Abstract
The MADS transcription factors (TF) constitute an ancient family of TF found in all eukaryotes that bind DNA as obligate dimers. Plants have dramatically expanded the functional diversity of the MADS family during evolution by adding protein–protein interaction domains to the core DNA-binding domain, allowing the formation of heterotetrameric complexes. Tetramerization of plant MADS TFs is believed to play a central role in the evolution of higher plants by acting as one of the main determinants of flower formation and floral organ specification. The MADS TF, SEPALLATA3 (SEP3), functions as a central protein–protein interaction hub, driving tetramerization with other MADS TFs. Here, we use a SEP3 splice variant, SEP3Δtet, which has dramatically abrogated tetramerization capacity to decouple SEP3 tetramerization and DNA-binding activities. We unexpectedly demonstrate that SEP3 heterotetramer formation is required for correct termination of the floral meristem, but plays a lesser role in floral organogenesis. The heterotetramer formed by SEP3 and the MADS protein, AGAMOUS, is necessary to activate two target genes, KNUCKLES and CRABSCLAW, which are required for meristem determinacy. These studies reveal unique and highly specific roles of tetramerization in flower development and suggest tetramerization may be required to activate only a subset of target genes in closed chromatin regions.
INTRODUCTION
Plants maintain a pool of pluripotent stem cells that give rise to new organs throughout their lifecycles. These stem cells are located at the root and shoot apical meristems from which all below ground and aerial tissues derive. During angiosperm reproduction, the shoot apical meristem is converted to an inflorescence meristem (IM) and finally to fully determinate floral meristems (FM), which terminate pluripotent cell production and commit meristematic cells to distinctive fates. From the FM, four types of floral organs are produced in whorls with the sepals, petals, stamen and carpel of the flower located in the outer-most to the inner-most whorls of the flower. The MADS family plays critical roles in all aspects of flowering, including the floral transition, floral meristem determinacy and floral organ development (for recent reviews (1,2)). One of the best-studied developmental networks involves the ABCE class MADS genes whose overlapping expression patterns control floral meristem identity, determinacy and the identity of the floral organs.
In Arabidopsis, the A gene, APETALA1 (AP1), is required for floral meristem identity, sepal and petal formation, the B genes, APETALA3 (AP3) and PISTILLATA (PI), are required for petal and stamen development, the C gene, AGAMOUS (AG), is required for determinacy of the floral meristem as well as male and female organogenesis and the E class genes, SEPALLATA1, SEPALLATA2, SEPALLATA3 and SEPALLATA4 (SEP1-4), are necessary for floral organ identity in all whorls (3–6). Single knock outs of any of the SEP genes do not have strong floral phenotypes, however in the sep1 sep2 sep3 (sep1/2/3) triple mutant all floral organs are converted to sepaloid-like structures with a lack of determinacy of the floral meristem resulting in the continuous generation of sepals where the carpel would exist in wild type (WT) flowers (3). Initial expression patterns of the class B and C genes in the triple mutant are unaffected, underscoring the critical role of the SEP genes in both floral organ formation and floral meristem identity. The sep1/2/3/4 quadruple mutant is further affected, with floral organogenesis abolished, resulting in leaf-like organs in all whorls and a reduction in the expression levels of the B and C class genes (7,8).
The A–E class MADS genes encode TFs that act in a combinatorial manner in an intricate protein–protein interaction network, forming both dimeric and tetrameric complexes and driving different developmental programs. During evolution, plants have progressively diversified the MADS TF family, which likely played a role in allowing for increased plant complexity. The SEPALLATA genes appear later in evolution than other MADS floral organ identity genes and have been postulated to be key players in the evolution of flowering plants from their non-flowering ancestors (9). The SEPALLATA proteins form a central organising hub for MADS complex formation, particularly MADS TFs involved in flowering and floral organ development (10). Based on extensive genetic and biochemical studies, SEP3 is both the most promiscuous and active member of the SEPALLATA clade, able to interact with all floral organ identity MADS TFs and, when ectopically expressed with the appropriate A, B or C genes, able to convert leaves into floral organs (6,11–13).
MADS TFs bind DNA as dimers through the MADS DNA-binding domain, a domain that is conserved across all eukaryotes. The addition of an alpha-helical keratin-like or ‘K’ domain allows plant MADS TFs to tetramerize, with the structure of the K domain of SEP3 recently characterized and the tetramerization determinants mapped at the amino acid level (14). A central question is whether tetramerization is required for floral organ development. One well-accepted hypothesis is that binding of tetrameric MADS TF complexes results in DNA-looping and that this change in the three-dimensional chromatin structure may be necessary for triggering expression of specific genes and therefore floral organogenesis (15,16). In order to determine the role of tetramerization in flower development independently of DNA-binding, we exploited a naturally occurring splice variant of SEP3, SEP3Δtet, which lacks a portion of the K domain (amino acids 161–174) shown to be important for homotetramerization in vitro (14,17). Here, we demonstrate that SEP3Δtet exhibits abrogated heterotetramerization capacity with different partner MADS TFs in vitro. However, the heterodimeric complexes are still able to form and are fully competent to bind DNA. Expression of SEP3 and SEP3Δtet in the sep1/2/3 mutant background under the native promoter demonstrates that while SEP3 is able to fully complement the floral phenotype, SEP3Δtet is unable to restore WT flowers. While the first three whorls (sepals, petals and stamens) harbor normal floral organs, the fourth whorl exhibits unfused carpels and an indeterminate flower-within-a-flower phenotype. The combined in vitro and in vivo experiments decouple the roles of DNA-binding from tetramer formation and demonstrate the absolute requirement of SEP3/AG tetramerization in activating genes required for floral meristem determinacy. In addition, these results suggest that tetramerization may be less crucial for floral organogenesis.
MATERIALS AND METHODS
Plant material and growth conditions
All experiments were performed using Arabidopsis thaliana in the Col-0 background. Seedlings mutated in the SEPALLATA genes (3), were grown in controlled growth chambers in long day conditions (16 h light/8 h dark) at 22°C for plant transformation. For in situ hybridization, western blots and qPCR analysis, plants were first grown 5 weeks in short days (8 h light/16 h dark) at 22°C, then transferred and grown for 2 weeks in long day conditions.
Plasmid construct for complementation analysis
A 6419 bp and a 6377 bp DNA fragment containing specific sequence encoding SEP3 WT (Atg24260.2) and SEP3Δtet (Atg24260.3) respectively, under the control of SEP3 native promoter were cloned into the BamHI site of the plant expression vector pFP100 which contains the GFP gene under control of a seed specific promoter for selection (18). Each construct, pSEP3::SEP3 (ABRC stock number CD3-2708) and pSEP3::SEP3Δtet (ABRC stock number CD3-2709) contained 4136 bp of the SEP3 promoter region, the first exon and first intron of SEP3, exons 2–8 of the cDNA sequence for the pSEP3::SEP3 construct or exons 2-3-4-5-7-8 for pSEP3::SEP3Δtet construct, and 740 bp of the SEP3 terminator. DNA templates and primers used to amplify the constructs are listed in Supplementary Table S2.
Plant transformation and floral phenotype analysis
Sep1/2/3 triple mutants do not produce seeds. Because mutations in SEP2 and SEP3 were generated by transposon insertions (3), complementation analysis was performed as follows to confirm stability of the transposon mutants. The progeny of two heterozygous sep1/2/3+/− seedlings (ABRC stock number CS69927) genotyped as described (3,7) and showing the expected segregation patterns with 25% of the plants exhibiting the triple mutant phenotype, were transformed by classical floral dipping methods using Agrobacterium tumefaciens-mediated gene transfer. Half of the plants were transformed with the pSEP3::SEP3 construct and the other half with the pSEP3::SEP3Δtet construct. Transformants were selected based on the fluorescence of GFP-positive seeds. More than 40 seeds carrying each transgene were put on soil and further genotyped to identify a minimum of 10 sep1/2/3 plants carrying the transgene. Primers (5′ TCAATAGGCAAGTGAC 3′ and 5′ CACTCTCTGAAGGTAGCTGAAG 3′) were used to discriminate between sep1/2/3 and sep1/2/3+/− plants carrying the construct. In parallel, untransformed control sep1/2/3 plants were genotyped from the same batch of seeds after sowing non-fluorescent (untransformed) seeds to validate the stability of the sep1/2/3 phenotype in the second generation of seeds. Floral phenotypic analyses were performed by light microscopy on flower numbers 10–19 based on their order of emergence on T1 plants genotyped sep1/2/3 for each construct and on control untransformed sep1/2/3 plants. Seeds of T1 lines expressing SEP3 or SEP3Δtet were deposited to the ABRC (stock number CS69928-31).
Protein extraction and western blot analysis
Proteins were extracted from Arabidopsis thaliana inflorescence meristems with small closed buds up to stage 10-11 (19). Between 20 and 40 mg of tissues were ground in liquid nitrogen and resuspended in 2X Coomassie loading buffer (50 μl per 100 mg tissue, diluted in 100 mM Tris buffer, pH 7.5, supplemented with an antiprotease cocktail (Roche)). After 5 min at 95°C, samples were centrifuged 5 min, and 8 μl of total soluble proteins were separated on a SDS 15% polyacrylamide gel and transferred to a Hybond LFP PVDF transfer membrane (GE Healthcare). Western blot analyses were performed using the SEP3 antibody (20) at a 1:1000 dilution. Detection was performed using the Clarity Western ECL Substrate kit (BIO-RAD). Equal loading of the samples was checked by Coomassie blue protein detection on a separate gel.
Environmental scanning electron microscopy
SEM experiments were performed at the Electron Microscopy facility of the ICMG Nanobio-Chemistry Platform (Grenoble, France). Untreated flowers were directly placed in the microscope chamber. Care was taken to maintain some humidity during the pressure decrease in the chamber in order to prevent tissue drying. Secondary electron images were recorded with a Quanta FEG 250 (FEI) microscope while maintaining the tissue at 2°C, under a pressure of 500 Pa and a 70% relative humidity. The accelerating voltage was 14 kV and the image magnification ranged from 100 to 800x. Flowers from 3 independent lines were observed for each genotype.
Gene expression analysis by real time PCR
RNA was isolated from Arabidopsis thaliana Col-0 inflorescence meristems with small closed buds up to stage 10–11 (19) using RNAeasy plant mini kit (Qiagen). Complementary DNA (cDNA) was primed with random hexamers on 1μg DNase-treated RNA using Superscript III Reverse Transcriptase (Thermo Fisher Scientific). Real time qRT-PCR was performed on a CFX Connect real-time system (BIO-RAD) using Sso Advanced Universal SYBR Green supermix (BIO-RAD) and specific primers listed in Supplementary Table S2. Three technical replicates were run in parallel per sample. Quantification of gene expression was performed using the comparative CT method with the CFX Connect Manager 3.1 software. The ACTIN2 (At3g18780) and the EF-1α (At1g09740) genes were used to normalize the qPCR data (21). Statistical analyses were performed by Student's t-test.
In situ hybridization
RNA in situ hybridization was performed as previously described (22). Briefly, inflorescences were harvested 2 weeks after bolting. DIG-labeled AG antisense probe was generated by T7 RNA polymerase activity from a 1-kb insert cloned into the pBS KS+ vector and digoxigenin-labeling mix (Roche).
EMSA experiments
AG (At4g18960.1), SEP3 WT (At1g24260.2) and SEP3Δtet (At1g24260.3) cDNAs were PCR amplified using specific primers (Supplementary Table S2), cloned into the pSP64 plasmid (Promega) and used for in vitro transcription translation (Promega SP6 High Yield Expression System). AP3 (At3g54340) and PI (At5g20240) were cloned into pSPUTK (23). EMSAs were performed as described (17) with 10 nM DNA labeled with Cy5 (Eurofins). Primers used to amplify DNA probes and probe sequences are listed in Supplementary Tables S2 and S3. For each EMSA, a negative control was run, labeled ‘DNA alone’ in which the in vitro translation assay was done with pSP64 vector without any insert and incubated with the DNA probe.
Yeast two-hybrid
The cDNAs encoding SEP3, SEP3Δtet, AP3, PI and AG were cloned into both pGBKT7 and pGADT7 vectors (Clontech), using the NdeI/PstI and NdeI/XhoI restriction sites, respectively. The bait and prey vectors were transformed into yeast strain Y187 and AH109, respectively. After mating, the resulting diploids were selected with medium lacking Leu and Trp (-LW). The selected strains were dotted at two different optical densities (OD600 of 2 and 20) onto two different selection plates containing -LW or -LWAH medium (lacking Leu, Trp, Ade and His), the latter used to select diploids presenting positive protein interaction. Screening was done in duplicate.
Homology modeling
Sequence alignments and secondary structure prediction of SEP3 and AG were performed with Clustal and JPred4, respectively, followed by homology modeling of the SEP3/AG heterotetramer using the structure of the SEP3 homotetramerization domain (4OX0) and threading with the AG sequence (24–26). The models were inspected in Coot and side chain conformations manually optimized for hydrogen bonding (27). All structural figures were generated with PyMOL (28).
Bioinformatics analysis
ChIP-seq data for SEP3 (29) and AG (30) were downloaded from Arrayexpress and mapped and normalized using bowtie1.2.1 (31) and MACS2.1.1 (32). MNase data (33) were mapped using bowtie1.2.1. For Segal scores (34), 10 000 bp were added to the flanking region of each gene and computed using the web server (https://genie.weizmann.ac.il/software/nucleo_prediction.html). Predicted binding sites for SEP3 were computed using the position weight matrices from JASPAR (35) and the genes scanned using FIMO from the MEME suite (36) with P-value < 10e–4.
RESULTS
SEP3 and SEP3Δtet bind DNA in vitro
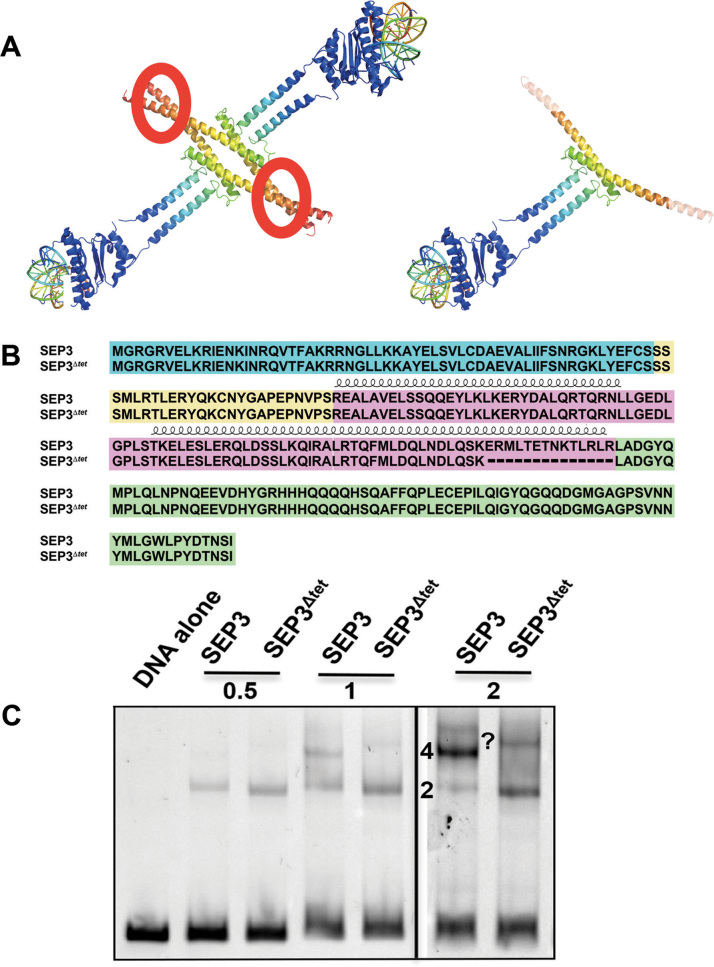
SEP3Δtet lacks residues 161–174 in the K domain, breaking the alpha helix that forms the major interaction surface necessary for stable tetramerization (Figure 1A and B). Using in vitro transcription-translation, the DNA-binding and oligomerization capabilities of the SEP3 and SEP3Δtet proteins were determined via electrophoretic mobility shift assays (EMSAs) (14,23,37). Similar production of SEP3 and SEP3Δtet proteins using the in vitro transcription-translation assay was confirmed by western blot analysis (Supplementary Figure S1A). As shown in Figure 1C, using a DNA fragment from the SEP3 promoter with two SEP3 binding sites (CArG boxes), the protein can bind DNA on a single site as a dimer or as a higher order complex with four molecules of SEP3 bound per DNA, suggesting cooperativity between the two dimers due to homotetramerization of the protein. This higher order complex was absent when only one CArG box was present (Supplementary Figure S1B). In contrast, the ability of SEP3Δtet to form this higher order complex is virtually abolished, demonstrating dramatically reduced cooperative binding by SEP3Δtet (Figure 1C). These data indicate that SEP3Δtet can efficiently bind DNA as a homodimer and recognizes the CArG box motif. However, cooperativity and tetramer formation is strongly impaired. SEP3Δtet was then used as a tool to study the impact of tetramerization on flower development in vivo.
Figure 1.
SEP3 dimers and tetramers bind DNA. (A) Composite model of SEP3 tetramer (left) and dimer (right) using the SEP3 K domain, (PDB 4OX0), and the homologous DNA-binding domain from MEF2A, (PDB 3KOV). The N-terminal DNA-binding domain is in blue with DNA shown as a cartoon and SEP3 K domain in rainbow with the tetramerization interface circled in red. The C-terminal domain is not shown. (B) Sequence alignment of SEP3 and SEP3Δtet with the M domain in blue, the I domain in yellow, the K domain in pink and the C-terminal domain in green. Alpha helices for the K domain are displayed schematically above the sequences. (C) EMSA with SEP3 and SEP3Δtet incubated at increasing protein concentrations (0.5, 1 and 2 μl of in vitro transcription-translation product) with a 103 bp fragment of the SEP3 promoter containing two CArG boxes. 2 and 4 indicate numbers of bound SEP3 or SEP3Δtet molecules per DNA. A higher molecular weight species of unknown composition, possibly corresponding to multiple bound dimers, is indicated by a question mark.
SEP3Δtet restores petals and stamen identity in the sep1/2/3 triple mutant background but not meristem determinacy
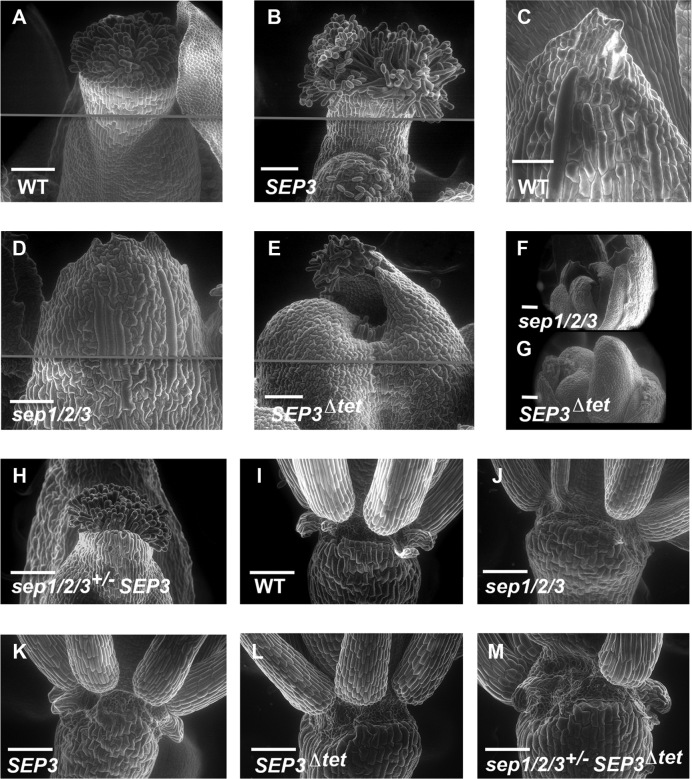
As the single and double sepallata mutants do not have strong phenotypes, the sep1/2/3 triple mutant background was chosen to investigate SEP3Δtet function. The sep1/2/3 mutant exhibited conversion of all floral organs to sepaloid-like structures and a lack of floral meristem determinacy (3). Sep1/2/3+/− Arabidopsis Col-0 plants were transformed with either SEP3 or SEP3Δtet under the control of the native promoter to generate sep1/2/3 pSEP3::SEP3 (hereafter referred to as sep1/2/3 SEP3) and sep1/2/3 pSEP3:: SEP3Δtet (hereafter referred to as sep1/2/3 SEP3Δtet) (Figure 2). Phenotypic complementation, defined by flowers having sepals, petals, stamens and carpels in whorls 1–4, respectively, with normal siliques harbouring seeds, was found in the overwhelming majority of T1 sep1/2/3 plants (15 of 18) expressing wild type SEP3 (Figure 2C, G, K). The remaining three T1 plants exhibited the classic sep1/2/3 phenotype. Conversely, 9 of 11 T1 sep1/2/3 plants expressing the SEP3Δtet construct showed incomplete flower development. While petals and stamens were clearly observed in these plants, two unfused carpeloid-like structures were produced instead of the WT gynoecium in whorl 4 (Figure 2D, H, L, M, O). Further inspection revealed that instead of containing ovules and seeds, whorl 4 exhibited supernumerary floral organs characteristic of the flower within-a-flower phenotype (Figure 2N, P). An extended gynophore was observed in these plants and all plants were sterile. The two remaining T1 plants carrying the vector SEP3Δtet exhibited the sep1/2/3 mutant phenotype. Petal number in both lines expressing either SEP3 or SEP3Δtet was identical to WT, while the number of stamens was slightly reduced for both lines compared to WT (5.8 ± 0.4 for WT versus 4.5 ± 0.2 for sep1/2/3 SEP3 lines and 4.5 ± 0.4 for sep1/2/3 SEP3Δtet lines). Among 18 plants genotyped sep1/2/3 in the pool of the untransformed T1 plants, 16 showed the characteristic triple mutant phenotype (Figure 2B, F and J) and none exhibited the sep1/2/3 SEP3Δtet phenotype. The remaining two plants showed the WT phenotype. These phenotypes were recapitulated in the T2 generation for the SEP3 and SEP3Δtet lines. In the case of plants expressing SEP3Δtet, sep1/2/3+/− plants were selected from the T1 and sep1/2/3 plants carrying the transgene SEP3Δtet were selected in the T2 generation. The T2 generation was used for all subsequent experiments.
Figure 2.
SEP3 tetramers are required for the normal development of the fourth whorl and flower determinacy in Arabidopsis. (A–D) show inflorescences in WT, sep1/2/3 and the sep1/2/3 triple mutant transformed with either SEP3 (C) or SEP3Δtet (D) construct. Flower indeterminacy (arrows) is observed in B and D, while normal siliques (arrows) are produced in A and C. (E–H) show close-ups of a young flower as per A to D. Sep1/2/3 flowers usually stay closed as in (F). (I–L) Close-ups after dissection of E to H. The fourth whorl of sep1/2/3 SEP3Δtet plants shows organ defects (L) and indeterminacy (M–P). Scale bar is 1 cm in A to D, 1 mm in E to O, and 0.5 mm in P.
In order to determine if the observed phenotypes were due to differences in protein levels, western blots using a SEP3-specific antibody was performed on independent lines for each construct. SEP3 and SEP3Δtet accumulate at similar levels to WT SEP3 in all transformed lines (Supplementary Figure S2). These data suggest that the differences in whorl 4 between sep1/2/3 expressing SEP3 or SEP3Δtet were unlikely to be due to protein accumulation level, but rather due to bona fide differences in protein function.
SEP3Δtet restores carpel cells in the sep1/2/3 mutant background but not the production of nectaries
To better characterize the floral organ identity in the triple mutant plants expressing SEP3Δtet, scanning electron microscopy experiments were performed. The cell surface morphology of all the floral organs in the sep1/2/3 mutant has a sepaloid-like character (Figure 3 and Supplementary Figure S3) as previously described (4). In contrast, the distinct cell surface morphology of sepals, petals and stamens were observed in WT and sep1/2/3 plants expressing SEP3 or SEP3Δtet (Supplementary Figure S3). Inspection of the cell surface in the fourth whorl in these lines reveals, similar to WT, stigmatic papillae, valve cells, style and replum (Figure 3A, B, E). This indicates that SEP3Δtet is able to launch carpel development. However, the gynoecium is unfused (Figure 3E). The interior of the carpel structure reveals a cell surface morphology characteristic of stamen and carpels in sep1/2/3 SEP3Δtet plants (Figure 3G). In addition, no nectaries were observed in SEP3Δtet plants (Figure 3L) but were present in the WT (Figure 3I) and SEP3 complemented lines (Figure 3K). These data show that the expression of SEP3Δtet fully restores petal and stamen development. In contrast, while the carpel development program was launched, gynoecium development is incomplete due to a lack of determinacy of the floral meristem. We questioned whether the indeterminate floral phenotype observed could be due to a downregulation of AG as this gene is a known regulator of floral meristem determinacy through the repression of the stem cell maintenance gene WUSCHEL (WUS) (38–40).
Figure 3.
Scanning electron micrographs of flowers for WT and mutants. (A and B) show normal carpels for WT and sep1/2/3 expressing SEP3 (labeled SEP3 for simplicity in the figure). (C) Normal sepal morphology in WT flowers. (D) In a sep1/2/3 triple mutant flower, the fourth whorl is replaced by sepaloid-like organs, as per C. (E) In sep1/2/3 SEP3Δtet plant (labeled SEP3Δtet) a swollen fourth whorl structure made of two unfused carpels is observed. (F) The indeterminate growth of flowers in sep1/2/3, with a succession of sepaloid-like organs. (G) In sep1/2/3 SEP3Δtet plants, the production of additional stamen and carpeloid-like organs is observed in the fourth whorl. (H) WT carpel fusion is restored with a single copy of SEP3. (I) Nectaries are present at the base of Arabidopsis WT stamens, absent in sep1/2/3 (J) and restored with the expression of SEP3 (K) but not SEP3Δtet (L). Nectary formation is dependent on SEP3 levels with a single copy of SEP3 sufficient to restore nectary formation (M). In panels A, B, D and E, the gray line indicates the junction between 2 separate images. This was required to achieve the resolution and complete view of the tissue or organ. Scale bar corresponds to 100 μm.
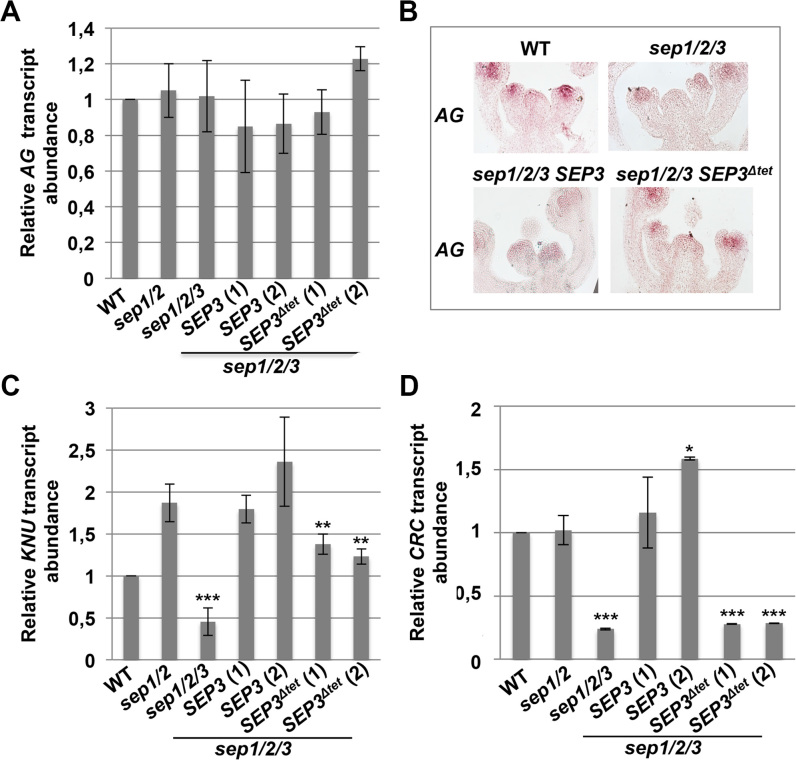
SEP3Δtet does not impact AG expression but misregulates KNU and CRC
SEP3 binds to the promoter of AG, suggesting the possibility that the lack of determinacy due to SEP3Δtet could be due to downregulation or misregulation of AG. As shown in Figure 4A, AG transcript levels in IMs and closed flower buds in WT, mutant and transformed lines were indistinguishable. In situ hybridization also confirmed that the early pattern of AG expression, in the centre of the floral meristem and in the primordia of whorls three and four, was similar in all lines (Figure 4B). This is consistent with previous reports for the sep1/2/3 mutant that also reported unaltered early AG expression with respect to WT (3). Thus, SEP3Δtet does not have a dramatic impact on AG expression levels and seems unlikely to account for the observed indeterminacy phenotype. Additional qRT-PCR experiments were performed on the floral MADS genes AP3, PI, AP1 and SEP4 to determine if there was misregulation of other floral organ identity genes, however no unexpected differences were observed (Supplementary Figure S4).
Figure 4.
Expression of SEP3 and SEP3Δtet does not affect AG, but deregulates CRC and KNU expression. (A and B) AG expression level and pattern was determined by qPCR and in situ hybridization, respectively, on IMs and closed flower buds up to stage 11, from WT, sep1/2, sep1/2/3, and the triple mutant expressing SEP3 or SEP3Δtet. (C) KNU and (D) CRC transcript abundance was analysed by qPCR. Data correspond to the mean of 3 independent biological replicates for WT, sep1/2 and sep1/2/3 plants, and 2 biological replicates for sep1/2/3 SEP3 and sep1/2/3 SEP3Δtet lines. Two independent lines were tested for sep1/2/3 SEP3 and sep1/2/3 SEP3Δtet. Expression was normalized to ACTIN2 and EF-1α. Asterisks indicate significant differences from sep1/2 (* P < 0.05, **P < 0.01, ***P < 0.001). Sep1/2 showed higher KNU expression levels than WT likely due to misregulation of KNU in the sep1/2 line. SEP3Δtet lines gave reduced KNU expression with respect to SEP3 complemented lines (P < 0.05).
Since AG expression was not significantly altered, we examined downstream targets of AG as possible candidates for the observed indeterminacy phenotype. AG has been previously shown to activate KNUCKLES (KNU) and CRABSCLAW (CRC) (40–46), known repressors of WUS. As shown in Figure 4C, KNU expression was significantly lower in sep1/2/3 compared to WT, and in sep1/2/3 expressing SEP3Δtet compared to either the double sep1/2 mutant or the triple mutant expressing SEP3. Interestingly, sep1/2 mutant and transformed lines showed higher expression levels of KNU compared to WT, suggesting a misregulation of KNU. KNU has significant expression in stamen and ovules in addition to its expression in the floral meristem (47). Different SEPALLATA proteins may play slightly different roles in regulating KNU in specific floral tissues, resulting in the observed KNU expression levels in sep1/2 mutants. CRC is required for nectary formation and, as expected by the absence of nectaries in sep1/2/3 and SEP3Δtet plants, CRC expression was strongly decreased in these lines and restored to WT and sep1/2 levels when the sep1/2/3 triple mutant was complemented with SEP3 but not when transformed with SEP3Δtet (Figure 4D). These experiments demonstrate that both KNU and CRC are specifically misregulated in the triple mutant expressing SEP3Δtet, with CRC showing the clearest downregulation.
Interaction patterns of SEP3Δtet and MADS floral organ identity TFs
To better understand the ability of SEP3Δtet to restore floral organ formation but not determinacy, we first compared the ability of SEP3 and SEP3Δtet to interact with AG by yeast-2-hybrid. Interactions for SEP3 and SEP3Δtet with AG were positive, indicating that SEP3Δtet can interact with AG in vivo (Supplementary Figure S5). The binary interactions of SEP3, AP3 and PI were also tested and were consistent with published reports (10) (Supplementary Table S1). The interaction of SEP3Δtet and AG was also validated by EMSA, using a single CArG box showing that heterodimers formed and bound DNA (Supplementary Figure S6).
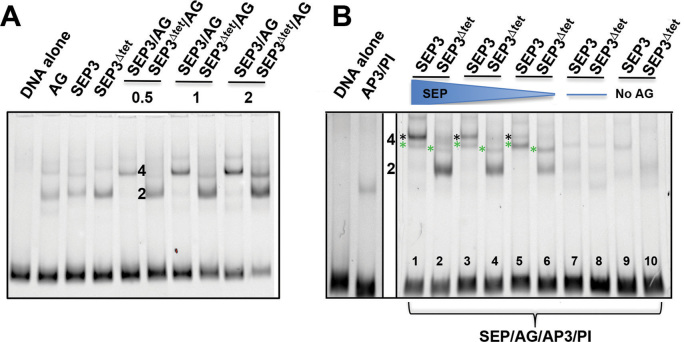
To further decipher the interaction patterns and oligomerization state of SEP3 and SEP3Δtet, EMSAs were performed with SEP3 or SEP3Δtet and the different floral organ identity interaction partners AG, AP3 and PI using a DNA fragment with two binding sites. As shown in Figure 5A, the tetrameric interaction of SEP3Δtet with AG was strongly impaired. At the lowest protein concentration used, SEP3Δtet/AG complexes were predominantly dimeric, resulting in the disappearance of the higher molecular weight band observed for SEP3/AG. Titrating increasing protein concentrations gave rise to a slight amount of higher molecular weight complex for SEP3Δtet/AG, however, the predominant species was a dimer under all conditions tested, indicating lack of cooperative binding by SEP3Δtet/AG (Supplementary Figure S7). In the presence of AP3 and PI, while SEP3/AG tetramers are favored (Figure 5B, lanes 3 and 5), the putative tetrameric complexes with SEP3/AP3/PI/AG were formed (Figure 5B, lane 7). In the presence of SEP3Δtet, SEP3Δtet/AG heterodimers predominate (Figure 5B, lane 4) with a decreased amount of SEP3Δtet/AG/AP3/PI complex formed as compared to SEP3/AG/AP3/PI, indicating reduced cooperative binding and impaired tetramerization. In the absence of SEP3 or SEP3Δtet, an AG/AP3/PI complex of higher molecular weight was observed, suggesting full occupancy of the available MADS binding sites and possible formation of a higher order complex even in the absence of SEP3. Overall, SEP3Δtet was strongly impaired in its ability to bind cooperatively and form tetrameric complexes with its MADS partners. As interactions with AG were most affected by the SEP3Δtet mutation and the observed developmental defects were all in the fourth whorl, the interaction patterns of SEP3Δtet/AG were further explored in the context of the KNU and CRC promoters.
Figure 5.
Analysis of tetramer formation between SEP3 and SEP3Δtet and floral organ identity partners. (A) EMSA of SEP3, SEP3Δtet and AG with increasing protein concentrations (0.5, 1 and 2 μl of in vitro transcription-translation product) with a 103 bp fragment of the SEP3 promoter as per Figure 1. Lanes are as indicated and labeled. For single proteins, 2 μl of in vitro transcription-translation product was used. (B) EMSA of SEP3, SEP3Δtet with floral organ identity partners AP3, PI and AG with the same DNA as in (A). Lanes are as marked with decreasing concentrations of SEP3 and SEP3Δtet with equivolumes of SEP3 or SEP3Δtet and AG present in lanes 5 and 6 and no SEP3 or SEP3 Δtet present in lanes 7 and 8. In lanes 9 and 10, no AG was present but equivolumes of AP3, PI and SEP3 or SEP3Δtet were added. A total of 4 μl of in vitro transcription-translation product was used in all assays. 2 and 4 indicate number of protein molecules bound with green asterisks indicating SEP3 (or SEP Δtet)/AG/AP3/PI tetramer and black asterisks indicating SEP3/AG tetramers.
In vitro binding of SEP/AG to the KNU and CRC promoters
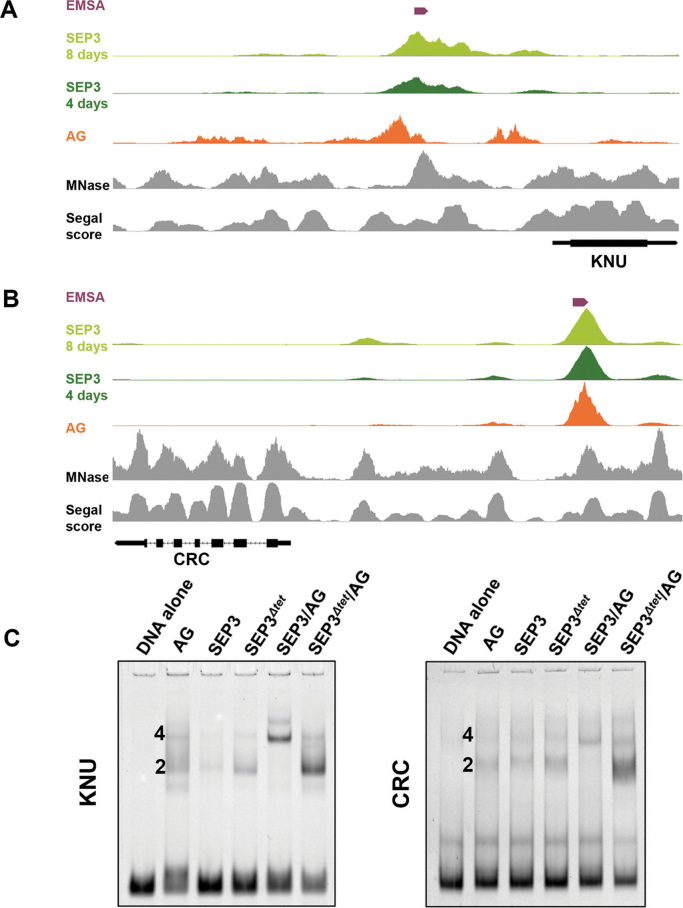
The promoters of both KNU and CRC contain multiple putative binding sites for AG and SEP3 based on ChIP-seq data and binding site prediction (http://jaspar.genereg.net/) (29,35,42,48) (Figure 6). Furthermore, extensive characterization of the KNU promoter has identified binding sites for AG that overlap with Polycomb response elements (PRE) (49) and with predicted SEP3 binding sites. As shown in Figure 6A and B, the AG and SEP3 ChIP-seq binding sites overlap in the KNU and CRC promoters, respectively. Based on these data, promoter sequences from KNU and CRC, each with two putative binding sites for SEP3 and AG, were used for EMSA experiments and the binding patterns shown in Figure 6C. For both the KNU and CRC promoters, SEP3/AG showed robust cooperativity and tetramerization, with the majority of the bound species exhibiting a higher molecular weight shift as compared to the dimeric species. In contrast, SEP3Δtet/AG exhibited dramatically reduced cooperativity and higher order complex formation, with the major species corresponding to a single SEP3Δtet/AG dimer. These results indicate that impaired tetramerization of SEP3Δtet with AG affects the in vitro binding of the proteins at the promoters of KNU and CRC and provides a molecular basis for the observed phenotypes for SEP3 and SEP3Δtet in the triple mutant background.
Figure 6.
Binding of SEP3, SEP3Δtet and AG to KNU and CRC promoters. (A and B) ChIP-seq peaks for SEP3 (light and dark green), AG (orange), MNase signal (33) (gray) and Segal score (34) (gray) for the KNU and CRC promoters, respectively. The DNA fragments used in EMSA assays in C, are indicated in purple. (C) EMSA of SEP3, SEP3Δtet and AG with a 87 bp DNA fragment from the promoter region of KNU and a 136 bp DNA fragment from the promoter region of CRC. 2 and 4 indicate number of protein molecules bound per DNA.
DISCUSSION
The quartet model of floral organ formation presupposes the formation of tetrameric MADS complexes as critical to the proper regulation of downstream target genes, putatively through DNA looping (10,15,16). Here, we begin to decouple the role of DNA-binding from tetramerization by investigating the activity of a naturally occurring splice variant, SEP3Δtet, which we predicted would be impaired in tetramerization based on structural and biochemical studies (14,17). While the physiological role of SEP3Δtet is not known, SEP3 undergoes temperature dependent alternative splicing, with the SEP3Δtet variant showing increased expression at lower temperatures (50). Other MADS genes also exhibit temperature dependent alternative splicing, including the flowering time genes FLOWERING LOCUS M (FLM) and MADS AFFECTING FLOWERING2 (MAF2) (37,51,52). The FLM and MAF2 isoforms expressed preferentially at lower ambient temperatures are able to form complexes with the MADS TF, SHORT VEGETATIVE PHASE (SVP), and delay flowering via repression of the downstream floral pathway integrators, SUPRESSOR OF CONSTANS OVEREXPRESSION 1 (SOC1) and FLOWERING LOCUS T (FT). However, increased ambient temperature results in FLM and MAF2 isoforms that are unable to repress FT and SOC1. Indeed, temperature dependent alternative splicing may be a mechanism used more generally by MADS TFs to tune their activity in response to temperature. One could speculate that the SEP3Δtet isoform may have a role in titrating SEP3 activity in a temperature dependent manner by favoring dimeric versus tetrameric complex formation. Our previous overexpression studies with SEP3Δtet in the WT background demonstrated early flowering and a floral phenotype characterized by perturbations in floral organ number (50), however interpreting the role of the SEP3Δtet isoform in the WT background was not possible due to the presence of the four SEP proteins and the role of tetramerization could not be fully addressed. In order to more fully explore the role of SEP3 tetramerization on flower development and differentiate the function of SEP3-containing dimeric versus tetrameric MADS complexes, transformation of SEP3Δtet in the sep1/2/3 background was required.
Surprisingly, sep1/2/3 plants expressing SEP3 or SEP3Δtet demonstrate a requirement for robust SEP3-driven tetramerization of MADS TF complexes in floral meristem determinacy but seemingly not petal, stamen and carpel identity. SEP3Δtet is able to fulfil the majority of regulatory functions of the SEP3 isoform, including activation of genes required for floral organ specification (4,6). However, SEP3Δtet did not complement the indeterminacy phenotype. Indeed, the continued proliferation of meristematic cells may point to a physiological function of the SEP3Δtet isoform in delaying premature floral meristem termination under low temperature conditions in which cell division is slower, however this remains to be determined. While the putative physiological role of the SEP3Δtet isoform is still speculative, sep1/2/3 plants expressing SEP3Δtet demonstrate a clear and reproducible phenotype with complementation of floral organ identity and defects limited to an indeterminacy phenotype. Complementation of floral organ identity defects may be due to the residual tetramerization capabilities of different combinations of MADS TF partners. Indeed, the floral organ identity tetramers for petal and stamen development putatively contain only a single SEP3 or SEP3Δtet and the presence of three other MADS partner proteins may exercise a compensatory effect on tetramerization and cooperative DNA-binding. We observed that, for the same DNA sequence, the formation of a higher molecular weight species corresponding to four MADS TFs (SEP3Δtet / AP3/PI/AG) was less affected than with two MADS TFs (SEP3Δtet /AG), (Figure 5). In addition, while the in vitro studies described suggest that tetramerization is impaired with SEP3Δtet/AG/AP3/PI, in vivo conditions may favor or stabilize these weak tetrameric MADS complexes or ternary factors may contribute to higher order complex formation. Conversely, tetramer formation may not be absolutely required for activation of target genes during floral organ development with simply the correct dimeric species formed between the A, B, C and E partners and occupancy of the appropriate CArG box binding sites. This second hypothesis is likely in the case of carpel organogenesis as tetramer formation with AG was strongly impaired with SEP3Δtet and cooperative DNA binding dramatically affected, yet the fourth whorl cells adopted carpel characteristics including clear development of the stigma, valve and replum. To fully understand whether dimeric species are sufficient to trigger floral organ development further mutational studies in vitro and in vivo of multiple MADS TFs will need to be performed with complete abrogation of tetramerization and cooperative DNA binding for all partner combinations.
Unfused carpel and extended gynophore together with lack of determinacy are phenotypes observed in sep1/2/3 expressing SEP3Δtet and described for crc and knu mutants, respectively (47,53). CRC and KNU are known to belong to two parallel pathways regulating carpel development and determinacy, are directly activated by AG and also have SEP3 binding sites in their promoter regions (40,41,49,54). Gene expression of CRC and KNU is downregulated in the sep1/2/3 triple versus the sep1/2 double mutant and is restored back to sep1/2 levels of expression in sep1/2/3 SEP3 lines, but not in sep1/2/3 SEP3Δtet lines. This shows a trend with the tetramerization competent SEP3 able to more strongly activate KNU and CRC versus SEP3Δtet. One cannot rule out the possibility that the 14-amino acid deletion in SEP3Δtet results in the loss of a ternary factor interface that impedes recruitment of additional proteins needed for activation of floral meristem determinacy genes. However, consistent with our results indicating SEP3/AG tetramerization is crucial for floral meristem determinacy (55,56), the examination of different ag mutant alleles, AG-Met205 and ag-4, weak and strong mutant alleles of AG, respectively, exhibit similar phenotypes to sep1/2/3 SEP3Δtet. AG-Met205 has meristem determinacy defects but no defects in stamen organogenesis or carpel specification (55). The ag-4 allele shows a stronger phenotype with stamen in the third whorl and indeterminacy in the fourth whorl with no carpel formation. Analysis of these mutant alleles reveals that AG-Met205 has a lysine to methionine mutation that breaks a putative salt bridge important for tetramerization with SEP3 (Supplementary Figure S8). For ag-4, a portion of the K domain is deleted due to skipping of exon 6, resulting in loss of the tetramerization interface while preserving dimerization and DNA-binding (Supplementary Figure S8). These data demonstrating that both AG and SEP3Δtet tetramerization interface mutants share similar phenotypes provides additional evidence that the SEP3/AG tetramer is absolutely required for floral meristem determinacy.
A fundamental question remains as to why SEP3/AG tetramers are required for KNU and CRC activation and seem to play a lesser role in floral organogenesis. One possible explanation is the strong repression of both these genes prior to flower development (Figure 6A and B). AG activation of KNU requires the eviction of the Polycomb repressive complex 2 (PRC2) from the KNU promoter. AG binding sites overlap with putative Polycomb response elements (PRE), and direct competition for these binding sites between AG and PRC2 is thought to play a role in KNU activation by AG (49). If AG must outcompete PRC2 for binding to the KNU promoter, robust tetrameric interactions with SEP3 may be required. Likewise, the CRC promoter shows strong nucleosome positioning based on MNase experiments (33) and is bound by the PRC2 interacting protein EARLY EMBRYONIC FLOWER1 (EMF1) in seedlings, indicating that this gene is in a closed region of chromatin (57). After SEP3 binding, this closed region was shown to open, triggering CRC expression (29). Thus, the inability of SEP3Δtet to fully restore CRC and KNU expression in the triple mutant background suggests that tetrameric SEP/AG complexes are needed to overcome strong repression. This may be due to an important cooperativity or avidity effect, with impaired tetramerization resulting in decreased overall occupancy of the SEP3Δtet/AG complex at its cognate sites. A second intriguing possibility for the misregulation of KNU and CRC by the SEP3Δtet /AG complex is the lack of DNA-looping (15,16,23). The abrogation of tetramerization eliminates SEP3 driven DNA-looping. DNA-looping is known to be important in gene regulation (58), although it is still controversial the extent of DNA-looping engendered by the MADS TF as few chromatin conformation capture studies with sufficient resolution have been performed in plants (59,60). Identification of transient short-range DNA loops by tetrameric MADS complexes will require further investigation to fully address this question.
These in vitro and in vivo experiments provide novel data as to the requirements of tetramerization for MADS TF function in flower development. The absolute requirement for strong tetrameric interactions may be limited to robustly repressed targets (such as CRC and KNU) as floral organ identity programs were successfully launched in the sep1/2/3 triple mutant expressing SEP3Δtet, including carpel development in the fourth whorl. The combination of experiments presented here refines the model for flower and floral organ development and sheds new light on the role of the SEPALLATA MADS TFs in triggering different developmental programs.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank K. Kaufmann and C. Smaczniak for plasmids, SEP3 antibody, helpful discussions and advice, B. Davies, M. Yanofsky and J. Ripoll for sep mutant seeds, and G. Tichtinsky, G. Vachon and R. Dumas for helpful discussions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Action Thématique et Incitative sur Programme (ATIP)-Avenir (to C.Z.); Grenoble Alliance for Integrated Structural Cell Biology (ANR-10-LABX-49-01 to V.H., A.J., C.Z., Q.C., A.S., V.C., F.P.); Agence Nationale de la Recherche (project Flopinet to C.Z., V.H., A.J., F.P., C.C.) and Centre National de la Recherche Scientifique (to S.J.C.). Funding for open access charge: ATIP-Avenir, ANR.
Conflict of interest statement. None declared.
REFERENCES
- 1. O’Maoileidigh D.S., Graciet E., Wellmer F.. Gene networks controlling Arabidopsis thaliana flower development. New Phytol. 2014; 201:16–30. [DOI] [PubMed] [Google Scholar]
- 2. Smaczniak C., Immink R.G., Angenent G.C., Kaufmann K.. Developmental and evolutionary diversity of plant MADS-domain factors: insights from recent studies. Development. 2012; 139:3081–3098. [DOI] [PubMed] [Google Scholar]
- 3. Pelaz S., Ditta G.S., Baumann E., Wisman E., Yanofsky M.F.. B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature. 2000; 405:200–203. [DOI] [PubMed] [Google Scholar]
- 4. Pelaz S., Tapia-Lopez R., Alvarez-Buylla E.R., Yanofsky M.F.. Conversion of leaves into petals in Arabidopsis. Curr. Biol. 2001; 11:182–184. [DOI] [PubMed] [Google Scholar]
- 5. Goto K., Kyozuka J., Bowman J.L.. Turning floral organs into leaves, leaves into floral organs. Curr. Opin. Genet. Dev. 2001; 11:449–456. [DOI] [PubMed] [Google Scholar]
- 6. Honma T., Goto K.. Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature. 2001; 409:525–529. [DOI] [PubMed] [Google Scholar]
- 7. Ditta G., Pinyopich A., Robles P., Pelaz S., Yanofsky M.F.. The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Curr. Biol. 2004; 14:1935–1940. [DOI] [PubMed] [Google Scholar]
- 8. Liu C., Xi W., Shen L., Tan C., Yu H.. Regulation of floral patterning by flowering time genes. Dev. Cell. 2009; 16:711–722. [DOI] [PubMed] [Google Scholar]
- 9. Zahn L.M., Kong H., Leebens-Mack J.H., Kim S., Soltis P.S., Landherr L.L., Soltis D.E., Depamphilis C.W., Ma H.. The evolution of the SEPALLATA subfamily of MADS-box genes: a preangiosperm origin with multiple duplications throughout angiosperm history. Genetics. 2005; 169:2209–2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Theissen G., Saedler H.. Plant biology. Floral quartets. Nature. 2001; 409:469–471. [DOI] [PubMed] [Google Scholar]
- 11. de Folter S., Immink R.G., Kieffer M., Parenicova L., Henz S.R., Weigel D., Busscher M., Kooiker M., Colombo L., Kater M.M. et al. Comprehensive interaction map of the Arabidopsis MADS Box transcription factors. Plant Cell. 2005; 17:1424–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Immink R.G., Tonaco I.A., de Folter S., Shchennikova A., van Dijk A.D., Busscher-Lange J., Borst J.W., Angenent G.C.. SEPALLATA3: the ‘glue’ for MADS box transcription factor complex formation. Genome Biol. 2009; 10:R24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pelaz S., Gustafson-Brown C., Kohalmi S.E., Crosby W.L., Yanofsky M.F.. APETALA1 and SEPALLATA3 interact to promote flower development. Plant J. 2001; 26:385–394. [DOI] [PubMed] [Google Scholar]
- 14. Puranik S., Acajjaoui S., Conn S., Costa L., Conn V., Vial A., Marcellin R., Melzer R., Brown E., Hart D. et al. Structural basis for the oligomerization of the MADS domain transcription factor SEPALLATA3 in Arabidopsis. Plant Cell. 2014; 26:3603–3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Melzer R., Verelst W., Theissen G.. The class E floral homeotic protein SEPALLATA3 is sufficient to loop DNA in ‘floral quartet’-like complexes in vitro. Nucleic Acids Res. 2009; 37:144–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mendes M.A., Guerra R.F., Berns M.C., Manzo C., Masiero S., Finzi L., Kater M.M., Colombo L.. MADS domain transcription factors mediate short-range DNA looping that is essential for target gene expression in Arabidopsis. Plant Cell. 2013; 25:2560–2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Silva C.S., Puranik S., Round A., Brennich M., Jourdain A., Parcy F., Hugouvieux V., Zubieta C.. Evolution of the plant reproduction master regulators LFY and the MADS transcription factors: the role of protein structure in the evolutionary development of the flower. Front. Plant Sci. 2015; 6:1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bensmihen S., To A., Lambert G., Kroj T., Giraudat J., Parcy F.. Analysis of an activated ABI5 allele using a new selection method for transgenic Arabidopsis seeds. FEBS Lett. 2004; 561:127–131. [DOI] [PubMed] [Google Scholar]
- 19. Smyth D.R., Bowman J.L., Meyerowitz E.M.. Early flower development in Arabidopsis. Plant Cell. 1990; 2:755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kaufmann K., Muino J.M., Jauregui R., Airoldi C.A., Smaczniak C., Krajewski P., Angenent G.C.. Target genes of the MADS transcription factor SEPALLATA3: integration of developmental and hormonal pathways in the Arabidopsis flower. PLoS Biol. 2009; 7:e1000090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Conn S.J., Gilliham M., Athman A., Schreiber A.W., Baumann U., Moller I., Cheng N.H., Stancombe M.A., Hirschi K.D., Webb A.A. et al. Cell-specific vacuolar calcium storage mediated by CAX1 regulates apoplastic calcium concentration, gas exchange, and plant productivity in Arabidopsis. Plant Cell. 2011; 23:240–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Carles C.C., Fletcher J.C.. The SAND domain protein ULTRAPETALA1 acts as a trithorax group factor to regulate cell fate in plants. Genes Dev. 2009; 23:2723–2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Smaczniak C., Immink R.G., Muino J.M., Blanvillain R., Busscher M., Busscher-Lange J., Dinh Q.D., Liu S., Westphal A.H., Boeren S. et al. Characterization of MADS-domain transcription factor complexes in Arabidopsis flower development. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:1560–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Higgins D.G. CLUSTAL V: multiple alignment of DNA and protein sequences. Methods Mol. Biol. 1994; 25:307–318. [DOI] [PubMed] [Google Scholar]
- 25. Thompson J.D., Higgins D.G., Gibson T.J.. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic. Acids. Res. 1994; 22:4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Drozdetskiy A., Cole C., Procter J., Barton G.J.. JPred4: a protein secondary structure prediction server. Nucleic. Acids. Res. 2015; 43:W389–W394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Emsley P., Lohkamp B., Scott W.G., Cowtan K.. Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr. 2010; 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. The PyMOL Molecular Graphics System, V., Schrödinger, LLC. [Google Scholar]
- 29. Pajoro A., Madrigal P., Muino J.M., Matus J.T., Jin J., Mecchia M.A., Debernardi J.M., Palatnik J.F., Balazadeh S., Arif M. et al. Dynamics of chromatin accessibility and gene regulation by MADS-domain transcription factors in flower development. Genome Biol. 2014; 15:R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. DS O.M., Wuest S.E., Rae L., Raganelli A., Ryan P.T., Kwasniewska K., Das P., Lohan A.J., Loftus B., Graciet E. et al. Control of reproductive floral organ identity specification in Arabidopsis by the C function regulator AGAMOUS. Plant Cell. 2013; 25:2482–2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Langmead B., Trapnell C., Pop M., Salzberg S.L.. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009; 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008; 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang T., Zhang W., Jiang J.. Genome-wide nucleosome occupancy and positioning and their impact on gene expression and evolution in plants. Plant Physiol. 2015; 168:1406–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kaplan N., Moore I.K., Fondufe-Mittendorf Y., Gossett A.J., Tillo D., Field Y., LeProust E.M., Hughes T.R., Lieb J.D., Widom J. et al. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2009; 458:362–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mathelier A., Fornes O., Arenillas D.J., Chen C.Y., Denay G., Lee J., Shi W., Shyr C., Tan G., Worsley-Hunt R. et al. JASPAR 2016: a major expansion and update of the open-access database of transcription factor binding profiles. Nucleic. Acids. Res. 2016; 44:D110–D115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Grant C.E., Bailey T.L., Noble W.S.. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011; 27:1017–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pose D., Verhage L., Ott F., Yant L., Mathieu J., Angenent G.C., Immink R.G., Schmid M.. Temperature-dependent regulation of flowering by antagonistic FLM variants. Nature. 2013; 503:414–417. [DOI] [PubMed] [Google Scholar]
- 38. Bowman J.L., Smyth D.R., Meyerowitz E.M.. Genes directing flower development in Arabidopsis. Plant Cell. 1989; 1:37–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mizukami Y., Huang H., Tudor M., Hu Y., Ma H.. Functional domains of the floral regulator AGAMOUS: characterization of the DNA binding domain and analysis of dominant negative mutations. Plant Cell. 1996; 8:831–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sun B., Xu Y., Ng K.H., Ito T.. A timing mechanism for stem cell maintenance and differentiation in the Arabidopsis floral meristem. Genes Dev. 2009; 23:1791–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Liu X., Kim Y.J., Muller R., Yumul R.E., Liu C., Pan Y., Cao X., Goodrich J., Chen X.. AGAMOUS terminates floral stem cell maintenance in Arabidopsis by directly repressing WUSCHEL through recruitment of Polycomb Group proteins. Plant Cell. . 2011; 3654–3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gomez-Mena C., de Folter S., Costa M.M., Angenent G.C., Sablowski R.. Transcriptional program controlled by the floral homeotic gene AGAMOUS during early organogenesis. Development. 2005; 132:429–438. [DOI] [PubMed] [Google Scholar]
- 43. Alvarez J., Smyth D.R.. CRABS CLAW and SPATULA, two Arabidopsis genes that control carpel development in parallel with AGAMOUS. Development. 1999; 126:2377–2386. [DOI] [PubMed] [Google Scholar]
- 44. Prunet N., Morel P., Thierry A.M., Eshed Y., Bowman J.L., Negrutiu I., Trehin C.. REBELOTE, SQUINT, and ULTRAPETALA1 function redundantly in the temporal regulation of floral meristem termination in Arabidopsis thaliana. Plant Cell. 2008; 20:901–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zuniga-Mayo V.M., Marsch-Martinez N., de Folter S.. JAIBA, a class-II HD-ZIP transcription factor involved in the regulation of meristematic activity, and important for correct gynoecium and fruit development in Arabidopsis. Plant J. 2012; 71:314–326. [DOI] [PubMed] [Google Scholar]
- 46. Yumul R.E., Kim Y.J., Liu X., Wang R., Ding J., Xiao L., Chen X.. POWERDRESS and diversified expression of the MIR172 gene family bolster the floral stem cell network. PLos Genet. 2013; 9:e1003218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Payne T., Johnson S.D., Koltunow A.M.. KNUCKLES (KNU) encodes a C2H2 zinc-finger protein that regulates development of basal pattern elements of the Arabidopsis gynoecium. Development. 2004; 131:3737–3749. [DOI] [PubMed] [Google Scholar]
- 48. Lee J.Y., Baum S.F., Oh S.H., Jiang C.Z., Chen J.C., Bowman J.L.. Recruitment of CRABS CLAW to promote nectary development within the eudicot clade. Development. 2005; 132:5021–5032. [DOI] [PubMed] [Google Scholar]
- 49. Sun B., Looi L.S., Guo S., He Z., Gan E.S., Huang J., Xu Y., Wee W.Y., Ito T.. Timing mechanism dependent on cell division is invoked by Polycomb eviction in plant stem cells. Science. 2014; 343:1248559. [DOI] [PubMed] [Google Scholar]
- 50. Conn V.M., Hugouvieux V., Nayak A., Conos S.A., Capovilla G., Cildir G., Jourdain A., Tergaonkar V., Schmid M., Zubieta C. et al. A circRNA from SEPALLATA3 regulates splicing of its cognate mRNA through R-loop formation. Nat Plants. 2017; 3:17053. [DOI] [PubMed] [Google Scholar]
- 51. Lee J.H., Ryu H.S., Chung K.S., Pose D., Kim S., Schmid M., Ahn J.H.. Regulation of temperature-responsive flowering by MADS-box transcription factor repressors. Science. 2013; 342:628–632. [DOI] [PubMed] [Google Scholar]
- 52. Airoldi C.A., McKay M., Davies B.. MAF2 is regulated by temperature-dependent splicing and represses flowering at low temperatures in parallel with FLM. PLoS One. 2015; 10:e0126516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bowman J.L., Smyth D.R.. CRABS CLAW, a gene that regulates carpel and nectary development in Arabidopsis, encodes a novel protein with zinc finger and helix-loop-helix domains. Development. 1999; 126:2387–2396. [DOI] [PubMed] [Google Scholar]
- 54. Lee J.Y., Baum S.F., Alvarez J., Patel A., Chitwood D.H., Bowman J.L.. Activation of CRABS CLAW in the nectaries and carpels of Arabidopsis. Plant Cell. 2005; 17:25–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sieburth L.E., Running M.P., Meyerowitz E.M.. Genetic separation of third and fourth whorl functions of AGAMOUS. Plant Cell. 1995; 7:1249–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sieburth L.E., Meyerowitz E.M.. Molecular dissection of the AGAMOUS control region shows that cis elements for spatial regulation are located intragenically. Plant Cell. 1997; 9:355–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kim S.Y., Lee J., Eshed-Williams L., Zilberman D., Sung Z.R.. EMF1 and PRC2 cooperate to repress key regulators of Arabidopsis development. PLos Genet. 2012; 8:e1002512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bulger M., Groudine M.. Functional and mechanistic diversity of distal transcription enhancers. Cell. 2011; 144:327–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Liu C., Weigel D.. Chromatin in 3D: progress and prospects for plants. Genome Biol. 2015; 16:170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wang C., Liu C., Roqueiro D., Grimm D., Schwab R., Becker C., Lanz C., Weigel D.. Genome-wide analysis of local chromatin packing in Arabidopsis thaliana. Genome Res. 2015; 25:246–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.