Abstract
Background and Aims
Flowers can be highly variable in nectar volume and chemical composition, even within the same plant, but the causes of this variation are not fully understood. One potential cause is nectar-colonizing bacteria and yeasts, but experimental tests isolating their effects on wildflowers are largely lacking. This study examines the effects of dominant species of yeasts and bacteria on the hummingbird-pollinated shrub, Mimulus aurantiacus, in California.
Methods
Wildflowers were inoculated with field-relevant titres of either the yeast Metschnikowia reukaufii or the bacterium Neokomagataea sp. (formerly Gluconobacter sp.), both isolated from M. aurantiacus nectar. Newly opened flowers were bagged, inoculated, harvested after 3 d and analysed for microbial abundance, nectar volume, and sugar and amino acid concentration and composition.
Key Results
Yeast inoculation reduced amino acid concentration and altered amino acid composition, but had no significant effect on nectar volume or sugar composition. In contrast, bacterial inoculation increased amino acid concentration, enhanced the proportion of nectar sugars comprised by monosaccharides, and reduced nectar volume.
Conclusions
The results presented suggest that microbial inhabitants of floral nectar can make nectar characteristics variable among flowers through divergent effects of yeasts and bacteria on nectar chemistry and availability, probably modifying plant–pollinator interactions.
Keywords: Flower longevity, flower microbiome, host–microbe, Mimulus aurantiacus, nectar chemistry, plant defence, plant-pollinator
INTRODUCTION
It is widely recognized that flowers vary greatly in nectar volume and chemical composition (Mitchell, 2004, and references therein), which can affect pollinator foraging and plant fitness (Cnaani et al., 2006). Some of this variation reflects genetic differences among plants (Mitchell, 2004), but nectar characteristics can be highly variable even within individual plants (Biernaskie and Cartar, 2004; Herrera et al., 2006). One factor that is gaining increased recognition as a major source of this variation is nectar-colonizing micro-organisms. Recent studies have linked microbial abundance to a variety of nectar characteristics, including temperature (Herrera et al., 2010), pH (Vannette et al., 2013; Tucker and Fukami, 2014), volatile profiles (Rering et al., 2018), and the concentration and composition of sugars (Herrera et al., 2008; Canto and Herrera, 2012; de Vega and Herrera, 2012; Canto et al., 2015; Vannette and Fukami, 2017), secondary metabolites (Vannette et al., 2013) and amino acids (Herrera et al., 2008; Herrera and Pozo, 2010; Peay et al., 2012; Vannette et al., 2013), as well as pollinator foraging (Good et al., 2014; Junker et al., 2014; Schaeffer and Irwin, 2014; Vannette and Fukami, 2016) and plant reproduction (Herrera et al., 2013; Vannette et al., 2013). In addition, some studies have reported taxon-specific effects, particularly between bacteria and yeasts, both of which are frequently found in floral nectar (Vannette et al., 2013; Good et al., 2014; Tucker and Fukami, 2014; Rering et al., 2018).
Most of these studies have used field observations examining correlational data, laboratory experiments using nectar extracted from plants (e.g. Peay et al., 2012; Vannette et al. 2013, 2016), or field experiments where entire microbial communities are added to the nectar of real flowers (Canto et al., 2007, 2008, 2011; de Vega and Herrera, 2013). It remains unknown whether bacteria and yeasts differentially influence nectar characteristics in real flowers. This knowledge is needed to understand fully microbial effects on nectar traits as microbial species composition can vary among flowers due to differences in microbial dispersal history and strong competitive interactions within flowers (Tucker and Fukami, 2014; Vannette and Fukami, 2017; Toju et al., 2018). Given that nectar microbial communities might be often dominated by either yeasts or bacteria (Tucker and Fukami, 2014) and that yeasts and bacteria may have contrasting effects on nectar chemistry and pollination visits (Vannette et al., 2013; Good et al., 2014; Rering et al., 2018), their separate effects in wildflowers are of particular interest. In this study, we inoculated flowers of wild Mimulus auranticus plants, a hummingbird-pollinated shrub in California, with a species of the dominant nectar yeast, Metschnikowia reukaufii, or that of the dominant nectar bacteria, Neokomagatea sp. (previously called Gluconobacter sp.), to quantify their effects on nectar characteristics.
MATERIALS AND METHODS
Study organisms
We conducted this study using micro-organisms isolated from the nectar of flowering plants at Jasper Ridge Biological Preserve (JRBP) in the foothills of the Santa Cruz mountains near Stanford, California. Yeasts and bacteria were isolated from the nectar of the flowering shrubs Mimulus aurantiacus (sticky monkeyflower) and Eriodictyon californicum (yerba santa). Both plant species are visited by hummingbirds, and bees including Bombus vosnesenskii, Ceratina acantha and Xylocopa micans, although M. aurantiacus is primarily hummingbird pollinated, whereas E. californicum is mainly insect pollinated. The butterfly Euphydryas chalcedona also visits E. californicum.
The nectar microbes of M. aurantiacus have been studied previously (Belisle et al., 2012; Dhami et al., 2016; Tsuji et al., 2016; Vannette and Fukami, 2017; Dhami et al., 2018), and the dominant micro-organisms are readily cultured and inoculated into live flowers (e.g. Vannette et al., 2013). In M. aurantiacus, both bacteria and yeasts appear to modify floral nectar traits including pH, sugar composition, H2O2 concentration, and amino acid composition, although this evidence comes from laboratory experiments using extracted nectar, not field experiments with nectar in real flowers (Vannette et al., 2013). In a field correlational study, microbial abundance explained 35 % of the variation in sugar composition of M. aurantiacus (Vannette and Fukami, 2017), suggesting that micro-organisms influence nectar traits under field conditions. Microbial diversity within flowers is relatively limited. Often, single or a few species of micro-organism dominate a nectar sample (Belisle et al., 2012; Vannette and Fukami, 2017). As a result, examining the effect of single microbial species is ecologically relevant for M. aurantiacus (Belisle et al., 2012) and other plant species (Herrera et al., 2009).
We chose microbial taxa that were common and dominant representatives of floral microbial communities in our study plants. The fungal species most frequently isolated from both plant species was the yeast Metschnikowia reukaufii. Given the considerable phenotypic diversity within this yeast species (Herrera et al., 2014; Dhami et al. 2016, 2018), we used multiple strains: one isolated from M. aurantiacus and two from E. californicum. No significant differences in growth or effects on nectar parameters were detected between the strains (P > 0.05), so strains are pooled for all analyses below. We also used a bacterial strain isolated from M. aurantiacus (NCBI JX437138.1), a bacterium in the Acetobacteraceae and a close BLAST match to the genus Neokomagatea sp. (previously called Gluconobacter sp.) identified using a region of the 16S rRNA gene. The Gram-negative Neokomagataea, an acetic acid bacterium, is a frequent colonist of floral nectar in many species of California plant species (R. L. Vannette, unpubl. res.) and some other biogeographic regions (Samuni-Blank et al., 2014; Reis and Teixeira, 2015). We used a single strain from the genus Neokomagataea. Yeasts were maintained on yeast malt agar (YMA) and Neokomagataea on Reasoner’s 2A (R2A) medium supplemented with 20 % sucrose (Oxoid formula).
Experimental design
In the same area of JRBP as used by Belisle et al. (2012), we chose 29 M. aurantiacus plants, which spanned a gradient of sun exposure and water availability. Five of the 29 plants received experimental water addition, mimicking 2-fold the annual rainfall. For this purpose, from January to May of 2014, 10 L of tap water was delivered once a week, using a 10 L container placed at the base of each plant. The containers had a spigot, so that the water was delivered slowly. No significant effect of watering was detected on any measured variables (P > 0.10), so we do not include this factor in the statistical models below. During May and June of 2014, unopened buds on each plant were marked using a small jewelry tag that had a unique identification number and then bagged using a small organza bag, which prevented hummingbirds and other floral visitors from accessing the flower, inhibiting microbial dispersal to flowers via the flower-visiting animals (Belisle et al., 2012; Vannette and Fukami, 2017). More flowers were marked and bagged than were used to ensure that experimental cohorts of flowers had opened on the same day. Bagged flowers were checked daily.
Microbial inoculation
Microbial suspensions were prepared using a sterile 20 % sucrose solution. On the morning of inoculation, suspensions of each yeast and bacterial strain were prepared at 1000 cells µL–1 from a colony formed on 3- or 4-day-old plates, which probably included both viable and non-viable cells. Into each newly opened flower, we pipetted 1 µL of suspension (1000 cells) of a single species or a sterile control solution comprised only of the 20 % sucrose solution. Flowers from all plants received each treatment, depending on flower availability. Most plants hosted between one and three replicates of each treatment (control, yeasts, and bacteria), which was limited by the number of flowers opening each day. Upon anthesis, M. aurantiacus flowers typically contain about 2–5 µL of nectar. Feeding by a hummingbird in artificial nectar solutions after banding (Vannette and Fukami, 2017) typically results in the deposition of, on average, 155 (range = 10–1180) colony-forming units (CFUs) on a YMA plate or, on average, 2000 (range = 10–10 000) CFUs on a R2A plate (unpubl. res.), so our experimentally introduced microbial titres were within the range of those probably deposited in nectar of M. aurantiacus.
Álvarez-Pérez and Herrera (2013) showed that bacteria and yeasts appeared to co-occur frequently in the flowers that they studied in Spain. However, this study analysed the presence and absence of bacteria and yeasts, not their abundance. Even if bacteria and yeasts co-occur frequently in terms of their presence/absence, it is possible that they rarely co-dominate, in the sense that bacteria are rarely abundant if yeasts are abundant, and vice versa. In fact, laboratory experiments that used bacterial and yeast strains isolated from M. aurantiacus flowers (Tucker and Fukami, 2014) indicated that bacteria and yeasts, in general, and Neokomagataea sp. (formerly Gluconobacter sp.) and M. reukaufii, in particular, would engage in strongly antagonistic interactions in nectar and, as a consequence, rarely co-dominate. This study suggests that whichever arrives in a flower earlier than the other would dominate in that flower, suppressing the other (Tucker and Fukami, 2014). Furthermore, a field experiment using wild M. aurantiacus flowers (Toju et al., 2018) yielded results that are consistent with those of Tucker and Fukami (2014). Given that bacteria and yeasts are likely to affect nectar traits most extensively when they are abundant, these two earlier studies suggesting that bacteria and yeasts rarely co-dominate provide a biological justification for the lack of co-inoculation of bacteria and yeasts in our experimental design. We note, however, that it may be worthwhile in future research to try co-inoculation to better understand the effect of bacteria and yeasts on nectar traits.
Immediately after the inoculation, flowers were re-bagged. Three days following inoculation, flowers were clipped at the pedicel and stored in a cooler until nectar could be extracted. Individual flowers are typically open between 4 and 7 (Peay et al., 2012). We chose to harvest after 3 d to maximize recovery of inoculated flowers. Storage in the cooler lasted no longer than 3 h. In total, 222 flowers were inoculated over 7 d, with the experiment spanning 2 weeks during peak flowering during 2014, from 22 May to 6 June. We did not observe stigma closure on any bagged flower, indicating that pollen was not deposited on the stigma (Fetscher and Kohn, 1999), and thus flowers were probably not visited by pollinators.
Laboratory analyses
In the laboratory, the corolla tube was separated from the calyx, and nectar collected from each flower using a 20-µL microcapillary tube. The volume of each sample was quantified and the nectar diluted in 15 µL of sterile water. Flower status, including corolla condition (turgid or wilted/senescent), was noted during nectar extraction. Serial dilutions of nectar samples were plated (0.1 and 0.01 µL of original sample plated) to determine CFU density in each flower. Yeast-inoculated samples were plated on YMA without the antibacterial chloramphenicol, and bacterial-inoculated samples were plated on R2A without the antifungal cycloheximide, to assess whether additional micro-organisms were present. Sucrose-inoculated flowers were plated on both media types to ensure that no detectable microbes were present. The number of CFUs on each plate was counted after 5 d. We analysed 123 flowers inoculated with M. reukaufii, 33 flowers inoculated with Neokomagataea sp., and 40 flowers inoculated with sucrose solution.
Sugars and amino acids were also quantified from the same nectar samples. For sugars, 1 µL of remaining nectar was diluted in 49 µL of 50:50 acetonitrile:water containing 0.5 µg/µL–1 maltose as an internal standard. For each sample, 1 µL of the above dilution was injected and separated using ultra performance liquid chromatography (UPLC). Sugars were separated on a Luna amide column (50 × 2 mm, 3 µm, Phenomenex, Torrance, CA, USA) and quantitated using evaporative light scattering detection (Waters, Milford, MA, USA). Mono- and disaccharides detected in each sample (with elution times matching fructose, glucose, and sucrose) were quantified using a series of external standards. The remaining nectar was used to quantify amino acid content using Waters AccQ-Tag derivatization and standards, separated on an AccQTag Ultra Column (2.1 × 100 mm, 1.7 µm, Waters) and analysed using UV detection. However, only a sub-set of total samples could be used for all analyses due to microbial effects on nectar presence and volume. Amino acid identity was based on the retention time of a series of external standards and concentration calculated from standard curves.
Statistical analyses
We examined whether microbial CFUs and nectar characteristics within individual flowers varied among inoculation treatments using a linear mixed model, where plant individual and inoculation date were included as random effects, using the R package nlme v3.1 (Pinheiro et al., 2012). In each model, microbial inoculation treatment (yeasts, bacteria, or control) was used as a predictor. Separate models examined inoculation effects on log10-transformed total CFUs per flower, nectar volume, the concentrations of sucrose, glucose, and fructose, the proportion of monosaccharides [i.e. (glucose + fructose)/(glucose + fructose + sucrose)], and the total concentration of amino acids. We also analysed raw CFU counts using a negative binomial generalized linear mixed model (GLMM) using the package lme4 (Bates et al., 2015) followed by a likelihood ratio test to assess the significance of inoculation treatment. To assess whether inoculation treatment influenced the probability that a flower contained nectar following inoculation (yes or no), we used logistic regression with inoculation treatment as a predictor. We examined sources of variation in the amino acid composition in nectar using a multivariate analysis of variance (MANOVA) including microbe inoculation treatment and plant identity as predictors.
All analyses were performed in the R statistical environment (v.3.3.1, R Core Team, 2013).
RESULTS
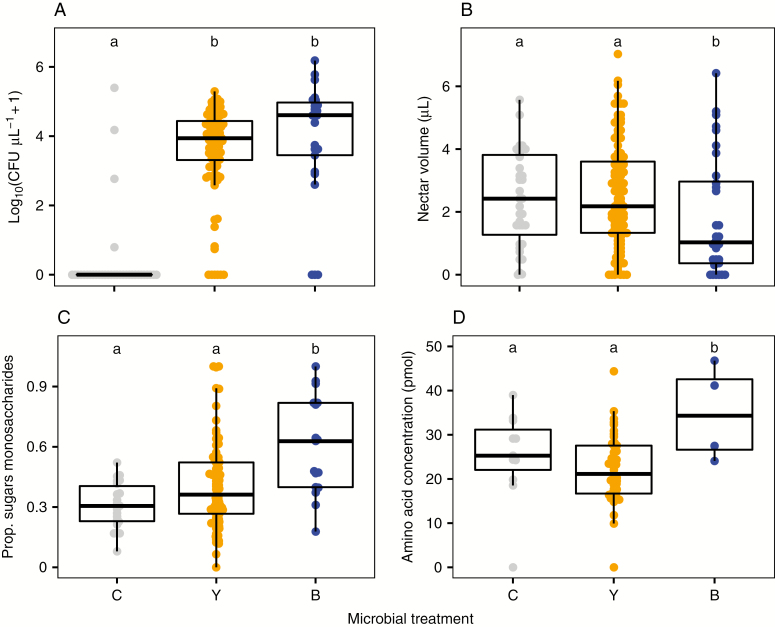
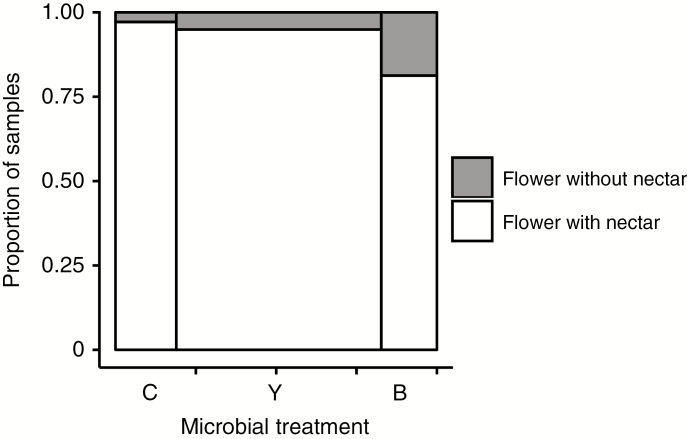
Treatments were effective, with focal microbes detected in 90 % of inoculated flowers, while <10 % of the uninoculated flowers had detectable microbial growth. When present, both yeasts and bacteria grew to high CFU densities in nectar (linear model Fig. 1A; F2,141 = 90.16, P < 0.0001; glmm χ2 = 21.5, P < 0.001) and influenced nectar characteristics (Fig. 1B–D), but the effects depended on the microbial species identity. Microbial inoculation influenced nectar volume (Fig. 1B; F2,156 = 3.8, P = 0.024). Specifically, flowers inoculated with Neokomagataea had, on average, 25 and 30 % lower nectar volume than flowers inoculated with yeast and sucrose control flowers, respectively, whereas yeasts did not significantly influence nectar volume (Fig. 1B). Microbial inoculation treatments did not significantly influence the concentration of sucrose (F2,87 = 1.62, P = 0.20), glucose (F2,87 = 1.80, P = 0.17) or fructose (F2,87 = 1.9, P = 0.16) in floral nectar. However, microbial inoculation influenced the ratio of nectar sugars (Fig. 1C; F2,91= 12.5, P < 0.001). Neokomagataea doubled the proportion of monosaccharides in nectar compared with the control, whereas yeast inoculation did not significantly affect the proportion of monosaccharides in nectar (Fig. 1C). Microbial inoculation also influenced amino acid concentration (F2,42 = 4.95, P = 0.01). Neokomagataea-inoculated flowers increased the concentration of amino acids in nectar compared with sucrose- or yeast-inoculated flowers (Fig. 1D). Finally, inoculation with Neokomagataea increased the probability that flowers contained no nectar [Fig. 2; χ2 = (2) = 6.26, P=0.044]. Flowers without nectar, particularly in the bacterial inoculation treatment, were also typically wilted and browning.
Fig. 1.
Effects of microbial inoculation treatment on nectar characteristics 3 d after inoculation with yeast (Y), bacteria (B) or a sucrose control (C). Measured characteristics included (A) log10 (microbial CFUs + 1) in nectar assessed on YM or R2A plates; (B) total nectar volume (µL) per flower; (C) the proportion of monosaccharides in nectar (fructose + glucose)/(fructose + glucose + sucrose); and (D) the total amino acid concentration measured in nectar (pmol). The midline in the box represents the median, and edges represent Q1 and Q3. Letters in each panel represent treatments that differed (P < 0.05 based on a Tukey HSD test).
Fig. 2.
Mosaic plot indicating the frequency of measurable nectar (yes >0 µL of nectar per flower) in flowers that were inoculated with sucrose control (C), yeast (Y) or bacteria (B) 3 d previously. Flowers without nectar were also wilted and browned. n = 185 recovered flowers (some were lost due to wind or florivory). Treatments differed at P = 0.015. Bar width indicates the relative number of replicates within each treatment group.
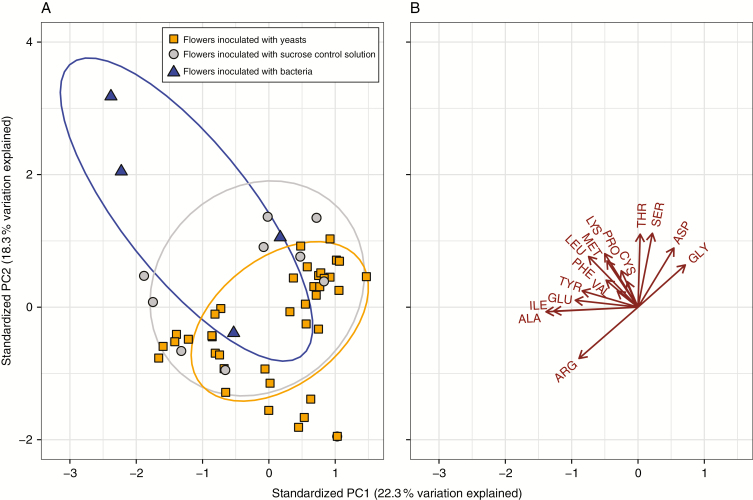
Inoculation treatment and plant identity were the main sources of variation in amino acid composition within floral nectar (Fig. 3; MANOVA Taxa Pillai’s trace2,42 = 1.16, P = 0.001; Plant ID Pillai’s trace15,42 = 5.38, P = 0.00028). The MANOVA indicated that proline, lysine, methionine, leucine, phenylalanine and isoleucine were reduced with yeast inoculation. Neokomagataea tended to increase the concentration of some amino acids in nectar, but the amino acid composition in Neokomagataea-inoculated flowers was not statistically distinguishable from that in sucrose-inoculated flowers, which may in part reflect the small sample size for Neokomagataea-inoculated flowers.
Fig. 3.
Effect of microbial inoculation on the composition of amino acids in nectar. (A) Ovals indicate the 95 % confidence interval for taxonomic groups (yeasts, bacteria, or sucrose control). MANOVA treatment P < 0.0001. (B) Letters indicate amino acid vectors, with arrows pointing towards samples with a higher abundance of amino acids.
DISCUSSION
Our results indicate that the dominant species of yeasts and bacteria in our nectar system cause contrasting changes in floral characteristics, thereby contributing to increased variation in nectar chemistry among flowers. For example, bacterial inoculation more strongly affected sugar composition and the presence of floral nectar than did yeast inoculation, similar to ex situ studies using M. aurantiacus nectar (Vannette et al., 2013). On the other hand, yeasts decreased the concentration of specific amino acids, while bacteria tended to increase amino acid concentration overall. Those amino acids that were reduced by yeasts were largely overlapping with those previously documented (Peay et al., 2012; Vannette and Fukami, 2014, Tucker and Fukami, 2014). Microbe-induced changes in nectar characteristics may result from microbial metabolism or plant response to microbial presence. For example, bacterial-induced increases in proline, among other amino acids, in floral nectar (Figs. 1D and 3) might suggest plant resorption of floral nectar and the concentration of solutes or, alternatively, plant stress response (Fabro et al., 2004). The observation of reduced nectar volume and increased floral senescence following bacterial inoculation (Fig. 2) may also suggest plant stress and subsequent floral senescence. This observation suggests that plants may respond to bacterial growth in nectar, and that at least some bacterial species can directly inflict biotic stress and damage on plant tissues, as has been previously documented in Malus–pathogen interactions (Venisse et al., 2002). Further work is required to examine whether other species of floral bacteria or yeasts elicit similar plant responses, and whether co-inoculation with two or more microbial species attenuates or increases these effects.
Microbe-driven divergence in nectar traits may introduce variation in floral attractiveness to particular floral visitors. For example, bumble-bees are attracted to yeast-colonized flowers (Herrera et al., 2013; Schaeffer and Irwin, 2014), while bacterial growth can reduce visitation by diverse floral visitors (Vannette et al., 2013; Good et al., 2014). Specifically, variation in amino acid composition (Inouye and Waller, 1984; Carter et al., 2006) and nectar presence and volume (or indicators of these properties) influence pollinator choice, even among individual flowers (von Arx et al., 2012). Because floral visitor species vary in attraction to particular nectar characteristics, visitor communities to flowers colonized by different microbial communities may diverge, even within the same plant species. The resulting heterogeneity in microbial communities and nectar characteristics among flowers of the same species may in turn increase the species diversity of floral visitors. These scenarios remain speculative, but suggest that it might be difficult to explain the abundance and distribution of plants, pollinators, or nectar microbes if interactions among the three groups were not considered simultaneously.
CONCLUSIONS
We found that micro-organisms could influence nectar characteristics and increase variability among individual flowers in the wild. Our results suggest that microbial communities that vary in species composition can also vary in their effects on plant traits. The ecological consequences of this variation on plants, pollinators, and micro-organisms remain to be fully explored.
DATA ACCESSIBILITY
Accession numbers for sequence data are included in the article.
ACKNOWLEDGEMENTS
We thank Nona Chiariello and other JRBP staff for logistical support, Robert Schaeffer and other members of the Vannette lab for comments, and Kaoru Tsuji for help with analysis. R.L.V. and T.F. designed the experiment. R.L.V. performed the experiment, collected and analysed data, and wrote the first draft of the manuscript. R.L.V. and T.F. revised the manuscript. We acknowledge support from the NSF (DEB 1149600, DEB 1737758) and the Terman Fellowship and the Department of Biology at Stanford University. R.L.V. was supported by the Gordon and Betty Moore Foundation through the Life Sciences Research Fellowship (GBMF 2550.02) and a USDA Hatch award (NE1501).
LITERATURE CITED
- Álvarez-Pérez S, Herrera C. 2013. Composition, richness and nonrandom assembly of culturable bacterial–microfungal communities in floral nectar of Mediterranean plants. FEMS Microbiol Ecol 83: 685–699. [DOI] [PubMed] [Google Scholar]
- von Arx M, Goyret J, Davidowitz G, Raguso RA. 2012. Floral humidity as a reliable sensory cue for profitability assessment by nectar-foraging hawkmoths. Proceedings of the National Academy of Sciences, USA 109: 9471–9476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates D, Maechler M, Bolker B, Walker S. 2015. Fitting linear mixed-effects models using lme4. Journal of Statistical Software 67: 1–48. [Google Scholar]
- Belisle M, Peay KG, Fukami T. 2012. Flowers as islands: spatial distribution of nectar-inhabiting microfungi among plants of Mimulus aurantiacus, a hummingbird-pollinated shrub. Microbial Ecology 63: 711–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biernaskie J, Cartar R. 2004. Variation in rate of nectar production depends on floral display size: a pollinator manipulation hypothesis. Functional Ecology 18: 125–129. [Google Scholar]
- Canto A, Herrera CM. 2012. Micro-organisms behind the pollination scenes: microbial imprint on floral nectar sugar variation in a tropical plant community. Annals of Botany 110: 1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canto A, Perez R, Medrano M, Castellanos MC, Herrera CM, García IM. 2007. Intra-plant variation in nectar sugar composition in two Aquilegia species (Ranunculaceae): Contrasting patterns under field and glasshouse conditions. Annals of Botany 99: 653–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canto A, Herrera CM, Medrano M, Perez R, García IM. 2008. Pollinator foraging modifies nectar sugar composition in Helleborus foetidus (Ranunculaceae): an experimental test. American Journal of Botany 95: 315–320. [DOI] [PubMed] [Google Scholar]
- Canto A, Herrera CM, García IM, Pérez R, Vaz M. 2011. Intraplant variation in nectar traits in Helleborus foetidus (Ranunculaceae) as related to floral phase, environmental conditions and pollinator exposure. Flora-Morphology, Distribution, Functional Ecology of Plants 206: 668–675. [Google Scholar]
- Canto A, Herrera CM, García IM, García M, Bazaga P. 2015. Comparative effects of two species of floricolous Metschnikowia yeasts on nectar. Anales del Jardín Botánico de Madrid 71: e019. [Google Scholar]
- Carter C, Shafir S, Yehonatan L, Palmer RG, Thornburg R. 2006. A novel role for proline in plant floral nectars. Naturwissenschaften 93: 72–79. [DOI] [PubMed] [Google Scholar]
- Cnaani J, Thomson JD, d Papaj DR. 2006. Flower choice and learning in foraging bumblebees: effects of variation in nectar volume and concentration. Ethology 112: 278–285. [Google Scholar]
- Dhami MK, Hartwig T, Fukami T. 2016. Genetic basis of priority effects: insights from nectar yeast. Proceedings of the Royal Society B: Biological Sciences 283: 20161455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhami MK, Hartwig T, Letten AD, Banf M, Fukami T. 2018. Genomic diversity of a nectar yeast clusters into metabolically, but not geographically, distinct lineages. Molecular Ecology (in press). [DOI] [PubMed] [Google Scholar]
- Fabro G, Kovács I, Pavet V, Szabados L, Alvarez ME. 2004. Proline accumulation and AtP5CS2 gene activation are induced by plant–pathogen incompatible interactions in Arabidopsis. Molecular Plant-Microbe Interactions 17: 343–350. [DOI] [PubMed] [Google Scholar]
- Fetscher AE, Kohn JR. 1999. Stigma behavior in Mimulus aurantiacus (Scrophulariaceae). American Journal of Botany 86: 1130–1135. [PubMed] [Google Scholar]
- Good AP, Gauthier M-PL, Vannette RL, Fukami T. 2014. Honey bees avoid nectar colonized by three bacterial species, but not by a yeast species, isolated from the bee gut. PLoS One 9: e86494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera CM, Pozo MI. 2010. Nectar yeasts warm the flowers of a winter-blooming plant. Proceedings of the Royal Society B: Biological Sciences 277: 1827–1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera CM, Perez R, Alonso C. 2006. Extreme intraplant variation in nectar sugar composition in an insect-pollinated perennial herb. American Journal of Botany 93: 575–581. [DOI] [PubMed] [Google Scholar]
- Herrera CM, Garcia IM, Perez RF. 2008. Invisible floral larcenies: microbial communities degrade floral nectar of bumble bee-pollinated plants. Ecology 89: 2369–2376. [DOI] [PubMed] [Google Scholar]
- Herrera CM, de Vega C, Canto A, Pozo ML. 2009. Yeasts in floral nectar: a quantitative survey. Annals of Botany 103: 1415–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera CM, Pozo MI, Medrano M. 2013. Yeasts in nectar of an early-blooming herb: sought by bumble bees, detrimental to plant fecundity. Ecology 94: 273–279. [DOI] [PubMed] [Google Scholar]
- Herrera CM, Pozo MI, Bazaga P. 2014. Nonrandom genotype distribution among floral hosts contributes to local and regional genetic diversity in the nectar-living yeast Metschnikowia reukaufii. FEMS Microbiology Ecology 87: 568–575. [DOI] [PubMed] [Google Scholar]
- Inouye DW, Waller GD. 1984. Responses of honey bees (Apis mellifera) to amino acid solutions mimicking floral nectars. Ecology 65: 618–625. [Google Scholar]
- Junker RR, Romeike T, Keller A, aLangen D. 2014. Density-dependent negative responses by bumblebees to bacteria isolated from flowers. Apidologie 45: 467–477. [Google Scholar]
- Mitchell RJ. 2004. Heritability of nectar traits: why do we know so little?Ecology 85: 1527–1533. [Google Scholar]
- Peay KG, Belisle M, Fukami T. 2012. Phylogenetic relatedness predicts priority effects in nectar yeast communities. Proceedings of the Royal Society B: Biological Sciences 279: 749–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinheiro J, Bates D, DebRoy S, Sarkar D.2012. https://CRAN.R-project.org/package=nlme nlme: linear and nonlinear mixed effects models.
- R Core Team 2013. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: URL http://www.R-project.org/ [Google Scholar]
- Reis VM, Teixeira KR. 2015. Nitrogen fixing bacteria in the family Acetobacteraceae and their role in agriculture. Journal of Basic Microbiology 55: 931–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rering CC, Beck JJ, Hall G, McCarthy M, Vannette RL, 2018. Nectar-inhabiting microorganisms influence nectar volatile composition and attractiveness to a generalist pollinator. New Phytologist (in press). [DOI] [PubMed] [Google Scholar]
- Samuni-Blank M, Izhaki I, Laviad S, Bar-Massada A, Gerchman Y, Halpern M. 2014. The role of abiotic environmental conditions and herbivory in shaping bacterial community composition in floral nectar. PLoS One 9: e99107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer R, Irwin RE. 2014. Yeasts in nectar enhance male fitness in a montane perennial herb. Ecology 95: 1792–1798. [DOI] [PubMed] [Google Scholar]
- Toju H, Vannette RL, Gauthier MPL, Dhami MK, Fukami T. 2018. Priority effects can persist across floral generations in nectar microbial metacommunities. Oikos (in press) [Google Scholar]
- Tsuji K, Dhami MK, Cross DJR, Rice CP, Romano NH, Fukami T. 2016. Florivory and pollinator visitation: a cautionary tale. AoB PLANTS 8: plw036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker CM, Fukami T. 2014. Environmental variability counteracts priority effects to facilitate species coexistence: evidence from nectar microbes. Proceedings of the Royal Society of London B: Biological Sciences 281: 20132637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannette RL, Fukami T. 2014. Historical contingency in species interactions: towards niche-based predictions. Ecology Letters 17:115–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannette RL, Fukami T. 2016. Nectar microbes can reduce secondary metabolites in nectar and alter effects on nectar consumption by pollinators. Ecology 97: 1410–1419. [DOI] [PubMed] [Google Scholar]
- Vannette RL, Fukami T. 2017. Dispersal enhances beta diversity in nectar microbes. Ecology Letters 20: 901–910. [DOI] [PubMed] [Google Scholar]
- Vannette RL, Gauthier M-PL, Fukami T. 2013. Nectar bacteria, but not yeast, weaken a plant–pollinator mutualism. Proceedings of the Royal Society B: Biological Sciences 280: e20122601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vega C, Herrera CM. 2012. Relationships among nectar-dwelling yeasts, flowers and ants: patterns and incidence on nectar traits. Oikos 121: 1878–1888. [Google Scholar]
- de Vega C, Herrera CM. 2013. Microorganisms transported by ants induce changes in floral nectar composition of an ant-pollinated plant. American Journal of Botany 100: 792–800. [DOI] [PubMed] [Google Scholar]
- Venisse J-S, Malnoy M, Faize M, Paulin J-P, Brisset M-N. 2002. Modulation of defense responses of Malus spp. during compatible and incompatible interactions with Erwinia amylovora.Molecular Plant-Microbe Interactions 15: 1204–1212. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Accession numbers for sequence data are included in the article.