Abstract
Purpose
Colorectal carcinoma (CRC) is among the most common causes of death. Recent studies have shown that both STAT3 and miR-572 contribute to CRC progression. STAT3 plays an important role in miRNA expression. Moreover, MOAP-1, which is a pro-apoptotic protein that induces cell death or apoptosis, has a direct correlation with miRNA. Therefore, the current study is designed to explore whether miR-572 and STAT3 are involved in a common pathway and the role of MOAP-1 in this process.
Patients and methods
The expressions of STAT3, miR-572, and MOAP-1 in human CRC tissues and multiple cell lines were estimated by qRT-PCR or Western blot. MTT, transwell migration, and invasion assays were used to assess cell growth, migration, and invasion, respectively. Dual-luciferase reporter assay was applied to examine the association between miR-572 and MOAP-1.
Results
Elevated STAT3 levels were accompanied by increased miR-572 and decreased MOAP-1 levels in primary CRC specimens and cell lines. STAT3 promoted CRC cell growth, migration, and invasion via the upregulated expression of miR-572. Subsequently, miR-572 inhibited MOAP-1 protein expression through an interaction with its 3′UTR.
Conclusion
Our study proposes a novel STAT3-miR-572-MOAP-1 pathway involved in the process of CRC progression, which might be a potential target for the development of new preventive and therapeutic approaches against human colorectal cancer.
Keywords: colorectal neoplasm, STAT3, MOAP-1, miR-572, tumor progression
Introduction
Around the world colorectal carcinoma (CRC) is one of the most common and deadly malignant tumors. It has been ranked as the third malignancies both in China and in the USA, according to recent cancer statistics.1,2 Despite medical advancements in the past decade, nearly 50% of CRC patients have tumor recurrence resulting in a poor prognosis. The 5-year survival rate is 64.9% so far.3 The recurrence of CRC is assumed to be as a result of tumor invasion and metastasis. Therefore, it is of great significance to understand the molecular mechanism of CRC progression in order to develop therapeutic strategies to improve the prognosis of patients with CRC.
Components of JAK/STAT3, which include the oncogenic transcription factor STAT3, play a crucial role in mutiple pathways of physiological and pathophysiological processes involved in tumor proliferation, invasion, and metastasis.4,5 In CRC, activation of STAT3 often positively correlates with tumor progression and a poor prognosis.6,7 miRNAs, a class of evolutionally conserved small non-coding RNA molecules (approximately 19–23 nucleotides), take part in post-transcriptional regulation of gene expression.8 The modification in miRNA expression profiles represents a shared feature in all human cancers, indicating the importance of such variations in carcinogenesis and tumor progression.9–11 Earlier reports revealed that STAT3 induces miRNA (eg, miR-182-5p and miR-21) expression.12,13 miR-572 contributes to human CRC cell proliferation by suppressing PPP2R2C expression and post-transcriptionally regulating suppressor of cytokine sig-naling 1 (SOCS1) and p21.14,15 Moreover, MOAP-1 is a pro-apoptotic protein that induces cell death or apoptosis. It can bind with Bax protein and trigger its activation.16 Recently, some studies have shown a direct correlation between miR-NAs and MOAP-1.17,18 However, the status of MOAP-1 expression in CRC has remained unknown up to now. Given the importance of miR-572 and STAT3 in CRC progression, we decided to explore whether they are involved in a common pathway and the role of MOAP-1 in this process.
In the current study, we described a putative miR-572 binding site at the 3′UTR of MOAP-1 in the proposal of a key mechanism behind STAT3-mediated positive impact on CRC progression. Additionally, we explored whether STAT3 inhibited MOAP-1 expression through the upregulation of miR-572, as well as the functional implication of this regulation.
Material and methods
CRC tissue specimens
The study protocol was approved by the Ethical Committee of Tangdu Hospital of The Fourth Military Medical University, and written informed consent was provided by all the participants prior to surgery. The specimens were obtained from 40 patients with CRC who underwent colorectal resection between January 1, 2016 and May 30, 2017. The tumor samples were confirmed by pathological examination, frozen in liquid nitrogen, and stored at −80°C until use.
Cell lines and culture
Human CRC cell lines LS174T, SW620, HT29, LOVO, HCT116, and SW480 were obtained from American Type Culture Collection (ATCC, Manassas, VA, USA). The cells were cultured in RPMI-1640 (Thermo Fisher Scientific, Waltham, MA, USA) as per the manufacturer’s recommendations, with 10% FBS and 1% streptomycin in a humidified atmosphere of 5% CO2 at 37°C.
qRT-PCR
Trizol reagent (Thermo Fisher Scientific) was used to extract total RNA for miR-572, STAT3 or MOAP-1 mRNA studies. miRNA-specific TaqMan MiRNA Assay Kit (Thermo Fisher Scientific) was used to reveal the expression levels of miR-572. qRT-PCR was achieved using the SYBR green reagent with an ABI Prism™ 7000HT sequence detection system (Thermo Fisher Scientific). The relative expression was assessed by a comparative CT method and standardized to the expression of GAPDH and U6 small RNA, respectively. The primer sequences were as follows: STAT3, 5′-CAGCAGCTTGACACACGGTA-3′ (forward primer) and 5′-AAACACCAAAGTGGCATGTGA-3′ (reverse primer); MOAP-1, 5′-ACATGAAAATGGCTCCT TAGAC-3′ (forward primer) and 5′-GACACGAATAACATCAAGTGCT-3′ (reverse primer); miR-572, 5′-GCCAGATCTCTGAGGAAAGCAGGAGGAGG-3′ (forward primer) and 5′-GCCGAATTCTCGGCACAAATCTTCAGAGC-3′ (reverse primer); GAPDH, 5′-CAAGGTCAT CCATGACAACTTTG-3′ (forward primer) and 5′-GTCCACCACCCTGTTGCTGTAG-3′ (reverse primer). U6 small RNA: 5′-CTCGCTTCGGCAGCACA-3′ (forward primer) and 5′-AACGCTTCACGAATTTGCGT-3′ (reverse primer).
miRNA transfection and siRNA treatment
Both hsa-miR-572 mimic (MISSION® microRNA Mimic) and hsa-miR-572 inhibitor (MISSION® Lenti microRNA Inhibitor) were purchased from Sigma-Aldrich Co. (St Louis, MO, USA). The miR control and anti-miR control were obtained from Shanghai GenePharma Co., Ltd. (Shanghai, China). LS174T, SW620, HCT116, and SW480 cells were transfected using X-tremeGENE™ HP-DNA transfection reagent (Hoffman-La Roche Ltd., Basel, Switzerland) according to the manufacturer’s guidelines. MOAP-1 siRNA (ID: 130305 and 30053) and STAT3 siRNA (ID: 116558 and 116558) were acquired from Thermo Fisher Scientific. LS174T and SW480 cells were transfected using Lipofectamine® RNAiMAX Reagent (Thermo Fisher Scientific).
Western blot assay
Cells were washed with PBS, collected, and lysed with RIPA lysis buffer (Thermo Fisher Scientific). Qubit® Protein Assay Kit and Qubit® 2.0 Fluorometer (Thermo Fisher Scientific) were used to determine protein concentrations. Approximately 50 μg of total protein was subjected to 10% SDS-PAGE gel and transferred to PVDF membranes according to the standard procedure. Membranes were blocked with 10% milk in TBST and incubated with primary antibodies against MOAP-1 ([Sigma-Aldrich Co.] SAB1407857, 1:1,000 dilution), STAT3 ([Cell Signaling Technology, Danvers, MA, USA] #12640, 1:1,000 dilution), p-STAT3 ([Cell Signaling Technology] #9145, 1:2,000 dilution), GAPDH ([Proteintech®, Rosemont, IL, USA] 60004-1-Ig, 1:10,000 dilution), and β-actin ([Sigma-Aldrich Co.] clone AC-15, 1:6,000 dilution) overnight, followed by corresponding HRP-linked secondary antibody ([Abbkine, Wuhan, China], 1:4 000 dilution) and visualized using a Pierce™ ECL Western blotting substrate (Thermo Fisher Scientific).
3′UTR luciferase reporter assay
Human wild-type MOAP-1 or its mutant with the putative miR-572-binding sites in the 3′UTR of MOAP-1 was synthesized and cloned downstream of firefly luciferase gene in the psiCHECK™-2 vector (Promega Corporation, Fitchburg, WI, USA). The LS174T and SW620 cells were cultured in 96-well plates and co-transfected with 400 ng of constructed luciferase reporter plasmids, 50 ng of renilla luciferase reporter vector (pRL-TK), and 20 nM of miR-572 or control mimic using Lipofectamine™ 2000 (Thermo Fisher Scientific) as per the manufacturer’s instructions. Cells were collected 48 h after transfection, and the luciferase activity was scrutinized using Dual Luciferase Reporter Assay System (Promega Corporation). Renilla luciferase activities were used as the internal control for the normalization of luciferase activity.
Cell proliferation assay
Cells were plated in 96-well plates. Cell growth was tested using a standard MTT assay and cells were counted with trypan blue (0.04%, Sigma-Aldrich Co.) by a hemocytometer.
Cell migration and invasion assay
Cell migration assay was performed using a transwell system according to the manufacturer’s instructions (Corning Incorporated, Corning, NY, USA). Briefly, transfected cells were trypsinized, and 1×105 cells were seeded into the upper chamber with serum-free option-MEM. The lower chamber was filled with 800 μL medium containing 10% FBS as a chemo-attractant. After incubation for 24 h, cells on the bottom side of the filter were fixed with 5% formaldehyde for 20 min and stained with 0.1% crystal violet solution. The numbers of cells in five randomly selected fields were calculated under a light microscope. For the invasion assay, the capacity of cell invasion was measured using Matrigel matrix-coated Boyden chambers (Corning Incorporated) according to the company’s instructions. The rest of the experiment was performed as transwell migration assay.
Statistical analysis
For statistical analyses, we used SPSS (version 16.0; SPSS Inc., Chicago, IL, USA). All of the data were depicted as the mean ± SD. Each error bar indicated the variation between the means of three independent experiments. The comparisons between two groups or among more than two groups were analyzed using Student’s t-test or ANOVA. Spearman’s correlation analysis was used to determine the relationship between STAT3 and MOAP-1 mRNA levels, and P-value <0.05 was considered to be statistically significant.
Results
Elevated STAT3 levels were accompanied by increased miR-572 and decreased MOAP-1 levels in primary CRC specimens and cell lines
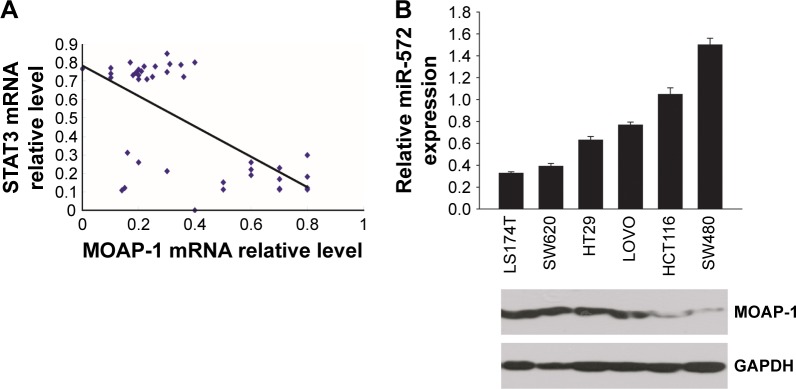
In exploring the relevance of this proposed regulation, 40 cases of frozen human CRC tissues were analyzed. As shown in Figure 1A, there was a negative correlation (P=0.0005) between STAT3 and MOAP-1 mRNA levels. Likewise, there was a markedly inverse expression of miR-572 and MOAP-1 protein in several human CRC cell lines (Figure 1B).
Figure 1.
Elevated STAT3 levels were accompanied by increased miR-572 and decreased MOAP-1 levels in human primary colorectal carcinoma (CRC) specimens and cell lines.
Notes: (A) The correlation between STAT3 and MOAP-1 mRNA levels was determined by qRT-PCR in 40 cases of frozen CRC tissues (P=0.0005). (B) Levels of miR-572 were inversely related to MOAP-1 protein in human CRC cell lines LS174T, SW620, HT29, LOVO, HCT116, and SW480. Relative miR-572 levels and MOAP-1 protein levels were determined by qRT-PCR and Western blot, respectively.
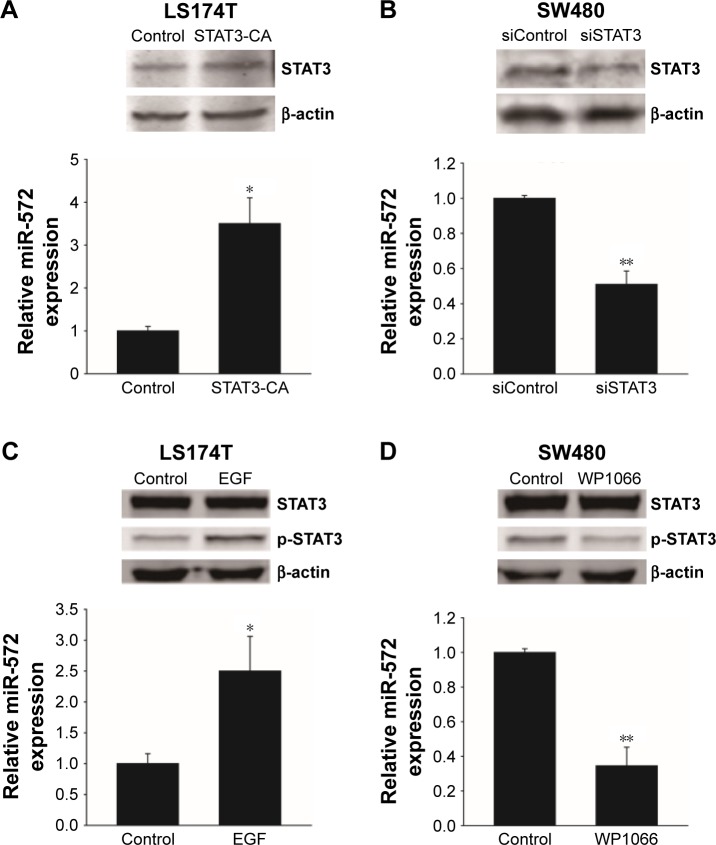
STAT3 upregulated miR-572 in CRC cell lines
When the LS174T cells were transfected with a constitutively activated mutant of STAT3 (STAT3-CA), the expression level of miR-572 was increased (Figure 2A). Additionally, when serum-starved LS174T cells were treated with a ligand for STAT3 activation, EGF (Sigma-Aldrich Co.), miR-572 expression was upregulated, as shown in Figure 2C, thus suggesting that STAT3 activation positively regulated miR-572 expression. In contrast, inhibition of STAT3 by siRNA or a STAT3 inhibitor, WP1066 (Sigma-Aldrich Co.), significantly reduced miR-572 expression levels in SW480 cells (Figure 2B and D).
Figure 2.
STAT3 upregulated miR-572 expression in colorectal carcinoma cell lines.
Notes: (A) The levels of miR-572 expression in LS174T cells transfected with a constitutively activated mutation of STAT3 (STAT3-CA) or empty vector (control). (B) The levels of miR-572 expression in SW480 cells transfected with siSTAT3 and siControl. (C) The levels of miR-572 expression in serum-starved LS174T cells treated with EGF (50 ng/mL) or without EGF (control). (D) The levels of miR-572 expression in SW480 cells treated with WP1066 (10 μM) or without WP1066 (control). The levels of miR-572 were determined by qRT-PCR after transfection with STAT3 or siSTAT3 for 24 h, or after treatment with EGF or WP1066 for 30 min. The protein levels of STAT3 (A and B), or STAT3 and p-STAT3 (C and D) are shown at the top center of corresponding figures. *P<0.05; **P<0.01.
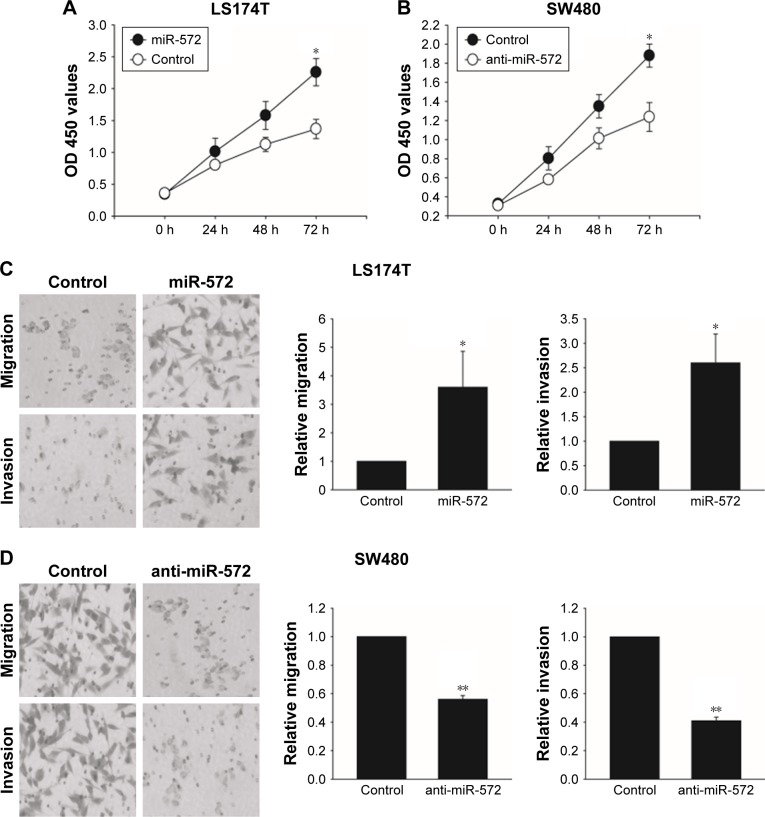
miR-572 promoted CRC cell growth, migration, and invasion
In order to find out whether miR-572 stimulated an oncogenic phenotype of tumor cells, we performed overexpressed or reduced function assays in CRC cells using an miR-572 mimic or inhibitor. Overexpression of miR-572 resulted in a significantly increased viability of LS174T cells in an MTT assay (Figure 3A). In contrast, as shown in Figure 3B, miR-572 inhibition significantly reduced cell growth compared with the control group, as measured by an MTT assay. Moreover, to determine whether miR-572 controlled the ability of tumor cell migration and invasion, transwell migration and invasion assays were performed. The results exhibited that miR-572 was able to promote LS174T cells’ migration and invasion (Figure 3C). Conversely, the ability of migration and invasion in SW480 cells was significantly reduced by the inhibition of miR-572, as demonstrated in Figure 3D.
Figure 3.
miR-572 promoted colorectal carcinoma cell growth, migration, and invasion.
Notes: The proliferation of LS174T cells (A) transfected with miR-572 mimic (20 nM) or with control and SW480 cells (B) transfected with miR-572 inhibitor (multiplicity of infection =5) or with control was analyzed by MTT assay. LS174T cells (C) transfected with miR-572 mimic (20 nM) or with control and SW480 cells (D) transfected with miR-572 inhibitor (multiplicity of infection =5) or with control were subjected to transwell migration or invasion assays. *P<0.05; **P<0.01.
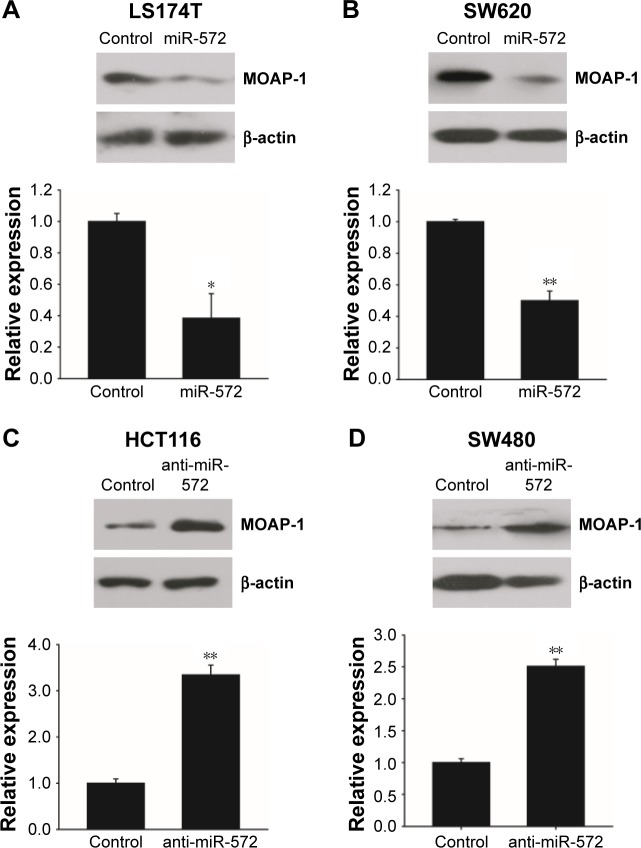
miR-572 downregulated MOAP-1 expression
The counter relationship between miR-572 and MOAP-1 mRNA levels implied that miR-572 might decrease MOAP-1 expression. To prove this speculation, we transfected LS174T and SW620 cells with miR-572 mimic and measured MOAP-1 expression levels by Western blot. The results indicated that MOAP-1 expression was decreased significantly in these two cell lines, as clearly shown in Figure 4A and B. Accordingly, miR-572 inhibitors increased MOAP-1 expression levels in HCT116 and SW480 cell lines, as revealed in Figure 4C and D.
Figure 4.
miR-572 downregulated MOAP-1 protein expression.
Notes: The protein levels of MOAP-1 were determined by Western blot after transfection with miR-572 mimic (20 nM) or control in LS174T (A) and SW620 (B) cells, or after transfection with miR-572 inhibitor (multiplicity of infection =5) or control in HCT116 (C) and SW480 (D) cells. β-actin was used as loading control. Scanning densitometry of immunoblotting for each panel was measured (lower). *P<0.05; **P<0.01.
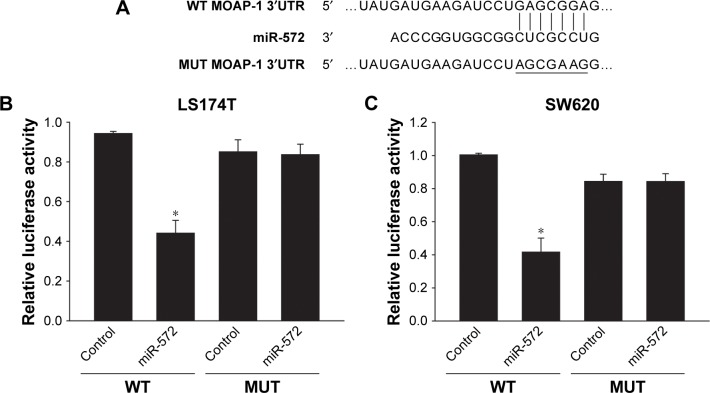
miR-572 silenced MOAP-1 by targeting its 3′UTR
To determine whether MOAP-1 was a target of miR-572, TargetScan prediction software was used to scan the 3′UTR of the human MOAP-1 gene in search of an miR-572 binding site. The seed sequence of miR-572 indicated complete complementarity to a target site of the MOAP-1 3′UTR (Figure 5A). By using 3′UTR luciferase reporter assay in LS174T (Figure 5B) and SW620 cells (Figure 5C), we validated that the activity of luciferase cloned with a wild-type MOAP-1 3′UTR was dramatically reduced by miR-572, while its activity was unaffected in the clone with a mutation in the putative 3′UTR target site (Figure 5B and C). These results proposed that miR-572 blocked MOAP-1 expression through binding to its 3′UTR sequence.
Figure 5.
miR-572 inhibited MOAP-1 expression.
Notes: (A) Sequence of miR-572 and the predicted miR-572-binding site at MOAP-1 3′UTR. Dual-luciferase reporter assay was applied in LS174T (B) and SW620 (C) cells after co-transfection with established luciferase reporter vectors (including wild-type [WT] or mutated [MUT] MOAP-1 3′UTR) and miR-572 or miR-control. *P<0.05.
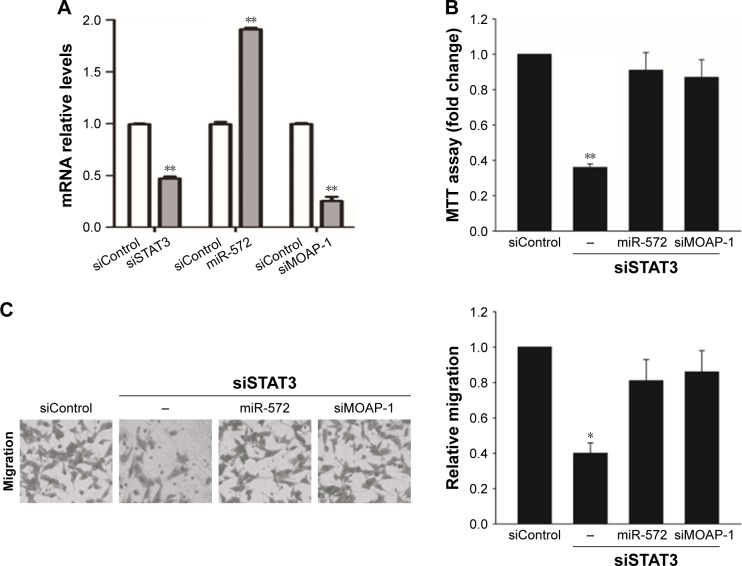
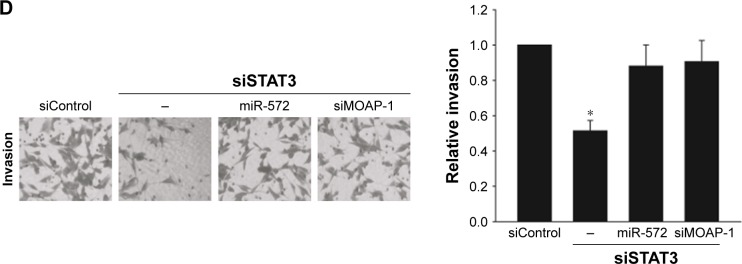
STAT3 induced CRC progression via an miR-572-MOAP-1 pathway
In order to estimate whether STAT3 induced CRC cell growth, migration, and invasion via miR-572-MOAP-1 pathway, we first assessed the consequences of targeting STAT3 in cell growth (Figure 6). Interestingly, although cell growth in SW480 cells with STAT3 knockdown was significantly reduced, the proliferative potential of these cells was restored after miR-572 mimics or MOAP-1 siRNA was transfected (Figure 6B). In a similar way, we observed that the abilities of cell migration and invasion in SW480 cells with STAT3 knockdown were notably reduced. Nevertheless, these inhibition effects were rescued after miR-572 mimics or MOAP-1 siRNA transfection (Figure 6C and D). Collectively, these results showed that there might be an miR-572-MOAP-1 pathway involved in STAT3-induced CRC cell growth, migration, and invasion.
Figure 6.
STAT3 induced colorectal carcinoma progression via an miR-572-MOAP-1 pathway.
Notes: After transfection with STAT3 siRNA for 24 h, SW480 cells were transfected with miR-572 mimic or MOAP-1 siRNA for 24 h. The relative mRNA levels of STAT3, miR-572, and MOAP-1 were determined by qRT-PCR (A). SW480 cells transfected with STAT3 siRNA or siRNA control alone or in combination with miR-572 mimic or MOAP-1 siRNA were analyzed by MTT assay (B), transwell migration (C) and invasion (D) assays. *P<0.05. **P<0.01.
Discussion
STAT3-induced transcription of protein-coding genes has been widely studied in human cancers. Nevertheless, the role of STAT3 in the transcription of non-protein-coding genes, such as miRNAs, is rarely studied. miRNAs are essential for the development of the malignant phenotype in CRC.19–21 Recent studies have provided increasing evidence that STAT3 plays an important role in miRNA expression. To name a few, STAT3 induces miR-182-5p to promote glioma tumorigenesis.12 STAT3-regulated exosomal miR-21 enhances angiogenesis and participates in neoplastic processes of transformed human bronchial epithelial cells.13 STAT3 regulates stemness, tumorigenesis, and migration of lung-derived brain metastasis cells through miR-21 activation.22 These findings propose that STAT3 and miRNAs are cooperatively involved in tumor initiation and progression.
In the present study, we provided proof that the activation of miR-572 is induced by STAT3. We speculated that STAT3 binds in miR-572 promoter regions, as it is important for transcriptional induction of miR-572. In support of this speculation, is the result that STAT3 activation by EGF treatment resulted in the upregulation of miR-572. On the other hand, inhibition of STAT3 by siRNA or WP1066 markedly reduced the expression levels of miR-572, signifying that STAT3 and miR-572 were necessary for CRC cell growth, migration, and invasion. Our study further supported that STAT3 performs a key role in the regulation of miRNAs in human CRC.
The roles of miR-572 in carcinogenesis and tumor progression remain controversial. Growing evidence suggests that miR-572 is an oncogene in ovarian cancer14,15 and nasopharyngeal carcinoma.23 Wang et al reported that the serum level of miR-572 is significantly increased in the patients with renal cell carcinoma.24 Whereas, in stage T2–T3N0 CRC patients who underwent resection and adjuvant chemotherapy, miR-572 is downregulated in patients with distant metastases.25 miR-572 is one of the commonly downregulated miRNAs in recurrent prostate cancer samples in comparison to non-recurrent tumors.26 Furthermore, it is significantly downregulated in basal cell carcinoma tissues compared with normal skin.27 In view of these contradictory results, we performed overexpressed or reduced function assays using miR-572 in multiple CRC cell lines. Our study showed that miR-572 promoted CRC cell growth, migration, and invasion, which proposed that miR-572 stimulated an oncogenic phenotype of CRC cells.
Initially, MOAP-1 was described as a pro-apoptotic protein that binds with multiple members of the Bcl-2 family, including Bax.28,29 It is involved in Bax mitochondrial translocation and activation.16 It is reported that the abilities of miR-25 to promote cell proliferation and to inhibit apoptosis are attributable to its suppressed effect on MOAP-1 in lung cancer.17 Moreover, miR-1228 supports cell growth after it is attenuated by overexpressed MOAP-1 in breast cancer.18 MOAP-1 is also a tumor suppressor protein linked to the RASSF1A protein.30 Our study first established that MOAP-1 expression is promoted by miR-572 inhibitor. Given the biological function and mechanism of miRNAs, we speculated that the mechanism in inhibition of MOAP-1 expression might be attributed to miRNA regulation at the post-transcriptional level. To this end, we performed 3′UTR luciferase reporter assay and validated that miR-572 blocked MOAP-1 expression through binding in its 3′UTR sequence.
Conclusion
This study proposed a STAT3-miR-572-MOAP-1 pathway involved in the process of CRC progression. The activation of STAT3 upregulates miR-572 expression and successively reduces MOAP-1 through interaction with its 3′UTR, resulting in a positive impact on CRC cell growth, migration, and invasion. Therefore, our study provides a potential novel target for the development of new therapeutic agents for human CRC.
Acknowledgments
The authors thank Jianfei Xue, MD, PhD, who works in the Department of Neurosurgery, The University of Texas MD Anderson Cancer Center, for his insightful manuscript editing and revision.
Footnotes
Disclosure
Dr Qing Qiao was supported by Scientific and Technological Project of Shanxi Province in China (No 2014K11-03-03-03); Dr Nan Wang was supported by Scientific and Technological Project of Shanxi Province in China (No 2014K11-03-03-14) and National Natural Science Foundation of China (NSFC, No 81200330). The authors report no other conflicts of interest in this work.
References
- 1.Siegel RL, Miller KD, Fedewa SA, et al. Colorectal cancer statistics, 2017. CA Cancer J Clin. 2017;67(3):177–193. doi: 10.3322/caac.21395. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Wu W, Guo F, Ye J, et al. Pre- and post-diagnosis physical activity is associated with survival benefits of colorectal cancer patients: a systematic review and meta-analysis. Oncotarget. 2016;7(32):52095–52103. doi: 10.18632/oncotarget.10603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Poli V, Camporeale A. STAT3-mediated metabolic reprograming in cellular transformation and implications for drug resistance. Front Oncol. 2015;5:121. doi: 10.3389/fonc.2015.00121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu H, Lee H, Herrmann A, Buettner R, Jove R. Revisiting STAT3 signalling in cancer: new and unexpected biological functions. Nat Rev Cancer. 2014;14(11):736–746. doi: 10.1038/nrc3818. [DOI] [PubMed] [Google Scholar]
- 6.Zhu X, Sun L, Lan J, et al. BRG1 targeting STAT3/VEGFC signaling regulates lymphangiogenesis in colorectal cancer. Oncotarget. 2016;7(24):36501–36509. doi: 10.18632/oncotarget.9038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bosch-Barrera J, Menendez JA. Silibinin and STAT3: A natural way of targeting transcription factors for cancer therapy. Cancer Treat Rev. 2015;41(6):540–546. doi: 10.1016/j.ctrv.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 8.Bracken CP, Scott HS, Goodall GJ. A network-biology perspective of microRNA function and dysfunction in cancer. Nat Rev Genet. 2016;17(12):719–732. doi: 10.1038/nrg.2016.134. [DOI] [PubMed] [Google Scholar]
- 9.Bracken CP, Khew-Goodall Y, Goodall GJ. Network-based approaches to understand the roles of miR-200 and other microRNAs in cancer. Cancer Res. 2015;75(13):2594–2599. doi: 10.1158/0008-5472.CAN-15-0287. [DOI] [PubMed] [Google Scholar]
- 10.Ishimoto T, Baba H, Izumi D, et al. Current perspectives toward the identification of key players in gastric cancer microRNA dysregulation. Int J Cancer. 2016;138(6):1337–1349. doi: 10.1002/ijc.29627. [DOI] [PubMed] [Google Scholar]
- 11.Lin S, Gregory RI. MicroRNA biogenesis pathways in cancer. Nat Rev Cancer. 2015;15(6):321–333. doi: 10.1038/nrc3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xue J, Zhou A, Wu Y, et al. miR-182-5p induced by STAT3 activation promotes glioma tumorigenesis. Cancer Res. 2016;76(14):4293–4304. doi: 10.1158/0008-5472.CAN-15-3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Y, Luo F, Wang B, et al. STAT3-regulated exosomal miR-21 promotes angiogenesis and is involved in neoplastic processes of transformed human bronchial epithelial cells. Cancer Lett. 2016;370(1):125–135. doi: 10.1016/j.canlet.2015.10.011. [DOI] [PubMed] [Google Scholar]
- 14.Wu AH, Huang YL, Zhang LZ, et al. MiR-572 prompted cell proliferation of human ovarian cancer cells by suppressing PPP2R2C expression. Biomed Pharmacother. 2016;77:92–97. doi: 10.1016/j.biopha.2015.12.005. [DOI] [PubMed] [Google Scholar]
- 15.Zhang X, Liu J, Zang D, et al. Upregulation of miR-572 transcriptionally suppresses SOCS1 and p21 and contributes to human ovarian cancer progression. Oncotarget. 2015;6(17):15180–15193. doi: 10.18632/oncotarget.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Foley CJ, Freedman H, Choo SL, et al. Dynamics of RASSF1A/MOAP-1 association with death receptors. Mol Cell Biol. 2008;28(14):4520–4535. doi: 10.1128/MCB.02011-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu T, Chen W, Kong D, et al. miR-25 targets the modulator of apoptosis 1 gene in lung cancer. Carcinogenesis. 2015;36(8):925–935. doi: 10.1093/carcin/bgv068. [DOI] [PubMed] [Google Scholar]
- 18.Lin L, Liu D, Liang H, et al. MiR-1228 promotes breast cancer cell growth and metastasis through targeting SCAI protein. Int J Clin Exp Pathol. 2015;8(6):6646–6655. [PMC free article] [PubMed] [Google Scholar]
- 19.Li T, Yang J, Lv X, et al. miR-155 regulates the proliferation and cell cycle of colorectal carcinoma cells by targeting E2F2. Biotechnol Lett. 2014;36(9):1743–1752. doi: 10.1007/s10529-014-1540-3. [DOI] [PubMed] [Google Scholar]
- 20.Fantini S, Salsi V, Reggiani L, Maiorana A, Zappavigna V. The miR-196b miRNA inhibits the GATA6 intestinal transcription factor and is upregulated in colon cancer patients. Oncotarget. 2017;8(3):4747–4759. doi: 10.18632/oncotarget.13580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang RY, Wang Z, Chen HQ, Zhu SB. Negative correlation between miR-200c and decorin plays an important role in the pathogenesis of colorectal carcinoma. Biomed Res Int. 2017;2017:1038984. doi: 10.1155/2017/1038984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singh M, Garg N, Venugopal C, et al. STAT3 pathway regulates lung-derived brain metastasis initiating cell capacity through miR-21 activation. Oncotarget. 2015;6(29):27461–27477. doi: 10.18632/oncotarget.4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yan L, Cai K, Liang J, et al. Interaction between miR-572 and PPP2R2C and their effects on proliferation, migration and invasion of nasopharyngeal carcinoma (NPC) cells. Biochem Cell Biol. 2017;95(5):578–584. doi: 10.1139/bcb-2016-0237. [DOI] [PubMed] [Google Scholar]
- 24.Wang C, Hu J, Lu M, et al. A panel of five serum miRNAs as a potential diagnostic tool for early-stage renal cell carcinoma. Sci Rep. 2015;5:7610. doi: 10.1038/srep07610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bobowicz M, Skrzypski M, Czapiewski P, et al. Prognostic value of 5-microRNA based signature in T2-T3N0 colon cancer. Clin Exp Metastasis. 2016;33(8):765–773. doi: 10.1007/s10585-016-9810-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pashaei E, Pashaei E, Ahmady M, Ozen M, Aydin N. Meta-analysis of miRNA expression profiles for prostate cancer recurrence following radical prostatectomy. PLoS One. 2017;12(6):e0179543. doi: 10.1371/journal.pone.0179543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sand M, Skrygan M, Sand D, et al. Expression of microRNAs in basal cell carcinoma. Br J Dermatol. 2012;167(4):847–855. doi: 10.1111/j.1365-2133.2012.11022.x. [DOI] [PubMed] [Google Scholar]
- 28.Vos MD, Dallol A, Eckfeld K, et al. The RASSF1A tumor suppressor activates Bax via MOAP-1. J Biol Chem. 2006;281(8):4557–4563. doi: 10.1074/jbc.M512128200. [DOI] [PubMed] [Google Scholar]
- 29.Fu NY, Sukumaran SK, Yu VC. Inhibition of ubiquitin-mediated degradation of MOAP-1 by apoptotic stimuli promotes Bax function in mitochondria. Proc Natl Acad Sci U S A. 2007;104(24):10051–10056. doi: 10.1073/pnas.0700007104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Law J, Salla M, Zare A, et al. Modulator of apoptosis 1 (MOAP-1) is a tumor suppressor protein linked to the RASSF1A protein. J Biol Chem. 2015;290(40):24100–24118. doi: 10.1074/jbc.M115.648345. [DOI] [PMC free article] [PubMed] [Google Scholar]