Abstract
We have synthesized three peptides from the mdm-2 binding domain of human p53, residues 12–26 (PPLSQETFSDLWKLL), residues 12–20, and 17–26. To enable transport of the peptides across the cell membrane and at the same time to maximize the active mdm-2 binding α-helical conformation for these peptides, each was attached at its carboxyl terminus to the penetratin sequence, KKWKMRRNQFWVKVQRG, that contains many positively charged residues that stabilize an α-helix when present on its carboxyl terminal end. All three peptides were cytotoxic to human cancer cells in culture, whereas a control, unrelated peptide attached to the same penetratin sequence had no effect on these cell lines. The same three cytotoxic peptides had no effect on the growth of normal cells, including human cord blood-derived stem cells. These peptides were as effective in causing cell death in p53-null cancer cells as in those having mutant or normal p53. Peptide-induced cell death is not accompanied by expression of apoptosis-associated proteins such as Bax and wafp21. Based on these findings, we conclude that the antiproliferative effects of these p53-derived peptides are not completely dependent on p53 activity and may prove useful as general anticancer agents.
Activation of the p53 protein results in the transcription of antiproliferative proteins and in induction of apoptosis, in part through expression of Bax and wafp21 proteins (1). Regulation of the lifetime of the p53 molecule is known to be a critical factor that affects its ability to control cell cycle events. Binding of p53 to the oncogene-encoded mdm-2 protein targets it for ubiquitination and degradation (2).
In a previous study, we found that the N-terminal kinase that activates the jun protein, jun N-terminal kinase (JNK), binds to p53 and promotes its phosphorylation (3). Consequently, p53 no longer interacts with mdm-2, a phenomenon that results in the prolongation of its half-life and its ability to reverse cell cycle events that lead to cell transformation.
The x-ray structure of the complex of the mdm-2 binding peptide domain of p53 with the mdm-2 protein has been elucidated (4). The region of human p53 from residues 12–26 (with the sequence PPLSQETFSDLWKLL) is the one that contacts the mdm-2 protein, which adopts an α-helical conformation when bound to mdm-2. This sequence is divided into two subdomains (1) consisting of an invariant segment (residues 12–20) and residues 17–26 containing all of the residues that actually contact the binding domain of mdm-2 (4).
Based on our results with the JNK-induced phosphorylation of p53, it occurred to us that blockade of p53-mdm-2 interactions can counter cell-transforming events. One manner to effect this blockade would be to use peptides from the p53 interaction site to compete for the binding of p53 to mdm-2.
Prior studies on the use of peptides from p53 that activate the mutant protein, resulting in apoptosis, have been performed on several different transformed cell lines. For example, we and others have found that a carboxyl terminal peptide, corresponding to p53 homotetramerization domain residues 361–382, introduced into cells either as a vector or as the peptide bound to a leader sequence from antennapedia protein enabling membrane transport of the peptide (5), induced apoptosis in transformed cell lines that contained either mutant or normal p53 (6, 7), but not in transformed cell lines that were p53 null (6, 7). That apoptosis was p53-dependent was further confirmed in studies that demonstrated that this peptide induced expression of Bax, wafp21, FAS, and other apoptosis-associated proteins induced by p53 (1, 6, 7).
In another study, a GST fusion peptide with the sequence MPRFMDYWEGLN was introduced into osteosarcoma cells, which overexpress mdm-2, and in other cell lines that express mdm-2 and p53 but are transformed by the HPV16 E6 oncogene (8). This peptide also induced apoptosis in p53-containing cells, but had no effect on cells with homozygous deletions of p53.
In this study, we have prepared several peptides from the p53 amino terminal domain corresponding to the region that directly binds to mdm-2, attached to a positively charged leader sequence of the antennapedia protein (5). This sequence was placed in such a way as to stabilize the α-helical conformation of each peptide (9, 10). We have introduced these peptides into a number of different human cancers and also untransformed cell lines to determine their efficacy and specificity in blocking cell proliferation.
Materials and Methods
Peptides.
Six peptides from the mdm-2 binding domain of p53 were synthesized by solid phase synthesis and purified by HPLC to >95% purity. Three of these contained the antennapedia leader sequence (KKWKMRRNQFWVKVQRG, designated as “Leader”) on their carboxyl terminal ends, and three were devoid of this sequence.
The three p53 peptides in peptide linkage to the leader sequence were (i) residues 12–20 (PPLSQETFS)-Leader, denoted as PNC-21 that contains the invariant domain; (ii) residues 12–26 (PPLSQETFSDLWKLL)-Leader, denoted as PNC-27 that comprises the mdm-2 binding domain (4); and (iii) residues 17–26 (ETFSDLWKLL)-Leader, denoted as PNC-28, which contains all of the residues that contact mdm-2 (4) and which overlaps the invariant sequence by four residues. A modified form of PNC-27 was synthesized in which a fluorescent rhodamine label was attached to its carboxyl terminus (of the leader sequence; Research Genetics, Huntsville, AL).
Of the three p53 peptides that were not connected to the leader sequence, we used one in this study, called PNC-26, whose sequence is identical to that for PNC-27 without the leader sequence. In addition to the above peptide, we synthesized an unrelated peptide from cytochrome p450, called X13 (MPFSTGKRIMLGE), which we have previously used as a control peptide (11) attached to the leader sequence on its carboxyl terminal end, called X13-Leader.
Cells.
BMRPA1 cells are untransformed, normal rat pancreatic acinar cells that have maintained, in addition to their epithelial cell phenotype and growth in culture, their differentiated cell functions such as continued enzyme production and activation of zymogen secretion by secretagogue. BMRPA1 cells have neither c-k-ras nor p53 mutations, are unable to grow in anchorage-independent conditions, and do not form tumors in Nu/Nu mice (ref. 12; L. Y. Bao and J.M., unpublished results). BMRPA1.TUC-3 or TUC-3 cells are BMRPA1 cells transformed by transfection with a plasmid containing an activated human K-ras oncogene [single base mutation at codon 12, valine substitution for the wild-type glycine in the ras protein (K-rasval12); a kind gift of Dr. M. Perucho (Burnham Institute, La Jolla, CA)] and a neomycin resistance gene (J.M. and L. Y. Bao, unpublished results). TUC-3 cell clones were selected on their basis of resistance to G418 and the overexpression of K-rasval12. TUC-3 cells no longer display an epithelial cell phenotype and acinar cell functions, but grow significantly faster than BMRPA1 cells and have a transformed spindle cell phenotype. In addition, we have found that these cells form colonies under anchorage-independent conditions in vitro and tumors in vivo in nude mice (S. Bradu, R. Huynh, H. O. Akman, T. Zheng, L. Y. Bao, M.R.P., and J.M., unpublished results).
E49 cells are rat brain capillary endothelial cells with the phenotypic characteristics of spontaneously transformed cells and exhibit the ability to form tumors in 50–60% of the Nu/Nu mice injected with 107 cells (kind gift of Dr. M. DelPiano, Max-Planck Institute, Dortmund, Germany).
A549 cells, human lung non-small-cell carcinoma cells, HeLa, human cervical carcinoma cells, p53-null SW 1417 cells, human metastatic colon adenocarcinoma cells, MDA-MB-453 (p53-null human metastatic breast carcinoma cells), H1299 (p53-null human non-small-cell lung cancer), and SAOS2 (p53-null osteosarcoma) were obtained from the American Type Culture Collection. Each cell line was grown in its specified culture medium containing 10% FBS. On reaching ≈80% confluence, cells were passaged [subjected to release by trypsin-EDTA (GIBCO)] for continued growth or collected, adjusted to a predetermined cell concentration, and reseeded into 24-well tissue culture dishes (TCD) or 25 cm2 TC flasks (TCF) according to the experimental design. All experiments were carried out with cells in exponential growth phase.
Incubation of Cells with PNC Peptides.
BMRPA1, TUC-3, and other transformed cells were collected, and 2 × 104 cells were seeded into each well of 24-well TCD containing 1 ml of culture medium. Cells were allowed to adhere for 24 h when the medium in sets of triplicate wells was replaced with media containing from 1 to 100 μg/ml of the peptide to be tested. The peptides were first dissolved at 10 mg/ml PBS from which the predetermined peptide concentrations were prepared in the specified media. The peptide-containing media were sonicated briefly before being added to the cells. Another set of triplicate wells (controls) in each experiment was processed identically but with peptide-free media. Cells were fed every 24 h with 1 ml of their respective peptide-containing media. The cultures were examined daily for changes in cell growth and morphology. Cells were released and collected after 5 days of treatment, and the number of viable cells was established in each of the triplicate wells in the presence of trypan blue.
Molecular Analysis of Peptide-Induced Cytotoxicity.
For immunofluorescence examination of the expression of apoptosis-related proteins, cells were allowed to attach to glass coverslips for 48 h. Then peptide-containing medium was added for another 24 h. At this time, the coverslips were removed, the cells fixed in ice-cold methanol, and the samples processed with antibody to c-myc, p53, Bax, bcl-2, and wafp21 according to the manufacturer's description (Oncogene Science), using ProLong Antifade to prevent rapid bleaching of the fluorescence image during microscopic examination in a Zeiss Fluorescent Photomicroscope equipped with epifluorescence.
For analysis of apoptosis-related proteins by immunoblotting, cells were grown in 25 cm2 TCF for 24 h when the medium was exchanged for peptide-containing medium. After another 48–72 h the cells were lysed in situ in cell lysis buffer [1% Triton X-100 in 0.05 M Tris⋅HCl (pH 8.0)/0.15 mM NaCl/0.02% Na azide/0.1 mg/ml PMSF/0.001 mg/ml aprotinin]. Cell proteins were separated on 10% SDS/PAGE followed by electrotransfer to nitrocellulose and immunoblotting with apoptosis-specific antibodies to Bax, waf-1p21, and p53 as directed (Oncogene Science; Santa Cruz Biotechnology). Antibody-labeled proteins were sidentified by chemiluminescence using ECL methodology (Amersham Pharmacia) and Kodak x-ray film.
Oocyte Microinjection.
Oocytes were obtained from Xenopus laevis frogs from Connecticut Valley Biological (Southampton, MA) as described (11). All microinjection experiments were performed at least six times on 30 oocytes, prepared from collagenase-digested ovarian follicles that were then incubated at 19°C for 12–24 h. Microinjected oocytes were incubated in Barth's medium or Barth's medium containing insulin, present at a concentration of 10 μg/ml, for various times at 25°C. Oocyte maturation was determined by observing germinal vesicle breakdown (GVBD). [Val12]Ha-ras-p21 was either injected into oocytes (100 μg/ml, 50 nl per oocyte) alone or coinjected with an inhibitory peptide (500 μg/ml).
Gamma Irradiation of Cells.
TUC-3 and BMRPA1 cells were irradiated with 10 Gy at room temperature in 5 ml of fresh growth medium with a Philips RT250 x-ray machine (250-kV peak, 15 mA, 0.39-mm Cu HVL) at a dose of 2.7 Gy/min.
Incubation of Cells with Actinomycin D.
Actinomycin D induces apoptosis in cells (13). To induce apoptosis in the cell lines used in this study, cell cultures described above were incubated in media containing 0.5 μg/ml actinomycin D (Sigma; ref. 13).
DNA Fragmentation Analysis for p53-Induced Apoptosis.
DNA was isolated from cells, which were lysed as described in the preceding paragraph, using the standard QIAamp DNA mini kit procedure (Qiagen). Isolated DNA was subjected to agarose gel electrophoresis and stained with ethidium bromide. As positive controls, we used lysate provided in the Suicide-Track DNA Ladder isolation kit (Oncogene Science) and TUC-3 cells treated with actinomycin D (13).
Colony Forming Unit Assays.
We have tested the effects of the peptides on the abilities of cultures of human stem cells to differentiate into erythroid or myeloid cell lines in the presence of growth factors. The technique, described in refs. 14 and 15, consists of culturing unfractionated cord blood samples of 2 and 4 μl diluted in 1 ml of semisolid Iscove's modified Dulbecco's medium (IMDM; GIBCO) containing 0.8% methyl cellulose and defined concentrations of hematopoietic growth factors (14, 15)—i.e., 1.5 units/ml erythropoietin (Epo), 2 × 10−10 M granulocyte colony-stimulating factor (GCSF), 100 ng/ml stem cell factor (SCF), 4.5 × 10−10 M granulocyte–macrophage colony-stimulating factor (GM-CSF), and 2 × 10−10 M interleukin-3 (IL-3). EPO, GCSF, and SCF were generous gifts from Amgen, and the Genetics Institute (Cambridge, MA) kindly donated both GM-CSF and IL-3. All cultures were established in duplicate dishes and visually evaluated at 12–14 days for the number and overall shape and size of erythroid bursts, GM colonies, and mixed colonies in the absence or presence of each peptide, present at a concentration of 100 μg/ml.
Computational Methods.
To ascertain the effects of adding the leader sequence to either the amino or carboxyl terminal end of the p53 12–26 peptide, we computed the α-helix probability profile for each peptide by using two different methods, one using helix probabilities from the protein database (16) and the other using the Ising model based on helix nucleation (σ) and growth (s) equilibrium constants determined experimentally from block copolymers for each of the twenty naturally occurring L amino acids (9), modified by inclusion of the effects of charges on these parameters as described (9, 10).
Results
Helix Probability Profiles for p53 Peptides.
Effect of the leader sequence.
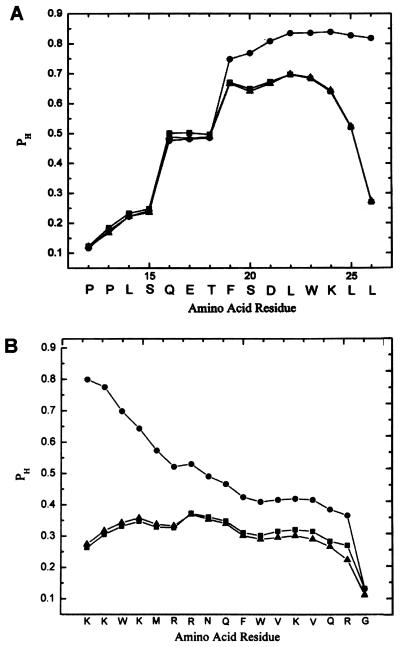
To compute whether the leader sequence would stabilize the α-helical conformation for the p53 peptide, we generated the α-helical probability profiles for the p53 12–26 peptide without the leader sequence (PNC-26) and for this peptide attached to the leader sequence at both amino and carboxyl terminal ends as shown in Fig. 1A, using the Ising model that includes the effects of charges on amino acids (9). This method has been used previously to compute the helical probability profiles for a number of different peptides including the S-peptide of ribonuclease A and a series of cytochrome C peptides that stimulate T-cell activation, with good agreement between computational and experimental results (10). Very similar results were obtained by using an independent method based on empirical probabilities derived from proteins with known x-ray structures (16). As shown in Fig. 1A, the amino acid residues of PNC-26 from Glu-17–Lys-24 (filled squares) have high helical probabilities; residues 25 and 26 have significantly lower helical probabilities. If the leader sequence containing high positive charge content is added to this sequence on its amino-terminal end, it has little effect on the probability profile (filled triangles). Reciprocally, as shown in Fig. 1B, addition of the leader sequence to the amino-terminal end of the p53 sequence has no effect on its residue helical probabilities.
Figure 1.
(A) Helix probability (PH) profiles for the p53 12–26 peptide with no leader sequence (filled squares), with positively charged antennapedia leader sequence (filled triangles) on its amino-terminal end and with the leader sequence on its carboxyl terminal end (filled circles). (B) Helix probability profile for the antennapedia leader sequence alone (filled triangles), attached to the amino-terminal end of the p53-12–26 peptide (filled squares) and attached to the carboxyl terminal end (filled circles). Profiles were computed by using experimentally determined helix–coil parameters including the effects of charges as described by Vasquez et al. (9, 10).
However, if the leader sequence is added to the carboxyl terminal end of the p53 peptide, the helix probabilities of residues 19–26 (filled circles, Fig. 1A) and of the residues of the leader sequence (filled circles, Fig. 1B) increase substantially. This is particularly significant for the p53 sequence, because these residues are the ones involved in the direct contacts with mdm-2 (4). Because our hypothesis was that this peptide might exert antiproliferative effects in transformed cells by binding to mdm-2, we chose to employ the peptide attached to the leader sequence on its carboxyl terminal end where the active conformation for the contact residues in the peptide would have a high probability for adopting the α-helical conformation. We therefore also added the leader sequence to the carboxyl terminal ends of the other two peptides for the same reason.
BMRPA1 and TUC-3 Cells.
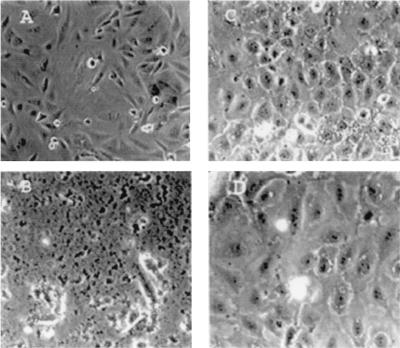
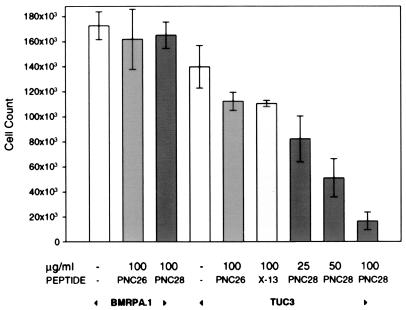
We have developed a line of rat pancreatic acinar cells in culture, called BMRPA1 (12). These cells form stable contact-inhibited monolayers as shown in Fig. 2C for untreated cells. These cells have been stably transfected with the k-ras oncogene to yield acinar pancreatic carcinoma cells, called TUC-3, that are not contact-inhibited and become spindle-shaped, many of them becoming multinucleated as shown in Fig. 2A. We have incubated these cells with each of the three p53 peptides attached to the leader sequence, PNC-21, -27, and -28 for 3 days. Fig. 2 B and D shows the effects of treatment of TUC-3 and BMRPA1 cells, respectively, with PNC-28. PNC-28 induces almost 100% cell death in TUC-3 cells (Fig. 2B), but has no effect on cell viability of BMRPA1 cells (Fig. 2D). Identical results were obtained with PNC-21 and -27 peptides. PNC-28 is cytotoxic to TUC-3 cells in a dose-related manner as shown in the last three bar graphs on the right side of Fig. 3. In these experiments, the cells were incubated with PNC-28 for 3 days. At 100 μg/ml (right-most bar in the figure), PNC-28 induces cell death in 90% of TUC-3 cells. The effective dose range for PNC-28 (and for PNC-21 and -27 peptides) is narrow for TUC-3 cells in that the effective doses occur over only about a 4-fold (25–100 μg/ml, IC50 = 40 μg/ml) increase in concentration (last three bars in Fig. 3). On the other hand, control peptides like X13-Leader and PNC-26, the p53 mdm-2 binding peptide with no leader sequence, had no effect on TUC-3 cell growth (fifth and sixth bars in Fig. 3). As can also be seen in Fig. 3, PNC-28 and control peptides had no effect on the viability of BMRPA1 cells (third and second bars, respectively).
Figure 2.
Effects of PNC-28-Leader on rat pancreatic acinar cells (BMRPA1) and the corresponding ras-transformed cell line (TUC-3). Untreated cells for the transformed and normal acinar cells are shown in A and C, respectively. After 3 days of treatment, the transformed cells are necrotic (B), whereas the acinar cells are the same as before treatment (D).
Figure 3.
Effects of different peptides on the growth of BMRPA1 and TUC-3 cells. Experiments with each of these cell lines are indicated by the arrowheads at the bottom of the figure. The specific peptides and, above them, their concentrations (μg/ml), are given along the abscissa. The number of viable cells (ordinate) was determined by using the trypan blue exclusion method.
From the results shown in Figs. 2 and 3, we conclude that the cytotoxic effect of this peptide is selective for the transformed cells and is specific to the amino acid sequence and is not a nonspecific effect of the peptide or of the leader sequence, because X13-Leader does not affect TUC-3 cells and because PNC-28 has no effect on the untransformed BMRPA1 cells.
We further explored the effects, after 3 days, of discontinuing incubation of TUC-3 cells with PNC-28 after 24 h of incubation with this peptide. At a dose of PNC-28 of 100 μg/ml, the cytotoxicity level was about 30% rather than almost 100% in cells receiving continuous treatment. This result suggests that the peptide is inactivated over a 24-h period and that full cytotoxicity requires continuous dosing and is compatible with our finding (data not shown) that, in several different transformed human cell lines, membrane penetration of PNC-27, containing a carboxyl terminal fluorescent rhodamine label, occurs in at least 50% of the cells within 15 min of exposure and can be detected within the cells for about 1 day subsequently. The peptides may be inactivated through such processes as degradation, extrusion, or inactivating binding to other molecules over a 24-h period.
Oocyte Microinjection.
Oncogenic ras-p21 (containing a G12V substitution) induces oocyte maturation that is blocked by agents, including p21 peptides that do not interfere with insulin-induced maturation (17), that depend on activation of normal cellular ras-p21. This finding suggests that oncogenic p21 activates a unique signaling pathway that is distinct from that used by the activated normal counterpart protein (17). Because PNC-28 is cytotoxic for TUC-3 cells, which are ras-transformed, but not to the normal counterpart BMRPA1 cells, we investigated whether the p53 peptides selectively block [Val12]p21- but not insulin-induced oocyte maturation.
Coinjection of 500 μg/ml of PNC-26 or -28 with 100 μg/ml of [Val12]p21 resulted in 30% inhibition of oncogenic p21-induced oocyte maturation after 36 h. Neither peptide had any effect on insulin-induced maturation, suggesting that these peptides induce specific inhibitory effects on oncogenic protein-initiated signal transduction.
However, unlike the inhibitory effects of other peptides on [Val12]p21-induced maturation, which reach 100% inhibition, these peptides reach a maximum level of 30% inhibition. This result is consistent with the results of the experiment described above in which PNC-28 was withdrawn after 24 h, resulting in a cytotoxicity level of about 30% rather than almost 100% in cells receiving continuous treatment and suggests that inactivation of the peptide occurs in oocytes by a similar mechanism.
Effect of p53 Peptides on Stem-Cell Proliferation.
To further test the specificity of these peptides in inducing cell death of transformed cells, we determined their effects on normally proliferating human stem cell lines, from the cord blood of five different donors, to test whether these peptides block normal stem cell differentiation, as in bone marrow, that would mitigate against their efficacy as potential chemotherapeutic agents.
Each set of stem cells was induced to differentiate into hematopoietic progenitor cells, which form colonies that contain both granulocytes and macrophages (GM) and erythroid bursts and mixed (GM plus erythroid bursts), under the effect of added growth factors without peptide or in the presence of 100 μg/ml of PNC-28 or X13-Leader. The results are summarized in Table 1.
Table 1.
Effects of PNC-28 and control X13-Leader peptides on growth factor-induced proliferation of human stem cells from five different donors
| Sample | Absolute colony counts
|
||
|---|---|---|---|
| Untreated | X13-Leader* | PNC-28* | |
| 1 | 2,000 | 3,000 | 2,500 |
| 2 | 22,667 | 17,500 | 17,667 |
| 3 | 11,167 | 11,000 | 14,000 |
| 4 | 6,667 | 7,000 | 6,333 |
| 5 | 5,333 | 8,000 | 5,333 |
Growth factors consisted of 1.5 units/ml erythropoietin (Epo), 2 × 10−10 M granulocyte colony-stimulating factor (GCSF), 100 ng/ml stem cell factor (SCF), 4.5 × 10−10 M granulocyte–macrophage colony-stimulating factor (GM-CSF), and 2 × 10−10 M interleukin-3 (IL-3).
All peptides were present at a concentration of 100 μg/ml.
As can be seen from this table, neither the negative control X13-leader nor PNC-28 peptide substantially affects the colony counts from the stem cells of any of the five donors. Although not shown in this table, the distribution and size of colonies for each experiment were the same in the absence or presence of each peptide. This finding correlates with our other findings that PNC-28 does not affect the normal growth of BMRPA1 cells and has no effect on the ability of insulin-activated c-ras p21 to induce maturation of oocytes by normal signal transduction.
Effects of Peptides on Human Cancer Cell Lines.
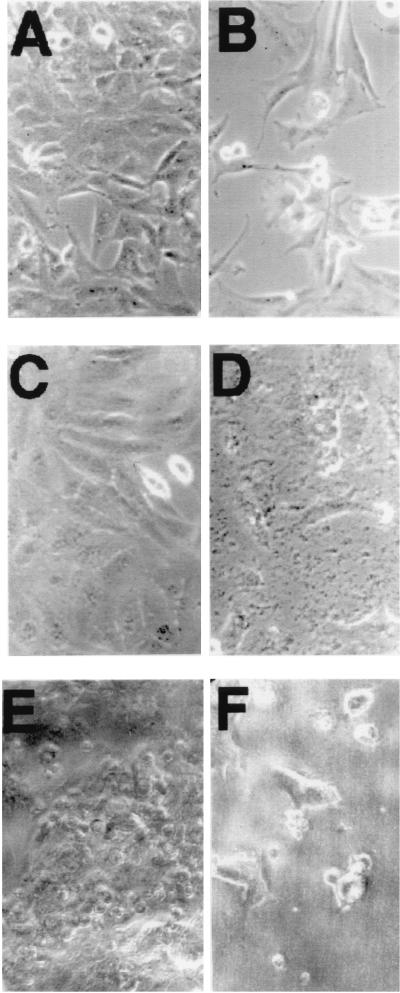
As shown in Fig. 4, we further studied the effects of each of these three p53 peptides on other transformed cell lines that include E49 (a transformed rat endothelial cell line; Fig. 4 A and B), HeLa (squamous carcinoma; C and D), and SW 1417 (colon adenocarcinoma; E and F). The results are summarized in Fig. 4 for untreated cells (A, C, and E) and cells incubated with 100 μg/ml of PNC-28 (B, D, and F). As can be seen in this figure, PNC-28 induces extensive cell death that reached 96, 100, and 86% in the three cell lines, respectively. Neither X13-Leader nor PNC-26, which does not contain the leader sequence, had any effect on cell viability (data not shown). Virtually identical effects were observed on other cell lines (data not shown) such as A549 (small-cell lung carcinoma) for which no viable cells were observed after 3 days.
Figure 4.
Effect of 3 days of treatment of three different cancer cell lines with 100 μg/ml of PNC-28 peptide. A, C, and E show untreated E-49, HeLa, and SW1417 cells, whereas B, D, and F show the corresponding frames of the same cells treated with 100 μg/ml of PNC-28.
A surprising finding from Fig. 4 is that PNC-28 induces extensive cell death in the SW1417 cell line, a human colon cancer cell line that is p53 null (Fig. 4F). Both PNC-21 and -27 also had the same effect on this cell line, whereas X13-Leader had no effect on cell viability, indicating that the cytotoxic effect of PNC-28, -21, and -27 is specific to these p53 peptides. Identical results were achieved with three other p53-null cell lines, namely MDA-MB-453 (human breast cancer), H1299 (human colon cancer), and SAOS2 (osteosarcoma) (data not shown). In contrast to these results, SAOS2 cells have been found to be unaffected by a carboxyl terminal peptide that specifically induces p53-dependent apoptosis (6–8). Our results suggest that the amino-terminal p53 peptides induce cytotoxic effects that appear to be independent of p53 function.
Absence of Markers for Apoptosis.
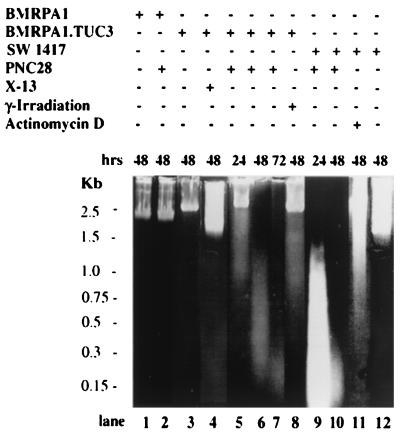
The above conclusion is reinforced by the results of experiments, shown in Fig. 5, in which cell lines treated with p53 peptides were subjected to lysis and the DNA extracted and stained with propidium bromide. Untransformed BMRPA1 cells have high molecular weight DNA whether untreated (lane 1) or treated with 100 μg/ml PNC-28 for 48 h (lane 2). A very similar pattern was likewise obtained for untreated TUC-3 cells and TUC-3 cells treated for 48 h with control X13-Leader peptide (lanes 3 and 4, respectively). However, DNA from TUC-3 cells treated for 24, 48, and 72 h with PNC-28 peptide distributes into fragments of progressively lower molecular weight, as shown in lanes 5–7, respectively. This pattern is not typical of apoptosis, which typically results in fragmentation of DNA into higher molecular forms, and is one that is obtained by treating cells with agents known to induce apoptosis such as γ-irradiation and actinomycin D (13). Lane 8 shows that irradiation of TUC-3 cells, after 48 h, results in fragmentation of DNA into higher molecular weight fragments, in contrast to lane 6, which shows DNA fragmentation into low molecular weight forms from TUC-3 cells incubated with PNC-28 for 48 h.
Figure 5.
Propidium iodide staining of total cell DNA revealing the fragmentation patterns for different cell lines [BMRPA1, BMRPA1.TUC-3 (TUC-3), and SW1417] subjected to different conditions (i.e., PNC-28, actinomycin D, or irradiation). Cell line and treatment agent are given above each lane of the gels; lane numbers (“lane”) are given at the bottom of the figure; the time of treatment is given immediately above each lane; and molecular weight markers are given to the left of the gel.
Similarly, DNA in SW1417 cells treated for 24 and 48 h with PNC-28 undergoes fragmentation into low molecular weight species, as shown in lanes 9 and 10, respectively, whereas DNA from SW1417 cells incubated for 48 h with actinomycin D, which induces apoptosis (13), fragments into high molecular weight species, as shown in lane 11. DNA from untreated SW1417 cells is shown in lane 12. To further confirm these findings, we probed for expression of protein markers of p53-induced apoptosis in TUC-3 and SW1417 cells incubated with PNC-28. These markers include p53 itself, wafp21, Bax, c-myc, and bcl-2. In both the immunostaining and Western blot analysis we were not able to detect any of the above mentioned apoptosis-specific proteins, in contrast to results obtained in several cell lines subjected to conditions that induce apoptosis. Thus it appears that the three p53 peptides induce cytotoxicity, in transformed cells, that does not involve apoptosis.
Discussion
Structure of the p53-Leader Sequence Peptide.
Peptides from the amino (residues 80–93; ref. 18) and carboxyl (residues 361–382; refs. 6 and 7) terminal domains of p53 have been found to induce p53-dependent apoptosis in transformed cells containing mutant p53, but not in cell lines that are homozygously p53 deleted. We have now identified a new set of peptides (PNC-21, -27, and -28) from the amino-terminal mdm-2-binding domain of p53 that also induce cell death, but not apoptosis, in transformed cells. We synthesized these peptides under the assumption that they would bind to mdm-2. Because the active structure of this region of p53 bound to mdm-2 is an α-helix (4), we constructed this peptide such that it would contain a high α-helical content by placing the positively charged leader sequence on the carboxyl terminal end of the peptide so as to stabilize this conformation (9, 19), as indicated by the results shown in Fig. 1A. A similar strategy was used in designing peptides that would induce a strong T cell proliferative response (10) and was also recently used in the design of the carboxyl terminal p53 peptide (7), corresponding to residues 361–382 of p53, that has a well defined α-helical structure as the isolated peptide (20). The latter peptide was active in promoting p53-dependent apoptosis when introduced into cells attached to either a positively charged leader or a positively charged tat sequence on its carboxyl terminal end. Placement of the positively charged tat sequence on the amino-terminal end of the p53 sequence, where it would be expected to destabilize the α-helical structure, resulted in loss of activity of the peptide (R.L.F., unpublished results).
Activity of the p53 Peptides.
All three p53 peptides, PNC-21, -27, and -28 induce cell death that occurs in a dose-related manner. In contrast, two control peptides, one that contains the p53-12–26 sequence but no penetratin sequence and one p53-unrelated sequence with the penetratin sequence attached (X13-Leader), had no effect on any of the tumor cell lines studied. The latter result suggests that the penetratin leader sequence itself has no effect on the cells, a conclusion we have directly confirmed by incubating the penetratin sequence alone with several different human tumor cell lines.
The antiproliferative effects of these peptides on a wide variety of tumor cells cannot be attributed to nonspecific toxicity because none of the p53-leader peptides had any effect on normal pancreatic acinar (BMRPA1) cells either in growth phase or in stable contact-inhibited monolayers. Furthermore, none of these peptides had any significant effect on the abilities of human stem cells derived from cord blood to form colonies in response to specific growth factors that induce differentiation of these cells into different hematopoietic cell lines. These results all suggest that the p53 peptides promote cell death only in abnormally proliferating cells.
Possible Mechanisms for Induction of Cell Death in Malignant Cells.
Selective cytotoxicity of the p53 peptides for transformed cells raises the question as to the mechanism by which this effect occurs. In contrast to cells treated with carboxyl terminal p53 peptides (6–8), none of the malignant cell lines treated with the amino-terminal peptides expressed elevated levels of proteins like Bax and wafp21 and, as shown in Fig. 5, the DNA rapidly fragmented into low molecular weight forms, in contrast to results with cells that were treated with agents that induce apoptosis—i.e., γ-irradiation and treatment with actinomycin D. These results suggest that the p53 peptides induce cell death by using mechanisms that are independent of p53-induced apoptosis or that are independent of p53 protein activation.
The latter conclusion is supported by our findings that the peptides in this study, also in contrast to the carboxyl terminal peptides, all induce cell death in transformed cells, such as SW1417 colon cancer cells, which contain homozygous p53 gene deletions. This result does not definitively exclude activation by the p53 peptides of other proteins, such as p73, that have p53-like effects (21). However, any such activation process would have to involve mechanisms that do not depend on p53-induced apoptotic proteins. The fact that these peptides induce cell death in p53-defective transformed cells as effectively as they induce it in cells with viable p53 indicates that these peptides have a general antiproliferative effect on transformed cells.
One target of the p53 peptides may be mdm-2 itself. Although a major function of this protein is to attenuate the functioning of p53, there is evidence that this protein can induce neoplastic effects by p53-independent mechanisms (22). Possibly, the p53 peptides in this study block interaction of mdm-2 with other protein targets that interact with it in the same p53-binding domain, but not necessarily with the same residues as p53 itself. Compatible with this view is our finding that all three p53 peptides, PNC-21, -27, and -28, which contain overlapping sequences from the p53 mdm-2 binding domain, were about equally effective in inducing cell death. The one sequence that lies in common to all three active peptides is residues 17–20 (Glu-Thr-Phe-Ser). This segment would appear to be important for the selective cytotoxic effects of the whole peptide containing residues 12–26. Of these four residues, two, Glu-17 and Phe-19, are contact residues for the binding of p53 to mdm-2. Both of these residues appear to be important additionally in stabilizing the α-helical conformation. If only two of the contact residues are contained in this common sequence, this leaves open the possibility that the p53 peptides interact with another, undetermined cellular target.
Acknowledgments
This work was supported in part by National Institutes of Health RO1 Grant CA 42500, a Veteran's Affairs Merit Review Grant, and a grant from the Lustgarten Foundation for Pancreatic Cancer Research (to M.R.P. and J.M.); Environmental Protection Agency Grants R8825361 and R826685 and National Institutes of Health Grant R01 OH04192 (to P.W.B.-R.); and National Institutes of Health RO1 Grant CA 82528 and a Herbert Irving Scholar Award (to R.L.F.).
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Haffner R, Oren M. Curr Opin Genet Dev. 1995;5:84–90. doi: 10.1016/s0959-437x(95)90058-6. [DOI] [PubMed] [Google Scholar]
- 2.Haupt Y, Maya R, Kazaz A, Oren M. Nature (London) 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 3.Fuchs S Y, Adler V, Pincus M R, Ronai Z. Proc Natl Acad Sci USA. 1998;95:10541–10546. doi: 10.1073/pnas.95.18.10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kussie P H, Gorina S, Marechal V, Elenbass B, Moreau J, Levine A, Pavletich N P. Science. 1996;174:948–953. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- 5.Derossi D, Chassaing G, Prochiantz A. Trends Cell Biol. 1998;8:84–87. [PubMed] [Google Scholar]
- 6.Selivanova G, Iotsova V, Okan I, Fritsche M, Strom M, Groner B, Grafstrom R C, Wiman K G. Nat Med. 1997;3:632–638. doi: 10.1038/nm0697-632. [DOI] [PubMed] [Google Scholar]
- 7.Kim A L, Raffo A J, Pincus M R, Brandt-Rauf P W, Monaco R, Abarzua P, Fine R L. J Biol Chem. 1999;274:34924–34931. doi: 10.1074/jbc.274.49.34924. [DOI] [PubMed] [Google Scholar]
- 8.Wasylyk C, Salcvi R, Argentini M, Demeuil C, Delumeau I, Abecassis I, Debussche L, Wasylyk B. Oncogene. 1999;18:1921–1934. doi: 10.1038/sj.onc.1202528. [DOI] [PubMed] [Google Scholar]
- 9.Vasquez M, Pincus M R, Scheraga H A. Biopolymers. 1987;26:351–372. doi: 10.1002/bip.360260305. [DOI] [PubMed] [Google Scholar]
- 10.Vasquez M, Pincus M R, Scheraga H A. Biopolymers. 1987;26:373–393. doi: 10.1002/bip.360260306. [DOI] [PubMed] [Google Scholar]
- 11.Amar S, Glozman A, Chung D L, Alder V, Ronai Z, Friedman F K, Robinson R, Brandt-Rauf P W, Yamaizumi Z, Pincus M R. Cancer Chemother Pharmacol. 1997;41:79–85. doi: 10.1007/s002800050711. [DOI] [PubMed] [Google Scholar]
- 12.Bao L Y, Thelmo W L, Somnay S, Madahar C, Michl J. FASEB J. 1994;8:64A. [Google Scholar]
- 13.Kleeff J, Kornmann M, Sawhney H, Korc M. Int J Cancer. 2000;86:399–407. doi: 10.1002/(sici)1097-0215(20000501)86:3<399::aid-ijc15>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 14.Migliaccio G, Migliaccio A R, Adamson J W. Blood. 1988;72:248–256. [PubMed] [Google Scholar]
- 15.Rubinstein P, Dobrila L, Rosenfield R E, Adamson J W, Migliaccio G, Migliaccio A R, Taylor P E, Stevens C E. Proc Natl Acad Sci USA. 1995;92:10119–10122. doi: 10.1073/pnas.92.22.10119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karplus K, Barrett C, Hughey R. Bioinformatics. 1998;14:846–856. doi: 10.1093/bioinformatics/14.10.846. [DOI] [PubMed] [Google Scholar]
- 17.Pincus M R, Brandt-Rauf P W, Michl J, Friedman F K. Cancer Invest. 2000;18:39–50. doi: 10.3109/07357900009023061. [DOI] [PubMed] [Google Scholar]
- 18.Muller-Tiffman B F, Halazonetis T D, Elting J J. Proc Natl Acad Sci USA. 1998;95:6079–6084. doi: 10.1073/pnas.95.11.6079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zimmerman S S, Scheraga H A. Biopolymers. 1977;16:811–843. doi: 10.1002/bip.1977.360160408. [DOI] [PubMed] [Google Scholar]
- 20.Clore E M, Omichinski J G, Sakaguchi K, Zambrano N, Sakamoto H, Appella E, Gronenborn A M. Science. 1994;265:386–391. doi: 10.1126/science.8023159. [DOI] [PubMed] [Google Scholar]
- 21.Yang A, McKeon F. Nat Rev Mol Cell Biol. 2000;1:199–207. doi: 10.1038/35043127. [DOI] [PubMed] [Google Scholar]
- 22.Freedman D A, Wu L, Levine A J. Cell Mol Life Sci. 1999;55:96–107. doi: 10.1007/s000180050273. [DOI] [PMC free article] [PubMed] [Google Scholar]