Abstract
Transfer RNA-derived small RNAs (tsRNAs) are an emerging class of small RNAs, yet their regulatory roles have not been well understood. Here we studied the molecular mechanisms and consequences of tsRNA-mediated regulation in Drosophila. By analyzing 495 public small RNA libraries, we demonstrate that most tsRNAs are conserved, prevalent and abundant in Drosophila. By carrying out mRNA sequencing and ribosome profiling of S2 cells transfected with single-stranded tsRNA mimics and mocks, we show that tsRNAs recognize target mRNAs through conserved complementary sequence matching and suppress target genes by translational inhibition. The target prediction suggests that tsRNAs preferentially suppress translation of the key components of the general translation machinery, which explains how tsRNAs inhibit the global mRNA translation. Serum starvation experiments confirm tsRNAs participate in cellular starvation responses by preferential targeting the ribosomal proteins and translational initiation or elongation factors. Knock-down of AGO2 in S2 cells under normal and starved conditions reveals a dependence of the tsRNA-mediated regulation on AGO2. We also validated the repressive effects of representative tsRNAs on cellular global translation and specific targets with luciferase reporter assays. Our study suggests the tsRNA-mediated regulation might be crucial for the energy homeostasis and the metabolic adaptation in the cellular systems.
INTRODUCTION
Small RNAs, such as microRNAs (miRNAs), small interfering RNAs (siRNAs) and PIWI-interacting RNAs (piRNAs), regulate gene expression via RNA interference (RNAi) in metazoans (1,2). The discovery of tRNA-derived small RNAs (tsRNAs, also named as tRNA-derived fragments or tRFs) has considerably expanded the small RNA repertoire (3–6). Multiple lines of evidence have suggested that tsRNAs are not simply the random products of tRNA degradation, but play important roles in biological processes (3,6–12). First, in yeasts and mammalian cells tRNAs are cleaved at specific sites by different endonucleases, which generates tsRNAs with precise cleavage signatures (13–15). Such cleavage either occurs in the anticodon loop of the tRNA to generate the ‘tRNA halves’ of ∼35 nucleotides (nt) in length, or in the D-loop or T-loop to produce short 5′-tsRNAs or 3′-tsRNAs, respectively (14–20). Second, some tsRNAs could be induced by stress, which might be an adaptive feature for the cellular systems (19–22). Third, the expression levels of tsRNAs can be regulated in animal tissues or cell types during development (5,8,23,24). Last, certain tsRNAs have been experimentally verified to play regulatory roles in the biological processes such as DNA damage response (25), cell proliferation and cancer progression (26–29), transposon silencing (30,31), sperm maturation (6,32) or epigenetic inheritance (12). In summary, tsRNAs might have important biological functions, yet their regulatory function deserves to be further elucidated.
It has been demonstrated that tsRNAs could mediate the global translational regulation of gene expression through diverse mechanisms (13,19,20,29,33–35). For example, in mammalian cells, the angiogenin-induced tsRNAs could promote eIF2α-independent translational arrest and stress granule assembly (19,20), or displace eIF4G/eIF4A from mRNAs to slow down translational initiation (13). Moreover, recent studies suggest that human 5′ tsRNAGln interacts with the multi-synthetase complex to affect translation (33,34). Recently, it was found that a 22-nt LeuCAG3′tsRNA from humans is essential for cell viability since it binds the mRNAs of ribosomal proteins (RPS28 and RPS15) to enhance their translation (29). It is also reported that a stress-dependent tsRNA in Halobacteria directly binds to ribosomes and reduces protein synthesis (35). In short, the above studies represent various scenarios in which the regulatory effects rely on the interactions between tsRNAs and the general translation machinery or high-order cytoplasmic structures such as polyribosomes, processing bodies, and stress granules. By contrast, other studies have demonstrated that complementarity between some tsRNAs and their target mRNAs is indispensable for efficient silencing (10,23,25,36,37). In support of these discoveries, in many organisms, tsRNAs are found associated with Argonaute (AGO) proteins which are essential for target recognition in an RNAi manner (10,23,25,37,38). In particular, a re-analysis of the human CLASH (cross-linking, ligation and sequencing of hybrids) data (39) has identified various AGO1-tsRNA-mRNA chimeras, further suggesting that tsRNAs regulate targets via RNAi (23). Therefore, as these discrepancies suggest, a unifying model to describe how tsRNAs recognize and regulate the target mRNAs has been lacking.
The discrepancy in findings related to the molecular mechanisms of tsRNA-mediated regulation might be (partly) caused by the diversity of tsRNA biogenesis and function across species. For instance, the stress-induced tsRNAs are cleaved by Rny1 (a member of the RNase T2 family) in yeasts (40) and angiogenin (a vertebrate-specific member of RNase A family) in human cells (19), while the biogenesis of tsRNAs is not clear in other organisms such as Drosophila. Moreover, the locations of tsRNAs in the tRNA precursors (5′, middles or 3′ ends) also exhibited distinct patterns across species (23,24). Hence, a thorough investigation in one model organism is necessary toward a general framework that unifies the mechanisms underlying tsRNA-mediated gene regulation.
Here we comprehensively examined the role of tsRNAs in modulating gene expression in Drosophila. We present evidence that tsRNAs are conserved, prevalent and abundant in Drosophila. With cellular transfection of tsRNA mimics, mRNA-seq, ribosome profiling (Ribo-seq) (41,42) and luciferase reporter assay, we show that tsRNAs inhibit the translational efficiency of specific genes with conserved antisense pairing. The target prediction and our experimental evidence suggest that tsRNAs preferentially suppress translation of the key components of the general translation machinery. We also found that tsRNA-mediated repression depends on AGO2, suggesting tsRNAs inhibit specific targets in an RNAi-like manner. Finally, we provide a unifying model that describes how tsRNAs participate in cellular starvation response by regulating translation of specific and general mRNAs.
MATERIALS AND METHODS
S2 cell culture, transfection, serum starvation and other treatments
Drosophila S2 cells were cultured in Schneider's Insect Medium (Sigma) containing 10% (by volume) heat-inactivated fetal bovine serum, 100 U/ml penicillin and 100 μg/ml streptomycin (Thermo Fisher) at 27°C without CO2 for 24 h to reach 2–4 × 106 cells/ml before further treatments. For serum starvation assays, the cells were subsequently cultured in serum-containing or serum-free medium for 24 additional hours before harvesting. For tsRNA transfections, the cells were bathed with the synthetic mimics or the single-stranded negative control RNA (ss-NC) at a final concentration of 100 nM using Lipofectamine 2000 transfection reagent according to the manufacturer’s instructions (Thermo Fisher) for 32 h. Mock-treated cells were also treated with Lipofectamine 2000 in the same manner, except that no synthetic mimic or NC RNA was used. For AGO2 knockdown, the cells were bathed with a siRNA duplex that specifically targets the AGO2 mRNA or the double-stranded negative control small RNA (ds-NC) at a final concentration of 30 nM for 24 h. See Supplementary Table S1 for the oligomer sequences. For rapamycin treatment, rapamycin (Sigma) was diluted with Dimethyl sulfoxide (DMSO), and the cells were treated at a final concentration of 274 nM for 2 h. As the control in the rapamycin-treatment experiment, we treated S2 cells with DMSO for 2 h.
Western blotting was performed as a conventional procedure using horseradish peroxidase-conjugated secondary antibody (1:8000, CWBiotech). Primary antibodies specific for mouse monoclonal β-Tubulin (1:1000, CWBiotech) and rabbit polyclonal AGO2 (1:1000, Abcam) were used.
Luciferase reporter assays
To monitor the repressive effects of tsRNAs on the global translational activities, we co-transfected the psiCHECK-2 vector (Promega, 100 ng) and a tsRNA mimic (or the tsRNA mimic cocktail) at different final concentration (12.5, 25, 50, 100 and 200 nM, respectively) into S2 cells with the Lipofectamine 2000 transfection reagent (Thermo Fisher). To validate the target site of tsRNA T16, we chose the target site sequence corresponding to the full-length synthetic tsRNA mimic (GenePharma), extending 5 bp at both ends to form a sequence around 30 nt, synthesized a 3 × repetition of this sequence (Sangon) and cloned it to the multiple cloning region (1640–1674 bp) of psiCHECK-2 vector. The empty or the target-site containing psiCHECK-2 plasmids (100 ng), together with NC or tsRNA mimic (final concentration of 100 nM), were co-transfected into S2 cells with Lipofectamine 2000. The luciferase activities of firefly and Renilla were measured according to the manual of the Dual-Luciferase Reporter Assay System 48 h after transfection.
Experimental and analytical procedures for RNA sequencing and ribosome profiling
The ribosome profiling procedures were performed according to a previous study (43), with some modifications. See Supplementary Methods for the detailed experimental and analytical procedures.
Public datasets
Small RNA sequencing datasets of Drosophila melanogaster, Drosophila simulans and Drosophila virilis were downloaded from the NCBI website. Supplementary Table S2 summarizes the accession numbers and other relevant information. The small RNAs were mapped on the reference of genomes or sequences with Bowtie2 (44). The miRNA annotations were downloaded from miRBase V21 (www.mirbase.org).
Prediction of small RNA target sites
The mRNA sequences and annotations were downloaded from www.FlyBase.org (r6.06). The conserved targets of AGO2-bound tsRNAs were predicted by requiring 7-mer antisense perfect match to the tsRNAs, and are conserved between D. melanogaster and D. virilis. The genome alignments between D. melanogaster and D. virilis were downloaded from UCSC Genome Browser (genome.ucsc.edu) (45). AGO1-bound or AGO2-bound tsRNAs information is based on previously published IP-seq results (46).
We defined the miRNAs that are preferentially bound with AGO2 rather than AGO1 in IP-seq experiments based on the following criteria: (i) their raw reads > 50, (ii) the ratio of normalized mapped AGO2-bound reads to AGO1-bound reads is ≥ 2-folds. For the miRNAs that are expressed in the normal or starved S2 cells, we employed TargetScan6.0 (47) to identify the seed (position 2–8) pairing target sites in the 3′ UTRs based on genome-wide alignments of 12 Drosophila species downloaded from UCSC Genome Browser. We tested the conservation pattern for each putative target site in D. melanogaster using phyloP (48). We treated the target sites in 3′ UTRs with phyloP P < 0.001 as bona fide target sites of miRNAs.
We retrieved the AUB-bound small RNAs (Sequence Read Archive accession numbers: SRR060648, SRR1434932, SRR1568760, SRR2147102 and SRR513394) sequenced by previous studies (49–52), characterized the 23–29 nt tsRNAs, and pooled the data after normalizing the library size. We predicted the target mRNAs of these AUB-bound tsRNAs with a similar pattern of piRNA target recognition (53,54). Briefly, we used RNAhybrid (55) to scan all the transcripts for the target sites with the following requirements: (i) perfect pairing between the nucleotides 2–11 of the tsRNAs and the target sites, and (ii) at most four mismatches between nucleotides 12–21 of the tsRNAs and target sites.
Analysis of the 5′TOP-like signature and the genes orthologous to known human TOP genes
For each protein-coding transcript annotated in FlyBase r6.06, the nucleic acid base composition within the first eight nucleotides was counted using a custom script in Perl. Totally 115 5′TOP genes were defined in Drosophila. Sixty seven genes are orthologous to the well-characterized human 5′TOP genes (56) based on the gene orthologous relationship retrieved from Ensembl BioMart (www.ensembl.org/biomart). Another 48 genes were defined as 5′TOP genes with the two criteria: (i) with eight pyrimidines or beginning with Cytosine and have seven pyrimidines out of the first eight nucleotides of the mRNA transcript, and (ii) with log2 (TE) < −0.5 in rapamycin treatment as revealed by the Ribo-seq results.
AGO2 CLIP-seq data analysis
The AGO2 cross-linking and immunoprecipitation sequencing (CLIP-seq) data of nuclear extract of S2 cells were taken from (57) and the Nascent-seq data of S2 cells (58) were downloaded from sequence read archive (SRA) under accession SRP008848. Both the CLIP-seq and Nascent-seq reads were mapped to reference genome of D. melanogaster with STAR (59) using default parameters. The RPKM for a CDS (or full-length mRNA) was calculated as (reads uniquely mapped to that region)/(length of that region) × 109/(total number of uniquely mapped reads). For each gene, the longest transcript was used in the analysis, and the fold enrichment (fe) score for that transcript is calculated as RPKMCLIP-seq/RPKMNascent-seq. To search the 7-mer motifs that are associated with preferential AGO2 binding, we examined the occurrences of all the possible 47 = 16 384 motifs in the CDS (or mRNA) of each gene. For each 7-mer motif, we grouped the expressed genes into 100 bins based on increasing densities of the motif and detected the correlation between the density of that motif and the mean fe score in each bin. We conducted the analyses using the CDS regions and full-length mRNAs separately, and obtained very similar results.
RESULTS
tsRNAs contribute significantly to the small RNA repertoire of Drosophila
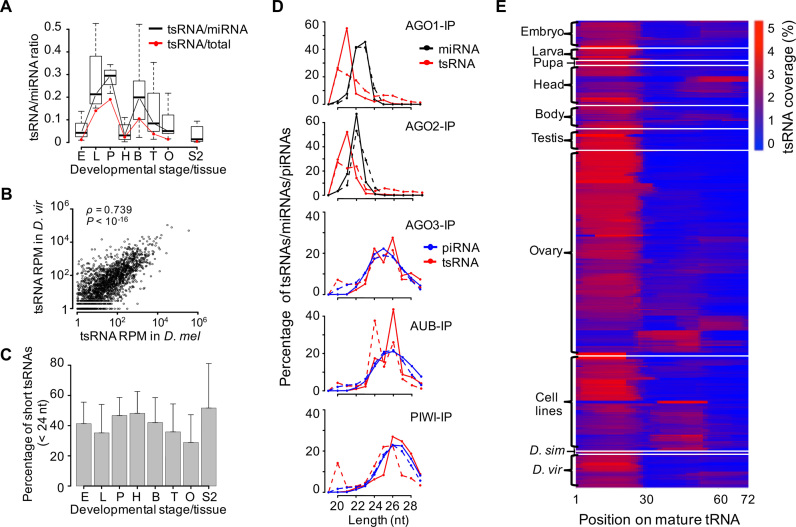
To identify experimentally detectable tsRNAs in Drosophila, we screened 495 publicly available deep-sequencing libraries of small RNAs, which encompassed three species (D. melanogaster, D. simulans and D. virilis; the number of libraries is N = 457, 2 and 36, respectively) and 21 cell lines/developmental stages (Supplementary Table S2). More than 5 billion small RNAs with a length of 20–29 nt were mapped on the reference genomes, and 169 850 445 of these reads were mapped to the mature tRNA sequences. The abundance ratio of tsRNAs to miRNAs varied widely across libraries, with a median ratio of 7.2, 7.5 and 5.5% in D. melanogaster, D. simulans and D. virilis, respectively. Interestingly, we found the numbers of tsRNA reads are even higher than those of miRNA reads in certain libraries of D. melanogaster (Supplementary Table S2), suggesting that tsRNAs are not uncommon in Drosophila. Among various developmental stages of D. melanogaster, the total abundance of tsRNAs was the highest in pupae and the lowest in embryos and adult heads (Figure 1A), which is very similar to the expression pattern of the mature tRNAs (60). We in total detected 3083 distinct tsRNA species (each having a unique sequence) located in the 5′ ends of tRNAs of D. virilis, and about 83.4% of these tsRNA species were also detected in the libraries of D. melanogaster (with identical sequences), suggesting the tsRNA sequences are overall highly conserved. The abundance of the 5′-tsRNAs was significantly correlated in D. melanogaster and D. virilis (Spearman’s ρ = 0.739, P < 10−16, Figure 1B), suggesting the expression levels of tsRNAs or the source-tRNAs are constrained by natural selection during evolution. Consistent with previous observations that tsRNAs are preferentially generated from certain tRNAs (23), we found tRNAGly, tRNAGlu, tRNALys and tRNAAsp contribute the most to the total tsRNAs (on average 15.3, 13.8, 9.9 and 8.1%, respectively).
Figure 1.
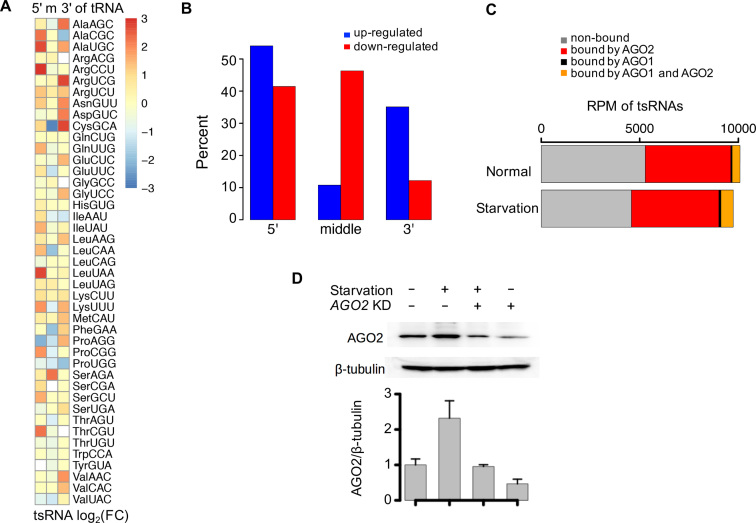
tsRNAs are conserved, abundant and prevalent in Drosophila. (A) Boxplots of the ratios of tsRNA reads to miRNA reads in individual developmental categories (embryos, larvae, pupae, heads, bodies, testes and ovaries, each represented by the initial uppercase letter) and S2 cells of Drosophila melanogaster, based on a total of 147 small RNA libraries that were prepared without any specific isolation step or treatments. In the boxplots throughout the manuscript, the inside bands represent the median values, the bottom and top of the boxes represent the first and third quantiles and the whiskers extend to the most extreme data points which are no more than 1.5 interquartile range. Also shown are the average ratios of tsRNA reads to all genome-mapped reads excluding rRNAs, snRNAs and snoRNAs (red line). (B) The normalized abundances of individual 20–22 nt tsRNA species (N = 1725 with RPM > 1) that were shared in D. melanogaster (x-axis) and Drosophila virilis (y-axis), measured as reads per million (RPM). (C) The percentage of short tsRNAs (20–23 nt) out of the total tsRNAs (20–29 nt) in the small RNA libraries of different developmental categories. For each developmental stage/tissue/cell-line (defined as in A), the mean and s.d. of the percentage were given. (D) The distributions of tsRNA lengths in AGO1, AGO2, AGO3, AUB and PIWI-IP libraries. The AGO1 and AGO2-IP results were extracted from two sets of studies in S2 cells (GSM280087, GSM280088; SRR768820, SRR768821); the AGO3, AUB and PIWI-IP results were extracted from two sets of studies in female ovaries (SRR060648, SRR060649, SRR060651; SRR2147101, SRR2147102, SRR2147103). The black lines represent the small RNA reads mapped to the primary miRNAs; the red lines represent the small RNA reads mapped to tRNAs; the blue lines represent the 23–29 nt small RNA reads mapped to the whole genome excluding miscRNAs, miRNAs and tRNAs. (E) The relative coverage of tsRNAs at each nucleotide position on the mature tRNAs in each library (N = 495).
In Drosophila, miRNAs, and siRNAs, which have the similar length (20–23 nt), are primarily bound by AGO1 and AGO2 respectively (2,61); while piRNAs, which are 24–29 nt in length, are bound by AGO3, PIWI and AUB primarily in germline cells and early embryos (46,62–70). Interestingly, the length of tsRNAs in S2 cells, heads and pupae were generally shorter than those in embryo, testes or ovaries (the mean proportion of reads shorter than 24 nt is 0.517, 0.481 and 0.467 versus 0.413, 0.359 and 0.288 for the former three versus the latter three categories of samples, Figure 1C; the length distribution is presented in Supplementary Figure S1A), which is analogous to the length comparison between siRNAs/miRNAs and piRNAs. To exclude the possibility that the RNA isolation procedures varying between different studies might result in the detection bias, we analyzed the distributions of tsRNAs that were associated with different AGO proteins based on previously published IP-seq libraries (46,52,67,71). The short tsRNAs (20–22 nt) were mainly detected in the AGO1 and AGO2 IP-seq of S2 cells (upper panels of Figure 1D), whereas the long tsRNAs (23–29 nt) were primarily enriched in the AGO3, AUB and PIWI IP-seq libraries of ovaries (lower panels of Figure 1D). Consistent with previous observations that tsRNAs can be preferentially bound by different AGO proteins (10,23,37), the diversity of tsRNA lengths might be shaped by their interactions with different AGO proteins across Drosophila tissues.
Similar to previous observations in mammals (14,23), Drosophila tsRNAs also have asymmetrical distributions in the tRNA precursors. On average, the 5′-tsRNAs that were located before the anticodon loop of the tRNAs accounted for 84.2% and 78.2% of the total tsRNA reads in the libraries of D. melanogaster and D. virilis, respectively. The enrichment of 5′-tsRNAs was well manifested when we calculated the normalized base coverage at each position along a generalized tRNA in each library (Figure 1E). Curiously, in 81 (16.4%) out of the 495 libraries either the middle-tsRNAs (i.e. overlapping with the anticodon loop) or 3′-tsRNAs (i.e. following the anticodon loop) were over-represented, although at this moment we are unable to attribute this pattern to specific tissues, genetic backgrounds or experimental procedures (such as AGOs-IP and β-elimination). Although the length of tsRNAs varied across the 495 libraries, the general trend is that in a library, the average lengths of the 5′-tsRNAs are significantly longer than those of the middle-tsRNAs (the relative ratio is 1.06 ± 0.05 across libraries; P < 10−10, paired t-test; Supplementary Figure S1B) and the middle-tsRNAs are significantly longer than the 3′-tsRNAs (the relative ratio is 1.02 ± 0.06 across libraries; P < 10−10, paired t-test; Supplementary Figure S1C).
Altogether, our results suggest that tsRNAs are conserved, prevalent and abundant in Drosophila. Moreover, these tsRNAs tend to be bound by different AGO proteins in somatic and germline cells, and biased toward 5′ end of tRNAs.
Drosophila tsRNAs potentially inhibit global mRNA translation
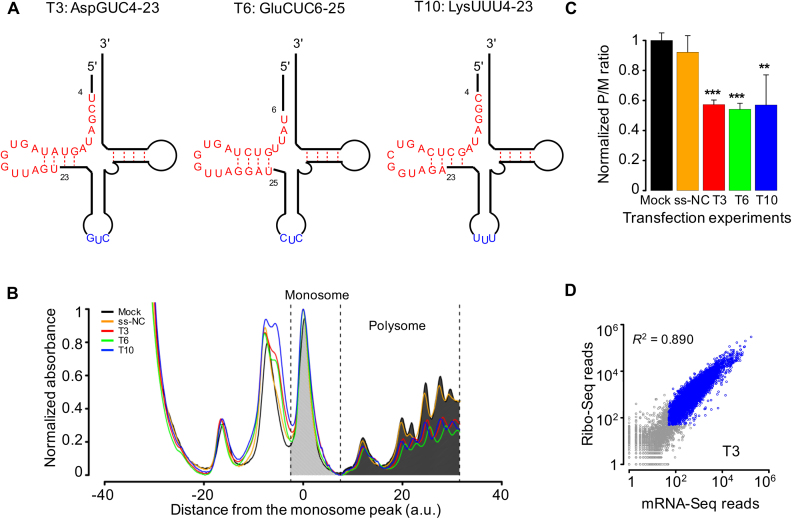
To probe their cellular functions, we chose 12 tsRNAs: AspGUC1-20, AspGUC1-29, AspGUC4-23, GlnUUG36-55, GluCUC1-20, GluCUC6-25, GluCUC27-46, GlyGCC3-22, GlyGCC4-23, LysUUU4-23, LysUUU8-27 and ValCAC35-54 (each tsRNA was denoted by the amino acid, anti-codon and the start and end positions of the tRNA; Figure 2A). These 12 tsRNAs, abbreviated as T1 through T12, respectively, were overall abundant in Drosophila, and most of them (11 out 12) were bound by AGO1 or AGO2 in the IP-seq experiments (Supplementary Table S3). For each of these tsRNAs, we transfected the single-stranded mimic into Drosophila S2 cells. We conducted two sets of control experiments side-by-side, either by treating the cells with only the transfection reagent (‘mock’) or by transfecting the cells with a ‘negative control’ single-stranded small RNA (‘ss-NC’) as previously described (72,73).
Figure 2.
Transfections of tsRNA mimics into S2 cells repress global translational activities. (A) Schematic representations of the structures of three tsRNAs tsRNAAspGUC (T3), tsRNAGluCUC (T6) and tsRNALysUUU (T10) in the mature tRNAs. tsRNAs are in red and the anti-codons in blue. (B) Absorbance profiles (at a UV wavelength of 254 nm) of RNAs partitioned using 10–45% sucrose gradients from S2 cells transfected without any RNA sequence (Mock, black), with a negative-control small RNA (ss-NC, orange), with tsRNA T3 (red), T6 (green) or T10 (blue). For each profile, both the absorbance and the position were normalized according to the monosome peak. Dashed lines indicate the boundaries of the monosome and polysome partitions for a measurement of the overall translational activity. (C) The ratios of the aggregate intensities within polysome partition (P) to the monosome partition (M) normalized by the median value of the control experiments. Error bars represent one standard deviation computed from three repeated experiments. Asterisks indicate significant different P/M ratios between transfection and mock: *P < 0.05; **P < 0.01; ***P < 0.001. (D) Scatter plots of the Ribo-seq read counts against the mRNA-seq read counts of individual genes in T3 transfection experiments. Blue circles represent genes that have >50 mRNA-seq reads and >50 Ribo-seq reads.
tsRNAs potentially suppress the global mRNA translational activities in mammals (13,19,20,33,34). To test whether Drosophila tsRNAs affect mRNA translation, we performed quantitative analysis of the polyribosome (polysome) fractionation profiles, which has been used to measure the global translational rate (74–77). Based on the centrifuge sedimentation patterns, we partitioned the mRNA–ribosome complex into two distinct fractions: a monosome fraction, in which the mRNA molecule is bound to a single ribosome and a polysome fraction, in which one mRNA molecule is translated by multiple ribosomes (Figure 2B and Supplementary Figure S2A). The polysome/monosome (P/M) ratio analysis suggests nearly all the examined tsRNAs potentially reduced the global translational activities in the transfection experiments (Figure 2C and Supplementary Figure S2B). In particular, relative to mock, transfection of T3 (AspGUC4-23), T6 (GluCUC6-25) or T10 (LysUUU4-23) resulted in considerably lower P/M ratios (mean ± s.d. was 67.4 ± 2.0%, 72.1 ± 2.9% and 57.3 ± 5.7%, respectively; Student’s t-test P < 0.01 in each comparison; Figure 2C). These results suggest that the endogenous tsRNAs of Drosophila we examined could suppress the global mRNA translational activities. Future studies are needed to examine whether the repressive effects of these tsRNAs are affected by their nucleotide compositions, lengths or their start and end positions on the source-tRNAs.
tsRNAs regulate translation of target genes through conserved antisense matching
To investigate the molecular mechanism by which tsRNAs repress the global translational activity, we carried out mRNA-seq and Ribo-seq to quantify the alterations in mRNA abundance and translational activities in S2 cells that were transfected with T3, T6 or T10 single-stranded mimic (‘Materials and Methods’ section). We also sequenced the mock and the ss-NC transfected S2 cells side by side as negative controls. We mapped the mRNA-seq and the RPF (ribosome protected fragment) reads on the reference genome of D. melanogaster using STAR (59) and summarized the gene expression levels with HTSeq-count (78) (see Table 1 and Supplementary Table S4 for the mapping statistics). Overall, in each sample we obtained significant correlations between the mRNA-seq and RPF read counts for the protein-coding genes (Figure 2D; Supplementary Figure S3A and B). We normalized the read counts and calculated the fold change (FC) of mRNAs (FCmRNA) or RPFs (FCRPF) in the tsRNA transfection experiments using DESeq2 (79). We measured the protein translational efficiency (TE = RPF/mRNA) as previously described (41,80–83), and contrasted TE in tsRNA-transfected versus mock or ss-NC transfected S2 cells (FCTE = FCRPF/FCmRNA) to evaluate the inhibitory efficacy of tsRNAs at the translational level.
Table 1. Gene expression changes for tsRNA target genes in S2 cells under different conditions.
| Experiment | Control | No. of target genes | log2 (FCmRNA) (mean ± s.d.) | P(FCmRNA) | log2 (FCTE) (mean ± s.d.) | P(FCTE) |
|---|---|---|---|---|---|---|
| T3 | ss-NC | 396 | 0.067 ± 0.21 | 1.37 × 10−7 | −0.28 ± 0.51 | 6.11 × 10−8 |
| T6 | ss-NC | 381 | 0.074 ± 0.20 | 5.77 × 10−8 | −0.14 ± 0.25 | < 2.20 × 10−16 |
| T10 | ss-NC | 646 | 0.026 ± 0.17 | 0.0006 | −0.14 ± 0.33 | < 2.20 × 10−16 |
| AGO2 KD | ds-NC | 351 | 0.053 ± 0.45 | 0.003 | 0.057 ± 0.35 | 2.22 × 10−4 |
| Starvation | Normal | 408 | 0.17 ± 0.55 | 1.91 × 10−10 | −0.17 ± 0.58 | 7.45 × 10−11 |
| Starvation + AGO2 KD | Starvation + ds-NC | 408 | 0.048 ± 0.38 | 0.42 | −0.040 ± 0.23 | 0.013 |
In total 6511 genes that were expressed in the S2 cells cultured under normal conditions were used in all the analyses.
In the T3, T6 and T10 experiments, the target mRNAs were expressed in S2 cells and had conserved 7-mer sites antisense pairing with T3, T6 and T10 tsRNAs, respectively.
In the AGO2 KD (knock-down) experiment, 351 mRNAs expressed in S2 cells had conserved target sites of AGO2-bound tsRNAs (target sites per kb >1.5).
In the serum starvation and serum starvation+AGO2 KD experiments, 408 target mRNAs expressed in S2 cells have UpSites >3.
P(FCmRNA) and P(FCTE) were calculated based on the comparisons (Kolmogorov–Smirnov tests) between target genes of tsRNAs and the remaining genes in the mRNA-seq and Ribo-seq data, respectively.
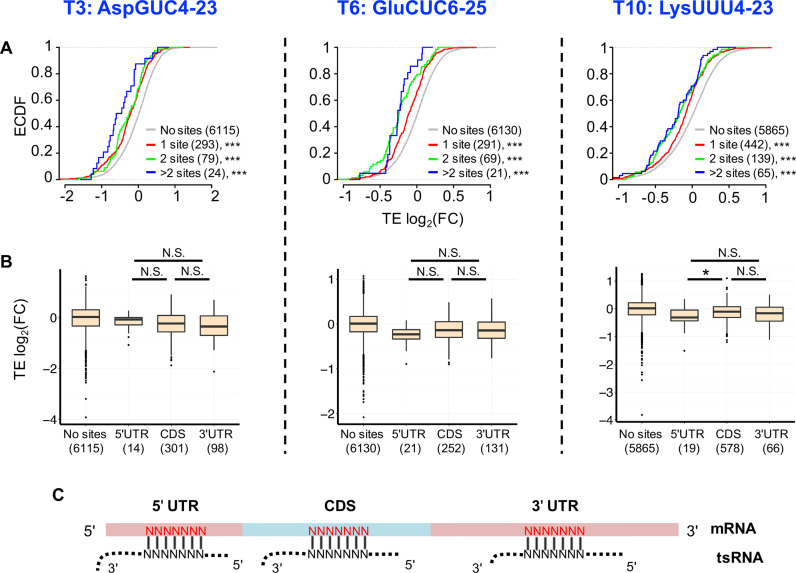
For each transfected tsRNA (T3, T6 or T10), we extensively searched for the motifs in the mRNAs that were associated with mRNA destabilization or translational inhibition (‘Materials and Methods’ section). Our results are summarized as follows. First, conserved 7-mer sites that are perfectly complementary to a tsRNA are significantly associated with reduced translational efficiency of mRNAs. For mRNAs that have 7-mer sites conserved between D. melanogaster and D. virilis and antisense paired to any part of a tsRNA, the mRNA abundances of the predicted target genes were slightly increased after T3, T6 or T10 transfection (Supplementary Figure S3C). Nevertheless, TEs were significantly decreased than the background mRNAs (no sites) in all the three transfection experiments (P < 0.001 in each comparison, KS tests, Figure 3A). These results suggest that tsRNAs repress translation of the target genes via antisense matching. Second, the number of conserved 7-mer target sites within a transcript was significantly associated with the translational repressiveness (ρ < 0 and P < 2.2 × 10−6 in each transfection experiment, Figure 3A), suggesting tsRNAs suppress translation of target mRNAs in an additive manner. [Here we dissected a target site with conserved n-mer motif (n > 7) antisense pairing to a tsRNA into (n-6) 7-mer target sites]. Third, the position of a target site in a mRNA does not affect the translational repressiveness. In each transfection experiment, the genes with conserved target sites at different locations of a mRNA (5′ UTR, CDS, 3′ UTR) yield nearly similarly decreased TEs compared to the mRNAs without conserved target sites (in the T3 experiment, the TE deduction was not statistically significant for genes with target sites in 5′ UTRs due to the small number of target genes; and in the T10 experiment, target sites in the 5′ UTR is associated with slightly stronger repression effects than those in CDSes, Figure 3B). Hence, unlike miRNAs that primarily recognize target sites in the 3′ UTRs (1,2), tsRNAs suppress target mRNAs by antisense-pairing the target sites in the whole mRNAs. Fourth, the conserved 7-mer target sites are preferentially complementary to the 5′ ends (starting from positions 1–3) of the tsRNAs (Supplementary Figure S4A), which is consistent with the pattern observed in human CLASH data (23,39). Nevertheless, we did not find the target sites complementary to the 5′ end of tsRNAs are associated with stronger inhibitory effects than the target sites matching to other locations of the tsRNAs (Supplementary Figure S4B).
Figure 3.
tsRNAs repress translation of target mRNAs via antisense pairing. (A) Cumulative distribution of changes in translational efficiency (TE) for all the genes. The x-axis is the log2(FCTE) in S2 cells transfected with the tsRNA mimic (T3, T6, T10 from left to right) versus transfection with ss-NC. The genes with 7-mer sites conserved between Drosophila melanogaster and Drosophila virilis and antisense paired to any part of a tsRNA are defined as tsRNA target genes. Red: genes with one conserved tsRNA target site, Green: genes with two conserved tsRNA target sites, Blue: genes with more than two conserved tsRNA target sites, Gray: genes without any target site of the transfected tsRNA in the entire mRNA. The number of genes in each category is indicated in parentheses. The asterisks indicate a significant difference in TE FC compared with the ss-NC control group. ***P < 0.001. (B) Repression efficiency of tsRNA is independent of the location of the tsRNA target sites in a mRNA transcript. The y-axis is the log2(FCTE) (comparing T3, T6 and T10 transfections with ss-NC from left to right). The x-axis is the location of the tsRNA mimic target sites in a mRNA transcript. ‘No sites’: mRNAs without conserved target sites for that tsRNA; ‘5′UTR’: mRNAs with conserved target sites in the 5′ UTRs, ‘CDS’: mRNAs with conserved target sites in CDSes, ‘3′UTR’: mRNAs with conserved target sites in the 3′ UTRs. The number of genes in each category is given in the parentheses. *P < 0.05; N.S. not statistically significant. (C) A scheme showing a tsRNA antisense pairing to evolutionarily conserved 7-mer target sites (in red) in different locations of a mRNA.
In summary, our experimental results suggest that tsRNAs modulate the expression of genes via conserved antisense sequence matching (Figure 3C), in an additive manner, and primarily at the translational level.
AGO2-bound tsRNAs preferentially repress components of the general translational machinery
In Drosophila, mutation of Dcr-1, a key component of the miRNA biogenesis pathway, does not significantly decrease the expression of tsRNAs (23), suggesting the biogenesis of tsRNA is independent of the miRNA pathway. Nevertheless, a previous study suggests that Dcr-2, a key effector in the siRNA pathway (1,2), cleaves tRNAs into tsRNAs (84). By examining the contents of short (20–22 nt) and long (23–29 nt) tsRNAs in the small RNA sequencing libraries in various mutants versus the matched wild-types (85–87), we found that mutations of Dicer-2 (Dcr-2) and r2d2, both of which are involved in the siRNA pathway (1,2), significantly reduce the abundance of the short tsRNAs (Supplementary Table S5). Since in Drosophila many tsRNAs (20–22 nt) are loaded onto AGO2 which is crucial for siRNA-mediated silencing (46), we hypothesize that the biogenesis of the short tsRNAs (20–22 nt) might rely on Dcr-2 and r2d2, and the functioning of tsRNA might depend on AGO2.
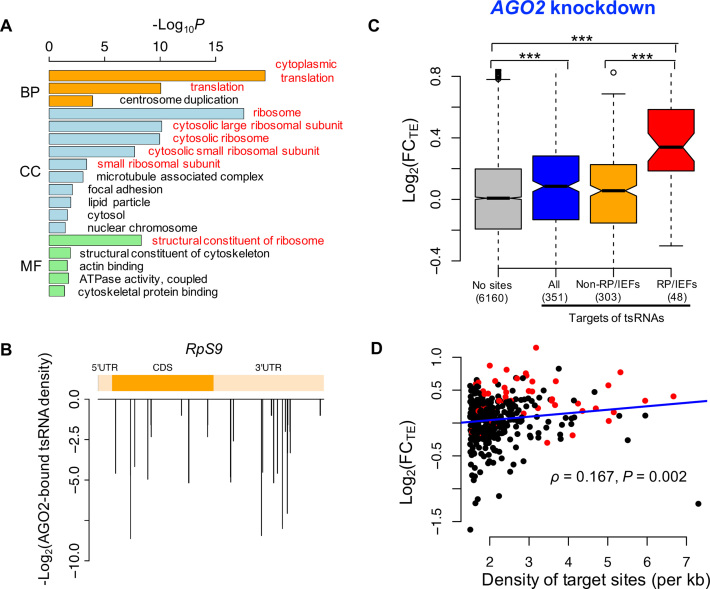
Next, we focused on the AGO2-bound tsRNAs in Drosophila S2 cells that were characterized previously (46) and pursued their regulatory function. We identified the 7-mer target sites that were conserved between D. melanogaster and D. virilis and antisense matching to at least one of the AGO2-bound tsRNAs in S2 cells. Then for each mRNA (the longest one was chosen in case one gene had many transcripts), we counted the number of conserved 7-mer target sites in that mRNA and calculated the density of target sites (per kb). The gene ontology (GO) analysis revealed the top target genes with the highest target densities were significantly enriched in the ribosomal proteins (RPs) and translational initiation or elongation factors (IEFs) (Figure 4A, see Figure 4B and Supplementary Figure S5 for some representative target genes), suggesting that the AGO2-bound tsRNAs might preferentially target the core components of the general translational machinery.
Figure 4.
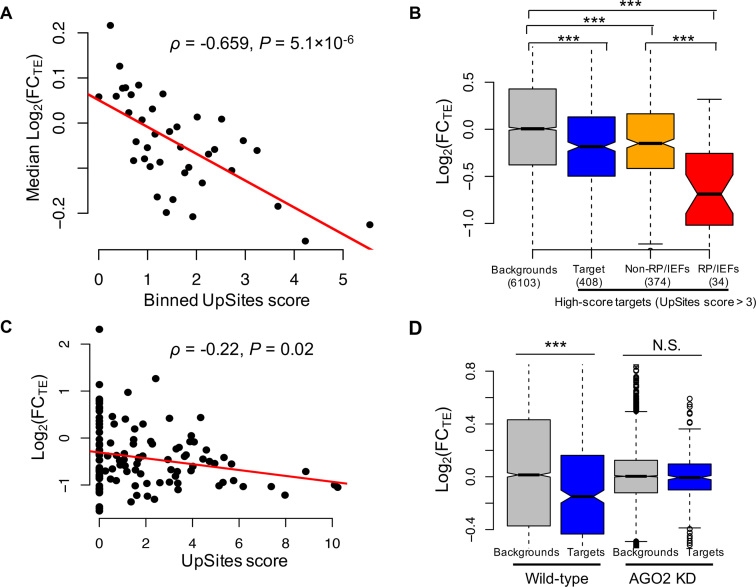
tsRNAs mediated translational repression in an AGO2-dependent manner. (A) GO analysis of the top 600 target genes of the AGO2-bound tsRNAs. The ontology related to translation is in red. (B) The target site density of AGO2-bound tsRNAs on RpS9 mRNA. The y-axis is the -log2 (reads of AGO2-bound tsRNAs) for each target site. (C) The log2(FC) for TEs in the AGO2 knock-down versus ds-NC transfected S2 cells that were cultured in normal condition. The targets of tsRNAs: mRNAs with conserved target sites of AGO2-bound tsRNAs with the density >1.5 per kb of mRNA (N = 351); ‘No sites": the remaining mRNAs expressed in S2 cells; RP/IEFs: the RPs or translational IEFs that were also targets of tsRNAs; non-RP/IEFs: the targets of tsRNAs that are not RPs or translational IEFs. The number of genes in each category is given in the parentheses. ***P < 0.001. (D) The density of tsRNA target sites in a mRNA (x-axis) is positively correlated with the extent of translational de-repression (y-axis) after AGO2 was knocked down. The top 351 targets of the AGO2-bound tsRNAs were used in this analysis. RP/IEFs were shown in red (N = 48). The Spearman’s correlation and P-value were shown.
To confirm that AGO2 is important for tsRNA-mediated regulation, we knocked down AGO2 mRNAs in S2 cells with RNAi oligos (‘Materials and Methods’ section). The polysome fractionation profile analysis suggests that global translational activity was slightly upregulated in AGO2 knock-down S2 cells compared to the cells transfected with a negative control siRNA duplex (ds-NC) (Supplementary Figure S6). We further carried out mRNA-seq and Ribo-seq of the AGO2 knock-down and the control S2 cells (Table 1 and Supplementary Table S4). We found TEs for the top targets of AGO2-bound tsRNAs (the density of target sites per kb > 1.5, N = 351) were significantly upregulated in the AGO2 knock-down S2 cells (P = 0.0002, Figure 4C), suggesting AGO2 is indispensable for tsRNA-mediated translational suppression (analogous results were obtained if we required the density of target sites per kb > 2). Strikingly, among the 351 strong target genes, the RPs and IEFs (N = 48) had significantly higher TE increases (0.340 ± 0.299, log2 scaled, mean ± s.d.) than the remaining target genes did (0.008 ± 0.328, log2 scaled; N = 303) (P < 10−8, KS test) after AGO2 knock-down, supporting our hypothesis that tsRNAs primarily inhibit the translation of the components of the general translational machinery. Moreover, after AGO2 was knocked down, the extent of de-repression (log2(FCTE)) was significantly positively correlated with the density of tsRNA target sites among the 351 strong targets (Spearman’s ρ = 0.167, P = 0.002, Figure 4D), suggesting that tsRNAs overall repress translation of the target mRNAs in an additive manner, as shown in our tsRNA transfection experiments (Figure 3A).
tsRNAs participate in cellular starvation response
tsRNAs have attracted intense interest due to their potential regulatory roles in cellular stress response (18–20,40). To examine whether tsRNAs participate in cellular starvation response in Drosophila, we sequenced the small RNAs in S2 cells under normal and serum-free medium (i.e. serum starvation) (‘Materials and Methods’ section). After normalizing the small RNA reads that were mapped to known miRNAs, transposable elements (siRNAs) and tRNAs (tsRNAs), we obtained similar numbers of total tsRNAs in the normal and starved S2 cells. After collapsing tsRNAs according to their locations in tRNAs (5′ end, middle or 3′ end), we found that under serum starvation, the upregulated tsRNA fragments (> 1.5-folds) were significantly enriched in the 5′ ends of tRNAs, while the downregulated tsRNA fragments tend to be located in the middle regions of tRNAs (P = 0.0012, t-test, Figure 5A and B). These results suggest that the 5′-tsRNAs and middle-tsRNAs might have different functional roles in cellular starvation response, although we could not exclude the possibility that the tRNA-degradation machinery is affected under stress conditions.
Figure 5.
tsRNAs participate in Drosophila cellular starvation response. (A) The log2 (FC) of tsRNA abundance from 5′, middle and 3′ end of each tRNA after serum starvation. Each tRNA type was denoted by the amino acid and anti-codon. (B) The changes in tsRNA abundances are different in tsRNAs from 5′, middle and 3′ ends of tRNAs. Upregulation under serum starvation is shown in blue, and downregulation under serum starvation is shown in red. The y-axis is the percent of upregulated/downregulated tsRNAs in the three types of tsRNAs. The upregulated tsRNAs were defined as tsRNAs with FC > 1.5 after serum starvation. The downregulated tsRNAs were defined as those tsRNAs with a FC < 0.66 after serum starvation. (C) The abundance of tsRNAs (RPM) in normal and starved S2 cells. tsRNA not bound by AGO1 nor AGO2 is in gray, tsRNA only bound by AGO2 is in red, tsRNA only bound by AGO1 is in black, tsRNA both bound by AGO1 and AGO2 is in orange. The tsRNA/AGO binding information was taken from previously published IP-seq results from Czech et al. (46). (D) Western blotting of AGO2 in the four different conditions of S2 cells. The four conditions of S2 cells: cultured under normal condition, serum starvation, AGO2 knockdown while serum deprived, and AGO2 knockdown. Error bars represent one standard deviation computed from two repeated experiments.
We found 53.3 and 59.4% of the tsRNAs in the normal and starved S2 cells, respectively, were overlapping with the AGO2-bound tsRNAs characterized previously (46). Meanwhile, only very few (∼1%) of the tsRNAs showed evidence of preferential association with AGO1 but not AGO2 (Figure 5C). The protein level of AGO1 was similar in the normal and starved S2 cells (Supplementary Figure S7). By contrast, AGO2 was upregulated at translation level after serum starvation [log2(FCRPF) = 0.635 and log2(FCmRNA) = -0.06] in the Ribo-seq and mRNA-seq results, which was further verified by western blotting (Figure 5D). These results suggest that AGO2 might participate starvation response in S2 cells. The polysome fractionation profile analyses suggest the global translational activities of S2 cells were significantly reduced in the serum starvation conditions (Supplementary Figure S6A and B, blue). Nevertheless, knock-down of AGO2 restored the global translational suppression in S2 cells (Supplementary Figure S6), suggesting that AGO2 participates in the global translation regulation. Altogether, these observations prompt us to hypothesize that the AGO2-bound tsRNAs might participate in Drosophila cellular starvation response.
Next, we questioned whether the changes in tsRNA abundances were associated with changes in TEs. For each tsRNA that is expressed in the normal or starved S2 cells and shows evidence of AGO2-binding in a previous study (46), we predicted the 7-mer target sites that were antisense matching to that tsRNA and conserved between D. melanogaster and D. virilis (‘Materials and Methods’ section). Since many tsRNAs are overlapping in sequences, for each target site in a mRNA, we calculated the total number of tsRNA reads matching that site in the normal and starved S2 cells, respectively. Then for each target mRNA, we only considered the target sites with the summed tsRNA reads differentially expressed between normal and starved conditions by at least 2-folds, and calculated the density of target sites (per kb) of the upregulated (UpSites) and downregulated (DnSites) summed tsRNAs. To evaluate the regulatory impact of the changes in tsRNAs on target genes, we conducted mRNA-seq and Ribo-seq of the starved S2 cells (Table 1 and Supplementary Table S4) and contrasted the gene expression profiles in the normal and starved conditions. Overall, under serum starvation, we found a significant negative correlation between FCTE (log2 scaled) and the UpSites score for a mRNA (ρ = −0.659, P = 5.1 × 10−6 when the mRNAs were binned based on increasing UpSites scores, Figure 6A; ρ = −0.113, P = 5.6 × 10−20 for the raw data). This result suggests that the upregulated tsRNAs in cellular starvation, in general, exerted stronger repressive effects on translation of the target genes in an additive manner. Moreover, the highest scoring target genes of the upregulated tsRNAs (defined as UpSites > 3, N = 408) have significantly reduced TEs comparing to the backgrounds under serum starvation (−0.174 ± 0.583 versus 0.033 ± 0.726, P = 7.5 × 10−11, KS test, Figure 6B). In particular, TEs for RPs and IEFs (N = 34) showed a further reduction (−0.622 ± 0.559, P = 3.2 × 10−7, KS test) compared to the backgrounds. Notably, the mRNAs of the highest scoring target genes were significantly upregulated compared to those of the background genes in the starved S2 cells [log2(FCmRNA) is 0.168 ± 0.548 versus −0.017 ± 0.566, P = 1.92 × 10−10, KS test, Supplementary Figure S8A], presumably due to the cellular intrinsic properties to compensate the reduced translation of the mRNAs (88–91). In short, these comparisons further confirm the model that tsRNAs repress target mRNAs at the translational level, as revealed in our transfection experiments (Figure 3A). Curiously, we observed a significant negative correlation, instead of a positive one, between the FCTE (log2 scaled) and the DnSites score for a mRNA (ρ = −0.075, P = 1.2 × 10−9, Supplementary Figure S8B), presumably because the UpSites and DnSites scores were highly correlated (ρ = 0.478, P < 10−16, Supplementary Figure S8C) and the effects of the downregulated tsRNAs were masked by the upregulated tsRNAs. Indeed, when we only focused on the mRNAs without any sites of the upregulated tsRNAs (UpSites = 0), the target mRNAs of the downregulated tsRNAs had significantly higher changes in TEs compared to the remaining mRNAs (0.247 ± 0.822 versus 0.065 ± 0.774, P = 0.02, N = 87 and 1862, respectively, KS test, Supplementary Figure S8D).
Figure 6.
tsRNAs repress translation of target genes in cellular starvation response. (A) mRNAs with a higher density of target sites by the upregulated AGO2-bound tsRNAs (UpSites score, x-axis) show stronger translational repression (y-axis) under cellular starvation. The genes with UpSites score of zero (N = 2765) were grouped together, and the remaining 3746 genes were evenly divided into 38 groups based on increasing UpSites scores. The Spearman’s correlation and P-value were shown. (B) Genes targeted by the upregulated AGO2-bound tsRNAs (UpSites score > 3 per kb, N = 408) have decreased TEs in the starved S2 cells. ‘RP/IEFs’: RPs or translational IEFs; ‘backgrounds’: the remaining genes (UpSites score < 3 per kb, N = 6103) expressed in S2 cells. ***P < 0.001. (C) A significant negative correlation between FCTE (log2 scaled, y-axis) and the UpSites score (x-axis) for the 5′TOP genes under serum starvation (N = 115). (D) The change of TEs (y-axis) for the highest scoring target genes of upregulated tsRNAs in the starved S2 cells (UpSites > 3, and 5′TOP genes were not included, N = 376) under normal (left) and AGO2 knock-down condition (right panel). ***P < 0.001; N.S. not significant.
One caveat in the above analysis is that the reduced TEs for RPs and IEFs under serum starvation might be confounded by the mTOR signaling pathway, which activates translation of 5′TOP (terminal oligopyrimidine) genes by inhibiting the inhibitory eIF4E-binding proteins (92–94). In mammals, RPs and IEFs are significantly over-represented with the 5′TOP genes (56,95). Thus we questioned whether the targets of tsRNAs and 5′TOP genes are significantly overlapped in Drosophila as well. By carrying out mRNA-seq and Ribo-seq of the S2 cells that were treated with rapamycin, which negatively affects translation by specifically inhibiting the mTOR signaling pathway (96) and slowed down the global translation (Supplementary Figure S6), we in total identified 115 5′TOP genes expressed in Drosophila S2 cells (67 were homologous to known 5′TOP genes in humans, and 48 were newly identified in this study, Materials and Methods, and Supplementary Table S6). These 5′TOP genes showed significantly reduced RPFs (P = 2.6 × 10−44, Wilcoxon rank-sum (WRS) test) and TEs (P = 1.5 × 10−65, WRS test) compared to the remaining genes in the rapamycin-treated S2 cells (Supplementary Figure S9). Consistent with previous observations that 5′TOP genes are involved in the cellular stress response (97–99), the 5′TOP genes overall showed significantly reduced TEs compared to the background genes in the starved S2 cells (−0.421 ± 0.647 versus 0.028 ± 0.719, P = 4.67 × 10−16, KS test). Notably, under serum starvation, we found a significant negative correlation between FCTE (log2 scaled) and the UpSites score for the 5′TOP genes (ρ = −0.22, P = 0.02, Figure 6C), suggesting some of the 5′TOP genes were also suppressed by tsRNAs at the translational level. Importantly, after removing all the 5′TOP genes, we still found the highest scoring target genes of the upregulated tsRNAs (N = 376) have significantly reduced TEs compared to the background genes under serum starvation (−0.140 ± 0.581 versus 0.038 ± 0.726, P = 1.2 × 10−8, KS test, Figure 6D). Similarly, after removing 5′TOP genes, we also observed a significant negative correlation between FCTE (log2 scaled) and the UpSites score for a mRNA (ρ = −0.103, P = 1.3 × 10−16, Supplementary Figure S10) after serum starvation. Therefore, the reduced TEs of the upregulated tsRNAs were not caused by the mTOR signaling pathway.
To further verify that AGO2 is indispensable in the tsRNA-mediated starvation response, we carried out mRNA-seq and Ribo-seq of the AGO2 knock-down S2 cells that were cultured under serum starvation. In parallel, we sequenced the ds-NC transfected S2 cells cultured under the same conditions as controls. Under serum starvation and AGO2 knock-down, the suppressive effects of the upregulated tsRNAs on the translation of the highest scoring target genes that did not include 5′TOP genes (UpSites > 3, N = 376) diminished (−0.019 ± 0.227 versus −0.003 ± 0.247, P = 0.07, KS test, Figure 6D), which further indicates that AGO2 is indispensable in the tsRNA-mediated translational repression under serum starvation.
Taken together, our results suggest that tsRNAs regulate translation of the target genes via antisense pairing and participate in cellular starvation response, and the tsRNA-mediated regulatory process is dependent on AGO2.
tsRNA- and miRNA-mediated regulation are largely independent
Although Drosophila AGO1 primarily bind miRNAs and AGO2 typically bind siRNAs (62,63,100), many miRNAs that are highly complementary to their miRNA* are bound by AGO2 (66,101,102). Here we questioned whether the target genes of AGO2-bound tsRNAs were also targeted by AGO2-bound miRNAs. We identified 27 miRNAs that were highly expressed in S2 cells cultured at normal conditions and preferentially bound with AGO2 rather than AGO1 as evidenced by the IP-seq experiments (46). We predicted 668 protein-coding genes expressed in Drosophila S2 cells as conserved targets of these AGO2-bound miRNAs (‘Materials and Methods’ section, Supplementary Figure S11). After AGO2 was knocked down, TEs of the predicted targets of most AGO2-bound miRNAs were significantly reduced, whereas the target mRNAs were upregulated at normal culture condition (Supplementary Figure S11), suggesting that the AGO2-bound miRNAs primarily repress targets by mRNA destabilization or cleavage. Among the predicted targets of AGO2-bound miRNAs and tsRNAs, 65 genes were targeted by both miRNAs and tsRNAs (tsR/miR), which is slightly higher than the expected number under the assumption of randomness (Supplementary Figure S12A and B). Moreover, 603 genes were only targeted by miRNAs (miR), and 286 genes were only targeted by tsRNAs (tsR). Consistent with the observation for individual miRNAs (Supplementary Figure S11), after AGO2 was knocked down, the TEs were significantly reduced for genes in the miR category compared to the background genes (Supplementary Figure S12C). By contrast, TEs for genes in the tsR category were significantly increased compared to the background genes (Supplementary Figure S12C). Taken together, our results suggest that only a small fraction (∼10%) of genes targeted by the AGO2-bound miRNAs were also targeted by tsRNAs. Our results also suggest that the AGO2-bound miRNAs primarily destabilize target mRNAs (Supplementary Figure S11) whereas the AGO2-bound tsRNAs mainly suppress targets by inhibiting translation.
It is reported that miRNAs are involved in starvation responses in mammalian cells (103–108). We identified 18 miRNAs that were upregulated by more than 1.2-folds in the starved S2 cells (these miRNAs were primarily preferentially bound by AGO1, Supplementary Figure S13). Under serum starvation, TEs for the conserved target genes of most of these miRNAs were decreased (Supplementary Figure S13), suggesting these AGO1-bound miRNAs were upregulated in serum starvation response to repress translation of the target genes. In total, we identified 646 conserved target genes for these 18 upregulated miRNAs, and under serum starvation, 55 of these target genes (tsR/miR) were overlapping with the target genes of the upregulated AGO2-bound tsRNAs (UpSites score > 3) (Supplementary Figure S14A). The observed number of tsR/miR genes (N = 55) is slightly higher than the expected number under the assumption of randomness (mean is 40.5 and 95% CI is (29,52), P = 0.01, Supplementary Figure S14B), suggesting most of genes targeted by the upregulated tsRNAs might not be affected by miRNAs. TEs for the target genes of the upregulated miRNAs were more significantly reduced compared to the background genes under serum starvation (Supplementary Figure S14C). Also, the genes only targeted by the upregulated tsRNAs but not by the upregulated miRNAs (tsR, N = 286) had significantly lower TEs compared to the background genes under serum starvation (Supplementary Figure S14C). These results suggest that under serum starvation both the tsRNAs and AGO1-bound miRNAs are upregulated to suppress translation of the target genes, although only a small fraction of the target genes are overlapping. Indeed, the genes targeted by both miRNAs and tsRNAs (tsR/miR, N = 55) had lower (but non-significant) TEs compared to the genes only targeted by miRNAs (miR, N = 591).
Overall, our results suggest that only a small number of target genes are commonly repressed by both tsRNAs and miRNAs, and the target repression between these two regulatory pathways are largely independent.
A model describing how tsRNAs suppress translation of specific targets and global mRNAs
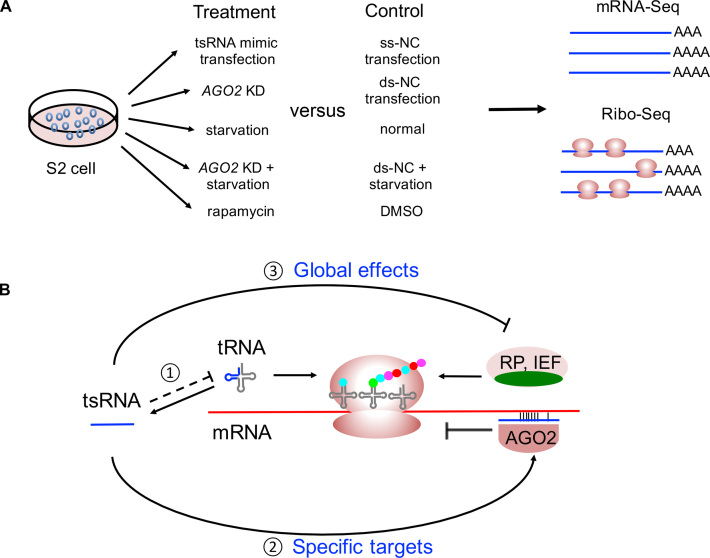
In the present study, we investigated the regulatory function of tsRNAs in Drosophila based on tsRNA mimic transfection, high-throughput sequencing, AGO2 knock-down, and cellular starvation response (Figure 7A). Our study provides a unifying model that describes how tsRNAs regulate specific and global mRNA translation via antisense matching (Figure 7B). Our study also supports previous results that tsRNAs may function as messengers to convey stress signals and ensure efficient attenuation of the genome-wide translational program (10,13,19,20,23,25,33–37). While the slowdown of translation has been hypothesized to be an optimized strategy organisms evolve to live under stressful conditions (104,109–111), the tsRNA-mediated regulation might have multiple advantages. First, since the abundance of tRNAs limits translational activities (112), the global translational activities would be reduced when a fraction of the full-length tRNAs is processed into tsRNAs. Second, the tsRNAs generated from tRNA cleavage preferentially repress the key components of the global translational machinery via antisense pairing, which further slows down the global mRNA translation (Figure 7B). Third, the stress-induced tsRNA-mediated regulation is accomplished in chained reactions, which is bio-energetically cost-effective and quick-responsive to environmental stress. Therefore, under our model, the tsRNA-mediated regulation would repress both the specific and global mRNA translation in a timely and efficient manner.
Figure 7.
A model describing how tsRNAs suppress translation of specific targets and global mRNAs. (A) An overview of the experimental treatments of S2 cells and sequencing workflow in the study. (B) tsRNAs preferentially target RPs and IEFs via antisense pairing to regulate the global translational activities. (1) AGO2 bound the tsRNAs cleaved from tRNAs, and those tsRNAs specifically bind to the mRNAs with partial complementarity and inhibit their translation. (2) Translation of some RPs and IEFs is repressed by tsRNAs, which in turn suppresses the global translational activities. Under starvation, translational repression is also regulated by mTOR pathway via 5′TOP genes, and some of the tsRNA targets are overlapping with the 5′TOP genes (in green).
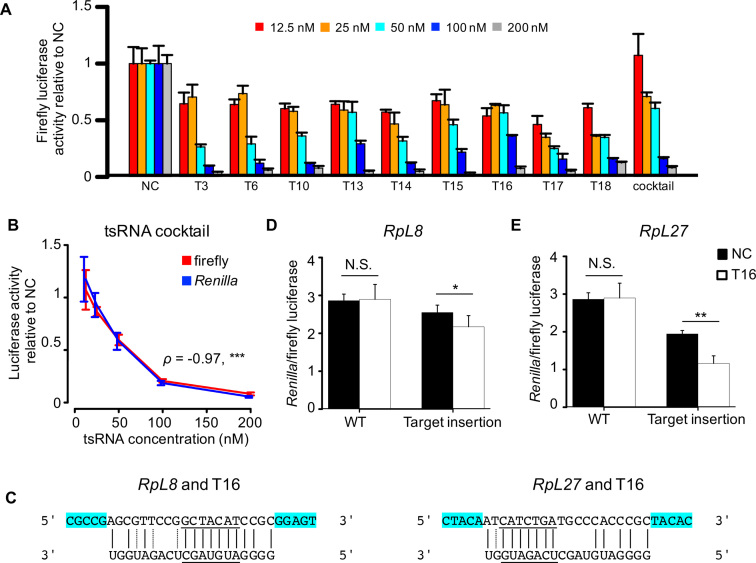
To further validate this model, we performed the luciferase reporter assay to test the repressive effects of individual tsRNAs on global translation and the specific target mRNAs through antisense pairing. Based on the results of our Ribo-seq experiments, we chose nine tsRNAs (Table 2, see Supplementary Table S7 for the detailed information of the predicted translational target genes) that were bound by AGO2 in the previous IP-seq study (46) and evaluated their regulatory impact on the global translational activities in S2 cells. Three of these tsRNAs (T3, T6 and T10) were used in our transfection experiments that were followed with mRNA-seq and Ribo-seq (Table 1). The other six tsRNAs were relatively abundant in Drosophila S2 cells, and five of them were upregulated in the serum starvation condition. Our target prediction suggests all the nine tsRNAs target mRNAs of RPs or other translational factors through conserved 7-mer antisense matching. To monitor the repressive effects of each tsRNA on the global translational activities, we transferred the single-strand tsRNA mimic at different concentrations into the S2 cells together with the psiCHECK-2 plasmids. As negative controls, we also transfected ss-NC at the same concentrations together with the psiCHECK-2 plasmids. Compared to the transfection of NC, the luciferase activities were lower after transfection with any of the nine tsRNAs (Figure 8A for firefly luciferase; see Supplementary Figure S15 for the Renilla luciferase). Remarkably, the luciferase activities were significantly reduced at higher concentrations of tsRNA mimics (Spearman’s ρ < -0.8, P < 0.0003 for each of the nine tsRNA transfection experiments), suggesting the tsRNAs transfected at higher concentrations exerted stronger repressive effects on the global translational activities. Moreover, we also mixed the nine tsRNA equally into a cocktail of tsRNA mimics, and transfected the cocktail of different concentrations into S2 cells. We also observed similar patterns of global translational repression by the tsRNA cocktails as in the single tsRNA mimic transfection experiments (ρ = −0.971, P < 1.83 × 10−9 when either the firefly or Renilla luciferase activity was used; Figure 8A and B). Noteworthy, these luciferase reporter assay results are well consistent with our polysome fractionation profiles analysis which revealed the P/M ratios were reduced after transfection of an individual single-strand tsRNA (Figure 2B). Overall, these results are in line with the notion that tsRNAs repress the global translational activities.
Table 2. The tsRNAs used in the luciferase reporter assay.
| ID | tsRNA | Sequence (5′-3′) | No. of predicted target genes | No. of target genes related to translation |
|---|---|---|---|---|
| T3 | AspGUC4-23 | UCGAUAGUAUAGUGGUUAGU | 396 | 3 |
| T6 | GluCUC6-25 | UAUUGUCUAGUGGUUAGGAU | 381 | 1 |
| T10 | LysUUU4-23 | CGGAUAGCUCAGUCGGUAGA | 646 | 4 |
| T13 | AspGUC32-53 | CUGUCACGCGGGAGACCGGGGU | 272 | 18 |
| T14 | LysCUU1-23 | GCCCGGCUAGCUCAGUCGGUAGA | 545 | 15 |
| T15 | LeuUAG1-20 | GGCAGCGUGGCCGAGCGGUC | 1228 | 42 |
| T16 | AlaAGC1-20 | GGGGAUGUAGCUCAGAUGGU | 786 | 27 |
| T17 | ArgUCU1-20 | GUCCCUUUGGCGCAGAGGAU | 1140 | 47 |
| T18 | GluCUC27-48 | UCCGGCUCUCACCCGGAAGGCC | 735 | 20 |
The target mRNAs were expressed in S2 cells and had conserved 7-mer sites antisense pairing with each tsRNA, respectively.
Figure 8.
Verifying the repressive effects of tsRNAs on global translational activities and specific target sites with dual luciferase reporter assay. (A) Co-transfection of tsRNA mimics and the psiCHECK-2 plasmids into S2 cells reduced the activities of firefly luciferase. The color key shows different final concentrations of tsRNA mimics. The tsRNA cocktail was made by mixing equal amount of the nine tsRNA mimics of the same concentrations. The luciferase activity of S2 cells transfected with the tsRNA mimics were normalized by those transfected with ss-NC at the same concentration (three replicates were performed for each assay). (B) Higher concentrations of tsRNA cocktail cause lower firefly (red) and Renilla (blue) luciferase activities on the psiCHECK-2 plasmid. ***P < 0.001 (three replicates were performed for each assay). (C) The base pairing between tsRNA T16 and the predicted targeting sites in RpL8 and PpL27 mRNAs. The targeting sites conserved between Drosophila melanogaster and Drosophila virilis were underlined. The flanking 5 bp on both sides of the target sites (highlighted in cyan) were included in the synthesized targeting sites. (D and E) Relative luciferase activities in S2 cells co-transfected with T16 and the psiCHECK-2 plasmids containing the target site (RpL8, D; RpL27, E) are significantly lower than those co-transfected with ss-NC and the same plasmid. As negative controls, no significant difference in the relative luciferase activities was observed in S2 cells when the empty psiCHECK-2 plasmid (WT) was co-transfected with T16 or with ss-NC (*P < 0.05, **P < 0.01, five replicates were performed).
Furthermore, we show evidence that one of the nine tsRNAs (T16) repress the translational activity of RpL8 and RpL27 mRNAs through antisense pairing. Under serum starvation, T16 was upregulated to 1.4-fold in S2 cells. Both RpL8 and RpL27 were predicted targets of T16 (Figure 8C), and RpL8 showed 2-fold decrease and RpL27 showed a 1.85-fold decrease in TE under the serum starvation. We synthesized a 3 × replication of each target site and separately cloned them into the 3′ UTR of the Renilla gene in the dual luciferase reporter psiCHECK-2 plasmid. We co-transfected the T16 tsRNA mimic together with the target-containing or the empty psiCHECK-2 plasmids into S2 cells. In parallel, we also co-transfected ss-NC together with the target-containing or the empty psiCHECK-2 plasmids into S2 cells. The dual luciferase assay results revealed a significant repression effect of T16 on the target site derived from RpL8 (P = 0.049, Student’s t-test, Figure 8D) and RpL27 (P = 0.002, t-test Figure 8E), suggesting that both RpL8 and RpL27 are authentic target genes of the T16 tsRNA. Altogether, our luciferase reporter assays provide further evidence to support our model that tsRNAs suppress the global translational activities by specifically targeting factors in the global translational machinery through antisense pairing.
DISCUSSION
In this study, we present evidence that tsRNAs are conserved, prevalent and abundant in Drosophila. We found Drosophila tsRNAs preferentially inhibit translation of RPs and IEFs via antisense pairing. Since RPs and IEFs are crucial components of the general translational machinery, our results for the first time well explain how tsRNAs inhibit the global mRNA translation. We also found that the tsRNA-mediated regulation in S2 cells is dependent on AGO2, largely independent from the miRNA-mediated regulation and overlapped with the mTOR signaling pathway. Our results improve our understanding of the molecular mechanisms by which tsRNAs participate in the cellular starvation response.
While we have primarily focused on the short tsRNAs (20–22 nt) that are evolutionarily conserved between D. melanogaster and D. virilis and bound with AGO2, a considerable fraction of the tsRNAs are the long (23–29 nt) tsRNAs that could be bound by AGO3, AUB or PIWI in Drosophila germlines (Figure 1). Recent studies have demonstrated that tsRNAs could be crucial for mammalian sperm maturation and fertilization (12,32). Hence, we questioned whether the long tsRNAs might be related to the proper functioning of Drosophila reproductive systems. In Drosophila ovaries, tsRNAs had a length distribution resembling that of piRNAs (Figure 1). Similar to piRNAs (51), 26.3, 60.3 and 75.8% of the AGO3, AUB, and PIWI-bound tsRNA reads begin with uridine, respectively (Supplementary Figure S16A), suggesting they might recognize their target sites in a piRNA-like manner. To probe the regulatory effects of these long tsRNAs, we retrieved the mRNA and Ribo-seq datasets of mature oocytes and activated eggs (113) and predicted the target sites of the AUB-bound tsRNAs (‘Materials and Methods’ section). Among the genes that were expressed in the mature and activated eggs, we predicted 780 mRNAs potentially targeted by the top 200 most abundant AUB-bound tsRNAs. The target mRNAs have significantly lower expression levels than the non-targets in both mature oocytes (P = 7.15 × 10−8, KS test; Supplementary Figure S16B, left) and activated eggs (P = 4.06 × 10−8, KS test; Supplementary Figure S16B, right), suggesting the target mRNAs might be suppressed by the AUB-bound tsRNAs. Interestingly, TEs of the target mRNAs were significantly lower than those of the non-targets in the mature oocytes (P = 0.0004, KS test; Supplementary Figure S16C, left), but the difference was not observed in the activated eggs (P = 0.11, KS test; Supplementary Figure S16C, right). Since translational control plays a critical role during the development of the Drosophila germlines and early embryos (113–116), here our results suggest that the long tsRNAs might play important roles in these processes.
Accumulating evidence indicates that Drosophila AGO2 not only functions as a slicer (63,117), but is also related to different types of small RNA loading (102,118,119) and involved in many other transcriptional or post-transcriptional regulatory processes either directly or through adaptor proteins (57,108,120). Our results suggest that Drosophila AGO2 is required for the tsRNA-mediated translational repression of target mRNAs, especially for the RPs/IEFs in the general translational machinery. However, how AGO2 participates in this process is not well understood. The gene silencing mechanisms mediated by the AGO-small RNA complex are determined not only by the bound small RNAs and co-factors but also by the specific mRNA targets (121). Here we analyzed the data of CLIP-seq on nuclear extracts of Drosophila S2 cells (57) and examined the profiles of mRNAs bound by AGO2 (‘Materials and Methods’ section). With the Nascent-seq data of S2 cells (58) as the input control, we calculated the fold enrichment (fe) score to measure the enrichment of an expressed mRNA in the AGO2 CLIP-seq data. A higher fe score means the mRNA is more enriched in the AGO2 CLIP-seq data. We found the fe scores are significantly higher for the RPs/IEFs (N = 89) than the remaining genes (N = 6,005) that are expressed in Drosophila S2 cells [P = 10−36, (WRS test, Supplementary Figure S17A], suggesting that mRNAs of RPs/IEFs are preferentially bound by AGO2. Next, we focused on the mRNAs that possess conserved target sites of AGO2-bound tsRNAs (with the density of target sites > 1 per kb of mRNA, N = 710). The RP/IEF targets (N = 42) have significantly higher fe scores than the background genes (P = 10−15, WRS test, Supplementary Figure S17B). However, the fe scores are very similar between the remaining targets (excluding RPs/IEFs) and the background genes (Supplementary Figure S17B). Overall, these results suggest that AGO2 preferentially bind mRNAs of RPs/IEFs in Drosophila S2 cells. Remarkably, we found eight 7-mer motifs whose densities in the CDS sequences are significantly positively correlated with the fe scores in the AGO2 CLIP-seq data (Supplementary Figure S18 and Table S8). Notably, the densities of the eight 7-mers are significantly higher for the RPs/IEFs than the remaining expressed genes (P = 10−34, WRS test, Supplementary Figure S17C). Moreover, the RP/IEF targets (N = 42) have significantly higher densities of these motifs than the background genes (P = 10−23, WRS test, Supplementary Figure S17D), whereas such a difference does not exist between the remaining targets (excluding RPs/IEFs) and the background genes (Supplementary Figure S17D). It should be noted that the patterns we reported in Supplementary Figures S17 and 18 and Table S8 were based on the analyses of the CDS regions. We observed very similar patterns when we performed the same analytical procedures on the full-length mRNAs. Altogether, our results suggest that AGO2 tends to preferentially bind mRNAs of RPs/IEFs that harbor high densities of these motifs. Since many tsRNAs are bound by AGO2, our results support the scenario that the preferential binding of AGO2 to mRNAs of RPs/IEFs would facilitate forming the complexes of tsRNAs and mRNAs of RPs/IEFs that have the target sites. Further experiments, such as CLASH (cross-linking, ligation and sequencing of hybrids) (39) or CLEAR (covalent ligation of endogenous AGO-bound RNAs)-CLIP (122) are required to directly detect the tsRNA–mRNA chimera in the cytoplasm.
Since tsRNAs are evolutionarily ancient and present in both prokaryotes and eukaryotes, it is not surprising that the biogenesis, regulatory mechanisms and functional consequences have evolved across species (23). It is possible that the tremendous diversity and functional divergence of tsRNAs across species would lead to inconsistent observations if one only focuses on a few tsRNAs. For example, previous studies in plants have demonstrated that tsRNAs are upregulated upon stress (123), and such tsRNAs suppress target genes or transposable elements by degrading mRNAs (30,124,125). Here we found that in Drosophila tsRNAs preferentially suppress translation of the mRNAs of the RPs and other translational factors via antisense pairing. Interestingly, it is reported that human RP RPL35A is among the most abundant in the tsRNA-mRNA chimeras in the human AGO1 CLASH data, and RPL35A mRNA can be targeted by different tsRNAs at different regions (23). Hence, the targeting of RP mRNAs by tsRNAs might be a conserved mechanism between Drosophila and animals. Our study also provides a unifying model that describes how tsRNAs regulate specific and global mRNA translation via antisense matching (Figure 7B). Cells employ multiple strategies to reprogram the global and specific mRNA translations upon environmental stresses (104,109–111). It is known that the tRNAs can be dynamically regulated concerning their abundance and nucleotide modifications during stress adaptation (126,127). Under our model, the tsRNA-mediated regulation may be crucial for the energy homeostasis and metabolic adaptation in the cellular systems. Further efforts are needed to explore whether such a model applies to other species such as humans.
DATA AVAILABILITY
All raw deep-sequencing data generated by this study have been submitted to SRA (PRJNA378597) and GEO (GSE106697). All other data supporting the findings of this study are available within the article and its supplementary files.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Qi Chen for valuable suggestions. We thank Dr Matthew Taliaferro for providing the AGO2 CLIP-seq data in nuclear extract of S2 cells. We thank Mr Hong Zhang and Ms Qi Zhang for technical assistance. We thank the Biodynamic Optical Imaging Centre from Peking University for sequencing services.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministry of Science and Technology of China [2016YFA0500800]; National Natural Science Foundation of China [91731301, 31771411, 31571333, 91431101]; Peking-Tsinghua Center for Life Science (to Ji.L.); Peking-Tsinghua Centre for Life Sciences Postdoctoral Fellowship (to F.H.) (in part); postdoctoral fellowship from China Postdoctoral Science Foundation (to F.H.) (in part). Funding for open access charge: National Natural Science Foundation of China; Ministry of Science and Technology of China.
Conflict of interest statement. None declared.
REFERENCES
- 1. Jacquier A. The complex eukaryotic transcriptome: unexpected pervasive transcription and novel small RNAs. Nat. Rev. Genet. 2009; 10:833–844. [DOI] [PubMed] [Google Scholar]
- 2. Ghildiyal M., Zamore P.D.. Small silencing RNAs: an expanding universe. Nat. Rev. Genet. 2009; 10:94–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Shigematsu M., Honda S., Kirino Y.. Transfer RNA as a source of small functional RNA. J. Mol. Biol. Mol. Imaging. 2014; 1:8. [PMC free article] [PubMed] [Google Scholar]
- 4. Keam S.P., Hutvagner G.. tRNA-derived fragments (tRFs): Emerging new roles for an ancient RNA in the regulation of gene expression. Life. 2015; 5:1638–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kumar P., Kuscu C., Dutta A.. Biogenesis and function of transfer RNA-related fragments (tRFs). Trends Biochem. Sci. 2016; 41:679–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Peng H., Shi J., Zhang Y., Zhang H., Liao S., Li W., Lei L., Han C., Ning L., Cao Y. et al. A novel class of tRNA-derived small RNAs extremely enriched in mature mouse sperm. Cell Res. 2012; 22:1609–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lee Y.S., Shibata Y., Malhotra A., Dutta A.. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev. 2009; 23:2639–2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Karaiskos S., Naqvi A.S., Swanson K.E., Grigoriev A.. Age-driven modulation of tRNA-derived fragments in Drosophila and their potential targets. Biol. Direct. 2015; 10:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Blanco S., Dietmann S., Flores J.V., Hussain S., Kutter C., Humphreys P., Lukk M., Lombard P., Treps L., Popis M. et al. Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders. EMBO J. 2014; 33:2020–2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Haussecker D., Huang Y., Lau A., Parameswaran P., Fire A.Z., Kay M.A.. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA. 2010; 16:673–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yeung M.L., Bennasser Y., Watashi K., Le S.Y., Houzet L., Jeang K.T.. Pyrosequencing of small non-coding RNAs in HIV-1 infected cells: evidence for the processing of a viral-cellular double-stranded RNA hybrid. Nucleic Acids Res. 2009; 37:6575–6586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen Q., Yan M., Cao Z., Li X., Zhang Y., Shi J., Feng G.H., Peng H., Zhang X., Zhang Y. et al. Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science. 2016; 351:397–400. [DOI] [PubMed] [Google Scholar]
- 13. Ivanov P., Emara M.M., Villen J., Gygi S.P., Anderson P.. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol. Cell. 2011; 43:613–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sobala A., Hutvagner G.. Transfer RNA-derived fragments: origins, processing, and functions. Wiley Interdiscip. Rev. RNA. 2011; 2:853–862. [DOI] [PubMed] [Google Scholar]
- 15. Cole C., Sobala A., Lu C., Thatcher S.R., Bowman A., Brown J.W., Green P.J., Barton G.J., Hutvagner G.. Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. RNA. 2009; 15:2147–2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lee S.R., Collins K.. Starvation-induced cleavage of the tRNA anticodon loop in Tetrahymena thermophila. J. Biol. Chem. 2005; 280:42744–42749. [DOI] [PubMed] [Google Scholar]
- 17. Li Y., Luo J., Zhou H., Liao J.Y., Ma L.M., Chen Y.Q., Qu L.H.. Stress-induced tRNA-derived RNAs: a novel class of small RNAs in the primitive eukaryote Giardia lamblia. Nucleic Acids Res. 2008; 36:6048–6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fu H., Feng J., Liu Q., Sun F., Tie Y., Zhu J., Xing R., Sun Z., Zheng X.. Stress induces tRNA cleavage by angiogenin in mammalian cells. FEBS Lett. 2009; 583:437–442. [DOI] [PubMed] [Google Scholar]
- 19. Yamasaki S., Ivanov P., Hu G.F., Anderson P.. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J. Cell Biol. 2009; 185:35–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Emara M.M., Ivanov P., Hickman T., Dawra N., Tisdale S., Kedersha N., Hu G.F., Anderson P.. Angiogenin-induced tRNA-derived stress-induced RNAs promote stress-induced stress granule assembly. J. Biol. Chem. 2010; 285:10959–10968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Thompson D.M., Lu C., Green P.J., Parker R.. tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA. 2008; 14:2095–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hsieh L.C., Lin S.I., Shih A.C., Chen J.W., Lin W.Y., Tseng C.Y., Li W.H., Chiou T.J.. Uncovering small RNA-mediated responses to phosphate deficiency in Arabidopsis by deep sequencing. Plant Physiol. 2009; 151:2120–2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kumar P., Anaya J., Mudunuri S.B., Dutta A.. Meta-analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets. BMC Biol. 2014; 12:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kumar P., Mudunuri S.B., Anaya J., Dutta A.. tRFdb: a database for transfer RNA fragments. Nucleic Acids Res. 2015; 43:D141–D145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Maute R.L., Schneider C., Sumazin P., Holmes A., Califano A., Basso K., Dalla-Favera R.. tRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:1404–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Goodarzi H., Liu X., Nguyen H.C., Zhang S., Fish L., Tavazoie S.F.. Endogenous tRNA-derived fragments suppress breast cancer progression via YBX1 displacement. Cell. 2015; 161:790–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Honda S., Loher P., Shigematsu M., Palazzo J.P., Suzuki R., Imoto I., Rigoutsos I., Kirino Y.. Sex hormone-dependent tRNA halves enhance cell proliferation in breast and prostate cancers. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:E3816–E3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pekarsky Y., Balatti V., Palamarchuk A., Rizzotto L., Veneziano D., Nigita G., Rassenti L.Z., Pass H.I., Kipps T.J., Liu C.G. et al. Dysregulation of a family of short noncoding RNAs, tsRNAs, in human cancer. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:5071–5076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kim H.K., Fuchs G., Wang S., Wei W., Zhang Y., Park H., Roy-Chaudhuri B., Li P., Xu J., Chu K. et al. A transfer-RNA-derived small RNA regulates ribosome biogenesis. Nature. 2017; 552:57–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Martinez G., Choudury S.G., Slotkin R.K.. tRNA-derived small RNAs target transposable element transcripts. Nucleic Acids Res. 2017; 45:5142–5152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schorn A.J., Gutbrod M.J., LeBlanc C., Martienssen R.. LTR-retrotransposon control by tRNA-derived small RNAs. Cell. 2017; 170:61–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sharma U., Conine C.C., Shea J.M., Boskovic A., Derr A.G., Bing X.Y., Belleannee C., Kucukural A., Serra R.W., Sun F. et al. Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science. 2016; 351:391–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Keam S.P., Sobala A., Ten Have S., Hutvagner G.. tRNA-derived RNA fragments associate with human multisynthetase complex (MSC) and modulate ribosomal protein translation. J. Proteome Res. 2017; 16:413–420. [DOI] [PubMed] [Google Scholar]
- 34. Sobala A., Hutvagner G.. Small RNAs derived from the 5′ end of tRNA can inhibit protein translation in human cells. RNA Biol. 2013; 10:553–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gebetsberger J., Zywicki M., Kunzi A., Polacek N.. tRNA-derived fragments target the ribosome and function as regulatory non-coding RNA in Haloferax volcanii. Archaea. 2012; 2012:260909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang Q., Lee I., Ren J., Ajay S.S., Lee Y.S., Bao X.. Identification and functional characterization of tRNA-derived RNA fragments (tRFs) in respiratory syncytial virus infection. Mol. Ther. 2013; 21:368–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Loss-Morais G., Waterhouse P.M., Margis R.. Description of plant tRNA-derived RNA fragments (tRFs) associated with argonaute and identification of their putative targets. Biol. Direct. 2013; 8:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hirano T., Iwasaki Y.W., Lin Z.Y., Imamura M., Seki N.M., Sasaki E., Saito K., Okano H., Siomi M.C., Siomi H.. Small RNA profiling and characterization of piRNA clusters in the adult testes of the common marmoset, a model primate. RNA. 2014; 20:1223–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Helwak A., Kudla G., Dudnakova T., Tollervey D.. Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell. 2013; 153:654–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Thompson D.M., Parker R.. The RNase Rny1p cleaves tRNAs and promotes cell death during oxidative stress in Saccharomyces cerevisiae. J. Cell Biol. 2009; 185:43–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ingolia N.T., Ghaemmaghami S., Newman J.R., Weissman J.S.. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009; 324:218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ingolia N.T., Lareau L.F., Weissman J.S.. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011; 147:789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dunn J.G., Foo C.K., Belletier N.G., Gavis E.R., Weissman J.S.. Ribosome profiling reveals pervasive and regulated stop codon readthrough in Drosophila melanogaster. Elife. 2013; 2:e01179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Langmead B., Salzberg S.L.. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012; 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rosenbloom K.R., Armstrong J., Barber G.P., Casper J., Clawson H., Diekhans M., Dreszer T.R., Fujita P.A., Guruvadoo L., Haeussler M. et al. The UCSC Genome Browser database: 2015 update. Nucleic Acids Res. 2015; 43:D670–D681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Czech B., Malone C.D., Zhou R., Stark A., Schlingeheyde C., Dus M., Perrimon N., Kellis M., Wohlschlegel J.A., Sachidanandam R. et al. An endogenous small interfering RNA pathway in Drosophila. Nature. 2008; 453:798–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Friedman R.C., Farh K.K., Burge C.B., Bartel D.P.. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009; 19:92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pollard K.S., Hubisz M.J., Rosenbloom K.R., Siepel A.. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010; 20:110–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Olivieri D., Senti K.A., Subramanian S., Sachidanandam R., Brennecke J.. The cochaperone shutdown defines a group of biogenesis factors essential for all piRNA populations in Drosophila. Mol. Cell. 2012; 47:954–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rozhkov N.V., Aravin A.A., Zelentsova E.S., Schostak N.G., Sachidanandam R., McCombie W.R., Hannon G.J., Evgen’ev M.B.. Small RNA-based silencing strategies for transposons in the process of invading Drosophila species. RNA. 2010; 16:1634–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wang W., Yoshikawa M., Han B.W., Izumi N., Tomari Y., Weng Z., Zamore P.D.. The initial uridine of primary piRNAs does not create the tenth adenine that is the hallmark of secondary piRNAs. Mol. Cell. 2014; 56:708–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Senti K.A., Jurczak D., Sachidanandam R., Brennecke J.. piRNA-guided slicing of transposon transcripts enforces their transcriptional silencing via specifying the nuclear piRNA repertoire. Genes Dev. 2015; 29:1747–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Goh W.S., Falciatori I., Tam O.H., Burgess R., Meikar O., Kotaja N., Hammell M., Hannon G.J.. piRNA-directed cleavage of meiotic transcripts regulates spermatogenesis. Genes Dev. 2015; 29:1032–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhang P., Kang J.Y., Gou L.T., Wang J., Xue Y., Skogerboe G., Dai P., Huang D.W., Chen R., Fu X.D. et al. MIWI and piRNA-mediated cleavage of messenger RNAs in mouse testes. Cell Res. 2015; 25:193–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Krüger J., Rehmsmeier M.. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006; 34:W451–W454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Thoreen C.C., Chantranupong L., Keys H.R., Wang T., Gray N.S., Sabatini D.M.. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature. 2012; 485:109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Taliaferro J.M., Aspden J.L., Bradley T., Marwha D., Blanchette M., Rio D.C.. Two new and distinct roles for Drosophila Argonaute-2 in the nucleus: alternative pre-mRNA splicing and transcriptional repression. Genes Dev. 2013; 27:378–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Khodor Y.L., Rodriguez J., Abruzzi K.C., Tang C.H., Marr M.T. 2nd, Rosbash M.. Nascent-seq indicates widespread cotranscriptional pre-mRNA splicing in Drosophila. Genes Dev. 2011; 25:2502–2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. White B.N., Tener G.M., Holden J., Suzuki D.T.. Analysis of tRNAs during the development of Drosophila. Dev. Biol. 1973; 33:185–195. [DOI] [PubMed] [Google Scholar]
- 61. Carthew R.W., Sontheimer E.J.. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009; 136:642–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Tomari Y., Du T., Zamore P.D.. Sorting of Drosophila small silencing RNAs. Cell. 2007; 130:299–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Okamura K., Ishizuka A., Siomi H., Siomi M.C.. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes Dev. 2004; 18:1655–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Vagin V.V., Sigova A., Li C., Seitz H., Gvozdev V., Zamore P.D.. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006; 313:320–324. [DOI] [PubMed] [Google Scholar]
- 65. Gunawardane L.S., Saito K., Nishida K.M., Miyoshi K., Kawamura Y., Nagami T., Siomi H., Siomi M.C.. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science. 2007; 315:1587–1590. [DOI] [PubMed] [Google Scholar]
- 66. Kawamura Y., Saito K., Kin T., Ono Y., Asai K., Sunohara T., Okada T.N., Siomi M.C., Siomi H.. Drosophila endogenous small RNAs bind to Argonaute 2 in somatic cells. Nature. 2008; 453:793–797. [DOI] [PubMed] [Google Scholar]
- 67. Brennecke J., Aravin A.A., Stark A., Dus M., Kellis M., Sachidanandam R., Hannon G.J.. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007; 128:1089–1103. [DOI] [PubMed] [Google Scholar]
- 68. Yin H., Lin H.. An epigenetic activation role of Piwi and a Piwi-associated piRNA in Drosophila melanogaster. Nature. 2007; 450:304–308. [DOI] [PubMed] [Google Scholar]
- 69. Li C., Vagin V.V., Lee S., Xu J., Ma S., Xi H., Seitz H., Horwich M.D., Syrzycka M., Honda B.M. et al. Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell. 2009; 137:509–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Brennecke J., Malone C.D., Aravin A.A., Sachidanandam R., Stark A., Hannon G.J.. An epigenetic role for maternally inherited piRNAs in transposon silencing. Science. 2008; 322:1387–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Han B.W., Wang W., Li C., Weng Z., Zamore P.D.. piRNA-guided transposon cleavage initiates Zucchini-dependent, phased piRNA production. Science. 2015; 348:817–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Wang Y., Luo J., Zhang H., Lu J.. microRNAs in the same clusters evolve to coordinately regulate functionally related genes. Mol. Biol. Evol. 2016; 33:2232–2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Zhao Y., Lin J., Xu B., Hu S., Zhang X., Wu L.. MicroRNA-mediated repression of nonsense mRNAs. Elife. 2014; 3:e03032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Arava Y., Wang Y., Storey J.D., Liu C.L., Brown P.O., Herschlag D.. Genome-wide analysis of mRNA translation profiles in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:3889–3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Gandin V., Sikstr M.K., Alain T., Morita M., McLaughlan S., Larsson O., Topisirovic I.. Polysome fractionation and analysis of mammalian translatomes on a genome-wide scale. J. Vis. Exp. 2014; 87:e51455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Chassé H., Boulben S., Costache V., Cormier P., Morales J.. Analysis of translation using polysome profiling. Nucleic Acids Res. 2017; 45:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Pospisek M., Valasek L.. Polysome profile analysis–yeast. Methods Enzymol. 2013; 530:173–181. [DOI] [PubMed] [Google Scholar]
- 78. Anders S., Pyl P.T., Huber W.. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015; 31:166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Tuller T., Carmi A., Vestsigian K., Navon S., Dorfan Y., Zaborske J., Pan T., Dahan O., Furman I., Pilpel Y.. An evolutionarily conserved mechanism for controlling the efficiency of protein translation. Cell. 2010; 141:344–354. [DOI] [PubMed] [Google Scholar]
- 81. Guo H., Ingolia N.T., Weissman J.S., Bartel D.P.. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010; 466:835–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Yang J.R., Chen X., Zhang J.. Codon-by-codon modulation of translational speed and accuracy via mRNA folding. PLoS Biol. 2014; 12:e1001910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Zhang Z., Presgraves D.C.. Drosophila X-Linked genes have lower translation rates than autosomal genes. Mol. Biol. Evol. 2016; 33:413–428. [DOI] [PubMed] [Google Scholar]
- 84. Durdevic Z., Mobin M.B., Hanna K., Lyko F., Schaefer M.. The RNA methyltransferase Dnmt2 Is required for efficient Dicer-2-dependent siRNA pathway activity in Drosophila. Cell Rep. 2013; 4:931–937. [DOI] [PubMed] [Google Scholar]
- 85. Fukunaga R., Han B.W., Hung J.H., Xu J., Weng Z., Zamore P.D.. Dicer partner proteins tune the length of mature miRNAs in flies and mammals. Cell. 2012; 151:533–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Ameres S.L., Horwich M.D., Hung J.H., Xu J., Ghildiyal M., Weng Z., Zamore P.D.. Target RNA-directed trimming and tailing of small silencing RNAs. Science. 2010; 328:1534–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Ghildiyal M., Xu J., Seitz H., Weng Z., Zamore P.D.. Sorting of Drosophila small silencing RNAs partitions microRNA* strands into the RNA interference pathway. RNA. 2010; 16:43–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Liu Y., Beyer A., Aebersold R.. On the dependency of cellular protein levels on mRNA abundance. Cell. 2016; 165:535–550. [DOI] [PubMed] [Google Scholar]
- 89. McManus C.J., May G.E., Spealman P., Shteyman A.. Ribosome profiling reveals post-transcriptional buffering of divergent gene expression in yeast. Genome Res. 2014; 24:422–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Artieri C.G., Fraser H.B.. Evolution at two levels of gene expression in yeast. Genome Res. 2014; 24:411–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Wang Z., Sun X., Zhao Y., Guo X., Jiang H., Li H., Gu Z.. Evolution of gene regulation during transcription and translation. Genome Biol. Evol. 2015; 7:1155–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Kang S.A., Pacold M.E., Cervantes C.L., Lim D., Lou H.J., Ottina K., Gray N.S., Turk B.E., Yaffe M.B., Sabatini D.M.. mTORC1 phosphorylation sites encode their sensitivity to starvation and rapamycin. Science. 2013; 341:1236566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Shimobayashi M., Hall M.N.. Making new contacts: the mTOR network in metabolism and signalling crosstalk. Nat. Rev. Mol. Cell Biol. 2014; 15:155–162. [DOI] [PubMed] [Google Scholar]
- 94. Laplante M., Sabatini D.M.. mTOR signaling in growth control and disease. Cell. 2012; 149:274–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Hsieh A.C., Liu Y., Edlind M.P., Ingolia N.T., Janes M.R., Sher A., Shi E.Y., Stumpf C.R., Christensen C., Bonham M.J. et al. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature. 2012; 485:55–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Hay N., Sonenberg N.. Upstream and downstream of mTOR. Genes Dev. 2004; 18:1926–1945. [DOI] [PubMed] [Google Scholar]
- 97. Demetriades C., Doumpas N., Teleman A.A.. Regulation of TORC1 in response to amino acid starvation via lysosomal recruitment of TSC2. Cell. 2014; 156:786–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Zhang H., Stallock J.P., Ng J.C., Reinhard C., Neufeld T.P.. Regulation of cellular growth by the Drosophila target of rapamycin dTOR. Genes Dev. 2000; 14:2712–2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Blenis J. TOR, the gateway to cellular metabolism, cell growth, and disease. Cell. 2017; 171:10–13. [DOI] [PubMed] [Google Scholar]
- 100. Förstemann K., Horwich M.D., Wee L., Tomari Y., Zamore P.D.. Drosophila microRNAs are sorted into functionally distinct Argonaute complexes after production by Dicer-1. Cell. 2007; 130:287–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Ghosh U., Adhya S.. Non-equivalent roles of AGO1 and AGO2 in mRNA turnover and translation of Cyclin D1 mRNA. J. Biol. Chem. 2016; 291:7119–7127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Seitz H., Ghildiyal M., Zamore P.D.. Argonaute loading improves the 5′ precision of both microRNAs and their miRNA∗ strands in flies. Curr. Biol. 2008; 18:147–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Detzer A., Engel C., Wünsche W., Sczakiel G.. Cell stress is related to re-localization of Argonaute 2 and to decreased RNA interference in human cells. Nucleic Acids Res. 2011; 39:2727–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Karginov F.V., Hannon G.J.. Remodeling of Ago2–mRNA interactions upon cellular stress reflects miRNA complementarity and correlates with altered translation rates. Genes Dev. 2013; 27:1624–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Garcia-Segura L., Abreu-Goodger C., Hernandez-Mendoza A., Dimitrova Dinkova T.D., Padilla-Noriega L., Perez-Andrade M.E., Miranda-Rios J.. High-throughput profiling of Caenorhabditis elegans starvation-responsive microRNAs. PLoS One. 2015; 10:e0142262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Leung A.K.L., Sharp P.A.. MicroRNA functions in stress responses. Mol. Cell. 2010; 40:205–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Xu P., Vernooy S.Y., Guo M., Hay B.A.. The Drosophila microRNA mir-14 suppresses cell death and is required for normal fat metabolism. Curr. Biol. 2003; 13:790–795. [DOI] [PubMed] [Google Scholar]
- 108. Wilczynska A., Bushell M.. The complexity of miRNA-mediated repression. Cell Death Differ. 2015; 22:22–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Buchan J.R., Parker R.. Eukaryotic stress granules: the ins and outs of translation. Mol. Cell. 2009; 36:932–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Balagopal V., Parker R.. Polysomes, P bodies and stress granules: states and fates of eukaryotic mRNAs. Curr. Opin. Cell Biol. 2009; 21:403–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Yamasaki S., Anderson P.. Reprogramming mRNA translation during stress. Curr. Opin. Cell Biol. 2008; 20:222–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Qian W., Yang J.R., Pearson N.M., Maclean C., Zhang J.. Balanced codon usage optimizes eukaryotic translational efficiency. PLoS Genet. 2012; 8:e1002603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Kronja I., Yuan B., Eichhorn S.W., Dzeyk K., Krijgsveld J., Bartel D.P., Orr-Weaver T.L.. Widespread changes in the posttranscriptional landscape at the Drosophila oocyte-to-embryo transition. Cell Rep. 2014; 7:1495–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Wong M.D., Jin Z., Xie T.. Molecular mechanisms of germline stem cell regulation. Annu. Rev. Genet. 2005; 39:173–195. [DOI] [PubMed] [Google Scholar]
- 115. Chen S., Wang S., Xie T.. Restricting self-renewal signals within the stem cell niche: multiple levels of control. Curr. Opin. Genet. Dev. 2011; 21:684–689. [DOI] [PubMed] [Google Scholar]
- 116. Horner V.L., Wolfner M.F.. Transitioning from egg to embryo: triggers and mechanisms of egg activation. Dev. Dyn. 2008; 237:527–544. [DOI] [PubMed] [Google Scholar]
- 117. Miyoshi K., Tsukumo H., Nagami T., Siomi H., Siomi M.C.. Slicer function of Drosophila Argonautes and its involvement in RISC formation. Genes Dev. 2005; 19:2837–2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Wee L.M., Flores-Jasso C.F., Salomon W.E., Zamore P.D.. Argonaute divides its RNA guide into domains with distinct functions and RNA-binding properties. Cell. 2012; 151:1055–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Miyoshi K., Miyoshi T., Siomi H.. Many ways to generate microRNA-like small RNAs: non-canonical pathways for microRNA production. Mol. Genet. Genomics. 2010; 284:95–103. [DOI] [PubMed] [Google Scholar]
- 120. Iwasaki S., Kawamata T., Tomari Y.. Drosophila argonaute1 and argonaute2 employ distinct mechanisms for translational repression. Mol. Cell. 2009; 34:58–67. [DOI] [PubMed] [Google Scholar]
- 121. Peters L., Meister G.. Argonaute proteins: mediators of RNA silencing. Mol. Cell. 2007; 26:611–623. [DOI] [PubMed] [Google Scholar]
- 122. Moore M.J., Scheel T.K.H., Luna J.M., Park C.Y., Fak J.J., Nishiuchi E., Rice C.M., Darnell R.B.. miRNA-target chimeras reveal miRNA 3′-end pairing as a major determinant of Argonaute target specificity. Nat. Commun. 2015; 6:8864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Hackenberg M., Huang P.J., Huang C.Y., Shi B.J., Gustafson P., Langridge P.. A comprehensive expression profile of microRNAs and other classes of non-coding small RNAs in barley under phosphorous-deficient and -sufficient conditions. DNA Res. 2012; 20:109–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Alves C.S., Vicentini R., Duarte G.T., Pinoti V.F., Vincentz M., Nogueira F.T.. Genome-wide identification and characterization of tRNA-derived RNA fragments in land plants. Plant Mol. Biol. 2017; 93:35–48. [DOI] [PubMed] [Google Scholar]
- 125. Cognat V., Morelle G., Megel C., Lalande S., Molinier J., Vincent T., Small I., Duchêne A.M., Maréchal-Drouard L.. The nuclear and organellar tRNA-derived RNA fragment population in Arabidopsis thaliana is highly dynamic. Nucleic Acids Res. 2017; 45:3460–3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Kirchner S., Ignatova Z.. Emerging roles of tRNA in adaptive translation, signalling dynamics and disease. Nat. Rev. Genet. 2015; 16:98–112. [DOI] [PubMed] [Google Scholar]
- 127. Orioli A. tRNA biology in the omics era: Stress signalling dynamics and cancer progression. Bioessays. 2017; 39:1600158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw deep-sequencing data generated by this study have been submitted to SRA (PRJNA378597) and GEO (GSE106697). All other data supporting the findings of this study are available within the article and its supplementary files.