ABSTRACT
Polyoxin (POL) is an unusual peptidyl nucleoside antibiotic, in which the peptidyl moiety and nucleoside skeleton are linked by an amide bond. However, their biosynthesis remains poorly understood. Here, we report the deciphering of PolG as an ATP-dependent ligase responsible for the assembly of POL. A polG mutant is capable of accumulating multiple intermediates, including the peptidyl moiety (carbamoylpolyoxamic acid [CPOAA]) and the nucleoside skeletons (POL-C and the previously overlooked thymine POL-C). We further demonstrate that PolG employs an ATP-dependent mechanism for amide bond formation and that the generation of the hybrid nucleoside antibiotic POL-N is also governed by PolG. Finally, we determined that the deduced ATP-binding sites are functionally essential for PolG and that they are highly conserved in a number of related ATP-dependent ligases. These insights have allowed us to propose a catalytic mechanism for the assembly of peptidyl nucleoside antibiotic via an acyl-phosphate intermediate and have opened the way for the combinatorial biosynthesis/pathway engineering of this group of nucleoside antibiotics.
IMPORTANCE POL is well known for its remarkable antifungal bioactivities and unusual structural features. Actually, elucidation of the POL assembly logic not only provides the enzymatic basis for further biosynthetic understanding of related peptidyl nucleoside antibiotics but also contributes to the rational generation of more hybrid nucleoside antibiotics via synthetic biology strategy.
KEYWORDS: polyoxin, peptidyl nucleoside antibiotic, ATP-dependent ligase, amide bond, synthetic biology
INTRODUCTION
Nucleoside antibiotics constitute a large family of important microbial secondary metabolites with broad-spectrum bioactivities, such as antibacterial, antifungal, antiviral, and anticancer activities (1–4). Peptidyl nucleoside antibiotics, including polyoxin (POL), nikkomycin (NIK), blasticidin S, and arginomycin, are well characterized and typically include two parts, the peptidyl moiety and the nucleoside skeleton, which are connected by an amide bond (Fig. 1A) (4). POLs, a group of structurally related peptidyl nucleoside antibiotics, are produced by Streptomyces cacaoi var. asoensis and Streptomyces aureochromogenes (5). Mechanistically, POL mimics the structure of UDP-N-acetylglucosamine, a building block for fungal chitin biosynthesis. Accordingly, it functions as a powerful competitive inhibitor of chitin synthetase, targeting fungal cell wall biosynthesis (6–8). As an effective and nontoxic fungicide, POL has been extensively used to control several important fungus-caused plant diseases in agriculture (9).
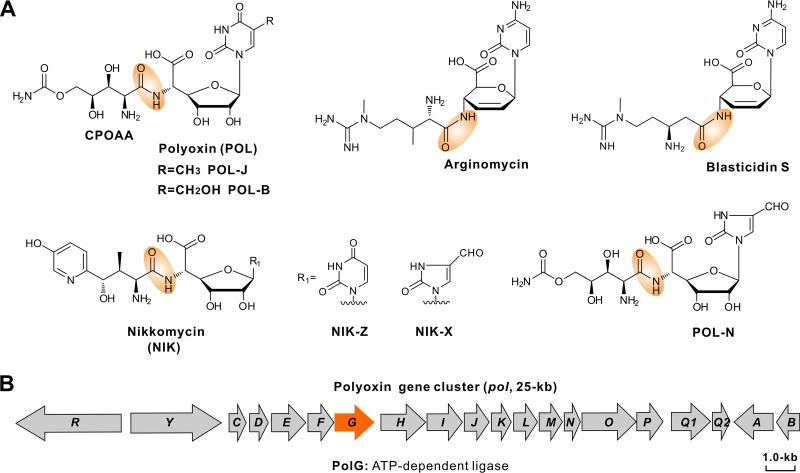
FIG 1.
Chemical structures of related peptidyl nucleoside antibiotics and genetic organization of the POL gene cluster. (A) Chemical structures of POL and related peptidyl nucleoside antibiotics. Amide bonds are highlighted by an orange shadow region. (B) Genetic organization of the POL biosynthetic gene cluster; the gene in orange is required for the assembly of POL (amide bond formation).
Earlier studies reported that the POL gene cluster consists of 20 genes (Fig. 1B). Genes polLMNOP were defined for the biosynthesis of the peptidyl moiety carbamoylpolyoxamic acid (CPOAA) (Fig. 1A), and genes polBADHIJK were identified for nucleoside skeleton (Fig. 1A) formation (5, 9). The CPOAA biosynthetic pathway contains an unusual acetylation cycle associated with tandem reduction and sequential hydroxylation (5). For the nucleoside skeleton, formation of the key intermediate octosyl acid requires a carbon-carbon bond construction catalyzed by a radical S-adenosyl-l-methionine (SAM)-dependent enzyme (10, 11). Chen et al. have determined the enzymatic mechanism of C-5 methylation for the nucleoside skeleton and further solved the crystal structure of the C-5 methylase, PolB (12). Previous works also initiated the production of polyoxin-nikkomycin (POL-NIK) hybrid nucleoside antibiotics through synthetic biology approaches (13–15).
In the present study, we have demonstrated that PolG functions as an ATP-dependent ligase for POL assembly, by first activating the peptidyl CPOAA moiety through activation of the carboxyl group via a direct phosphorylation. We have also dissected the enzymatic basis for the generation of the hybrid nucleoside antibiotic POL-N, potentially allowing for the combinatorial biosynthesis of this group of antibiotics.
RESULTS
In vivo reconstitution of the POL-B pathway in S. aureochromogenes CXR14.
To reconstitute the POL-B pathway in S. aureochromogenes CXR14 (CXR14 here) (Table 1), the cosmid pCHW201 (containing the whole POL gene cluster but with deletion of polF, which encodes a hypothetic oxidoreductase for polyoximic acid biosynthesis) (Table 1) (9) was introduced into the strain via conjugation. After validation, the recombinant strain CXR14::pCHW201 was grown in fermentation medium and the metabolites were analyzed by high-pressure liquid chromatography (HPLC). The results showed HPLC peaks at 10.4 min and 14.5 min (Fig. 2A; see also Fig. S1A in the supplemental material), which were absent from a control strain lacking the gene cluster (CXR14::pJTU2463b) (Fig. 2A). Further liquid chromatography-mass spectrometry (LC-MS) analysis showed that the two peaks generate [M+H]+ ions at m/z 508.1521 and m/z 492.1542, respectively, whose fragmentation patterns are correspondingly matched to those of the theoretical mass of POL-B and POL-J (Fig. S1B and C). To confirm these assignments, both compounds were purified and analyzed by 1H and 13C nuclear magnetic resonance (NMR) (see Fig. S2 and S3 in the supplemental material). These NMR signals are in agreement with those of POL-B (16) and POL-J (17) as reported previously. Taken together, these data establish that the target recombinant CXR14::pCHW201 is conferred with the capability of POL-B and POL-J production.
TABLE 1.
Strains, plasmids, and cosmids used in this study
| Strain, plasmid, or cosmid | Description | Reference or source |
|---|---|---|
| S. aureochromogenes | ||
| CXR14 | YB172 (industrial POL producer) derivative with the entire POL gene cluster deleted | 13 |
| CXR14::pCHW201 | Strain CXR14 containing pCHW201 | This study |
| CXR14::pCHW201/ΔpolG | Strain CXR14 containing pCHW201/ΔpolG | This study |
| CXR14::pJTU2463b | Strain CXR14 containing pJTU2463b | This study |
| E. coli | ||
| DH10B | F− mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80dlacZΔM15 ΔlacX74 deoR recA1 endA1 araΔ139 D(ara leu)7697 galU galK λ− rpsL nupG | Gibco-BRL |
| BW25113/pIJ790 | λ Red (gamma beta exo) cat araC rep101 | 18 |
| Rosetta(DE3)/pLysS | F− ompT hsdSB(rB− mB−) gal dcm λ(DE3 [lacI lacUV5-T7 gene1 ind1 sam7 nin5]) pLysS (Cmlr) | Novagen |
| ET12567(pUZ8002) | F− dam-13::Tn9 dcm-6 hsdM hsdR recF143 zjj-202::Tn10 galK2 galT22 ara14 pacY1 xyl-5 leuB6 thi-1 pUZ8002 | 31 |
| Plasmids | ||
| pJTU2463b | int aac(3)IV oriT RK2 phiC31 attP | This study |
| pET28a | neo reppMB1 T7 promoter | Novagen |
| pJTU4774 | The cosmid with XbaI and SpeI sites infilled, containing the entire POL gene cluster | This study |
| pCHW201 | pJTU4774 derivative with polF in frame deleted via PCR-targeting technology | This study |
| pCHW201/ΔpolG | pCHW201 derivative with polG in frame deleted via PCR-targeting technology | This study |
| pET28a/polG | pET28a derivative carrying an NdeI-EcoRI fragment containing the optimized polG | This study |
| pET28a/polGK110A | pET28a/polG derivative with the K110A mutation | This study |
| pET28a/polGK150A | pET28a/polG derivative with the K150A mutation | This study |
| pET28a/polGE188A | pET28a/polG derivative with the E188A mutation | This study |
| pET28a/polGE267A | pET28a/polG derivative with the E267A mutation | This study |
| pET28a/polGE280A | pET28a/polG derivative with the E280A mutation | This study |
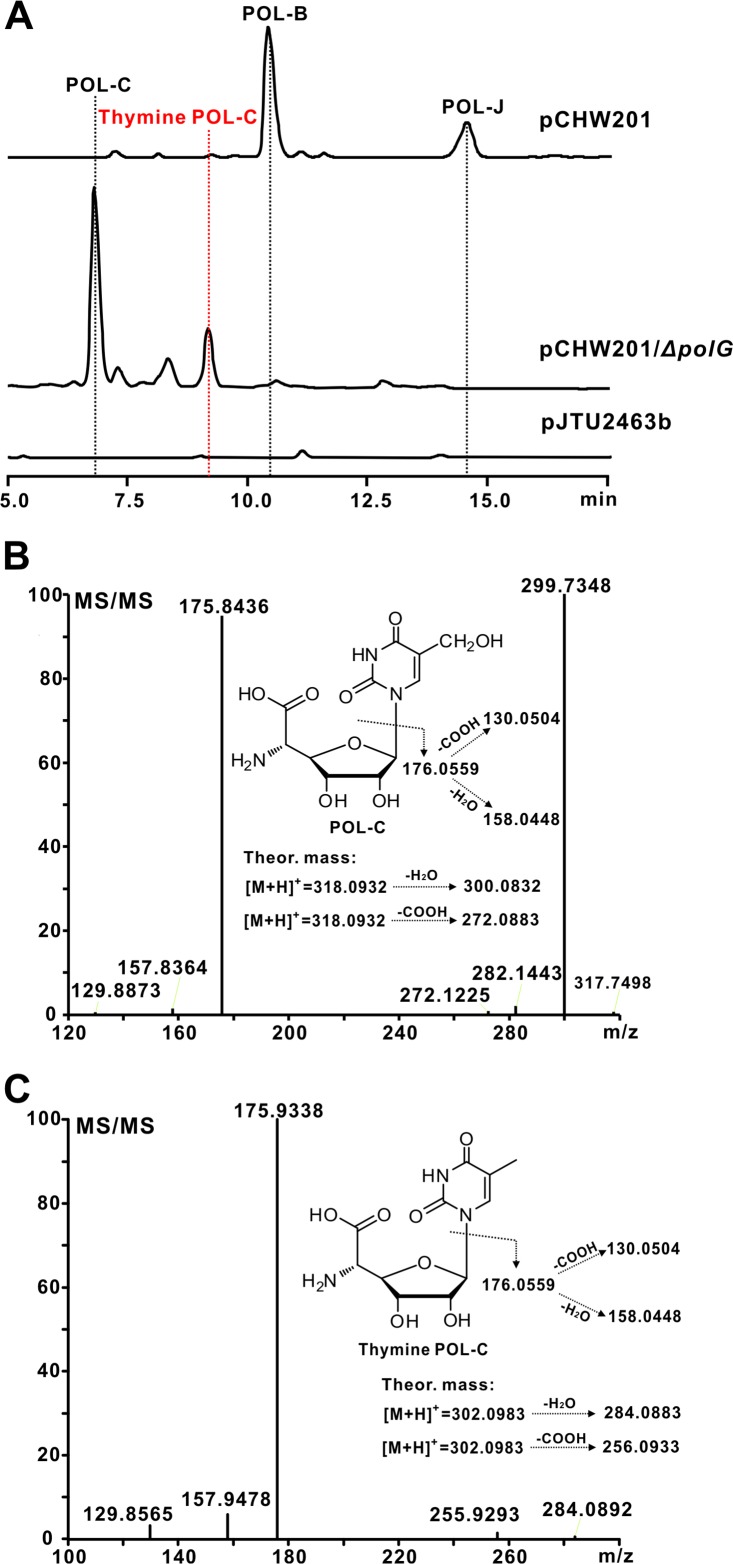
FIG 2.
HPLC and HRMS analysis of the targeted metabolites accumulated by the target recombinants. (A) HPLC analysis of the metabolites produced by strains CXR14::pCHW201 and CXR14::pCHW201/ΔpolG. Line labels: pCHW201, metabolites produced by S. aureochromogenes CXR14::pCHW201; pCHW201/ΔpolG, metabolites produced by S. aureochromogenes CXR14::pCHW201/ΔpolG; pJTU2463b, metabolites produced by S. aureochromogenes CXR14::pJTU2463b, used as a negative control. (B) HRMS/MS analysis of the target metabolite POL-C produced by strain pCHW201/ΔpolG. The theoretical fragmentation pattern of POL-C is listed as a control. (C) HRMS/MS analysis of the target metabolite thymine POL-C produced by pCHW201/ΔpolG strain. The theoretical fragmentation pattern of thymine POL-C is also shown as a control. Theor., theoretical.
Genetic assignment of polG as a candidate gene responsible for the assembly of POL.
To define the functional role of polG in POL biosynthesis, we mutated the target gene in the cosmid pCHW201 via a PCR-targeting technology (see Fig. S4A and B in the supplemental material) (18). After confirmation, the pCHW201/ΔpolG variant was conjugated into strain CXR14 and fermented for production of metabolites. The HPLC analysis indicated that strain pCHW201/ΔpolG is able to generate new peaks at 6.8 min and 9.1 min but its production of POL-B and POL-J is abolished (Fig. 2A and S4C and D). We then isolated the two new peaks for further LC-MS and NMR analysis. The results showed that the peak at 6.8 min is characterized by an [M+H]+ ion at m/z 318.0918, which could be fragmented into m/z 299.7348 and m/z 175.8436, consistent with those of the intermediate POL-C (Fig. 2B and S4E). The identity of this compound was further confirmed by 1H and 13C NMR analysis (see Fig. S5 in the supplemental material) (19). The second LC peak (9.1 min) gave rise to a strong [M+H]+ ion at m/z 302.0969, 16 Da (an oxygen atom loss) less than the mass of POL-C. Tandem mass spectrometry (MS/MS) analysis indicated that this ion could give rise to the main fragment ions at m/z 129.8565, 157.9478, and 175.9338, resembling the fragmentation pattern of the POL-C-related molecule (Fig. 2C and S4F). For validation of this metabolite, we purified it for 1H, 13C, and two-dimensional (2D) NMR analysis. The 1H and 13C chemical shifts were similar to those of POL-C, except for the C-7 position signals at Hδ 1.71 ppm (s, 3H) and Cδ (11.53 ppm). These are characteristic for the methyl group, which was confirmed by the heteronuclear multiple-bond correlation (HMBC) correlations of H-7 with C-4, C-5, and C-6. The detailed assignments of thymine POL-C are supported by correlation spectroscopy (COSY), HMBC, and heteronuclear multiple-quantum correlation (HMQC) spectra (see Fig. S6 in the supplemental material). These results indicate that this compound is a previously unidentified component of the POL pathway, thymine POL-C (Fig. S6).
To confirm that the CPOAA biosynthesis remains intact in the polG mutant, we reanalyzed the metabolites of the polG mutant (CXR14::pCHW201/ΔpolG) by LC-MS. A characteristic [M+H]+ ion is present at m/z 209.0762, whose fragmentation pattern is matched to that of the authentic CPOAA standard (see Fig. S7 in the supplemental material). These data show that polG is responsible for the assembly of POL-B and POL-J.
Biochemical characterization of PolG as an ATP-dependent ligase.
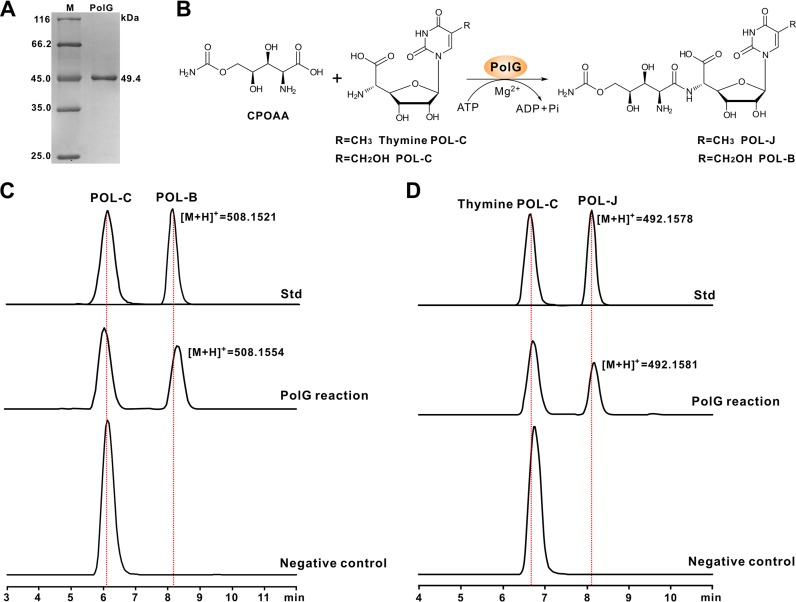
PolG shows homology to a large number of proteins containing an ATP-grasp domain (58% identities to NikS) (see Fig. S8 in the supplemental material), although very few of them have been functionally assigned. To correlate the functional role of PolG in vitro to the assembly of POL-B, we initially sought to overexpress it in Escherichia coli. This was unsuccessful, likely due to the codon bias of E. coli, and we therefore optimized the polG codons according to the E. coli preference (see Table S1 in the supplemental material), which resulted in the overexpression of PolG (Fig. 3A). Subsequently, we purified the PolG protein and tested its activity in vitro. The LC-MS results (using POL-C as the substrate) showed a characteristic [M+H]+ ion at m/z 508.1554 (Fig. 3B and C), whose fragmentation pattern is fully consistent with that of the authentic POL-B standard (see Fig. S9A in the supplemental material). This MS ion was not detected in the PolG-negative control or in reaction mixtures in the absence of ATP/Mg2+ (Fig. 3C and Fig. S9C). We further tested to see if ATP could be substituted for GTP to maintain PolG activity, and the results indicated that CPOAA is selectively activated by ATP during the assembly of POL-B (Fig. S9D). We then tested the substrate specificity of PolG, and as expected, this enzyme could also convert thymine POL-C as the substrate to POL-J (Fig. 3B and D and Fig. S9B). Hence, the results verify that PolG functions as an ATP-dependent ligase (amide synthetase) for POL-B/POL-J assembly.
FIG 3.
Biochemical characterization of PolG as an ATP-dependent ligase. (A) SDS-PAGE analysis of PolG (5 μg per lane) purified from E. coli; (B) scheme of the PolG-catalyzed reaction. CPOAA, carbamoylpolyoxamic acid; (C) LC-MS analysis of the PolG-catalyzed reaction with POL-C as the substrate; (D) LC-MS analysis of the PolG-catalyzed reaction with thymine POL-C as the substrate. Std, the authentic standards of POL-C and POL-B (thymine PoL-C and POL-J); PolG reaction, the PolG-catalyzed reaction using POL-C/thymine POL-C as the individual substrate; negative control, the reaction (POL-C/thymine POL-C added as the substrate) without the PolG enzyme added.
Enzymatic basis for the assembly of the hybrid peptidyl nucleoside antibiotic POL-N.
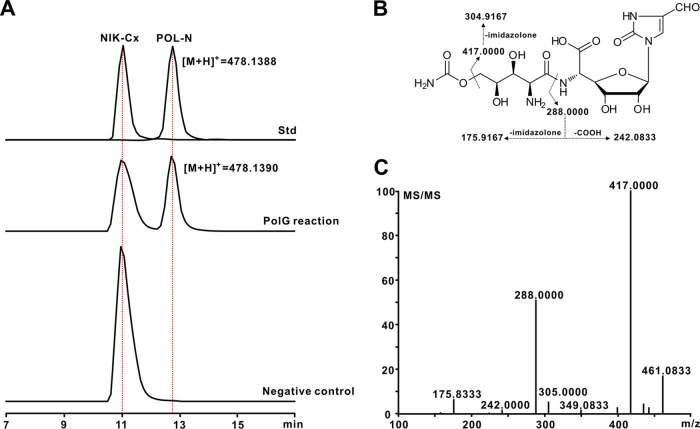
POL-N was previously reported as a POL-NIK hybrid nucleoside antibiotic (1, 14, 15), but the enzymatic mechanism for its biosynthesis is as yet undefined. POL-N is structurally similar to POL-B, except for the difference in the bases (Fig. 1A), suggesting that PolG is perhaps also the candidate required for POL-N assembly (see Fig. S10A in the supplemental material). To assess this, the PolG activity was reconstituted in vitro using NIK-Cx and CPOAA as the substrates (Fig. S10B to D). The results obtained by LC-MS showed that the PolG reaction is able to generate the characteristic [M+H]+ ion at m/z 478.1390, which is absent for the negative control (Fig. 4A). MS/MS analysis generated main fragment ions at m/z 288.0000 and m/z 417.0000, identical to those from an authentic POL-N standard (Fig. 4B and C and S10E). Hence, these data show that the construction of the hybrid nucleoside antibiotic POL-N is governed by the PolG enzyme, which possesses considerable substrate flexibility.
FIG 4.
Enzymatic basis for the generation of the hybrid peptidyl nucleoside antibiotic POL-N. (A) LC-MS analysis of the PolG-catalyzed reaction with NIK-Cx as the substrate; (B) fragmentation pattern of the authentic POL-N standard; (C) HRMS/MS analysis of the fragment ions of the target PolG-catalyzed product.
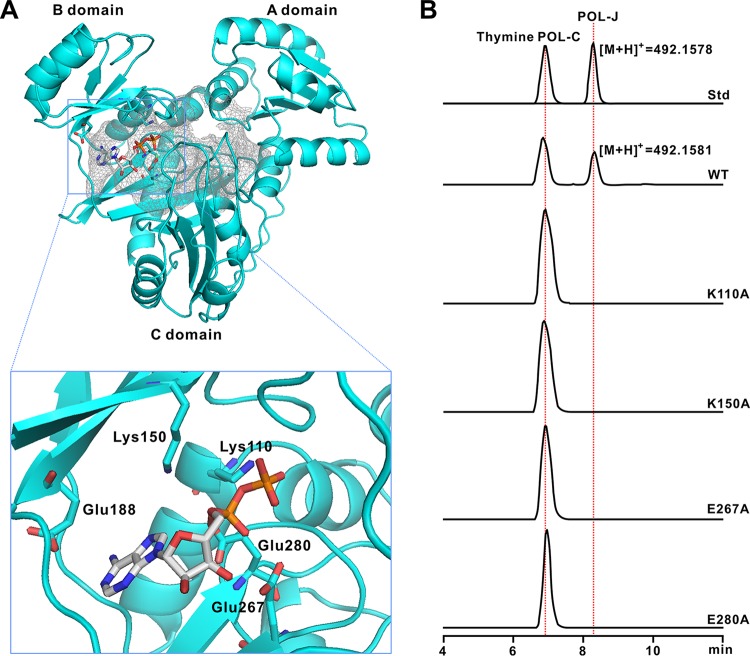
Determination of the conserved sites of PolG by a combined homology structure model and biochemical investigation.
To investigate the ATP-binding pocket of PolG in more detail, we constructed a homology structure (Fig. 5A; see also Fig. S11A to C in the supplemental material) based on that of RizA (PDB 4WD3), a well-defined ligase in the rhizociticin pathway from Bacillus subtilis (20). According to this model, PolG contains three independent domains (designated A, B, and C domains) (Fig. 5A), which are conserved in the L-amino acid ligases, including RizA, BL0025, and BacD (20). Furthermore, in silico analysis showed that the five conserved residues deduced for ATP binding in RizA could also be discerned in PolG (Lys110, Lys150, Glu267, Glu188, and Glu280) (Fig. 5A; see also Fig. S8 in the supplemental material). While the conserved ATP-binding sites are shared by a number of ATP-dependent ligases, their function is as yet undefined (20). To determine if these sites are essential for PolG activity, we individually mutated them and purified the resultant PolG variants. The exception was E188A, which unaccountably did not express in E. coli (see Fig. S12A in the supplemental material). In vitro assays with the mutated PolG were unable to generate the characteristic [M+H]+ ion of POL-J (m/z 492.1581) yet (Fig. 5B). The PolG variants were also tested on the substrate POL-C, again without showing activity (Fig. S12B). Hence, the conserved ATP-binding sites are essential for the activity of PolG enzyme.
FIG 5.
A structural model for PolG and in vitro characterization of its variants. (A) Homology structure model of PolG. This structure is constructed on the basis of RizA from Bacillus subtilis (PDB 4WD3), and the active ATP-binding sites are indicated in the rectangular region. These sites are proposed to be essential for the binding of ATP. (B) LC-MS analysis of the reactions of PolG and its variants. Std, authentic standards of thymine POL-C and POL-J. WT, the reaction with the intact PolG added. Lines labeled K110A, K150A, E267A, and E280A show the individual reactions of PolG variants.
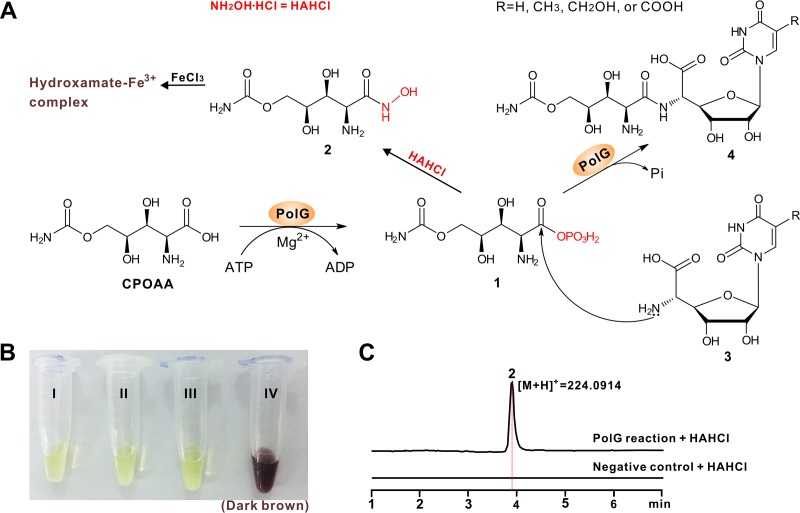
The acyl-phosphorylated CPOAA (compound 1) is an activated intermediate during the assembly of POL.
ATP-dependent ligase-catalyzed reactions typically proceed through an unstable acyl-adenylate or acyl-phosphate intermediate (21). We therefore attempted to trap the deduced compound (labeled with boldface “1” in Fig. 6A) by derivatizing it to form the hydroxamate compound (compound 2), which is readily identified by a chromogenic reaction in the presence of Fe3+ (Fig. 6B). The products of the PolG reaction plus NH2OH·HCl were submitted for LC-MS analysis, generating an [M+H]+ ion of compound 2 at m/z 224.0914, which is totally absent from the negative control (Fig. 6C). MS/MS analysis of the target ion generated a major fragment ion series that is well matched to that of compound 2 (see Fig. S13A and B in the supplemental material). In addition, the LC-MS analysis verified that the predicted product AMP could not be detected (Fig. S13C), thereby excluding the possibility of an acyl-adenylate as the intermediate. Hence, the phosphorylated CPOAA (compound 1) is an activated intermediate in the process of POL assembly.
FIG 6.
Proposed catalytic mechanism for PolG. (A) Proposed mechanism for the PolG-catalyzed reaction and scheme for the chromogenic reaction of the hydroxamate complex. Compound 1 is converted to compound 2, which might react with Fe3+ to form the hydroxamate-Fe3+ complex (dark brown). (B) Detection of compound 2 by chromogenic reaction. I, Fe3+ solution only; II, negative control of the PolG reaction with NH2OH·HCl and Fe3+ added; III, PolG reaction with Fe3+ added; IV, PolG reaction with NH2OH·HCl and Fe3+ added (dark brown). (C) LC-MS analysis of the PolG reaction with NH2OH.
DISCUSSION
POL is an environmentally friendly fungicide with an important role in agriculture in combating a variety of plant diseases, including tobacco brown spot disease, caused by Alternaria alternata, and apple Alternaria leaf spot, caused by Alternaria mali (9). Amide bond-containing biomolecules are usually derived from the templated nonribosomal peptide synthetase (NRPS) pathways or the nontemplated pathways (21, 22), often involving ATP-dependent ligases for assembling natural product scaffolds (23). In the present study, a mechanism is proposed for the PolG-catalyzed reaction, such that PolG first activates the nonproteinogenic amino acid CPOAA to form an acyl-phosphate intermediate (compound 1), and then the amino group of the nucleoside skeleton (compound 3) attacks the carbonyl carbon (compound 1) to form an amide bond (compound 4) by displacing the phosphate group (Fig. 6A).
Previous precursor (uracil analog)-directed biosynthetic studies afforded several POL derivatives with modified nucleoside skeletons (24, 25), and this implicated that the enzyme for the assembly of POL harbors a flexible specificity for the substrate, especially for the nucleoside skeleton. In the present study, this seems to be certainly true, and we demonstrated that PolG is able to recognize diverse nucleoside motifs (POL-C, thymine POL-C, and NIK-Cx). More interestingly, thymine POL-C was previously available only as an artificial POL-C analog prior to its discovery as a natural product (26, 27).
The ATP-dependent ligase family enzymes are widely distributed in the microbial metabolic pathways, including de novo purine biosynthesis, gluconeogenesis, and fatty acid synthesis (23); however, their roles have been significantly underappreciated for natural product biosynthesis (21). In this study, PolG and its related homologs, different from those of ATP-dependent ligases in natural product biosynthesis (28–30), are situated in an independent clade (see Fig. S14 in the supplemental material). In this respect, PolG represents a group of distinctive ATP-dependent ligases with considerable substrate promiscuity, and this group of enzymes are of great potential as catalysts for future biomanufacturing purposes to synthesize rationally designed peptidyl nucleoside antibiotics. As a consequence, it will be of significance to elucidate the precise enzymatic mechanism of PolG by solving its crystal structure, and a related study is now in progress.
In summary, we have dissected the assembly logic of POL, which follows an ATP-dependent strategy for amide bond formation via an activated acyl-phosphate intermediate. We have clarified the chemical structure of the previously overlooked thymine POL-C and describe the enzymatic basis for the biosynthesis of the hybrid nucleoside antibiotic POL-N. This work will contribute to the targeted discovery of novel peptidyl nucleoside antibiotics using PolG as a probe.
MATERIALS AND METHODS
Strains, reagents, and protocols.
S. aureochromogenes strains are cultivated either on a plate of mannitol soya flour (MS) medium for sporulation or in tryptone soya broth (TSB) medium (31). Strains and plasmids used in this study are described in Table 1; PCR primers are listed in Table S2 in the supplemental material. All chemical reagents were purchased from J&K Scientific Ltd., Sigma-Aldrich (USA), or Sinopharm unless otherwise indicated. The general methods performed in this study were according to the standard protocols of Green and Sambrook (32) or Kieser et al. (31).
Analysis of the metabolites by strain CXR14::pCHW201 and its derivative.
For analysis of the target metabolites accumulated by CXR14::pCHW201 and its derivatives, HPLC (Shimadzu LC-20A) was performed on a C18 column (Inertsil ODS-3, 4.6 by 250 mm, 5 μm). The conditions for HPLC analysis were according to the method of Chen et al. (4), and the elution was monitored at UV of 263 nm by a diode array detector (DAD).
Model building and analysis of PolG structure.
The homology structural model of PolG was constructed according to the X-ray structure of RizA from Bacillus subtilis (PDB 4WD3) and further refined using Discovery Studio 4.1 (accelrys) and Pymol softwares.
Enzymatic assays of PolG and its variants.
For activity of PolG and its variants, the reaction mixture (100 mM Tris-Cl buffer, pH 8.0; 1 mM substrate; 2 mM ATP; 4 mM MgCl2; 25 μg protein) was incubated at 30°C for 8 h, and then the reaction was terminated by the immediate addition of an equivalent volume of methanol. After centrifugation to remove protein, the supernatant was filtered by a 0.22-μm filter. Subsequent liquid chromatography-high resolution mass spectrometry (LC-HRMS) analysis was performed on a C18 column (Inertsil ODS-3, 4.6 by 250 mm, 5 μm) with the elution gradient of 5% to 25% methanol–0.15% trifluoroacetic acid (TFA) over 20 min at a flow rate of 0.5 ml/min. LC-HRMS was conducted on an electrospray ionization (ESI)-ion trap mass spectrometer (Thermo LTQ-Obitrap) in a positive mode with drying gas (275°C, 10 liters/ml) and a nebulizer pressure of 30 lb/in2.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants of the National Natural Science Foundation of China (31770041, 21402146), Hubei Provincial Natural Science Foundation of China (2016CFB458), and the Open Funding Projects of the State Key Laboratory of Bioactive Substance and Function of Natural Medicines (GTZK201701) and the State Key Laboratory of Microbial Metabolism (MMLKF16-03).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00501-18.
REFERENCES
- 1.Isono K. 1988. Nucleoside antibiotics: structure, biological activity, and biosynthesis. J Antibiot (Tokyo) 41:1711–1739. doi: 10.7164/antibiotics.41.1711. [DOI] [PubMed] [Google Scholar]
- 2.Chen S, Kinney WA, Van Lanen S. 2017. Nature's combinatorial biosynthesis and recently engineered production of nucleoside antibiotics in Streptomyces. World J Microbiol Biotechnol 33(4):66. doi: 10.1007/s11274-017-2233-6. [DOI] [PubMed] [Google Scholar]
- 3.Niu G, Tan H. 2015. Nucleoside antibiotics: biosynthesis, regulation, and biotechnology. Trends Microbiol 23:110–119. doi: 10.1016/j.tim.2014.10.007. [DOI] [PubMed] [Google Scholar]
- 4.Chen W, Qi J, Wu P, Wan D, Liu J, Feng X, Deng Z. 2016. Natural and engineered biosynthesis of nucleoside antibiotics in Actinomycetes. J Ind Microbiol Biotechnol 43:401–417. doi: 10.1007/s10295-015-1636-3. [DOI] [PubMed] [Google Scholar]
- 5.Qi J, Wan D, Ma H, Liu Y, Gong R, Qu X, Sun Y, Deng Z, Chen W. 2016. Deciphering carbamoylpolyoxamic acid biosynthesis reveals unusual acetylation cycle associated with tandem reduction and sequential hydroxylation. Cell Chem Biol 23:935–944. doi: 10.1016/j.chembiol.2016.07.011. [DOI] [PubMed] [Google Scholar]
- 6.Benitez T, Villa TG, Acha IG. 1976. Effects of polyoxin D on germination, morphological development and biosynthesis of the cell wall of Trichoderma viride. Arch Microbiol 108:183–188. doi: 10.1007/BF00428949. [DOI] [PubMed] [Google Scholar]
- 7.Hector RF, Pappagianis D. 1983. Inhibition of chitin synthesis in the cell wall of Coccidioides immitis by polyoxin D. J Bacteriol 154:488–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Becker JM, Covert NL, Shenbagamurthi P, Steinfeld AS, Naider F. 1983. Polyoxin D inhibits growth of zoopathogenic fungi. Antimicrob Agents Chemother 23:926–929. doi: 10.1128/AAC.23.6.926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen W, Huang T, He X, Meng Q, You D, Bai L, Li J, Wu M, Li R, Xie Z, Zhou H, Zhou X, Tan H, Deng Z. 2009. Characterization of the polyoxin biosynthetic gene cluster from Streptomyces cacaoi and engineered production of polyoxin H. J Biol Chem 284:10627–10638. doi: 10.1074/jbc.M807534200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He N, Wu P, Lei Y, Xu B, Zhu X, Xu G, Gao Y, Qi J, Deng Z, Tang G, Chen W, Xiao Y. 2017. Construction of an octosyl acid backbone catalyzed by a radical S-adenosylmethionine enzyme and a phosphatase in the biosynthesis of high-carbon sugar nucleoside antibiotics. Chem Sci 8:444–451. doi: 10.1039/C6SC01826B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lilla EA, Yokoyama K. 2016. Carbon extension in peptidylnucleoside biosynthesis by radical SAM enzymes. Nat Chem Biol 12:905–907. doi: 10.1038/nchembio.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen W, Li Y, Li J, Wu L, Li Y, Wang R, Deng Z, Zhou J. 2016. An unusual UMP C-5 methylase in nucleoside antibiotic polyoxin biosynthesis. Protein Cell 7:673–683. doi: 10.1007/s13238-016-0289-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qi J, Liu J, Wan D, Cai YS, Wang Y, Li S, Wu P, Feng X, Qiu G, Yang SP, Chen W, Deng Z. 2015. Metabolic engineering of an industrial polyoxin producer for the targeted overproduction of designer nucleoside antibiotics. Biotechnol Bioeng 112:1865–1871. doi: 10.1002/bit.25594. [DOI] [PubMed] [Google Scholar]
- 14.Zhai L, Lin S, Qu D, Hong X, Bai L, Chen W, Deng Z. 2012. Engineering of an industrial polyoxin producer for the rational production of hybrid peptidyl nucleoside antibiotics. Metab Eng 14:388–393. doi: 10.1016/j.ymben.2012.03.006. [DOI] [PubMed] [Google Scholar]
- 15.Li J, Li L, Tian Y, Niu G, Tan H. 2011. Hybrid antibiotics with the nikkomycin nucleoside and polyoxin peptidyl moieties. Metab Eng 13:336–344. doi: 10.1016/j.ymben.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 16.Uchida K, Kato K, Yamaguchi K, Akita H. 2000. First total syntheses of (+)-polyoxin B and (+)-polyoxin D. Heterocycles 53:2253–2262. doi: 10.3987/COM-00-8998. [DOI] [Google Scholar]
- 17.Fujino H, Nagatomo M, Paudel A, Panthee S, Hamamoto H, Sekimizu K, Inoue M. 2017. Unified total synthesis of polyoxins J, L, and fluorinated analogues on the basis of decarbonylative radical coupling reactions. Angew Chem Int Ed Engl 56:11865–11869. doi: 10.1002/anie.201706671. [DOI] [PubMed] [Google Scholar]
- 18.Gust B, Challis GL, Fowler K, Kieser T, Chater KF. 2003. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc Natl Acad Sci U S A 100:1541–1546. doi: 10.1073/pnas.0337542100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barrett AGM, Lebold SA. 1990. (Phenylthio)nitromethane in the total synthesis of polyoxin-C. J Org Chem 55:3853–3857. doi: 10.1021/jo00299a030. [DOI] [Google Scholar]
- 20.Kagawa W, Arai T, Ishikura S, Kino K, Kurumizaka H. 2015. Structure of RizA, an L-amino-acid ligase from Bacillus subtilis. Acta Crystallogr F Struct Biol Commun 71:1125–1130. doi: 10.1107/S2053230X15012698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goswami A, Van Lanen SG. 2015. Enzymatic strategies and biocatalysts for amide bond formation: tricks of the trade outside of the ribosome. Mol Biosyst 11:338–353. doi: 10.1039/C4MB00627E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Giessen TW, Marahiel MA. 2012. Ribosome-independent biosynthesis of biologically active peptides: application of synthetic biology to generate structural diversity. FEBS Lett 586:2065–2075. doi: 10.1016/j.febslet.2012.01.017. [DOI] [PubMed] [Google Scholar]
- 23.Fawaz MV, Topper ME, Firestine SM. 2011. The ATP-grasp enzymes. Bioorg Chem 39:185–191. doi: 10.1016/j.bioorg.2011.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Isono K, Suhadolnik RJ. 1976. The biosynthesis of natural and unnatural polyoxins by Streptomyces cacaoi. Arch Biochem Biophys 173:141–153. doi: 10.1016/0003-9861(76)90244-7. [DOI] [PubMed] [Google Scholar]
- 25.Isono K, Crain PF, Odiorne TJ, McCloskey JA, Suhadolnik RJ. 1973. Biosynthesis of the 5-fluoropolyoxins, aberrant nucleoside antibiotics. J Am Chem Soc 95:5788–5789. doi: 10.1021/ja00798a072. [DOI] [PubMed] [Google Scholar]
- 26.Harding KE, Southard JM. 2005. A formal synthesis of thymine polyoxin C. Tetrahedron Asymmetry 16:1845–1854. doi: 10.1016/j.tetasy.2005.03.034. [DOI] [Google Scholar]
- 27.Nishiyama T, Mohile SS, Kajimoto T, Node M. 2007. Synthesis of thymine polyoxin C by using L-threonine aldolase-catalyzed aldol reaction. Heterocycles 71:1397–1402. doi: 10.3987/COM-07-11055. [DOI] [Google Scholar]
- 28.Feng J, Wu J, Gao J, Xia Z, Deng Z, He X. 2014. Biosynthesis of the beta-methylarginine residue of peptidyl nucleoside arginomycin in Streptomyces arginensis NRRL 15941. Appl Environ Microbiol 80:5021–5027. doi: 10.1128/AEM.01172-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hollenhorst MA, Clardy J, Walsh CT. 2009. The ATP-dependent amide ligases DdaG and DdaF assemble the fumaramoyl-dipeptide scaffold of the dapdiamide antibiotics. Biochemistry 48:10467–10472. doi: 10.1021/bi9013165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Z, Du L, Zhang W, Zhang X, Jiang Y, Liu K, Men P, Xu H, Fortman JL, Sherman DH, Yu B, Gao S, Li S. 2017. Complete elucidation of the late steps of bafilomycin biosynthesis in Streptomyces lohii. J Biol Chem 292:7095–7104. doi: 10.1074/jbc.M116.751255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kieser T, Bibb MJ, Chater KF, Butter MJ, Hopwood DA. 2000. Practical Streptomyces genetics, 2nd ed John Innes Foundation, Norwich, United Kingdom. [Google Scholar]
- 32.Green MR, Sambrook J. 2012. Molecular cloning: a laboratory manual, 4th ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.