Abstract
The use of proteins that bind and catalyze reactions with DNA alongside DNA nanostructures has broadened the functionality of DNA devices. DNA binding proteins have been used to specifically pattern and tune structural properties of DNA nanostructures and polymerases have been employed to directly and indirectly drive structural changes in DNA structures and devices. Despite these advances, undesired and poorly understood interactions between DNA nanostructures and proteins that bind DNA continue to negatively affect the performance and stability of DNA devices used in conjunction with enzymes. A better understanding of these undesired interactions will enable the construction of robust DNA nanostructure-enzyme hybrid systems. Here, we investigate the undesired disassembly of DNA nanotubes in the presence of viral RNA polymerases (RNAPs) under conditions used for in vitro transcription. We show that nanotubes and individual nanotube monomers (tiles) are non-specifically transcribed by T7 RNAP, and that RNA transcripts produced during non-specific transcription disassemble the nanotubes. Disassembly requires a single-stranded overhang on the nanotube tiles where transcripts can bind and initiate disassembly through strand displacement, suggesting that single-stranded domains on other DNA nanostructures could cause unexpected interactions in the presence of viral RNA polymerases.
INTRODUCTION
Nucleic acid nanotechnology takes advantage of the reliable nature of Watson-Crick base pairing and our knowledge of nucleic acid structure to build molecular devices and sensors and direct the self-assembly of DNA and RNA materials. Numerous one-, two- and three-dimensional DNA nanostructures have been assembled (1–7) and these structures have been functionalized with a variety of proteins (8), nanoparticles (9–12), and other small molecules (13–15). Additionally, chemical logic circuits, timers, and spatial patterns have been constructed via DNA strand-displacement reactions (16–18) and these systems have been used to control DNA devices (16,19).
Often, the integration of proteins that interact with nucleic acids can lead to more functional nucleic acid systems. Recombinases, topoisomerases, helicases, ligases, and relaxases have been utilized to induce structural and mechanical changes in nucleic acid devices (20–26). DNA polymerases have been used to induce structural changes in DNA nanostructures (20,27), replicate nanostructures (28), and build a range of dynamic chemical reaction networks (29–35). RNA polymerases, such as T7 RNA polymerase, have been used to transcribe self-assembling RNA nanostructures (36–39) and materials (28), drive molecular motion and structural changes in DNA nanostructures (40,41), and engineer dynamic chemical reaction networks (42–44). These applications take advantage of specific reactions that occur between nucleic acids and proteins, but often unintended interactions arise when coupling DNA devices and DNA binding proteins which can hamper device functionality (20,27,40). To continue to expand the functionality of nucleic acid devices, an understanding of the non-specific or unintended interactions between DNA nanostructures and DNA binding proteins is imperative.
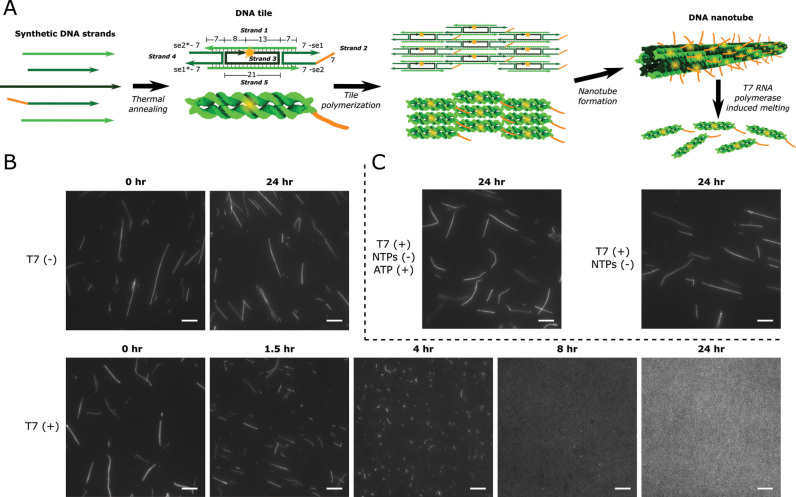
Here, we elucidate non-specific and unintended interactions that arise between viral RNA polymerases and DNA nanostructures. These interactions lead to the disassembly of the DNA nanotubes in the presence of viral RNAPs. Viral RNAPs are highly processive, monomeric enzymes that require no accessory factors to conduct transcription (45). Given the relative simplicity of viral RNAPs and the knowledge of their structure and function (46), these enzymes are utilized for many in vitro and in vivo applications, many pertaining to nucleic acid nanotechnology (36,40,42,47). DNA nanotubes are model nanostructures for understanding DNA self-assembly (15,48–50) and mechanics (51,52) that self-assemble from DNA monomers termed tiles. The tiles are composed of two parallel DNA double helices rigidly connected by double crossover junctions; tile polymerization occurs via hybridization of single-stranded domains termed sticky ends (Figure 1A) (1).
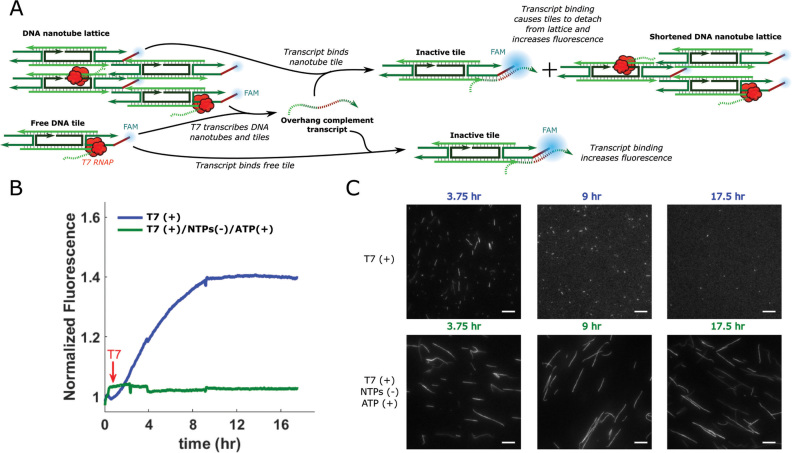
Figure 1.
DNA nanotube design and disassembly in the presence of 8.6 U/μl T7 RNAP in transcription conditions. (A) DNA nanotube structure and assembly. Left: DNA tiles are DAE-E double crossover molecules (71) that assemble from 5 strands of synthetic DNA. Tiles consist of two DNA helices that are rigidly bound by two crossover motifs with four single-stranded sticky end (se) regions on the two helix ends. Sticky ends are complementary across the diagonal of the tile (complementary sequences denoted by *) (1). Tiles also contain a single-stranded overhang domain on strand 2 (orange) and a Cy3 fluorophore (yellow) at the 5′ end of strand 3. Numbers next to domains indicate domain lengths in number of bases. Middle: The complementary sticky ends specifically program the DNA tiles to form the lattice shown as an abstract line representation (top) and a 3D rendering (bottom). Right: The lattice cyclizes to form nanotubes (1). (B) Fluorescence micrographs of DNA nanotubes (var1_7—SI Section 1) before and after incubation with (T7 (+)) or without (T7 (–)) T7 RNAP in transcription conditions. (C) Nanotube stability (var1_7) in the presence of T7 RNAP with ATP (ATP (+)) as the only NTP or without any NTPs (NTPs (–)). Scale bars: 10 μm.
Here we observe that DNA nanotubes disassemble when incubated with either T7 RNAP or SP6 RNAP. To understand why this unintended disassembly occurs, we characterize how viral RNAPs and DNA nanotubes interact and find that DNA nanotubes and tiles are transcribed non-specifically by T7 RNAP. Furthermore, we show that RNA transcripts produced from the DNA nanotubes induce nanotube disassembly even in the absence of active T7 RNAP, likely through a toehold-mediated branch migration process initiated at single-stranded overhangs on the tiles. In addition, we show that nanotube disassembly is not particularly dependent on the sequence of the DNA nanotubes but does depend on the relative concentrations of RNAP, DNA tiles, and double-stranded DNA containing RNAP promoter sequences. Viral RNAPs have been shown to non-specifically bind (53) and transcribe (54) a variety of DNA sequences, but non-specific transcription of DNA nanostructures has not been previously reported. Off-target viral RNAP transcription could significantly affect the structure and function of many other synthetic DNA devices as many DNA nanostructures necessarily contain single-stranded domains as sites for binding other DNA structures, DNA-functionalized nanoparticles, or small molecules (9,12,55–57). Our elucidation of the mechanism underlying RNAP-induced nanotube disassembly should enable the development of targeted design strategies to reduce these unintended reactions for DNA nanostructures, a number of which are discussed.
MATERIALS AND METHODS
DNA oligonucleotides and enzymes
All DNA oligonucleotides were purchased from Integrated DNA Technologies, Inc (IDT). Modified DNA was ordered HPLC purified. Non-modified DNA was ordered PAGE purified. Sequences are listed in SI Section 1. T7 RNAP was purchased from Cellscript (200 U/μl, Catalog # C-T7300K), SP6 RNAP from ThermoFisher Scientific (100 U/μl, Catalog # EP0133), yeast inorganic pyrophosphatase from New England Biolabs (NEB) (0.1 U/μl, Catalog # M2403S) and DNase I from NEB (2 U/μl, Catalog # M0303S). RNase A was taken from a Qiagen mini-prep kit (10 mg/ml). All reactions (unless otherwise stated) were conducted using NEB RNAPol Reaction Buffer (Catalog # M0251S).
Measurement of nanotube stability
To measure DNA nanotube stability in the presence of T7 RNAP, the nanotubes were assembled, incubated with T7 RNAP, and imaged via fluorescence microscopy. DNA nanotubes were assembled by annealing 5 μM of each tile strand in NEB RNAPol Reaction Buffer and following the nanotube annealing protocol (SI Section 2). The nanotubes were then incubated at 37°C with T7 RNAP in NEB RNAPol Reaction Buffer, which was supplemented with ribonucleotide triphosphates (NTPs) and additional MgCl2 (these reaction conditions are referred to as ‘transcription conditions’ throughout the text). The final concentrations of each component in the transcription conditions were as follows: 1 μM DNA tiles, 20 mM MgCl2, 8.6 U/μl (4.3% v/v) T7 RNAP and 5 mM of each NTP (ATP, UTP, GTP and CTP). Control experiments without T7 RNAP were performed under otherwise identical conditions. Fluorescence micrographs were obtained by diluting an aliquot of a sample 20-fold in NEB RNAPol Reaction Buffer and pipetting onto an untreated glass coverslip (Fisher Scientific—Catalog # 12-545-E). Fluorescence micrographs were scaled such that the minimum intensity pixel in the image is shown in black and the maximum intensity pixel is shown in white. The grayscale intensity of the other pixels was scaled linearly between the minimum and maximum intensity values. Due to this conversion, images of disassembled nanotubes may appear to have a higher background.
Characterization of RNA transcripts
RNA was identified by incubating DNA nanotubes with T7 RNAP in transcription conditions (except T7 RNAP was at a concentration of 4.3 U/μl (2.1% v/v), and yeast inorganic pyrophosphatase and biotinylated bovine serum albumin (BSA) (58) were present at concentrations of 0.00135 U/μl (1.35% v/v) and 0.1 mg/ml, respectively), digesting nanotube DNA with DNase I, and detecting RNA products via non-denaturing PAGE. Samples were incubated at 37°C for 20 h, heated to 65°C for 30 min to denature T7 RNAP, and cooled to 37°C after which each sample was split into two aliquots. DNase I was added to one aliquot to a final concentration of 0.21 U/μl (10.5% v/v); the samples were then both incubated for 2 h at 37°C. For samples also digested with RNase A, RNase A was added to a final concentration of 0.12 U/μl (17.4% v/v) after DNase I digestion. The samples were then incubated at 37°C for 2.5 h (SI Section 2 for PCR protocol). Samples were subsequently run on a 10% non-denaturing polyacrylamide gel (Tris-acetate-EDTA buffer with 12.5 mM magnesium acetate) at 100 V for 2 h. Gels were run with a 10 bp DNA ladder (ThermoFisher Scientific) and stained with SYBR Gold. The gel was imaged using a Syngene EF2 G:Box gel imager equipped with a blue light transilluminator (emission max ∼450 nm) and a UV032 filter (bandpass 572–630 nm).
Detecting tile-specific sequences in RNA with fluorescent DNA probes
To determine if the RNA produced from DNA nanotube incubation with T7 RNAP contained DNA tile sequences, a fluorescent probe containing tile sequences was annealed with the RNA and the mobility of the probe was assessed with non-denaturing PAGE. RNA was generated and T7 RNAP was denatured using the same incubation protocol described for Characterization of RNA transcripts. After T7 RNAP denaturation, two aliquots were prepared with DNase I added to both as described in Characterization of RNA transcripts. Additionally, RNase A was added to one of the aliquots to a final concentration of 0.12 U/μl (17.4% v/v). Samples were incubated at 37°C for 24 h, then held at 75°C for 30 min to denature DNase I. Cy3-labeled DNA probes were then added to the samples, followed by cooling from 90°C to 25°C at a rate of –1°C/min (SI Section 2 contains PCR protocol details). Samples were analyzed with PAGE as described above in Characterization of RNA transcripts. Gels were imaged prior to staining with SYBR Gold to detect only the Cy3 labeled probe, and then again after staining to detect all nucleic acids. For samples that served as size markers on the gels (strand 1:strand 3 and strand 5:strand 3 complexes), the same incubation procedures were followed but T7 RNAP or other enzymes were not added to the samples.
Detecting binding between the RNA transcripts and the DNA nanotube single-stranded overhang domain
To detect hybridization between the RNA transcripts produced from DNA nanotubes and the single-stranded overhang domain on the nanotubes over time, var1_7-3 nanotubes (which have a FAM modification at the 5′ end of the single-stranded overhang) were incubated at 37°C with T7 RNAP in transcription conditions. During the incubation, the fluorescence of the sample was measured in a quantitative PCR machine (Stratagene MxPro3000, FAM filter: 492–516 nm). To avoid saturating the fluorescence signal, 25% of the strands with the single-stranded overhang domain had a FAM modification at their 5′ end while the other 75% of these strands were the same sequence but did not have a FAM modification. Fluorescence readings were taken every minute. Aliquots were taken periodically from the samples to measure nanotube stability using fluorescence microscopy as described in Measurement of nanotube stability.
RESULTS
DNA nanotubes disassemble in the presence of active viral RNA polymerases
To characterize the interaction between viral RNAPs and DNA nanotubes, we first tested the stability of DNA nanotubes with nanotubes we called var1_7 (see SI Section 1 for sequences). To ensure thermal stability at 37°C, these nanotubes were designed with longer sticky ends than previously developed versions (Figure 1A) (1). The nanotubes also contained a single-stranded overhang on strand 2 of the DNA tiles for use with another project relating to dynamic activation and deactivation of tiles (59). We also found that the single-stranded overhang domain aided in the efficient assembly of nanotubes in transcription buffer (Supplementary Figure S1 and SI Section 3).
The assembled nanotubes were incubated at 37°C in transcription conditions with and without T7 RNAP. Without T7 RNAP, the nanotubes were still intact after 24 h (Figure 1B). However, when these nanotubes were incubated under the same conditions with T7 RNAP, no nanotubes were visible in fluorescence micrographs after 24 h (Figure 1B). The addition of heat-denatured T7 RNAP did not disassemble nanotubes over 24 h (Supplementary Figure S2), suggesting active T7 RNAP is required to induce nanotube disassembly. Nanotubes also disassembled in the presence of SP6 RNAP, another viral RNA polymerase (SI Section 5). Additionally, RNAP-induced disassembly was dependent on the concentrations of the DNA tiles (Supplementary Figure S3) and RNAP used (Supplementary Figure S4).
T7 RNAP-induced disassembly of DNA nanotubes is not sequence specific
We hypothesized that viral RNA polymerases might induce DNA nanotube disassembly because the polymerases bind to specific sequences within the DNA tiles due to a similarity of these sequences to the RNAP promoter sequences. T7 RNAP has been shown to melt 6–7 bases of promoter region DNA to create a pre-initiation complex (60), so T7 RNAP could conceivably bind to nanotubes and cause disassembly by creating these transcriptional bubbles near the hybridized sticky ends of the nanotubes. To reduce the possibility of a specific binding interaction between viral RNA polymerases and the nanotubes, we re-designed the tile sequences to produce a new variant (var2_7) that had less overlap with the promoter sequences of the viral RNAPs than the original tile sequences (See SI Section 7 and Supplementary Figure S5 for the design process and SI Section 1 for tile sequences). However, we found that the var2_7 nanotubes were no more stable than the var1_7 nanotubes in the presence of T7 or SP6 RNAP (Supplementary Figure S6). These results suggest that RNAP-induced nanotube disassembly is not due to specific recognition of promoter-like sequences in the DNA tiles.
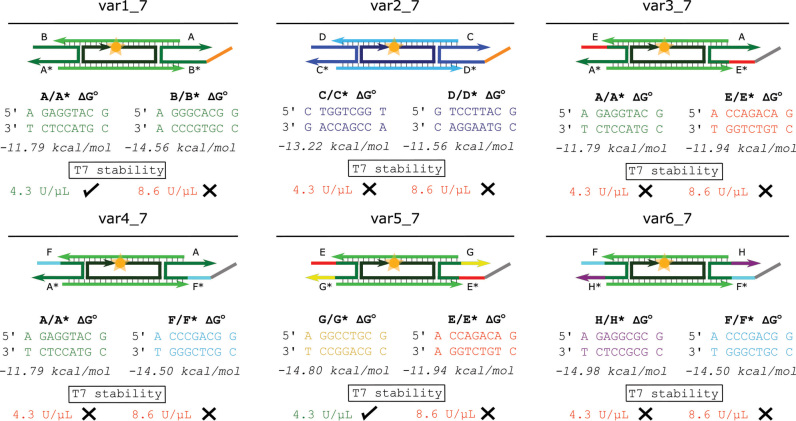
We next hypothesized that non-specific binding might be inducing disassembly. T7 RNAP, for example, binds to its preferred promoter sequence only slightly more strongly than to other sequences (53,61) so off-target binding of T7 RNAP to the nanotubes might cause sticky end melting and subsequent nanotube disassembly. If this were the case, increasing the hybridization energy of the tile sticky ends might prevent polymerases from melting hybridized sticky ends along the nanotubes, thus preventing disassembly. To test whether sticky end hybridization energy influenced the stability of nanotubes in the presence of T7 RNAP, we designed four new tile variants where the sticky end sequences had different hybridization energies (var3_7–var6_7—Figure 2 and SI Section 1) and incubated each of these nanotube variants for 24 h at two different concentrations of T7 RNAP. Fluorescence micrographs of the nanotubes taken after 24 h showed that each of the new variants disassembled when incubated with the higher RNAP concentration and only var5_7 nanotubes were still intact after incubation with the lower RNAP concentration (Figure 2 and Supplementary Figure S7). These tests suggested that increasing sticky end strength does not significantly increase nanotube stability in the presence of T7 RNAP.
Figure 2.
The sticky end sequences, sticky end energies, and the stability in the presence of T7 RNAP in transcription conditions of nanotubes with six different 7 bp sticky end sequences. The domains of variants var2_7 to var6_7 whose sequences are different from the sequences of the respective domains in var1_7 are shown in different colors than the domains in var1_7 (SI Section 1 for sequences). Free energies were estimated for comparison using NUPACK (72) (SI Section 8.1). Nanotubes were deemed stable (check) at the enzyme unit concentration listed if nanotubes at 1 μM tile concentration were present after 24 h of incubation with T7 RNAP in transcription conditions and were deemed unstable (X) if no nanotubes were present after this incubation. Fluorescence micrographs of the nanotubes after 24 h of incubation with and without T7 RNAP are shown in Supplementary Figure S7.
To further test the influence of the hybridization energy of nanotube sticky ends on nanotube stability in the presence of active T7 RNAP, we developed another set of nanotubes variants based on the var1_7 and var3_7 variants. These variants were altered so that the tiles contained either 6 or 8 bp sticky ends (Supplementary Figure S8). We characterized the stability of each of these variants and found no clear relationship between sticky end length and stability in the presence of T7 RNAP (Supplementary Figures S8 and S9). Taken together, these results suggest that changing the sequences of tiles is unlikely to increase the stability of nanotubes in the presence of viral RNAPs.
NTPs are required for T7 RNAP-induced DNA nanotube disassembly
To further investigate the mechanism underlying nanotube disassembly in the presence of active viral RNA polymerases, we next examined which aspects of RNAP activity were required to induce nanotube disassembly, using var1_7 nanotubes and T7 RNAP as a model system.
Since T7 RNAP has been shown to non-specifically transcribe DNA at high enzyme and template concentrations (54), we investigated whether transcription is involved in T7 RNAP-induced DNA nanotube disassembly. We first tested whether NTPs are required for T7 RNAP-induced nanotube disassembly to occur. Since not including NTPs in the solution would significantly change the free magnesium concentration of the samples, which could also influence nanotube stability (Supplementary Figure S10), we compared nanotube stability in the presence of T7 RNAP with all four NTP types present at 5 mM each to nanotube stability with only one type of NTP present at 20 mM. This comparison kept the free magnesium concentration the same between the samples, but only allowed transcription in the sample with all of the NTP types.
While we had previously observed that var1_7 nanotubes disassembled after 24 h of incubation with all four NTPs and 8.6 U/μl of T7 RNAP, these same nanotubes did not disassemble after 24 h with either 20 mM of only ATP (Figure 1C), only CTP, only UTP, or only GTP (Supplementary Figure S11 and SI Section 10) and 8.6 U/μl of T7 RNAP. Similarly, nanotubes did not disassemble after 24 h with T7 RNAP when NTPs were not present (Figure 1C). These results suggest that transcription is required for T7 RNAP-induced nanotube disassembly.
RNA is produced when DNA nanotubes and tiles are incubated with T7 RNAP
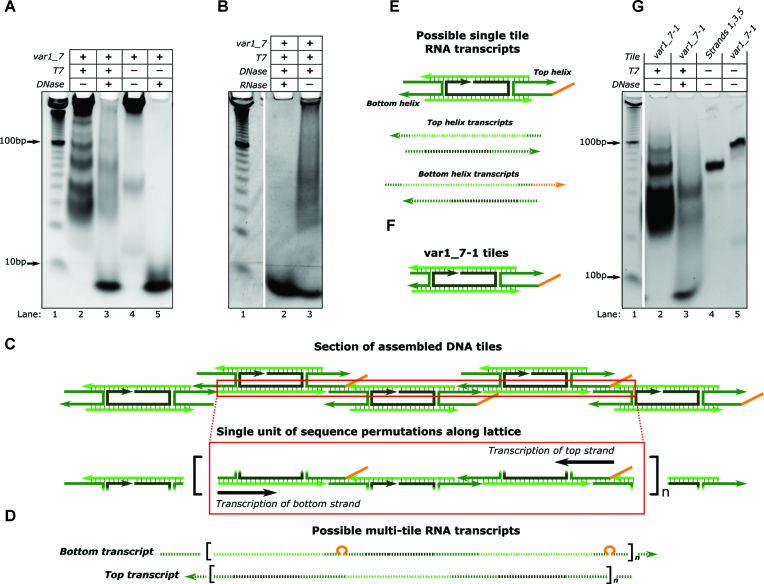
Since T7 RNAP-induced DNA nanotube disassembly was only observed when all four NTP types were present, we hypothesized that DNA nanotubes might act as transcription substrates for T7 RNAP and that some aspect of the transcription process might be causing disassembly. To begin to test this hypothesis, we investigated whether RNA was produced when T7 RNAP and DNA nanotubes were incubated together in transcription conditions. We incubated DNA nanotubes with T7 RNAP in transcription conditions for 20 h and heat denatured the sample to remove any enzyme bound to the DNA. We then split the sample in half and digested one aliquot with DNase I to remove all nanotube DNA (62). The samples were analyzed via polyacrylamide gel electrophoresis and stained with SYBR Gold to visualize both RNA and DNA.
A range of products, migrating more slowly than 10 bp DNA migrates, were observed in the stained gel for both aliquots (Figure 3A—lanes 2 and 3), but not for a control sample of nanotubes incubated without T7 RNAP (Figure 3A—lane 4). The complete absence of products in the lane containing DNA nanotubes and DNase I but no T7 RNAP (Figure 3A—lane 5) suggested that the products in Figure 3A—lane 3 were not leftover nanotube DNA. These products were not present if DNA nanotubes, active T7 RNAP, or NTPs were left out of the incubation mixture (Supplementary Figure S12), suggesting that all of these components are required for the products in lane 3 of Figure 3A to be present. Additionally, the products were not observed after nanotubes incubated with T7 RNAP in transcription conditions were treated with DNase I and then RNase A (Figure 3B). This treatment should degrade all DNA and RNA species (63). Together, these controls verified that the products observed in Figure 3A—lane 3 were RNA.
Figure 3.
RNA is produced when DNA nanotubes are incubated with T7 RNAP in transcription conditions. (A and B) Non-denaturing PAGE results showing samples of nanotubes incubated with or without T7 RNAP (T7) and treated with or without DNase I (DNase) and RNase A (RNase) as shown. (C–F) DNA: solid lines, RNA: dashed lines. (C) Schematic of part of a DNA nanotube lattice. The red box encloses a single DNA duplex along the lattice that T7 RNAP could transcribe. This duplex is repeated along the length of the lattice. (D) Possible multi-tile transcripts that could be produced via transcription of the DNA duplex highlighted in C. Transcripts of the bottom strand could contain the complement of the single-stranded overhang domain (orange) (67). (E) Transcripts that could be produced by transcription of single tiles. (F) The single tile variant (var1_7-1) used in G. (G) Non-denaturing PAGE results using var1_7-1 tiles that cannot form nanotubes. Individual tiles form during annealing (lanes 4 and 5). Tiles were incubated with T7 RNAP in transcription conditions and treated with DNase I as labeled. Bands at the bottom of the gels (migrating more quickly than 10 bp DNA migrates) for samples incubated with DNase I are likely short oligonucleotides from DNase I digestion.
We next asked what RNA products might be produced by T7 RNAP in the presence of nanotubes. The RNA products observed in Figure 3A span a wide range of molecular weights. Long transcripts (migrating more slowly than 100 bp ladder DNA migrates) could be produced from DNA nanotubes, which contain DNA helices that extend for micrometers along the nanotube lattice (Figure 3C and D). Additionally, since some free tiles remain after nanotube assembly, free tiles might also serve as transcription templates (Figure 3E). To investigate whether free tiles could serve as templates for transcription, we designed tiles that cannot polymerize into nanotubes by removing two of the sticky ends from the var1_7 tiles (var1_7-1 in Figure 3F and SI Section 1). Formation of these tiles was confirmed using PAGE. The top and bottom tile strands (1 and 5) both bind to the middle (strand 3) to form a complex (Figure 3G—lane 4) while the 5 strands that comprise the var1_7-1 tile variant form a single larger complex (Figure 3G—lane 5). We used the same gel to characterize the RNA produced when these tiles were incubated with T7 RNAP for 20 h in transcription conditions. RNA was produced but fewer high molecular weight products (products migrating more slowly than 100 bp ladder DNA migrates) were produced during the transcription of single tiles (Figure 3G—lane 3) than during the transcription of DNA nanotubes and T7 RNAP (Figure 3A—lane 3). Thus, it appears that both nanotubes and free tiles can serve as substrates for T7 RNAP transcription.
The RNA produced during T7 RNAP incubation with DNA nanotubes specifically binds DNA tile sequences
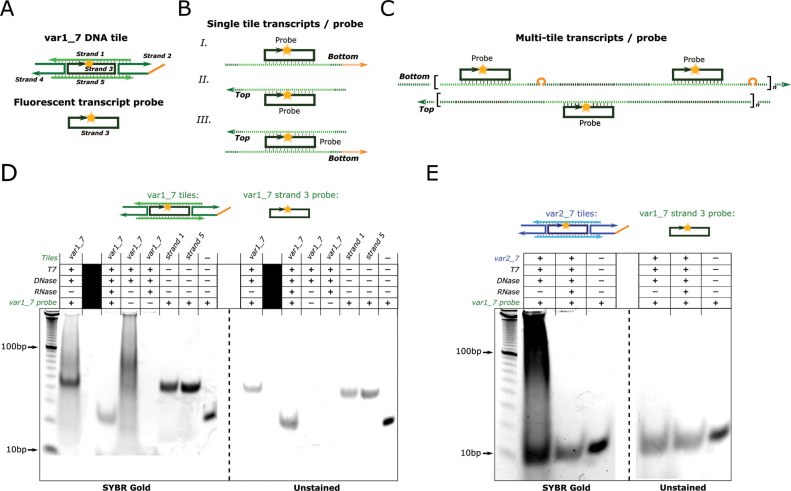
Since DNA nanotubes and tiles are presumably the only transcriptional templates for T7 RNAP in the experiments presented in Figure 3, we would expect the RNA produced during these experiments to have some of the same sequences as the DNA tiles. To test if this were the case, we annealed the RNA products produced from incubation of var1_7 nanotubes and T7 RNAP with the Cy3-modified strand 3 of these tiles (Figure 4A). This strand should be able to bind to a variety of possible transcripts produced from single var1_7 tile or nanotube templates (Figure 4B and C, respectively). As such, this strand served as a probe that could detect specific tiles sequences within RNA transcripts. To prepare RNA transcripts to probe for tile sequences, we incubated var1_7 nanotubes with T7 RNAP in transcription conditions for 24 h, heat denatured the RNAP, and digested the nanotube DNA with DNase I. At this point, the sample should have contained RNA produced during the incubation but no nanotube DNA. After heat denaturing the sample to inactivate the DNase I, we added the Cy3-modified strand 3 of the var1_7 tiles. We then annealed the sample to allow the most stable pairings of RNA and the probe to form. To determine whether the probe bound to the RNA, we analyzed the sample with non-denaturing PAGE. We captured a fluorescence image of the unstained gel to locate the Cy3-labeled probe on the gel and then stained the gel with SYBR Gold to identify the locations of all the nucleic acids. As a control, we conducted the same experiment but also included RNase A during the DNase I digestion step. In this control, the solution to which the probe was added should not have contained any intact nucleic acids, so a mobility shift of the fluorescent probe should not be observed.
Figure 4.
RNA produced from DNA nanotube incubation with T7 RNAP specifically binds DNA tile sequences. (A–C) DNA: solid lines, RNA: dashed lines. (A) Schematic of the fluorescent DNA probe used to detect the presence of tile sequences in RNA. The probe is strand 3 of the var1_7 tiles and contains a Cy3 fluorophore at its 5′ end. It can bind to a range of possible transcription products (B and C). (B) Schematics of the probe bound to possible single tile transcripts. (C) Schematics of the probe bound to possible RNA transcribed from assembled nanotubes. Aggregation of many such possible transcripts and a probe are also possible. (D–E) Non-denaturing PAGE results showing samples of nanotubes incubated with or without T7 RNAP (T7) and treated with or without DNase I (DNase) and RNase A (RNase) as shown. Nanotube tile variants used during the T7 RNAP incubation and the fluorescent probe for each experiment are shown above the gels.
The probe strand migrated more slowly in the sample containing the RNA produced during T7 RNAP incubation with DNA nanotubes than in the sample treated with both DNase I and RNase A (Figure 4D - unstained gel), suggesting that the probe binds to some of the RNA produced when nanotubes are incubated with T7 RNAP. To verify that this mobility shift was the result of the probe's sequence, we repeated these experiments using a fluorescent probe of the same length that did not have sequence overlap with the var1_7 tile strands (orthogonal probe—SI Section 1.2). This probe migrated at the same rate in the presence of the RNA products as the individual probe (Supplementary Figure S13). Likewise, no shift of the probe was observed when the probe for var1_7 tiles was mixed with the RNA produced during an incubation of the var2_7 nanotubes and T7 RNAP (Figure 4E). But the probe for the var2_7 tiles did shift when mixed with the var2_7 derived RNA (Supplementary Figure S14). Thus, the RNA produced by a set of tiles binds specifically to the DNA from those tiles.
The sizes of the complexes formed when each of the fluorescent probes bound to their respective RNA products were similar to the sizes of the complexes formed by the probe and either strand 1 or strand 5 of the DNA tiles (Figure 4D). This suggests that the majority of the RNA products bound to the probe are transcripts from either a single tile or a region of the DNA nanotube lattice that corresponds to a single tile. Either these transcripts are the main transcription products or the probe is unable to bind larger transcripts, possibly because these longer transcripts hybridize to themselves or other RNAs rather than to the probe during the annealing process.
RNA produced during the incubation of DNA nanotubes with T7 RNAP interacts with DNA tiles and disassembles nanotubes
We next explored how the transcription of DNA nanotubes and tiles might lead to nanotube disassembly. One possibility is that the process of transcription itself (i.e. melting of the DNA helix along the length of the nanotubes during transcription) causes the nanotubes to disassemble. Alternatively, RNA transcripts could cause disassembly by displacing DNA within the tiles or nanotubes via branch migration (64).
To determine whether the transcription process alone was responsible for nanotube disassembly, we incubated nanotubes with both T7 RNAP and RNase A to prevent RNA from accumulating. In the presence of RNase A, nanotubes incubated with T7 RNAP in transcription conditions did not disassemble (Supplementary Figure S15). These results indicate that RNA transcripts are necessary for nanotube disassembly. To assess whether the presence of transcripts (as opposed to the production of transcripts) was sufficient to induce nanotube disassembly, we first prepared an RNA transcript solution by incubating nanotubes with T7 RNAP in transcription conditions, denaturing T7 RNAP, digesting the nanotube DNA with DNase I, and heat denaturing DNase I (Supplementary Figure S16). We refer to this process as the preparation of RNA transcript solution. We prepared RNA transcript solution using var1_7 DNA nanotubes and tested whether the RNA transcripts in this solution could disassemble nanotubes by adding intact var1_7 nanotubes to the solution after DNA nanotube digestion with DNase I and subsequent DNase I heat denaturation. The var1_7 nanotubes that were added to the RNA transcript solution disassembled to a significant degree after 8 h (Supplementary Figure S16).
To ensure that disassembly was caused specifically by RNA, we prepared the RNA transcript solution with 20 mM of ATP during the T7 RNAP incubation rather than 5 mM of each NTP type. This change should produce a solution that does not contain RNA but still contains DNA degradation products and inactivated enzymes. DNA nanotubes added to this solution did not disassemble (Supplementary Figure S16). To test if any RNA, rather than RNA containing sequences complementary to the nanotubes, could induce nanotube disassembly, we again prepared the RNA transcript solution using var1_7 nanotubes but added var2_7 nanotubes, which have completely different sequences than the var1_7 nanotubes, to the RNA transcript solution. The var2_7 nanotubes did not disassemble in these conditions (Supplementary Figure S16). Thus, the RNA transcripts produced by a set of DNA nanotubes is sufficient to disassemble DNA nanotubes of the same sequence.
Having established that RNA transcripts produced in the presence of DNA nanotubes and T7 RNAP are necessary and sufficient for DNA nanotube disassembly, we next asked how disassembly might occur. Nucleic acid branch migration processes are accelerated by the availability of a single-stranded toehold region that a complementary displacing strand can bind to (64). The tiles we studied contained a 7 nucleotide single-stranded domain on strand 2. This domain might act as a binding site for a complementary RNA (Figure 5A). Hybridization of an RNA transcript at this site could allow the RNA to efficiently initiate displacement of the adjacent sticky end, causing tile or nanotube disassembly. In support of this hypothesis, we found that a 14 nucleotide DNA strand consisting of the complement of the 7-base single-stranded overhang domain and the complement of the 7-base sticky end domain adjacent to the overhang was able to disassemble nanotubes (Supplementary Figure S17).
Figure 5.
RNA transcripts produced when var1_7-3 DNA nanotubes are incubated with T7 RNAP in transcription conditions bind to the single-stranded overhang domain of the DNA tiles during nanotube disassembly. (A) Schematic showing the process of RNA hybridization and fluorescence increase. RNA transcripts bind to the single-stranded overhang and hybridization of the RNA and overhang increases the fluorescence of the FAM molecule at the 5′ end of the overhang. This hybridization can also disassemble tiles, inactivating them for nanotube assembly and/or causing them to detach from nanotubes. (B) Normalized FAM fluorescence of nanotubes in transcription conditions with a full set of NTPs or just ATP before and after the addition T7 RNAP. Fluorescence values for each sample are normalized to the fluorescence of the sample at t = 0. T7 RNAP was added at the time indicated by the red arrow. (C) Fluorescence micrographs of the samples in B after different incubation times. Scale bars: 10 μm.
If nanotube disassembly in the presence of T7 RNAP occurred via toehold-mediated strand displacement in which the single-stranded overhang domain served as a toehold, disassembly should only occur when the RNA can bind to the overhang domain to initiate displacement. To determine if the ability of the RNA transcripts to bind to the single-stranded overhang domain was required for nanotube disassembly, we created a new set of tiles, the var1_7-2 tiles (for sequences see SI Section 1), by changing the overhang sequence of the var1_7 tiles. We then determined whether RNA produced from nanotubes containing the original overhang sequence (var1_7) could disassemble the var1_7-2 nanotubes. We prepared the RNA solution using var1_7 nanotubes, then added var1_7-2 nanotubes to the RNA solution and found that the var1_7-2 nanotubes did not disassemble (Supplementary Figure S16). These results suggest that disassembly requires a single-stranded overhang domain that matches that of the nanotubes from which the RNA was transcribed.
To verify that RNA transcripts bind to the single-stranded overhang domains during the process of disassembly, we designed an experiment to characterize RNA-overhang binding using fluorescence spectroscopy. We designed a tile variant where the overhang domain had a FAM fluorophore at its 5′ end (var1_7-3 in SI Section 1). Nucleic acid hybridization near a FAM fluorophore has been shown to induce a change in FAM fluorescence intensity (65,66), so our goal was to measure hybridization at the overhang by looking for changes in the FAM fluorescence of our samples. To test that hybridization at the overhang would result in a measurable change of fluorescence signal we confirmed that the fluorescence of samples containing var1_7-3 nanotubes changed significantly after the addition of the overhang complement strand (Supplementary Figure S18).
To determine whether the RNA transcripts produced when DNA nanotubes are incubated with T7 RNAP bind to the single-stranded overhang domain of the tiles (Figure 5A), we incubated var2_7-3 nanotubes with T7 RNAP in a quantitative PCR machine with either 5 mM of each NTP type or 20 mM of only ATP and measured FAM fluorescence over time. In the sample containing NTPs (where transcription can occur), a steady increase in fluorescence was observed for 10 h while the ATP-only control (where transcription cannot occur) showed no fluorescence change (Figure 5B). Fluorescence micrographs of the samples throughout the experiment show that nanotubes in the sample with all NTP types disassemble but those in the ATP-only control do not (Figure 5C). Furthermore, the fluorescence of the sample containing all NTP types plateaus when nanotube disassembly is nearly complete, suggesting that RNA binding to the single-stranded overhang domain coincides with disassembly.
Since our results indicated that the single-stranded overhang on the DNA tiles is directly involved in the nanotube disassembly process, we would not expect nanotubes without this domain to disassemble when incubated with T7 RNAP in transcription conditions. To test this hypothesis, we annealed var3_7 nanotubes with and without single-stranded overhangs in transcription buffer without spermidine (Supplementary Figure S1), as we observed that nanotubes without overhangs did not assemble with spermidine present. We then incubated each nanotube variant with T7 RNAP in transcription conditions. We found the nanotubes without the single-stranded overhang were stable with T7 RNAP for 24 h while those with the overhang disassembled after 24 h (Supplementary Figure S19). We also observed that tiles with a double-stranded (rather than single-stranded) overhang assembled in transcription buffer containing spermidine (Supplementary Figure S20) and did not disassemble after 24 h when incubated with T7 RNAP in transcription conditions (Supplementary Figure S21). Thus the overhang domain on the sticky end of the tiles must be single-stranded for disassembly to occur. These results provide further evidence for our model of toehold-mediated nanotube disassembly.
Nanotubes are more stable in the presence of T7 RNAP when DNA containing the T7 RNAP promoter sequence is present
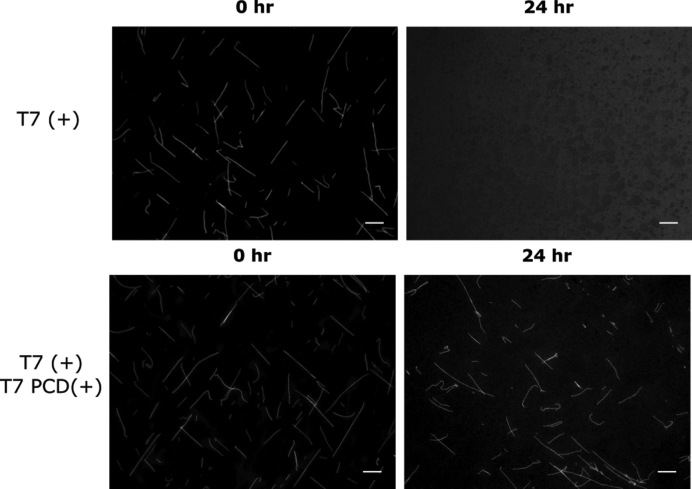
The above results establish that T7 RNAP can induce the disassembly of DNA nanotubes through non-specific production of RNA that interacts with the DNA tiles. Such non-specific interactions by an enzyme are often suppressed if a preferred substrate is available for binding. We would therefore expect the native T7 RNAP promoter sequence, which T7 RNAP should prefer as a substrate over the DNA nanotube sequences, to slow down disassembly. To test this hypothesis, we incubated var3_7 nanotubes and 50 nM of a DNA duplex containing the T7 RNAP promoter sequence (see SI Section 1.4 for sequence) with T7 RNAP in transcription conditions. Addition of 50 nM of this T7 RNAP promoter containing duplex (PCD) to a 1 μM solution of DNA nanotubes was sufficient to prevent nanotube disassembly for at least 24 h (Figure 6). The availability of T7 RNAP promoter sites presumably decreases the net activity of T7 RNAP on DNA nanotubes, which is in line with our observation that lower concentrations of T7 RNAP could prevent nanotube disassembly for some tile variants (Supplementary Figure S7).
Figure 6.
DNA nanotubes are stable when incubated with T7 RNAP in transcription conditions in the presence of a T7 RNAP promoter containing DNA duplex (PCD). Fluorescence micrographs of var3_7 nanotubes incubated with T7 RNAP in transcription conditions with (bottom) or without (top) 50 nM of a T7 PCD (SI Section 1.4 for sequences). Scale bars: 10 μm.
DISCUSSION
The results we presented here elucidate a mechanism by which DNA nanotubes, well-characterized DNA nanostructures, can disassemble in the presence of viral RNA polymerases commonly used to control gene expression. DNA nanotubes and tiles serve as substrates for non-specific transcription by T7 RNAP. The RNA produced during this non-specific transcription interacts directly with the DNA nanotubes, leading to nanotube disassembly. Disassembly appears to occur via a toehold-mediated branch migration process involving a single-stranded overhang domain on the tiles.
The DNA nanotubes studied in this work contain structural motifs, such as nicks, helical crossovers, and single-stranded overhangs that are not commonly observed in genomic DNA. While it would be reasonable to expect that these motifs could inhibit T7 RNAP transcription, we observed transcripts with high molecular weight (migrating more slowly than 100bp ladder DNA migrates in PAGE), suggesting that T7 RNAP has the capacity to traverse these potential roadblocks during transcription. Indeed, T7 RNAP has been shown to transcribe single-stranded overhangs in the template strand of a transcription substrate (67), past nicks and gaps with high efficiency (67,68), and through immobile double Holliday junctions (69). Thus, T7 RNAP’s transcription of tiles or across many tiles along a nanotube lattice seems plausible. Furthermore, the rate of non-specific transcription of DNA nanotubes might be higher compared to other non-specific substrates as nicks in transcription templates have been shown to promote T7 RNAP transcription (70).
Given the ability of T7 RNAP to non-specifically transcribe DNA nanotubes with many different sequences, it seems reasonable to assume that T7 RNAP would also non-specifically transcribe many other DNA nanostructures. Further, single-stranded domains on DNA nanotubes and origami have served as attachment sites for metals (9,12,55), enzymes (56), and fluorescence tags (15). In the presence of a viral transcriptional system, these nanostructures could disassemble or lose functionality via a mechanism similar to the mechanism elucidated for nanotubes in this work. Our results also suggest that DNA strand-displacement circuits might lose functionality in the presence of viral RNAPs. For example, non-specific transcription of strand-displacement components could trigger unintended actuation (40). Given the widespread use of viral RNA polymerases across synthetic biology, both in vitro and in vivo, our results should be taken into consideration when combining these enzymes (T7 RNAP in particular) with DNA nanostructures or other DNA devices.
Our careful elucidation of the mechanisms underlying DNA nanotube disassembly by T7 RNAP also suggest methods for reducing the rate of non-specific transcription and viral RNAP-induced nanostructure disassembly. Addition of a competitor for T7 RNAP binding mitigates nanotube disassembly (Figure 6), and disassembly could also be mitigated by the degradation of RNA by RNase A (Supplementary Figure S15) or by lowering the amount of RNAP used (Supplementary Figure S7). Eliminating single-stranded domains on the nanotubes (by either removing or sequestering them in dsDNA) also prevents most nanotube disassembly (Supplementary Figures S19 and S21). Together these results serve as a set of basic design principles for designing robust DNA nanostructures for use alongside viral RNAPs.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Dr Abdul Mohammed and Dr Joshua Fern for insightful conversations.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Science Foundation Graduate Research Fellowship [DGE-1232825 to S.W.S., DGE-1326120 to L.G.]; Department of Energy [DE-SC001 0426]. Funding for open access charge: Department of Energy [DE-SC001 0426].
Conflict of interest statement. None declared.
REFERENCES
- 1. Rothemund P.W.K., Ekani-Nkodo A., Papadakis N., Kumar A., Fygenson D.K., Winfree E.. Design and Characterization of Programmable DNA Nanotubes. J. Am. Chem. Soc. 2004; 126:16344–16352. [DOI] [PubMed] [Google Scholar]
- 2. Wei B., Dai M., Yin P.. Complex shapes self-assembled from single-stranded DNA tiles. Nature. 2012; 485:623–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rothemund P.W.K. Folding DNA to create nanoscale shapes and patterns. Nature. 2006; 440:297–302. [DOI] [PubMed] [Google Scholar]
- 4. Andersen E.S., Dong M., Nielsen M.M., Jahn K., Subramani R., Mamdouh W., Golas M.M., Sander B., Stark H., Oliveira C.L.P. et al. . Self-assembly of a nanoscale DNA box with a controllable lid. Nature. 2009; 459:73–76. [DOI] [PubMed] [Google Scholar]
- 5. Han D., Pal S., Nangreave J., Deng Z., Liu Y., Yan H.. DNA origami with complex curvatures in three-dimensional space. Science. 2011; 332:342–346. [DOI] [PubMed] [Google Scholar]
- 6. Dietz H., Douglas S.M., Shih W.M.. Folding DNA into twisted and curved nanoscale shapes. Science. 2009; 325:725–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Douglas S.M., Dietz H., Liedl T., Högberg B., Graf F., Shih W.M.. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature. 2009; 459:414–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wilner O.I., Shimron S., Weizmann Y., Wang Z.-G., Willner I.. Self-assembly of enzymes on DNA scaffolds: en route to biocatalytic cascades and the synthesis of metallic nanowires. Nano Lett. 2009; 9:2040–2043. [DOI] [PubMed] [Google Scholar]
- 9. Pearson A.C., Liu J., Pound E., Uprety B., Woolley A.T., Davis R.C., Harb J.N.. DNA origami metallized site specifically to form electrically conductive nanowires. J. Phys. Chem. B. 2012; 116:10551–10560. [DOI] [PubMed] [Google Scholar]
- 10. Hung A.M., Micheel C.M., Bozano L.D., Osterbur L.W., Wallraff G.M., Cha J.N.. Large-area spatially ordered arrays of gold nanoparticles directed by lithographically confined DNA origami. Nat. Nanotechnol. 2010; 5:121–126. [DOI] [PubMed] [Google Scholar]
- 11. Bui H., Onodera C., Kidwell C., Tan Y., Graugnard E., Kuang W., Lee J., Knowlton W.B., Yurke B., Hughes W.L.. Programmable periodicity of quantum dot arrays with DNA origami nanotubes. Nano Lett. 2010; 10:3367–3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kuzyk A., Schreiber R., Fan Z., Pardatscher G., Roller E.-M., Högele A., Simmel F.C., Govorov A.O., Liedl T.. DNA-based self-assembly of chiral plasmonic nanostructures with tailored optical response. Nature. 2012; 483:311–314. [DOI] [PubMed] [Google Scholar]
- 13. Zhang Z., Yang Y., Pincet F., Llaguno M.C., Lin C.. Placing and shaping liposomes with reconfigurable DNA nanocages. Nat. Chem. 2017; 9:653–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Langecker M., Arnaut V., Martin T.G., List J., Renner S., Mayer M., Dietz H., Simmel F.C.. Synthetic lipid membrane channels formed by designed DNA nanostructures. Science. 2012; 338:932–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mohammed A.M., Šulc P., Zenk J., Schulman R.. Self-assembling DNA nanotubes to connect molecular landmarks. Nat. Nanotechnol. 2017; 12:312–316. [DOI] [PubMed] [Google Scholar]
- 16. Zhang D.Y., Seelig G.. Dynamic DNA nanotechnology using strand-displacement reactions. Nat. Chem. 2011; 3:103–113. [DOI] [PubMed] [Google Scholar]
- 17. Fern J., Scalise D., Cangialosi A., Howie D., Potters L., Schulman R.. DNA strand-displacement timer circuits. ACS Synth. Biol. 2017; 6:190–193. [DOI] [PubMed] [Google Scholar]
- 18. Zenk J., Scalise D., Wang K., Dorsey P., Fern J., Cruz A., Schulman R.. Stable DNA-based reaction-diffusion patterns. RCS Adv. 2017; 7:18032–18040. [Google Scholar]
- 19. Zhang D.Y., Hariadi R.F., Choi H.M.T., Winfree E.. Integrating DNA strand-displacement circuitry with DNA tile self-assembly. Nat. Commun. 2013; 4:1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schiffels D., Liedl T., Fygenson D.K.. Nanoscale structure and microscale stiffness of DNA nanotubes. ACS Nano. 2013; 7:6700–6710. [DOI] [PubMed] [Google Scholar]
- 21. Corbett S.L., Sharma R., Davies A.G., Wälti C.. Enhancement of RecA-mediated self-assembly in DNA nanostructures through basepair mismatches and single-strand nicks. Sci. Rep. 2017; 7:41081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Du S.M., Wang H., Tse-Dinh Y.-C., Seeman N.C.. Topological transformations of synthetic DNA knots. Biochemistry. 1995; 34:673–682. [DOI] [PubMed] [Google Scholar]
- 23. Bertrand O.J.N., Fygenson D.K., Saleh O.A.. Active, motor-driven mechanics in a DNA gel. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:17342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. O’Neill P., Rothemund P.W.K., Kumar A., Fygenson D.K.. Sturdier DNA Nanotubes via Ligation. Nano Lett. 2006; 6:1379–1383. [DOI] [PubMed] [Google Scholar]
- 25. Li T., Zhang H., Hu L., Shao F.. Topoisomerase-based preparation and AFM imaging of multi-interlocked circular DNA. Bioconj. Chem. 2016; 27:616–620. [DOI] [PubMed] [Google Scholar]
- 26. Sagredo S., Pirzer T., Aghebat Rafat A., Goetzfried M.A., Moncalian G., Simmel F.C., de la Cruz F.. Orthogonal Protein Assembly on DNA Nanostructures Using Relaxases. Angew. Chem. Int. Ed. 2016; 55:4348–4352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Agarwal N.P., Matthies M., Joffroy B., Schmidt T.L.. Structural transformation of wireframe DNA origami via DNA polymerase assisted gap-filling. ACS Nano. 2018; 12:2546–2553. [DOI] [PubMed] [Google Scholar]
- 28. Mohsen M.G., Kool E.T.. The discovery of rolling circle amplification and rolling circle transcription. Acc. Chem. Res. 2016; 49:2540–2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Padirac A., Fujii T., Rondelez Y.. Bottom-up construction of in vitro switchable memories. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:E3212–E3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Montagne K., Gines G., Fujii T., Rondelez Y.. Boosting functionality of synthetic DNA circuits with tailored deactivation. Nat. Commun. 2016; 7:13474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kishi J.Y., Schaus T.E., Gopalkrishnan N., Xuan F., Yin P.. Programmable autonomous synthesis of single-stranded DNA. Nat. Chem. 2017; 10:155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jung C., Ellington A.D.. A primerless molecular diagnostic: phosphorothioated-terminal hairpin formation and self-priming extension (PS-THSP). Anal. Bioanal. Chem. 2016; 408:8583–8591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gines G., Zadorin A.S., Galas J.-C., Fujii T., Estevez-Torres A., Rondelez Y.. Microscopic agents programmed by DNA circuits. Nat. Nanotechnol. 2017; 12:351. [DOI] [PubMed] [Google Scholar]
- 34. Meijer L.H.H., Joesaar A., Steur E., Engelen W., van Santen R.A., Merkx M., de Greef T.F.A.. Hierarchical control of enzymatic actuators using DNA-based switchable memories. Nat. Commun. 2017; 8:1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhao W., Ali M.M., Brook M.A., Li Y.. Rolling circle amplification: applications in nanotechnology and biodetection with functional nucleic acids. Angew. Chem. Int. Ed. 2008; 47:6330–6337. [DOI] [PubMed] [Google Scholar]
- 36. Geary C., Rothemund P.W.K., Andersen E.S.. A single-stranded architecture for cotranscriptional folding of RNA nanostructures. Science. 2014; 345:799–805. [DOI] [PubMed] [Google Scholar]
- 37. Geary C., Chworos A., Verzemnieks E., Voss N.R., Jaeger L.. Composing RNA nanostructures from a syntax of RNA structural modules. Nano Lett. 2017; 17:7095–7101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jepsen M.D.E., Sparvath S.M., Nielsen T.B., Langvad A.H., Grossi G., Gothelf K.V., Andersen E.S.. Development of a genetically encodable FRET system using fluorescent RNA aptamers. Nat. Commun. 2018; 9:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Stewart J.M., Subramanian H.K.K., Franco E.. Self-assembly of multi-stranded RNA motifs into lattices and tubular structures. Nucleic Acids Res. 2017; 45:5628–5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Franco E., Friedrichs E., Kim J., Jungmann R., Murray R., Winfree E., Simmel F.C.. Timing molecular motion and production with a synthetic transcriptional clock. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:E784–E793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pomerantz R.T., Ramjit R., Gueroui Z., Place C., Anikin M., Leuba S., Zlatanova J., McAllister W.T.. A tightly regulated molecular motor based upon T7 RNA polymerase. Nano Lett. 2005; 5:1698–1703. [DOI] [PubMed] [Google Scholar]
- 42. Kim J., White K.S., Winfree E.. Construction of an in vitro bistable circuit from synthetic transcriptional switches. Mol. Syst. Biol. 2006; 2:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kim J., Winfree E.. Synthetic in vitro transcriptional oscillators. Mol. Syst. Biol. 2011; 7:465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Subsoontorn P., Kim J., Winfree E.. Ensemble Bayesian analysis of bistability in a synthetic transcriptional switch. ACS Synth. Biol. 2012; 1:299–316. [DOI] [PubMed] [Google Scholar]
- 45. McAllister W.T. Eckstein F, Lilley DMJ. Transcription by T7 RNA Polymerase. Mechanisms of Transcription. 1997; Berlin, Heidelberg: Springer; 15–25. [Google Scholar]
- 46. Choi K.H. Viral polymerases. Adv. Exp. Med. Biol. 2012; 726:267–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Green A.A., Kim J., Ma D., Silver P.A., Collins J.J., Yin P.. Complex cellular logic computation using ribocomputing devices. Nature. 2017; 548:117–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mohammed A.M., Schulman R.. Directing Self-Assembly of DNA Nanotubes Using Programmable Seeds. Nano Lett. 2013; 13:4006–4013. [DOI] [PubMed] [Google Scholar]
- 49. Jorgenson T.D., Mohammed A.M., Agrawal D.K., Schulman R.. Self-Assembly of Hierarchical DNA Nanotube Architectures with Well-Defined Geometries. ACS Nano. 2017; 11:1927–1936. [DOI] [PubMed] [Google Scholar]
- 50. Hariadi R.F., Yurke B., Winfree E.. Thermodynamics and kinetics of DNA nanotube polymerization from single-filament measurements. Chem. Sci. 2015; 6:2252–2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hariadi R.F., Winfree E., Yurke B.. Determining hydrodynamic forces in bursting bubbles using DNA nanotube mechanics. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:E6086–E6095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hariadi R.F., Yurke B.. Elongational-flow-induced scission of DNA nanotubes in laminar flow. Phys. Rev. E. 2010; 82:046307. [DOI] [PubMed] [Google Scholar]
- 53. Sousa R., Patra D., Lafer E.M.. Model for the mechanism of bacteriophage T7 RNAP transcription initiation and termination. J. Mol. Biol. 1992; 224:319–334. [DOI] [PubMed] [Google Scholar]
- 54. Martin C.T., Coleman J.E.. Kinetic analysis of T7 RNA polymerase-promoter interactions with small synthetic promoters. Biochemistry. 1987; 26:2690–2696. [DOI] [PubMed] [Google Scholar]
- 55. Liu W., Halverson J., Tian Y., Tkachenko A.V., Gang O.. Self-organized architectures from assorted DNA-framed nanoparticles. Nat. Chem. 2016; 8:867–873. [DOI] [PubMed] [Google Scholar]
- 56. Yan H., Park S.H., Finkelstein G., Reif J.H., LaBean T.H.. DNA-Templated Self-Assembly of Protein Arrays and Highly Conductive Nanowires. Science. 2003; 301:1882–1884. [DOI] [PubMed] [Google Scholar]
- 57. Liu D., Park S.H., Reif J.H., LaBean T.H.. DNA nanotubes self-assembled from triple-crossover tiles as templates for conductive nanowires. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:717–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Mohammed A.M., Velazquez L., Chisenhall A., Schiffels D., Fygenson D.K., Schulman R.. Self-assembly of precisely defined DNA nanotube superstructures using DNA origami seeds. Nanoscale. 2017; 9:522–526. [DOI] [PubMed] [Google Scholar]
- 59. Green L.N., Amodio A., Subramanian H.K.K., Ricci F., Franco E.. pH-driven reversible self-assembly of micron-scale DNA scaffolds. Nano Lett. 2017; 17:7283–7288. [DOI] [PubMed] [Google Scholar]
- 60. Bandwar R.P., Patel S.S.. The energetics of consensus promoter opening by T7 RNA polymerase. J. Mol. Biol. 2002; 324:63–72. [DOI] [PubMed] [Google Scholar]
- 61. Gunderson S.I., Chapman K.A., Burgess R.R.. Interactions of T7 RNA polymerase with T7 late promoters measured by footprinting with methidiumpropyl-EDTA-iron(II). Biochemistry. 1987; 26:1539–1546. [DOI] [PubMed] [Google Scholar]
- 62. Sutton D.H., Conn G.L., Brown T., Lane A.N.. The dependence of DNase I activity on the conformation of oligodeoxynucleotides. Biochem. J. 1997; 321:481–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ausubel F.M., et al. Current Protocols in Molecular Biology. 1994; NY: John Willey & Sons, Inc. [Google Scholar]
- 64. Zhang D.Y., Winfree E.. Control of DNA strand displacement kinetics using toehold exchange. J. Am. Chem. Soc. 2009; 131:17303–17314. [DOI] [PubMed] [Google Scholar]
- 65. Padirac A., Fujii T., Rondelez Y.. Quencher-free multiplexed monitoring of DNA reaction circuits. Nucleic Acids Res. 2012; 40:e118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Behrens S., Fuchs B.M., Amann R.. The effect of nucleobase-specific fluorescence quenching on in situ hybridization with rRNA-targeted oligonucleotide probes. Syst. Appl. Microbiol. 2004; 27:565–572. [DOI] [PubMed] [Google Scholar]
- 67. Rong M., Durbin R.K., McAllister W.T.. Template strand switching by T7 RNA polymerase. J. Biol. Chem. 1998; 273:10253–10260. [DOI] [PubMed] [Google Scholar]
- 68. Zhou W., Reines D., Doetsch P.W.. T7 RNA polymerase bypass of large gaps on the template strand reveals a critical role of the nontemplate strand in elongation. Cell. 1995; 82:577–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Pipathsouk A., Belotserkovskii B.P., Hanawalt P.C.. When transcription goes on Holliday: double Holliday junctions block RNA polymerase II transcription in vitro. BBA. Gene. Regul. Mech. 2017; 1860:282–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Gong P., Martin C.T.. Mechanism of instability in abortive cycling by T7 RNA polymerase. J. Biol. Chem. 2006; 281:23533–23544. [DOI] [PubMed] [Google Scholar]
- 71. Fu T.J., Seeman N.C.. DNA double-crossover molecules. Biochemistry. 1993; 32:3211–3220. [DOI] [PubMed] [Google Scholar]
- 72. Zadeh J.N., Steenberg C.D., Bois J.S., Wolfe B.R., Pierce M.B., Khan A.R., Dirks R.M., Pierce N.A.. NUPACK: analysis and design of nucleic acid systems. J. Comput. Chem. 2011; 32:170–173. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.