicon sequencing of water samples collected at two well-studied locations in the Neuse River Estuary, NC. Samples were collected between May and December 2016 with enhanced sampling efforts in response to two named storms. Using hsp60 sequences, 21 Vibrio species were identified, including the potential human pathogens V. cholerae, V. parahaemolyticus, and V. vulnificus. Changes in the Vibrio community mirrored seasonal and storm-related changes in the water column, especially in response to an influx of nutrient-rich freshwater to the estuary after Hurricane Matthew, which initiated dramatic changes in the overall Vibrio community. Individual species dynamics were wide ranging, indicating that individual Vibrio taxa have unique ecologies and that total Vibrio abundance predictors are insufficient for risk assessments of potentially pathogenic species. Positive relationships between Vibrio, dinoflagellates, and Cyanobacteria were identified, as were intraspecies associations, which further illuminated the interactions of cooccurring Vibrio taxa along environmental gradients.
IMPORTANCE The objectives of this research were to utilize a novel approach to improve sequence-based surveys of Vibrio communities and to demonstrate the usefulness of this approach by presenting an analysis of Vibrio dynamics in the context of environmental conditions, with a particular focus on species that cause disease in humans and on storm effects. The methods presented here enabled the analysis of Vibrio dynamics with excellent taxonomic resolution and could be incorporated into future ecological studies and risk prediction strategies for potentially pathogenic species. Next-generation sequencing of hsp60 and other innovative sequence-based approaches are valuable tools and show great promise for studying Vibrio ecology and associated public health risks.
KEYWORDS: Vibrio, hsp60, amplicon sequencing, microbial ecology, public health
INTRODUCTION
The genus Vibrio encompasses a diverse group of heterotrophic bacteria which are ubiquitous and abundant members of native microbial assemblages in open ocean, estuarine, and freshwater ecosystems. Vibrio spp. are Gram-negative rods belonging to Gammaproteobacteria and are chemoorganotrophic with facultative fermentative metabolisms (1). The distribution and dynamics of Vibrio populations are strongly influenced by their occurrence along environmental gradients (2, 3) as well as ecosystem-level interactions controlled by resource availability, predation, and host abundance (4–6). Through obligate heterotrophic growth on organic substrates, Vibrio species contribute to nutrient cycling within their diverse habitats and are involved in both the uptake and remineralization of carbon, phosphorus, and nitrogen (7–9). As free-living members of the bacterioplankton, Vibrio spp. have been shown to proliferate quickly and can even form blooms in response to environmental changes due to their motility, ability to metabolize diverse substrates, and high rates of population turnover (10).
Although many Vibrio species associated with human and animal hosts are benign commensals, several are human and animal pathogens and pose major international threats to public health. Potentially pathogenic species include Vibrio cholerae, the causative agent of cholera, and Vibrio vulnificus and Vibrio parahaemolyticus, which are associated with seafood poisoning and wound infections and cause the majority of Vibrio-related illnesses (vibriosis) in developed nations (1). These organisms are common in estuarine waters and bioaccumulate in oysters and other filter feeders during the warm summer months. Like other members of the genus, potentially pathogenic species are most abundant in warm waters and exhibit strong seasonality, with most infections occurring during the summer months (11, 12). When conditions are optimal, virtually 100% of oysters carry Vibrio, and while some strains of these species are not virulent, the consumption of raw shellfish is an important mode of infection for those which are pathogenic to humans (13–15). Recreational water contact, especially secondary contact such as from fishing and boating activities, is also an important mode of transmission for Vibrio pathogens (16).
The presence and distribution of Vibrio species in coastal habitats is largely dependent on environmental conditions. Salinity and temperature have been frequently correlated with Vibrio and have been widely utilized to determine the risk of illness from pathogenic species (2, 17–19). Many risk assessment models for Vibrio rely almost entirely on water temperature, which explains only about half of the interannual variation in total Vibrio abundance quantified using culture-based methods (20). Efforts to model specific species of Vibrio have been hampered by the fact that many risk assessment methodologies are incapable of delving into species-level variation in Vibrio communities. This is problematic both because of the unique ecologies of individual species and because models for the entire genus are inadequate predictors of potential pathogens. Another shortfall of current methods is their failure to account for shifts in the Vibrio community in response to storm events. Vibrio dynamics in shallow coastal and estuarine systems are strongly affected by storm-related high winds and enhanced flow, which can reintroduce particle-attached cells to the water column (21). Storm-associated precipitation and increased freshwater river discharge can dramatically influence salinity, nutrient availability, phytoplankton abundance, and other factors in coastal estuaries which may impact Vibrio abundance and community composition (21, 22). Other environmental factors, including dissolved oxygen (DO), chlorophyll a, and turbidity, have been shown to affect Vibrio abundances in water and shellfish in various studies, but these relationships are not well resolved, and opposite trends have sometimes been demonstrated (19). Continued research efforts are necessary to define the relationship between the environment and Vibrio, especially for species that are important from a public health perspective, in order to identify consistent patterns associated with the ecology of specific species within the genus.
The gene for the small subunit of 16S rRNA has been used to identify and study prokaryotes for decades. This marker is an excellent tool for surveying bacterial assemblages in marine and coastal environments and has been frequently used to define bacterial responses to environmental factors (10, 23). Unfortunately, the relatedness and rapid evolution of Vibrio species makes them difficult to identify and differentiate using approaches which rely on the 16S rRNA gene (24–26). Culture-based methods have also been problematic due to the prevalence of false positives, the need for molecular testing to confirm species identity, and the time to result (27). Despite these challenges, the prevalence of Vibrio in populated coastal areas, as well as the economic value of recreational waters and shellfish fisheries, underscores the importance of excellent methods for the study of potential Vibrio pathogens and Vibrio communities as a whole (28). Next-generation sequencing (NGS) technologies enable the simultaneous analysis of DNA sequences for all microbes in a given sample, permitting the study of complex environmental microbial communities (29, 30). As NGS technologies become less expensive, quicker, and more widely available, it is essential to consider how they can be used to detect and study Vibrio and other groups that are important from both ecological and public health perspectives. The heat shock protein 60 gene (hsp60) is an excellent marker for differentiating closely related taxa, since it is protein encoding and thus more variable than 16S rRNA markers (31). This gene has been used to identify and differentiate Vibrio and other clinically relevant bacteria with high taxonomic resolution (32, 33) and is a promising target for NGS of PCR amplicons, since reads as short as 200 bp can be used for species-level identification (31).
The objectives of this research were twofold. First, we aimed to evaluate the use of NGS of hsp60 amplicons for environmental surveys of Vibrio. To demonstrate the utility of this approach, our second objective was to conduct an analysis of Vibrio communities in the context of environmental parameters within both microbial and phytoplankton assemblages. To achieve these goals, we initiated a 7-month biweekly sampling campaign at two well-studied sampling stations in the Neuse River Estuary (NRE), a eutrophic estuary in eastern North Carolina. Samples were collected from May through December 2016 with enhanced sampling efforts immediately following two named storm events, Tropical Storm Colin and Hurricane Matthew. We conducted NGS of both 16S rRNA and hsp60 amplicons. Sequencing both amplicons enabled us to compare hsp60 and 16S rRNA results for Vibrio populations, analyze Vibrio dynamics with improved taxonomic resolution using hsp60, and better understand Vibrio in the context of the larger microbial community using the 16S rRNA marker. Using hsp60, we identified specific species of Vibrio that are important from a human health perspective (V. vulnificus, V. parahaemolyticus, and V. cholerae) and observed how individual species dynamics impacted the entire Vibrio community. We also collected environmental, chemical, and phytoplankton photopigment data and associated these variables with Vibrio abundances and community structure using a range of informative statistical approaches.
RESULTS
Vibrio sequences.
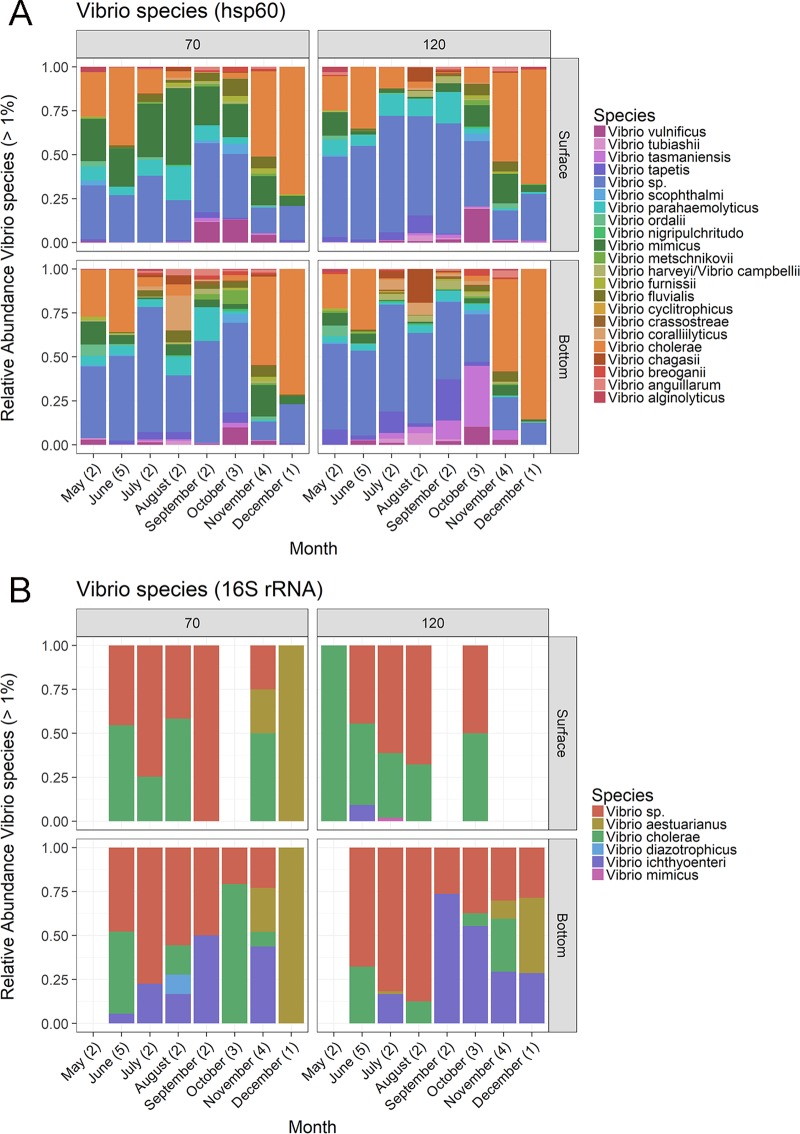
There were 21 Vibrio species identified using hsp60 amplicons, including the potential pathogens V. vulnificus, V. parahaemolyticus, and V. cholerae (Fig. 1A). There were only 5 species of Vibrio identified using 16S rRNA amplicons, and there were multiple samples in which there were no 16S rRNA Vibrio sequences (Fig. 1B). The 16S rRNA analysis identified three species, V. ichthyoenteri, V. aestuarianus, and V. diazotrophicus, which did not appear in the hsp60 data because there were no representatives of these species in the curated cpn60 database used to assign taxonomy to the hsp60 reads. In total, annotated Vibrio reads made up 0.02% of 16S rRNA reads and 0.5% of hsp60 reads across the study (n = 184). The percentages of Vibrio reads in individual samples ranged from 0 to 0.2% for 16S rRNA and from 0.35 to 3.2% for hsp60. Observed, Shannon, and chao1 alpha diversity metrics for hsp60 and 16S rRNA amplicons were correlated but were significantly higher for the 16S rRNA gene (see Fig. S1 and Tables S3 and S4 in the supplemental material). The hsp60 amplicon exhibited higher diversity for the genus Vibrio than the 16S rRNA amplicon (Fig. S1). hsp60 and 16S rRNA alpha diversity metrics for Vibrio were not significantly correlated (Table S3). Only two Vibrio taxa, V. cholerae and V. mimicus, were observed in both amplicon data sets. See the supplemental material for summaries of the Proteobacteria, Gammaproteobacteria, Vibrionales, and Vibrionaceae in the 16S rRNA and hsp60 data (see Fig. S2 to S5).
FIG 1.
Relative abundances of Vibrio species in the NRE from May through December 2016 using the hsp60 gene (A) and 16S rRNA gene (B). Panels designate sampling site (stations 70 or 120) and depth (surface or bottom water). Numbers in parentheses indicate the number of sampling events per month. There were no 16S reads assigned to the Vibrio genus in several samples, as indicated by blank data slots.
Vibrio and the environment.
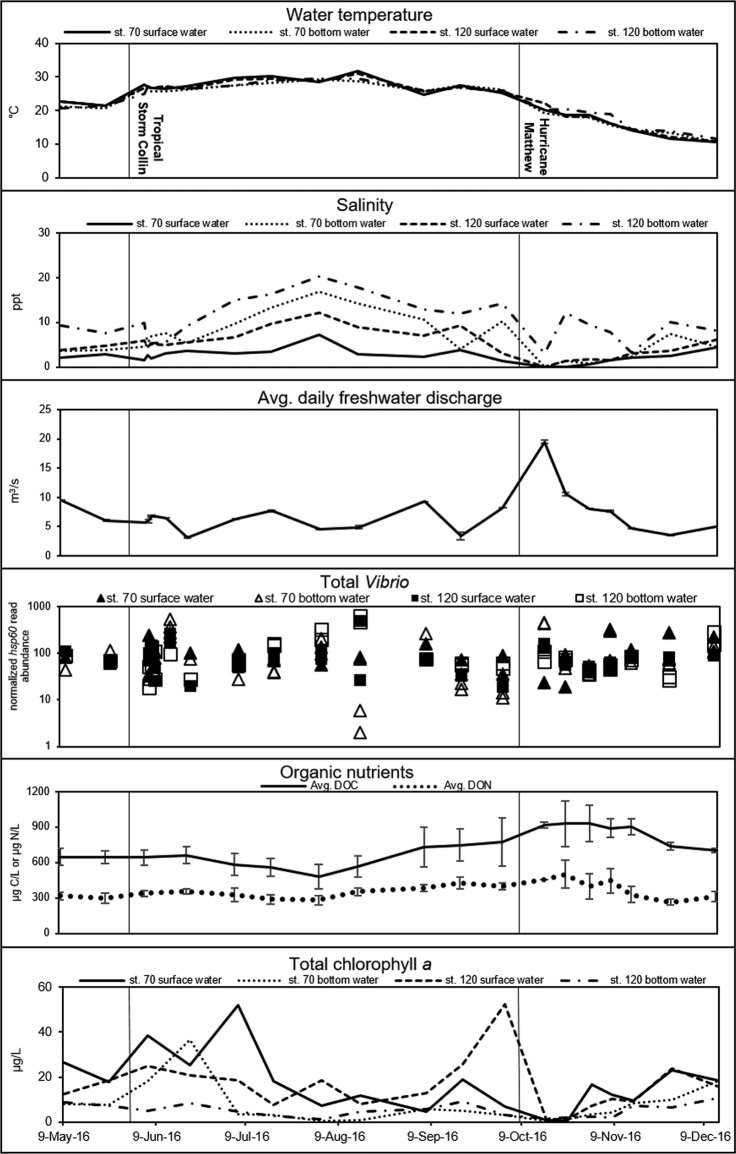
Temperatures in the NRE during the study period ranged from 10.5 to 31.6°C. Salinities ranged from 0.02 to 20.2 ppt, with higher salinities observed during the summer months and in the bottom water. The average daily freshwater discharge remained relatively constant and averaged 6.3 m3/s from May through early October 2016 and increased to 19.5 m3/s on the first sampling day following Hurricane Matthew. A corresponding decrease in salinity to near freshwater levels (0.02 ppt) was observed immediately following the hurricane. On average, the normalized abundance of total Vibrio reads increased 9% in the surface waters and 39% in the bottom waters during the summer months. Normalized Vibrio read abundances following Tropical Storm Colin and Hurricane Matthew increased in the bottom waters by 9% and decreased in the surface waters by 12.6% on average. Dissolved organic nitrogen (DON) increased from an average of 340 μg/liter in samples taken prior to Hurricane Matthew to an average of 449 μg/liter in storm-associated samples. Dissolved organic carbon (DOC) increased from 640 μg/liter on average before the storm to 917 μg/liter in Hurricane Matthew-associated samples. The total chlorophyll a concentration, which is a proxy for phytoplankton abundance, was highly variable during the study period, with values ranging from 0.5 to 52 μg/liter, and the highest numbers were observed in response to two phytoplankton blooms in the surface waters. Chlorophyll a concentrations associated with both bloom events were 52 μg/liter, and each bloom was captured at a single time point, the first in July at station 70 and the second in October at station 120. These blooms were likely associated with dinoflagellates based on high concentrations of the dinoflagellate-related phytoplankton photopigment peridinin (data not shown). These results are summarized in Fig. 2 and are typical of the highly variable conditions in the NRE, with the exception of those associated with Hurricane Matthew, which can be considered an extreme event on the basis of the observed influx of freshwater discharge and corresponding changes in salinity, chlorophyll a, and nutrient concentrations. Storm-related freshwater pulses in the NRE were previously shown to decrease residence times and salinity and contribute to elevated nutrient concentrations (34).
FIG 2.
Temperature, salinity, average daily river discharge, total Vibrio abundance, organic nutrients, and total chlorophyll a for all sampling dates, stations, and depths in the NRE during the study period. Organic nutrient data are averaged across stations and depths for each sampling date. Named storm events Tropical Storm Colin (June 2016) and Hurricane Matthew (October 2016) are marked by vertical lines.
We investigated how the full suite of measured temporal and environmental parameters impacted Vibrio communities by using an accepted nonparametric modeling approach (distLM), which can be used to assess the contributions of a range of variables to variations in multivariate abundance data (35). All fitted variables except particulate organic carbon (POC), pH, and particulate nitrogen (PN) were significant in the marginal tests. In the sequential tests, 26 of 34 total variables were fit to the model (see Table S2 for a list of all tested variables). Three factors, month, salinity, and days post-Hurricane Matthew, explained 49% of the observed variation in the Vibrio community, with 67% of observed variation explained by the full model. For Tropical Storm Colin, only the categorical (storm/no storm) variable, which explained <1% of the total variation, was fit in the sequential distLM test; the temporal variable (days poststorm) was not fit by the model. For Hurricane Matthew, the categorical variable was included in the model, though it was not significant, and explained <1% of the total variation. However, the temporal variable for days post-Hurricane Matthew was the third variable fit in the sequential model and explained 6.7% of all observed variation in the Vibrio community. DistLM statistics for the variables included in the sequential model are listed in Table 1. Analysis of similarity (ANOSIM) testing indicated that Tropical Storm Colin did not significantly explain the observed variation in Vibrio communities between samples (R = −0.1222, P = 0.986) but that Hurricane Matthew did (R = 0.164, P = 0.009). Both storms impacted the water column and Vibrio communities in the NRE, but Hurricane Matthew was the larger storm and had a much more dramatic impact on both biotic and abiotic conditions in the water column. The observed Δsalinity, which is defined as the difference between bottom and surface water salinities (36), is one example of the effect Hurricane Matthew had on the abiotic conditions in the estuary. The average Δsalinity was ∼5 ppt during the sample period across all samples. For Hurricane Matthew-associated samples, the average Δsalinity dropped to 0.5 ppt due to enhanced freshwater discharge, confirming that the hurricane had a large impact on salinity and stratification in the estuary.
TABLE 1.
Distance-based linear modeling for normalized and square root-transformed hsp60 Vibrio read abundances and environmental and temporal predictor variables
| Predictor variablea | Marginal testsb |
Sequential testsc |
|||
|---|---|---|---|---|---|
| P valued | Proportion of variation explained | Adj. R2 | P valued | Proportion of variation explained | |
| Month | 0.0001 | 0.338 | 0.308 | 0.0001 | 0.338 |
| Salinity | 0.0001 | 0.151 | 0.393 | 0.0001 | 0.086 |
| Days post-Hurricane Matthew | 0.0001 | 0.251 | 0.445 | 0.0001 | 0.067 |
| PO4 | 0.0001 | 0.080 | 0.462 | 0.0001 | 0.019 |
| NO3/NO2 | 0.0001 | 0.083 | 0.477 | 0.0001 | 0.017 |
| Rainfall | 0.0013 | 0.024 | 0.489 | 0.0002 | 0.014 |
| C:N | 0.0001 | 0.045 | 0.498 | 0.0007 | 0.011 |
| DON | 0.0001 | 0.057 | 0.508 | 0.0001 | 0.012 |
| River discharge | 0.0001 | 0.090 | 0.517 | 0.0002 | 0.011 |
| DO | 0.0001 | 0.104 | 0.522 | 0.0082 | 0.008 |
| Station | 0.0001 | 0.055 | 0.528 | 0.004 | 0.008 |
| NH4 | 0.0078 | 0.017 | 0.532 | 0.0204 | 0.006 |
| Temperature | 0.0001 | 0.117 | 0.536 | 0.0351 | 0.006 |
| Chlorophyll a | 0.0003 | 0.027 | 0.538 | 0.0939 | 0.005 |
| SiO2 | 0.0001 | 0.036 | 0.540 | 0.1885 | 0.004 |
| Turbidity | 0.0001 | 0.067 | 0.541 | 0.2271 | 0.004 |
| DIC | 0.0001 | 0.105 | 0.542 | 0.275 | 0.004 |
| TSS | 0.5504 | 0.005 | 0.543 | 0.239 | 0.004 |
| BP | 0.0001 | 0.035 | 0.545 | 0.1523 | 0.004 |
| Tropical Storm Colin | 0.0001 | 0.034 | 0.549 | 0.0212 | 0.006 |
| Average wind speed | 0.0001 | 0.033 | 0.562 | 0.0001 | 0.013 |
| Hurricane Matthew | 0.0001 | 0.070 | 0.565 | 0.1149 | 0.008 |
| Depth | 0.0001 | 0.043 | 0.566 | 0.1948 | 0.004 |
| Storm sampling | 0.0001 | 0.055 | 0.567 | 0.2411 | 0.004 |
| DOC | 0.0001 | 0.054 | 0.569 | 0.2132 | 0.004 |
Only variables which were included in the sequential model are listed. C:N, carbon-to-nitrogen molar ratio; DON, dissolved organic nitrogen; DO, dissolved oxygen; DIC, dissolved inorganic carbon; TSS, total suspended solids; BP, barometric pressure; DOC, dissolved organic carbon.
Predictor variables were taken individually.
Stepwise tests began with a null model and fit predictor variables to achieve the highest adjusted (adj.) R2.
P values from 9,999 model permutations. Bold typeface indicates significance at P < 0.05.
Relating Vibrio dynamics to extreme events.
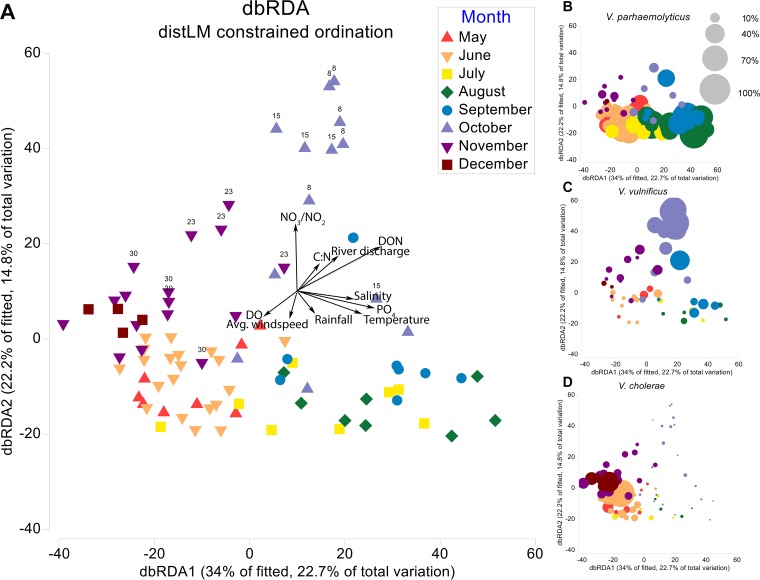
Samples taken following Hurricane Matthew grouped at the top of the y axis of the distance-based redundancy analysis (dbRDA) ordination (Fig. 3A), which was positively correlated with increased nitrate/nitrite (NO3/NO2) concentrations, as well as decreased salinity and increased freshwater discharge and DON as floodwaters from the storm made their way through the estuary. As freshwater discharge returned to baseline (see Fig. 2), the Vibrio community composition shifted as V. vulnificus and cooccurring taxa, which had increased in concentration immediately after the storm, began to subside (see Fig. 3C and 4). The communities sampled in November and December were more similar to those observed in May and June than to those taken immediately before the storm in late September and early October based on their positions in the ordination. Samples taken in July, August, and September grouped together and were positively associated with temperature, phosphate (PO4), and salinity. Samples associated with Tropical Storm Colin grouped with the other samples collected during the month of June. The unconstrained principal components (PCO) plot is available in the supplemental material (see Fig. S6).
FIG 3.
(A) Distance-based redundancy analysis (dbRDA) plot of the fitted distLM model for Vibrio communities in the NRE. Each point in the ordination represents the Vibrio community in a given water sample. The distance between points is the Bray-Curtis distance (dissimilarity) between Vibrio communities. Vectors denote significant environmental gradients (P < 0.05) in the distLM. Points associated with increased river discharge after Hurricane Matthew are labeled according to the number of days post-Matthew the sample was taken. The ordination is also presented as bubble plots in which point size is proportional to the relative abundances of V. parahaemolyticus (B), V. vulnificus (C), and V. cholerae (D). The bubble key indicates bubble size for 100, 70, 40, and 10% of the range for each species.
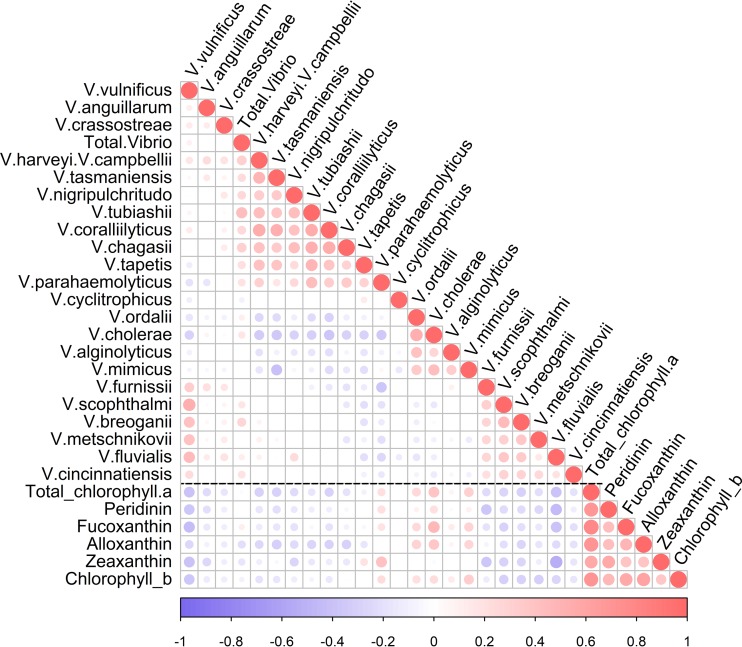
FIG 4.
Spearman rank correlation plot showing correlations between Vibrio taxa and between Vibrio taxa and phytoplankton photopigments. The black dotted line distinguishes Vibrio-Vibrio relationships from Vibrio-pigment relationships. Only significant correlations (P < 0.05) are plotted.
Dynamics of species with public health implications.
The dbRDA bubble plots (Fig. 3B to D) illustrate how the abundances of potential human pathogens V. parahaemolyticus, V. vulnificus, and V. cholerae changed during the study period. V. parahaemolyticus normalized read abundances were highest in the spring and summer months and were positively correlated with water temperature (r = 0.64). V. cholerae normalized read abundances were highest in December and May and were significantly correlated with chlorophyll a (r = 0.39) and temperature (r = −0.66). V. vulnificus rapidly responded to the changes in the water column after Hurricane Matthew and had the highest normalized read abundances in samples taken immediately after the storm. The V. vulnificus normalized read abundance was also significantly correlated with DOC (r = 0.36) and DON (r = 0.42). Only a subset of correlations between V. vulnificus, V. parahaemolyticus, V. cholerae, and quantitative environmental parameters are listed here, see Fig. S7 for a full summary of significant correlations. The presence of V. parahaemolyticus, V. vulnificus, and V. cholerae in the water column was confirmed by digital droplet PCR using previously published primer sets (37–39) (data not shown).
Inter- and intraspecies relationships.
Potential species-level interactions between Vibrio taxa were investigated using correlation analyses (Fig. 4). Significant relationships were identified between Vibrio species, which were grouped based on the angular order of eigenvectors (AOE) into clusters of cooccurring taxa. Total Vibrio abundance, which has been suggested as a proxy for estimating the concentrations of potential human pathogens (40), was only weakly correlated to V. parahaemolyticus (r = 0.19), V. vulnificus (r = 0.09), and V. cholerae (r = 0.16). We also used correlations to test for interactions between Vibrio and phytoplankton photopigments, which represent distinct classes of primary producers in the NRE (41) (Fig. 4; see Table 2 for a list of the dominant phytoplankton classes associated with each photopigment). V. parahaemolyticus was positively correlated with nearly every photopigment measured, with the exception of alloxanthin. V. parahaemolyticus, V. ordalii, V. cholerae, V. mimicus, and V. alginolyticus also had positive correlations with many photopigments, with the strongest between V. cholerae and fucoxanthin (r = 0.47) and total chlorophyll a (r = 0.40). The photopigment peridinin, which is associated with dinoflagellates and was the major contributor to both phytoplankton blooms observed during this study, was significantly positively correlated with this group of Vibrio taxa.
TABLE 2.
Dominant phytoplankton classes associated with accessory photopigments in the NREa
Significant correlational relationships with r > 0.4 between Vibrio species and the top 15 most abundant phyla identified in the 16S rRNA data are plotted for all samples (Fig. 5A) and for Hurricane Matthew samples only (Fig. 5B). Figure 5A shows a correlation between V. parahaemolyticus and the Cyanobacteria (r = 0.53). This relationship was correspondingly observed in the phytoplankton photopigment data, where the Cyanobacteria-associated photopigment zeaxanthin was also correlated with V. parahaemolyticus (r = 0.41). The Cyanobacteria were negatively associated with V. fluvialis (r = −0.49) and V. vulnificus (r = −0.46). Total Vibrio spp. and several individual Vibrio taxa were not included in the overall network, as their correlations fell below the threshold of an r value >0.4, though total Vibrio spp. did correlate with the Acidobacteria (r = −0.41). The network analysis presented in Fig. 5B, which includes only samples associated with Hurricane Matthew, had a greater complexity and contained a higher number of correlational relationships above the r > 0.4 threshold. V. vulnificus and cooccurring Vibrio species abundances increased rapidly immediately after the storm (Fig. 3C, Fig. 4). This corresponded to decreases in V. cholerae and several other Vibrio and 16S rRNA taxa.
FIG 5.
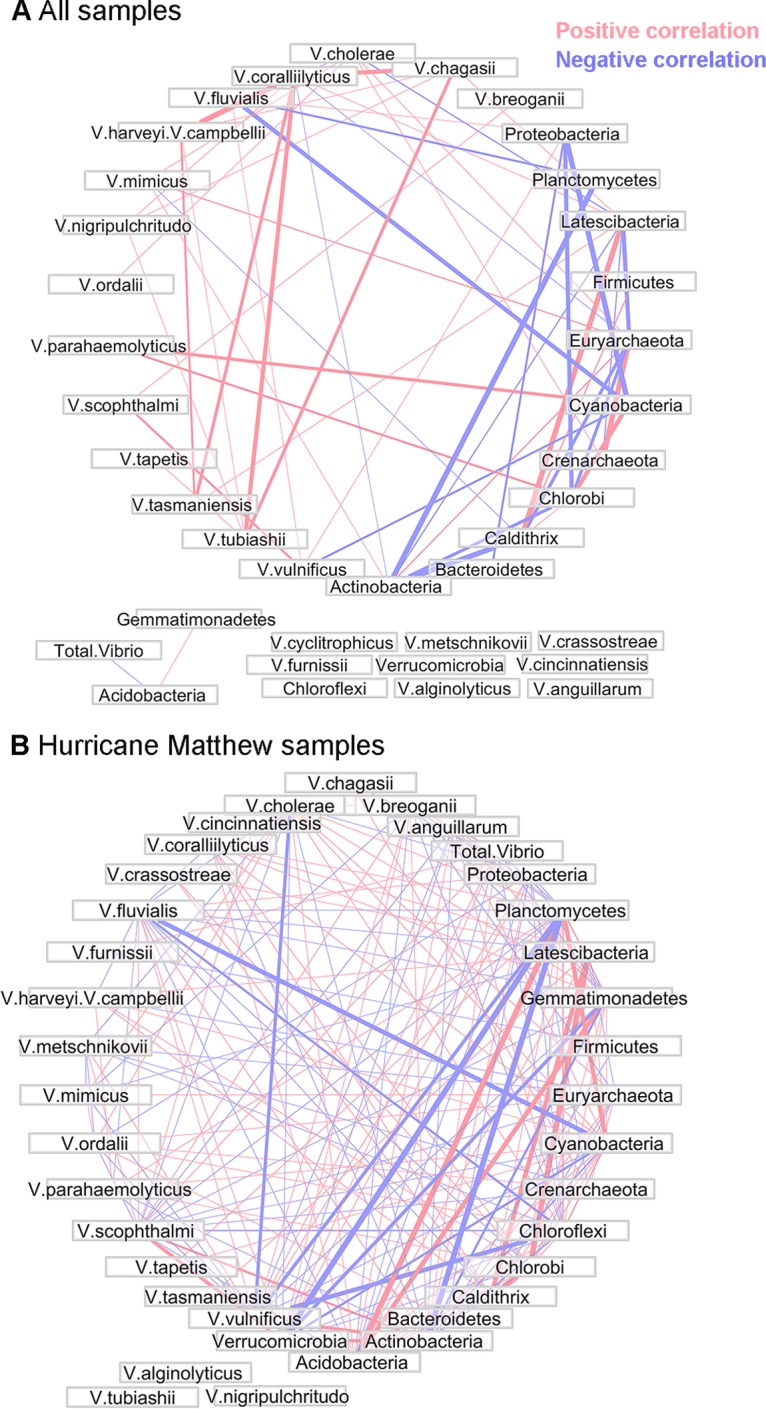
Network plots showing positive and negative Spearman rank correlations (Spearman's r > 0.4) between Vibrio species identified using hsp60 and the top 15 most abundant bacterial phyla identified using 16S rRNA for all samples (A) and for samples associated with Hurricane Matthew (B). Vector width is proportional to the strength of the correlation.
DISCUSSION
Next-generation sequencing of hsp60 amplicons enabled us to accomplish our objective of achieving enhanced taxonomic resolution for the Vibrio in the NRE, which provides an important advantage for studying the genus. This approach also enabled us to meet our second objective of examining Vibrio dynamics in the context of the environment. We were able to characterize and more deeply understand how both the Vibrio community as a whole and individual species relate to a wide range of ecological factors. Here, we present a number of the prominent relationships observed as a result of our nonparametric modeling effort. In addition, we highlight a series of correlational associations across Vibrio species, phytoplankton groups, and 16S rRNA taxa, particularly focusing on storm effects and potentially virulent taxa.
Total Vibrio dynamics.
The shifts in total Vibrio abundance mirrored the changes in the abiotic conditions in the water column (Fig. 2). We saw total Vibrio increase in the bottom water and decrease in the surface water following Tropical Storm Colin and Hurricane Matthew. This was caused by fluctuations in the overall Vibrio community in response to changing environmental conditions, which were especially prominent after Hurricane Matthew. Because it was primarily an inland rain event in eastern North Carolina, Hurricane Matthew caused major flooding upstream of the NRE, resulting in a notable increase in freshwater discharge and persistently low salinities for weeks after the storm (Fig. 2). This storm-related increase in discharge also resulted in elevated concentrations of organic matter as runoff and floodwaters entered the estuary (Fig. 2).
Total Vibrio abundance increased during the summer months, as has been seen in numerous studies (42–46). However, concentrations remained relatively high in the winter months, even as water temperatures dipped to <11°C. This may be tied to organisms which had entered the viable but nonculturable (VBNC) state, which would have still been detected using NGS and other molecular methods. The VBNC state has been well documented for Vibrio species under temperature stress (47). However, recent findings of a culture-based monitoring effort which ran in the NRE from 2003 to 2013 found increasing total culturable Vibrio over time and determined that the largest monthly increases were in the winter months when Vibrio had previously been undetectable (B. A. Froelich, R. Gonzalez, A. D. Blackwood, K. Lauer, and R. T. Noble, submitted for publication). The storm seemed to interrupt this downward trend in concentrations, and Vibrio numbers rose >100-fold at some stations following Hurricane Matthew. This storm-related increase may have contributed to persistent Vibrio presence in the water column through the end of the study period in December 2016 despite winter water temperatures, corresponding to previous observations that the impact of a storm can persist for weeks to months after the event (21).
Vibrio reads represented a maximum of 3.24% of total hsp60 reads in a given sample, with an average of 0.5% Vibrio reads across all samples. This result was not unexpected given that, despite their prevalence, Vibrio populations typically represent <1% of bacterioplankton (1). The development of Vibrio-specific primer sets which have been tested for a wide array of species would ensure greater sequencing depth for Vibrio taxa than was achieved in the current study and might be useful in future amplicon sequencing surveys. Vibrio spp. have been shown to proliferate rapidly in response to nutrient pulses and other environmental changes and can form blooms, in which Vibrio taxa become dominant members of the bacterioplankton (10, 48). We did not observe this phenomenon during the course of our study, perhaps because Vibrio bloom conditions are transient and can occur and resolve within 48 h (48), and our approximately biweekly sampling schedule did not have sufficient temporal resolution to capture such an event.
Relationships to environmental parameters.
The month in which samples were taken explained the greatest proportion of variation in Vibrio community composition as revealed by the distLM analysis (Table 1). This result was not surprising, since seasonal shifts in temperature, salinity, and other factors in the NRE are well documented, as is the seasonality of Vibrio populations (19, 42, 49). Temperature and nutrient concentrations significantly impacted Vibrio communities in the distLM marginal tests but explained relatively little variation in the sequential model after month, salinity, and days post-Hurricane Matthew were considered. This is because variations caused by shifts in temperature and nutrient concentrations were autocorrelated with month, salinity, and days poststorm, which were already included in the model and accounted for more variation overall. Salinity, which also varies month to month in the NRE as the water column becomes stratified during periods of moderate freshwater discharge or long intervals when there is minimal wind-driven vertical mixing (49), explained the second largest proportion of variation in Vibrio abundance (Table 1). This salinity effect was not surprising, since salinity is often correlated with Vibrio concentrations, though previous studies found that temperature accounted for more variation (19). In 2016, the salinity in the NRE was hugely affected by Hurricane Matthew, which had a particularly dramatic impact on the hydrographic structure of the estuary based on the shift to near freshwater salinities following the storm (Fig. 2). This shift in salinity post-hurricane caused striking changes in the Vibrio community, and the magnitude of this change is likely why salinity accounted for an overall greater proportion of variance than temperature in the current study.
Despite the aforementioned link between Hurricane Matthew and low salinities, days post-Hurricane Matthew accounted for an additional 6.7% of variation (Table 1) even after salinity and month were fitted to the model. This lead us to hypothesize that the increase in organic matter after the storm (Fig. 2) had a large impact on Vibrio community structure despite the fact that DON and DOC were not substantial contributors to the fitted sequential model. While the immediate storm-related changes in the Vibrio community did not persist, there were distinct differences between the pre- and poststorm communities (Fig. 3). Additional studies with longer time series are needed to see if these storm-related changes persist, as they could potentially change Vibrio dynamics in the future.
V. parahaemolyticus, V. vulnificus, and V. cholerae.
There was substantial variation in the abundances of V. parahaemolyticus, V. vulnificus, and V. cholerae over time and in relation to environmental gradients (Fig. 3B to D). Since salinity was the most important environmental factor associated with changes in the Vibrio community as a whole, it is likely that much of the species-level differences we observed were also tied to salinity. V. parahaemolyticus, which causes most human vibriosis in the United States and early mortality syndrome in shrimp, was most abundant in the spring and summer months, as has been observed in many other studies (50–52). V. parahaemolyticus almost completely disappeared after Hurricane Matthew when salinities dropped to near freshwater levels (Fig. 2). This was not surprising, since the optimal salinity range for V. parahaemolyticus is between 25 and 35 ppt (53), and V. parahaemolyticus likely could not tolerate the near-freshwater salinities observed after the storm. The V. parahaemolyticus decline could also have been caused by temperature, which had been gradually decreasing from peak summer temperatures when the storm hit in early October 2016 (Fig. 2). However, V. parahaemolyticus reemerged, albeit at lower concentrations, in November and December despite falling water temperatures. This may be because DNA sequencing methods, as mentioned previously, can detect VBNC cells under temperature stress or because V. parahaemolyticus in the NRE is able to cope with lower temperatures than previously thought. Despite reports that V. parahaemolyticus is seldom isolated from waters <15°C (54), there is some evidence for growth at a minimum temperature of 8.3°C (55), which is colder than our observed December temperatures of ∼10°C (Fig. 2). Continued winter monitoring efforts are needed to understand the effects of cold temperatures on V. parahaemolyticus in the environment.
V. vulnificus, another potential pathogen which causes both seafood-related and wound infections, was present in nearly all samples and became much more prolific after Hurricane Matthew (Fig. 3C). This is an interesting finding, especially since one of the largest single outbreaks of V. vulnificus, which included 22 total cases and 5 confirmed deaths, was reported after Hurricane Katrina in 2005 due to contact with estuarine flood waters (56). V. vulnificus has been shown to have the greatest abundances at salinities between 5 and 10 ppt (57) and, as a Vibrio species that can tolerate lower salinities, likely benefitted from the drop in salinity poststorm, which may have negatively impacted competitors just as nutrient-rich storm waters flooded the system. The correlations between V. vulnificus abundances and organic matter (see Fig. S7 in the supplemental material), which noticeably increased after Hurricane Matthew (Fig. 2), suggest that V. vulnificus was able to exploit the increasing availability of nutrients in the estuary. This relationship between V. vulnificus and the availability of organic matter was demonstrated in previous microcosm experiments in which researchers showed that V. vulnificus abundances increased following inputs of organic matter (9). The results of the present study add to a growing body of literature showing that V. vulnificus populations can respond rapidly to changes in salinity and enhanced nutrient concentrations associated with floodwaters, enabling rapid poststorm proliferation and contributing to higher disease risk (3, 56, 58).
V. cholerae, the causative agent of cholera, had the highest abundances in the spring and winter months. On the basis of salinity alone, V. cholerae, which has a salinity range of 1 to 10 ppt (19), should have been able to take advantage of the low salinities after Hurricane Matthew. However, there was no increase in V. cholerae levels after Hurricane Matthew, and V. cholerae was negatively correlated with V. vulnificus and other taxa which increased in abundance after the storm (Fig. 4). This may be because V. cholerae abundances in the water column were relatively low in the months prior to the storm (Fig. 3D), indicating that perhaps some other environmental factor was controlling V. cholerae before the storm hit. The negative relationship we observed between V. cholerae and temperature (Fig. 3; Fig. S7) was surprising, and seemed to be driven by high December numbers. V. cholerae has been shown to grow at temperatures between 10 and 15°C, but it becomes VBNC below that temperature range (59, 60). Despite this, the presence of V. cholerae was confirmed in these winter samples by digital droplet PCR; so, it is possible that a cold-tolerant population of V. cholerae emerged. Continued research efforts are necessary to determine whether the winter reemergence of V. cholerae is a recurring or persistent trend and, if so, to further investigate the causes.
Ecology of Vibrio associations.
As ubiquitous members of the bacterioplankton, Vibrio taxa constantly intermingle with each other, with other microbes, and with phytoplankton communities. There is evidence in the present study (Fig. 4) and in the scientific literature that Vibrio taxa are associated with phytoplankton generally; Turner et al. (42) found that the abundances of culturable Vibrio were significantly correlated with phytoplankton abundance, and numerous other studies have identified links between Vibrio and chlorophyll a concentrations (19). The dinoflagellate-associated phytoplankton photopigment peridinin was the major contributor to the bloom events observed in July and October 2016 and was positively correlated with V. parahaemolyticus, V. ordalii, V. cholerae, and V. mimicus (Fig. 4). While the precise nature of the positive correlations between these specific Vibrio taxa and dinoflagellates is currently unknown, these results contribute to existing evidence that dinoflagellate-dominated phytoplankton assemblages may encourage Vibrio survival or growth in coastal waters (61–63).
The same Vibrio taxa that were correlated with peridinin/dinoflagellates, V. parahaemolyticus, V. ordalii, V. cholerae, and V. mimicus, had positive associations with several other photopigments. Of these, three species, V. ordalii, V. cholerae, and V. mimicus, were also highly correlated with each other. Although correlations between Vibrio species may not elucidate the details of direct interactions, they do reflect overlapping ecologies (40), and we observed similar correlational patterns between distinct groups of Vibrio species across our entire investigation, providing evidence that there are ecological drivers of cooccurring species. All three of the aforementioned species have pathogenic potential: V. cholerae is the causative agent of cholera, V. mimicus is so named because it can cause gastrointestinal illness with symptoms and biochemical characteristics that mimic cholera (64), and V. ordalii can cause vibriosis in fish (65). V. mimicus, like V. cholerae, has been shown to thrive at relatively low salinities (1 to 10 ppt, 4 ppt optimal salinity for V. mimicus) compared to other Vibrio, many of which have higher salinity requirements (19, 66). On the other hand, relatively little is known about the ecology of V. ordalii, though previous studies of diseased fish have recorded salinities of >33 ppt (67). Since the bulk of academic literature has focused on Vibrio that are pathogenic to humans, identifying ecological similarities between Vibrio that have been less thoroughly studied, such as V. ordalii, and those for which we have more comprehensive academic knowledge, such as V. cholerae, can help us to define the ecologies of lesser-known Vibrio taxa.
There were several correlations across all samples (Fig. 5A) between hsp60 Vibrio taxa and 16S rRNA bacterial phyla. Notably, V. parahaemolyticus was associated with Cyanobacteria, a result which was confirmed in the phytoplankton photopigment data (see relationship between V. parahaemolyticus and Cyanobacteria-associated zeaxanthin in Fig. 4). Cyanobacteria have been associated with Vibrio species in a variety of contexts, perhaps most notably in studies which have identified relationships between toxin-producing Cyanobacteria blooms and V. cholerae (68) and identified Cyanobacteria as contributors to V. cholerae persistence or growth (9, 69, 70). Very little research has been done to investigate specific linkages between V. parahaemolyticus and Cyanobacteria, though there is evidence that some strains of V. parahaemolyticus may nonrandomly associate with cyanobacterial mats (71). Physical interactions between V. parahaemolyticus and Cyanobacteria could have important implications for the growth and survival of V. parahaemolyticus, as has been shown for V. cholerae (68–70). Additional studies investigating the possible interactions between V. parahaemolyticus and Cyanobacteria are warranted based on these results.
The correlation network for Hurricane Matthew samples (Fig. 5B) was different and more complex than that for all samples (Fig. 5A), illustrating the shifting dynamics in both the Vibrio and entire bacterioplankton communities as the water column freshened and nutrient concentrations increased. In the context of Vibrio species succession, this heightened complexity highlights a need to further understand the relative importance of temperature, salinity, nutrients, and phytoplankton populations as drivers of specific Vibrio species in response to disruptive changes in the water column associated with extreme events.
Conclusions.
NGS sequencing of hsp60 and other protein-coding marker genes paired with information generated from sequencing the more widely used 16S rRNA gene can dramatically improve the study of Vibrio and/or other closely related microbes and provide important information about the potential emergence of pathogens. This study demonstrates the utility of hsp60 amplicon sequencing for Vibrio populations, reveals the complexity of Vibrio dynamics in the NRE, and illustrates how those dynamics can shift seasonally and in response to storm events. Although they represented only a small percentage of the total diversity in both our 16S rRNA and hsp60 data, our results indicate that members of the Vibrio genus are themselves a complex and ever-changing community. The associations between cooccurring subgroups of species reveals cohorts of Vibrio taxa with similar ecologies, and relationships with various phytoplankton groups provide additional evidence for interactions between Vibrio and phytoplankton. A major outcome of this study is that there may be considerable benefit from the Vibrio research community adapting to the use of species-specific methods, since total Vibrio abundance was not strongly correlated with the presence of potential pathogens. We suggest that sequence-based approaches are useful in combination with quantitative and digital PCR-based assays that have the benefit of tracking Vibrio species and pathogenic subgroups of those species with high specificity. This type of approach would yield information that can be pitted against environmental and phytoplankton abundance data to more completely understand the ecology of this complex genus.
MATERIALS AND METHODS
Study site and environmental sampling.
The NRE is a shallow, eutrophic density-stratified estuary in eastern North Carolina. It is an ecologically and economically important tributary of Pamlico Sound, the second largest lagoonal estuary in the United States. The estuary has minimal tidal influence due to buffering by North Carolina's extensive barrier island system, and flow variability is largely governed by wind and river inflow (72). The NRE is subject to climatic shifts, including storm and drought events, which affect freshwater inflow and residence times in the estuary. These events have been shown to affect Vibrio densities in the NRE due to changes in salinity, temperature, and the resuspension of shallow bottom sediments during storms, which can reintroduce Vibrio cells to the water column (21, 22). Both recreational water use and oyster harvesting are actively conducted in the NRE and its feeder creeks, and the watershed includes communities that depend on aquaculture and tourism.
The water quality in the NRE has been monitored for more than 40 years through multiple projects, most notably, the ongoing NRE Modeling and Monitoring program (ModMon), which has been in operation since the mid-1990s. In coordination with ModMon, water samples for this study were collected from the surface and bottom waters at ModMon stations 70 (28 km downstream from the head of the estuary) and 120 (43 km downstream). The locations of the stations are shown in Fig. 6. Samples were collected biweekly May through December 2016, with additional sampling efforts after two named storm events. Tropical Storm Colin passed over eastern North Carolina on 6 June 2016, and additional storm-associated samples were collected daily from 6 to 8 June and on 13 June 2016. Hurricane Matthew, which brought heavy rainfall, catastrophic flooding, strong winds, and moderate storm surge to eastern North Carolina, passed over on 8 to 9 October 2016. In response to Hurricane Matthew, sampling was increased from biweekly to weekly from the first poststorm sampling date on 17 October through the end of November 2016. Freshwater discharge from the Neuse River was elevated following both storm events, and samples collected on dates after Tropical Storm Colin and Hurricane Matthew were considered “storm samples” if they were taken before freshwater discharge returned to or below the average for the sampling period. In total, samples were collected on 23 trips, 3 of which were associated with Tropical Storm Colin and 4 of which were associated Hurricane Matthew. Samples were also categorized by month and season, with samples collected in May designated “spring samples,” samples collected June to August designated “summer samples,” samples collected September to November designated “fall samples,” and samples collected in December designated “winter samples.” All water samples were collected using a diaphragm pump and a weighted hose, with the exception of the post-Tropical Storm Colin samples, which were collected using a Van Dorn bottle. Surface samples were collected at approximately 0.2 m below the water surface, and bottom samples were collected approximately 0.5 m above the bottom sediments. Measurements of depth, salinity, water temperature, DO, pH, and turbidity were measured in situ using a fully calibrated YSI 6000 multiprobe sonde (Yellow Springs Instruments, Yellow Springs, OH). Bottles containing water samples were insulated from outside temperatures in a cooler in the shade and were returned to the laboratory for sample processing within 6 h of collection for water quality, phytoplankton photopigment, and molecular analyses. A previous study of the effect of storage time on total Vibrio, V. parahaemolyticus, and V. vulnificus abundances found no significant change in concentrations for up to 24 h (73). The field and laboratory methods for all water quality and phytoplankton photopigment analyses are summarized in Table 3.
FIG 6.
Sampling locations in the Neuse River Estuary (NRE) in eastern North Carolina.
TABLE 3.
Summary of measurement methods for environmental parameters
| Measured parameter | Abbreviation | Unit | Method, instrument, or source for measurement | Reference(s) |
|---|---|---|---|---|
| Salinity | ppt | Measured in situ using water quality sonde | ||
| Temperature | °C | Measured in situ using water quality sonde | ||
| pH | Measured in situ using water quality sonde | |||
| Dissolved oxygen | DO | mg/liter | Measured in situ using water quality sonde | |
| Turbidity | NTU | Measured in situ using water quality sonde | ||
| Total suspended solids | TSS | mg/liter | Dry mass captured by filtering 200 ml of sample water through 0.7-μm glass fiber filters | 3 |
| Extracted chlorophyll a | μg C/liter | Fluorometric measurement using acetone extract | 94 | |
| Particulate organic carbon | POC | μg C/liter | Costech ECS 4010 analyzer | 36 |
| Particulate nitrogen | PN | μg N/liter | Costech ECS 4010 analyzer | 36 |
| Dissolved organic carbon | DOC | mg C/liter | Shimadzu TOC 5000-A | 36 |
| Dissolved inorganic carbon | DIC | mg C/liter | Shimadzu TOC 5000-A | 95 |
| Nitrate/nitrite | NO3/NO2 | μg N/liter | Lachat QuikChem 8000 automated ion analyzer | 36 |
| Ammonium | NH4 | μg N/liter | Lachat QuikChem 8000 automated ion analyzer | 36 |
| Dissolved organic nitrogen | DON | μg N/liter | Lachat QuikChem 8000 automated ion analyzer | 34 |
| Dissolved inorganic nitrogen | DIN | μg N/liter | Sum of NO3/NO2 and NH4 | 34 |
| Total dissolved nitrogen | TDN | μg N/liter | Lachat QuikChem 8000 automated ion analyzer | 34 |
| Carbon-to-nitrogen molar ratio | C:N | Ratio of POC to PN | 36 | |
| Phosphate | PO4 | μg P/liter | Lachat QuikChem 8000 automated ion analyzer | 34 |
| Silicate | SiO4 | μM | Lachat QuikChem 8000 automated ion analyzer | 96 |
| Primary productivity | PPR | mg C/m3/h | Light/dark [14C]bicarbonate incorporation | 97, 98 |
| River discharge | m/s | Daily avg from USGS gauging station 02089500 at Ft. Barnwell, 30 km upstream from the head of the NRE | ||
| Rainfall | cm | Daily sum from NWS station at New Bern, NC, airport | ||
| Daily avg wind speed | m/s | Daily avg from NWS station at New Bern, NC, airport | ||
| Phytoplankton photopigments | μg/liter | HPLCa | 41 |
HPLC, high-pressure liquid chromatography.
gDNA extraction and amplicon sequencing.
Undiluted water was filtered through 47-mm 0.4-μm-pore-size polycarbonate filters (HTTP; Millipore) in 100-ml aliquots and stored at −80°C until extraction. Genomic DNA (gDNA) was extracted from duplicate polycarbonate filters (2 filters per sample) using the PowerSoil DNA isolation kit (MoBio Laboratories, Carlsbad, CA) according to the manufacturer's protocol. A mini bead beater (BioSpec Products) was used at speed 3 for 2 min to optimize cellular lysis, and gDNA was eluted into 10 mM Tris buffer (pH 8). A Qubit fluorometer (Thermo Fisher) was used to determine the concentrations of all nucleic acid samples. Duplicate gDNA samples were not pooled and were PCR amplified and sequenced individually.
Extracted gDNA was submitted to the UNC Chapel Hill High-Throughput Sequencing Facility for PCR amplification of the hsp60 universal target (UT) and V4V5 region of the 16S rRNA genes (see Table 4 for primer sequences), library preparation, and amplicon sequencing. The hsp60 primers we used are universal for chaperonin sequences found in bacteria, some archaea, mitochondria, and plasmids and have been used as targets for the detection and identification of Vibrio and other clinically relevant bacteria (26, 32, 33, 74). For the hsp60 PCR, primers were mixed at a 1:3 ratio of H279/H280 to H1612/H1613 to improve template representation of amplicon sequences as described by Hill et al. (75). A total of four PCR-barcoded and pooled amplicon libraries for each gene target were created; two libraries were created for each gene target, one for water samples collected from May to August 2016 and one for samples collected September to December 2016. The barcode sequences are listed in Table S1 in the supplemental material. Two-step library preparation was performed according to a modified system from Lundberg et al. (76). The library preparation did not include peptide nucleic acid (PNA) probes for blocking unwanted amplicons, and we did not use the molecular barcoding protocol, since the first thermocycling step involved more than 2 cycles of PCR amplification. For 16S rRNA PCR amplification (step 1), samples were thermocycled using a program of denaturing at 95°C for 1 min, 10 cycles of primer annealing at 50°C for 2 min and extension at 72°C for 2 min, followed by a cooldown to 4°C. For hsp60 PCR amplification (step 1), samples were thermocycled using a program of denaturing at 95°C for 1 min, 2 cycles of primer annealing at 42°C for 2 min and extension at 72°C for 2 min, 2 cycles of primer annealing at 46.5°C for 2 min and extension at 72°C for 2 min, 2 cycles of primer annealing at 50°C for 2 min and extension at 72°C for 2 min, and 6 cycles of primer annealing at 56°C for 2 min and extension at 72°C for 2 min, followed by a cooldown to 4°C. Multiple annealing temperatures were used for hsp60 PCR amplification to reduce bias at any one temperature (75). The final (step 2) thermocycling conditions were the same for both amplicon reactions: denaturing at 95°C for 45 s followed by 4 cycles of denaturation at 95°C for 15 s with primer annealing at 63°C for 30 s, and 17 cycles of denaturation at 95°C for 15 s with primer annealing at 65°C for 30 s, a final extension at 72°C for 30 s, and a cooldown to 4°C. Magnetic bead size selection was used after each step of amplification. Each pooled library was sequenced in a separate Illumina MiSeq (San Diego, CA) run using 2 × 300 nucleotide (nt) chemistry. All water samples were sequenced in duplicate, and a mock community obtained through BEI Resources, NIAID, NIH as part of the Human Microbiome Project (genomic DNA from microbial mock community B [staggered, low concentration], V5.2L, for 16S rRNA gene sequencing, HM-783D) and negative controls were included in each sequencing run.
TABLE 4.
Amplicon sequencing primers
| Gene target | Primer name | Primer sequencea | Reference |
|---|---|---|---|
| 16S SSU rRNA (V4-V5) | 515F | GCCTCCCTCGCGCCATCAGAGATGTGTATAAGAGACAGNNNNGTGYCAGCMGCCGCGGTAA | 99 |
| 926R | GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTNNNNCCGYCAATTYMTTTRAGTTT | 99 | |
| hsp60 UT | H279 | GCCTCCCCTCGCGCCATCAGAGATGTGATAAGAGACAGNNNNGAIIIIGCIGGIGAYGGIACIACIAC | 100 |
| H1612 | GCCTCCCCTCGCGCCATCAGAGATGTGATAAGAGACAGNNNNAGAIIIIGCIGGYGACGGYACSACSAC | 75 | |
| H280 | GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTNNNNYKIYKITCICCRAAICCIGGIGCYTT | 100 | |
| H1613 | GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTNNNNACGRCGRTCRCCGAAGCCSGGIGCCTT | 75 |
Underlined sequences indicate the molecular adaptor.
Sequence QC and processing.
Sequences for the 16S rRNA gene target and the hsp60 gene target were analyzed independently but using the same data analysis pipeline unless otherwise noted. Illumina MiSeq software was implemented for base calling and initial quality filtering to generate demultiplexed paired FastQ files containing forward and reverse sequencing reads and quality scores. Residual primer and adapter content was trimmed using the JGI BBDuk tool (k = 20, ktrim = r, hdist = 1, minlength = 200). Read quality was assessed before and after trimming using FastQC (77). Trimmed sequences were analyzed using the Qiime pipeline (78) under default parameters unless otherwise indicated. Forward and reverse reads were joined based on overlapping regions and both joined and forward unjoined reads were used for further analysis as described in previous amplicon studies (79, 80). UCLUST (81) as implemented in Qiime was used for de novo operational taxonomic unit (OTU) picking at 97% nucleotide identity, and representative sequences were selected for each OTU. The 97% nucleotide identity threshold is commonly used for 16S rRNA amplicons (82) and has previously been used for hsp60 in a clinical bacterial survey (74). Taxonomy for 16S rRNA sequences was assigned to the Greengenes reference database (http://greengenes.secondgenome.com, accessed February 2017) to the highest similarity score using BLAST (83) in Qiime. For hsp60 sequences, a reference database was created by downloading all nonredundant nucleotide sequences for the hsp60 UT from the manually curated chaperonin database (http://www.cpndb.ca, accessed February 2017). All Vibrio sequences from this database were aligned using MUSCLE (84), and a phylogenetic tree was created using RAxML v8.2.11 (85) in Geneious v10 (86) to ensure sufficient phylogenetic resolution in the reference database (data not shown). An hsp60 taxonomy mapping file was created using a python script for generating Qiime input files (87), and taxonomy was assigned to the curated chaperonin database using the highest BLAST similarity score in Qiime. OTUs that did not return significant hits at >90% sequence similarity and E values <1e−5 were labeled as “no blast hit.” Chimeric sequences were detected using the BLAST fragments method in Qiime. All chimeric sequences, chloroplast sequences, mitochondrial sequences, and OTUs that appeared in fewer than two samples were filtered from downstream analyses.
Data analysis.
Alpha diversity metrics were obtained using the estimate_richness function of phyloseq (88) in R v.3.4.0 (89) for both whole-sequence libraries and for subsetted Vibrio taxa. The observed (number of OTUs), chao1 (richness), and Shannon entropy (OTU-based diversity) metrics were plotted using the plot_richness function of phyloseq. Nonparametric Wilcoxon signed-rank tests in SPSS v24 (IBM, Armonk, NY) were used to test whether alpha diversity estimates for hsp60 versus 16S rRNA amplicons were significantly different. Pearson correlations were used to test for correlations between diversity metrics calculated for both amplicons. Singleton OTUs were included for alpha diversity estimates, as many of these metrics are dependent on the number of singletons, but were filtered prior to further analyses. After alpha diversity calculations, the data were normalized by random subsampling to the lowest number of sequences per sample for each amplicon (20,053 for hsp60 and 22,139 for 16S rRNA). The abundances of Vibrio taxa in both the 16S rRNA and hsp60 libraries were determined by subsetting Vibrio reads at the genus level in phyloseq. Taxa comprising <1% of the Vibrio population were removed from the analysis. Vibrio reads not assigned to the species level were categorized as “Vibrio spp.” Vibrio harveyi and Vibrio campbellii reference sequences for hsp60 were phylogenetically indistinguishable and are presented as a single taxonomic group. Total Vibrio abundance was calculated as the sum of normalized reads for each sample. Because Vibrio taxa identified in the hsp60 sequence data were more abundant, diverse, and of greater clinical relevance than those identified using the 16S rRNA marker, we used hsp60 for all subsequent analyses of Vibrio taxa.
Normalized Vibrio hsp60 read counts were imported into Primer-e v7 (35) for multivariate analyses using the Permanova+ add-on package. Data were square root transformed to preserve subsampled abundances, allowing abundant species to play a greater role while taking into account contributions from less-dominant taxa, as suggested by Nguyen et al. (90) for ecological analysis of read abundance data. Distance-based linear modeling (distLM), a distance-based regression tool for the analysis of multivariate data, was used to assess the relationship between the multivariate cloud, generated using a Bray-Curtis dissimilarity matrix, and environmental, temporal, or categorical predictor variables based on stepwise adjusted R2 selection criteria and 9,999 model permutations. Each variable was initially analyzed separately in the marginal test and then subjected to a stepwise sequential selection procedure, where the amount of multivariate variability explained by each variable added to the model is conditional on the variables already included in the model. Both named storms were included in the distLM in two ways: first, as categorical variables, where samples were labeled either storm samples (samples associated with increased freshwater discharge into the estuary after a storm event) or nonstorm samples, and second, as temporal variables in which the number of days after the storm was considered. A full list of tested variables is in Table S2. A dbRDA plot was used to visualize the ordination of the fitted distLM analysis. An unfitted ordination (principle components ordination) was also plotted and is available in the supplemental material. The relative abundances of potentially pathogenic species V. vulnificus, V. parahaemolyticus, and V. cholerae are presented in the dbRDA ordination using the bubble plot function in Primer-e. Quantitative environmental variables that were significant in the distLM were projected as vectors on the dbRDA ordination and indicate both the strength and direction of significant environmental gradients. ANOSIM tests with 9,999 permutations were used to test whether the Vibrio communities in samples associated with Tropical Storm Colin and Hurricane Matthew were more similar to each other than to communities not associated with a storm.
To investigate the potential links between Vibrio, phytoplankton, and the dominant bacterial phyla in the water column, the following Spearman rank correlation matrices were calculated in R: (i) for Vibrio taxa and phytoplankton photopigments and (ii) for Vibrio taxa and the top 15 most abundant 16S rRNA phyla. Significant (P < 0.05) Vibrio-Vibrio and Vibrio-phytoplankton photopigment correlations were plotted using the package corrplot (91) in R. Vibrio species were ordered into cooccurring groups within the plot using the AOE. Vibrio-16S rRNA phyla correlations (P < 0.05, r > 0.4) were plotted as networks using the Metscape plugin (92) for Cytoscape v3.5.1 (93). Spearman rank correlations were also used to assess the relationships between V. vulnificus, V. parahaemolyticus, V. cholerae, and various environmental parameters and were plotted in corrplot.
Accession numbers.
Raw sequence data are available in NCBI's SRA database by sample under accession numbers SRR6843512 to SRR6843673 (16S rRNA) and SRR6873873 to SRR6874034 (hsp60).
Supplementary Material
ACKNOWLEDGMENTS
We thank Hans Paerl and the members of his research group at the UNC Institute of Marine Sciences for allowing us to access the NRE Modeling and Monitoring Project (ModMon) data and for collecting water samples. This project would not have been possible without the invaluable assistance we received during sample collection and processing from members of the Noble lab, especially Justin Hart, Denene Blackwood, Brett Froelich, Joe Purifoy, and Rachel Canty. We also thank Amy Perou and Piotr Mieczkowski at the UNC High-Throughput Sequencing Facility, Hemant Kelkar at the UNC Center for Bioinformatics, and Jeff Roach at UNC Research Computing for their guidance during sequencing and data analyses.
This work was supported by grants from the UNC Research Opportunities Initiative and NSF AIR (award number 1602023) to R.T.N.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00333-18.
REFERENCES
- 1.Thompson JR, Randa MA, Marcelino LA, Tomita-Mitchell A, Lim E, Polz MF. 2004. Diversity and dynamics of a North Atlantic coastal Vibrio community. Appl Environ Microbiol 70:4103–4110. doi: 10.1128/AEM.70.7.4103-4110.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Froelich BA, Williams TC, Noble RT, Oliver JD. 2012. Apparent loss of Vibrio vulnificus from North Carolina oysters coincides with a drought-induced increase in salinity. Appl Environ Microbiol 78:3885–3889. doi: 10.1128/AEM.07855-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wetz JJ, Blackwood AD, Fries JS, Williams ZF, Noble RT. 2008. Trends in total Vibrio spp. and Vibrio vulnificus concentrations in the eutrophic Neuse River Estuary, North Carolina, during storm events. Aquat Microb Ecol 53:141–149. doi: 10.3354/ame01223. [DOI] [Google Scholar]
- 4.DePaola A, McLeroy S, McManus G. 1997. Distribution of Vibrio vulnificus phage in oyster tissue and other estuarine habitats. Appl Environ Microbiol 63:2464–2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee K, Ruby EG. 1994. Effect of the squid host on the abundance and distribution of symbiotic Vibrio fischeri in nature. Appl Environ Microbiol 60:1565–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hornák K, Masin M, Jezbara J, Bettarel Y, Nedoma J, Sime-Ngando T, Simek K. 2005. Effects of decreased resource availability, protozoan grazing and viral impact on a structure of bacterioplankton in a canyon-shaped reservoir. FEMS Microbiol Ecol 52:315–327. doi: 10.1016/j.femsec.2004.11.013. [DOI] [PubMed] [Google Scholar]
- 7.Urdaci MC, Stal LJ, Marchand M. 1988. Occurrence of nitrogen fixation among Vibrio spp. Arch Microbiol 150:224–229. doi: 10.1007/BF00407784. [DOI] [Google Scholar]
- 8.Criminger JD, Hazen TH, Sobecky PA, Lovell CR. 2007. Nitrogen fixation by Vibrio parahaemolyticus and its implications for a new ecological niche. Appl Environ Microbiol 73:5959–5961. doi: 10.1128/AEM.00981-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eiler A, Gonzalez-Rey C, Allen S, Bertilsson S. 2007. Growth response of Vibrio cholerae and other Vibrio spp. to cyanobacterial dissolved organic matter and temperature in brackish water. FEMS Microbiol Ecol 60:411–418. doi: 10.1111/j.1574-6941.2007.00303.x. [DOI] [PubMed] [Google Scholar]
- 10.Gilbert JA, Steele JA, Caporaso JG, Steinbruck L, Reeder J, Temperton B, Huse S, McHardy AC, Knight R, Joint I, Somerfield P, Fuhrman JA, Field D. 2012. Defining seasonal marine microbial community dynamics. ISME J 6:298–308. doi: 10.1038/ismej.2011.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daniels NA, MacKinnon L, Bishop R, Altekruse S, Ray B, Hammond RM, Thompson S, Wilson S, Bean NH, Griffin PM, Slutsker L. 2000. Vibrio parahaemolyticus infections in the United States, 1973–1998. J Infect Dis 181:1661–1666. doi: 10.1086/315459. [DOI] [PubMed] [Google Scholar]
- 12.Givens CE, Bowers JC, DePaola A, Hollibaugh JT, Jones JL. 2014. Occurrence and distribution of Vibrio vulnificus and Vibrio parahaemolyticus—potential roles for fish, oyster, sediment, and water. Lett Appl Microbiol 58:503–510. doi: 10.1111/lam.12226. [DOI] [PubMed] [Google Scholar]
- 13.Pruzzo C, Gallo G, Canesi L. 2005. Persistence of vibrios in marine bivalves: the role of interactions with haemolymph components. Environ Microbiol 7:761–772. doi: 10.1111/j.1462-2920.2005.00792.x. [DOI] [PubMed] [Google Scholar]
- 14.Letchumanan V, Chan K, Lee L. 2014. Vibrio parahaemolyticus: a review on the pathogenesis, prevalence, and advance molecular identification techniques. Front Microbiol 5:705. doi: 10.3389/fmicb.2014.00705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shen X, Cai Y, Liu C, Liu W, Hui Y, Su Y. 2009. Effect of temperature on uptake and survival of Vibrio parahaemolyticus in oysters (Crassostrea plicatula). Int J Food Microbiol 136:129–132. doi: 10.1016/j.ijfoodmicro.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 16.Dechet AM, Yu PA, Koram N, Painter J. 2008. Nonfoodborne Vibrio infections: an important cause of morbidity and mortality in the United States, 1997–2006. Clin Infect Dis 46:970–976. doi: 10.1086/529148. [DOI] [PubMed] [Google Scholar]
- 17.Constantin de Magny G, Long W, Brown CW, Hood RR, Huq A, Murtugudde R, Colwell RR. 2009. Predicting the distribution of Vibrio spp. in the Chesapeake Bay: a Vibrio cholerae case study. Ecohealth 6:378–389. doi: 10.1007/s10393-009-0273-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davis BJ, Jacobs JM, Davis MF, Schwab KJ, DePaola A, Curriero FC. 2017. Environmental determinants of Vibrio parahaemolyticus in the Chesapeake Bay. Appl Environ Microbiol 83:e01147-17. doi: 10.1128/AEM.01147-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takemura AF, Chien DM, Polz MF. 2014. Associations and dynamics of Vibrionaceae in the environment, from the genus to the population level. Front Microbiol 5:38. doi: 10.3389/fmicb.2014.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martinez-Urtaza J, Bowers JC, Trinanes J, DePaola A. 2010. Climate anomalies and the increasing risk of Vibrio parahaemolyticus and Vibrio vulnificus illnesses. Food Res Int 43:1780–1790. doi: 10.1016/j.foodres.2010.04.001. [DOI] [Google Scholar]
- 21.Fries JS, Characklis GW, Noble RT. 2008. Sediment-water exchange of Vibrio spp. and fecal indicator bacteria: implications for persistence and transport in the Neuse River Estuary, North Carolina. Water Res 42:941–950. doi: 10.1016/j.watres.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 22.Hsieh JL, Fries JS, Noble RT. 2008. Dynamics and predictive modelling of Vibrio spp. in the Neuse River Estuary, North Carolina, USA. Environ Microbiol 10:57–64. doi: 10.1111/j.1462-2920.2007.01429.x. [DOI] [PubMed] [Google Scholar]
- 23.Steele JA, Countway PD, Xia L, Vigil PD, Beman JM, Kim DY, Chow CT, Sachdeva R, Jones AC, Schwalback MS, Rose JM, Hewson I, Patel A, Sun F, Caron DA, Fuhrman JA. 2011. Marine bacterial, archaeal and protistan association networks reveal ecological linkages. ISME J 5:1414–1425. doi: 10.1038/ismej.2011.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cano-Gomez A, Hoj L, Owens L, Andreakis N. 2011. Multilocus sequence analysis provides basis for fast and reliable identification of Vibrio harveyi-related species and reveals previous misidentification of important marine pathogens. Syst Appl Microbiol 34:561–565. doi: 10.1016/j.syapm.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 25.Pascual J, Macian MC, Arahal DR, Garay E, Pujalte MJ. 2010. Multilocus sequence analysis of the central clade of the genus Vibrio by using the 16S rRNA, recA, pyrH, rpoD, gyrB, rctB, and toxR genes. Int J Syst Evol Microbiol 60:154–165. doi: 10.1099/ijs.0.010702-0. [DOI] [PubMed] [Google Scholar]
- 26.Szabo G, Preheim SP, Kauffman KM, David LA, Shapiro J, Alm EJ, Polz MF. 2013. Reproducibility of Vibrionaceae population structure in coastal bacterioplankton. ISME J 7:509–519. doi: 10.1038/ismej.2012.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Froelich BA, Noble RT. 2014. Factors affecting the uptake and retention of Vibrio vulnificus in oysters. Appl Environ Microbiol 80:7454–7459. doi: 10.1128/AEM.02042-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Broberg CA, Calder TJ, Orth K. 2011. Vibrio parahaemolyticus cell biology and pathogenicity determinants. Microbes Infect 13:992–1001. doi: 10.1016/j.micinf.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ininbergs K, Bergman B, Larsson J, Ekman M. 2015. Microbial metagenomics in the Baltic Sea: recent advancements and prospects for environmental monitoring. AMBIO 44(Suppl 3):439–450. doi: 10.1007/s13280-015-0663-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shaw JLA, Monis P, Weyrich LS, Sawade E, Drikas M, Cooper AJ. 2015. Using amplicon sequencing to characterize and monitor bacterial diversity in drinking water distribution systems. Appl Environ Microbiol 81:6463–6473. doi: 10.1128/AEM.01297-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kwok AYC, Wilson JT, Coulthart M, Ng L, Mutharia L, Chow AW. 2002. Phylogenetic study and identification of human pathogenic Vibrio species based on partial hsp60 gene sequences. Can J Microbiol 48:903–910. doi: 10.1139/w02-089. [DOI] [PubMed] [Google Scholar]
- 32.Silvester R, Alexander D, Antony AC, Hatha M. 2017. GroEL PCR-RFLP—an efficient tool to discriminate closely related pathogenic Vibrio species. Microb Pathog 105:196–200. doi: 10.1016/j.micpath.2017.02.029. [DOI] [PubMed] [Google Scholar]
- 33.Schellenberg JJ, Links MG, Hill JE, Dumonceaux TJ, Kimani J, Jaoko W, Wachihi C, Mungai JN, Peters GA, Tyler S, Graham M, Severini A, Fowke KR, Ball TB, Plummer FA. 2011. Molecular definition of vaginal microbiota in East African commercial sex workers. Appl Environ Microbiol 77:4066–4074. doi: 10.1128/AEM.02943-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peierls B, Paerl HW. 2010. Temperature, organic matter, and the control of bacterioplankton in the Neuse River Estuary and Pamlico Sound estuarine system. Aquat Microb Ecol 60:139–149. doi: 10.3354/ame1415. [DOI] [Google Scholar]
- 35.Clarke KR, Gorley RN. 2015. PRIMER v7: user manual/tutorial. PRIMER-E, Plymouth, United Kingdom. [Google Scholar]
- 36.Peierls BL, Christian RR, Paerl HW. 2003. Water quality and phytoplankton as indicators of hurricane impacts on a large estuarine system. Estuaries 26.5:1329–1343. doi: 10.1007/BF02803635. [DOI] [Google Scholar]
- 37.Garrido-Maestu A, Chapela M, Vieites Cabado AG. 2015. lolB gene, a valid alternative for qPCR detection of Vibrio cholerae in food and environmental samples. Food Microbiol 46:535–540. doi: 10.1016/j.fm.2014.09.012. [DOI] [PubMed] [Google Scholar]
- 38.Campbell MS, Wright AC. 2003. Real-time PCR analysis of Vibrio vulnificus from oysters. Appl Environ Microbiol 69:7137–7144. doi: 10.1128/AEM.69.12.7137-7144.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taiwo M, Baker-Austin C, Powell A, Hodgson E, Natas OB, Walker DI. 2017. Comparison of toxR and tlh based PCR assays for Vibrio parahaemolyticus. Food Control 77:116–120. doi: 10.1016/j.foodcont.2017.02.009. [DOI] [Google Scholar]
- 40.Blackwell KD, Oliver JD. 2008. The ecology of Vibrio vulnificus, Vibrio cholerae, and Vibrio parahaemolyticus in North Carolina estuaries. J Microbiol 46:146–153. doi: 10.1007/s12275-007-0216-2. [DOI] [PubMed] [Google Scholar]
- 41.Paerl HW, Hall NS, Peierls BL, Rossignol KL, Joyner AR. 2013. Hydrologic variability and its control of phytoplankton community structure and function in two shallow, coastal, lagoonal ecosystems: the Neuse and New River Estuaries, North Carolina, USA. Estuaries Coasts 37(Suppl 1):31–45. doi: 10.1007/s12237-013-9686-0. [DOI] [Google Scholar]
- 42.Turner JW, Good B, Cole D, Lipp EK. 2009. Plankton composition and environmental factors contribute to Vibrio seasonality. ISME J 3:1082–1092. doi: 10.1038/ismej.2009.50. [DOI] [PubMed] [Google Scholar]
- 43.DePaola A, Nordstrom JL, Bowers JC, Wells JG, Cook DW. 2003. Seasonal abundance of total and pathogenic Vibrio parahaemolyticus in Alabama oysters. Appl Environ Microbiol 69:1521–1526. doi: 10.1128/AEM.69.3.1521-1526.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parveen S, Hettiarachchi KA, Bowers JC, Jones JL, Tamplin ML, McKay R, Beatty W, Brohawn K, DaSilva LV, DePaola A. 2008. Seasonal distribution of total and pathogenic Vibrio parahaemolyticus in Chesapeake Bay oysters and waters. Int J Food Microbiol 128:354–361. doi: 10.1016/j.ijfoodmicro.2008.09.019. [DOI] [PubMed] [Google Scholar]
- 45.O'Neill KR, Jones SH, Grimes DJ. 1992. Seasonal incidence of Vibrio vulnificus in the Great Bay estuary of New Hampshire and Maine. Appl Environ Microbiol 58:3257–3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Froelich BA, Bowen J, Gonzalez R, Snedeker A, Noble R. 2013. Mechanistic and statistical models of total Vibrio abundance in the Neuse River Estuary. Water Res 47:5783–5793. doi: 10.1016/j.watres.2013.06.050. [DOI] [PubMed] [Google Scholar]
- 47.Oliver JD. 2010. Recent findings on the viable but nonculturable state in pathogenic bacteria. FEMS Microbiol Rev 34:415–425. doi: 10.1111/j.1574-6976.2009.00200.x. [DOI] [PubMed] [Google Scholar]
- 48.Westrich JR, Ebling AM, Landing WM, Joyner JL, Kemp KM, Griffin DW, Lipp EK. 2016. Saharan dust nutrients promote Vibrio bloom formation in marine surface waters. Proc Natl Acad Sci U S A 113:5964–5969. doi: 10.1073/pnas.1518080113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rudek J, Paerl HW, Mallin MA, Bates PW. 1991. Seasonal and hydrological control of phytoplankton nutrient limitation in the lower Neuse River Estuary, North Carolina. Mar Ecol Prog Ser 75:133–142. [Google Scholar]
- 50.Chowdhury MAR, Yamanaka H, Miyoshi S, Shinoda S. 1990. Ecology and seasonal distribution of Vibrio parahaemolyticus in aquatic environments of a temperate region. FEMS Microbiol Lett 74:1–9. doi: 10.1111/j.1574-6968.1990.tb04046.x. [DOI] [Google Scholar]
- 51.Deepanjali A, Kumar HS, Karunasagar I. 2005. Seasonal variation in abundance of total and pathogenic Vibrio parahaemolyticus bacteria in oysters along the southwest coast of India. Appl Environ Microbiol 71:3575–3580. doi: 10.1128/AEM.71.7.3575-3580.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cheng W, Juang F, Chen J. 2004. The immune response of Taiwan abalone Haliotis diversicolor supertexta and its susceptibility to Vibrio parahaemolyticus at different salinity levels. Fish Shellfish Immunol 16:295–306. doi: 10.1016/S1050-4648(03)00111-6. [DOI] [PubMed] [Google Scholar]
- 53.Kaneko T, Colwell RR. 1974. Distribution of Vibrio parahaemolyticus and related organisms in the Atlantic Ocean off South Carolina and Georgia. Appl Microbiol 28:1009–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Horie S, Okuzumi M, Kato N. 1966. Comparative observations on the range of growth temperature among three biotypes of Vibrio parahaemolyticus in plankton and fish in the open sea. Bull Jap Soc Sci Fish 30:786–791. (In Japanese) doi: 10.2331/suisan.30.786. [DOI] [Google Scholar]
- 55.Miles DW, Ross T, Olley J, McMeekin TA. 1997. Development and evaluation of a predictive model for the effect of temperature and water activity on the growth rate of Vibrio parahaemolyticus. Int J Food Microbiol 38:133–142. doi: 10.1016/S0168-1605(97)00100-1. [DOI] [PubMed] [Google Scholar]
- 56.Centers for Disease Control and Prevention. 2005. Vibrio illnesses after Hurricane Katrina—multiple states, Aug-Sep 2005. MMWR Morb Mortal Wkly Rep 54:928–931. [PubMed] [Google Scholar]
- 57.Randa MA, Polz MF, Lim E. 2004. Effects of temperature and salinity on Vibrio vulnificus population dynamics as assessed by quantitative PCR. Appl Environ Microbiol 70:5469–5476. doi: 10.1128/AEM.70.9.5469-5476.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shaw KS, Jacobs JM, Crump BC. 2014. Impact of Hurricane Irene on Vibrio vulnificus and Vibrio parahaemolyticus concentrations in surface water, sediment, and cultured oysters in Chesapeake Bay, MD, USA. Front Microbiol 5:204. doi: 10.3389/fmicb.2014.00204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Datta PP, Bhadra RK. 2003. Cold shock response and major cold shock proteins of Vibrio cholerae. Appl Environ Microbiol 69:6361–6369. doi: 10.1128/AEM.69.11.6361-6369.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Singleton FL, Attwell R, Jangi S, Colwell RR. 1982. Effects of temperature and salinity on Vibrio cholerae growth. Appl Environ Microbiol 44:1047–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mouriño-Pérez RR, Worden AZ, Azam F. 2003. Growth of Vibrio cholerae O1 in red tide waters off California. Appl Environ Microbiol 69:6923–6931. doi: 10.1128/AEM.69.11.6923-6931.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Seong KA, Jeong HJ. 2011. Interactions between the pathogenic bacterium Vibrio parahaemolyticus and red-tide dinoflagellates. Ocean Sci 46:105–115. doi: 10.1007/s12601-011-0010-2. [DOI] [Google Scholar]
- 63.Eiler A, Langenheder S, Bertilsson S, Tranvik LJ. 2003. Heterotrophic bacterial growth efficiency and community structure at different natural organic carbon concentrations. Appl Environ Microbiol 69:3701–3709. doi: 10.1128/AEM.69.7.3701-3709.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Davis BR, Fanning GR, Madden JM, Steigerwalt AG, Bradford HB, Smith HL, Brenner DJ. 1981. Characterization of biochemically atypical Vibrio cholerae strains and designation of a new pathogenic species, Vibrio mimicus. J Clin Microbiol 14:631–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ransom DP, Lannan CN, Rohovec JS, Fryer JL. 1984. Comparison of histopathology caused by Vibrio anguillarum and Vibrio ordalii in three species of Pacific salmon. J Fish Dis 7:107–115. doi: 10.1111/j.1365-2761.1984.tb00913.x. [DOI] [Google Scholar]
- 66.Chowdhury MA, Yamanaka H, Miyoshi S, Aziz KM, Shinoda S. 1989. Ecology of Vibrio mimicus in aquatic environments. Appl Environ Microbiol 55:2073–2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Colquhoun DJ, Aase IL, Wallace C, Barklien A, Gravningen K. 2004. First description of Vibrio ordalii from Chile. Bull Eur Assoc Fish Pathol 24:185–188. [Google Scholar]
- 68.Chaturvedi P, Agrawal MK, Bagchi SN. 2015. Microcystin-producing and non-producing cyanobacterial blooms collected from the Central India harbor potentially pathogenic Vibrio cholerae. Ecotoxicol Environ Saf 115:67–74. doi: 10.1016/j.ecoenv.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 69.Islam M, Rahim Z, Alam MJ, Begum S, Moniruzzaman SM, Umeda A, Amako K, Albert MJ, Sack RB, Huq A, Colwell RR. 1999. Association of Vibrio cholerae O1 with the cyanobacterium, Anabaena sp., elucidated by polymerase chain reaction and transmission electron microscopy. Trans R Soc Trop Med Hyg 93:36–40. doi: 10.1016/S0035-9203(99)90171-2. [DOI] [PubMed] [Google Scholar]
- 70.Epstein PR. 1993. Algal blooms in the spread and persistence of cholera. Biosystems 31:209–221. doi: 10.1016/0303-2647(93)90050-M. [DOI] [PubMed] [Google Scholar]
- 71.Ward CE. 2014. Investigating the interactions between Cyanobacteria and Vibrio parahaemolyticus. Undergraduate honors thesis. University of New Hampshire, Durham, NH. [Google Scholar]
- 72.Luettich RA, Carr SD, Reynolds-Fleming JV, Fulcher CW, McNinch JE. 2002. Semi-diurnal seiching in a shallow, micro-tidal lagoonal estuary. Cont Shelf Res 22:1669–1681. doi: 10.1016/S0278-4343(02)00031-6. [DOI] [Google Scholar]
- 73.Ghazaleh MN, Froelich BA, Noble RT. 2014. The effect of storage time on Vibrio spp. and fecal indicator bacteria in an Isco autosampler. J Microbiol Methods 104:109–116. doi: 10.1016/j.mimet.2014.06.021. [DOI] [PubMed] [Google Scholar]
- 74.Peterson SW, Knox NC, Golding GR, Tyler SD, Tyler AD, Mabon P, Embree JE, Fleming F, Fanella S, Van Domselaar G, Mulvey MR, Graham MR. 2016. A study of the infant nasal microbiome development over the first year of life and in relation to their primary adult caregivers using cpn60 universal target (UT) as a phylogenetic marker. PLoS One 11:e0152493. doi: 10.1371/journal.pone.0152493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hill JE, Town JR, Hemmingsen SM. 2006. Improved template representation in cpn60 polymerase chain reaction (PCR) product libraries generated from complex templates by application of a specific mixture of PCR primers. Environ Microbiol 8:741–746. doi: 10.1111/j.1462-2920.2005.00944.x. [DOI] [PubMed] [Google Scholar]
- 76.Lundberg DS, Yourstone S, Mieczkowski P, Jones CD, Dangl JL. 2013. Practical innovations for high-throughput amplicon sequencing. Nat Methods 10:999–1002. doi: 10.1038/nmeth.2634. [DOI] [PubMed] [Google Scholar]
- 77.Andrews S. 2010. FastQC, a quality control tool for high throughput sequence data. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- 78.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Gonzalez Pena A, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mrozinska S, Radkowski P, Gosiewski T, Szopa M, Bulanda M, Ludwig-Galezowska HL, Morawsak I, Sroka-Oleksiak A, Matejko B, Kapusta P, Salamon D, Malecki MT, Wolkow PP, Klupa T. 2016. Qualitative parameters of the colonic flora in patients with HNF1A-MODY are different from those observed in type 2 diabetes mellitus. J Diabetes Res 2016:3876764. doi: 10.1155/2016/3876764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gosiewski T, Ludwig-Galezowska AH, Huminska K, Sroka-Oleksiak A, Radkowski P, Salamon D, Wojciechowicz J, Kus-Slowinska M, Bulanda M, Wolkow PP. 2017. Comprehensive detection and identification of bacterial DNA in the blood of patients with sepsis and healthy volunteers using a next-generation sequencing method—the observation of DNAemia. Eur J Clin Microbiol Infect Dis 36:329–336. doi: 10.1007/s10096-016-2805-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Edgar EC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 82.Konstantinidis KT, Tiedje JM. 2005. Genomic insights that advance the species definition for prokaryotes. Proc Natl Acad Sci U S A 102:2567–2572. doi: 10.1073/pnas.0409727102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 84.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Mentijies P, Drummond A. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Baker CM. 7 October 2016. Entrez qiime: a utility for generating QIIME input files from the NCBI databases, v2.0. https://github.com/bakerccm/entrez_qiime.
- 88.McMurdie PJ, Holmes S. 2013. Basic storage, access, and manipulation of phylogenetic sequencing data with phyloseq. https://github.com/joey711/phyloseq.
- 89.R Core Team. 2013. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: http://www.R-project.org. [Google Scholar]
- 90.Nguyen NH, Smith D, Kabir P, Kennedy P. 2014. Parsing ecological signal from noise in next generation amplicon sequencing. New Phytol 205:1389–1393. doi: 10.1111/nph.12923. [DOI] [PubMed] [Google Scholar]
- 91.Wei T, Simko V. 2017. R package “corrplot”: visualization of a correlation matrix, v0.84. https://github.com/taiyun/corrplot.
- 92.Basu S, Duren W, Evans CR, Burant C, Michailidis G, Karnovsky A. 2017. Sparse network modelling and Metscape-based visualization methods for the analysis of large-scale metabolomics data. Bioinformatics 33:1545–1553. doi: 10.1093/bioinformatics/btx012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2489–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Arar EJ, Collins GB. 1997. Method 445.0: in vitro determination of chlorophyll a and pheophytin a in marine and freshwater algae by fluorescence. United States Environmental Protection Agency, Office of Research and Development, National Exposure Laboratory, Cincinnati, OH. [Google Scholar]
- 95.Crosswell JR, Wetz MS, Hales B, Paerl HW. 2012. Air-water CO2 fluxes in the microtidal Neuse River Estuary, North Carolina. J Geophys Res Oceans 117:1–12. doi: 10.1029/2012JC007925. [DOI] [Google Scholar]
- 96.Grasshof K, Erhardt M, Kremling K (ed). 1976. Methods of seawater analysis, 2nd ed Verlag Chemie, Weinheim, Germany. [Google Scholar]
- 97.Mallin MA, Paerl HW. 1992. Effects of variable irradiance on phytoplankton productivity in shallow estuaries. Limnol Oceanogr 37:54–62. doi: 10.4319/lo.1992.37.1.0054. [DOI] [Google Scholar]
- 98.Wetz MS, Hutchinson EA, Lunetta RS, Paerl HW, Taylor JC. 2011. Severe droughts reduce estuarine primary productivity with cascading effects on higher trophic levels. Limnol Oceanogr 56:627–638. doi: 10.4319/lo.2011.56.2.0627. [DOI] [Google Scholar]
- 99.Parada AE, Needham DM, Fuhrman JA. 2016. Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ Microbiol 18:1403–1414. doi: 10.1111/1462-2920.13023. [DOI] [PubMed] [Google Scholar]
- 100.Goh SH, Potter S, Wood JO, Hemmingsen SM, Reynolds RP, Chow AW. 1996. HSP60 gene sequences as universal targets for microbial species identification: studies with coagulase-negative staphylococci. J Clin Microbiol 34:818–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.