Abstract
Background
Eukaryotic translation initiation factor 4E (eIF4E) is a key regulator of protein synthesis. Changes in eIF4E activity disproportionally affect the translation of a subset of oncogenic mRNAs in some cancers.
Materials and methods
We have assessed the expression levels of vascular endothelial growth factor C (VEGFC), eIF4E, eIF4E-binding proteins (4E-BPs) and phospho-4E-BP1 in clear cell renal carcinoma (ccRCC; n=101) using immunohistochemistry and analyzed the relevant mRNA levels and survival using online databases.
Results
The protein levels of VEGFC, an eIF4E-regulated gene, were upregulated in ccRCC tissues compared with adjacent normal renal tissues, indicating an enhanced eIF4E activity in ccRCC. The expression of eIF4E had no significant changes in ccRCC tissues. However, 4E-BP1 and phospho-4E-BP1 were found to be overexpressed in ccRCC tissues (P<0.05), and the high mRNA and protein levels of 4E-BP1 and phospho-4E-BP1 correlated with an unfavorable clinical outcome in ccRCC patients. Meanwhile, the mRNA expression of PIK3CD and PIK3CG were enhanced in ccRCC.
Conclusion
From these results, we could infer that the increase in eIF4E activity may be caused by the increased phospho-4E-BP1 level, which was probably due to the activation of phosphoinositide 3-kinase (PI3K) pathway.
Keywords: eIF4E, 4E-BP1, phospho-4E-BP1, VEGFC, PI3K, ccRCC
Introduction
Kidney cancer is one of the most common tumors in the urinary system. In the past 2 decades, the amount of kidney cancer has been increasing. Kidney cancer consists of multiple types, including transitional cell carcinoma of kidney, renal cell carcinoma (RCC), inverted papilloma and kidney lymphoma. Among the four types of kidney cancer, RCC accounts for the largest proportion. RCC contains multiple pathological categories, including chromophobe RCC, clear cell renal carcinoma (ccRCC) and papillary RCC. Among them, ccRCC accounts for the highest proportion witĥ75% of cases.1
The translations of mRNAs need to be tightly controlled in cells as the imbalance of translation leads to cancer occurrence and development.2–4 At the transcription elongation step, nuclear-transcribed mRNAs have 5′-cap added. Translation in eukaryotic cells begins with binding between 5′-cap and the eukaryotic translation initiation factor 4F (eIF4F).5,6 eIF4F complex includes eIF4A, eukaryotic translation initiation factor 4E (eIF4E) and eIF4G. eIF4E is able to recognize and bind to 5′-cap of mRNA, allowing the translation initiation.7,8 Changes in eIF4E activity only have a tiny influence on the mRNA translation of house-keeping genes such as glyceraldehyde-3-phosphate dehydrogenase (GAPDH).9 Most house-keeping mRNAs have short and low complexity 5′-untranslated regions (UTRs). Unlike them, some 5′-cap-dependent mRNAs own long 5′-UTRs with a high complexity, whose protein translation is tightly controlled by eIF4E including the mRNAs of cyclin D3, phosphoribosyl pyrophosphate synthetase 2 (PRPS2) and vascular endothelial growth factor (VEGF).3,10–13
Besides the amplification of eIF4E, eIF4E activity is also controlled by eIF4E-binding proteins (4E-BPs) such as 4E-BP1, 4E-BP2 and 4E-BP3.14 4E-BP1 is the most common isoform of 4E-BPs. 4E-BPs and eIF4G compete with eIF4E for the same binding site.15–17 The phosphoinositide 3-kinase (PI3K) pathway can phosphorylate 4E-BPs, causing the separation of 4E-BPs and eIF4E.6 Therefore, the phosphorylated 4E-BP1 is able to promote protein translation. PI3K can phosphorylate phosphatidylinositol 3,4 (PIP2) and activate the AKT.18,19 The activity of AKT can be inhibited by phosphatase and tensin homolog (PTEN).6,20,21 PI3Ks belong to three distinct categories called Class I, II and III.22 PI3K class I consists of multiple genes including PIK3CA, PIK3CB, PIK3CD and PIK3CG.
The expression level of eIF4E or 4E-BP1 is deregulated in many cancers such as bladder, ovary and prostate.23–33 In the present study, we focused on the expression and active levels of 4E-BP1 and eIF4E to confirm the prognostic values of these factors in ccRCC.
Materials and methods
Expression and clinical data obtained from Gene Expression Omnibus (GEO), The Cancer Genome Atlas (TCGA) and The Cancer Proteome Atlas (TCPA) databases
In the research, ccRCC datasets were extracted from the GEO database (http://www.ncbi.nlm.nih.gov/geo/). Four datasets with >400 specimens, GSE40435,34 GSE46699,35 GSE1564136 and GSE66272,37 were obtained. The probe ID was converted into a gene symbol first. When a gene was mapped to different probes, the genic expression value was calculated by the average expression value. Next, the data were translated into log2 logarithms, and the median normalization was performed using the robust multichip averaging method.38 We compared the normal control vs cancer datasets using the Student’s t-test. A P value of <0.05 was considered as statistically significant. The clinical data and mRNA expression data of eIF4E and 4E-BP1 in ccRCC were extracted from the TCGA database.39,40 The survival analysis of eIF4E and 4E-BP1 protein expression was directly obtained from the TCPA database (http://www.tcpaportal.org/tcpa/index.html).41–44 According to the median value of gene or protein expression, samples were divided into low- and high-expression groups.
Patients and specimens
The research consisted of 202 samples from 101 patients with ccRCC. All the patients had a renal resection at the Fujian Provincial Hospital between January 2016 and June 2017. The standard requirements for patients included in the study were 1) histologically confirmed ccRCC; 2) no history of other malignancy, and 3) no prior neoadjuvant chemotherapy. The study was performed with the approval of the ethics committee of Fujian Provincial Hospital. Written informed consent was given by the patients for their information, and specimens were stored in the hospital database and used for research.
Immunohistochemical staining
Paraffin blocks that contained sufficient formalin-fixed tumor specimens were serial sectioned at 3 μm and mounted on silane-coated slides for immunohistochemical staining analysis. Dimethylbenzene rehydrated through 100% ethanol, 100% ethanol, 95% ethanol, 85% ethanol and 75% ethanol were applied to deparaffinize. In all, 0.01 mol/L of sodium citrate buffer (pH 6.0) was used to the progress of antigen retrieval treatment (autoclaved at 121°C, 2 min). Then, 3% H2O2 was applied to block endogenous peroxidase at room temperature for 10 min. The sections were washed in PBS solution subsequently and blocked with 10% goat serum (ZhongShan Biotechnology, Beijing, China) for 30 min and incubated with anti-eIF4E (ab33766, 1:100 dilution, monoclonal; Abcam, Cambridge, MA, USA) or anti-eIF4E-BP1 (ab32024, 1:100 dilution, monoclonal; Abcam) antibody or anti-vascular endothelial growth factor C (VEGFC; ab83905, 1:100 dilution, polyclonal; Abcam) or anti-phospho-4E-BP1 (Thr37/46) (236B4) (2855, 1:200 dilution, monoclonal; Cell Signaling Technology, Boston, MA, USA) at 4°C for 12 h. The sections were washed in PBS solution three times and incubated with HRP-conjugated secondary antibody for 30 min at room temperature. All slides were counterstained with diaminobenzidine (DAB) solution and 20% hematoxylin and dehydrated. The primary antibody diluent was regarded as negative control.
Evaluation of immunostaining intensity
Immunohistochemical staining tissue sections were reviewed and scored by two independent pathologists. The score was calculated according to the proportion of stained tumor cells and intensity of cellular staining. The intensity of cellular staining was scored between 0 and 3: 0, no staining; 1, weak staining; 2, moderate staining; and 3, strong staining. The proportion of stained tumor cells was scored between 1 and 4: 1, 0–25%; 2, 26–50%; 3, 51–75% and 4, 75–100%. The multiplication of these two variables was calculated as the final score. The staining was divided into five grades according to the final score as follows: 0 score, 0; 1 score, 1–2; 2 score, 3–4; 3 score, 6–8 and 4 score, 9–12.
Statistical analyses
In the research, the student’s t-test was used to calculate the mRNA expression level in ccRCC tissues and adjacent normal renal tissues by using GraphPad Prism 5 (GraphPad Software, Inc., La Jolla, CA, USA). Log-rank test was used to calculate the survival analysis by using IBM SPSS version 19.0 (IBM Corporation, Armonk, NY, USA). Multivariate survival analysis was performed by using stepwise Cox proportional hazards regression model. P value of <0.05 was considered as statistically significant.
Results
Expression levels of eIF4E were not changed in ccRCC
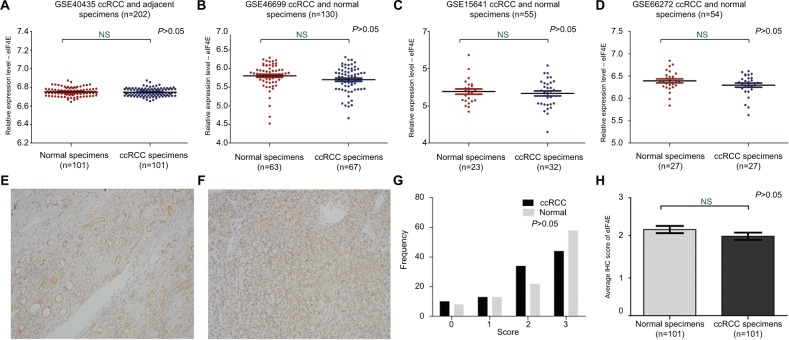
In order to explore the expression of eIF4E in ccRCC patients, a total of four related publications were used. Based on these datasets (GSE40435, GSE46699, GSE15641, GSE66272; Table S1), no change in eIF4E expression was observed in the tissues of ccRCC compared with adjacent normal renal tissues (Figure 1A–D). These results suggested that the mRNA of eIF4E expression did not change in ccRCC. Moreover, the expression of eIF4E protein in 101 tissues of ccRCC and their adjacent normal renal tissues was detected by immunohistochemical staining. Representative immunohistochemical-stained tissue sections and the frequency distributions of the immunohistochemical scores are presented (Figure 1E–G). The mean scores of eIF4E proteins in ccRCC and adjacent normal renal tissues were 2.11 and 2.29, respectively (Figure 1H), consistent with the conclusion from mRNA analysis that the level of eIF4E did not change in ccRCC.
Figure 1.
The mRNA and protein level of eIF4E in ccRCC.
Notes: The mRNA expression level of eIF4E in ccRCC was measured. Four mRNA datasets were used including GSE40435 (A), GSE46699 (B), GSE15641 (C) and GSE66272 (D). The protein expression level of eIF4E in 101 ccRCCs was measured using immunohistochemical staining. Representative adjacent normal renal tissues’ staining (E), ccRCC tissues’ staining (F), frequency distributions of protein expression across the cohort (G) and the average score of immunohistochemical staining for eIF4E (H) are shown. (E and F) Magnification ×100.
Abbreviations: ccRCC, clear cell renal carcinoma; eIF4E, eukaryotic translation initiation factor 4E; IHC, immunohistochemistry; NS, not significant.
Protein expression level of VEGFC was upregulated in ccRCC
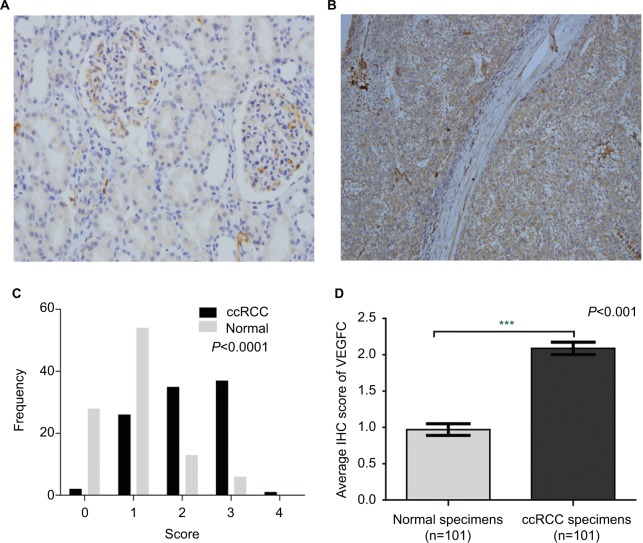
VEGFC, an important downstream target of eIF4E, is regulated by eIF4E activity. In order to further investigate the activity of eIF4E in ccRCC, we set out to evaluate the expression of VEGFC protein in ccRCC. The expression of VEGFC protein in 101 tissues of ccRCC and their adjacent normal renal tissues was detected by immunohistochemical staining. Representative immunohistochemical-stained tissue sections and the frequency distributions of the immunohistochemical scores are presented (Figure 2A–C). The mean scores of VEGFC proteins in ccRCC and adjacent normal renal tissues were 2.09 and 0.97, respectively(Figure 2D), suggesting that although eIF4E expression levels were unchanged, the activity of eIF4E in ccRCC tissues was enhanced.
Figure 2.
The protein expression of VEGFC in ccRCC.
Notes: The protein expression level of VEGFC in ccRCC was measured using immunohistochemical staining. Representative adjacent normal renal tissues’ staining (A), ccRCC tissues’ staining (B), frequency distributions of protein expression across the cohort (C) and the average score of immunohistochemical staining (D) are shown. (A and B) Magnification ×100. ***P<0.001.
Abbreviations: VEGFC, vascular endothelial growth factor C; ccRCC; clear cell renal carcinoma; IHC, immunohistochemistry.
Expression levels of phospho-4E-BP1 and 4E-BP1 were enhanced in ccRCC
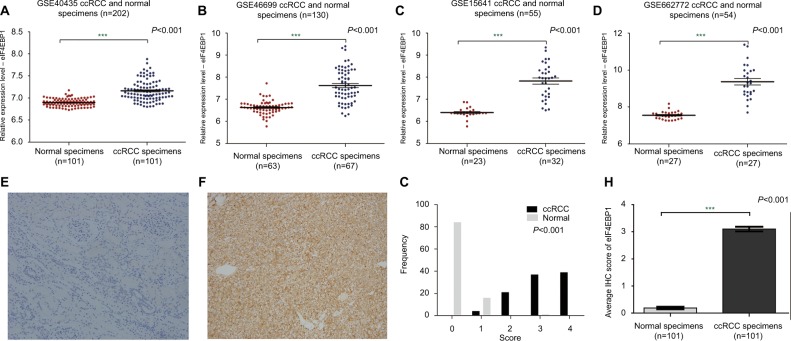
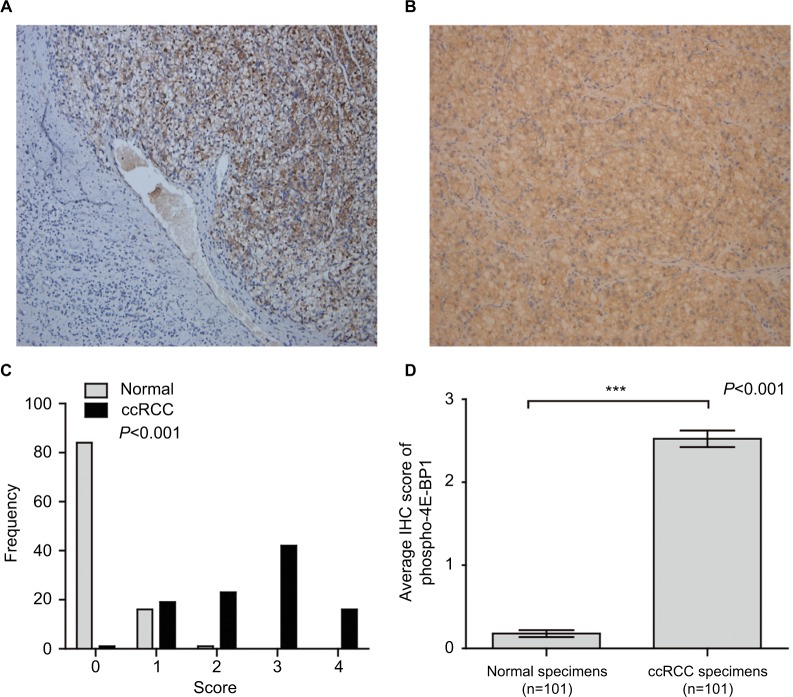
In order to investigate how the activity of eIF4E was enhanced in ccRCC, we next investigated the expression and activity of 4E-BP1. To our surprise, the mRNA and protein expression of 4E-BP1 were overexpressed in ccRCC tissues based on four datasets (GSE40435, GSE46699, GSE15641, GSE66272; Figure 3A–D and Table S1) and immunohistochemical staining of 4E-BP1 in 101 ccRCC tissues and their adjacent normal renal tissues. Representative immunohistochemical-stained tissue sections of 4E-BP1 and the frequency distributions of these scores are presented (Figure 3E–G). The mean scores of 4E-BP1 proteins in ccRCC and adjacent normal renal tissues were 3.10 and 0.19, respectively (Figure 3H). Then, we detected the protein expression of phospho-4E-BP1 in the 101 sample cohort. Representative immunohistochemical-stained tissue sections of phospho-4E-BP1 and the frequency distributions of these scores are presented (Figure 4A–C). The mean scores of phospho-4E-BP1 in ccRCC and adjacent normal renal tissues were 2.52 and 0.18, respectively (Figure 4D), suggesting that both 4E-BP1 and phospho-4E-BP1 levels were much higher in ccRCC tissues.
Figure 3.
The mRNA and protein expression of 4E-BP1 in ccRCC.
Notes: The mRNA expression level of 4E-BP1 in ccRCC was measured. Four mRNA datasets were used including GSE40435 (A), GSE46699 (B), GSE15641 (C) and GSE66272 (D). The protein expression level of 4E-BP1 in ccRCC was measured using immunohistochemical staining. Representative adjacent normal renal tissues’ staining (E), ccRCC tissues’ staining (F), frequency distributions of protein expression across the cohort (G) and the average score of immunohistochemical staining (H) are shown. (E and F) Magnification ×100. ***P<0.001.
Abbreviations: ccRCC; clear cell renal carcinoma; eIF4E, eukaryotic translation initiation factor 4E; BP, binding protein.
Figure 4.
The protein expression of phospho-4E-BP1 in ccRCC.
Notes: The protein expression level of phospho-4E-BP1 in ccRCC was measured using immunohistochemical staining. Representative adjacent normal renal tissues’ staining (A), ccRCC tissues’ staining (B), frequency distributions of protein expression across the cohort (C) and the average score of immunohistochemical staining (D) are shown. (A and B) Magnification ×100. ***P<0.001.
Abbreviations: BP, binding protein; ccRCC; clear cell renal carcinoma; IHC, immunohistochemistry.
Expression levels of PIK3CD and PIK3CG were upregulated in ccRCC
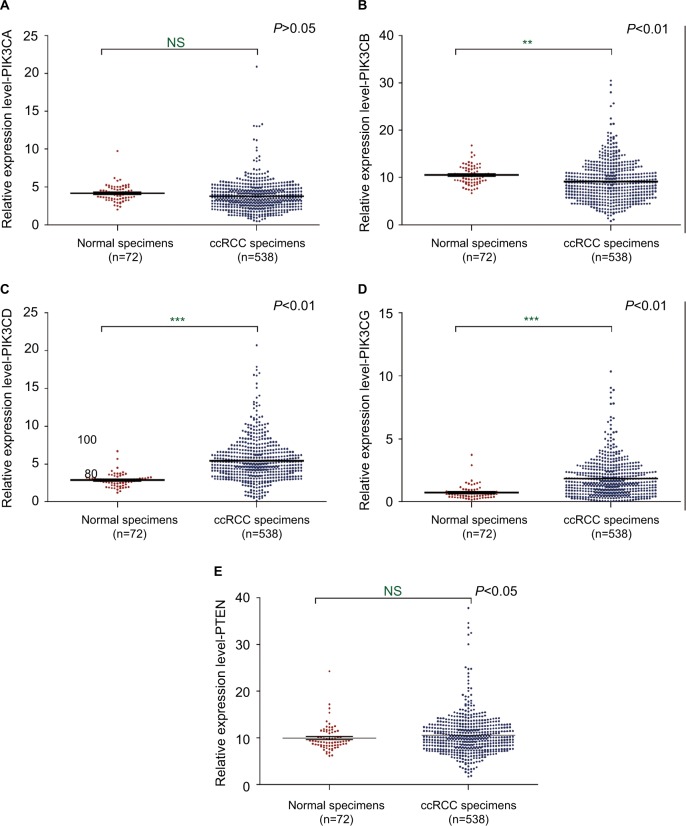
4EBPs could be phosphorylated due to the activation of PI3K pathway. Phosphorylated 4EBPs enable the assembly of eIF4F by dislodging 4EBPs from eIF4E. In order to further investigate the mechanisms underlying eIF4E activation in ccRCC, the expression levels of PIK3CA, PIK3CB, PIK3CD, PIK3CG and PTEN were examined (Table S2). The mRNAs of PIK3CD and PIK3CG were upregulated in ccRCC tissues based on the TCGA database (Figure 5).
Figure 5.
The mRNA expression of PIK3s and PTEN in ccRCC.
Notes: The mRNA expression levels of PIK3CA (A), PIK3CB (B), PIK3CD (C), PIK3CG (D) and PTEN (E) in ccRCC were analyzed using the TCGA database. **P<0.01; ***P<0.001.
Abbreviations: PTEN, phosphatase and tensin homolog; ccRCC, clear cell renal carcinoma; TCGA, The Cancer Genome Atlas; NS, not significant.
Expression levels of 4E-BP1 and phospho-4E-BP1 were strongly associated with ccRCC survival
In the TCGA ccRCC cohort, we observed that patients with advanced stage and grade were at significantly increased risk of death. Patients with age >60 years, laterality=left, pharmaceutical therapy and radiation therapy also have a high risk of death in ccRCC (Table 1).
Table 1.
Univariate analysis of the correlation between clinicopathological parameters and survival of ccRCC patients in the TCGA cohort
| Variables | Patients (n) | MST (days) | Log-rank test | P-value |
|---|---|---|---|---|
| Age (years) | ||||
| ≤60 | 261 | NA | ||
| >60 | 261 | 2415 | 13.256 | 0.000* |
| Gender | ||||
| Female | 182 | 2564 | ||
| Male | 340 | 2830 | 0.04 | 0.841 |
| Location | ||||
| Left | 246 | 2343 | ||
| Right | 275 | 3695 | 4.657 | 0.031* |
| Race | ||||
| White | 463 | 2752 | ||
| Other | 59 | 1913 | 0.284 | 0.594 |
| Tumor stage | ||||
| I/II | 314 | NA | ||
| III/IV | 205 | 1133 | 76.153 | 0.000* |
| Histologic grade | ||||
| G1/G2 | 242 | NA | ||
| G3/G4 | 280 | 1913 | 34.145 | 0.000* |
| Pharmaceutical therapy | ||||
| Yes | 73 | 1378 | ||
| No | 85 | 2830 | 11.489 | 0.001* |
| Radiation therapy | ||||
| Yes | 45 | 927 | ||
| No | 116 | 2764 | 17.13 | 0.000* |
| Surgery locoregional procedure | ||||
| Yes | 16 | 1724 | ||
| No | 146 | 1964 | 0.103 | 0.748 |
| Surgery metastatic procedure | ||||
| Yes | 31 | 1567 | ||
| No | 132 | 1964 | 0.996 | 0.318 |
Note:
P<0.05, statistical significance.
Abbreviations: ccRCC, clear cell renal carcinoma; TCGA, The Cancer Genome Atlas; MST, median survival time, NA, not available.
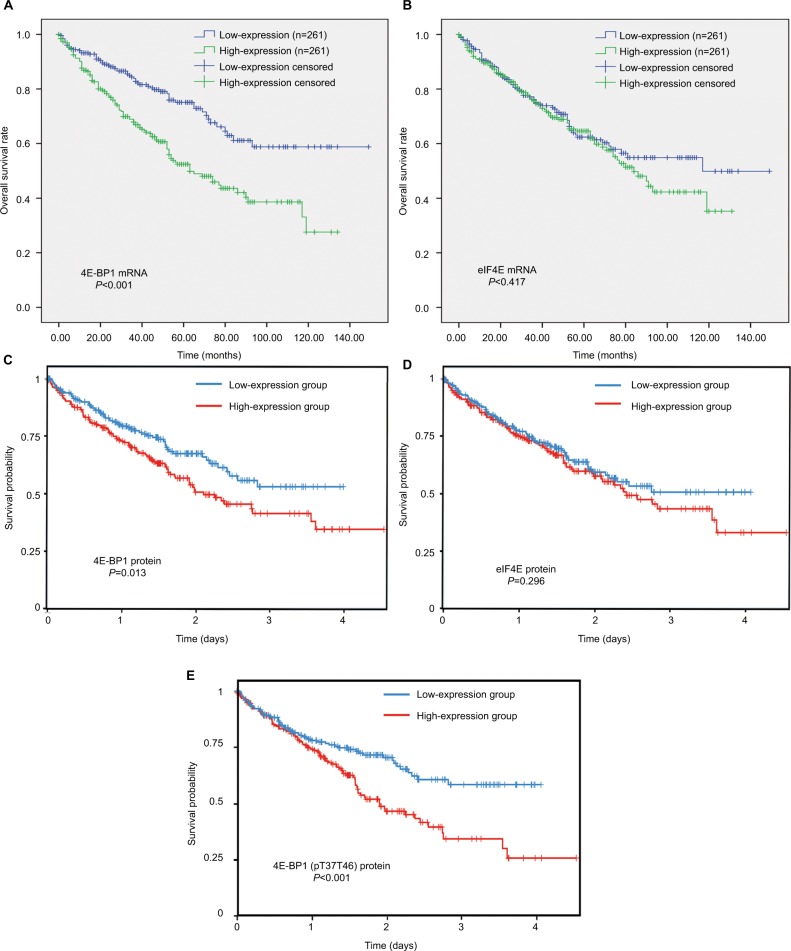
The survival analyses of 4E-BP1and eIF4E mRNA are shown in Figure 6A and B. We found that the mRNA expression of 4E-BP1 (Figure 6A), but not eIF4E (Figure 6B), was associated with the clinical outcomes of ccRCC patients (Table S3), after adjusting for tumor location, grade, stage and patients’ age, gender and race (Table 2). The survival analyses of 4E-BP1, eIF4E and phospho-4E-BP1 protein were obtained from the TCPA database. With the cutoff value set at the median, we found that the protein expression of 4E-BP1 (Figure 6C) and phospho-4E-BP1 (Figure 6E), but not eIF4E (Figure 6D), was also associated with the clinical outcomes of 445 ccRCC patients.
Figure 6.
The prognostic value of 4E-BP1, eIF4E and phospho-4E-BP1 in ccRCC.
Notes: The Kaplan–Meier survival analyses of 4E-BP1 (A) and eIF4E (B) mRNA expression of the overall survival time of ccRCC patients using the OncoLnc database. The Kaplan–Meier survival analyses of 4E-BP1 (C), eIF4E (D) and phospho-4E-BP1 (E) protein expression of the overall survival time of ccRCC patients using the TCPA database.
Abbreviations: BP, binding protein; eIF4E, eukaryotic translation initiation factor 4E; ccRCC; clear cell renal carcinoma; TCPA, The Cancer Proteome Atlas.
Table 2.
Multivariate analysis of the correlation between clinicopathological parameters and survival of ccRCC patients in the TCGA cohort
| Covariates | Coefficient | Standard error | HR | 95% CI for HR | P-value |
|---|---|---|---|---|---|
| Gender (female vs male) | 0.067 | 0.161 | 1.070 | 0.780–1.467 | 0.676 |
| Age (≤60 vs >60 years) | 0.425 | 0.159 | 0.654 | 0.479–0.893 | 0.008* |
| Race (white vs other) | 0.100 | 0.301 | 1.105 | 0.613–1.993 | 0.739 |
| Location (left vs right) | 0.372 | 0.152 | 1.450 | 1.073–1.959 | 0.015* |
| Tumor stage (I/II vs III/IV) | 1.118 | 0.301 | 0.532 | 0.372–0.762 | 0.001* |
| Histologic grade (G1/G2 vs G3/G4) | 0.631 | 0.183 | 1.105 | 0.613–1.993 | 0.000* |
| eIF4E expression (low vs high) | 0.212 | 0.154 | 0.809 | 0.598–1.093 | 0.168 |
| 4E-BP1 expression (low vs high) | 0.463 | 0.167 | 1.589 | 1.146–2.204 | 0.006* |
Note:
P<0.05, statistical significance.
Abbreviations: ccRCC, clear cell renal carcinoma; HR, hazard ratio; TCGA, The Cancer Genome Atlas; eIF4E, eukaryotic translation initiation factor 4E; BP, binding protein.
Discussion
In this study, the expression and prognostic relevance of eIF4E, 4E-BP1 and phospho-4E-BP1 in ccRCC were examined. Our analysis included four datasets with >400 specimens from the GEO database, one web tool for interactively exploring survival correlations performed by the TCPA database and one ccRCC cohort (n=101) with immunohistochemistry (IHC) analyses.
eIF4E is highly elevated and deregulated in many cancers such as lung and breast.45,46 Surprisingly, no association was found between eIF4E expression and survival (Figure 6B and D), whereas the protein level of VEGFC (Figure 2) was upregulated in ccRCC tissues compared with adjacent normal renal tissues. The protein translation of VEGFC mRNA was regulated by eIF4E. Our results indicated that eIF4E activity is upregulated in ccRCC.
In a simple model to calculate the regulatory effect toward eIF4E activity, one phospho-4E-BP1 protein could eliminate the inhibitory effect of two 4E-BP1 proteins on eIF4E activity,47,48 suggesting that phospho-4E-BP1 plays a greater role than unphosphorylated 4E-BP1 in the regulation of eIF4E activity. In our study, high mRNA and protein expression level of 4E-BP1 were observed in ccRCC tissues compared with adjacent normal renal tissues (3.10 vs 0.19, tumor vs normal; Figure 3). At the same time, a strong phosphorylation status of the 4E-BP1 in ccRCC was also observed (2.52 vs 0.18, tumor vs normal; Figure 4). Therefore, the increased eIF4E activity observed in our study may be due to an enhanced proportion of phospho-4E-BP1 in ccRCC compared with adjacent normal renal tissues (Figure 7). As we know, eIF4E activity can be controlled by the phospho-4E-BP1 and the expression levels of eIF4E, and the increased eIF4E activity drives cancer progression. Therefore, phosphorylated 4E-BP1 and eIF4E overexpression synchronously drive disease progression in ccRCC.49
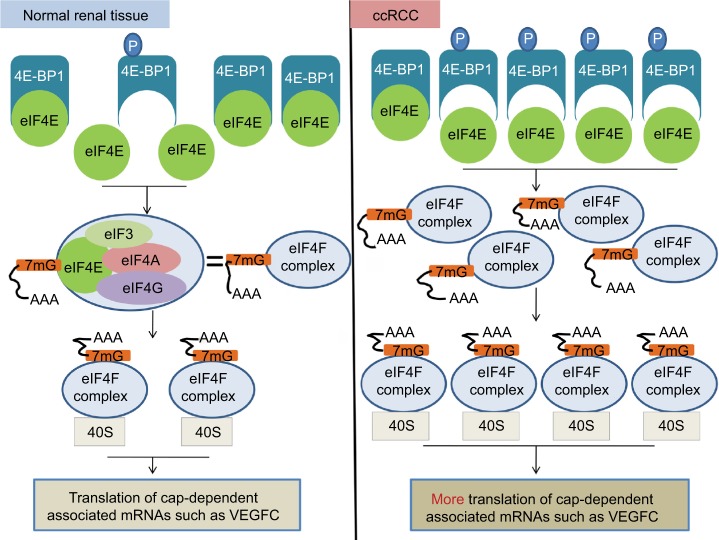
Figure 7.
A diagram illustrating the potential molecular mechanisms underlying the regulation of eIF4E activity by 4E-BP1 and phospho-4E-BP1 in ccRCC.
Notes: In normal renal tissue, most eIF4E activity is inhibited due to the combination of eIF4E and 4E-BP1. This ensures that the translations of cap-dependent associated mRNA are tightly controlled. In ccRCC, the mRNA level of eIF4E does not change. However, the PI3K pathway was activated and 4E-BP1 was phosphorylated, leading to the release of 4E-BP1 from eIF4E and the increased activity of eIF4E. The increased activity of eIF4E actively promoted cap-dependent translation. eIF4F complex, eIF4E+eIF3+eIF4A+eIF4G.
Abbreviations: eIF4E, eukaryotic translation initiation factor 4E; BP, binding protein; ccRCC; clear cell renal carcinoma; PI3K, phosphoinositide 3-kinase; VEGFC, vascular endothelial growth factor C.
The simultaneous increase in 4E-BP1 protein expression and phosphorylation may be due to the malfunction of the feedback loop between 4E-BP1 and eIF4E. When eIF4E is overactivated, the enhanced 4E-BP1 is supposed to suppress its activity. However, due to the upregulation of the PI3K pathway, i.e., overexpression of PIK3CD and PIK3CG mRNA (Figure 5), most of newly synthesized 4E-BP1 proteins were phosphorylated and lost the ability to suppress eIF4E activity. Thus, the protein expression of 4E-BP1 cannot be restored and remained high in ccRCC; albeit most of the 4E-BP1 protein were phosphorylated and inactive.
Collectively, our data displayed enhanced activity of eIF4E in ccRCC, which is probably due to the increased ratio of phospho-4E-BP1 against 4E-BP1 (Figure 7). In addition, there was a significantly unfavorable influence of 4E-BP1 and phospho-4E-BP1 expression on the survival (Figure 6). These results suggested that 4E-BP1, eIF4E and phospho-4E-BP1 are important determinants of diagnosis and disease progression in ccRCC.
Acknowledgments
This work was funded by the Youth Scientific Research Project of Fujian Provincial Heath Department, China (2015-1-2); the International S&T Cooperation Program of China (2016YFE0121900) and the Scientific Research Innovation Team Construction Program of Fujian Normal University (IRTL1702).
Footnotes
Author contributions
Feng Li and Qingshui Wang performed the experiments, wrote the paper and prepared figures and/or tables.
Xiaoxue Xiong and Chenyi Wang performed the experiments and analyzed the data.
Xiaohua Liu, Ziqiang Liao, and Bifeng Xie contributed to reagents/analysis tools.
Yao Lin conceived and designed the experiments, wrote the paper and prepared figures and/or tables.
All authors contributed toward data analysis, drafting and revising the paper and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Nickerson ML, Jaeger E, Shi Y, et al. Improved identification of von Hippel-Lindau gene alterations in clear cell renal tumors. Clin Cancer Res. 2008;14(15):4726–4734. doi: 10.1158/1078-0432.CCR-07-4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ruggero D. Translational control in cancer etiology. Cold Spring Harb Perspect Biol. 2013;5(2):152–158. doi: 10.1101/cshperspect.a012336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Silvera D, Formenti SC, Schneider RJ. Translational control in cancer. Nat Rev Cancer. 2010;10(4):254–266. doi: 10.1038/nrc2824. [DOI] [PubMed] [Google Scholar]
- 4.Feoktistova K, Tuvshintogs E, Do A, Fraser CS. Human eIF4E promotes mRNA restructuring by stimulating eIF4A helicase activity. Proc Natl Acad Sci U S A. 2013;110(33):13339. doi: 10.1073/pnas.1303781110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Topisirovic I, Svitkin YV, Sonenberg N, Shatkin AJ. Cap and cap-binding proteins in the control of gene expression. Wiley Interdiscip Rev RNA. 2011;2(2):277–298. doi: 10.1002/wrna.52. [DOI] [PubMed] [Google Scholar]
- 6.Siddiqui N, Sonenberg N. Signalling to eIF4E in cancer. Biochem Soc Trans. 2015;43(5):763–772. doi: 10.1042/BST20150126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sonenberg N. eIF4E, the mRNA cap-binding protein: from basic discovery to translational research. Biochem Cell Biol. 2008;86(2):178. doi: 10.1139/O08-034. [DOI] [PubMed] [Google Scholar]
- 8.And NKG, Wickens M. Control of translation initiation in animals. Annu Rev Cell Dev Biol. 1998;14:399. doi: 10.1146/annurev.cellbio.14.1.399. [DOI] [PubMed] [Google Scholar]
- 9.Cunningham JT, Moreno MV, Lodi A, Ronen SM, Ruggero D. Protein and nucleotide biosynthesis are coupled by a single rate-limiting enzyme, PRPS2, to drive cancer. Cell. 2014;157(5):1088. doi: 10.1016/j.cell.2014.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rosenwald IB, Kaspar R, Rousseau D, et al. Eukaryotic translation initiation factor 4E regulates expression of cyclin D1 at transcriptional and post-transcriptional levels. J Biol Chem. 1995;270(36):21176–21180. doi: 10.1074/jbc.270.36.21176. [DOI] [PubMed] [Google Scholar]
- 11.Kevil CG, De Benedetti A, Payne DK, Coe LL, Laroux FS, Alexander JS. Translational regulation of vascular permeability factor by eukaryotic initiation factor 4E: implications for tumor angiogenesis. Int J Cancer. 1996;65(6):785–790. doi: 10.1002/(SICI)1097-0215(19960315)65:6<785::AID-IJC14>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 12.Benedetti AD, Graff JR. eIF-4E expression and its role in malignancies and metastases. Oncogene. 2004;23(18):3189. doi: 10.1038/sj.onc.1207545. [DOI] [PubMed] [Google Scholar]
- 13.Bhat M, Robichaud N, Hulea L, Sonenberg N, Pelletier J, Topisirovic I. Targeting the translation machinery in cancer. Nat Rev Drug Discov. 2015;14:261–278. doi: 10.1038/nrd4505. [DOI] [PubMed] [Google Scholar]
- 14.Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem. 1999;68:913. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- 15.Mader S, Lee H, Pause A, Sonenberg N. The translation initiation factor eIF-4E binds to a common motif shared by the translation factor eIF-4 gamma and the translational repressors 4E-binding proteins. Mol Cell Biol. 1995;15(9):4990–4997. doi: 10.1128/mcb.15.9.4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pause A, Belsham GJ, Gingras AC, et al. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5′-cap function. Nature. 1994;371(6500):762. doi: 10.1038/371762a0. [DOI] [PubMed] [Google Scholar]
- 17.Poulin F, Gingras AC, Olsen H, Chevalier S, Sonenberg N. 4E-BP3, a new member of the eukaryotic initiation factor 4E-binding protein family. J Biol Chem. 1998;273(22):14002–14007. doi: 10.1074/jbc.273.22.14002. [DOI] [PubMed] [Google Scholar]
- 18.Hay N, Sonenberg N. Upstream and downstream of mTOR. Genes Dev. 2004;18(16):1926. doi: 10.1101/gad.1212704. [DOI] [PubMed] [Google Scholar]
- 19.Cantley LC. The phosphoinositide 3-kinase pathway. Science. 2002;296:1655–1657. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- 20.Yuan TL, Cantley LC. PI3K pathway alterations in cancer: variations on a theme. Oncogene. 2008;27(41):5497. doi: 10.1038/onc.2008.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pearce LR, Huang X, Boudeau J, et al. Identification of Protor as a novel Rictor-binding component of mTOR complex-2. Biochem J. 2007;405(3):513–522. doi: 10.1042/BJ20070540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Faes S, Dormond O. PI3K and AKT: unfaithful partners in cancer. Int J Mol Sci. 2015;16(9):21138–21152. doi: 10.3390/ijms160921138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nathan CO, Carter P, Liu L, et al. Elevated expression of eIF4E and FGF-2 isoforms during vascularization of breast carcinomas. Oncogene. 1997;15(9):1087–1094. doi: 10.1038/sj.onc.1201272. [DOI] [PubMed] [Google Scholar]
- 24.Nathan CO, Liu L, Li BD, Abreo FW, Nandy I, De Benedetti A. Detection of the proto-oncogene eIF4E in surgical margins may predict recurrence in head and neck cancer. Oncogene. 1997;15(5):579. doi: 10.1038/sj.onc.1201216. [DOI] [PubMed] [Google Scholar]
- 25.De BA, Harris AL. eIF4E expression in tumors: its possible role in progression of malignancies. Int J Biochem Cell Biol. 1999;31(1):59–72. doi: 10.1016/s1357-2725(98)00132-0. [DOI] [PubMed] [Google Scholar]
- 26.Crew JP, Fuggle S, Bicknell R, Cranston DW, de Benedetti A, Harris AL. Eukaryotic initiation factor-4E in superficial and muscle invasive bladder cancer and its correlation with vascular endothelial growth factor expression and tumour progression. Br J Cancer. 2000;82(1):161–166. doi: 10.1054/bjoc.1999.0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berkel HJ, Turbat-Herrera EA, Shi R, de Benedetti A. Expression of the translation initiation factor eIF4E in the polyp-cancer sequence in the colon. Cancer Epidemiol Biomarkers Prev. 2001;10(6):663–666. [PubMed] [Google Scholar]
- 28.Li BD, Gruner JS, Abreo F, et al. Prospective study of eukaryotic initiation factor 4E protein elevation and breast cancer outcome. Ann Surg. 2002;235(5):738–739. doi: 10.1097/00000658-200205000-00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seki N, Takasu T, Mandai K, et al. Expression of eukaryotic initiation factor 4E in atypical adenomatous hyperplasia and adenocarcinoma of the human peripheral lung. Clin Cancer Res. 2002;8(10):3046–3053. [PubMed] [Google Scholar]
- 30.Lee JW, Choi JJ, Lee KM, et al. eIF-4E expression is associated with histopathologic grades in cervical neoplasia. Hum Pathol. 2005;36(11):1197–1203. doi: 10.1016/j.humpath.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 31.Matthews-Greer J, Caldito G, de Benedetti A, et al. eIF4E as a marker for cervical neoplasia. Appl Immunohistochem Mol Morphol. 2005;13(4):367–370. doi: 10.1097/01.pai.0000170625.98446.3e. [DOI] [PubMed] [Google Scholar]
- 32.Culjkovic B, Borden KL. Understanding and targeting the eukaryotic translation initiation factor eIF4E in head and neck cancer. J Oncol. 2009;2009:981679. doi: 10.1155/2009/981679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pettersson F, Yau C, Dobocan MC, et al. Ribavirin treatment effects on breast cancers overexpressing eIF4E, a biomarker with prognostic specificity for luminal B-type breast cancer. Clin Cancer Res. 2011;17(9):2874. doi: 10.1158/1078-0432.CCR-10-2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wozniak MB, Le Calvez-Kelm F, Abedi-Ardekani B, et al. Integrative genome-wide gene expression profiling of clear cell renal cell carcinoma in Czech Republic and in the United States. PLoS One. 2013;8(3):e57886. doi: 10.1371/journal.pone.0057886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eckel-Passow JE, Serie DJ, Bot BM, et al. Somatic expression of ENRAGE is associated with obesity status among patients with clear cell renal cell carcinoma. Carcinogenesis. 2014;35(4):822. doi: 10.1093/carcin/bgt485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jones J, Otu H, Spentzos D, et al. Gene signatures of progression and metastasis in renal cell cancer. Clin Cancer Res. 2005;11:5730–5739. doi: 10.1158/1078-0432.CCR-04-2225. [DOI] [PubMed] [Google Scholar]
- 37.Wotschofsky Z, Gummlich L, Liep J, et al. Integrated microRNA and mRNA signature associated with the transition from the locally confined to the metastasized clear cell renal cell carcinoma exemplified by miR-146-5p. PLoS One. 2016;11(2):e0148746. doi: 10.1371/journal.pone.0148746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Irizarry RA, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 39.Cancer Genome Atlas Research Network. Weinstein JN, Collisson EA, et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet. 2013;45(10):1113. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anaya J. OncoLnc: linking TCGA survival data to mRNAs, miRNAs, and lncRNAs. Peerj Comput Sci. 2016;2:e67. [Google Scholar]
- 41.Uhlen M, Oksvold P, Fagerberg L, et al. Towards a knowledge-based Human Protein Atlas. Nat Biotechnol. 2010;28(12):1248–1250. doi: 10.1038/nbt1210-1248. [DOI] [PubMed] [Google Scholar]
- 42.Uhlen M, Zhang C, Lee S, et al. A pathology atlas of the human cancer transcriptome. Science. 2017;357(6352):eaan2507. doi: 10.1126/science.aan2507. [DOI] [PubMed] [Google Scholar]
- 43.Li J, Lu Y, Akbani R, et al. TCPA: a resource for cancer functional proteomics data. Nat Methods. 2013;10(11):1046. doi: 10.1038/nmeth.2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yoshihara K, Shahmoradgoli M, Martínez E, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang R, Geng J, Wang JH, Chu XY, Geng HC, Chen LB. Overexpression of eukaryotic initiation factor 4E (eIF4E) and its clinical significance in lung adenocarcinoma. Lung Cancer. 2009;66(2):237–244. doi: 10.1016/j.lungcan.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 46.Heikkinen T, Korpela T, Fagerholm R, et al. Eukaryotic translation initiation factor 4E (eIF4E) expression is associated with breast cancer tumor phenotype and predicts survival after anthracycline chemotherapy treatment. Breast Cancer Res Treat. 2013;141(1):79–88. doi: 10.1007/s10549-013-2671-2. [DOI] [PubMed] [Google Scholar]
- 47.Millican-Slater RA, Sayers CD, Hanby AM, Hughes TA. Expression of phosphorylated eIF4E-binding protein 1, but not of eIF4E itself, predicts survival in male breast cancer. Br J Cancer. 2016;115(3):339–345. doi: 10.1038/bjc.2016.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Coleman LJ, Peter MB, Teall TJ, et al. Combined analysis of eIF4E and 4E-binding protein expression predicts breast cancer survival and estimates eIF4E activity. Br J Cancer. 2009;100(9):1393. doi: 10.1038/sj.bjc.6605044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Campbell L, Jasani B, Griffiths DF, Gumbleton M. Phospho-4e-BP1 and eIF4E overexpression synergistically drives disease progression in clinically confined clear cell renal cell carcinoma. Am J Cancer Res. 2015;5(9):2838. [PMC free article] [PubMed] [Google Scholar]