ABSTRACT
In this study, we identified a P450 enzyme (STH10) and an oxidoreductase (POR) from Thanatephorus cucumeris NBRC 6298 by a combination of transcriptome sequencing and heterologous expression in Pichia pastoris. The biotransformation of 11-deoxycortisol was performed by using Pichia pastoris whole cells coexpressing sth10 and por, and the product analysis indicated that the STH10 enzyme possessed steroidal 19- and 11β-hydroxylase activities. This is a novel fungal P450 enzyme with 19-hydroxylase activity, which is different from the known steroidal aromatase cytochrome P450 19 (CYP19) and CYP11B families of enzymes.
IMPORTANCE Hydroxylation is one of the most important reactions in steroid functionalization; in particular, C-19 hydroxylation produces a key intermediate for the synthesis of 19-nor-steroid drugs without a C-19 angular methyl group in three chemoenzymatic steps, in contrast to the current industrial process, which uses 10 chemical reactions. However, hydroxylation of the C-19 angular methyl group remains a very challenging task due to the high level of steric resistance to the C-19 methyl group between the A and B rings. The present report describes a novel fungal P450 enzyme with 19-hydroxylase activity. This opens a new venue for searching effective biocatalysts for the useful process of steroidal C-19 hydroxylation, although further studies for better understanding of the structural basis of the regioselectivity and substrate specificity of this fungal steroidal 19-hydroxylase are warranted to facilitate the engineering of this enzyme for industrial applications.
KEYWORDS: 11β-hydroxylase, 19-hydroxylase, steroid hydroxylation, cytochrome P450
INTRODUCTION
Steroidal compounds are widely used as pharmaceuticals such as anti-inflammatory, immunosuppressive, and anticancer agents (1). Steroid-based drugs currently represent the second largest category of marketed drugs after antibiotics (2). The physiological activity and pharmaceutical value of steroids vary with the different functional group modifications of the steroidal core such as hydroxylation, dehydrogenation, esterification, and so on (3). Hydroxylation is considered one of the most important reactions in steroid functionalization. This reaction introduces an oxygen molecule into the inactivated C-H bond on the steroid core and remains a great challenge with respect to steroidal transformation for steroid chemistry. Steroid hydroxylation may result in profound changes in physicochemical and pharmaceutical properties such as bioactivity, solubility, and absorption, and the hydroxylated steroids can be used as high-value drugs or as intermediates for further chemical synthesis (2). For example, 11β,17,21-trihydroxypregn-4-ene-3,20-dione (hydrocortisone [cortisol]) is used as anti-inflammatory steroid drug (4), and 11α-hydroxylated pregn-4-ene-3,20-dione represents an commercially important intermediate in the production of contraceptive drugs (5). A 7α-hydroxy derivative of 3β-hydroxyandrost-5-en-17-one (dehydroepiandrosterone) exhibits properties that are more highly immunoprotective than those exhibited by dehydroepiandrosterone (6). The 16α-hydroxylated steroids have increased glucocorticoid activity (7).
Among the various hydroxylation reactions of steroids, hydroxylation at C-19 produces a very useful hydroxysteroid intermediate for the synthesis of 19-nor-steroids without the C-19 angular methyl group in three chemoenzymatic steps (see, for example, 19-nor-androstenedione in Fig. 1), while they are currently prepared in a series of 10 chemical reactions (8). The 19-nor-steroids are widely used key intermediates for the production of highly effective contraceptives, such as norethindrone (9), mifepristone (10), and tibolone (11). However, hydroxylation of the C-19 angular methyl group remains a very difficult task due to the fact that the C-19 methyl group is located in the middle of the A and B rings of the steroid, with high levels of steric resistance.
FIG 1.
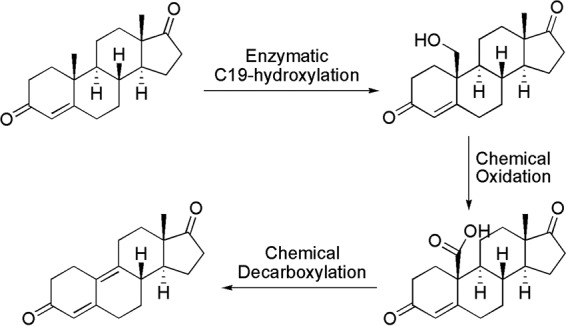
Transformation of androstenedione to 19-nor-androstenedione via a three-step chemoenzymatic process.
The best-known biocatalyst that leads to the hydroxylation at C-19 of steroids is a P450 enzyme, i.e., aromatase. However, it catalyzes the conversion of androgens in a complicated three-step reaction through the formation of 19-hydroxy and 19-aldehyde intermediates, followed by the aromatization (step III) of the A ring to produce estrogens (12). Until now, the ideal biocatalyst that can catalyze 19-hydroxylation of steroids without steroidal aromatization has remained unknown.
In 1961, a fungus (Thanatephorus cucumeris) that catalyzed the hydroxylation of 17,21-dihydroxypregn-4-ene-3,20-dione (11-deoxycortisol, RSS, cortexolone, or cortodoxone) to produce 19-hydroxy and 11β-hydroxy 11-deoxycortisol was discovered (13). To the best of our knowledge, this is the only microorganism that hydroxylates the C-19 of steroids. In 1982, Clark and colleagues further investigated the 11β- and 19-hydroxylation of 11-deoxycortisol by T. cucumeris and confirmed that the 11β- and 19-hydroxylation enzyme(s) of T. cucumeris was inducible by 11-deoxycortisol. They showed that the ratio of 11β- and 19-hydroxylation products remained 1:0.84 (14). Recently, we have found that the 11β- and 19-hydroxylation of 11-deoxycortisol by T. cucumeris was affected by the initial pH of the cell culture (15). Therefore, we envisioned the possibility that T. cucumeris contains one or two unique P450 enzymes responsible for 11β- and 19-hydroxylation of 11-deoxycortisol.
In this study, de novo RNA sequencing was performed after exposure of the T. cucumeris to 11-deoxycortisol. On the basis of the bioinformatics analysis, a few inducible cytochrome P450 (CYP) genes were identified in the sequence of T. cucumeris. Among them, a highly induced CYP gene was cloned and expressed in Pichia pastoris. The recombinant enzyme was functionally characterized with both C-19 and 11β-hydroxylase activities.
RESULTS
Transcriptome assembly of T. cucumeris.
On the basis of the fact that the 19- and 11β-hydroxylases were inducible by 11-deoxycortisol under the T. cucumeris biotransformation conditions, we inferred that the target genes responsible for 11β- and 19-hydroxylation of 11-deoxycortisol would be upregulated at the transcriptional level. The total amounts of RNAs from the cultures with or without 11-deoxycortisol were isolated. More than 19 million reads were obtained from sequencing for each sample, generating 2.3 billion nucleotides on average (Table 1). De novo assembly using Trinity produced 45,065 transcripts (mean length, 1,228 bp; N50, 1,939 bp), corresponding to 31,466 unigenes (mean length, 1,004 bp; N50, 1,653 bp) (Table 2). The length distribution of contigs assembled from all clean reads is shown in Fig. 2A. As shown in Fig. 2B, 88.2% of unigenes have over 80% similarity to entries in the public database.
TABLE 1.
Overview of the RNA-seq outcome, including numbers for total raw, high-quality reads and nucleotides and statistical Q20, Q30, and GC percentagesa
| Transcriptome | No. of raw reads | No. of clean reads | No. of clean bases | Q20 (%) | Q30 (%) | GC (%) |
|---|---|---|---|---|---|---|
| Sample A | 20,106,548 | 19,791,168 | 2.47 billion | 96.18 | 92.03 | 51.82 |
| Sample B | 18,486,306 | 18,139,002 | 2.27 billion | 96.02 | 91.92 | 50.86 |
Q20 and Q30 represent the percentages of nucleotide bases with Phred quality scores (Q) of 20 and 30, respectively.
TABLE 2.
An overview of annotated unigenes in seven databasea
| Database(s) | No. of unigenes annotated | % of total unigenes annotated |
|---|---|---|
| nr | 24,871 | 79.04 |
| nt | 6,574 | 20.89 |
| KO | 7,623 | 24.22 |
| SwissProt | 12,676 | 40.28 |
| Pfam | 14,280 | 45.38 |
| GO | 14,306 | 45.46 |
| KOG | 9,069 | 28.82 |
| All | 2,585 | 8.21 |
| At least one | 25,619 | 81.41 |
| Total | 31,466 | 100 |
nr, NCBI nonredundant nucleotide sequence database; nt, NCBI nonredundant protein sequence database; KO, Eukaryotic Ontology database; SwissProt, Swiss-Prot protein database; Pfam, Pfam database; GO, Gene Ontology database; KOG, Eukaryotic Ortholog Group database.
FIG 2.
Overview of de novo transcriptome assembly in T. cucumeris. (A) Length distribution of contigs assembled from all cleaned reads from two samples of T. cucumeris as determined using Trinity software. (B) Similarity distribution of the top BLAST hits for each unigene.
Annotation and classification and recombinant strains for expression of candidate CYP and P450 oxidoreductase (POR) genes.
A total of 233 candidate genes of p450 were identified by searching for transcripts possessing the cytochrome p450 domain (PF00067) against a Pfam-A database. Differential transcript analysis revealed that 8 candidate cytochrome P450 genes were transcribed at levels that were from 2-fold higher (sth3) to 10-fold higher (sth10) higher in the 11-deoxycortisol-treated sample than in the control (Table 3). Since the 19-hydroxylase activity was enhanced under conditions of 11-deoxycortisol induction, the upregulated genes might encode the 19-hydroxylase. Therefore, the most highly upregulated gene, sth10, was further analyzed. Sequence analysis of the amino acid sequence of STH10 revealed that it contains a common conserved heme-binding region spanning residues 514 to 532. Online alignment showed that T. cucumeris steroid hydroxylase STH10 exhibits significant identity with two cytochrome P450 proteins (NCBI accession numbers CUA72706.1 and CUA72679.1) of unknown function from Rhizoctonia solani, sharing 73% and 72% identity. Primary structure sequence alignment of STH10 with two related known P450 enzymes revealed that STH10 has 14.14% and 18.20% identity with human CYP19A1 (16) and hamster CYP11B1 (17), respectively. Human CYP19A1 catalyzes 19-hydroxylation of steroids, while hamster CYP11B1 possesses both 11β-hydroxylase and 19-hydroxylase activities.
TABLE 3.
The upregulated putative P450 genes
| Candidate gene | No. of FPKM for samplea: |
log2FCb | |
|---|---|---|---|
| A | B | ||
| sth10 | 2.28 | 2,645.9 | 10.18 |
| sth6 | 3.05 | 1,037.15 | 8.41 |
| sth30 | 5.03 | 84.01 | 4.06 |
| sth26 | 1.42 | 12.16 | 3.10 |
| sth44 | 3.5 | 26.66 | 2.93 |
| sth11 | 0.83 | 4.46 | 2.43 |
| sth5 | 3.29 | 13.21 | 2.01 |
| sth3 | 1.97 | 7.9 | 2.00 |
| por | 176.52 | 174.68 | −0.02 |
Sample A, T. cucumeris cells cultured without 0.5 g/liter 11-deoxycortisol; sample B, T. cucumeris cells cultured with 0.5 g/liter 11-deoxycortisol.
Log2FC, log2 fold change.
P450 catalysis also needs a suitable redox protein such as P450 oxidoreductase (POR) to transfer electrons, so a candidate P450 oxidoreductase gene was identified by searching performed with the POR conservative domain (PF000175/PF000258/PF00667) against the Pfam-A database. The transcriptional level of the por gene was nearly unchanged between the 11-deoxycortisol-treated sample and the control. The amino acid sequence of POR shares 99% identity with the cytochrome P450 oxidoreductase (sequence identifier [ID] ELU36991.1) from Rhizoctonia solani AG-1 IA.
The full-length sth10 and por genes were amplified and ligated into the expression plasmid together, and the product was designated picZ-STH10-POR. The resulting vector was used to transform P. pastoris X33 cells with sth10 and por genes. The corresponding recombinant strain was named X33-STH10-POR. A recombinant plasmid harboring the sth10 gene alone, picZ-STH10, was also constructed and used to generate P. pastoris strain X33-STH10 as a control.
11-Deoxycortisol biotransformation and product identification.
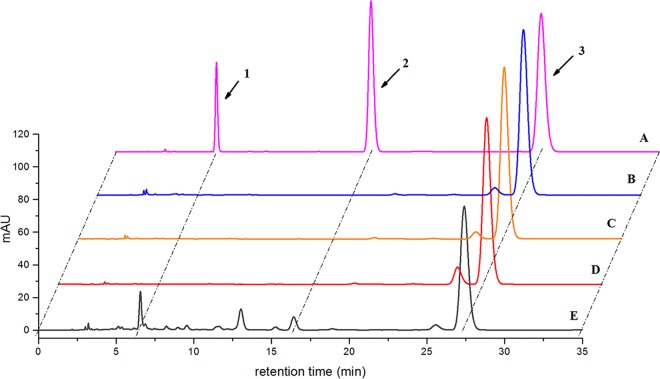
When the biotransformation of 11-deoxycortisol (RSS, cortexolone, or cortodoxone) was performed with recombinant strain X33-STH10-POR, two new main products with same retention times as 19-hydroxy-11-deoxycortisol (6.7 min) and 11β-hydroxy-11-deoxycortisol (cortisol or hydrocortisone; 16.5 min) were detected by high-performance liquid chromatography (HPLC) (Fig. 3, line E). In contrast, none of them was detected in the control reactions performed with yeast strains containing the empty vector expressing STH10 or POR alone (Fig. 3). In addition to the two main products (Fig. 4), several minor products were also detected by HPLC. These minor products were not detected in the control experiments (Fig. 3, lines B to D). Both product 1 and product 2 were purified and subjected to nuclear magnetic resonance (NMR) analyses.
FIG 3.
HPLC analysis of the biotransformation products of 11-deoxycortisol produced by recombinant strain X33-STH10-POR. (A) Standard references of 19-hydroxy-11-deoxycortisol (peak 1), 11β-hydroxy-11-deoxycortisol (peak 2), and 11-deoxycortisol (peak 3). (B) Transformation with strain X33 harboring empty vector. (C) Transformation with strain X33 expressing sth10 alone. (D) Transformation with strain X33 expressing por alone. (E) Transformation with strain X33 coexpressing sth10 and por. mAU, milli-absorbance units.
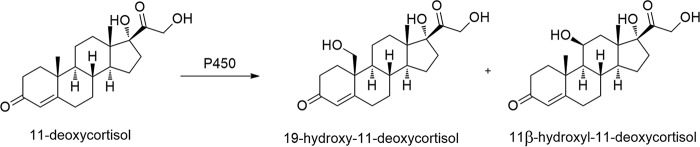
FIG 4.
Hydroxylation of 11-deoxycortisol catalyzed by recombinant strain X33-STH10-POR.
The molecular formula of product 1 was determined to be C21H30O5 on the basis of its positive mass spectrometry (MS) [M+Na]+ ion results at m/z 385.1996 (calculated value, 385.1991). The 1H NMR data were as follows: (CD3OD, 100 MHz) δ (ppm), 5.88 (s, 1H), 4.47 (dd J = 52, 79 Hz, 2H), 3.95 (dd J = 26, 42 Hz, 2H), 2.61 to 2.83 (m, 2H), 2.48 to 2.59 (m, 1H), 2.21 to 2.45 (m, 3H), 1.90 to 2.01 (m, 1H), 1.69 to 1.86 (m, 6H), 1.24 to 1.58 (m, 4H), 0.98 to 1.21 (m, 2H), and 0.68 (s, 3H); 13C NMR (CD3OD, 100 MHz) δ (ppm), 213.4, 203.2, 171.7, 126.7, 90.3, 67.8, 65.9, 55.2, 52.1, 49.3, 45.3, 37.5, 35.8, 34.9, 34.6, 34.4, 33.7, 32.1, 24.5, 22.3, and 15.4. On the basis of the analysis of the data described above, product 1 was identified as 19-hydroxy-11-deoxycortisol.
The molecular formula of product 2 was determined to be C21H30O5 on the basis of its positive MS [M+Na]+ ion results at m/z 385.1996 (calculated value, 385.1991). The 1H NMR data were as follows: (CD3OD, 100 MHz) δ (ppm), 5.66 (s, 1H), 4.45 (dd J = 52, 79 Hz, 2H), 4.40 (q J = 4, 16 Hz, 2H), 2.64 to 2.78 (m, 1H), 2.42 to 2.61 (m, 2H), 2.16 to 2.37 (m, 3H), 1.96 to 2.11 (m, 3H), 1.69 to 1.92 (m, 3H), 1.57 to 1.64 (m, 1H), 1.34 to 1.53 (m, 2H), 1.46 (s, 3H), 1.05 to 1.18 (m, 1H), 0.96 to 1.03 (m, 1H), and 0.88 (s, 3H); 13C NMR (CD3OD, 100 MHz) δ (ppm), 213.2, 202.7, 176.8, 122.6, 90.4, 68.8, 67.8, 57.7, 53.5, 50.0, 48.3, 40.9, 40.8, 35.9, 34.7, 34.4, 33.4, 33.0, 24.7, 21.5, and 17.9. On the basis of these data, product 2 was identified as 11β-hydroxy-11-deoxycortisol.
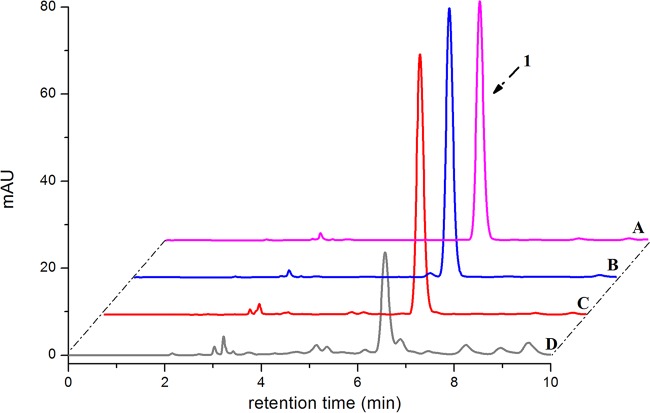
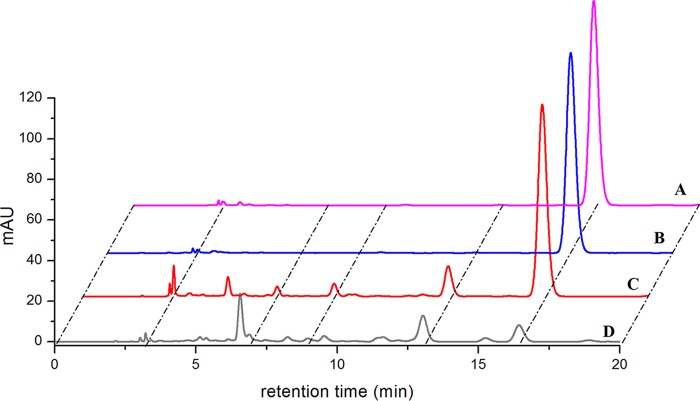
The biotransformation of 11β-hydroxy-11-deoxycortisol and 19-hydroxy-11-deoxycortisol was also performed with X33-STH10-POR resting cells. The reaction mixture was extracted with ethyl acetate, and the extract was analyzed by HPLC. No obvious product was detected from the reaction performed with 19-hydroxy-11-deoxycortisol as the substrate (Fig. 5). For the reaction performed with 11β-hydroxy-11-deoxycortisol as the substrate, three products were detected and showed the same retention times as the minor products from the reaction of 11-deoxycortisol, while no product was detected in the control experiments (Fig. 6).
FIG 5.
HPLC analysis of the biotransformation products of 19-hydroxy-11-deoxycortisol produced by recombinant strain X33-STH10-POR. (A) Standard reference 19-hydroxy-11-deoxycortisol. (B) Biotransformation with strain X33 harboring empty vector. (C) Biotransformation with strain X33-STH10-POR resting cells. (D) Biotransformation of 11-deoxycortisol with strain X33-STH10-POR resting cells.
FIG 6.
HPLC analysis of the biotransformation products of 11β-hydroxy-11-deoxycortisol produced by recombinant strain X33-STH10-POR. (A) Standard reference 11β-hydroxy-11-deoxycortisol. (B) Biotransformation with strain X33 harboring empty vector. (C) Biotransformation with strain X33-STH10-POR resting cells. (D) Biotransformation of 11-deoxycortisol with strain X33-STH10-POR resting cells.
DISCUSSION
Formation and transformation of secondary metabolites of many fungi, including antibiotics (18), immune suppressants (5), and hormones (19), are catalyzed by a vast amount of endogenous P450 (20). Fungi provide a rich source of P450 enzyme catalysts. Searching for the ideal P450 catalysts from fungi has attracted increasing attention. However, due to the presence of large amounts of P450 in fungi, to devise a universal and effective way to determine the required fungal strains of CYP450 orphans remains a major challenge (21). In this study, we used analysis of differential transcripts as the primary parameter for screening candidate P450s according to the fact that fungal P450s are inducible by many critical metabolites such as steroids and terpenes (22). A few candidate P450 genes were identified from T. cucumeris NBRC 6298. Among them, sth10 (which showed the highest induced transcript level) and por were amplified and coexpressed in P. pastoris X33. The recombinant whole cells catalyzed the hydroxylation of 11-deoxycortisol to give 19-hydroxy-11-deoxycortisol and 11β-hydroxy-11-deoxycortisol (Fig. 4). As such, sth10 is the hydroxylase gene responsible for both 19- and 11β-hydroxylation of 11-deoxycortisol. To the best of our knowledge, STH10 is a novel fungal P450 with steroidal 19-hydroxylase activity. Several minor products were also detected in the reaction of 11-deoxycortisol with X33-STH10-POR whole cells, while none of them was found in the control experiments, suggesting that the minor products were also produced by the cells coexpressing sth10 and por. On the basis of the fact that some P450s can catalyze the successive oxidation events seen on the substrates, we inferred that STH10 might further react with the hydroxylated products to produce these minor products. This inference is supported by the results from the biotransformation of 11β-hydroxy-11-deoxycortisol with X33-STH10-POR resting cells. The results are understandable because steroid hydroxylases can be induced by diverse steroids and because the inducible P450s can further modify the inducer substrate for detoxification or defense for adaptation to the ecological environment (23).
Two cytochrome P450 proteins (sequence ID CUA72706.1 and CUA72679.1) from R. solani showed high (73% and 72%) identity with T. cucumeris steroid hydroxylase STH10. We synthesized the two corresponding genes and heterologously expressed them in P. pastoris. Unfortunately, the recombinant strains coexpressing each of these two P450 genes with por did not hydroxylate 11-deoxycortisol to give 19-hydroxyl-11-deoxycortisol or 11β-hydroxyl-11-deoxycortisol (data not shown). These results suggest that POR may not match with these two P450 enzymes as the electron transfer partner or that they have catalytic properties that differ from the steroidal 19-hydroxylase and 11β-hydroxylase activities, although they have relatively high sequence identity. Further studies are needed to identify the structural determinants of STH10 with respect to its steroid 19-hydroxylase activity.
Introduction of oxygen at the C-19 methyl group is difficult because of the steric hindrance of the steroid core (24). As we know, the only well-studied P450 enzyme that catalyzes 19-hydroxylation of steroids is CYP19, or aromatase (12). However, this enzyme catalyzes a rather complex three-step process consisting of 19-hydroxylation, oxidation of the 19-hydroxy product to an aldehyde, and the loss of the 19-formyl group as formic acid, resulting in the aromatization of the steroid A ring (24). The reaction does not stop at the 19-hydroxylation step (16). In this study, no aldehyde or aromatized product was detected when 19-hydroxy-11-deoxycortisol was treated with X33-STH10-POR resting cells. This suggests that STH10 catalyzes only 19-hydroxylation without further oxidation with respect to the aldehyde. Therefore, STH10 is distinct from CYP19A1, consistent with the fact that the amino acid sequence identity between STH10 and CYP19A1 is rather low (14.14%).
In animals, glucocorticoid hormones such as cortisol and corticosterone are mainly produced by mitochondrial cytochrome P450 enzyme CYP11B1 (25), which shows diverse characteristics with respect to regio- and stereoselectivity among different species (26, 27). Human CYP11B1 is a pure desoxycortone 11β-hydroxylase without 19-hydroxylase or 18-oxidase activity (28). Rat CYP11B1 displays 11β- and 18-hydroxylase activities toward desoxycortone, but it does not carry out 19-hydroxylation (29). In contrast, hamster CYP11B1 hydroxylated desoxycortone at positions C-11 and C-19 and at nearly equal levels (17). It is believed that the minor differences in their primary CYP11B1 sequences account for the observed substrate and reaction specificities based on the high primary sequence similarity of examples of CYP11B1 from different species. It is surprising that STH10 and hamster CYP11B1 exhibit similar catalytic activities in spite of their low primary sequence identity.
Conclusion.
Transcriptome sequencing (RNA-seq) was used to identify a few cytochrome P450 genes from the fungus T. cucumeris which were inducible by 11-deoxycortisol. Among them, the highly induced sth10 gene and a redox partner, por, were coexpressed in the P. pastoris X33 strain. The biotransformation seen with the recombinant whole cells demonstrated that STH10 catalyzed the 19- and 11β-hydroxylations of 11-deoxycortisol. The purified products were identified by high-resolution mass spectrometry (HR-MS) and NMR. To the best of our knowledge, STH10 is a unique fungal P450 enzyme with steroidal 19-hydroxylase activity which is distinct from both the aromatase CYP19 and CYP11B families of enzymes. Further studies are needed to understand the structural basis of the regioselectivity and substrate specificity of this fungal enzyme to facilitate its engineering, with the aim of searching for effective steroidal 19-hydroxylases for use in industrial applications.
MATERIALS AND METHODS
Materials.
11-Deoxycortisol (purity, >98%) and 11β-hydroxy-11-deoxycortisol (purity, >98%) were obtained from Toronto Research Chemicals. 19-Hydroxy-11-deoxycortisol (purity, >95%) was produced with T. cucumeris NBRC 6298 as described previously (15). Peptone and yeast extract were purchased from Oxoid Ltd. All other chemicals were of analytical grade and bought from Merck.
Microorganisms, plasmids, and culture conditions.
Fungal strain T. cucumeris NBRC 6298 was obtained from the NITE Biological Resource Center (http://www.nbrc.nite.go.jp/NBRC2/NBRCCatalogueDetailServlet?ID=NBRC&CAT=00006298) and routinely maintained on potato dextrose agar (PDA) slants. Escherichia coli DH5α was grown at 37°C in Luria-Bertani (LB) medium (10 g/liter tryptone; 5 g/liter yeast extract; 10 g/liter NaCl; 15 g/liter agar; pH 7.0) and used as a host for molecular cloning of candidate genes. Pichia pastoris X33 (Invitrogen) was used as a host organism for the expression of candidate CYP genes and grown at 30°C in YPD medium (20 g/liter peptone; 10 g/liter yeast extract; 20 g/liter dextrose; 15 g/liter agar), buffered glycerol complex medium (BMG), or buffered minimal methanol medium (BMM) with Zeocin.
Fungal sample preparation and RNA sequencing.
For RNA sequencing, T. cucumeris was cultured with or without 0.5 g/liter 11-deoxycortisol for 24 h. The cells were collected, and the total RNA was isolated using TRIzol reagent (Promega, USA) followed by purification using RNeasy separation columns (RNeasy kit; Qiagen). The purified total RNA was sequenced with a HiSeq 2000 system (Illumina, USA) at Novogene Co., Ltd. (Tianjin, China).
Transcriptome assembly and annotation.
Clean data from the two samples were obtained from raw sequences by removing reads containing adapters or homopolymers or with low quality. The assembly of clean data was performed using Trinity (30). The resulting unigenes were annotated by searching against seven databases, including the NCBI nonredundant protein sequence (nr) database, the NCBI nonredundant nucleotide sequence (nt) database, the Pfam database (31), the Eukaryotic Ortholog Groups (KOG) database, the Swiss-Prot protein (Swiss-Prot) database, Kyoto Encyclopedia of Genes and Genomes (KEGG) databases (32), and the Gene Ontology (GO) database, using blastx (33) with an E value cutoff value of 10−5.
Gene expression analysis.
Gene expression levels were estimated according to the number of fragments per kilobase per million (FPKM) (34). Differential expression analysis of two samples was performed using the bioconductor edgeR package (35). The read counts were adjusted by the use of edgeR and one scaling normalized factor. P values were adjusted using false-discovery-rate (q) values (36). q values of <0.005 and fold change values of >2 were set as the thresholds for significantly differential expression levels (37).
Construction and transformation of recombinant plasmids.
The sth10 putative CYP gene with the highest level of induction by 11-deoxycortisol was amplified by PCR using cDNAs as the templates and the primers in Table 4 under the following conditions: initial denaturation at 95°C for 2 min, followed by 30 cycles of denaturation at 95°C for 45 s, annealing at 59°C for 45 s, and extension at 72°C for 1 min 30 s and a final extension step at 72°C for 10 min. The amplified gene was ligated to the KpnI/NotI-digested sites of E. coli/P. pastoris shuttle vector pPICZ A to form pPICZ-STH10. The identity of the gene in the plasmid was verified by sequencing (BGI, Beijing, China). The cytochrome P450 oxidoreductase (POR) from T. cucumeris NBRC 6298 was amplified by the use of the primers listed in Table 4. The coexpressing pPICZ-STH10-POR plasmid was constructed in accordance with data from a previous study (38). Briefly, the por cDNA was amplified and then ligated to the pUC57 vector at the KpnI and NotI restriction enzyme site to form pUC57-POR. One silent mutation at the EcoRI restriction enzyme site was created for elimination of the restriction site in the por gene using PCRs. The resulting sequence was ligated into pPICZ A at EcoRI and ApaI restriction sites to form pPICZ-MPOR recombination vector. To construct the sth10 expression cassette, the original PmeI site in the pPICZ A vector was mutated and the resulting construct was named dePmeI-pPICZA. Binary vector binary-pPICZ-MPOR was constructed by ligating the pPICZA-MPOR BamHI-digested fragment with a BglII-BamHI-digested dePmeI-pPICZA fragment. A clone positive for tail-to-head orientation (Pichia expression manual; Invitrogen) was selected. The sth10 target gene was cloned into the por-containing binary vector at KpnI-NotI restriction sites to build the coexpressing pPICZ-STH10-POR plasmid. The recombinant pPICZ-STH10-POR plasmid was linearized with PmeI and then electrotransformed into P. pastoris X33 cells according to the manufacturer's instructions for the Pichia expression kit. Cells of the recombinant P. pastoris X33 strain harboring pPICZ-STH10-POR were selected from YPD medium supplemented with 1 mg ml−1 Zeocin, and the integration of the sth10-por expression cassette was verified by PCR techniques. The correct strains were named pPICZ-STH10-POR. The pPICZ A empty vector, pPICZ-STH10, and pPICZ-MPOR were also transformed into P. pastoris X33 as control strains and named X33-CK, X33-STH10, and X33-POR, respectively. The names of all of the strains and vectors created and used in this work are listed in Table 5.
TABLE 4.
Primers of PCRs in this study
| Name | Sequence (5′–3′)a |
|---|---|
| F-KpnI-sth10 | CATCCGGTACCATGTCCAACTCAACTCTCGTTTCTTTTG |
| R-NotI-sth10 | CGCGGCGGCCGCTTATTCCTCCACGAGAGTGACTTTCAT |
| F-KpnI-por | CATCCGGTACCATGGCTCCTGCTCTCTCGAC |
| R-NotI-por | CGCGGCGGCCGCCTATGACCAGACATCCAACAACAAC |
| F-MEcoRI-por | ACCGATAATGCAGTCGAGTTCATGAATAACATCAAC |
| R-MEcoRI-por | GTTGATGTTATTCATGAACTCGACTGCATTATCGGT |
| F-EcoRI-por | CATCCGAATTCATGGCTCCTGCTCTCTCGAC |
| R-ApaI-por | GTTCGGGCCCCTATGACCAGACATCCAACAACAAC |
| F-dePmeI-pICZA | GGCCCAAAACTGACAGTTGAAACGCTGTCTTGGAACCT |
| R-dePmeI-pICZA | AGGTTCCAAGACAGCGTTTCAACTGTCAGTTTTGGGCC |
Restriction sites in primers are underlined, and mutations are bold.
TABLE 5.
Strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Source |
|---|---|---|
| Strains | ||
| Thanatephorus cucumeris NBRC 6298 | Wild type | NBRC |
| Pichia pastoris X33 | Wild type | Invitrogen |
| Escherichia coli TOP 10 | F− mcrA Δ(mrr-hsdRMS-mcrBC) Φ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara leu) 7697 galU galK rpsL (Strr) endA1 nupG | Invitrogen |
| Pichia pastoris X33-CK | Pichia pastoris X33 harboring pICZA empty vector | This work |
| Pichia pastoris X33-POR | Pichia pastoris X33 expressing por alone | This work |
| Pichia pastoris X33-STH10 | Pichia pastoris X33 expressing sth10 alone | This work |
| Pichia pastoris X33-STH10-POR | Pichia pastoris X33 coexpressing sth10 and por | This work |
| Plasmids | ||
| pPICZ A | Yeast expression vector (Pichia pastoris), Zeor | Invitrogen |
| pUC57 | Cloning vector, Ampr | Invitrogen |
| pPICZ-STH10 | T. cucumeris sth10 cDNA cloned in pPICZ A vector at KpnI-NotI site; Zeor | This work |
| pUC57-POR | T. cucumeris P450 reductase gene por cloned in pUC57 vector at KpnI-NotI site; Ampr | This work |
| pPICZ-MPOR | pPICZ A-por vector containing por gene mutated (silent mutation) at EcoRI; Zeor | This work |
| dePmeI-pPICZA | pPICZ A vector mutated at PmeI site; Zeor | This work |
| binary-pPICZ-MPOR | BglII-BamHI digested dePmeI-pPICZ A fragment cloned in pPICZ-MPOR at BamHI site; Zeor | This work |
| pPICZ-STH10-POR | sth10 gene cloned in binary-pPICZ-MPOR at KpnI-NotI site; Zeor | This work |
Ampr, ampicillin resistance; Strr, streptomycin resistance; Zeor, zeocin resistance.
Heterologous expression of sth10 and por in P. pastoris and steroid biotransformation.
Individual pPICZ-STH10-POR colonies with robust growth on YPD plates containing 1 mg ml−1 Zeocin were inoculated into 25 ml BMG and cultured until the optical density at 600 nm (OD600) reached 10. The cells were collected by centrifugation (4,000 × g, 5 min) and diluted to an OD600 of 1.0 in BMM containing aminolevulenic acid (2 mM), and 1% (vol/vol) methanol was added to induce expression of candidate genes every 12 h for 5 days at 20°C on a rotary shaker (250 rpm). The colonies of X33-CK, X33-STH10, and X33-POR were used as controls.
The whole-cell transformation of 11-deoxycortisol, 11β-hydroxy-11-deoxycortisol, or 19-hydroxy-11-deoxycortisol was performed at 30°C on a rotary shaker (200 rpm). The methanol-induced recombinant strains were first collected by centrifugation (4,000 × g, 5 min) and resuspended in 30 ml potassium phosphate buffer (50 mM, pH 7.5) containing aminolevulenic acid (2 mM) in 250-ml shake flasks. Then, the substrates were added to the respective reaction mixtures at a final concentration of 1 mM. Methanol (1% [vol/vol]) was added at each 24-h time interval. After 72 h, all of the samples were taken from the reaction mixtures and extracted with ethyl acetate for high-performance liquid chromatography (HPLC) analysis.
Preparative reaction and product identification.
P. pastoris X33-STH10-POR colonies were inoculated into 500 ml BMG in 2-liter shake flasks and incubated at 30°C and 250 rpm until the OD600 reached 10.0. Then, 5 ml of 11-deoxycortisol solution–methanol was added to the culture with a final substrate concentration of 1 mM. Methanol (1% [vol/vol]) was added to induce expression of the sth10 and por genes every 12 h for 5 days.
The transformation mixture was analyzed by HPLC on an Eclipse XDB C18 column (250 mm by 4.6 mm by 5 μm), using methanol-water (45/55 [vol/vol]) as the mobile phase. The flow rate was maintained at 0.6 ml/min, and the column temperature was 30°C. The substrate and the hydroxylation products were detected by absorbance at 254 nm.
To isolate the products, the reaction mixture was extracted twice with equivalent volumes of ethyl acetate. The extract was concentrated under conditions of reduced pressure, and the residue was resuspended in appropriate volumes of dichloromethane. The products were isolated by the use of a thin-layer chromatography (TLC) plate (Anhui Liangchen Silicon Material Co., Ltd., Anhui, China) (100 mm by 200 mm by 0.5 mm). The plate was developed with dichloromethane-methanol (15:1 [vol/vol]). Each band was scraped and extracted with dichloromethane, and further purification was performed by the use of preparative reverse-phase recycling HPLC, an XDB C18 column, and methanol-water (28/72 [vol/vol] for 0 to 60 min and 50/50 [vol/vol] for 60 to 90 min) as the eluent. The flow rate was maintained at 11 ml/min at a column temperature of 30°C. The 1H NMR and 13C NMR spectra were recorded at 400 MHz with a Bruker Avance III device using CD3OD as the solvent.
Accession number(s).
The RNA-seq data reported here were deposited in the NCBI Sequence Read Archive (SRA) under accession number SRX3599177. The nucleotide sequences of sth10 and por from T. cucumeris were deposited in the GenBank database under accession numbers MF818017 and MH006689, respectively.
ACKNOWLEDGMENTS
This work was financially supported by the Youth Innovation Promotion Association (to J.F.), the Key Research Program of Chinese Academy of Sciences (KFZD-SW-212), and the National High-Tech Research & Development Program of China (863 Program, no. 2011AA02A211-04).
We declare that there is no conflict of interest.
REFERENCES
- 1.Shahidi NT. 2001. A review of the chemistry, biological action, and clinical applications of anabolic-androgenic steroids. Clin Ther 23:1355–1390. doi: 10.1016/S0149-2918(01)80114-4. [DOI] [PubMed] [Google Scholar]
- 2.Donova MV, Egorova OV. 2012. Microbial steroid transformations: current state and prospects. Appl Microbiol Biotechnol 94:1423–1447. doi: 10.1007/s00253-012-4078-0. [DOI] [PubMed] [Google Scholar]
- 3.Tong WY, Dong X. 2009. Microbial biotransformation: recent developments on steroid drugs. Recent Pat Biotechnol 3:141–153. doi: 10.2174/187220809788700157. [DOI] [PubMed] [Google Scholar]
- 4.Ahluwalia A. 1998. Topical glucocorticoids and the skin-mechanisms of action: an update. Mediators Inflamm 7:183–193. doi: 10.1080/09629359891126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hogg JA. 1992. Steroids, the steroid community, and Upjohn in perspective: a profile of innovation. Steroids 57:593–616. doi: 10.1016/0039-128X(92)90013-Y. [DOI] [PubMed] [Google Scholar]
- 6.Janeczko T, Dmochowska-Gladysz J, Kostrzewa-Suslow E, Bialonska A, Ciunik Z. 2009. Biotransformations of steroid compounds by Chaetomium sp. KCH 6651. Steroids 74:657–661. doi: 10.1016/j.steroids.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 7.Bhatti HN, Khera RA. 2012. Biological transformations of steroidal compounds: a review. Steroids 77:1267–1290. doi: 10.1016/j.steroids.2012.07.018. [DOI] [PubMed] [Google Scholar]
- 8.Windholz TB, Windholz M. 1964. Recent advances in synthesis of 19-norsteroids. Angew Chem Int Ed Engl 3:353–361. doi: 10.1002/anie.196403531. [DOI] [PubMed] [Google Scholar]
- 9.Shenfield GM, Griffin JM. 1991. Clinical pharmacokinetics of contraceptive steroids - an update. Clin Pharmacokinet 20:15–37. doi: 10.2165/00003088-199120010-00002. [DOI] [PubMed] [Google Scholar]
- 10.Sun Y, Fang M, Davies H, Hu Z. 2014. Mifepristone: a potential clinical agent based on its anti-progesterone and anti-glucocorticoid properties. Gynecol Endocrinol 30:169–173. doi: 10.3109/09513590.2013.856410. [DOI] [PubMed] [Google Scholar]
- 11.Pinto-Almazán R, Segura-Uribe JJ, Farfán-García ED, Guerra-Araiza C. 2017. Effects of tibolone on the central nervous system: clinical and experimental approaches. Biomed Res Int 2017:8630764. doi: 10.1155/2017/8630764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sohl CD, Guengerich FP. 2010. Kinetic analysis of the three-step steroid aromatase reaction of human cytochrome P450 19A1. J Biol Chem 285:17734–17743. doi: 10.1074/jbc.M110.123711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takahashi T. 1964. Studies on microbiological transformation of steroids. 9. Transformation of C19-steroids by Pellicularia filamentosa f. sp. microsclerotia IFO 6298. Agric Biol Chem 28:38–47. doi: 10.1080/00021369.1964.10858205. [DOI] [Google Scholar]
- 14.Clark TA, Chong R, Maddox IS. 1982. The effect of dissolved oxygen tension on 11β- and 19-hydroxylation of Reichstein's substance S by Pellicularia filamentosa. Eur J Appl Microbiol Biotechnol 14:131–135. doi: 10.1007/BF00497887. [DOI] [Google Scholar]
- 15.Wang Y, Zhang F, Chen X, Feng J, Wu Q, Zhu D, Ma Y. 2016. Optimization of the transformation conditions for the 19-hydroxylation of cortexolone by Thanatephorus cucumeris. Chin J Appl Environ Biol 22:860–864. [Google Scholar]
- 16.Hong Y, Yu B, Sherman M, Yuan YC, Zhou D, Chen S. 2007. Molecular basis for the aromatization reaction and exemestane-mediated irreversible inhibition of human aromatase. Mol Endocrinol 21:401–414. doi: 10.1210/me.2006-0281. [DOI] [PubMed] [Google Scholar]
- 17.Véronneau S, Bernard H, Cloutier M, Courtemanche J, Ducharme L, Lefebvre A, Mason JI, Lehoux JG. 1996. The hamster adrenal cytochrome P450C11 has equipotent 11β-hydroxylase and 19-hydroxylase activities, but no aldosterone synthase activity. J Steroid Biochem Mol Biol 57:125–139. doi: 10.1016/0960-0760(95)00249-9. [DOI] [PubMed] [Google Scholar]
- 18.Amsterdam A, Sasson R. 2002. The anti-inflammatory action of glucocorticoids is mediated by cell type specific regulation of apoptosis. Mol Cell Endocrinol 189:1–9. doi: 10.1016/S0303-7207(01)00722-5. [DOI] [PubMed] [Google Scholar]
- 19.Fernandes P, Cruz A, Angelova B, Pinheiro HM, Cabral JMS. 2003. Microbial conversion of steroid compounds: recent developments. Enzyme Microb Technol 32:688–705. doi: 10.1016/S0141-0229(03)00029-2. [DOI] [Google Scholar]
- 20.van den Brink HM, van Gorcom RF, van den Hondel CA, Punt PJ. 1998. Cytochrome P450 enzyme systems in fungi. Fungal Genet Biol 23:1–17. doi: 10.1006/fgbi.1997.1021. [DOI] [PubMed] [Google Scholar]
- 21.Syed K, Shale K, Pagadala NS, Tuszynski J. 2014. Systematic identification and evolutionary analysis of catalytically versatile cytochrome p450 monooxygenase families enriched in model basidiomycete fungi. PLoS One 9:e86683. doi: 10.1371/journal.pone.0086683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kristan K, Rizner TL. 2012. Steroid-transforming enzymes in fungi. J Steroid Biochem Mol Biol 129:79–91. doi: 10.1016/j.jsbmb.2011.08.012. [DOI] [PubMed] [Google Scholar]
- 23.Deshmukh R, Khardenavis AA, Purohit HJ. 2016. Diverse metabolic capacities of fungi for bioremediation. Indian J Microbiol 56:247–264. doi: 10.1007/s12088-016-0584-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoshimoto FK, Guengerich FP. 2014. Mechanism of the third oxidative step in the conversion of androgens to estrogens by cytochrome P450 19A1 steroid aromatase. J Am Chem Soc 136:15016–15025. doi: 10.1021/ja508185d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nonaka Y, Fujii T, Kagawa N, Waterman MR, Takemori H, Okamoto M. 1998. Structure/function relationship of CYP11B1 associated with Dahl's salt-resistant rats—expression of rat CYP11B1 and CYP11B2 in Escherichia coli. Eur J Biochem 258:869–878. doi: 10.1046/j.1432-1327.1998.2580869.x. [DOI] [PubMed] [Google Scholar]
- 26.Momoi K, Okamoto M, Fujii S, Kim CY, Miyake Y, Yamano T. 1983. 19-Hydroxylation of 18-hydroxy-11-deoxycorticosterone catalyzed by cytochrome-P45011β of bovine adrenocortex. J Biol Chem 258:8855–8860. [PubMed] [Google Scholar]
- 27.Bureik M, Lisurek M, Bernhardt R. 2002. The human steroid hydroxylases CYP1B1 and CYP11B2. Biol Chem 383:1537–1551. doi: 10.1515/BC.2002.174. [DOI] [PubMed] [Google Scholar]
- 28.Schiffer L, Anderko S, Hobler A, Hannemann F, Kagawa N, Bernhardt R. 2015. A recombinant CYP11B1 dependent Escherichia coli biocatalyst for selective cortisol production and optimization towards a preparative scale. Microb Cell Fact 14:25. doi: 10.1186/s12934-015-0209-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gomez-Sanchez CE, Gomez-Sanchez EP, Shackleton CH, Milewich L. 1982. Identification of 19-hydroxydeoxycorticosterone, 19-oxo-deoxycorticosterone, and 19-oic-deoxycorticosterone as products of deoxycorticosterone metabolism by rat adrenals. Endocrinology 110:384–389. doi: 10.1210/endo-110-2-384. [DOI] [PubMed] [Google Scholar]
- 30.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, Mauceli E, Hacohen N, Gnirke A, Rhind N, di Palma F, Birren BW, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, Sonnhammer EL, Tate J, Punta M. 2014. Pfam: the protein families database. Nucleic Acids Res 42:D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, Yamanishi Y. 2008. KEGG for linking genomes to life and the environment. Nucleic Acids Res 36:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robinson MD, McCarthy DJ, Smyth GK. 2010. EdgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Storey JD, Tibshirani R. 2003. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A 100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Genome Biol 11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Syed K, Doddapaneni H, Subramanian V, Lam YW, Yadav JS. 2010. Genome-to-function characterization of novel fungal P450 monooxygenases oxidizing polycyclic aromatic hydrocarbons (PAHs). Biochem Biophys Res Commun 399:492–497. doi: 10.1016/j.bbrc.2010.07.094. [DOI] [PMC free article] [PubMed] [Google Scholar]